Significance
All organisms contain transposable DNA elements (TEs) that can be seriously deleterious. The dominant TE in mammals, the L1 (LINE-1) retrotransposon, has generated approximately 40% of the genome. L1 encodes two proteins, ORF1p and ORF2p, that are required for L1 retrotransposition. We show here that phosphorylation of ORF1p is required for this process. These results significantly advance our understanding of retrotransposition and indicate that L1 activity is integrated with, and thus potentially can perturb, host cellular signaling pathways. Thus, the effects of L1 may extend well beyond those of genome alteration as is currently thought.
Keywords: proline-directed protein kinase, LINE-1, peptidyl prolyl isomerase 1, retrotransposon, Pin1
Abstract
Although members of the L1 (LINE-1) clade of non-LTR retrotransposons can be deleterious, the L1 clade has remained active in most mammals for ∼100 million years and generated almost 40% of the human genome. The details of L1–host interaction are largely unknown, however. Here we report that L1 activity requires phosphorylation of the protein encoded by the L1 ORF1 (ORF1p). Critical phospho-acceptor residues (two serines and two threonines) reside in four conserved proline-directed protein kinase (PDPK) target sites. The PDPK family includes mitogen-activated protein kinases and cyclin-dependent kinases. Mutation of any PDPK phospho-acceptor inhibits L1 retrotransposition. The phosphomimetic aspartic acid can restore activity at the two serine sites, but not at either threonine site, where it is strongly inhibitory. ORF1p also contains conserved PDPK docking sites, which promote specific interaction of PDPKs with their targets. As expected, mutations in these sites also inhibit L1 activity. PDPK mutations in ORF1p that inactivate L1 have no significant effect on the ability of ORF1p to anneal RNA in vitro, an important biochemical property of the protein. We show that phosphorylated PDPK sites in ORF1p are required for an interaction with the peptidyl prolyl isomerase 1 (Pin1), a critical component of PDPK-mediated regulation. Pin1 acts via isomerization of proline side chains at phosphorylated PDPK motifs, thereby affecting substrate conformation and activity. Our demonstration that L1 activity is dependent on and integrated with cellular phosphorylation regulatory cascades significantly increases our understanding of interactions between L1 and its host.
L1 (or LINE-1) activity over the last ∼100 million years of primate evolution has generated ∼40% of the human genome (1, 2); thus, succeeding families of L1 elements are the main drivers of genetic expansion. These autonomously replicating elements convert their RNA transcripts and those of other genetic elements, particularly SINEs, into genomic DNA (3). A generic L1 element is 6–7 kb and contains the following: a 5′ UTR; ORF1, which encodes the coiled-coil mediated trimeric nucleic acid chaperone protein ORF1p; ORF2, which encodes a DNA endonuclease and reverse-transcriptase ORF2p; and a 3′ UTR terminated in a polyA sequence (reviewed in refs. 3 and 4). ORF1p, ORF2p, and L1 RNA form ribonucleoprotein particles (RNPs) that are likely intermediates in L1 retrotransposition (5–8). The L1-encoded proteins ORF1p and ORF2p are essential in cell culture-based retrotransposition assays (9) and in vitro assays using RNPs from cells transfected with L1 retrotransposition vectors (7). Although the role of ORF1p in retrotransposition is not known, mutations that affect its nucleic acid binding and chaperone activities can inactivate L1 (7, 9, 10).
L1 activity can damage DNA (11), can generate genetic diversity and rearrangements (12–16), and is activated in certain tumors (17–19) and other somatic cells (20), including neuronal cells (21–23). Despite being deleterious (24, 25), with at times catastrophic effects (26, 27), novel L1 families continue to evolve in modern mammals (15, 28–30), at least in some cases in response to evolving mammalian defensive measures (31). The existence of strong negative selection (24, 25) and robust host repressive mechanisms, which include methylation of L1 DNA (26), inhibition by APOBEC cytosine deaminases (32–35), and repression by Argonaute protein-mediated RNAi (36), support a parasitic nature of L1 elements (37); however, the overall effect of L1 on mammalian evolution and biology, and how L1 interacts with and persists in the host, remain unanswered questions.
Early reports suggested that ORF1p was phosphorylated (6, 38). Recent studies have shown that phosphorylation-related proteins can be coimmunoprecipitated with ORF1p or L1 RNPs (39, 40). In addition, the mitogen-activated protein kinase (MAPK) p38 has been implicated in L1 activation by environmental toxins (41, 42), and its expression can be increased by exogenous ORF1p (43). Although large-scale proteomic studies can only identify a subset of phospho sites (44), a phosphoproteomic study of human embryonal stem cells, in which L1 is active, identified an ORF1p fragment phosphorylated on S18 (45). This finding suggested to us that ORF1p phosphorylation might be required for L1 activity.
To investigate the role of ORF1p phosphorylation in retrotransposition, we used LC-MS/MS to determine the phosphorylation state of ORF1p purified from insect and HeLa cells and identified a total of 14 high-confidence phospho residues. Mutational analysis showed that highly conserved proline-directed protein kinase (PDPK) target sites and docking motifs are critical for L1 retrotransposition. PDPKs specifically phosphorylate serines or threonines with proline in the +1 position (S/T-P motifs) (46). Docking motifs on PDPK substrates ensure efficient kinase targeting, and phosphorylation of docking motifs by protein kinase A (PKA) can regulate PDPK binding (46, 47). Two PDPK docking motifs in ORF1p contain a predicted PKA site, and we show that mutation of either of these sites also inhibits L1 retrotransposition. Finally, we show that the serine PDPK sites in ORF1p mediate an interaction with the proline isomerase Pin1, an essential component of PDPK-mediated regulatory pathways. Pin1 binds to phosphorylated S/T-P motifs and, via proline isomerization, induces significant conformational changes, which can affect activity, stability, protein–protein interactions, phosphorylation state, and susceptibility to further posttranslational modifications (48, 49). Taken together, these results demonstrate a role for PDPK(s) in L1 retrotransposition and indicate that L1 activity is integrated into host kinase pathways, with potentially far-reaching effects on cellular function.
Results
ORF1p Is Phosphorylated on Multiple Residues.
Analysis of the primary amino acid sequence of ORF1p with ELM, the database of eukaryotic linear motifs (50), revealed a number of highly conserved kinase motifs, some of which are shown in Fig. 1. These include four S/T-P PDPK target sites: two serine sites, S18 and S27, lying in the N-terminal region, and two threonine sites, T203 and T213, in the RNA recognition motif (RRM) of ORF1p (51). ORF1p also contains multiple PDPK docking motifs, two of which contain predicted PKA sites at highly conserved T241 and T250. In addition, ORF1p contains a docking site for protein phosphatase 1 (PP1). Mouse ORF1p contains potential phosphorylation sites that correspond to positions S18, S27, S119, T203, T250, S254, and S287 in human ORF1p (Fig. 1 and Table 1), suggesting that these sites have been conserved for at least 120 million years, the estimated time at which rodents and primates diverged (52).
Fig. 1.
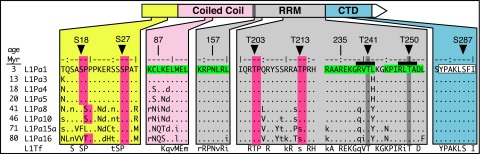
PDPK motifs are conserved in ORF1p. ELM identified in the modern L1Pa1 ORF1p consensus sequence (28): four PDPK S/T-P targets (dark-pink columns); four PDPK docking motifs approximated by (R/K)••••h•h, where • is any residue and h is a hydrophobic residue (green); within PDPK docking motifs, two PKA motifs, R•(S/T)-not-P (black bars) with the target sites T241 and T250; and a putative PP1 docking site (white). The consensus sequences of the ancestral L1Pa families and mouse L1Tf family were reported previously (30, 73, 74). RRM, RNA recognition motif (light gray); CTD, C-terminal domain (teal) (75).
Table 1.
MS/MS-detected phospho residues in ORF1p
| Residue | Insect | HeLa | L1Pa* | circa My |
| S16 | X | 6 | 27 | |
| S18† | X | X | 11 | 53 |
| S27† | X | X | 16 | 80 |
| S50 | X | X | 6 | 27 |
| S53 | X | X | 8 | 41 |
| S119† | X | 16 | 80 | |
| S145 | X | X | 8a | 42 |
| S166 | X | X | 8 | 41 |
| T203† | X | X | 16 | 80 |
| T250† | X | 16 | 80 | |
| S254† | X | 16 | 80 | |
| S281 | X | 16 | 80 | |
| S287† | X | 16 | 80 | |
| S290 | X | 16 | 80 |
X indicates phosphorylated L1Pa1 residues in peptides identified with ≥95% confidence, except T203, which was independently confirmed by manual inspection of the MS/MS spectra in HeLa cells. Only phospho residues with a rounded Mascot Delta Score ≥10 are shown, which corresponds to ≥91% confidence in the assignment of the phosphate to the listed residue, vs. ≤9% confidence for an alternate site in the peptide.
The column labeled “L1Pa” indicates the particular ancestral L1Pa family in which the indicated residue can first be detected. For example, the S16 ortholog of L1Pa1 is present in L1Pa6, which was active ∼27 Mya.
Homologs identified in mouse.
LC-MS/MS analysis of ORF1p purified from High Five insect or HeLa cells recovered peptides corresponding to ∼80% of ORF1p (SI LC-MS/MS data). Table 1 lists the high-confidence or manually confirmed phospho residues identified in ORF1p purified from each cell type. Confidence is based on two scores: the Mowse score, which ranks the confidence with which the peptide sequences match those of a given protein database (53), in our case NCBInr, and the Mascot Delta (MD) score, which ranks the confidence of phosphate assignment to a particular residue within a given peptide (54).
Three of the four PDPK target sites (S18, S27, and T203) were phosphorylated in both cell types and further verified for ORF1p-Flag by manual inspection of MS/MS spectra to confirm peptide identification and phospho site assignments (Fig. S1). Peptides containing the fourth PDPK site, T213, were either not recovered (HeLa cells) or not unambiguously identified (insect cells; SI Results) so the phosphorylation state of T213 remains unknown. Phosphorylation of only one of the two predicted PKA sites in the PDPK docking motifs—T250—was detected by LC-MS/MS, and only in insect cells.
In addition to the high-confidence sites listed in Table 1, LC-MS/MS identified other phosphorylation sites that could represent accurate assignments, as was the case with T203 (SI Results). Thus, our mutational analysis included the following actual or potential phosphorylation sites (those in bold are listed in Table 2): T14, S16, S18, S25, S26, S27, T30, S33, S50, Y52, S53, S119, S145, S166, T203, T250, S254, S281, S287, and S290, as well as T213 and T241, although the latter two were not identified as potential phosphorylation sites by LC-MS/MS.
Table 2.
Effects of ORF1p mutations on retrotransposition
| Relative colony area* | Average | ||
| PDPK target | |||
| S18A | 0.21 | 0.23 | 0.22 |
| S18D | 0.74 | 0.71 | 0.73 |
| S27A | 0.40 | 0.36 | 0.38 |
| S27D | 0.83 | 0.88 | 0.85 |
| S18A/S27A | 0.01 | 0.02 | 0.02 |
| S18D/S27D | 0.40 | 0.41 | 0.41 |
| T203G | 0.01 | 0.01 | 0.01 |
| T203S | 1.06 | 1.01 | 1.03 |
| T203D | 0.00 | 0.00 | 0.00 |
| T213G | 0.29 | 0.26 | 0.27 |
| T213S† | 0.67 | 0.66 | 0.66 |
| T213D | 0.00 | 0.01 | 0.01 |
| PDPK docking | |||
| T241A | 0.00 | 0.00 | 0.00 |
| T250G | 0.14 | 0.14 | 0.14 |
| PDPK proline | |||
| P19A | 0.26 | 0.26 | 0.26 |
| P28A | 0.60 | 0.54 | 0.57 |
| P204A | 0.00 | 0.00 | 0.00 |
| P214A | 0.40 | 0.29 | 0.30 |
Colony area is the percentage of well area covered by G418-resistant foci. All of these data, except that for T213S, were obtained at the same time along with eight determinations of WT, which yielded a mean % colony area of 48.6 ± 3.6.
T213S was assayed at a different time, and the mean % colony area for its WT (four determinations) was 39.5 ± 3.9. The WT mean was set at 1.00 for each dataset.
PDPK Sites in ORF1p Are Required for Retrotransposition.
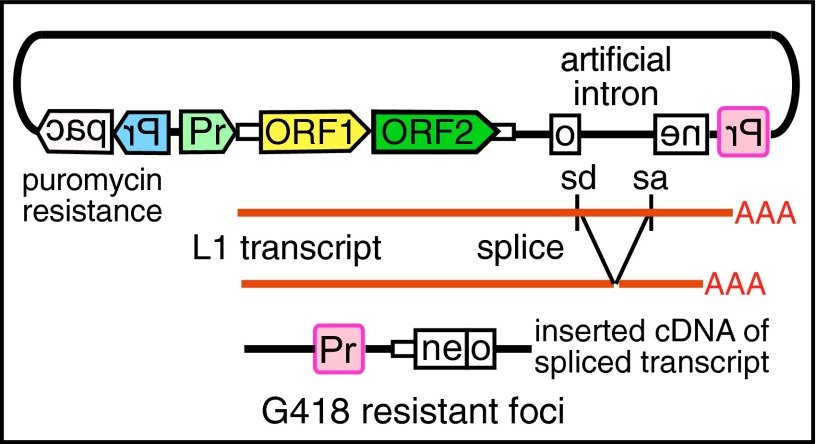
We tested the ORF1p mutants in L1 retrotransposition in HeLa cells using a previously described assay (9) with the engineered L1 construct pRTC-puro (Fig. 2). This vector contains a neomycin reporter gene (neo), interrupted by an intron, in the reverse orientation to L1. The neo gene becomes functional only after a cDNA copy of an appropriately spliced RNA is inserted into the genome, that is, a retrotransposition event, which rendered the cells resistant to G418, an analog of neomycin. After selection with G418, cells were fixed, stained, and quantified using the ImageJ plugin ColonyArea (55). We determined transfection efficiency by puromycin selection to kill all nontransfected cells, with surviving cells fixed and stained as described above (Fig. S2).
Fig. 2.
The retrotransposition vector contains a full-length modern L1.3 element, an active member of the L1Pa1 family (not to scale) (76) driven by the SV40 early promoter (Pr, light green); a neomycin-resistance gene (neo) in reverse orientation interrupted by a sense artificial intron with splice donor (sd) and acceptor (sa) sites and driven by the Rous sarcoma virus LTR promoter (Pr, pink); and a puromycin-resistance gene (pac) driven by the CMV promoter (Pr, blue).
We separately mutated the 22 actual or potential ORF1p phosphorylation sites listed above and found that mutations in all six PDPK-relevant sites (four target, two docking) seriously inhibited retrotransposition (Tables 1 and 2 and Fig. 3). In contrast, of the 16 non-PDPK sites, only S119, located in the first trimerization motif in the coiled coil (56), and S287 in the PP1 site produced >70% inhibition, despite the fact that some of these residues are highly conserved and were found to be phosphorylated in HeLa cells with high confidence, i.e., S50, S53, S145, and S166 (Table 1 and Fig. S3). The bias of inhibitory mutations toward the PDKP-relevant sites was highly significant (P = 0.0004, Fisher exact test).
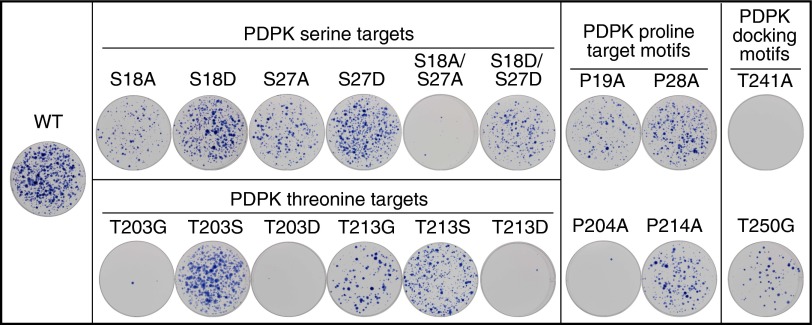
Fig. 3.
Mutation of PDPK target and docking sites in ORF1p inhibit retrotransposition. Wells show Giemsa-stained foci generated from HeLa cells transfected with L1 retrotransposition vectors that express WT or the indicated mutant ORF1p. These assays were carried out in duplicate; Table 2 presents the quantified data.
Regarding the serine PDPK target site mutations, alanine substitution of S18 or S27 decreased retrotransposition by 60–80%, but the double mutant S18A/S27A was barely active. Phosphomimetic substitutions S18D and S27D restored L1 function to >70% and 85% of WT, respectively, and S18D/S27D also rescued S18A/S27A, but to a lesser extent (Table 2). These results suggest that phosphorylation of both S18 and S27 are required for maximal retrotransposition activity, a result consistent with the known cooperative and synergistic effects of multisite phosphorylation; i.e., functional outcomes can be scaled based on gradients of phosphorylation events (57–59).
Mutation of either of the two threonine PDPK sites located in the RRM of ORF1p also strongly inhibited L1 activity. Mutation of T203 to glycine almost abolished retrotransposition, and although the phosphorylation state of T213 was not determined, T213G inhibited retrotransposition by ∼75%. The conservative T203S and T213S mutations, which preserve a PDPK target motif, exhibited WT and 65% of WT activity, respectively; however, in contrast to the serine sites, phosphomimetic substitutions at either threonine site eliminated retrotransposition. Thus, whereas phosphorylatable residues at these sites appear to be essential, a permanently acidic moiety at either threonine inactivates the protein, raising the possibility that reversible phosphorylation at these sites may be necessary during retrotransposition.
Reversible phosphorylation at the C-terminal PDPK threonines would necessitate phosphatase activity, and, as shown in Fig. 1, ELM identified a putative PPI docking motif (P283-I289) that overlaps the highly conserved sequence 282-YPAKLS-287. Previous studies have shown that mutations of this site to 282-AAALA-287 inhibit retrotransposition and the formation of L1 RNPs (7–9). Phosphorylation is not known to regulate PPI binding, but S287 was phosphorylated in insect cells, and S287A decreased retrotransposition by ∼80% (Fig. S3). Whether S287 phosphorylation affects PPI binding remains to be clarified.
Proline residues at PDPK S/T-P target sites are essential for substrate recognition and kinase activity. Therefore, if phosphorylation of these motifs is required for L1 activity, then mutating the proline, a critical component of the motif, should also inhibit retrotransposition. Fig. 3 and Table 2 show that P19A, P204A, and P214A parallel the inhibitory effects of S18A, T203G, and T213G, respectively; however, P28A was somewhat less inhibitory than S27A. Of note, in the absence of P28, the arginine at −3 relative to S27 creates a canonical motif for an AGC kinase, which does not tolerate proline in the +1 position (60–62). Thus, it is possible that S27 was phosphorylated by a non-PDPK kinase when P28 was mutated.
Mutations of PDPK Docking Motifs Inhibit L1 Activity.
As described earlier, ELM identified multiple PDPK docking motifs in ORF1p, two of which are potentially regulated by putative PKA sites T241 and T250 (Fig. 1). PDPK docking sites are thought to reside in close spatial proximity to target motifs, and the docking sites containing T241 and T250 lie very close to T203 and T213 in the ORF1p crystal structure (56). Although neither T241 nor T250 was found to be phosphorylated in HeLa cells, T250 was phosphorylated in insect cells. Because LC-MS/MS can miss low-abundance and reversible phospho residues (63), we determined the effect of mutations at each site. Substitution of T241 with alanine eliminated L1 activity, and glycine substitution at T250 reduced L1 activity by ∼85% (Fig. 3 and Table 2). Whether the decrease in L1 retrotransposition was related to altered phosphorylation or structural perturbation of the PDPK docking sites, or to some other cause, is not known.
ORF1p Mutants Are Expressed in HeLa Cells.
PDPK target site mutations did not prevent the expression of ORF1p in HeLa cells. ORF1p mutants that were inactive in retrotransposition could be expressed and purified for further analysis in vitro (Fig. 4 and Fig. S4). In addition, the proline and docking site mutant proteins were readily detected by Western blot analysis (Fig. S5).
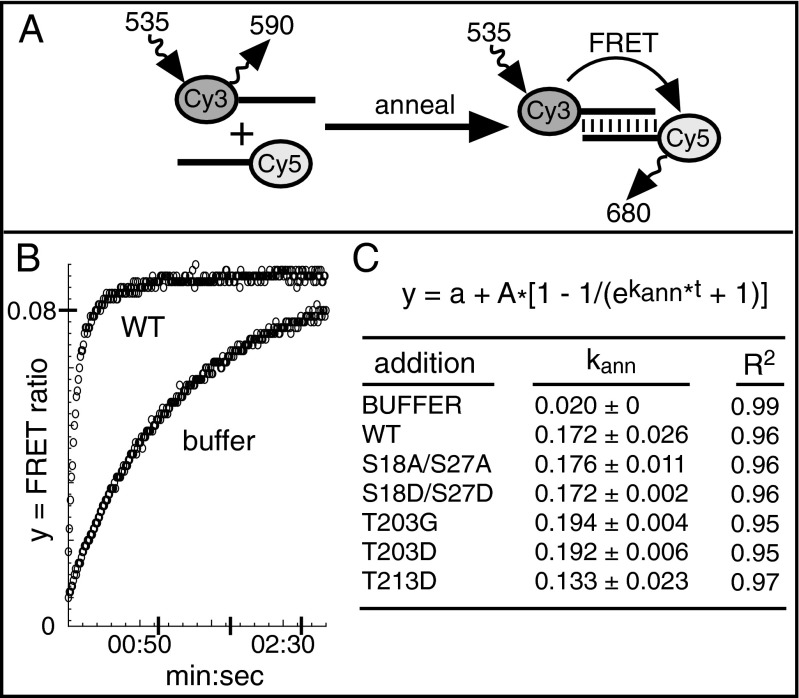
Fig. 4.
ORF1p PDPK mutants can anneal RNA. (A) Schematic of the FRET reaction showing complementary oligonucleotides labeled with the donor (Cy3) and the acceptor (Cy5) fluorophores and their respective excitation and emission wavelengths, as described in the text. (B) Curves for the annealing reaction with buffer or purified WT ORF1p. The FRET ratio (y axis) changes as a function of time (x axis). All reactions were carried out in triplicate. (C) Mean ± SD rate constants, kann, were derived from a least squares fit of the FRET ratio to the indicated rate equation. Pairwise t tests showed that the kann values of WT and ORF1p mutants were not statistically different; P values: S18A/S27A, 0.825; S18D/S27D, 0.984; T023G, 0.274; T203D, 0.301; T213D, 0.124.
RNA Annealing Activity of ORF1p Is Unaffected by PDPK Mutations.
RNA binding and nucleic acid chaperone activity are important biochemical properties of ORF1p, and facilitated annealing of RNA is an essential component of chaperone activity (64); therefore, we compared this activity for WT and mutant ORF1p using FRET (65). In this assay, RNA annealing allows transfer of emission energy from an excited donor fluorophore on one RNA strand to an acceptor fluorophore on its complementary strand (Fig. 4A). As annealing progresses, acceptor emission increases, with a concomitant decrease in donor emission. The FRET ratio (acceptor emission/donor emission) is plotted against time and fit to the exponential equation shown in Fig. 4C to obtain the annealing rate constant, kann. The kann of WT and S18D/S27D ORF1p, both active in retrotransposition, were not statistically different from those of the retrotransposition incompetent mutants S18A/S27A, T203G, T203D, and T213D (Fig. 4C). Thus, the ability of ORF1p to anneal RNA in vitro is not dependent on phosphorylation of its PDPK motifs.
ORF1p PDPK Target Sites Mediate an Interaction with Pin1.
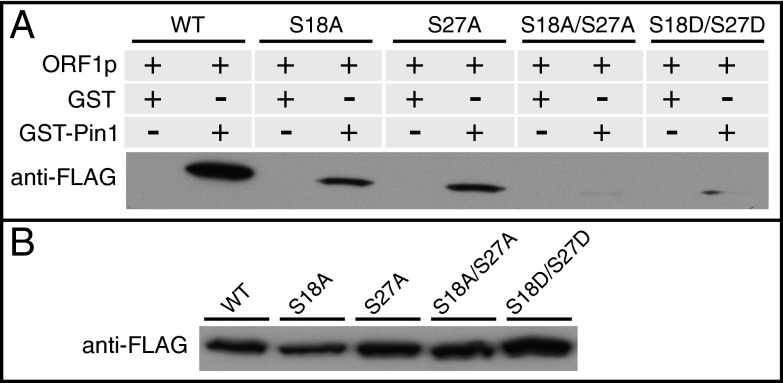
Pin1 is an essential component of numerous PDPK-mediated regulatory pathways (48, 49). This highly conserved protein specifically binds phosphorylated PDPK sites and catalyzes the cis/trans conversion of the proline side chains of S/T-P motifs, leading to conformational changes of its substrates with significant and varied functional outcomes (48, 49). To determine whether Pin1 could target ORF1p, we performed GST pull-down assays using lysates obtained from HeLa cells transfected with expression plasmids for WT ORF1p-Flag and the indicated mutants (Fig. 5). WT ORF1p bound to GST-Pin1, but not GST, and mutation of either the S18 or S27 PDPK sites (phosphorylated in WT ORF1p) strongly impaired Pin1 binding. The double mutant S18/27A almost completely eliminated the interaction with Pin1. These results indicate that in whole-cell lysates, the majority of Pin1 binding is mediated by S18 and S27. The phosphomimetic S18D/S27D, which restored approximately 40% of retrotransposition activity, barely if at all increased Pin1 binding. This finding is not surprising, given the central role of the phosphate group in Pin1 binding (66), and is consistent with failure of a phosphomimetic to restore Pin1 binding to mutated serine PDPK target sites on the transcription factor Nanog (67).
Fig. 5.
PDPK sites in ORF1p mediate binding to Pin1. (A) GST or GST-Pin1 were incubated with whole-cell lysates from HeLa cells transfected with expression plasmids for ORF1p WT or indicated mutants. (B) Relative amount of ORF1p-Flag per 125 µg of whole-cell lysate for each ORF1p construct.
Discussion
Given the dominant role of L1 retrotransposons in the structure and composition of most mammalian genomes, our lack of knowledge of the interface between L1 elements and their hosts represents a major void in our knowledge of mammalian biology. Therefore, our findings establishing that phosphorylation of ORF1p by PDPKs is essential for L1 retrotransposition constitute a major advance in our understanding of L1–host interactions.
Mutation of any of the six PDPK sites seriously inhibited or eliminated retrotransposition, whereas mutations of only two of the 16 non-PDPK sites were inhibitory. This strong bias (P = 0.0004, Fisher’s exact test) supports our conclusion that phosphorylation of ORF1p by PDPKs is necessary for L1 activity. In addition, mutating the prolines of each S/T-P motif, a critical element required for effective PDPK recognition, generally recapitulated the inhibitory effects of the phospho site mutations. Moreover, the rescue of activity by phosphomimetic substitutions at the N-terminal serines indicates that the relevant biochemical property at these PDPK sites is a negative charge, not an unphosphorylated serine side chain. Finally, the activity of the PDPK mutants in vitro indicates that PDPK phosphorylation is not required for RNA annealing, and that such mutations do not perturb the structural competence of the protein to perform this function. Taken together, these results strongly suggest that inhibition of retrotransposition is related to a defect of phosphorylation, not to structural changes caused by replacement of an unphosphorylated serine or threonine.
The prolyl isomerase Pin1 is a critical downstream modulator of phosphorylated PDPK sites that binds phosphorylated S/T-P motifs and catalyzes cis/trans isomerization of the prolyl bond. Our finding that the serine PDPK motifs in ORF1p mediate both L1 activity and an interaction with Pin1 suggests a mechanistic role for Pin1 in L1 retrotransposition. The functional consequences of the ORF1p/Pin1 interaction could be any one of multiple known effects induced by Pin1 isomerization, including altered kinetics of phosphorylation and dephosphorylation (48, 49). Given the effects of the S18D/S27D phosphomimetics, perhaps Pin1 protects the phosphorylated state of S18 and S27, possibly by inhibiting cis/trans prolyl-sensitive phosphatases (48, 49). Although L1 activity was partially restored by S18D/S27D, Pin1 binding was not. However, Pin1 binding at these sites would not be essential for L1 activity in the context of constitutive mimicked phosphorylation if its function at S18 and S27 was to protect phosphorylation. On the other hand, it is also possible that the stringent conditions of the GST pull-down, combined with the nonquantitative nature of Western blot analysis, failed to capture a weakened interaction between Pin1 and the S18D/S27D mutant that was nonetheless sufficiently stable within the cell to permit retrotransposition, albeit at a reduced level.
Our findings that ORF1p is a substrate for protein kinases indicates that L1 has appropriated a major regulatory cascade of the host, as is the case for numerous pathogens (68). In addition to its normal and evolutionary relevant replication niche in germline and early embryonic cells (26, 27, 69–71), L1 also can be active in some somatic cells, including certain tumors and neuronal progenitor cells (19, 20, 23), as well as a consequence of aging (72). ORF1p competition for kinases in any of these cells could perturb signaling cascades. In germline and early embryonic cells, even slightly deleterious effects of this competition could provide selective pressure for adaptive evolutionary changes in components of the phosphorylation-based regulatory pathways. Although the recent attention given to potential effects of L1 on cancer progression, neuronal development, and aging have focused mainly on retrotransposition or the effects of ORF2p, increased expression of ORF1p in these cells may dramatically alter their signaling and metabolic pathways, with consequences extending far beyond those of L1-induced genetic change. Our findings thus open areas for L1 research focused not only on the interplay between ORF1p and host factors necessary for retrotransposition, but also on questions regarding the overall effects of L1 protein expression on cellular function.
Materials and Methods
Retrotransposition Assays.
In this previously described tissue culture-based retrotransposition assay (9), HeLa cells were seeded in six-well plates at 2 × 105 cells per well, transfected with 1 µg of pRTC2-puro, and selected with G418 at 400 µg/mL beginning at 72 h posttransfection for ∼10 d before staining with KaryoMAX Giemsa (Gibco). Transfection efficiencies were assessed in parallel duplicate wells with 10 µg puromycin/mL for 24 h, starting at 1 d posttransfection, which was sufficient to kill all untransfected cells.
LC-MS/MS Analysis.
ORF1p-Flag purified from HeLa cells and untagged ORF1p purified from insect cells were analyzed for phosphorylation by the Mass Spectrometry and Proteomics Resource of the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University.
RNA Annealing Assay.
RNA annealing was measured using a FRET assay (64). Twenty nM ORF1p-Flag constructs, 100 nM Cy3-RNA, and 2× FRET buffer were incubated at 30 °C for 5 min, after which 20 µL of 100 nM Cy5-RNA was injected for final reaction concentrations of 50 nM of each RNA oligonucleotide, 10 nM ORF1p, 50 mM Tris pH 7.4, 150 mM NaCl, 3 mM MgCl2, and 1 mM DTT. Cy3 was excited at 535 nm, and emissions were read at 590 nm and 680 nm every 0.7 s for 3 min.
GST Pull-Down Assay.
GST or GST-Pin1 (500 nM) was immobilized on glutathione agarose before the addition of 1.5 mg of whole-cell extracts obtained from HeLa cells transfected with WT or mutant ORF1p-Flag expression plasmids. Pull-downs were washed four times, resolved via electrophoresis, transferred to nitrocellulose membranes, and probed with ANTI-FLAG M2 antibody (Sigma-Aldrich).
Supplementary Material
Acknowledgments
We thank Cesar Perez-Gonzalez for help in constructing the precursor of the pRTC2-puro plasmid, Dr. John Moran for the generous gifts of JM102 (the L1.3-containing retrotransposition cassette) and HeLa cells, Dr. Sandy Martin for advice, and Deborah Hinton for useful comments on the paper. We also thank TuKiet T. Lam, Jean E. Kanyo, and Mary LoPresti from the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University for the LC-MS/MS phospho site verification, data collection, and sample preparation, respectively. This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: MS/MS ORF1p phosphorylation data are available at yped.med.yale.edu/repository under project name ORF1p.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416869112/-/DCSupplemental.
References
- 1.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat Rev Genet. 2012;13(1):36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7(6):706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11(9):4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15(3):630–639. [PMC free article] [PubMed] [Google Scholar]
- 7.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14(21):3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 8.Doucet AJ, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6(10):e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran JV, et al. High-frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 10.Martin SL, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol. 2005;348(3):549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357(5):1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254(5039):1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 14.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283(5407):1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 15.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14(7):1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski JR. Retrotransposition and structural variation in the human genome. Cell. 2010;141(7):1110–1112. doi: 10.1016/j.cell.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Miki Y, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–645. [PubMed] [Google Scholar]
- 18.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141(7):1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tubio JMC, et al. ICGC Breast Cancer Group; ICGC Bone Cancer Group; ICGC Prostate Cancer Group Mobile DNA in cancer: Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345(6196):1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazazian HH., Jr Mobile DNA transposition in somatic cells. BMC Biol. 2011;9(1):62. doi: 10.1186/1741-7007-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 22.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460(7259):1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479(7374):534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1–containing loci in the human lineage. Mol Biol Evol. 2001;18(6):926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 25.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci USA. 2006;103(25):9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 27.Soper SFC, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15(2):285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17(6):915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat Genet. 1998;20(3):288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- 30.Naas TP, et al. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17(2):590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs FM, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 34.Muckenfuss H, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 35.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meister G. Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet. 2013;14(7):447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 37.Bestor TH. Cytosine methylation mediates sexual conflict. Trends Genet. 2003;19(4):185–190. doi: 10.1016/S0168-9525(03)00049-0. [DOI] [PubMed] [Google Scholar]
- 38.Kolosha VO, Martin SL. Polymorphic sequences encoding the first open reading frame protein from LINE-1 ribonucleoprotein particles. J Biol Chem. 1995;270(6):2868–2873. doi: 10.1074/jbc.270.6.2868. [DOI] [PubMed] [Google Scholar]
- 39.Taylor MS, et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell. 2013;155(5):1034–1048. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodier JL, Cheung LE, Kazazian HH., Jr Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 2013;41(15):7401–7419. doi: 10.1093/nar/gkt512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishizaka Y, Okudaira N, Okamura T. Regulation of retrotransposition of long interspersed element-1 by mitogen-activated protein kinases. In: Xavier DGDS, editor. Protein Kinases. InTech; Rijeka, Croatia: 2012. [Google Scholar]
- 42.Okudaira N, et al. Induction of long interspersed nucleotide element-1 (L1) retrotransposition by 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan photoproduct. Proc Natl Acad Sci USA. 2010;107(43):18487–18492. doi: 10.1073/pnas.1001252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuchen S, et al. The L1 retroelement-related p40 protein induces p38delta MAP kinase. Autoimmunity. 2004;37(1):57–65. doi: 10.1080/08916930310001637977. [DOI] [PubMed] [Google Scholar]
- 44.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11(6):427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 45.Rigbolt KT, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4(164):rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 46.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 47.Mayor F, Jr, Jurado-Pueyo M, Campos PM, Murga C. Interfering with MAP kinase docking interactions: Implications and perspective for the p38 route. Cell Cycle. 2007;6(5):528–533. doi: 10.4161/cc.6.5.3920. [DOI] [PubMed] [Google Scholar]
- 48.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 2002;12(4):164–172. doi: 10.1016/s0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- 49.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36(10):501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dinkel H, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42(Database issue) D1:D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci USA. 2009;106(3):731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adkins RM, Walton AH, Honeycutt RL. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol. 2003;26(3):409–420. doi: 10.1016/s1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- 53.Pappin DJC, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3(6):327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 54.Savitski MM, et al. Confident phosphorylation site localization using the Mascot Delta Score. Mol Cell Proteomics. 2011;10(2):M110.003830. doi: 10.1074/mcp.M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzmán C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE. 2014;9(3):e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khazina E, et al. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat Struct Mol Biol. 2011;18(9):1006–1014. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]
- 57.Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci USA. 2010;107(45):19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salazar C, Höfer T. Multisite protein phosphorylation—from molecular mechanisms to kinetic models. FEBS J. 2009;276(12):3177–3198. doi: 10.1111/j.1742-4658.2009.07027.x. [DOI] [PubMed] [Google Scholar]
- 59.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460(7252):274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kemp BE, Graves DJ, Benjamini E, Krebs EG. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977;252(14):4888–4894. [PubMed] [Google Scholar]
- 61.Zhu G, et al. Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J Biol Chem. 2005;280(11):10743–10748. doi: 10.1074/jbc.M413159200. [DOI] [PubMed] [Google Scholar]
- 62.Zhu G, et al. A single pair of acidic residues in the kinase major groove mediates strong substrate preference for P-2 or P-5 arginine in the AGC, CAMK, and STE kinase families. J Biol Chem. 2005;280(43):36372–36379. doi: 10.1074/jbc.M505031200. [DOI] [PubMed] [Google Scholar]
- 63.Alghamdi WM, Gaskell SJ, Barber J. Detection of low-abundance protein phosphorylation by selective 18O labeling and mass spectrometry. Anal Chem. 2012;84(17):7384–7392. doi: 10.1021/ac301038u. [DOI] [PubMed] [Google Scholar]
- 64.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13(12):2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajkowitsch L, Schroeder R. Coupling RNA annealing and strand displacement: A FRET-based microplate reader assay for RNA chaperone activity. Biotechniques. 2007;43(3):304–310, 306, 308 passim. doi: 10.2144/000112530. [DOI] [PubMed] [Google Scholar]
- 66.Schutkowski M, et al. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry. 1998;37(16):5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- 67.Moretto-Zita M, et al. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci USA. 2010;107(30):13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alto NM, Orth K. Subversion of cell signaling by pathogens. Cold Spring Harb Perspect Biol. 2012;4(9):a006114. doi: 10.1101/cshperspect.a006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92(5):1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brouha B, et al. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71(2):327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23(11):1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Meter M, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18(12):2186–2194. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- 74.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16(1):78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Januszyk K, et al. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem. 2007;282(34):24893–24904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- 76.Dombroski BA, Scott AF, Kazazian HHJ., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci USA. 1993;90(14):6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.