Abstract
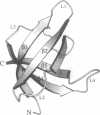
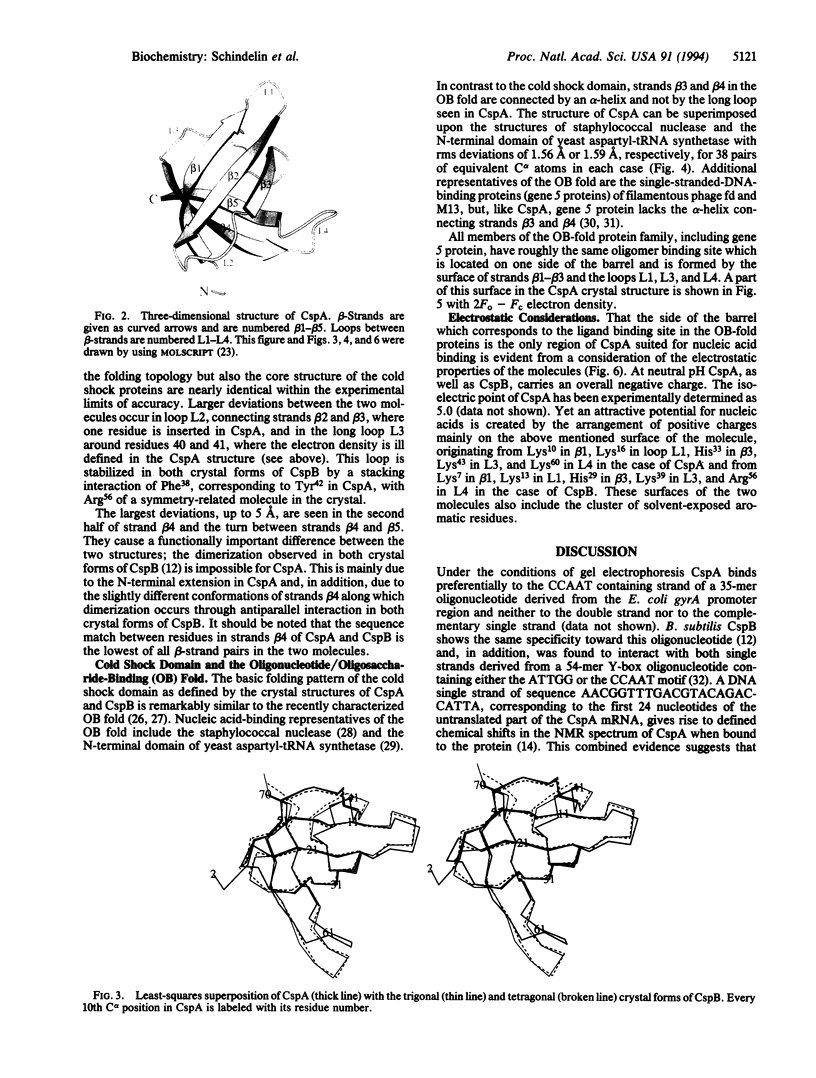
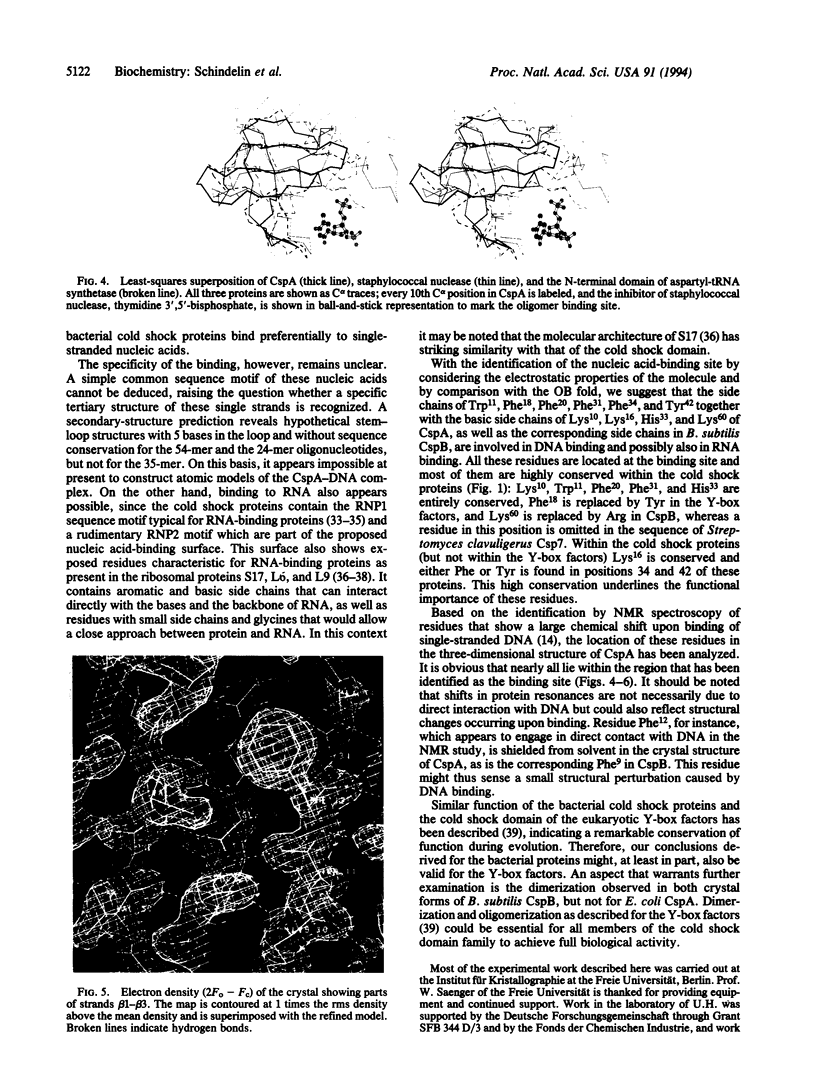
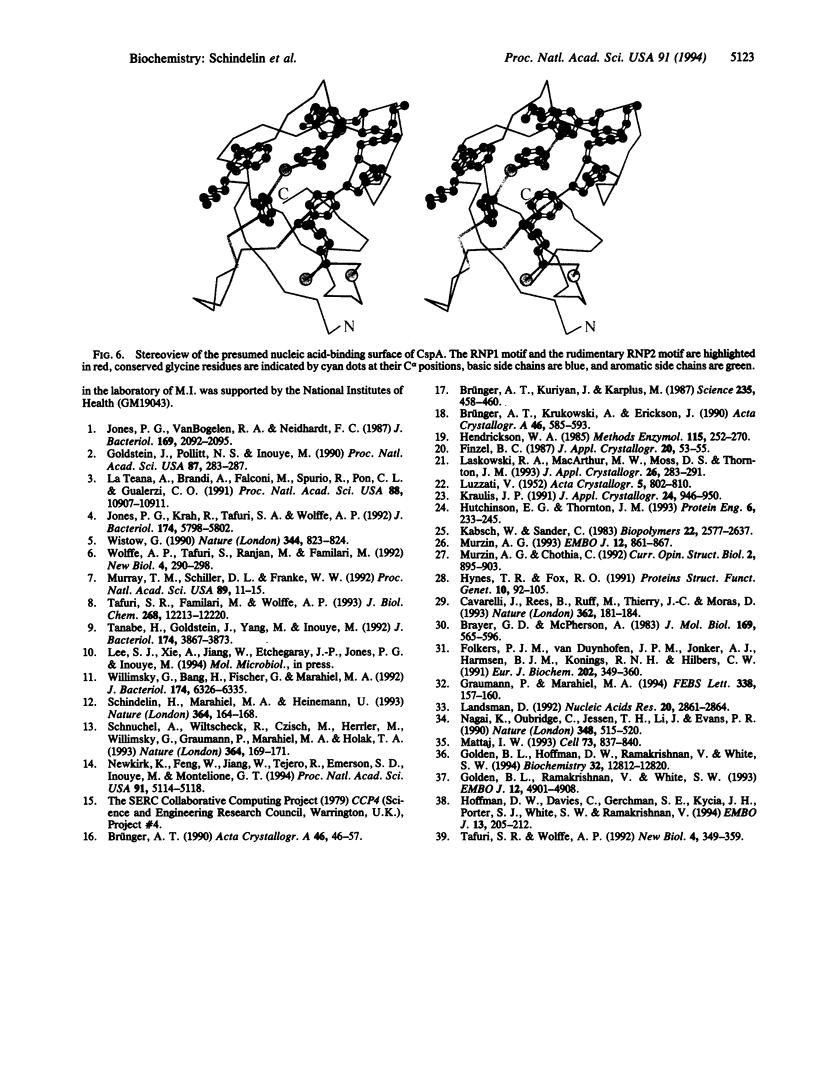
The major cold shock protein of Escherichia coli, CspA, produced upon a rapid downshift in growth temperature, is involved in the transcriptional regulation of at least two genes. The protein shares high homology with the nucleic acid-binding domain of the Y-box factors, a family of eukaryotic proteins involved in transcriptional and translational regulation. The crystal structure of CspA has been determined at 2-A resolution and refined to R = 0.187. CspA is composed of five antiparallel beta-strands forming a closed five-stranded beta-barrel. The three-dimensional structure of CspA is similar to that of the major cold shock protein of Bacillus subtilis, CspB, which has recently been determined at 2.45-A resolution. However, in contrast to CspB, no dimer is formed in the crystal. The surface of CspA is characteristic for a protein interacting with single-stranded nucleic acids. Due to the high homology of the bacterial cold shock proteins with the Y-box factors, E. coli CspA and B. subtilis CspB define a structural framework for the common cold shock domain.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brayer G. D., McPherson A. Refined structure of the gene 5 DNA binding protein from bacteriophage fd. J Mol Biol. 1983 Sep 15;169(2):565–596. doi: 10.1016/s0022-2836(83)80065-5. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Krukowski A., Erickson J. W. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr A. 1990 Jul 1;46(Pt 7):585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Folkers P. J., van Duynhoven J. P., Jonker A. J., Harmsen B. J., Konings R. N., Hilbers C. W. Sequence-specific 1H-NMR assignment and secondary structure of the Tyr41----His mutant of the single-stranded DNA binding protein, gene V protein, encoded by the filamentous bacteriophage M13. Eur J Biochem. 1991 Dec 5;202(2):349–360. doi: 10.1111/j.1432-1033.1991.tb16382.x. [DOI] [PubMed] [Google Scholar]
- Golden B. L., Hoffman D. W., Ramakrishnan V., White S. W. Ribosomal protein S17: characterization of the three-dimensional structure by 1H and 15N NMR. Biochemistry. 1993 Nov 30;32(47):12812–12820. doi: 10.1021/bi00210a033. [DOI] [PubMed] [Google Scholar]
- Golden B. L., Ramakrishnan V., White S. W. Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein. EMBO J. 1993 Dec 15;12(13):4901–4908. doi: 10.1002/j.1460-2075.1993.tb06184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J., Pollitt N. S., Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jan;87(1):283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P., Marahiel M. A. The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG- and CCAAT sequences in single stranded oligonucleotides. FEBS Lett. 1994 Jan 31;338(2):157–160. doi: 10.1016/0014-5793(94)80355-2. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Hoffman D. W., Davies C., Gerchman S. E., Kycia J. H., Porter S. J., White S. W., Ramakrishnan V. Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein. EMBO J. 1994 Jan 1;13(1):205–212. doi: 10.1002/j.1460-2075.1994.tb06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson E. G., Thornton J. M. The Greek key motif: extraction, classification and analysis. Protein Eng. 1993 Apr;6(3):233–245. doi: 10.1093/protein/6.3.233. [DOI] [PubMed] [Google Scholar]
- Hynes T. R., Fox R. O. The crystal structure of staphylococcal nuclease refined at 1.7 A resolution. Proteins. 1991;10(2):92–105. doi: 10.1002/prot.340100203. [DOI] [PubMed] [Google Scholar]
- Jones P. G., Krah R., Tafuri S. R., Wolffe A. P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992 Sep;174(18):5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. G., VanBogelen R. A., Neidhardt F. C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987 May;169(5):2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- La Teana A., Brandi A., Falconi M., Spurio R., Pon C. L., Gualerzi C. O. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman D. RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 1992 Jun 11;20(11):2861–2864. doi: 10.1093/nar/20.11.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. RNA recognition: a family matter? Cell. 1993 Jun 4;73(5):837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- Murray M. T., Schiller D. L., Franke W. W. Sequence analysis of cytoplasmic mRNA-binding proteins of Xenopus oocytes identifies a family of RNA-binding proteins. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):11–15. doi: 10.1073/pnas.89.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A. G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993 Mar;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990 Dec 6;348(6301):515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Newkirk K., Feng W., Jiang W., Tejero R., Emerson S. D., Inouye M., Montelione G. T. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5114–5118. doi: 10.1073/pnas.91.11.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin H., Marahiel M. A., Heinemann U. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature. 1993 Jul 8;364(6433):164–168. doi: 10.1038/364164a0. [DOI] [PubMed] [Google Scholar]
- Schnuchel A., Wiltscheck R., Czisch M., Herrler M., Willimsky G., Graumann P., Marahiel M. A., Holak T. A. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature. 1993 Jul 8;364(6433):169–171. doi: 10.1038/364169a0. [DOI] [PubMed] [Google Scholar]
- Tafuri S. R., Familari M., Wolffe A. P. A mouse Y box protein, MSY1, is associated with paternal mRNA in spermatocytes. J Biol Chem. 1993 Jun 5;268(16):12213–12220. [PubMed] [Google Scholar]
- Tafuri S. R., Wolffe A. P. DNA binding, multimerization, and transcription stimulation by the Xenopus Y box proteins in vitro. New Biol. 1992 Apr;4(4):349–359. [PubMed] [Google Scholar]
- Tanabe H., Goldstein J., Yang M., Inouye M. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J Bacteriol. 1992 Jun;174(12):3867–3873. doi: 10.1128/jb.174.12.3867-3873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willimsky G., Bang H., Fischer G., Marahiel M. A. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J Bacteriol. 1992 Oct;174(20):6326–6335. doi: 10.1128/jb.174.20.6326-6335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistow G. Cold shock and DNA binding. Nature. 1990 Apr 26;344(6269):823–824. doi: 10.1038/344823c0. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Tafuri S., Ranjan M., Familari M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 1992 Apr;4(4):290–298. [PubMed] [Google Scholar]