Significance
RecQ DNA helicases catalyze critical genome maintenance reactions in nearly all organisms. This study describes the crystal structure of a bacterial RecQ helicase bound in a productive complex with DNA. Together with biochemical experiments, the structure reveals a conserved coupling mechanism that links DNA binding to ATP hydrolysis in RecQ enzymes. These findings also help explain how structural dynamics could facilitate RecQ’s noted ability to process diverse DNA substrates. A model explaining the physical basis for RecQ substrate binding and unwinding is proposed.
Keywords: helicase, RecQ, mechanism, aromatic-rich loop, DNA bending
Abstract
RecQ helicases unwind remarkably diverse DNA structures as key components of many cellular processes. How RecQ enzymes accommodate different substrates in a unified mechanism that couples ATP hydrolysis to DNA unwinding is unknown. Here, the X-ray crystal structure of the Cronobacter sakazakii RecQ catalytic core domain bound to duplex DNA with a 3′ single-stranded extension identifies two DNA-dependent conformational rearrangements: a winged-helix domain pivots ∼90° to close onto duplex DNA, and a conserved aromatic-rich loop is remodeled to bind ssDNA. These changes coincide with a restructuring of the RecQ ATPase active site that positions catalytic residues for ATP hydrolysis. Complex formation also induces a tight bend in the DNA and melts a portion of the duplex. This bending, coupled with translocation, could provide RecQ with a mechanism for unwinding duplex and other DNA structures.
Helicases are motor proteins that convert the chemical energy of nucleoside triphosphate (NTP) hydrolysis into the mechanical energy needed to separate nucleic acid strands (1). The largest and most diverse helicase superfamilies, SF1 and SF2, use conserved sequence motifs (I, Ia, II–VI) within their helicase domains to couple NTP hydrolysis to conformational changes that mediate DNA translocation and unwinding (1, 2). Although the DNA-unwinding mechanisms of SF1 helicases have been examined extensively, far less is known about SF2 enzymes, in part because of the smaller number of available helicase/substrate complex structures.
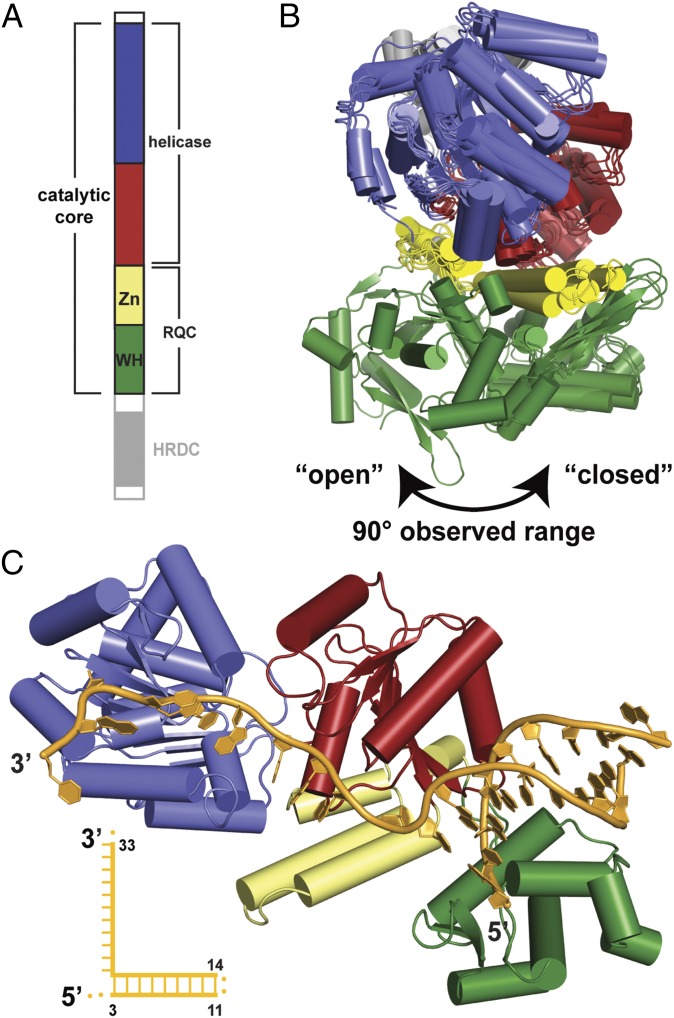
RecQ DNA helicases are SF2 enzymes with broad roles in promoting genomic stability in eubacterial and eukaryotic species (3). Their importance is underscored by the multiple genomic instability diseases caused by mutations in human recQ genes (4–8). At a structural level, most RecQ helicases share a similar domain architecture that includes a helicase domain, a RecQ C-terminal (RQC) element comprised of Zn2+-binding and winged-helix (WH) domains, and a helicase and RNaseD C-terminal (HDRC) domain (Fig. 1A) (9, 10). The helicase and RQC domains combine to form a catalytic core that is sufficient for DNA-unwinding activity in many RecQ proteins. Crystal structures of several RecQ catalytic cores have been determined, including Escherichia coli RecQ (EcRecQ), human RecQ1, and human Bloom syndrome protein (BLM) [refs. 10–12 and unpublished structures (4CDG, 2WWY, and 4CGZ) available through the Protein Data Bank (PDB)]. A comparison of these structures reveals strong similarities among domains within the catalytic core but also differences in the relative positioning of these domains among RecQ proteins. The EcRecQ and antibody-bound BLM structures form an open arrangement in which the WH domain is centered relative to the helicase and Zn2+-binding domains, but in RecQ1 and DNA-bound BLM a closed arrangement is observed with the WH domain positioned laterally to the helicase domain (Fig. 1B and Fig. S1) (10–12). The functional relevance of the open and closed arrangements is unknown.
Fig. 1.
Structure of the DNA-bound CsRecQ catalytic core. (A) Schematic diagram of RecQ DNA helicase. Coloring denotes the helicase (blue and red), Zn2+-binding (yellow), WH (green), and HRDC (gray) domains. (B) Overlay of RecQ structures highlights the mobility of the WH domain. (C) Ribbon diagram of the CsRecQ/DNA crystal structure. Domains are colored as in A with DNA colored orange. A labeled schematic of the DNA is shown with disordered nucleotides as dots.
Although several RecQ structures have been defined, insights into the mechanisms by which this important helicase family recognizes and unwinds DNA substrates have been hampered by limitations in reported RecQ/DNA complex structures. One outstanding question is how RecQ enzymes can unwind diverse DNA substrates. Most RecQ enzymes, including EcRecQ, unwind a very broad array of substrates that include duplex, triplex, quadruplex, and branched DNA (3). Human RecQ1, which unwinds Holliday junction but not triplex or quadruplex DNA, is a rare exception in its narrow substrate specificity (13). The mechanisms that confer functional diversity in most, but not all, RecQ enzymes remain unknown. A second question is how RecQ enzymes couple DNA binding with DNA-dependent ATPase functions. A chemo-mechanical coupling element called the “aromatic-rich loop” (ARL) is encoded C-terminal to motif II in RecQ proteins (10, 14), but how DNA binding at the ARL is linked to ATPase functions is unclear because current RecQ/DNA structures have not resolved the DNA/ARL interface. ARL-coupling elements have been examined in several SF1 helicases, but they map to different positions within the helicase domain and rely on structural features that are not found in RecQ enzymes (15–21), leaving their mechanistic similarity with ARLs in RecQ enzymes unclear. A third puzzling feature of RecQs is their varied use of a WH domain β-hairpin in DNA unwinding. In eukaryotic RecQ helicases, this β-hairpin projects away from the WH domain, forming a wedge that appears to separate DNA strands (11, 12, 22). However, the equivalent β-hairpin in EcRecQ is much shorter and is dispensable for helicase activity (10, 11), indicating that the bacterial and human RecQ proteins use different strand-separation mechanisms. This finding is even more surprising given that putative wedge elements have been identified in all other SF1 and SF2 DNA helicases of known structure (23). Understanding how bacterial RecQs function without a wedge element could offer new insights into the mechanisms by which helicases can unwind DNA.
In this report, the structure of a bacterial RecQ catalytic core bound to duplex DNA with a 3′ single-stranded extension provides insights into physical mechanisms of RecQ enzymes. DNA binding induces a rearrangement of the RecQ catalytic core in which the WH pivots ∼90° from an open to a closed conformation to secure duplex DNA against a surface of the helicase domain. Flexibility in the WH domain position could provide the structural plasticity needed for RecQ to accommodate diverse substrates in its DNA-binding site. A second conformational change remodels the ARL to allow ssDNA binding. This DNA-dependent conformational shift triggers reorganization of the ATP-binding pocket in a manner that positions active-site residues for catalysis, explaining how the ARL couples DNA binding and ATPase activities in RecQ enzymes. The DNA within the complex is bent at a 90° angle at the unwinding junction, and this sharp bend could explain the ability of bacterial RecQs to unwind DNA without the use of a wedge.
Results
X-Ray Crystal Structure of the RecQ/DNA Complex.
A screen of bacterial RecQ/DNA complexes yielded diffraction-quality crystals of the catalytic core domain of Cronobacter sakazakii RecQ (CsRecQ, 86% identical to EcRecQ) bound to a hairpin DNA comprised of a 10-bp duplex region flanked by a four-base hairpin sequence (GTAA) on one end and a 10-base 3′ extension on the other. The 2.4-Å resolution CsRecQ/DNA complex structure was determined by molecular replacement using the EcRecQ catalytic core structure (10) as a search model (Fig. 1C and Table S1). The asymmetric unit contained a single DNA-bound CsRecQ catalytic core monomer in which residues 4–514 of the helicase and nucleotides 3–11 and 14–33 of the hairpin DNA were resolved. The DNA-binding surface of CsRecQ was highly electropositive and was well conserved among RecQ proteins, especially within the ssDNA-binding region (Fig. S2).
The RecQ WH Domain Closes to Bind Duplex DNA.
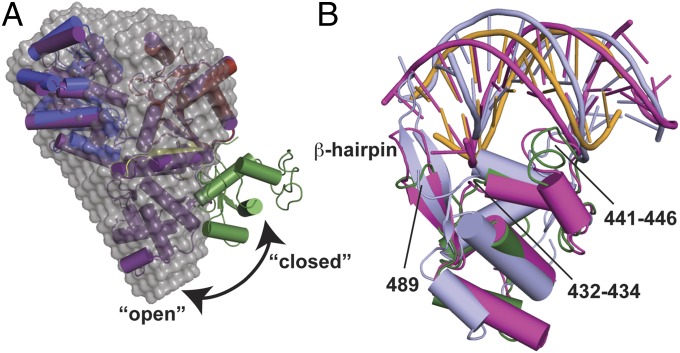
The CsRecQ/DNA domain arrangement differed significantly from EcRecQ. In contrast to the open conformation of the EcRecQ structure in which the WH domain was centered relative to the helicase and Zn2+-binding domains (10), the DNA-bound CsRecQ catalytic core was in a closed state with the WH domain positioned laterally to these domains (Fig. 2A). To test whether the WH domain of the free CsRecQ catalytic core is in an open position, similar to that of the EcRecQ catalytic core structure, small-angle X-ray scattering (SAXS) was used to determine the low-resolution shape of the CsRecQ catalytic core domain in the absence of DNA. The open RecQ form fits the SAXS-derived molecular envelope with a χ2 value of 1.50, whereas the closed form fits more poorly (χ2 = 1.99). The calculated radius of gyration (Rg) for the open form (25.1 Å) also is more consistent with the experimentally determined value for apo CsRecQ (25.3 ± 0.2 Å) than is the closed form (27.0 Å). These data show that the CsRecQ catalytic core exists in an open state in the absence of DNA and that the WH domain pivots to close onto DNA upon complex formation (Fig. 2A and Movie S1).
Fig. 2.
Dynamics of the WH domain in DNA binding. (A) Fits of closed CsRecQ (colored as in Fig. 1) and open EcRecQ (purple) (10) catalytic core protein structures to the averaged SAXS dummy atom model (gray). (B) Structural alignment of CsRecQ (green with DNA in orange), BLM (slate) (12), and WRN (magenta) (22) WH domains. Loops that contact dsDNA and the β-hairpin are labeled.
The CsRecQ WH domain structure closely resembled those from other RecQ proteins [Werner syndrome protein (WRN), RecQ1, and BLM; rmsds of 1.8 Å, 2.3 Å, and 2.5 Å, respectively], and it bound DNA using the same overall surface (Fig. 2B) (11, 12, 22). Loops from the WH domain (residues 432–434 and 441–446) and His489 from the WH β-hairpin contacted the phosphodiester backbone on either side of the DNA major groove (Fig. 2B and Figs. S3 and S4). This interface does not rely on sequence-specific major groove interactions to bind duplex DNA, as has been observed in many other WH domains (24); instead, interactions are made exclusively with the DNA backbone. Because previous studies have shown that non–duplex-folded (e.g., quadruplex) DNA structures compete with duplex structures for binding to a single site in the EcRecQ WH domain (25), the backbone-binding arrangement observed here most likely is used to bind other structures as well. Indeed, triplex and quadruplex DNAs appear to dock onto the WH DNA-binding surface with good steric and electrostatic complementarity (Fig. S4). The helicase domain also contacted the duplex, with interactions formed between residues 220–225 and the phosphodiester backbone. As was anticipated from biochemical experiments (11), the β-hairpin wedge found in human RecQ proteins (11, 12, 22, 26) was shorter in CsRecQ and made only limited contacts with the DNA, and no other wedge element was observed (Fig. 2B).
Electron density for the double-stranded portion of the crystallized DNA was notably poorer than for the ssDNA region (Fig. S4). Accordingly, the refined temperature factors for atoms within the duplex region were higher than those for ssDNA. The same parameters also were weaker for the WH domain relative to the rest of the RecQ catalytic core. These differences suggest that the WH domain and dsDNA could be more structurally dynamic than the remainder of the complex. This feature, combined with the noted DNA-binding promiscuity of the WH domain (25), could provide RecQ helicases with the structural tolerance required to bind diverse DNA structures (Discussion).
ssDNA Binding Alters the Structure of the RecQ ARL and the ATPase Active Site.
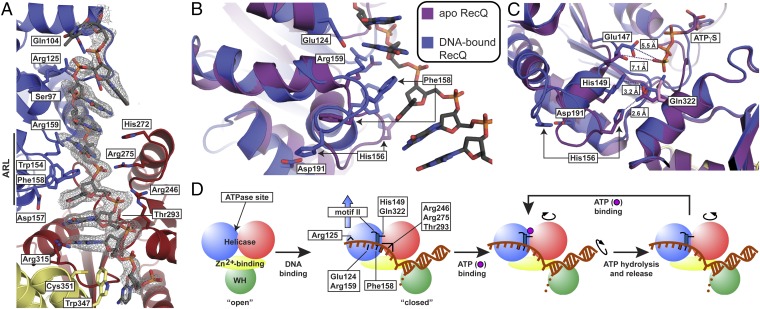
As part of a mixed electropositive/hydrophobic ssDNA-binding groove on CsRecQ, the ARL undergoes a major rearrangement to bind ssDNA (Fig. 3 A–C and Movie S2). The side chains of Phe158 and Arg159 move ∼10 Å and ∼5 Å, respectively, to contact ssDNA directly, with the Phe158 side chain flipping to base stack with the DNA. The DNA-bound ARL conformation also was stabilized by the formation of new intramolecular bonds (His149/Gln322, His156/Asp191, and Arg159/Glu124). Formation of the His156/Asp191 interaction required His156 to move ∼15 Å from its position in apo EcRecQ. In addition to ARL/ssDNA contacts, several other RecQ residues interacted with ssDNA (Fig. 3 A and B and Fig. S3). Arg315 contacted the base of G24, and Ser97, Arg125, Arg246, Arg275, and Thr293 contacted the phosphodiester backbone. Ser97 and Arg125 make the 3′-most ssDNA contacts as the DNA peels away from the helicase domain. All these residues are highly conserved among bacterial RecQs.
Fig. 3.
CsRecQ ssDNA-binding and DNA-translocation/unwinding model. (A) CsRecQ/ssDNA interface. An Fo−Fc DNA omit electron density map (contoured to 2σ) is shown in gray with the refined DNA model overlaid. Side chains of residues within 3.3 Å of ssDNA or that form new interactions in the DNA-bound form of RecQ are labeled. Domain coloring is as in Fig. 1. (B) Superposition of the free EcRecQ (purple) (10) and DNA-bound CsRecQ (blue) structures reveal DNA-dependent ARL rearrangement. Residues within the ARL that engage ssDNA or that appear to stabilize the DNA-bound form of the ARL are labeled. (C) Realignment of motifs II and VI by DNA binding-induced ARL remodeling in CsRecQ. EcRecQ and CsRecQ catalytic core structures are colored as in B, and the region around Gln322 was used to align the structures. In the free state, the side chain of Gln322 (pink) is oriented toward the empty ATP-binding site, whereas in the ATPγS-bound structure, Gln322 (purple) rotates to interact with His156 (10). In the DNA-bound state, Gln322 (blue) forms a new interaction with His149 of motif II, and His156 interacts with Asp191. Distances between Glu322 and His156 or His149 and between the carboxyl group of Glu147 and the gamma phosphate position observed in the EcRecQ/ATPγS (10) structure are shown. (D) Cartoon schematic of the RecQ DNA-binding and -translocation/unwinding mechanism. DNA binding (first step) induces changes described in the presented structure; key residues with roles in DNA binding and/or unwinding are depicted as black lines. Subsequent steps linking ATPase activities to domain movements involved in translocation are based on inchworm models from SF1 and SF2 helicases (20, 21, 33, 44). An ssDNA base is boxed to demonstrate its movement during translocation.
Beyond its direct binding role, DNA-dependent reorientation of the ARL was accompanied by two structural changes within the ATP-binding pocket: Motif II shifted toward the ATP-binding site and a new motif II/motif VI interaction was formed (Fig. 3C and Fig. S5). The first change caused the carboxyl group of Glu147 from motif II, the presumed general base for ATP hydrolysis, to close to within ∼5.5 Å of the binding site for the gamma phosphate of ATP. This distance contrasts with the 7.1-Å distance observed in the ATPγS-bound EcRecQ structure (Fig. 3C) (10). The second change formed a new interaction between His149 of motif II and Gln322 of motif VI, establishing a connection between the two lobes of the helicase domain. In apo EcRecQ, Gln322 points toward the ATP-binding site, whereas in the ATPγS-bound EcRecQ structure Gln322 binds to His156 of the ARL (10). Upon DNA binding, however, the altered ARL conformation displaces His156 away from Gln322 and aligns His149 with Gln322 (Fig. 3C). These observations thus reveal changes in the ATPase active site that could help link DNA binding and ATPase functions in RecQ.
Structure of DNA in the Binary Complex.
Examination of the CsRecQ/DNA structure provided several insights into the consequences of complex formation for the DNA. First, the duplex portion of the DNA was roughly B-form, but it included only 8 of the 10 anticipated base pairs along with a sheared G-A base pair formed between G11 and A14 of the hairpin [the G-A base pair was anticipated from previous studies of the GTAA hairpin structure (27)]. This finding indicates that, even in the absence of ATP, the energy of binding to the CsRecQ catalytic core was sufficient to unwind two base pairs at the ss/ds junction. Next, the C23 and G24 bases, which would be paired with G2 and C1 in the absence of RecQ, are in an ∼90° turn at the ss/ds junction. Finally, bases 24–26 stack together and are pointed into a pocket formed at the interface between the helicase domain and the Zn2+-binding platform (Fig. 3A). This stacking is capped by interaction between T26 and Phe158 from the ARL. The ssDNA bases 3′ of Phe158 are oriented away from the protein, and the remaining interactions are made with the phosphodiester backbone. The 3′ ssDNA end extends to an adjacent symmetrically related molecule within the crystal lattice, and the A32 base docks into the ATP active site at the position that ordinarily would bind the base in ATP (Fig. S6). This interaction appears to facilitate crystal packing because crystallization attempts with DNA containing shorter ssDNA or in which A32 is altered to thymine failed to produce crystals.
Mutational Analysis of Individual Residues in EcRecQ.
To determine the roles of interactions identified in the CsRecQ/DNA structure, variants of full-length EcRecQ with single alanine substitutions were purified, and their activities were measured in DNA-binding, ATPase, and DNA helicase assays. The variants were designed to disrupt interactions that appeared to stabilize the DNA-bound ARL conformation (Glu124Ala, His156Ala, and Asp191Ala), facilitate communication between the helicase lobes (His149Ala and Gln322Ala), bind ssDNA (Ser97Ala, Arg125Ala, Arg246Ala, Arg275Ala, Thr293Ala, Arg315Ala, and Trp347Ala), and bind dsDNA (Arg446Ala). The data were further correlated with a previous study examining EcRecQ ARL variants (Trp154Leu, Phe158Leu, and Arg159Leu) (14).
A fluorescence anisotropy assay was used to assess the equilibrium DNA-binding properties of the EcRecQ variants. RecQ concentration-dependent changes in fluorescence anisotropy were measured for a helicase substrate (F-3′overhang) comprised of a fluorescein-labeled 18-bp duplex DNA flanked by a 12-base 3′ ssDNA extension (Table 1). Wild-type EcRecQ binds the F-3′overhang substrate with an apparent dissociation constant (Kd, app) of 1.3 ± 0.2 nM. Most of the variants differed by less than onefold in Kd, app, indicating that the altered residues did not reduce DNA-binding affinity and that the variants were properly folded. However, two variants, Thr293Ala and Arg246Ala, had measurable DNA-binding defects (Kd, app values = 4.1 ± 0.4 nM and >50 nM, respectively). The side chains from both of these residues were within 3 Å of the phosphodiester backbone of ssDNA at the ss/ds junction in the CsRecQ/DNA structure (Fig. 3), highlighting the importance of this region in stabilizing the RecQ/DNA complex.
Table 1.
DNA-binding, DNA-dependent ATPase, and DNA-unwinding properties of EcRecQ, CsRecQ, and EcRecQ variants
| Variant | Function | DNA binding, Kd, app, nM | DNA-dependent ATPase activity | Helicase activity | ||||
| K1/2, nM | kmin⋅min−1 | kmax⋅min−1 | kmax /kmin | Midpoint*, nM | Maximum unwound, % | |||
| EcRecQ | 1.3 ± 0.2 | 9.1 ± 0.8 | 6.4 ± 0.0 | 1,430 ± 30 | 222 | 1.25 ± 0.07 | 100.1 ± 1.2 | |
| CsRecQ | 1.5 ± 0.1 | 32.5 ± 2.8 | 5.4 ± 0.2 | 994 ± 23 | 185 | 5.17 ± 0.17 | 101.7 ± 1.1 | |
| His156Ala | In ARL | 1.0 ± 0.1 | 11.8 ± 0.8 | 42.0 ± 5.4 | 1,220 ± 18 | 29 | 7.68 ± 2.4 | 26.0 ± 1.6 |
| Glu124Ala | ARL movement | 1.3 ± 0.2 | 3.0 ± 0.3 | 7.9 ± 0.2 | 782 ± 14 | 99 | NA | ∼0 |
| Asp191Ala | ARL movement | 1.4 ± 0.2 | 2.8 ± 0.4 | 56.8 ± 1.9 | 626 ± 17 | 11 | 1.56 ± 0.20 | 77.0 ± 2.1 |
| His149Ala | His–Gln interaction | 1.0 ± 0.2 | 4.0 ± 0.5 | 5.9 ± 1.7 | 311 ± 7.6 | 53 | 1.54 ± 0.07 | 79.6 ± 1.1 |
| Gln322Ala | His–Gln interaction | 1.5 ± 0.1 | NA | ∼0 | 5.2 ± 0.1 | NA | NA | ∼0 |
| Ser97Ala | ssDNA binding | 0.9 ± 0.1 | 9.9 ± 0.7 | 8.0 ± 0.6 | 1,289 ± 17 | 160 | 0.83 ± 0.07 | 97.8 ± 1.6 |
| Arg125Ala | ssDNA binding | 1.5 ± 0.2 | 20.7 ± 1.4 | 7.6 ± 0.2 | 882 ± 14 | 116 | NA | ∼0 |
| Arg246Ala | ssDNA binding | >50 | 128 ± 6.9 | 5.9 ± 0.2 | 452 ± 8.1 | 77 | 39.7 ± 3.62 | 41.6 ± 1.0 |
| Arg275Ala | ssDNA binding | 1.2 ± 0.1 | 96.3 ± 8.1 | 11.2 ± 0.2 | 451 ± 12 | 40 | 11.8 ± 0.57 | 88.5 ± 1.0 |
| Thr293Ala | ssDNA binding | 4.1 ± 0.4 | 133 ± 8.2 | 1.8 ± 0.1 | 86.0 ± 1.8 | 47 | NA | ∼0 |
| Arg315Ala | ssDNA binding | 2.6 ± 0.2 | 37.1 ± 5.7 | 6.3 ± 0.7 | 774 ± 33 | 122 | 2.02 ± 0.11 | 89.2 ± 1.2 |
| Trp347Ala | ssDNA binding | 2.1 ± 0.2 | 23.7 ± 4.2 | 9.4 ± 4.7 | 733 ± 32 | 78 | 1.85 ± 0.12 | 80.9 ± 1.3 |
| Arg446Ala | dsDNA binding | 1.2 ± 0.1 | 35.3 ± 1.5 | 6.8 ± 0.2 | 1,360 ± 15 | 198 | 2.76 ± 0.20 | 79.7 ± 1.6 |
NA, not applicable.
RecQ required for 50% maximal DNA unwinding.
The DNA-dependent ATPase activities of the EcRecQ variants were measured using a coupled spectrophotometric ATPase assay (Table 1) (14). In the absence of DNA, wild-type EcRecQ had a very weak ATPase activity (kmin ∼6⋅min−1) that was stimulated >200-fold by the addition of dT28 (kmax 1,430 ± 30⋅min−1). A dT28 concentration of 9.1 ± 0.8 nM was required for 50% maximal stimulation. Two of the variants (His156Ala and Asp191Ala) had elevated ATPase activity in the absence of DNA (kmin 42.0 ± 5.4⋅min−1 and 56.8 ± 1.9⋅min−1, respectively), which was similar to that observed for another ARL variant [Phe158Leu EcRecQ, kmin = 90⋅min−1 (14)]. Thus, multiple residues that help stabilize the DNA-bound ARL conformation appear to be important for limiting ATP hydrolysis in the absence of DNA.
The Gln322Ala variant was nearly devoid of ATPase activity even in the presence of DNA (kmax 5.2 ± 0.1⋅min−1). Interestingly, changing His149 (which interacts with Gln322 in the DNA-bound structure) to Ala also resulted in a reduced DNA-stimulated ATPase rate (kmax 311 ± 7.6⋅min−1), although the magnitude of the reduction was much less than that of the Gln322Ala variant. These data are consistent with a major catalytic role for Gln322 and a more minor role for His149 in supporting RecQ ATPase activity (Discussion).
The remaining EcRecQ variants had differences in DNA-dependent ATPase activities that correlated well with DNA-binding roles for each residue derived from the CsRecQ/DNA structure. Two variants (Arg246Ala and Arg275Ala) required much higher concentrations of dT28 for ATPase stimulation (11- and 14-fold higher K1/2) than wild-type EcRecQ and had reduced (threefold lower) kmax values. A third variant, Thr293Ala, also had a similarly elevated K1/2 but had a far greater decrease in its kmax value (17-fold lower), which could be linked to a role for the residue in translocation (Discussion). Other predicted DNA-binding variants (Arg125Ala, Arg315Ala, Trp347Ala, and Arg446Ala) displayed more modest defects, with two-to fourfold higher K1/2 values. Overall, these data are consistent with roles for several RecQ residues in DNA binding and highlight the particular importance of residues in the second helicase lobe (Arg246, Arg275, and Thr293) that contact ssDNA adjacent to the ss/dsDNA junction.
Finally, EcRecQ-variant helicase activities were tested using a substrate similar to F-3′overhang in a fluorescence-based assay (Table 1 and Fig. S7). The Glu124Ala variant lacked helicase activity; the absence of helicase activity was surprising, given the variant’s nearly wild-type ATPase levels. However, a similar pattern also had been observed previously for an Arg159Leu EcRecQ variant (14), and Glu124 and Arg159 interact in the DNA-bound structure, indicating the importance of this interaction for stabilizing the ARL during DNA unwinding. Gln322Ala was unable to unwind DNA, whereas His149Ala had nearly wild-type activity, paralleling the DNA-dependent ATPase activities of the variants. Two variants with proposed roles in translocation (Discussion), Arg125Ala and Thr293Ala, failed to unwind the substrate. Arg246Ala, which alters a residue that cooperates with Thr293 in ssDNA binding, also required ∼30-fold higher enzyme concentrations for half-maximal unwinding and unwound only 42% of the substrate. When combined with the CsRecQ/DNA structure, these data define molecular features that are critical for RecQ biochemical functions.
Discussion
RecQ enzymes are SF2 DNA helicases with central genome maintenance roles in organisms ranging from bacteria to humans (3). Although the domain architectures of several RecQ proteins have been defined, insights into the mechanisms of substrate recognition and unwinding by this important helicase family have been hampered by the limitations of reported RecQ/DNA complex structures. The structural and biochemical studies described here have identified DNA-dependent conformational changes that are essential for RecQ motor functions and provide an explanation for the unusual ability of RecQ to unwind diverse substrates. Moreover, the effect of RecQ on the conformation of bound DNA presents a possible helicase mechanism that relies on DNA bending to drive unwinding.
The largest DNA-dependent movement observed in CsRecQ was a 90° rotation of the WH domain that secures dsDNA against the helicase domain (Figs. 1 and 2 and Movie S1). This closed CsRecQ WH domain position is very similar to that observed in the structure of BLM/DNA (12) and RecQ1/DNA (2WWY, structure available through PDB), suggesting that the domain arrangement is conserved within the RecQ family. Similar DNA-dependent domain movements that help to grip duplex DNA have been observed in SF1 enzymes as well (20, 21). Interestingly, although the CsRecQ and BLM WH domain positions are dependent on DNA binding, the RecQ1 WH domain is fixed in a closed state regardless of whether DNA is bound (Fig. 2 and Fig. S1) (10–12). The structure-specific DNA-unwinding properties of these enzymes also vary: BLM and EcRecQ process diverse DNA structures, including duplex, triplex, and quadruplex DNA, but RecQ1 is far more restricted, unwinding only duplex and Holliday junction DNA (13, 28–31). This parallel suggests a model in which WH domain dynamics in BLM and bacterial RecQs could allow the flexibility needed to accommodate diverse DNA substrates, whereas the fixed position of the WH domain in RecQ1 limits its activity to a narrower range of substrates. This model further suggests that additional RecQ family members, such as WRN, that also can unwind diverse DNA structures (31, 32) would have dynamic WH domains as well.
Examination of the CsRecQ/DNA complex also showed that ssDNA binding restructures the ARL in a manner that is coupled with two changes in the ATPase active site. The first change causes the carboxyl group of Glu147 from motif II, the presumed general base used in ATP hydrolysis, to close to within 5.5 Å of the binding site for the gamma phosphate of ATP (Fig. 3C). Given that this closure correlates well with distances observed in structures of SF2 hepatitis C virus helicase NS3/nucleic acid complexes bound to ATPase transition state mimics (5.0–5.5 Å) (33, 34), this change could be critical for activating RecQ ATPase function in response to DNA binding. The second change alters the position of Gln322 (motif VI), shifting it away from His156 of the ARL and toward His149 of motif II. Similar motif II/VI interactions have been found to be important for coupling DNA binding with ATPase activity in other SF2 helicases (33, 35–37). In particular, this shift parallels that of a Gln from motif VI in NS3 helicase that also has conformations that alternate in a nucleotide- and nucleic acid-dependent manner (33, 35, 38). The primary role for the Gln in NS3 appears to be in positioning a water molecule for deprotonation by the general base (motif II Glu); the loss of ATPase and helicase functions in the Gln322Ala variant are consistent with a similar role for Gln322 in RecQ.
A coupling model emerges from these observations in which ssDNA binding to the ARL allosterically remodels the ATPase site to create a catalytically competent state. Placement of the RecQ ARL directly C-terminal to motif II allows a direct link between DNA binding and the observed active-site remodeling. In particular, when the Phe158 side chain flips to base stack with ssDNA, the remainder of the ARL is remodeled in a coordinated fashion that pushes the helix supporting motif II closer to the ATPase active site (Fig. 3 B and C and Fig. S5). The same helix motion also aligns His149 with Gln322 to create the motif II–VI linkage described above. Interestingly, nucleic acid-dependent ATPase active-site changes have been noted in other helicases as well (18, 34), but, to our knowledge, RecQ presents the first example in which movement of motif II is key (Fig. S5). This feature appears to arise from the juxtaposition of motif II and the ARL within RecQ enzymes. A similar motif II–ARL arrangement is present in the SF2 PriA DNA helicase (39), indicating that the DNA-dependent changes observed in RecQ could apply to other DNA helicase families as well.
The data presented here also help define residues within RecQ that are critical for translocation on ssDNA. In a model for NS3, two conserved Thr residues contact the phosphate backbone to facilitate helicase translocation (33, 40–42). These residues bind phosphates that are three bases apart in the absence of ATP, but when ATP binds, the gap is reduced to two bases (33, 40, 41). Cycling between these states with alternating ssDNA binding at each site is thought to drive helicase translocation via an inchworm mechanism. Similar mechanisms have been proposed for SF1 helicases (20, 21, 43). In EcRecQ and CsRecQ, Thr293 provides one of the equivalent Thr residues, whereas Arg125 replaces the second Thr. These residues bind DNA backbone phosphates that are separated by four bases in the CsRecQ/DNA structure, and each is required for helicase activity (Fig. 3 and Table 1). Our structural and biochemical data are consistent with a mechanism in which ATP-dependent positioning of RecQ ssDNA-binding sites centered on Arg125 and Thr293 could act to coordinate translocation (Fig. 3D). In this mechanism, ATPase-dependent conformational changes in the helicase domain would be coupled to an inchworm-style movement along ssDNA, analogous to the models proposed for translocation in many other SF1 and SF2 helicases (20, 21, 33, 44). Additional mechanistic and structural studies of RecQ/DNA complexes bound to ATPase cycle nucleotide mimics will be required to dissect further the physical mechanisms of translocation.
Finally, our structure uncovered a possible explanation for how bacterial RecQ proteins can unwind DNA without the use of a wedge element. Structural and mutagenesis data have revealed differences between eukaryotic and bacterial RecQ unwinding in their reliance on a β-hairpin wedge within the WH domain. The β-hairpin in RecQ1, BLM, and WRN projects away from the WH to split DNA at the ss/dsDNA junction, and mutations in DNA-binding residues within the hairpin essentially eliminate DNA unwinding (11, 12, 22). In contrast, the β-hairpin in the bacterial RecQ WH domain is much shorter, and, although the CsRecQ/DNA structure shows that it also associates with the DNA backbone (Fig. 2), mutations that substitute or delete the β-hairpin residue that binds DNA (His489) have negligible effects on DNA unwinding (11, 22). How bacterial RecQs unwind DNA in the absence of an apparent wedge element therefore was unknown.
CsRecQ/DNA complex formation induces an ∼90° bend in the DNA at the ss/ds junction, and this bend is sufficient to separate 2 bp of DNA in the absence of ATP (Figs. 1 and 2). This angle is larger than that in RecQ1, BLM, and other SF2 helicase/DNA complexes determined to date (20–60°) and instead is within the range observed in single-subunit SF1/DNA complexes (90–110°) (Fig. S8) (23). In the case of SF1 enzymes PcrA and UvrD, the angle at the ss/dsDNA bend has been proposed to help drive DNA unwinding by allowing the enzyme to peel ssDNA away from the duplex, although these enzymes also use wedge elements to assist in DNA unwinding (20, 21). We propose that CsRecQ and other bacterial RecQ enzymes that lack functional hairpin wedges have adapted to rely heavily on the DNA bend angle to facilitate DNA unwinding. In this model, the RecQ translocase activity would drive unwinding by peeling ssDNA away from the duplex (or other folded structures such as triplex or quadruplex) as it moves in a 3′–5′ direction. Further biophysical experiments will be required to test this model in RecQ and to assess the importance of nucleic acid bending in the unwinding mechanisms of other helicases. In sum, our structural and biochemical observations have provided key insights into the DNA-binding and -unwinding mechanisms of RecQ enzymes and help to define the physical basis for SF2 helicase function.
Materials and Methods
Detailed experimental procedures used in this study can be found in SI Materials and Methods. A summary of the experimental procedures follows.
Structural Studies.
CsRecQ was crystallized in complex with hairpin DNA using the hanging-drop vapor diffusion method, and the structure was determined by molecular replacement with apo EcRecQ (10) as a search model. SAXS data were collected at the National Magnetic Resonance Facility at Madison, WI using a Bruker NanoStar instrument for CsRecQ catalytic core at two concentrations. Bacterial catalytic core crystal structures were docked manually into dummy atom models generated from the SAXS data, and the scattering of the resulting all-atom model was predicted.
Biochemical Experiments.
Equilibrium DNA binding was measured as RecQ-dependent changes in fluorescence anisotropy of fluorescein-labeled DNA. ATPase activity was measured as previously described (14), except a Synergy 2 plate reader (BioTek) was used to monitor changes in A340nm over time. Helicase activity was tested using a fluorescence assay in which the fluorophore is placed opposite a quencher and unwinding is measured as an increase in fluorescence.
Supplementary Material
Acknowledgments
We thank the staff of the Advanced Photon Source (LS-CAT beamline) and Ken Satyshur for assistance with data collection, and members of the J.L.K. laboratory for critical reading of this manuscript. This work was funded by National Institutes of Health (NIH) Grant GM098885 (to J.L.K.). K.A.M. was supported in part by NIH Training Grant in Molecular Biosciences GM07215. The SAXS instrument was funded by NIH Grant S10RR027000 (to S.E.B.). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH Grant P41GM103399 (NIGMS). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by Grant 085P1000817 from the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor.
Footnotes
Conflict of interest statement: J.L.K. is a cofounder of Replisoma, Inc.
This article is a PNAS Direct Submission. K.R. is a guest editor invited by the Editorial Board.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4TMU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416746112/-/DCSupplemental.
References
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Gorbalenya AE, Koonin EV. Helicases: Amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3(3):419–429. [Google Scholar]
- 3.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.German J. Bloom syndrome: A mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72(6):393–406. [PubMed] [Google Scholar]
- 5.Yu CE, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 6.Kitao S, et al. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22(1):82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 7.Van Maldergem L, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43(2):148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siitonen HA, et al. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12(21):2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 9.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem Sci. 1997;22(11):417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J. 2003;22(19):4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pike AC, et al. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA. 2009;106(4):1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swan MK, et al. Structure of human Bloom’s syndrome helicase in complex with ADP and duplex DNA. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 5):1465–1475. doi: 10.1107/S139900471400501X. [DOI] [PubMed] [Google Scholar]
- 13.Popuri V, et al. The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283(26):17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 14.Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: Role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33(22):6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384(6607):379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 16.Dillingham MS, Soultanas P, Wiley P, Webb MR, Wigley DB. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc Natl Acad Sci USA. 2001;98(15):8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27(16):3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soultanas P, Dillingham MS, Velankar SS, Wigley DB. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J Mol Biol. 1999;290(1):137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- 19.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90(4):635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 20.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97(1):75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 21.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127(7):1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18(2):177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharyya B, Keck JL. Grip it and rip it: Structural mechanisms of DNA helicase substrate binding and unwinding. Protein Sci. 2014;23(11):1498–1507. doi: 10.1002/pro.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10(1):110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 25.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol. 2006;358(4):1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 26.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophys Chem. 2010;149(3):67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 27.van Dongen MJ, et al. Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): Formation of a G-A base pair in the loop. Nucleic Acids Res. 1997;25(8):1537–1547. doi: 10.1093/nar/25.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Maizels N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001;29(8):1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon BP, Lu L, Chu A, Bissler JJ. RecQ and RecG helicases have distinct roles in maintaining the stability of polypurine.polypyrimidine sequences. Mutat Res. 2008;643(1-2):20–28. doi: 10.1016/j.mrfmmm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273(42):27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 31.Brosh RM, Jr, et al. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J Biol Chem. 2001;276(5):3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- 32.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274(18):12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 33.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107(2):521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo D, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27(23):3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JL, et al. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: The crystal structure provides insights into the mode of unwinding. Structure. 1998;6(1):89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 36.Machius M, Henry L, Palnitkar M, Deisenhofer J. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc Natl Acad Sci USA. 1999;96(21):11717–11722. doi: 10.1073/pnas.96.21.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa N, et al. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J Biochem. 1999;126(6):986–990. doi: 10.1093/oxfordjournals.jbchem.a022566. [DOI] [PubMed] [Google Scholar]
- 38.Yao N, et al. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4(6):463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharyya B, et al. Structural mechanisms of PriA-mediated DNA replication restart. Proc Natl Acad Sci USA. 2014;111(4):1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317(5837):513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myong S, Ha T. Stepwise translocation of nucleic acid motors. Curr Opin Struct Biol. 2010;20(1):121–127. doi: 10.1016/j.sbi.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: Role of the 1B domain in SF1B helicase activity. EMBO J. 2008;27(16):2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.