Significance
Reproductive isolation is a key event leading to speciation as suggested by the observation of living populations in nature. This paper shows that novel reproductive populations of Schizosaccharomyces pombe, which are reproductively isolated from the WT population, can be created by genetically altering the primary structure of a mating pheromone and its receptor. Based on the biological concept of species, this reproductive group should be regarded as a new species. This is the first report, to our knowledge, of the artificial creation of a new species of any living organism in the history of evolutional research.
Keywords: fission yeast, mating pheromone, G-protein–coupled receptor, reproductive isolation, speciation
Abstract
The diversification of sex pheromones is regarded as one of the causes of prezygotic isolation that results in speciation. In the fission yeast Schizosaccharomyces pombe, the molecular recognition of a peptide pheromone by its receptor plays an essential role in sexual reproduction. We considered that molecular coevolution of a peptide-mating pheromone, M factor, and its receptor, Map3, might be realized by experimentally diversifying these proteins. Here, we report the successful creation of novel mating-type pairs by searching for map3 suppressor mutations that rescued the sterility of M-factor mutants that were previously isolated. Several strong suppressors were found to also recognize WT M factor. The substituted residues of these Map3 suppressors were mapped to F204, F214, and E249, which are likely to be critical residues for M-factor recognition. These critical residues were systematically substituted with each of the other amino acids by in vitro mutagenesis. Ultimately, we successfully obtained three novel mating-type pairs constituting reproductive groups. These novel mating-type pairs could not conjugate with WT maters. Furthermore, no flow of chromosomally integrated drug-resistance genes occurred between the novel and the WT mating pairs, showing that each experimentally created reproductive group [e.g., M factor(V5H) and Map3(F214H)] was isolated from the WT group. In conclusion, we have succeeded in creating an artificial reproductive group that is isolated from the WT group. In keeping with the biological concept of species, the artificial reproductive group is a new species.
Speciation is the most critical step in evolution (1). A new species branches off from an original species when a group of individuals is isolated reproductively (termed “reproductive isolation”) (2). Chemical communication between the two sexes is important in both attracting individuals of the opposite sex and the courtship reaction. Pheromone diversification may be a possible mechanism underlying reproductive isolation.
Female-attracting peptide pheromones of newts are providing a promising means to explore this mechanism. A decapeptide called sodefrin was first identified during the analysis of a cDNA library of the abdominal gland of the red-bellied newt Cynops pyrrhogaster (3, 4). A closely related newt, the sword-tailed newt Cynops ensicauda, produces a similar peptide pheromone named Silefrin (5). Interestingly, a sodefrin variant, aonirin, was found in the Nara area of Japan; 1 of 10 amino acids in aonirin differs from those in sodefrin, the prototype peptide (Table 1) (6). This variant peptide was found to not be effective in attracting females in the Niigata and Chiba areas of Japan (7). It was, thus, speculated that altering the primary structure of the female-attracting peptide of the red-bellied newt and coevolution of the corresponding receptor protein may lead to reproductive isolation. To verify this speculation, we are interested in artificially altering a pheromone and its receptor, thereby mimicking coevolution in nature, by using a genetically amenable model organism, the fission yeast Schizosaccharomyces pombe.
Table 1.
Primary structure of peptide pheromones in some red-bellied newts (Cynops sp.) and fission yeasts (Schizosaccharomyces sp.)
| Name | Species | Amino acid sequence |
| Cynops sp. | ||
| Sodefrin (prototype) | pyrrhogaster | SIPSKDALLK |
| Aonirin (Nara area) | pyrrhogaster | SIPSKDAVLK |
| Silefrin | ensicauda | SILSKDAQLK |
| Schizosaccharomyces sp. | ||
| M factor | pombe | YTPKVPYMC |
| M factor | octosporus | YQPKPPAMC (presumed) |
S. pombe has two sexes, which are usually termed mating-type h+ [plus (P)] and mating-type h− [minus (M)] (8–10). On nitrogen starvation, two haploid cells of opposite mating type mate to form a diploid zygote (11), which then commences meiosis and finally, culminates in an ascus containing four newly born ascospores. The mating pheromones of S. pombe are small peptides that play essential roles in the courtship reaction. The M-factor pheromone, YTPKVPYMCFar-OCH3, is a C-terminally farnesylated nonapeptide secreted by M cells (12–14) that is specifically recognized by a G-protein–coupled receptor, Map3, on the surface of P cells (15). P factor, the mating pheromone secreted by P cells, is a simple peptide composed of 23 amino acids that activates the corresponding receptor, Mam2, on M cells (16, 17). Meiosis also depends on the action of mating pheromone signals (17). Pheromones of S. pombe, thus, play important roles in sexual reproduction, mating, and meiosis.
The specificity of mating-type recognition is primarily determined by molecular recognition of the peptide pheromone by its cognate receptor. Mating pheromones play essential roles in sexual maturation, attraction of opposite mating-type cells (cell agglutination), copulation (cell fusion), and mate choice (14, 17–19). For S. pombe, all of the genes encoding the mating pheromones, receptors, and components of the signal transduction cascade emanating from the activated receptors have been identified and thoroughly investigated (11). The primary structures of both mating pheromones and their receptors can be easily altered by in vitro mutagenesis. Because mating competence depends on signaling by both the M- and P-type pheromones, complete impairment of M-factor signaling should prevent the mating reaction (13, 15–17).
Activation of the mating pheromone receptor is the initial event during the course of the mating process; thus, structural alteration of the pheromone peptides may affect the downstream signaling pathway. We reasoned that mutational alterations of either the pheromone peptides or the pheromone receptors might result in reproductive isolation from WT cells. If the modified pheromone (a ligand) and the receptor protein are structurally fit and ligand-induced activation of the receptor can be attained, the resulting modified versions of mating-competent cells might constitute a novel reproductive group. Owing to the small size of M factor (9 aa), a full set of single residue-substituted missense mutants of the mfm1 gene (coding for M factor) was previously successfully generated (18). Thorough screening of the 152 mfm1 mutants identified 35 sterile ones that might produce nonfunctional M-factor peptides. These mutant peptides were detected in culture filtrates, indicating that they are likely to be defective in molecular interaction with their specific receptor, Map3 (18).
The aim of this study was to identify mutated receptor proteins that could accept any of the mutated M-factor peptides and thereby, create novel mating-type pairs constituting an isolated reproductive group. Here, we report the successful creation of such new reproductive groups isolated from normal mating-type cells. Strict genetic evidence indicates that virtually no gene transfer occurs between the WT and the novel reproductive groups. Our success in prezygotic isolation in the fission yeast population by manipulating mating pheromone recognition systems represents a further advance toward the artificial creation of new species.
Results
Mating Pheromone System Functions as a Reproductive Barrier.
Mating pheromones are known to be required for successful copulation, but there are no quantitative data on the frequency of gene transfer between a pair of opposite mating-type strains carrying mutations in various pheromone-related genes. We first assayed the frequency of recombination of genetic markers between WT pairs of opposite mating types. When WT P- and M-type cells genetically marked by different drug-resistance genes (natR and hygR) were mixed to mate, the recombination frequency after a single conjugation cycle was approximately on the order of 10−1. As a negative control, a pair of strains of the same mating type showed a recombination frequency of less than 10−7. Next, several combinations of pheromone-related mutants were tested in the same way. For example, an h− strain harboring the deletion allele of the triple M-factor pheromone genes (mfm1+, mfm2+, and mfm3+) exhibited virtually no genetic recombination with WT h+ cells (<10−7). Furthermore, the recombination frequency was ∼10−7 for the null mutant of the M-factor receptor gene of the h+ strain mixed with the WT h− strain. These results summarized in Table 2 indicate that a deficiency in the mating pheromone systems of S. pombe might function as an efficient barrier to reproductive gene flow. We, therefore, reasoned that manipulations of the pheromone peptide and its receptor might produce novel reproductive groups that might be reproductively isolated from the WT group.
Table 2.
Recombination frequency in the cross between different mating-type strains carrying mutations in the pheromone and its receptor genes
| Combination | Frequency of recombinants | |
| Parent A (hygR) | Parent B (natR) | |
| M-type strain (WT) | P-type strain (WT) | 5 × 10−1 |
| M-type strain (WT) | M-type strain (WT) | <10−7 |
| P-type strain (WT) | P-type strain (WT) | <10−7 |
| M-type strain (pheromoneless) | P-type strain (WT) | <10−7 |
| P-type strain (receptorless) | M-type strain (WT) | 3 × 10−7 |
| M-type strain (pheromoneless) | P-type strain (receptorless) | <10−7 |
The same numbers of cells of parent A and parent B were mixed and cultured on malt extract medium (MEA) for 3 d. Cell suspensions were serially diluted, and aliquots were spread on YEA plates containing the appropriate drug. Colony numbers were counted after several days of incubation.
Identification of Suppressor Mutations in the map3 Gene That Rescue the Sterility of Mutated M Factor.
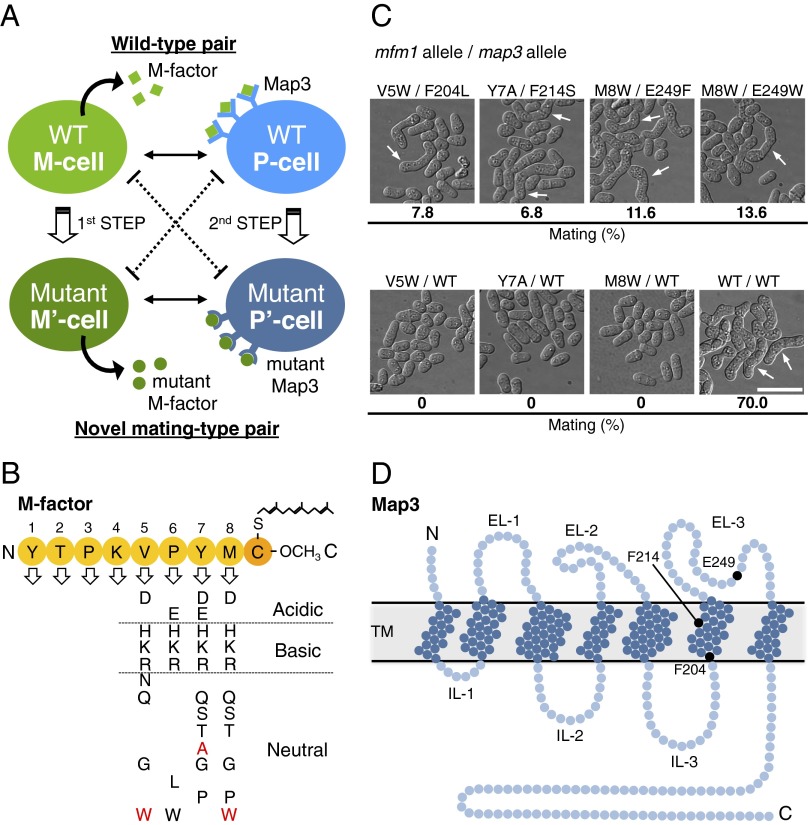
This study aimed to create novel mating-type pairs by genetically altering a mating pheromone and its receptor. As the first step of the strategy (Fig. 1A), we previously generated 35 different kinds of mfm1 mutants producing inactive M factor by comprehensively substituting each of the first eight of nine residues in M factor with the other 19 amino acids (18) (Fig. 1B). Most of these M-factor mutants are not likely to be recognized by the receptor, Map3. As the second step, here, we undertook random mutagenesis of the map3+ receptor gene to isolate suppressor mutants that recovered conjugation with the sterile M-factor mutants. Because the ligand binding domain of Map3 has not been determined, we introduced random mutations into the whole ORF of 365 amino acids by error-prone PCR. A high-quality map3 mutant library (prepared from ∼5.0 × 104 independent Escherichia coli transformants) constructed on a multicopy plasmid, pAL-KR(map3+) (Fig. S1A), was introduced into 35 sterile missense mutants of M factor (Fig. 1B). Approximately 6.5 × 105 colonies were inspected for their mating capability using efficient colony staining with iodine vapor (8). This large-scale screening gave 1,308 putative mating-positive colonies, which were then microscopically inspected to identify suppressor mutants of high mating efficiency (173 clones in total). Next, each suppressor map3 mutant gene was integrated into the map3+ locus on the chromosome I; map3 mutants showing significant mating ability, even in single copy number, were selected, and the map3 and mfm1 genes of these candidate mutants were sequenced. Ultimately, four genuine suppressor Map3 mutations (F204L, F214S, E249F, and E249W) coupled with the corresponding suppressed mfm1 mutations were identified (Fig. 1C). The map3 suppressor mutations were mapped to the 204th Phe (F204), 214th Phe (F214), and 249th Glu (E249) residues (Fig. 1D). These residues are likely to be critical for ligand recognition. According to domain assignments based on hydropathy analysis (15), F204 and F214 are located in the sixth transmembrane domain, and E249 is located in the third extracellular loop (Fig. 1D), suggesting that these domains might be implicated in ligand recognition.
Fig. 1.
Creation of novel mating-type pairs by screening for map3 mutants that suppress mating-deficient M-factor mutants. (A) Strategy for creating a novel mating-type pair. The first step was the comprehensive amino acid substitution of mating pheromone M factor by in vitro site-directed mutagenesis as described previously (18). Among 152 single residue-substituted missense mutants, 35 sterile M-factor mutants were selected (shown in B). The second step was random mutagenesis of the map3+ gene encoding M-factor receptor by error-prone PCR. We screened for suppressor map3 mutants that rescued the sterility of each of the M-factor mutants. In this way, novel mating-type pairs were successfully isolated. (B) List of sterile M-factor mutants. Nonfunctional substitutions are indicated by one-letter codes. The substitutions shown in red were suppressed by mutant Map3. (C) Mating ability of four genetically modified mating-type pairs isolated by screening. The mfm1 (encoding M factor) allele/map3 allele in each pair is indicated above the microphotographs, which show typical cell morphology and mating frequency for that pair. White arrows indicate typical zygotes. (Scale bar: 10 µm.) (D) The structure of Map3 predicted by hydropathy analysis (15), in which the positions of three possibly important residues for ligand recognition are indicated as black circles. EL, extracellular loop; IL, intracellular loop; TM, transmembrane.
Successful Creation of Novel Mating-Type Pairs.
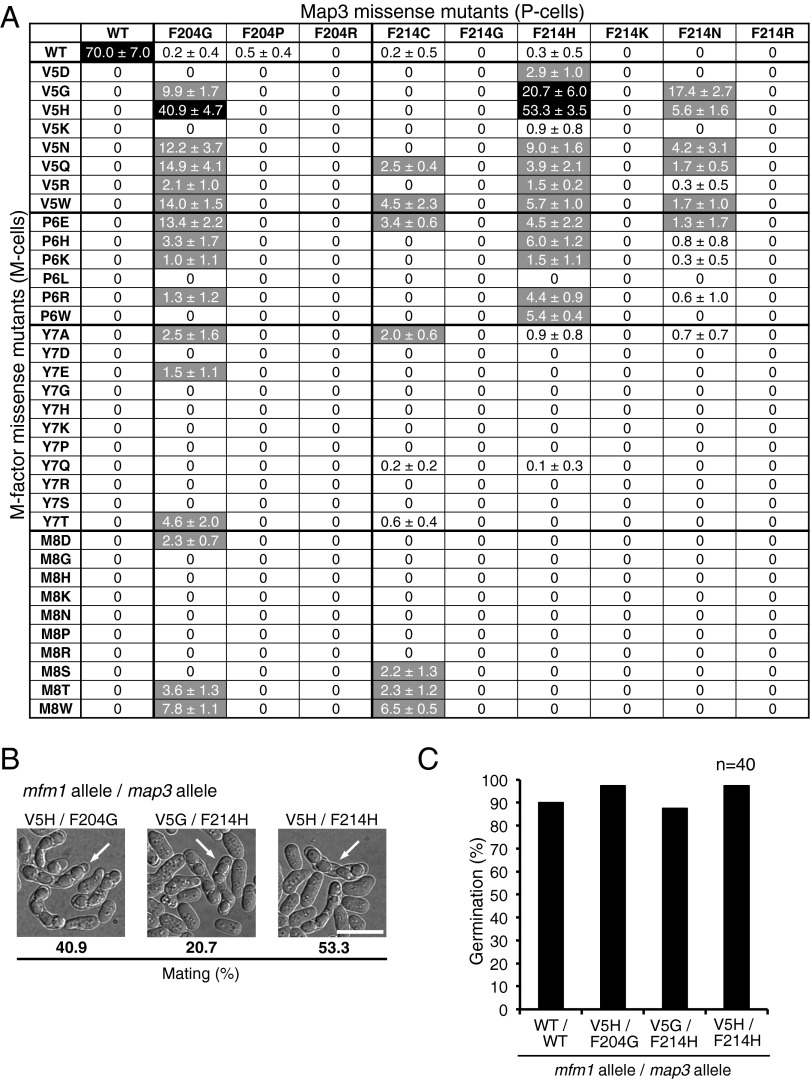
The four identified suppressor mutations of Map3 (F204L, F214S, E249F, and E249W) seemed to have reduced specificity for M-factor pheromone, because these map3 mutants retained high ability to mate with the WT strain (30–46%) (Table S1). We, therefore, attempted to generate novel mating-proficient pairs that were unable to conjugate with the WT. Three target residues (F204, F214, and E249) were systematically substituted to generate 57 missense map3 mutants, in which each of three residues was substituted with each of 19 other amino acids. Nine of these mutations (F204G, F204P, F204R, F214C, F214G, F214H, F214K, F214N, and F214R) showed sterility or extremely low mating proficiency (<1%) in the cross with WT M cells (Table S1).
These nine map3 mutants were crossed with the 35 sterile M-factor mutants (315 crosses) (Fig. 2A). This comprehensive mating assay showed that some intersex combinations had significant mating ability (black or gray in Fig. 2A). In particular, a pair comprising mfm1-V5H and map3-F214H showed fairly high mating frequency (53%), comparable with that of the WT (70%), although the mfm1-V5H mutant was unable to mate with WT P cells. [The mfm1+ gene produces precursor polypeptides composed of 42 amino acids, which are then processed to a mature M-factor peptide composed of nine residues from Tyr31 to Cys39. For simplification, hereafter, residue numbers are based on the mature peptide (i.e., mfm1-V5H instead of mfm1-V35H).] Intriguingly, some variant M factors of V5 and P6 residues were recognized by mutant Map3 proteins to some extent (Fig. 2A). These data also imply that the V5 and P6 residues of M factor are important for recognition by Map3. The diploid zygotes culminated in an ascus containing four haploid spores (Fig. 2B). Because these spores germinated normally and outgrew to vegetative cells, the descendants were fertile (Fig. 2C). Therefore, we concluded that three pairs, mfm1-V5H/map3-F204G, mfm1-V5G/map3-F214H, and mfm1-V5H/map3-F214H, were novel mating-type pairs that copulated at high frequency (20–55%). The map3 mutant harboring the double-mutation F204G and F214H recognized neither the WT M factor nor the sterile-type M factor(V5H) (Fig. S2), suggesting that these two residues, F204 and F214, are independently involved in M-factor recognition.
Fig. 2.
Mating ability of the cross between map3 missense mutants and sterile M-factor mutants. (A) Efficiency of mating between 35 M-type M-factor mutants and 9 P-type map3 mutants. The M-factor mutants are shown on the left, and the Map3 mutants are shown on the top. The map3 missense mutants (P-type cells) and M-factor missense mutants (M-type cells) were crossed in all combinations. The percentage of zygotes plus asci was determined after incubation for 2 d on malt extract medium agar. Three independent samples were inspected (at least 200 cells each) (SI Materials and Methods). Means with SDs were calculated. Mating frequency is also indicated as a three-level grading: black, >20%; gray, 1–20%, and white, <1% (almost sterile). (B) Microphotographs of three novel mating-type pairs. White arrows indicate typical asci. (Scale bar: 10 µm.) (C) Germination of spores produced by mating between novel mating-type pairs. Spores were isolated and inoculated on nutrient medium (YEA) by micromanipulation. Spore-derived colonies were observed after incubation for 3 d. The percentage of germination with the number of tested spores (n = 40) is shown.
Demonstration of Reproductive Isolation of the Novel Mating-Type Pairs.
Our microscopic observation showed that the conjugation frequency between the mutant strains and WT maters was extremely low. To verify reproductive isolation between the newly produced reproductive group and the WT group, a more sensitive genetic recombination test was applied. In this test, two drug-resistance markers, natR and hygR, are integrated into each of the different chromosomes (Fig. S3). If two strains carrying different drug-resistance markers successfully conjugate, then recombinant descendants harboring both resistance markers will appear. If no double-resistant cells are observed, then these strains must be reproductively isolated. This sensitive test facilitates the detection of extremely rare genetic recombination events (<10−7).
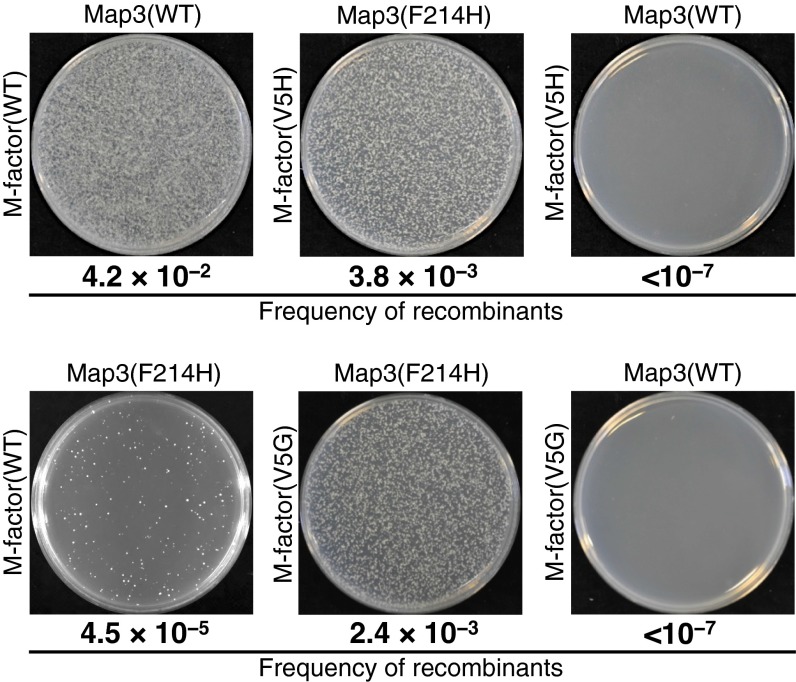
Equal numbers of cells carrying different drug-resistance markers were mixed and spotted onto solid conjugation medium (malt extract medium). After 2 d of incubation, an aliquot was spread onto nutrient agar plates containing the appropriate drugs, nourseothricin (Nat) and hygromycin B (Hyg). The isogeneic cross between opposite mating-type strains within the artificially generated reproductive group (e.g., mfm1-V5H and map3-F214H) or within the WT group produced abundant colonies (Fig. 3). Although the cross of WT P cells with M cells secreting mutant M factor(V5G) or M factor(V5H) produced virtually no double-resistant colonies (<10−7) (Fig. 3), these sterile M-factor mutants were able to mate with mutant P cells expressing mutant Map3(F214H), generating abundant double-resistant colonies (10−3). Thus, sterility caused by mutant M factors (such as V5H) was markedly recovered by a suppressor Map3 mutant, such as F214H. In addition, when P cells expressing mutant map3-F214H genes and WT M cells were mixed, only limited numbers of double-resistant colonies (4.5 × 10−5) appeared. Other mating combinations between mutant strains and WT strains were examined for genetic recombination of drug-resistance markers (Fig. S4). In all cases, gene flow was severely inhibited between different reproductive groups.
Fig. 3.
Reproductive isolation of novel mating-type pairs from the WT population as revealed by a sensitive genetic recombination assay. Results of the genetic recombination assay. The same numbers of P and M cells were mixed and cultured on conjugation medium (malt extract medium) for 2 d. Cells were spread onto YEA plates containing 100 µg/mL appropriate drugs: YEA + Nat, YEA + Hyg, and YEA + Nat + Hyg. After 3 d of incubation, colony numbers were counted. The number of Nat/Hyg double-resistant colonies was normalized by the colony numbers on plates containing single Nat or Hyg. High recombination frequencies were observed in crosses between WT maters as well as between the mutant pair mfm1-V5H and map3-F214H. Note that the mutant M factors were completely rejected by the WT Map3 (recombinant frequency, <10−7); similarly, the mutant receptor Map3(F214H) barely accepted WT M factor (recombination frequency, 4.5 × 10−5).
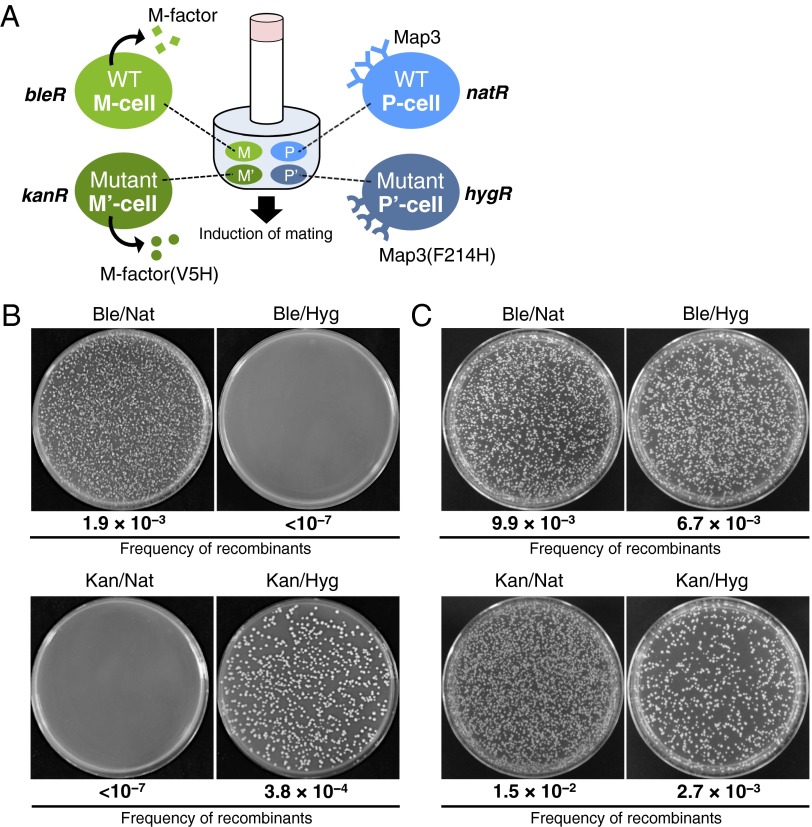
To further verify the prezygotic isolation of the WT group and novel reproductive groups, M- and P-type strains of both WT and mutant reproductive groups were mixed to mate (sympatric condition), and the flow of genetic markers between the reproductive groups was examined. The WT M strain, WT P strain, mfm1-V5H M strain, and map3-F214H P strain were differentially marked by the drug-resistance markers bleR, natR, kanR, and hygR, respectively (20) (Fig. S3). Equal numbers of cells of these four strains were mixed and cultured on malt extract medium to induce mating (Fig. 4A). After incubation for 2 d, the cell suspension was spread onto yeast extract medium (YEA) plates containing the appropriate drugs: phleomycin (Ble), G418 (Kan), Nat, and Hyg. Fig. 4B shows that isogeneic crosses between the two WT strains and between the two mutant strains gave plenty of Ble/Nat double-resistant and Kan/Hyg double-resistant colonies; in contrast, allogeneic crosses between the WT and the novel mating-type strains generated virtually no recombinant colonies (<10−7). As a positive control, two different pairs of WT strains were used in the genetic recombination test. Double-resistant clones appeared at roughly equal frequency for all combinations of strains (Fig. 4C). Taken together, these data show that virtually no gene flow occurred between the WT group and the novel reproductive group, indicating that there is almost complete reproductive isolation between the two groups. We conclude that genetic changes in sex pheromone systems generate a novel reproductive group that is reproductively isolated from the WT group, indicating that sympatric speciation can occur in a test tube for the model microorganism S. pombe.
Fig. 4.
Evidence for reproductive isolation of the novel mating-type group from the WT group in mixed mating cultures. (A) Experimental design. The novel reproductive group comprising M cells secreting M factor(V5H) and P cells expressing Map3(F214H) was cocultivated with WT cells of both mating types. Four heterothallic strains [WT M cells (FS476; heterothallic M), WT P cells (TS551; heterothallic P), mfm1-V5H M cells (TS655; heterothallic M′), and map3-F214H P cells (TS458; heterothallic P′)] were marked with the drug-resistance markers bleR, natR, kanR, and hygR, respectively (Fig. S3) and mixed in equal cell numbers. Mating was induced by incubation on malt extract medium plates. After incubation for 2 d, an aliquot was spread onto YEA plates containing the appropriate drugs. (B) Experimental results. Where crossing was achieved, double-resistant clones were observed. Colony numbers were counted on plates containing the four different drugs either singly or doubly in different combinations. (C) As a positive control, four WT strains [WT M cells (FS476; heterothallic MWT1), WT P cells (TS551; heterothallic PWT1), WT M cells (TS550; heterothallic MWT2), and WT P cells (TS452; heterothallic PWT2)] were marked with the drug-resistance markers bleR, natR, kanR, and hygR, respectively, and also mixed in equal cell numbers. Ble, phleomycin; Hyg, hygromycin B; Kan, G418; Nat, nourseothricin.
Discussion
The mechanism underlying speciation is a fundamental issue in evolutionary biology. Reproductive groups sharing a common gene pool constitute a species. Pre- and postzygotic reproductive isolation mechanisms are involved in the process of speciation. For insects and some vertebrates, the specificity of pheromones and receptors determines the reproductive group (21, 22). Mutational alterations of the mating pheromone system affect male/female recognition, resulting in prezygotic isolation. It is thought that such changes in pheromonal recognition generate novel reproductive groups, which trigger speciation. This hypothesis has not been proven, however, because artificial manipulations are not yet possible for the insects for which pheromone actions have been studied. We, therefore, considered that this hypothesis might be tested with unicellular organisms, such as fission yeast, because the genes encoding mating pheromones and their cognate receptors have been identified and are available for mutagenesis. This study has shown that artificial alterations of the primary structure of both a peptide pheromone and its receptor resulted in efficient reproductive isolation. Our results substantiate the hypothesis that the coevolution of pheromones and receptors may be one of the mechanisms underlying prezygotic isolation in nature.
These results raise the question of whether speciation mechanisms based on the coevolution of a sex pheromone and its receptor are really acting in natural populations of any higher organisms. Some male amphibians produce peptidyl pheromones that attract female individuals. The female-attracting pheromone from the male red-bellied newt C. pyrrhogaster is a decapeptide called sodefrin (Table 1) (3, 4). In relation to this study, a variant of sodefrin (Val8-sodefrin; also called aonirin) has been found in the Nara district (6). The Val8-sodefrin peptide is effective in attracting females in the Nara area but shows little or no activity toward females of the Niigata and Chiba areas (7), suggesting that the corresponding receptor has evolved to accept Val8-sodefrin among some newts. Although the receptor remains to be identified, these observations suggest that at least two different pheromone/receptor systems coexist in natural populations of newts in the Nara district. This observation seems to exemplify ongoing speciation in nature caused by mutational changes of peptide pheromones, similar to the artificial variants of S. pombe observed in this study.
The M-factor peptides of two fission yeast species, S. pombe and Schizosaccharomyces octosporus, differ in three of their nine residues (Table 1). A similar difference is observed in the peptide pheromones of the two newt species (3 among 10 residues). It seems plausible that S. pombe and S. octosporus may have branched away from their common ancestor because of mutational alterations in M factor and its receptor. This hypothesis would be a very attractive explanation for speciation, although at present, it is impossible to prove it. The Map3 receptors of S. pombe and S. octosporus are also conserved, showing 67% identity.
Yeasts have both asexual and sexual reproduction phases in their lifecycle. As a result, even sterile mutants harboring defective mutations in genes of the pheromone system survive and multiply. In contrast to yeasts and fungi, higher eukaryotes will fail to produce offspring if sexual reproduction is impaired. Loss of pheromone activity results in extinction of the lineage for these organisms. We speculate that changes in the activity of pheromones and receptors may not be marked but are probably very gradual, such that sexual reproduction is not prevented. The coevolution of pheromones and their corresponding receptors is likely to proceed little by little, and a mutational alteration may cause only a slight depression of pheromone activity or pheromone receptor activity. Before such a mutant is completely lost, a second suppressor mutation may occur to recover the first defect. Multiple subtle changes might take place repeatedly during the actual course of the coevolution of sex pheromones and receptor proteins before speciation occurs. This gradual coevolution is more likely to be the actual mechanism underlying the process of pheromone system-related prezygotic isolation.
We notice that M factor is encoded by three redundant genes (mfm1, mfm2, and mfm3) (13), and a single structure gene for P factor (map2) encodes a precursor protein that is processed to four mature peptides (16). An alternative mechanism for evolution of a pheromone system leading to reproductive isolation relies partly on such duplication of the pheromone-encoding genes. Redundancy of the genes may allow unrestricted alterations of pheromone structures, while still retaining the original version of the pheromone gene.
Materials and Methods
S. pombe strains used are listed in Table S2. Random mutagenesis of the map3 ORF was performed by error-prone PCR (23). Site-directed mutagenesis was conducted using in vitro DNA replication. Mutagenesis procedures are described in detail in SI Materials and Methods. Screening of mutants and quantitative mating assay are also described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported, in part, by Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Publication of Scientific Research Results (to T.N.). T.S. was supported by Japan Society for the Promotion of Science Exploratory Research Grant 11J02671.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501661112/-/DCSupplemental.
References
- 1.Coyne JA, Orr HA. Speciation. Sinauer; Sunderland, MA: 2004. [Google Scholar]
- 2.Smadja C, Butlin RK. On the scent of speciation: The chemosensory system and its role in premating isolation. Heredity (Edinb) 2009;102(1):77–97. doi: 10.1038/hdy.2008.55. [DOI] [PubMed] [Google Scholar]
- 3.Kikuyama S, et al. Sodefrin: A female-attracting peptide pheromone in newt cloacal glands. Science. 1995;267(5204):1643–1645. doi: 10.1126/science.7886452. [DOI] [PubMed] [Google Scholar]
- 4.Kikuyama S, Toyoda F. Sodefrin: A novel sex pheromone in a newt. Rev Reprod. 1999;4(1):1–4. doi: 10.1530/ror.0.0040001. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto K, et al. Silefrin, a sodefrin-like pheromone in the abdominal gland of the sword-tailed newt, Cynops ensicauda. FEBS Lett. 2000;472(2-3):267–270. doi: 10.1016/s0014-5793(00)01455-1. [DOI] [PubMed] [Google Scholar]
- 6.Nakada T, et al. Isolation, characterization and bioactivity of a region-specific pheromone, [Val8]sodefrin from the newt Cynops pyrrhogaster. Peptides. 2007;28(4):774–780. doi: 10.1016/j.peptides.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Iwata T, et al. Regionally specific occurrence of an active sodefrin variant in the red-bellied newt. Ann N Y Acad Sci. 2005;1040(1):351–353. doi: 10.1196/annals.1327.059. [DOI] [PubMed] [Google Scholar]
- 8.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics. Vol 1. Plenum; London: 1974. pp. 395–446. [Google Scholar]
- 9.Egel R. Mating-type genes, meiosis and sporulation. In: Nasim A, Young P, Johnson BF, editors. Molecular Biology of the Fission Yeast. Academic; San Diego: 1989. pp. 31–73. [Google Scholar]
- 10.Egel R. Fission yeast in general genetics. In: Egel R, editor. The Molecular Biology of Schizosaccharomyces Pombe. Springer; Heidelberg: 2004. pp. 1–12. [Google Scholar]
- 11.Nielsen O. Mating-type control and differentiation. In: Egel R, editor. The Molecular Biology of Schizosaccharomyces Pombe. Springer; Heidelberg: 2004. pp. 281–296. [Google Scholar]
- 12.Davey J. Mating pheromones of the fission yeast Schizosaccharomyces pombe: Purification and structural characterization of M-factor and isolation and analysis of two genes encoding the pheromone. EMBO J. 1992;11(3):951–960. doi: 10.1002/j.1460-2075.1992.tb05134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kjaerulff S, Davey J, Nielsen O. Analysis of the structural genes encoding M-factor in the fission yeast Schizosaccharomyces pombe: Identification of a third gene, mfm3. Mol Cell Biol. 1994;14(6):3895–3905. doi: 10.1128/mcb.14.6.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey J. Isolation and quantitation of M-factor, a diffusible mating factor from the fission yeast Schizosaccharomyces pombe. Yeast. 1991;7(4):357–366. [Google Scholar]
- 15.Tanaka K, Davey J, Imai Y, Yamamoto M. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol Cell Biol. 1993;13(1):80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: Its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8(3):328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- 17.Kitamura K, Shimoda C. The Schizosaccharomyces pombe mam2 gene encodes a putative pheromone receptor which has a significant homology with the Saccharomyces cerevisiae Ste2 protein. EMBO J. 1991;10(12):3743–3751. doi: 10.1002/j.1460-2075.1991.tb04943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seike T, Yamagishi Y, Iio H, Nakamura T, Shimoda C. Remarkably simple sequence requirement of the M-factor pheromone of Schizosaccharomyces pombe. Genetics. 2012;191(3):815–825. doi: 10.1534/genetics.112.140483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seike T, Nakamura T, Shimoda C. Distal and proximal actions of peptide pheromone M-factor control different conjugation steps in fission yeast. PLoS ONE. 2013;8(7):e69491. doi: 10.1371/journal.pone.0069491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22(13):1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- 21.Leary GP, et al. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA. 2012;109(35):14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng SH, et al. Pheromone evolution and sexual behavior in Drosophila are shaped by male sensory exploitation of other males. Proc Natl Acad Sci USA. 2014;111(8):3056–3061. doi: 10.1073/pnas.1313615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA fragment using a modified polymerase chain reaction. Biotechniques. 1989;1(1):11–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.