Significance
Paramyxoviruses, the cause of many important human and animal diseases, constitute a large family of enveloped negative-stranded RNA viruses including parainfluenza virus 5 (PIV5). The virion RNA is associated with ∼2,600 protomers of N-protein in the form of a helical ribonucleoprotein (RNP) (nucleocapsid). The RNP serves as the template for the viral polymerase in vivo. When expressed, N forms a 13 member N-ring that resembles the building block of the RNP. We have determined the atomic structure of the N-ring from PIV5 with 78 bound RNA residues. Precisely, six nucleotides of RNA are associated with each N protomer. Modeling PIV5-N in an “open conformation” in the ring structure indicates how transcription/replication could occur with minimal changes to the nucleocapsid structure.
Keywords: nucleoprotein, nucleocapsid ring, atomic structure, paramyxovirus, ribonucleoprotein
Abstract
Parainfluenza virus 5 (PIV5) is a member of the Paramyxoviridae family of membrane-enveloped viruses with a negative-sense RNA genome that is packaged and protected by long filamentous nucleocapsid-helix structures (RNPs). These RNPs, consisting of ∼2,600 protomers of nucleocapsid (N) protein, form the template for viral transcription and replication. We have determined the 3D X-ray crystal structure of the nucleoprotein (N)-RNA complex from PIV5 to 3.11-Å resolution. The structure reveals a 13-mer nucleocapsid ring whose diameter, cavity, and pitch/height dimensions agree with EM data from early studies on the Paramyxovirinae subfamily of native RNPs, indicating that it closely represents one-turn in the building block of the RNP helices. The PIV5-N nucleocapsid ring encapsidates a nuclease resistant 78-nt RNA strand in its positively charged groove formed between the N-terminal (NTD) and C-terminal (CTD) domains of its successive N protomers. Six nucleotides precisely are associated with each N protomer, with alternating three-base-in three-base-out conformation. The binding of six nucleotides per protomer is consistent with the “rule of six” that governs the genome packaging of the Paramyxovirinae subfamily of viruses. PIV5-N protomer subdomains are very similar in structure to the previously solved Nipah-N structure, but with a difference in the angle between NTD/CTD at the RNA hinge region. Based on the Nipah-N structure we modeled a PIV5-N open conformation in which the CTD rotates away from the RNA strand into the inner spacious nucleocapsid-ring cavity. This rotation would expose the RNA for the viral polymerase activity without major disruption of the nucleocapsid structure.
The Paramyxoviridae family of nonsegmented negative-strand RNA viruses includes many serious pathogens of humans and animals including mumps virus, measles virus, parainfluenza viruses 1–5, Nipah virus, Hendra virus, and Newcastle disease virus, all of which are in the Paramyxovirinae subfamily, and respiratory syncytial virus (RSV) and human metapneumovirus, which are in the subfamily Pneumoviridae. The Paramyxoviridae belong in the order Mononegavirales of membrane enveloped negative-strand RNA viruses (NSV), an order that includes the Rhabdoviridae (rabies virus and vesicular stomatitis virus; VSV), Orthomyxoviridae (influenza virus) and Filoviridae (Ebola virus) (1). Parainfluenza virus 5 (PIV5) is a paramyxovirus that was initially isolated from rhesus monkey-kidney cell cultures (2) and although PIV5 is not known to cause human disease (3), it is used as a prototype in the study of paramyxoviruses.
PIV5 has a nonsegmented single-stranded RNA genome of negative polarity of 15,246 nucleotides that encodes eight proteins (1): Three integral membrane proteins, fusion protein F and attachment protein hemagglutinin-neuraminidase (HN) and a small hydrophobic protein (SH). Inside the virion envelope lies a helical nucleocapsid core containing the RNA genome and the nucleocapsid (N), phosphoprotein (P), an innate immune system suppressor (V), and the large RNA-dependent RNA polymerase (L). Residing between the envelope and the core lies the viral matrix protein (M). The N protein and genome RNA together form a core structure, the ribonucleoprotein (RNP). The RNP of the NSVs share several functions essential to the virus life cycle: (i) N protein encapsidates the RNA genome forming the helical nucleocapsid structure necessary for the virus assembly and budding; (ii) it protects the genome from being recognized by the host innate immune response and prevents the RNA from degradation by nucleases (1); (iii) it serves as the template for the polymerase complex (P-L) during genomic replication and transcription. NSVs are a unique group of viruses where the nucleocapsid-associated RNA structure, rather than the naked RNA, is the only biologically active substrate for the viral polymerase complex activity (4). The structures of N proteins from different NSV families share common features despite low sequence identity across different families. Crystal structures of the N proteins of RSV (5), VSV (6), and rabies (7) have been obtained and have shown various oligomeric nucleocapsid-ring structures made by lateral contacts of single N protein molecules encapsidating an RNA strand that nestles in a long groove between the N-terminal domain (NTD) and the C-terminal domain (CTD) of successive N protomers. These nucleocapsid rings are most likely the building blocks of the long viral nucleocapsid-helical filaments encapsidating the viral RNA in vivo. Each ring could form one turn in the nucleocapsid helix after modeling-in a pitch in the ring formation based on early studies of tobacco mosaic virus (8). The pitch corresponds roughly to the height/thickness of the ring structure and can be modeled by introducing a slight slippage in the N–N protomer lateral contacts (5). The RNA encapsidated in the nucleocapsid rings is nuclease resistant (1), suggesting it would be inaccessible to the viral polymerase. For the L polymerase to gain access to the RNA it necessitates some flexibility in the N structure around the RNA binding groove, a dilemma that was partially answered in a recent study by Yabukarski and colleagues (9). They solved the structure of the RNA-free Nipah virus nucleoprotein in an open conformation showing that the hinge between the NTD and CTD domains is a flexible hinge allowing the NTD–CTD domains of N to assume different angles, thus hiding/or presenting the RNA in the groove as needed.
Here we describe the structure of a nucleocapsid-ring structure from PIV5 at 3.11-Å resolution. The structure shows that six nucleotides bind per N protein, consistent with the “rule of six” (10). The structural dimensions agree with data from electron microscopy (EM) (11). The structure shows high amino acid conservation in the RNA-binding pocket and for the scheme of RNA packaging. Based on the Nipah virus-N open conformation we propose a model where the RNA could become accessible to the viral polymerase with minimal changes to the nucleocapsid structure.
Results
PIV5-N Nucleocapsid-Ring Structure.
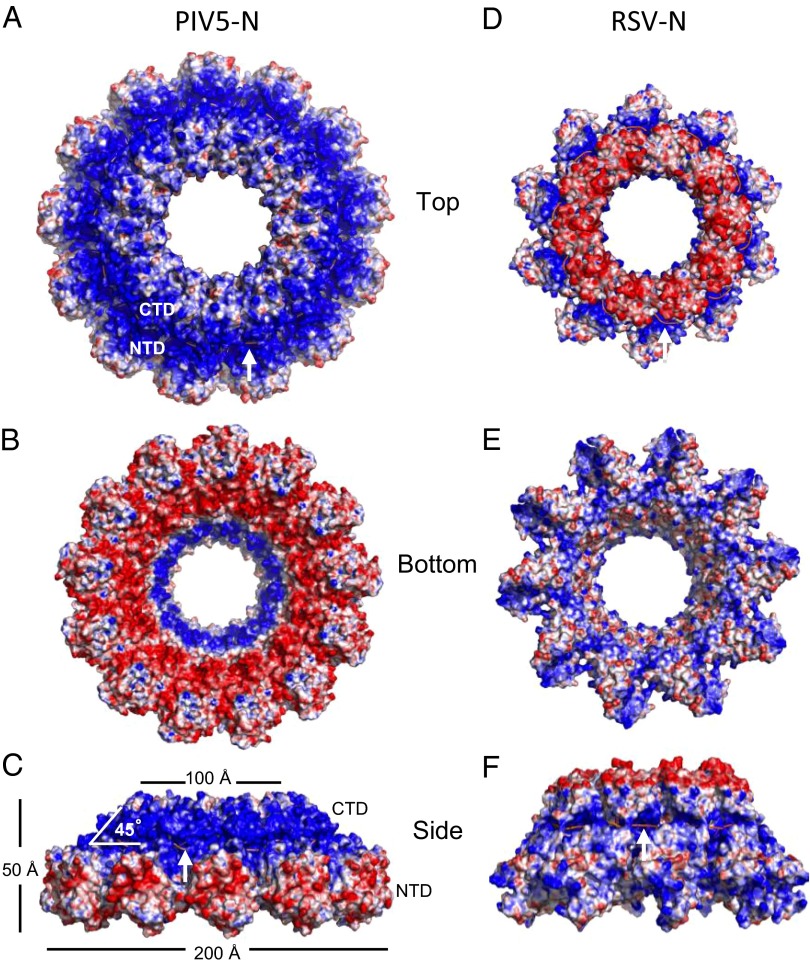
Recombinant PIV5-N was expressed in Escherichia coli and purified as described in Materials and Methods. An important step was the treatment of the N protein with RNase. At the beginning of purification the N protein was proteolyzed from the C terminus yielding a stable fragment of 48 kDa (Fig. 1A), with an absorbance of A260/A280 ∼1.0 suggesting that the protein bound nonspecific RNA. Electron microscopy imaging of the PIV5-N preparation showed ring structures with 20 nm diameters (Fig. 1B). PIV5-N was crystallized and its X-ray structure solved to 3.11 Å resolution with a space group of C2221 (Table 1). It had one large ring structure of 600 kDa in the asymmetric unit consisting of an RNA strand wrapped in a groove around 13 PIV5-N molecules (Fig. 2A, see also Fig. 4A). The 13-mer ring of PIV5-N is 20 nm wide at the bottom and 10 nm wide at the top with a cone-like cavity that runs in the middle of the ring structure with a narrow opening at the top of the ring of ∼5 nm (Figs. 2A and 3C). This narrow-top (positively charged), wide-bottom (negatively charge) structure (Fig. 3) owes to the fact that each N protomer rotates 45° relative to the plane of the RNA ring, with CTD (Fig. 2B, salmon) pointing toward the inside of the ring and the NTD (Fig. 2B, light blue) toward the outside (Fig. 3C), an orientation optimal for the packing of the nucleocapsid rings on top of each other to form the long nucleocapsid helix.
Fig. 1.
(A) Western blot of purified PIV5-N after proteolysis. Lane 1, molecular weight ladder. Lane 2, 2 μg of PIV5-N. (B) Electron microscopy of the purified PIV5-N protein shown in A. Top left inset magnifies one of the observed ring structures. (Scale bar, 50 nm for big box and 10 nm for Inset).
Table 1.
Crystallographic Statistics for Pb1 set
| Data collection | |
| Space group | C 2 2 21 |
| Cell dimensions | |
| a, b, c, Å | 205.64, 309.44, 233.24 |
| α, β, Υ, ° | 90 |
| Wavelength, Å | 0.95041 |
| Resolution limit, Å* | 3.11 (3.17–3.11) |
| Rmerge, %* | 17.8 (86.7) |
| (I/σI)* | 11.8 (2.05) |
| Completeness, %* | 99.9 (100) |
| Redundancy* | 7.0 (6.3) |
| Refinement | |
| Resolution, Å* | 45.01–3.11 (3.14–3.11) |
| Unique reflections | 132,287 |
| Rwork/Rfree, % | 22.71/ 26.25 |
| Number of atoms | |
| Protein | 40,755 |
| Nucleic Acid | 1,560 |
| Lead(II) | 14 |
| rmsd | |
| Bond lengths, Å | 0.01 |
| Bond angles, ° | 1.449 |
| Ramachandran plot | |
| Favored | 95.04% |
| Generously allowed | 3.19% |
| Molprobity score | |
| Overall | 2.24 (99th percentile) |
| Clashcore | 13.99 (96th percentile) |
*Data for highest resolution shell are indicated in parenthesis.
Fig. 2.
(A) Cartoon representation of PIV5-N nucleocapsid ring 13-mer structure (top view) with chains colored differently and the RNA strand shown in black. (B) Side view (from outside of ring) of PIV5-N monomer with domains colored accordingly: Narm (3–31) in dark blue, N-terminal domain (NTD) (32–263) in light blue, C-terminal domain (CTD) (264–372) in salmon, and Carm (373–401) in red. The bound RNA is shown in black. (C) RSV-N monomer structure with RNA (black) bound in the RNA binding pocket. (D) Nipah-N monomer structure missing both Narm and Carm. (E) View of the protomer–protomer interface from inside of ring, showing the interaction of the Narm and Carm with neighboring protomers N − 1 and N + 1. The CTD α11–η2–α12 hydrophobic pocket is labeled in blue font.
Fig. 4.
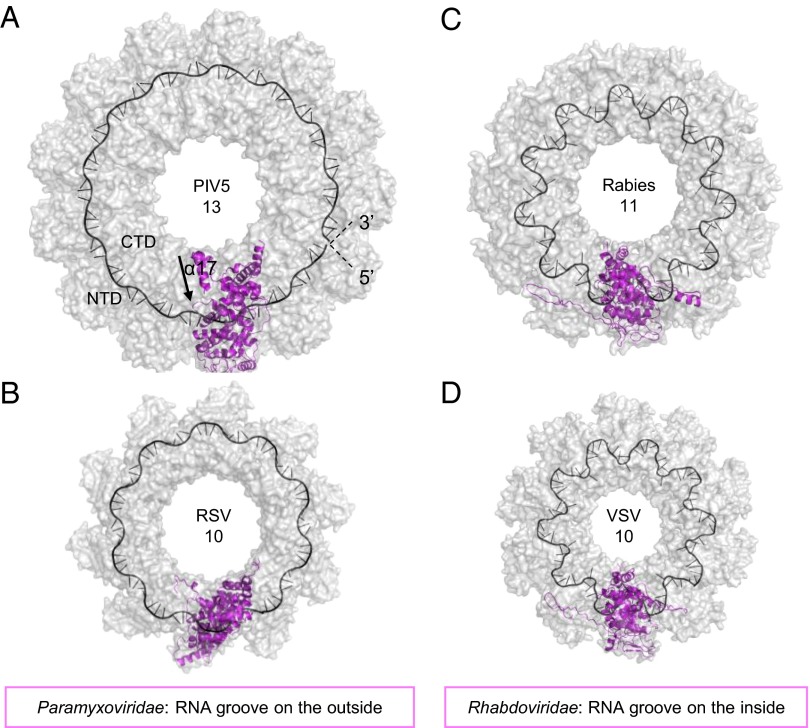
Surface representation (gray) of the nucleocapsid ring structures from different negative-strand RNA viruses. Paramyxoviridae virus nucleocapsid like PIV5-N 13-mer (A) and RSV 10-mer (B) with RNA (black cartoon) bound from the outside of the ring. Rhabdoviridae viruses like rabies 11-mer (C) and VSV 10-mer (D) with the RNA bound on the inside of the ring. One N-protomer in each ring is displayed as cartoon (magneta). Arrow in A indicates direction of helix α17.
Fig. 3.
Molecular surface representation of PIV5-N (A–C) and RSV-N (D–F) nucleocapsid rings colored by the electrostatic potential, ranging from −10 kT/e (red) to +10 kT/e (blue). RNA strand (salmon) is indicated by white arrow for clarity. PIV5-N charge distribution is more polarized than RSV-N with positive charges on the outside-top of the ring and negative charges on the inside-bottom of the ring.
N-Protomer Structure.
Each PIV5-N protomer had unambiguous electron density such that amino acids could be traced from residues 3–401 with the exception of residues 183–186. Similar to the RSV-N structure (Fig. 2C), PIV5-N protein could be separated into several subdomains: the Narm (3–31), the NTD (32-263), the CTD (264-372), and the Carm (373-401), where the NTD and CTD form the core of the protomer and the Narm and Carm extend out of the core toward the neighboring protomers to form tight lateral connections (Fig. 2 B and E). Nipah virus-N, which was crystallized in a monomeric open conformation (9), has a similar domain organization to PIV5-N and RSV-N. However, it lacks both the Narm and the Carm, and it did not bind RNA (Fig. 2D). PIV5-N core is mostly helical with the NTD formed by helices α2 to α10, one 3/10 helix η1, and two small parallel β-strands at the bottom of the molecule. The CTD is formed by helices α11 to α15, and five 3/10 helices η2 to η6. Both the Narm and the Carm are helices connected to the core by long flexible loops (Fig. 2B and Fig. S1). Based on the orientation of helix α17, which forms the last structured region in the C terminus, the missing C-terminal 108 amino acid residues from the PIV5-N structure would point toward the outside of the ring (Fig. 4A), similar to the “wheat head” structure described for RSV-N (5).
Nucleoprotein Interface in Ring Structure.
The protomer–protomer interface in RSV (5), VSV (6), and rabies (7) nucleocapsid rings consists of the Narm/Carm and the CTD domains of neighboring protomers, with the NTD making minimal domain contact. In contrast, in PIV5-N the interfaces make hydrogen-bonds, salt bridges, and hydrophobic interactions that span the whole length of two neighboring protomers with all of the domains contributing to the interface (Table 2). Each N has ∼2900 Å2 buried surface area with each of its 5′-neighbor (N − 1) and 3′-neighbor (N + 1), and the NTD contributes 46% to the N:N + 1 interface and 12% to that of N:N − 1 (Fig. 2E and Table 2). The Narm forms 37% of the N:N − 1 interface where it lies in a hydrophobic pocket formed by N − 1 CTD α11-η2-α12 motif, and the Carm forms 23% of the N:N + 1 interface where it lies on the top of a hydrophobic groove formed by the same α11–η2–α12 motif on N + 1 (Fig. 2E) (Table 2). PIV5-N amino acid alignment with N sequences of the Paramyxovirinae subfamily showed a level of amino acid identity ranging from 22% (Sendai virus-N) to 58% (mumps virus-N) (Fig. S1). However, lower levels of identity were found for other NSV N proteins (15.7% identity RSV-N, 13.9% rabies-N, and 15.3% VSV-N). Most of the conservation is concentrated in the RNA binding pocket and the N–N interface residues (such as α11–η2–α12 motif) suggesting that nucleocapsid-ring structures from other Paramyxovirinae would adopt a similar 13-mer conformation and not an RSV-like 10-mer structure, which can also be purified as an 11-mer (5).
Table 2.
Contribution of each domain to N–N interface in Å2
| Protein | N:N + 1 | N:N− 1 |
| Total | 2,946 | 2,908 |
| N-arm | 0 | 1,079 (37%) |
| NTD | 1,363 (46%) | 353 (12%) |
| CTD | 825 (28%) | 1,244 (42%) |
| C-arm | 666 (23%) | 80 (3%) |
Values in parenthesis indicate percentage of total interface.
RNA Binding.
The RNA strand in the PIV5-N nucleocapsid ring is 78 nucleotides (nt) long (6 nt per N protein protomer) in the 5′–3′ clockwise orientation (Fig. 4) and thus consistent with the “rule of six” in which the genome length of the Paramyxovirinae has to be a number of nucleotides that is divisible by six for efficient viral replication to occur. All of the bases were modeled as uracil despite the fact some electron densities/geometry suggested a bigger purine base. PIV5-N belongs to the Paramyxovirinae subfamily members of which obey the rule of six, in contrast to other NSVs where RSV-N binds to seven nucleotides, and VSV and rabies-N bind to nine nucleotides. Because PIV5 has to pack fewer nucleotides into a similar size RNA binding pocket than other NSVs, PIV5-N associated RNA has a less convoluted structure compared with the other NSV RNAs (Fig. 4) where the structure is known. The PIV5 N-associated RNA is nestled in a positively charged groove that is formed by the continuity of the pocket between the NTD–CTD domains of N protomers in the ring (Fig. 3). Whereas the RNA groove of Rhabdoviridae-Ns face to the inside of the ring cavity, those of PIV5-N and RSV-N face to the outside of the ring (Fig. 4).
RNA Binding Pocket Is Conserved.
Each N-protomer binds to six nucleotides and extends into the pockets of its N + 1 and N − 1 neighbors (Fig. 5A). Structural superposition of the PIV5-N and RSV-N monomers shows that the RNA configuration is highly conserved with a three-base stack (1–2–3) pointing in toward the protein (Fig. 5 B, C, and F), and with a three-base stack (4–5–6) oriented outwards to the solvent (Fig. 5 D–F), although RSV packs an additional nucleotide/ protomer compared with PIV5-N. The structure and amino acid sequence identity of the RNA binding pocket is highly conserved among paramyxoviruses (Fig. 5), and it can be divided into two subpockets. Pocket 1 consists of conserved residues from the NTD domain (K194, R195, Q202, Y260) and they bind to the bases facing inwards (Fig. 5 B and F). Pocket 2 consists of less conserved residues from the CTD domain (M266, G267, Y350, A351, R354, S355) and they bind bases facing outwards (Fig. 5 D and F). In pocket 1, RNA hydrogen bonding occurs through amino acid side chains, whereas in pocket 2 (with the exception of R354) hydrogen bonds are through main chain amide atoms. All of the above interactions occur via the phosphate–ribose backbone of the RNA with the exception of Q202 in pocket 1, which interacts with the O2 group of the base (Fig. 5 B and F).
Fig. 5.
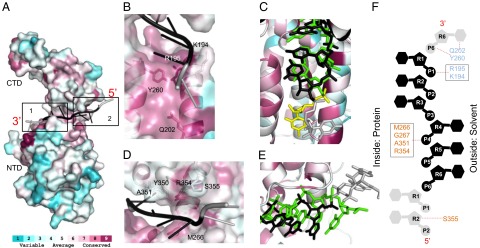
Conserved RNA binding pocket. (A) Consurf figure mapping amino acid conservation on PIV5-N structure based on alignment in Fig. S1 with conservation color scale shown at bottom. Each monomer binds to six nucleotides (black) and extends into the binding pocket of neighboring N molecules (gray). Two major binding pockets. (B) Close-up view of pocket 1: constitutes of conserved residues from NTD and binds the bases facing inward. (C) Close-up view of pocket 1: superposing PIV5-N RNA (black) and RSV-N RNA (green). Nucleotides from neighboring protomers are shown in gray (PIV5-N) and yellow (RSV-N). RNA is shown in stick representation. (D) Close-up view of pocket 2: constitutes of conserved residues from CTD and binds bases facing outward. (E) Same as C but for pocket 2. (F) Schematic representation figure showing the interaction of PIV5-N residues from pocket 1 (colored in light blue) and pocket 2 (colored in salmon) with RNA nucleotides (same color scheme as in A). The RNA is shown in 5′–3′ direction from bottom-to-top. Circles represent phosphates, pentagons represent the ribose sugars, hexagons represent the bases.
Open vs. Closed Conformation.
Superposition of the PIV5-N and Nipah-N structures (9) showed a poor overlap with a root-mean-square deviation (rmsd) of 3.6 Å. However, when the CTD and NTD domains were superposed separately they showed a much better overlay with rmsd values of 0.659 Å and 1.112 Å respectively, suggesting that they are nearly identical in structure. RSV CTD domain has an rmsd of 1.587 Å with that of PIV5-N CTD; however, its NTD is more divergent with an rmsd of 3.2 Å (Fig. 6A). Nipah-N was crystallized in complex with a phosphoprotein (P50) N-terminal fragment, where Nipah-N is in an open configuration. The only structural difference it has with PIV5-N is the angle between the NTD–CTD domains formed at the hinge region between PIV5-N helices α10 (NTD) and α11 (CTD) indicated in Fig. 6 B and C by an arrow. Modeling the binding of Nipah-P50 fragment onto the PIV5-N structure shows that it nestles almost perfectly into the PIV5-N CTD α11–η2–α12 hydrophobic pocket, interfering with the binding of Carm from the N − 1 protomer (Fig. 6D, box2) and the Narm from the N + 1 protomer (Fig. 6D, box 1). This domain exchange disruption results in the N-CTD losing 62% of its interface with N + 1 and 40% of its interface with N − 1. The CTD domain would now be free to rotate almost 20° counterclockwise around the hinge region and be able to assume a Nipah-like open conformation as modeled in Fig. 6 B and C. Modeling this open conformation of PIV5-N onto its nucleocapsid-ring structure shows the CTD rotating toward the center of the ring cavity (Fig. 6E) exposing the RNA in the RNA-binding groove (Fig. 6 E and F) making it accessible for the viral polymerase during genome transcription and replication.
Fig. 6.
Open vs. closed conformation. (A) Superposition of PIV5-N (gray) CTD (Left) and NTD (Right) domains with the corresponding domains from Nipah-N (blue, Upper) and RSV-N (green, Lower). (B) Superposing full-length PIV5-N (gray) and Nipah-N (blue) at their NTD domains shows their respective CTDs at a 20° angle. The (arrow) indicates PIV5-N flexible hinge region between NTD and CTD. (C) Overlay of PIV5-N open conformation model (gray) on Nipah-N (blue). (D) Modeling of Nipah-P fragment (green) on PIV5-N structure overlaps with the Narm from N + 1 (box1) and Carm from N − 1 (box2). (E) Modeling the PIV5-N open conformation in the nucleocapsid ring structure shows CTD tilting toward the central-cavity of the ring away from the RNA (black cartoon). The N-protomer in open conformation is colored according to Fig. 2. (F) Close-up view of the exposed RNA groove in the PIV5-N open conformation.
Discussion
Here we report a high-resolution X-ray crystal structure of the PIV5-N nucleocapsid ring from the Paramyxovirinae subfamily. The finding that six nucleotides bind to each N protomer is consistent with the “rule of six” whereby it was determined that the Sendai virus genome was only replicated efficiently when the total genome nucleotide length was a number divisible by six (10). This finding led to the prediction that each N protomer bound six nucleotides (10). Subsequently, it was found that all members of the Paramyxovirinae had a genome length that was a number divisible by six (12).
Nucleocapsid-ring structures can be thought of as representing one turn of a nucleocapsid helix that encapsidates the viral RNA in vivo (8). Our X-ray data shows a 13-mer ring with a diameter of 20 nm, an inner hole cavity of 5 nm in diameter, and a pitch of 5 nm, which agrees closely with previous measurements performed on native measles virus (13) and Sendai virus nucleocapsid-helix structures (11). It was calculated that there were 13.2 N molecules per helical turn and the helix was 20 nm wide with a 5.3-nm pitch (11). PIV5-N rings are narrow and positively charged on the outside-top, and wide and negatively charged on the inside-bottom (Fig. 3). This charge distribution makes the rings ideal for stacking on top of each other to build a nucleocapsid helix. The resulting PIV5-N helix would be mostly negatively charged on the outside, but that could be different among Paramyxovirus family members as RSV has a less polarized surface charge distribution in comparison (Fig. 3). This difference in charge distribution might explain why different Paramyxovirus nucleocapsid helices react differently to different salt concentrations: more coiled, rigid, with smaller helix-pitch evident at high salt concentrations, whereas low salt induces less coiled, more flexible, higher pitch structures (14). Nucleocapsid-ring structures can assume different pitch dimensions depending on the angle N protomers assume with respect to the RNA-strand ring (Fig. 3C), which in PIV5-N structure is 45°. Rotating the N protomer clockwise or counterclockwise relative to the RNA plane can result in lower or higher pitches respectively. Treating nucleocapsid helices with trypsin, which results in a 48-kDa N protein, results in a more coiled and rigid structure with a lower pitch (5.3 nm) (15) (16) implicating a role for the missing 108 amino acid residues from the PIV5-N atomic structure, due to proteolysis, in regulating pitch dimensions and hence the nucleocapsid flexibility.
Based on the electron density we could trace from residues 3–401, which, similar to the RSV-N structure (5), can be separated into four subdomains: Narm, NTD, CTD, and Carm. The missing residues 183–186 form an unstructured loop between helices α6 and α7 at the edge of the RNA binding groove and seem to be displaced by the binding of the RNA strand. The RSV-N equivalent loop (K170–S177) is structured and forms the edge of the RNA-binding groove by reaching up and forming hydrogen bonds with RSV-N (R338) in the CTD domain (5). The fact that RNA affects these equivalent loops differently in PIV5-N and RSV-N might indicate a difference in sizes in their RNA binding pockets.
The nucleocapsid ring encapsidates a 78-nt RNA strand which nestles in a positively charged groove on the outside of the ring. The RNA was derived from E. coli cells where N protein was expressed and as there was no specificity to the RNA binding, all of the bases were modeled as uracil. The Arp-Warp (17) program first built a single stranded DNA polynucleotide strand in the electron density of the initial solved phases; however, inspection of the refinement mFo − DFc difference maps at 3 σ contour level resulted in a positive electron density at all positions corresponding to the ribose 2′-OH group, suggesting indeed that it was an RNA rather than a DNA strand. The fact that the PIV5–RNA groove faces the outside of the ring doesn’t suggest that the RNA is more exposed or less protected, because the RNA was not cleaved by treatment with 0.1 mg/mL of RNase A for 30 min at room temperature during protein preparation. On the other hand, this orientation gives the advantage of an RNA strand being accessible by the viral polymerase machinery without major disruption to the nucleocapsid structure.
The RNA binding pocket of PIV5-N shares a lot of similarities with that of RSV-N both at the protein level and the RNA packing level. Both PIV5-N and RSV-N share the three-base-in three-base-out RNA conformation, with the extra seventh nucleotide of RSV being accommodated by the larger protomer–protomer interspace in the RSV-N 10-mer ring. In both PIV5-N and RSV-N, the RNA pocket could be divided into two subpockets: pocket 1 and pocket 2 consisting of highly conserved residues from NTD and CTD, respectively. Although, pocket 1–RNA interactions are through the amino acid side chains and those in pocket 2 are through backbone amide atoms, the side chain identities of pocket 2 residues still play an important role as mutations in VSV targeting Ser290 (A351 in PIV5-N) resulted in ring formations unable to encapsidate RNA (18), and mutating VSV-Y289 (Y350 in PIV5-N) affects RNA encapsidation and P binding, and consequently viral transcription and replication (19).
Because PIV5-N and Nipah-N (Fig. 6) NTD and CTD domains are almost identical when superimposed separately, we modeled a PIV5-N open structure, based on that of Nipah-N, by rotating the CTD domain 20° away from the RNA groove. Yabukarski and colleagues (9) suggested that P50 keeps the Nipah-N structure in the open conformation by rigidifying the helices in its CTD domain. However, VSV-N was stabilized in the closed conformation by its P60 protein fragment (20), which also replaces its RNA in the VSV-N–binding pocket, implying that P does not induce open RNA–groove conformation. Our data suggests that N could exist naturally in an open conformation, regardless of the P fragment, due to charge repulsion between the positively charged roof and ceiling of the RNA binding pocket (Fig. 3), resulting in the CTD and the NTD rotating away from each other. RNA binding in the groove then induces the closed conformation of N by bringing together the CTD and NTD domains supported by the finding that VSV nucleocapsid rings, which were defective in RNA binding, were in a slightly open conformation. The closed conformation of the nucleocapsid ring is then stabilized by the lateral arm connections between adjacent N-protomers (18), because the removal of RNA from already formed nucleocapsid rings doesn’t result in the opening of the RNA groove (21).
It was previously hypothesized that the NTD is the domain that bends downward when the open conformation is induced. This assumption was based on the fact that NTD made minimal lateral contacts in the RSV and VSV nucleocapsid rings (5, 6, 9, 18). The NTD domain in the PIV5-N ring contributes significantly to the lateral interface (Table 2). Moreover, when the phosphoprotein P50 fragment of Nipah-P is modeled on the PIV5-N structure, it binds to the CTD domain α11–η2–α12 hydrophobic motif, disrupting the arm exchange with neighboring protomers, resulting in CTD losing 60% and 40% of its contacts with its neighbors, whereas the NTD interface remains unaffected. This rearrangement of the CTD interface supports our hypothesis that the CTD is the domain that rotates upon P50 binding and not the NTD. Once CTD rotates 20° toward the spacious center ring-cavity, pocket 2, which harbors the three-bases-out stack, is exposed and the viral polymerase could start transcription from the 3′ end of the RNA strand. In PIV5, this open N subunit will remain bound to other subunits in the nucleocapsid via NTD–NTD interactions. However, in RSV where NTD forms minimal contact with the neighboring protomers, P could play a role in maintaining the open N subunit attached to the polymerase holo-complex, such that N could recover its original location on the nucleocapsid as the polymerase complex moves on along the RNA. The paramyxovirus P protein, which is a flexible tetramer (22), could induce up to four N-protomers to be in the open conformation at the site of readout, exposing up to 24 nucleotides, which would give more space for the polymerase to accommodate the RNA in its active site. The fact that each N-protomer extends into its neighboring RNA pockets could be indicative that once polymerase activity starts, and the first N molecule snaps open, it could start a chain reaction downstream on the RNA strand (3′ to 5′ direction) where consecutive RNA pockets are destabilized and induced to assume the open conformation.
Materials and Methods
PIV5-N Expression and Purification.
The coding sequence of PIV5-N (PIV5-W3A strain) was cloned into the pET28b expression vector (Novagen); the construct included an N-terminal His-tag followed by a thrombin cleavage site. PIV5-N was expressed from this vector in Escherichia coli (T7 Express lysY/Iq). Bacteria were grown at 37 °C to an OD600 of 0.6, at which point expression of PIV5-N was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Bacteria were grown for a further 18 h at 18 °C, harvested by centrifugation (20 min, 3,752 × g, 4 °C), resuspended in 40 mL per L of original bacterial culture of lysis buffer [1 M NaCl, 50 mM Tris pH 7.4, 1 M Urea, 5% (vol/vol) glycerol]. The lysis buffer was supplemented with EDTA-free complete protease inhibitors (Roche) (1 tablet per 40 mL of lysis buffer), 0.2 mg/mL lysozyme (Sigma), DNase I (Roche) and RNase A (Sigma) at 100 μg/mL concentrations each. Resuspended bacteria were lysed by five rounds of 2-min-pulse sonications with alternate 2-min incubations on ice. The lysate was centrifuged (20 min, 27,000 × g, 4 °C), and the supernatant fraction, which contained soluble PIV5-N, was purified by applying it to a 5-mL Ni2+-nitrilotriacetic (NTA) agarose column (Qiagen), which had been equilibrated with 3 column volumes of lysis buffer. The column was washed with 5 column volumes of lysis buffer containing 5 mM imidazole, followed by 5 column volumes of lysis buffer containg 20 mM imidazole. PIV5-N was eluted with 10 mM Tris pH 7.4, 250 mM NaCl, 250 mM imidazole, concentrated to 10 mg/mL, and further purified by gel filtration chromatography (Superdex 200 10/300, GE Healthcare) in 250 mM NaCl, 10 mM Tris, pH 8.0. PIV5-N was concentrated to 27 mg/mL in this same buffer, flash frozen in liquid N2, and stored at −80 °C. The concentration of PIV5-N was determined using a calculated ε280 of 46,300 M−1.
Structure Determination.
Crystals of PIV5-N were grown at 25 °C using the vapor diffusion sitting drop method with 0.6 μL of PIV5-N (23–35 mg/mL) mixed with a precipitant solution consisting of 0.3 μL of 30% (vol/vol) PEG 400, 0.1 M Mes sodium salt pH 6.5, 0.1 M MgCl2. Crystals were harvested and soaked in the above precipitant condition supplemented with 2.5 mM lead acetate trihydrate, with PEG 400 acting as the cryoprotectant. Three (180°, 1° oscillation range) datasets of Pb- derivatives (Pb1, Pb2, and Pb3) were collected at the Pb l-III edge λ = 0.95041 at the Life Science Collaborative Access Team (LSCAT) beamline 21-ID-D at the Advanced Photon Source.
PIV5-N lead (Pb) derivative datasets were processed and scaled separately using HKL2000 (23) to a resolution of 3.11 Å (Table 1). The crystals belonged to space group C2221 and had 13 molecules per asymmetric unit with a solvent content of 58%. To obtain accurate phases the three Pb datasets were merged in SCALA (24), and the resulting Pb-merge dataset was used in SHELX C/D (25) to determine a substructure solution of 8 Pb atoms in a half-disk geometry. The Pb-merge heavy metal solution was used in PROFESS (17) to determine noncrystallographic symmetry (NCS) operators which were used for density averaging in DM (17) to improve the quality of the SAD phases (FOM = 0.2) obtained from SHARP (26). The DM output map showed an outline of a ring structure and its map coefficients were used as an input to determine more accurate anomalous substructure solution and phases in PHENIX-EP (SAD-MR) program (27), this time using a single Pb dataset (Pb1) as input-reflection. The resulting phases had a figure of merit (FOM) of 0.286 and were density modified in RESOLVE (27) using extreme density modification. The resulting map was used in PHASER-EP (SAD-MR) again to improve initial phase quality, which resulted in accurate interpretable phases with a FOM of 0.855. NAUTILUS (17) was used to build a single continuous ring-shaped RNA strand, which was used as a starting model in BUCCANEER (17) to build 13 molecules of PIV5-N around the RNA structure with an initial Rwork/Rfree of 29.38/34.38. The model was manually adjusted and completed using COOT (28) and RCrane (29). Refinement of the model was carried out to 3.11-Å resolution limit in Phenix (27), with a random 5% of the data being omitted from refinement for determination of Rfree. Each round was followed by manual model building and adjustment as guided by inspection of σA-weighted 2mFo − DFc and mFo − DFc difference maps using COOT. Each round of refinement consisted of bulk solvent correction, followed by refinement of coordinates, individual B-factors, occupancies and automatic NCS restraints.
Structure validation was carried out with Molprobity (30), which showed that 95.04% and 98.23% of PIV5-N residues were in the favored and generously allowed regions of the Ramachandran plot, respectively. The structure had a Molprobity score of 2.24 (99th percentile) and a clashscore of 13.99 (96th percentile). Molecular figures were generated with PyMol (pymol.sourceforge.net).
Electron Microscopy.
NP complexes at a total protein concentration of ∼5 μg/mL were absorbed onto 300-mesh copper grids covered with a carbon film that had been freshly glow discharged. After wicking off excess solution, grids were stained with a freshly prepared 2% (wt/vol) aqueous solution of uranyl formate that was filtered immediately before use. Samples were observed in a JEOL 1400 electron microscope operated at 120kV, and images were acquired with a Gatan Ultrascan 4000 4k by 4k CCD camera at the Cryo-Electron Microscopy facility, Northwestern University, Evanston, IL. The diameters of the NP complexes were measured using Image J (US National Institutes of Health, Bethesda).
Western Blot.
Purified PIV5-N (2 μg) was electrophoresed on a 15% SDS/PAGE for 1 h at 180 V. The gel was blotted on PVDF membrane for 5 h at 30 V at 4 °C. The membrane was then blocked for 30 min with 5% milk, then treated with 1:100 dilution of PIV5-N primary monoclonal antibody (NP 214mAb), followed by a FITC-conjugated goat anti-mouse secondary antibody with extensive washes in between. The protein was detected on a Fuji FLA-5100 image reader with Multi Gauge v3.0 software (Fuji Medical Systems).
Supplementary Material
Acknowledgments
M.A. and G.P.L. are Specialists and R.A.L. is an Investigator of the Howard Hughes Medical Institute. We acknowledge use of the High Throughput Analysis Laboratory at Northwestern University. Our use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CHI1357. Our work at Life Sciences Collaborative Access Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor program (Grant 085PI000817).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4XJN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503941112/-/DCSupplemental.
References
- 1.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Wolters Kluwer/Lippincott, Williams and Wilkins; Philadelphia: 2013. pp. 957–995. [Google Scholar]
- 2.Hull RN, Minner JR, Smith JW. New viral agents recovered from tissue cultures of monkey kidney cells. I. Origin and properties of cytopathogenic agents S.V.1, S.V.2, S.V.4, S.V.5, S.V.6, S.V.11, S.V.12 and S.V.15. Am J Hyg. 1956;63(2):204–215. doi: 10.1093/oxfordjournals.aje.a119804. [DOI] [PubMed] [Google Scholar]
- 3.Goswami KKA, Lange LS, Mitchell DN, Cameron KR, Russell WC. Does simian virus 5 infect humans? J Gen Virol. 1984;65(Pt 8):1295–1303. doi: 10.1099/0022-1317-65-8-1295. [DOI] [PubMed] [Google Scholar]
- 4.Arnheiter H, Davis NL, Wertz G, Schubert M, Lazzarini RA. Role of the nucleocapsid protein in regulating vesicular stomatitis virus RNA synthesis. Cell. 1985;41(1):259–267. doi: 10.1016/0092-8674(85)90079-0. [DOI] [PubMed] [Google Scholar]
- 5.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326(5957):1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 6.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313(5785):357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 7.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 8.Klug A. From macromolecules to biological assemblies. Nobel Lecture, 8 December 1982. Biosci Rep. 1983;3(5):395–430. doi: 10.1007/BF01121953. [DOI] [PubMed] [Google Scholar]
- 9.Yabukarski F, et al. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol. 2014;21(9):754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 10.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67(8):4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egelman EH, Wu SS, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63(5):2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolakofsky D, et al. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72(2):891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakker SE, et al. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J Gen Virol. 2013;94(Pt 8):1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heggeness MH, Scheid A, Choppin PW. Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: Reversible coiling and uncoiling induced by changes in salt concentration. Proc Natl Acad Sci USA. 1980;77(5):2631–2635. doi: 10.1073/pnas.77.5.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mountcastle WE, Compans RW, Caliguiri LA, Choppin PW. Nucleocapsid protein subunits of simian virus 5, Newcastle disease virus, and Sendai virus. J Virol. 1970;6(5):677–684. doi: 10.1128/jvi.6.5.677-684.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mountcastle WE, Compans RW, Lackland H, Choppin PW. Proteolytic cleavage of subunits of the nucleocapsid of the paramyxovirus simian virus 5. J Virol. 1974;14(5):1253–1261. doi: 10.1128/jvi.14.5.1253-1261.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Green TJ, Tsao J, Qiu S, Luo M. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J Virol. 2008;82(2):674–682. doi: 10.1128/JVI.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayak D, Panda D, Das SC, Luo M, Pattnaik AK. Single-amino-acid alterations in a highly conserved central region of vesicular stomatitis virus N protein differentially affect the viral nucleocapsid template functions. J Virol. 2009;83(11):5525–5534. doi: 10.1128/JVI.02289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leyrat C, et al. Structure of the vesicular stomatitis virus N⁰-P complex. PLoS Pathog. 2011;7(9):e1002248. doi: 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green TJ, et al. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J Virol. 2011;85(6):2714–2722. doi: 10.1128/JVI.01927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol. 2000;7(9):777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter JCW, Sweet RM, editors. Methods in Enzymology. Academic Press; San Diego: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 24.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: Recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 11):2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Keating KS, Pyle AM. RCrane: Semi-automated RNA model building. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 8):985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.