Significance
Leprosy was thought to be exclusively caused by infection of humans by Mycobacterium leprae. In 2008, Han et al. proposed that Mycobacterium lepromatosis, a separate unculturable species, might be responsible for a rare yet severe form of the disease called diffuse lepromatous leprosy. Here, by using comparative genomics, we show that the two species are very closely related and derived from a common ancestor that underwent genome downsizing and gene decay. Since their separation 13.9 Mya, the two species have continued to lose genes, but from different regions of the genome, and M. leprae appears to be more recent. In a phylogeographic survey, by using differential PCR, we found that M. lepromatosis was scarce and restricted to patients from Mexico.
Keywords: Mycobacterium lepromatosis, genome sequencing, Mycobacterium leprae, comparative genomics, reductive evolution
Abstract
Mycobacterium lepromatosis is an uncultured human pathogen associated with diffuse lepromatous leprosy and a reactional state known as Lucio's phenomenon. By using deep sequencing with and without DNA enrichment, we obtained the near-complete genome sequence of M. lepromatosis present in a skin biopsy from a Mexican patient, and compared it with that of Mycobacterium leprae, which has undergone extensive reductive evolution. The genomes display extensive synteny and are similar in size (∼3.27 Mb). Protein-coding genes share 93% nucleotide sequence identity, whereas pseudogenes are only 82% identical. The events that led to pseudogenization of 50% of the genome likely occurred before divergence from their most recent common ancestor (MRCA), and both M. lepromatosis and M. leprae have since accumulated new pseudogenes or acquired specific deletions. Functional comparisons suggest that M. lepromatosis has lost several enzymes required for amino acid synthesis whereas M. leprae has a defective heme pathway. M. lepromatosis has retained all functions required to infect the Schwann cells of the peripheral nervous system and therefore may also be neuropathogenic. A phylogeographic survey of 227 leprosy biopsies by differential PCR revealed that 221 contained M. leprae whereas only six, all from Mexico, harbored M. lepromatosis. Phylogenetic comparisons indicate that M. lepromatosis is closer than M. leprae to the MRCA, and a Bayesian dating analysis suggests that they diverged from their MRCA approximately 13.9 Mya. Thus, despite their ancient separation, the two leprosy bacilli are remarkably conserved and still cause similar pathologic conditions.
Nearly a quarter million new cases of leprosy (Hansen's disease) are still recorded annually worldwide despite a remarkable decrease in prevalence in the past decade (1). Leprosy primarily affects the skin, peripheral nerves, and eyes, and manifests as a spectrum of diverse clinical forms varying in bacillary load and often accompanied by painful immunological reactions (2–4). A severe form of leprosy known as diffuse lepromatous leprosy (DLL) that is common in western Mexico and the Caribbean region, first described by Lucio and Alvarado in 1852 (5), is referred to as Lucio's leprosy. Such cases account for a sizable proportion (more than 20%) of all leprosy cases in western Mexico (5–8), Cuba (9), and Costa Rica (10) but are rarely reported elsewhere. In 1948, Latapi and Zamora noted that DLL cases had no dermal nodules and were characterized by a generalized and diffuse infiltration of the skin by histiocytes and acid-fast bacilli causing an appearance of swollen or “spotted” skin, which they termed “pure and primitive diffuse lepromatosis.” In addition, they reported that some patients developed acute necrotic skin reactions, “erythema necroticans,” and differentiated this condition as Lucio's phenomenon (6). The most notable clinical feature of DLL and Lucio's phenomenon is the diffuse mycobacterial invasion of endothelial cells surrounding small vessels, often leading to vascular occlusion (8). The initial cyanotic lesions, caused by poor blood supply and ischemia, gradually evolve into black necrotic lesions (11, 12). Hence, these cases are often associated with long-term morbidity (8) as well as a higher number of fatalities if not managed adequately (5, 13).
Lucio's phenomenon is usually observed among untreated or inadequately treated nonnodular DLL cases 1–3 y after the appearance of their symptoms. Although rare, Lucio's phenomenon has also been reported among other forms of lepromatous leprosy (6). Multidrug therapy (MDT) is currently the treatment recommended by the World Health Organization for all forms of leprosy, including DLL and Lucio's phenomenon (14, 15). Until recently, Mycobacterium leprae was considered the sole causative agent of all forms of leprosy, including Lucio's phenomenon, which is often referred to as a “form of leprosy reaction” (13, 16). The genome of M. leprae has undergone reductive evolution, with approximately half occupied by pseudogenes (17), and also displays remarkably low levels of genetic diversity (18–20).
In 2008, a new mycobacterial species named Mycobacterium lepromatosis was identified in a liver autopsy specimen from a homeless Mexican who died with DLL in Arizona (21). Since then, this species has been identified by PCR-based sequencing in several Mexican patients (22) as well as in individual cases from Singapore (23) and Canada (24). In addition, clinical presentations resembling Lucio's phenomenon have been reported elsewhere, i.e., Brazil (25, 26), India (27–29), Iran (30), and Malaysia (31); however, molecular confirmation of the mycobacterial agent was not carried out. In addition, several cases of mixed infection have been reported whereby both M. lepromatosis and M. leprae were detected (22), which undermines confidence in M. lepromatosis being the causative agent of DLL.
Knowledge about the biology and pathogenesis of M. lepromatosis is limited because this species remains uncultivated (21). To date, the DNA sequences of 22.8 kb of selected PCR fragments from M. lepromatosis are known, and these were sufficient to reveal striking sequence similarity to M. leprae and a close phylogenetic relationship, but this preliminary analysis provided little biological insight. Thus, at this stage, genome sequencing is the most efficient approach to investigate M. lepromatosis.
After the first description of M. lepromatosis by Han et al. in 2008 (21), we reported independent confirmation of this species in a biopsy specimen from a DLL case (Mx1-22A) from Monterrey, Mexico (32). Here, we combined various DNA enrichment approaches and deep sequencing to unveil the genome of M. lepromatosis directly from the archived biopsy specimen from this patient. Genome-wide comparison of M. lepromatosis and M. leprae provides deeper insight into the biology of M. lepromatosis and discloses the evolutionary history of these two closely related but clearly distinct species.
Results and Discussion
Genome Sequencing, Assembly, and Analysis.
Because M. lepromatosis cannot be cultured in vitro and an animal model is not yet available, the only source of its DNA is infected human tissue. Biopsy specimen Mx1-22A was used for DNA extraction and Illumina library preparation as described previously (20). To overcome the problem of host DNA, we used two methods to enrich M. lepromatosis DNA: whole-genome array capture using the M. leprae genome as bait (20) and removal of human DNA by hybridization with a human genomic DNA bait library (Materials and Methods). Illumina sequencing of the enriched as well as the original libraries provided 55-fold coverage, which was more than sufficient for the de novo whole-genome assembly (Table S1).
The genome assembly was obtained by relaxing the assembler program’s parameters (Materials and Methods) to account for the extremely biased read coverage from the array capture library. Contigs were considerably longer compared with those obtained with default assembly parameters, but at the cost of a higher chance of misassemblies. To avoid assembly errors, we split those contigs showing disrupted synteny with M. leprae so that, from the initial 110 contigs of the de novo assembly, we obtained a final set of 126 contigs. Most inconsistencies in the contigs were observed around areas of repetitive DNA, with some exceptions. One such exception was confirmed by PCR sequencing (as detailed later), proving that there is at least one instance of genome rearrangement between M. leprae and M. lepromatosis. However, we consider that the overall level of rearrangement is low, given that only a few contigs showed breaks in synteny with the reference genome and the GC skew of the “syntenic” version of the M. lepromatosis genome is virtually identical to that of M. leprae (Fig. S1).
A total of 3,206,741 bases of the M. lepromatosis Mx1-22A genome were represented in the 126 contigs, and, with one exception, these all aligned to the 3.27-Mb circular genome sequence of the Tamil Nadu (TN) reference strain of M. leprae (17). The exception was a 2.3-kb contig bearing five mycobacterial pseudogenes. A graphical comparison revealing genome-wide synteny and key features of the two genomes is presented in Fig. 1. In brief, the genome of M. lepromatosis appears to harbor at least 1,477 genes encoding proteins [i.e., coding DNA sequence (CDS)] and 1,334 pseudogenes.
Fig. 1.

Genome synteny and salient features of M. leprae and M. lepromatosis. The 126 contigs of M. lepromatosis are distinguished with red and orange colors. Links between M. leprae and M. lepromatosis are BLAST hits, with two shades of gray to distinguish individual contigs. White stripes indicate no BLAST hits and account for 5–6% of each genome’s specific sequences. Light blue stripes indicate dispersed repeats in M. leprae. Black line indicates a confirmed structural variation between M. leprae and M. lepromatosis.
Repetitive DNA.
Dispersed repeats were the major cause of breaks in our de novo assembly. Interestingly, in most cases, the locations of these repeats correspond to those of the four main families of repetitive DNA in M. leprae: RLEP (37 copies), REPLEP (15 copies), LEPREP (8 copies), and LEPRPT (5 copies) (33). These shared 75–90% sequence identity, in segments as much as 350 nt in length, with the most conserved corresponding to LEPREP and LEPRPT repeats and the lowest homology found for the REPLEP repeats. From this analysis, it is evident that the four repeat families were present in the most recent common ancestor (MRCA) of M. leprae and M. lepromatosis and that the levels of conservation between the repeats in the two species are proportional to their copy number. No additional repetitive DNA was detected in M. lepromatosis.
A Second Draft Genome.
Using the M. lepromatosis genome sequence, we designed specific PCR primers and used them to screen ∼260 biopsies from our collection, thereby identifying a second case of M. lepromatosis in another Mexican patient (Mx177) who presented with Lucio's phenomenon. This sample, which contained no M. leprae DNA, was shotgun-sequenced, without any enrichment, and the resultant sequences were mapped against the Mx1-22A genome assembly. Despite shallow coverage of the Mx177 sample (80% of the genome at an average coverage of 5× after excluding duplicate reads), we found only 12 SNPs in a 2-Mb alignment where SNP calling was feasible. This very low SNP frequency (1 in 167 kb) is reminiscent of the similarly low genetic diversity of M. leprae strains from the same geographical area (34) and in a set of worldwide M. leprae genomes in general (20).
Synteny and Conservation of the M. lepromatosis and M. leprae Genomes.
Ninety-four percent of the M. lepromatosis genome assembly could be aligned to the M. leprae TN genome, and there appears to be near-perfect collinearity and synteny, with 92% of the genes and pseudogenes shared (Fig. 1). Details of (pseudo)genes that have been deleted are provided in Dataset S1. In M. leprae, there are tandemly arranged asparagine permease genes (ML1304c ML1305c; ansP1 ansP2), but only one of these is present in M. lepromatosis (MLPM_1304). Likewise, a duplicated cluster of four genes in M. leprae (ML1053-ML1056; ML1180c-ML1183c) is present as a single copy in M. lepromatosis (MLPM_1053-MLPM_1056). This cluster encodes a member of the PE- and PPE-protein families and the ESAT-6 proteins, EsxL and EsxK.
M. lepromatosis has intact orthologs for 95% of the CDS present in M. leprae, but a further 132 CDS appear to have been pseudogenized in M. lepromatosis (Table S2). It is noteworthy that four of them (ilvX, proA, cysE, cysK) once encoded enzymes required for amino acid biosynthesis. Twenty-six M. leprae pseudogenes appear to have functional counterparts in M. lepromatosis (Table S3).
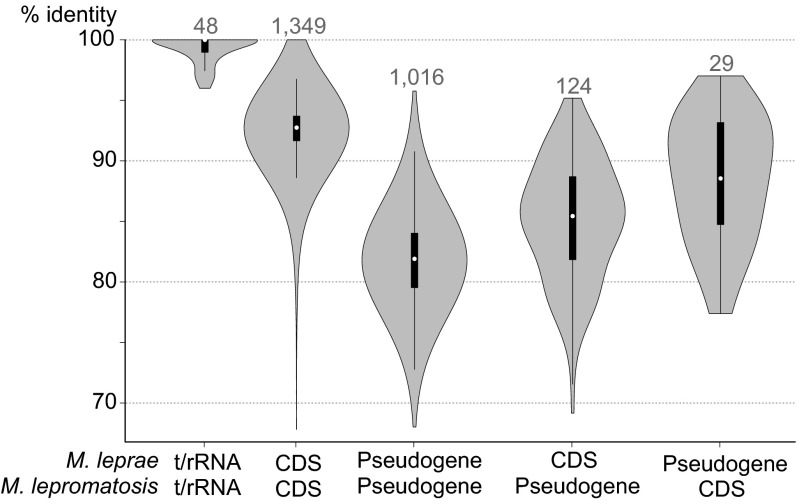
Levels of nucleotide sequence conservation for orthologous CDS and pseudogenes from M. lepromatosis and M. leprae varied between the functional categories, as may be seen from the violin plot (Fig. 2). The most conserved genes code for the rRNA and tRNA (sequence identity >95%), whereas the least conserved were those for the PE/PPE proteins, as these display the hallmarks of selective pressure (as detailed later). On average, CDS shared 93% nucleotide sequence identity between the two species, but this value was only 82% for the pseudogenes. A wide distribution of sequence conservation was seen among pseudogenes (Fig. 2), and this may reflect their respective dates of pseudogenization, as older pseudogenes will have had longer to diverge.
Fig. 2.
Nucleotide sequence identity between M. leprae and M. lepromatosis orthologs. Numbers above violins represent the number of ortholog pairs for that gene category. The most conserved are tRNA and rRNA genes, followed by the CDS. Pseudogenes are the least conserved. Identity of genes that are functional in one species but not in the other is between CDS and pseudogenes.
Species-Specific Sequences in M. lepromatosis and M. leprae.
We identified 84 genomic regions of M. lepromatosis larger than 500 nt that have no counterparts in M. leprae, accounting for a total of 166 kb, or ∼5% of the genome. These regions (range, 0.5–9.6 kb in size) consist essentially of pseudogenes (n = 163) except for three intact coding sequences: a hypothetical gene (MLPM_5094), a putative lipoprotein gene (MLPM_5098), and coproporphyrinogen III oxidase (hemN) (discussed later). Truncated remnants of 57 of these 163 pseudogenes remain in M. leprae, revealing that some of the reductive evolution in this leprosy bacillus stemmed from deletions within, or encompassing, pseudogenes since divergence from the MRCA.
Similarly, M. leprae has a total of 199 kb of sequences (>500 nt) in its genome with no counterpart in M. lepromatosis. These comprise 172 pseudogenes, of which 54 were partially present in M. lepromatosis, and 24 protein-coding genes, all annotated as encoding hypothetical or conserved hypothetical proteins, except for ML0398c (possible d-ribose–binding protein), which is truncated in M. lepromatosis.
Horizontally Acquired Genes.
Several M. leprae genes have no orthologs in other mycobacteria and appear to have been acquired by horizontal gene transfer. Among those with predicted functions are proS, encoding a eukaryotic-like prolyl tRNA synthetase, and ML2177, coding for a uridine phosphorylase that shows similarity to insect enzymes. Both these genes are conserved in sequence and location in M. lepromatosis, indicating that they were present in the MRCA. Likewise, there are two pseudogenes (MLPM_5100 and MLPM_5101 encoding a β-lactamase and LysR family transcriptional regulator) in a genomic island restricted to M. lepromatosis that appears to have been horizontally acquired by the MRCA and then lost by M. leprae.
Insight into Pathogenesis.
The higher morbidity and mortality reported to be associated with infection by M. lepromatosis and the resulting DLL suggest the presence of new virulence functions possibly borne by plasmids. For example, the pathogenesis of Mycobacterium ulcerans has been attributed to the horizontally acquired virulence plasmid encoding the mycolactone toxin (35, 36). However, despite intensive investigation of Illumina sequence reads with no matches in M. leprae and database searches, we found no evidence for plasmid or bacteriophage sequences.
To cope with iron limitation, intracellular pathogens often scavenge heme from host tissue or produce and release siderophores, such as mycobactin, to capture iron (37, 38). Both leprosy bacilli have retained the ESX-3 gene cluster that is involved in iron and zinc uptake in M. tuberculosis. The mycobactin (mbt) gene cluster is essential for the in vivo growth and virulence of M. tuberculosis (39, 40), but this cluster is missing from M. lepromatosis and M. leprae. In M. tuberculosis, hemN is located downstream of the mbt cluster, and, with the hemABCDEKLYZ genes, is required for heme biosynthesis. Interestingly, the hemN gene is present in M. lepromatosis but not in M. leprae (Fig. 3), indicating its loss occurred after separation from the MRCA and suggesting that M. leprae may be limited for heme production.
Fig. 3.
Synteny around the M. lepromatosis hemN locus and comparison with M. leprae and M. tuberculosis. Links between the genomes are BLAST hits; hemN and sirA are present in M. lepromatosis and M. tuberculosis (gray links) but have been deleted in M. leprae. The mbt cluster, present in M. tuberculosis, is deleted in M. leprae and M. lepromatosis. Scale is in kilobases. Note the genomic rearrangement between M. lepromatosis (first 10 kb of the contig) and M. leprae. Additionally, the sequence corresponding to the M. leprae genome between 780 and 795 kb is almost entirely deleted in M. lepromatosis.
Inspection of the genes least conserved between M. lepromatosis and M. leprae (Fig. 2) revealed that these correspond mainly to members of the PE and PPE protein families, characteristic of pathogenic mycobacteria, and to the ESX (type 7) protein secretion systems. Compared with M. tuberculosis and Mycobacterium marinum, there are very few PE and PPE proteins in leprosy bacilli. In M. leprae, ML0411 encodes a PPE protein that acts as an immunodominant serine-rich antigen, whereas the neighboring gene, ML0410, codes for a PE family protein. Both genes are present in M. lepromatosis but have diverged extensively: there is only 68% nucleotide sequence identity between MLPM_0411 and ML0411 (note that only half the genes could be aligned) and 73% between MLPM_0410 and ML0410. Interestingly, sequence comparison of ML0411 in a range of isolates of M. leprae of different geographical origin revealed this to be the most polymorphic gene in the genome, with the highest number of nonsynonymous substitutions (20, 41). It appears that the marked divergence in this locus is the consequence of selective pressure imparted by the host’s immune system (20).
The ability to invade the endothelium distinguishes M. lepromatosis from M. leprae. Because we found no evidence for the presence of a novel virulence gene that could account for this phenotype, we examined the gene clusters required to produce the five type 7 ESX secretion systems (T7Ss) for unusual features. Although not essential for growth in vitro, the ESX-1 system is the major virulence determinant in M. tuberculosis and M. marinum. ESX-1 mediates escape of the bacilli from the phagosome, thus allowing further replication, cytolysis, necrosis, and intercellular spread (42). The EsxA protein, a major substrate of ESX-1, ruptures the phagosomal membrane, thus acting as a principal virulence factor. Despite extensive conservation of the ESX-1 system among other mycobacterial pathogens, several of its genes encoding ESX-1 secreted proteins (Esp, Esx) are missing or nonfunctional in M. leprae and M. lepromatosis. These include espE, espB, espF, espG1, espH, espJ, espK, and pe35 in both, and, additionally, ppe68 in M. lepromatosis. Of the remaining genes in this locus, esxB, esxA, and espH are the least conserved (69–73% protein identity) between M. leprae and M. lepromatosis, and a similar trend was observed for the unlinked espACD operon (espA, 78%; espC, 77%; and espD, 86% protein identity) that regulates the expression and secretion of EsxA in a mutually dependent manner in M. tuberculosis (43–45).
Of the four other T7Ss, ESX-2 and ESX-4 are predicted to be nonfunctional in M. leprae and M. lepromatosis, whereas the ESX-5 locus is highly conserved. However, compared with M. tuberculosis, only the ESX-5 core genes (eccABCDE5 and mycP5) remain intact in both leprosy bacilli, there are no esx genes, and the PE protein gene (ML2534c/MLPM_2534) is nonfunctional. On the contrary, ESX-3 is the most conserved T7S system in mycobacteria and seems to fulfill an essential function in metal homeostasis, although its role in virulence is less clear (45).
Neuropathogenesis.
The ability to invade the Schwann cells of the peripheral nervous system is a hallmark of M. leprae, and this leads to the neuropathy and nerve damage associated with leprosy. Adherence to Schwann cells (46) has been proposed to be mediated by two cell wall components: the laminin-binding protein (ML1683c) and the terminal trisaccharide moiety of phenolic glycolipid 1 (PGL-1). To produce the trisaccharide, several enzymes are required, namely a rhamnosyl transferase (ML0128), a glucosyltransferase (ML2348), and four methyltransferases (ML0126, ML0127, ML23246c, and ML2347) (47). The genes encoding both these adhesin systems are highly conserved in M. lepromatosis, so invasion of Schwann cells is to be expected. Given the paucity of well-defined cases of infection with M. lepromatosis, studies of nerve involvement have not yet been conducted, but this will be facilitated by the tools arising from our investigation of the M. lepromatosis genome.
Disease Management and New Interventions.
Until very recently, recognition of M. lepromatosis as a separate species was questioned. Currently, M. lepromatosis can be identified by a nested PCR technique that targets the 16S rDNA (22). Given the 98% identity of the M. lepromatosis and M. leprae 16S rDNA sequences, a possible source of confusion, it may be advisable to establish a new method of identification that exploits sequences confined to M. lepromatosis such as the species-specific PCR templates and primers described here (Table 1 and Table S4). Immunodiagnostic approaches for leprosy are being pursued by using the highly specific trisaccharide from PGL-1 and LID1 (48), a fusion protein that includes sequences from genes ML0405 (espA) and ML2331. PGL-1-based tests should also detect M. lepromatosis, but tests involving LID-1 may be less sensitive because of the extensive variation in EspA reported earlier.
Table 1.
Geographical survey of leprosy bacilli by differential PCR analysis
| Country of origin | Sample size (suspicion of DLL/Lucio's) | M. lepromatosis | M. leprae |
| Venezuela | 77 | 0 | 77 |
| Mexico | 64 (4) | 6 | 58 |
| Mali | 48 | 0 | 48 |
| Brazil | 33 | 0 | 33 |
| Others | 5 (2) | 0 | 5 |
PCR was performed using primers specific for each species: LPM244F and LPM44R for M. lepromatosis; RLEP-7 and RLEP-8 for M. leprae. Full details are provided in SI Materials and Methods.
DLL cases resulting from infection with M. lepromatosis have responded favorably to the standard MDT (49), and, on examination of the drug resistance determining regions in the genes coding for the targets of rifampin, dapsone, and fluoroquinolones, only drug-susceptible sequences were found. New drugs developed to treat tuberculosis may also find application in leprosy. Inspection of the M. lepromatosis and M. leprae gene sequences for the targets of the experimental drugs bedaquiline, benzothiazinones, and Q203 suggest that these should be active, whereas the nitroimidazole prodrugs PA-824, TBA354, and delamanid will not be effective because the ddn gene, coding for the nitroreductase required for their activation, is missing.
Geographical Survey for M. lepromatosis.
To gain more insight into the global distribution of M. lepromatosis, a differential PCR test was implemented by using species-specific primers targeting hemN and RLEP (SI Materials and Methods). A total of 227 specimens from patients with leprosy were chosen (Table 1), including some with a history or suspicion of DLL or Lucio's reaction (n = 6). The largest patient groups were from Venezuela (n = 77), Mexico (n = 64), Mali (n = 48), and Brazil (n = 33); a small number of samples suspected of presenting with Lucio's reaction were obtained from elsewhere (Table 1). This analysis revealed the presence of M. leprae DNA in 221 cases and M. lepromatosis in only six, with no evidence for mixed infections. All six M. lepromatosis cases were of Mexican origin. Among the samples from DLL or Lucio's reaction cases, two were positive for M. lepromatosis and four for M. leprae.
On the Origin and Evolution of M. lepromatosis.
The results from the genome-wide comparison of M. leprae and M. lepromatosis indicate that pseudogenization took place in their ancestral forms and that the MRCA itself likely had a genome of reduced size compared with all other known mycobacteria. Based on the rate of nonsynonymous substitution in pseudogenes, it was estimated that a single massive pseudogenization event took place approximately 20 Mya (50). Since their separation from the MRCA, deletions and more pseudogenes have appeared in both species. M. lepromatosis shares the same repeat families with M. leprae, including their genomic locations, but these have diverged extensively in sequence. In M. leprae, genome reduction and gene truncation have been attributed to recombinational events between different repeat copies, but these events likely occurred in the MRCA (17, 33).
To estimate the divergence times of M. lepromatosis and the currently available M. leprae strains, substitution rates were calculated in a Bayesian framework by using the software package BEAST (Bayesian Evolutionary Analysis Sampling Trees). For phylogenetic and divergence time analysis, the M. lepromatosis genome was aligned with 18 modern and ancient M. leprae genomes. On average, there were 90 nucleotide substitutions between two M. leprae strains and 275,518 substitutions between M. leprae and M. lepromatosis. A substitution rate of 7.67−9 substitutions per site per year was estimated (Fig. S2A), similar to previous estimates using M. leprae genomes only (20). The resulting divergence time from the MRCA (TMRCA) for all M. leprae strains was calculated as 3,607 y ago [2,204–5,525 y ago 95% highest probability density (HPD)], comparable to previous results (20) (Fig. S2B). The TMRCA for M. leprae and M. lepromatosis was estimated to be 13.9 Mya (8.2–21.4 Mya 95% HPD; Fig. S2C). In this respect, the two leprosy bacilli differ quite markedly from the species comprising the M. tuberculosis complex (MTBC), as, from a recent paleomicrobiogical investigation that used two independent dating approaches, it was concluded that the maximal TMRCA was <6,000 y for the MTBC (51).
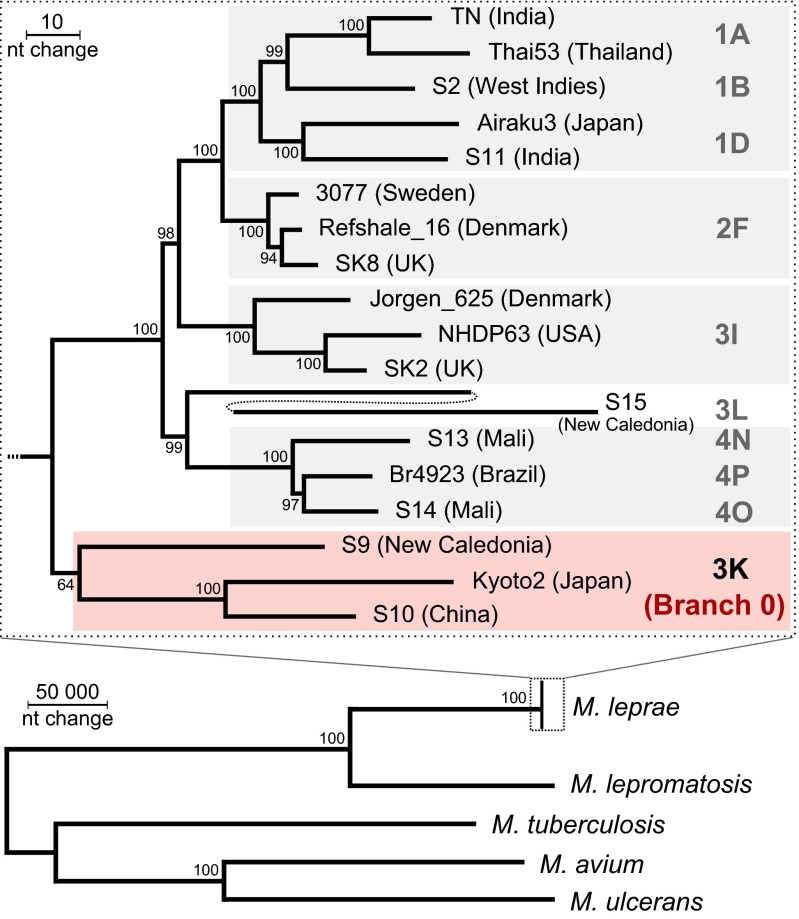
A phylogenetic comparison by maximum parsimony of M. lepromatosis with various M. leprae strains of different geographic origins and SNP subtypes is shown in Fig. 4 and by neighbor joining in Fig. S3. To gain further insight into the phylogenetic placement of M. lepromatosis among other mycobacterial species, additional phylogenetic trees were generated based on the concatenated amino acid alignments of GyrB, RpoB, and RpoC (Fig. S4), as well as the alignments of the 16S genes (Fig. S5). In all four phylogenetic trees, M. lepromatosis is positioned between the mycobacteria used as outgroups and M. leprae. The M. lepromatosis branch is closest to the M. leprae SNP-type 3K strains, consistent with the very recent report of the 3K strains (branch 0) being the most ancestral lineage of M. leprae known to date (20). The geographic distribution of type 3K strains is diverse, but sampling is insufficiently broad to predict a tentative origin. On the contrary, the predominance of reported DLL and confirmed cases of infection with M. lepromatosis in Mexico suggest that this pathogen may have evolved in Central America. More extensive investigation is required to explore this possibility and to retrace its origin.
Fig. 4.
Phylogeny of M. lepromatosis and M. leprae strains. Phylogenetic relationship of M. lepromatosis genome using a maximum parsimony tree, including M. tuberculosis, M. avium, and M. ulcerans as outgroups. SNP type is given at branch tips. Bootstrap support is indicated for each node. The long branch of S15 was split to reduce space.
Environmental Sources.
M. leprae has long been considered an obligate human pathogen with its primary route of transmission being interhuman. In the past 30 y, the nine-banded armadillo has been identified as a natural reservoir of M. leprae in the southern United States, and evidence for zoonotic transmission to humans is accumulating (34). The existence of nonhuman hosts or natural reservoirs of M. lepromatosis has not yet been investigated, but, recently, an apparent outbreak of M. lepromatosis infection in red squirrels (Sciurus vulgaris) was reported in the United Kingdom (52). Examination of infected squirrel tissue using the PCR-based sequencing procedures for the hemN gene outlined here has confirmed this finding and thus provides evidence for a nonhuman reservoir or intermediate host for M. lepromatosis. To date, the most convincing human cases of M. lepromatosis infection, particularly those with DLL, came from Mexico. One of the regional culinary traditions in rural Mexico is consumption of field rats (Rattus rattus) (53). Because M. lepromatosis seems to naturally infect rodents, such as squirrels, it is conceivable that field rats are a host and may serve as a disease reservoir. When a retrospective survey of confirmed cases of M. lepromatosis was conducted, two of the six Mexican subjects admitted having consumed meat from field rats. Availability of the M. lepromatosis genome sequence will enable us to search systematically for its presence in other potential animal reservoirs as well as in extant cases of leprosy. As a result, deeper understanding will be obtained of the incidence, etiology, epidemiology, clinical features, and pathogenesis of leprosy caused by M. lepromatosis.
Materials and Methods
DNA Extraction and Sequencing.
DNA was extracted from the biopsy specimen Mx1-22A (32), and the Illumina library was prepared as described elsewhere (20). To overcome the problem of the high level of host DNA in the library, we used two methods for enrichment of M. lepromatosis DNA: (i) whole-genome array capture using an M. leprae tiling array (20) and (ii) removal of human DNA by hybridization with a human genomic DNA bait library.
Genome Assembly and Annotation.
Reads mapping to the human genome were discarded and duplicate reads were removed before the assembly. De novo genome assembly was done in MIRA version 4.0rc4 (54). Contigs were anchored to the M. leprae TN genome, and those contigs that did not match were screened for contaminants and the presence of mycobacterial genes. Genome sequence synteny between M. lepromatosis and M. leprae was visualized in ACT (Artemis Comparison Tool) (55) (Fig. 1).
PCR Procedures.
To confirm the presence of the hemN gene and its flanking sequence, including a genomic rearrangement, we designed PCR primers (Table S4) to amplify overlapping genomic targets. Details of the procedures used for the geographical survey are provided in SI Materials and Methods.
SNP Calling and Phylogeny.
Genome sequences of Mycobacterium avium K10, M. lepromatosis, and the published M. leprae strains were aligned against the M. leprae TN genome. SNP alignments were analyzed in MEGA6 (Molecular Evolutionary Genetics Analysis version 6.0) (56). Further details of bioinformatics procedures, Bayesian dating analysis, and phylogeny are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Keith Harshman, Johann Weber, Richard Truman, Rahul Sharma, and Masanori Kai for helpful discussions and technical assistance. This work was supported by grants from the Fondation Raoul Follereau and the Swiss National Science Foundation (Brazilian Swiss Joint Research Program).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.B. is a guest editor invited by the Editorial Board.
All raw read files have been deposited in the trace archive of the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP047206). The de novo assembly of M. lepromatosis genome has been deposited at DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank (accession no. JRPY01000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421504112/-/DCSupplemental.
References
- 1.World Health Organization Global leprosy: Update on the 2012 situation. Wkly Epidemiol Rec. 2013;88(35):365–379. [PubMed] [Google Scholar]
- 2.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34(3):255–273. [PubMed] [Google Scholar]
- 3.Britton WJ, Lockwood DN. Leprosy. Lancet. 2004;363(9416):1209–1219. doi: 10.1016/S0140-6736(04)15952-7. [DOI] [PubMed] [Google Scholar]
- 4.Scollard DM, et al. The continuing challenges of leprosy. Clin Microbiol Rev. 2006;19(2):338–381. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucio R, Alsarado Y. Opusculo sobre el mal de San Làzaro, ó elefanciasis de los griegos, escrito por los profesores de medicina y cirugia. Imprento de M. Murguia y Compañia; Mexico City, Mexico: 1852. [Google Scholar]
- 6.Latapi F, Chevez-Zamora A. The “spotted” leprosy of Lucio: An introduction to its clinical and histological study. Int J Lepr. 1948;16:421–437. [Google Scholar]
- 7.Rea TH. Lucio’s phenomenon: An overview. Lepr Rev. 1979;50(2):107–112. doi: 10.5935/0305-7518.19790016. [DOI] [PubMed] [Google Scholar]
- 8.Rea TH, Jerskey RS. Clinical and histologic variations among thirty patients with Lucio’s phenomenon and pure and primitive diffuse lepromatosis (Latapi’s lepromatosis) Int J Lepr Other Mycobact Dis. 2005;73(3):169–188. [PubMed] [Google Scholar]
- 9.Moschella SL. The lepra reaction with necrotizing skin lesions. A report of six cases. Arch Dermatol. 1967;95(6):565–575. [PubMed] [Google Scholar]
- 10.Romero A, Brenes, Ibarra A, Fallas M. Clinical study of lepromatous leprosy in Costa Rica. Int J Lepr. 1949;17(1-2):27–33. [PubMed] [Google Scholar]
- 11.Vargas-Ocampo F. Diffuse leprosy of Lucio and Latapí: A histologic study. Lepr Rev. 2007;78(3):248–260. [PubMed] [Google Scholar]
- 12.Sehgal VN. Lucio’s phenomenon/erythema necroticans. Int J Dermatol. 2005;44(7):602–605. doi: 10.1111/j.1365-4632.2005.02567.x. [DOI] [PubMed] [Google Scholar]
- 13.Ang P, Tay Y-K, Ng S-K, Seow C-S. Fatal Lucio’s phenomenon in 2 patients with previously undiagnosed leprosy. J Am Acad Dermatol. 2003;48(6):958–961. doi: 10.1067/mjd.2003.326. [DOI] [PubMed] [Google Scholar]
- 14.Peixoto AB, Portela PS, Leal FRP de C, Brotas AM, Rodrigues NC dos S. Lucio’s phenomenon. Case study of an exceptional response to treatment exclusively with multibacillary multidrug therapy. An Bras Dermatol. 2013;88(6) Suppl 1:93–96. doi: 10.1590/abd1806-4841.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization (2014) WHO Recommended MDT Regimens. Available at www.who.int/lep/mdt/regimens/en/. Accessed September 24, 2014.
- 16.Helmer KA, Fleischfresser I, Kucharski-Esmanhoto LD, Fillus Neto J, Santamaria JR. The Lucio’s phenomenon (necrotizing erythema) in pregnancy. An Bras Dermatol. 2004;79(2):205–210. [Google Scholar]
- 17.Cole ST, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409(6823):1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 18.Williams DL, Gillis TP, Portaels F. Geographically distinct isolates of Mycobacterium leprae exhibit no genotypic diversity by restriction fragment-length polymorphism analysis. Mol Microbiol. 1990;4(10):1653–1659. doi: 10.1111/j.1365-2958.1990.tb00542.x. [DOI] [PubMed] [Google Scholar]
- 19.Monot M, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41(12):1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 20.Schuenemann VJ, et al. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science. 2013;341(6142):179–183. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- 21.Han XY, et al. A new Mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol. 2008;130(6):856–864. doi: 10.1309/AJCPP72FJZZRRVMM. [DOI] [PubMed] [Google Scholar]
- 22.Han XY, Sizer KC, Velarde-Félix JS, Frias-Castro LO, Vargas-Ocampo F. The leprosy agents Mycobacterium lepromatosis and Mycobacterium leprae in Mexico. Int J Dermatol. 2012;51(8):952–959. doi: 10.1111/j.1365-4632.2011.05414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han XY, Sizer KC, Tan H-H. Identification of the leprosy agent Mycobacterium lepromatosis in Singapore. J Drugs Dermatol. 2012;11(2):168–172. [PubMed] [Google Scholar]
- 24.Jessamine PG, et al. Leprosy-like illness in a patient with Mycobacterium lepromatosis from Ontario, Canada. J Drugs Dermatol. 2012;11(2):229–233. [PubMed] [Google Scholar]
- 25.Monteiro R, et al. Lucio’s phenomenon: Another case reported in Brazil. An Bras Dermatol. 2012;87(2):296–300. doi: 10.1590/s0365-05962012000200017. [DOI] [PubMed] [Google Scholar]
- 26.Souza CS, Roselino AM, Figueiredo F, Foss NT. Lucio’s phenomenon: Clinical and therapeutic aspects. Int J Lepr Other Mycobact Dis. 2000;68(4):417–425. [PubMed] [Google Scholar]
- 27.Saoji V, Salodkar A. Lucio leprosy with lucio phenomenon. Indian J Lepr. 2001;73(3):267–272. [PubMed] [Google Scholar]
- 28.Thappa DM, Karthikeyan K, Kumar BJ. Is it Lucio leprosy with Lucio phenomenon or something else? Indian J Lepr. 2002;74(2):161–166. [PubMed] [Google Scholar]
- 29.Kumari R, Thappa DM, Basu D. A fatal case of Lucio phenomenon from India. Dermatol Online J. 2008;14(2):10. [PubMed] [Google Scholar]
- 30.Golchai J, Zargari O, Maboodi A, Maboodi A, Granmayeh S. Lepromatous leprosy with extensive unusual ulcerations and cachexia. Is it the first case of Lucio’s phenomenon from Iran? Int J Lepr Other Mycobact Dis. 2004;72(1):56–59. doi: 10.1489/1544-581X(2004)072<0056:LLWEUU>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 31.Choon SE, Tey KE. Lucio’s phenomenon: A report of three cases seen in Johor, Malaysia. Int J Dermatol. 2009;48(9):984–988. doi: 10.1111/j.1365-4632.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 32.Vera-Cabrera L, et al. Case of diffuse lepromatous leprosy associated with “Mycobacterium lepromatosis”. J Clin Microbiol. 2011;49(12):4366–4368. doi: 10.1128/JCM.05634-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole ST, Supply P, Honoré N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr Rev. 2001;72(4):449–461. [PubMed] [Google Scholar]
- 34.Truman RW, et al. Probable zoonotic leprosy in the southern United States. N Engl J Med. 2011;364(17):1626–1633. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stinear TP, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17(2):192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demangel C, Stinear TP, Cole ST. Buruli ulcer: Reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat Rev Microbiol. 2009;7(1):50–60. doi: 10.1038/nrmicro2077. [DOI] [PubMed] [Google Scholar]
- 37.Chu BC, et al. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals. 2010;23(4):601–611. doi: 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- 38.Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15(2):328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 39.De Voss JJ, et al. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci USA. 2000;97(3):1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy PV, et al. Disruption of mycobactin biosynthesis leads to attenuation of Mycobacterium tuberculosis for growth and virulence. J Infect Dis. 2013;208(8):1255–1265. doi: 10.1093/infdis/jit250. [DOI] [PubMed] [Google Scholar]
- 41.Kai M, et al. Characteristic mutations found in the ML0411 gene of Mycobacterium leprae isolated in Northeast Asian countries. Infect Genet Evol. 2013;19:200–204. doi: 10.1016/j.meegid.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 42.Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8(2):e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortune SM, et al. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USA. 2005;102(30):10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garces A, et al. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS Pathog. 2010;6(6):e1000957. doi: 10.1371/journal.ppat.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoop EJM, et al. Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis Model Mech. 2011;4(4):526–536. doi: 10.1242/dmm.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rambukkana A. Molecular basis for the peripheral nerve predilection of Mycobacterium leprae. Curr Opin Microbiol. 2001;4(1):21–27. doi: 10.1016/s1369-5274(00)00159-4. [DOI] [PubMed] [Google Scholar]
- 47.Tabouret G, et al. Mycobacterium leprae phenolglycolipid-1 expressed by engineered M. bovis BCG modulates early interaction with human phagocytes. PLoS Pathog. 2010;6(10):e1001159. doi: 10.1371/journal.ppat.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Souza MM, Netto EM, Nakatani M, Duthie MS. Utility of recombinant proteins LID-1 and PADL in screening for Mycobacterium leprae infection and leprosy. Trans R Soc Trop Med Hyg. 2014;108(8):495–501. doi: 10.1093/trstmh/tru093. [DOI] [PubMed] [Google Scholar]
- 49.Han XY, Jessurun J. Severe leprosy reactions due to Mycobacterium lepromatosis. Am J Med Sci. 2013;345(1):65–69. doi: 10.1097/MAJ.0b013e31826af5fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han XY, et al. Comparative sequence analysis of Mycobacterium leprae and the new leprosy-causing Mycobacterium lepromatosis. J Bacteriol. 2009;191(19):6067–6074. doi: 10.1128/JB.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514(7523):494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meredith A, et al. Leprosy in red squirrels in Scotland. Vet Rec. 2014;175(11):285–286. doi: 10.1136/vr.g5680. [DOI] [PubMed] [Google Scholar]
- 53. Chacon R (October 8, 1999) In Mexico, field rats are secret delicacy. Miami Herald. Available at www.latinamericanstudies.org/mexico/rats.htm. Accessed October 16, 2014.
- 54.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB), Vol 99, pp 45–56. Available at www.bioinfo.de/isb/gcb99/talks/chevreux/main.html.
- 55.Carver TJ, et al. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21(16):3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.