Significance
Much of the mammalian genome recently was shown to be transcribed to long noncoding RNAs, one class of which is the enhancer RNAs (eRNAs) whose levels largely correlate with the mRNA levels of the target gene but whose functions are not yet clear. We examined the eRNA produced from a functional enhancer that directs cell-specific expression of the gonadotropin hormone α-subunit gene, chorionic gonadotropin alpha, in the pituitary. We show that this eRNA plays a crucial role in facilitating DNA looping between the enhancer and promoter and directs histone modifications that are essential for transcription initiation and without which the chromatin becomes repressive to transcription. In this way, the eRNA mediates the function of the enhancer in directing basal gene expression.
Keywords: enhancer, chromatin, ncRNA, gonadotropin, pituitary
Abstract
Since the discovery that many transcriptional enhancers are transcribed into long noncoding RNAs termed “enhancer RNAs” (eRNAs), their putative role in enhancer function has been debated. Very recent evidence has indicted that some eRNAs play a role in initiating or activating transcription, possibly by helping recruit and/or stabilize binding of the general transcription machinery to the proximal promoter of their target genes. The distal enhancer of the gonadotropin hormone α-subunit gene, chorionic gonadotropin alpha (Cga), is responsible for Cga cell-specific expression in gonadotropes and thyrotropes, and we show here that it encodes two bidirectional nonpolyadenylated RNAs whose levels are increased somewhat by exposure to gonadotropin-releasing hormone but are not necessarily linked to Cga transcriptional activity. Knockdown of the more distal eRNA led to a drop in Cga mRNA levels, initially without effect on the forward eRNA levels. With time, however, the repression on the Cga increased, and the forward eRNA levels were suppressed also. We demonstrate that the interaction of the enhancer with the promoter is lost after eRNA knockdown. Dramatic changes also were seen in the chromatin, with an increase in total histone H3 occupancy throughout this region and a virtual loss of histone H3 Lys 4 trimethylation at the promoter following the eRNA knockdown. Moreover, histone H3 Lys 27 (H3K27) acetylation, which was found at both enhancer and promoter in wild-type cells, appeared to have been replaced by H3K27 trimethylation at the enhancer. Thus, the Cga eRNA mediates the physical interaction between these genomic regions and determines the chromatin structure of the proximal promoter to allow gene expression.
Transcriptional enhancers comprise regions of DNA that act in cis to increase basal transcription by elevating promoter activity and often facilitate or determine the gene’s cell-specific expression. Their function is characteristically location and direction independent because of DNA looping between the enhancer and promoter of the target gene. It has long been proposed that the activity of enhancers is via sequence-specific DNA-binding proteins, often lineage-determining transcription factors, which interact with and stabilize the general transcription machinery to facilitate cell-specific gene expression (1). However, additional possibilities for enhancer function have arisen since the discovery that these regions often are transcribed to long noncoding RNAs (lncRNAs) or enhancer RNAs (eRNAs), many of whose levels correlate with those of their target genes (2–5). eRNAs are usually 800- to 2,000-bp long and have some distinctive characteristics; e.g., many of them are transcribed bidirectionally on both DNA strands from a central nontranscribed region, and they often lack polyadenylated [poly(A)] tails (2–6).
Since their initial identification and characterization as being commonly expressed (2, 3, 5, 6), it has been debated whether eRNAs play a role in transcriptional activation or are consequential, perhaps a by-product of the enhancer’s proximity with active promoters, resulting in the association of RNA polymerase II and thus also their transcription (7–9). However, recent evidence showing that knockdown of specific eRNAs resulted in reduced expression of their target genes supports a functional role for these RNAs in transcription (e.g., refs. 10–13), although the mechanisms through which eRNAs operate remain unclear.
The gene encoding the common α-subunit chorionic gonadotropin alpha (Cga) of the pituitary gonadotropins (luteinizing and follicle-stimulating hormones) and thyroid-stimulating hormone was reported to contain an enhancer located −4.6 to −3.7 kbp upstream of the transcriptional start site (TSS). This enhancer was shown to play a crucial role in the cell-specific expression of the gene in gonadotropes and thyrotropes in transgenic mice and was seen to increase the activity of the proximal promoter in pituitary cells (14, 15). In the current study we characterized this enhancer region and the eRNA that is expressed and show that it plays a major role in directing the activity of the proximal promoter through histone modifications, chromatin remodeling, and DNA looping.
Results
The Cga Enhancer Is Transcribed to Two Bidirectional eRNAs Whose Levels Increase Following GnRH Treatment but Are Not Intrinsically Linked to Cga Transcriptional Activity.
Based on the previous report of the functional Cga enhancer, we sought first to determine whether RNA is transcribed from this region. Using quantitative PCR (qPCR) on DNase-treated reverse-transcribed RNA from a gonadotrope cell line, we amplified fragments between −5300 and −3872 bp upstream of the Cga TSS, which includes most of the reported enhancer. RNA transcribed from the region further upstream, between −6309 and −5630 bp, also was amplified successfully and was expressed at a higher level than at the more proximal region (Fig. 1A). However, RNA was barely detected using primers targeting or spanning the region −5616 to −5377. To confirm the start and termination sites of these RNAs (hereafter, “eRNA-opposite” and “eRNA-forward,” respectively) and also their directionality, 5′ and 3′ RACE reactions were performed. These reactions revealed that the RNAs are transcribed bidirectionally from opposite strands starting at −5377 for the eRNA-forward and at −5616 for the eRNA-opposite. These RNAs could not be amplified from poly(A) RNA. Northern analysis using a labeled probe from the region encoding the eRNA-opposite confirmed the presence of a single-sized RNA containing this sequence (Fig. 1B).
Fig. 1.
The Cga enhancer is transcribed to two bidirectional eRNAs, whose levels increase after GnRH treatment but that are not intrinsically linked to Cga transcriptional activity. (A) Total RNA from αT3-1 cells was reverse-transcribed with random primers before qPCR, using the primer sets shown. RNA levels were quantitated using a standard curve of sonicated DNA normalized with Gapdh mRNA and presented relative to the highest levels. Data are shown as mean ± SEM, n = 4–6. Also shown is the average transcript ratio for the opposite and forward eRNAs (n = 17 or 21; ***P < 0.0001). The 5′ and 3′ RACE reactions confirmed the eRNA start and termination sites and their bidirectionality. (B) Northern analysis was carried out on total RNA from αT3-1 cells, using digoxigenin (DIG)-labeled eRNA (−6000 to −5630) as a probe. (C) ChIP for H3K4me1 was performed in untreated and GnRH-treated (100 nM, 2 h) αT3-1 cells. Immunoprecipitated DNA was measured by qPCR, and data are presented relative to levels in the input. Negative and positive controls are regions upstream of the Lhβ gene; the latter is an enhancer. Data are shown as mean ± SEM, n = 3–6. A Student’s t test comparing levels of each fragment in treated and untreated cells did not detect significant differences (P > 0.05). (D) After overexpression of HA-Pitx1, ChIP was performed using HA-antisera, and data were analyzed and presented similarly, but are shown relative to the highest levels, with Crystallin as a negative control and the Cga promoter as a positive control. Data are shown as mean ± SEM, n = 3–4. ANOVA followed by a Bonferroni t test revealed statistically different means (P < 0.05) designated by the letters “A” and “B”; bars with the same letter are similar (P > 0.05). (E and F) Cga mRNA and eRNA levels in untreated cells or in GnRH-treated (100 nM, 8 h) or forskolin-treated (10 μM, 4 h) cells were measured by qPCR, normalized to Gapdh mRNA, calibrated according to standard curves from cDNA samples for Cga and Gapdh or sonicated genomic DNA for the eRNAs, and are shown relative to levels in untreated cells. Data are shown as mean ± SEM, n = 3–6. **P < 0.01; ***P < 0.001; NS, P > 0.05, Student’s t test as in C.
Because enhancers characteristically carry H3K4 monomethylation (H3K4me1), we performed ChIP for this modification and found it substantially enriched between −4559 and −4460 bp upstream of the Cga TSS. ChIP also was carried out in cells treated with gonadotropin-releasing hormone (GnRH), which is the primary regulator of Cga expression in the gonadotropes; however, H3K4me1 levels were not affected by the treatment (Fig. 1C).
We looked for binding of various gene-specific transcription factors to this region and found that paired-like homeodomain 1 (Pitx1), which is required for expression of the Cga gene (16), has four putative binding motifs that match those of various Pitx-1–bound enhancers [T/CTAAT/GCC (17) at −6186, −5604, −4218, and −4168 bp], as well as an additional sequence (CAATCC at −4650 bp) matching a functional Pitx1-binding site on the luteinizing hormone beta (Lhb) gene promoter (16). ChIP analysis after overexpression of HA-Pitx1 or FLAG-Pitx1 (ChIP-grade Pitx1 antibodies were not available) revealed that Pitx1 is detected at similar levels at the region of the enhancer marked by H3K4me1 and at the proximal promoter (Fig. 1D). To test Pitx1 functionality, we transiently transfected shRNAs targeting Pitx1, which, although reducing Pitx1 mRNA levels by only ∼30%, caused a similar reduction (30–40%) in the levels of Cga mRNA and the eRNAs, without affecting the control (Fig. S1).
Treatment of the gonadotrope cells with GnRH leads to an increase in Cga mRNA levels and also slightly increased the levels of the eRNAs (Fig. 1E). However, forskolin, which activates Cga expression more strongly, did not affect the eRNA levels (Fig. 1F). Thus, the transcription of the eRNAs is not necessarily linked to that of Cga, nor is it triggered by activity of the promoter, although it is possible that forskolin-activated Cga transcription involves the activation of other enhancers at distinct genomic loci.
Stable Transfection of shRNA Targeting the eRNA-Opposite Reduces Cga Expression, and This Effect both Intensifies and Spreads.
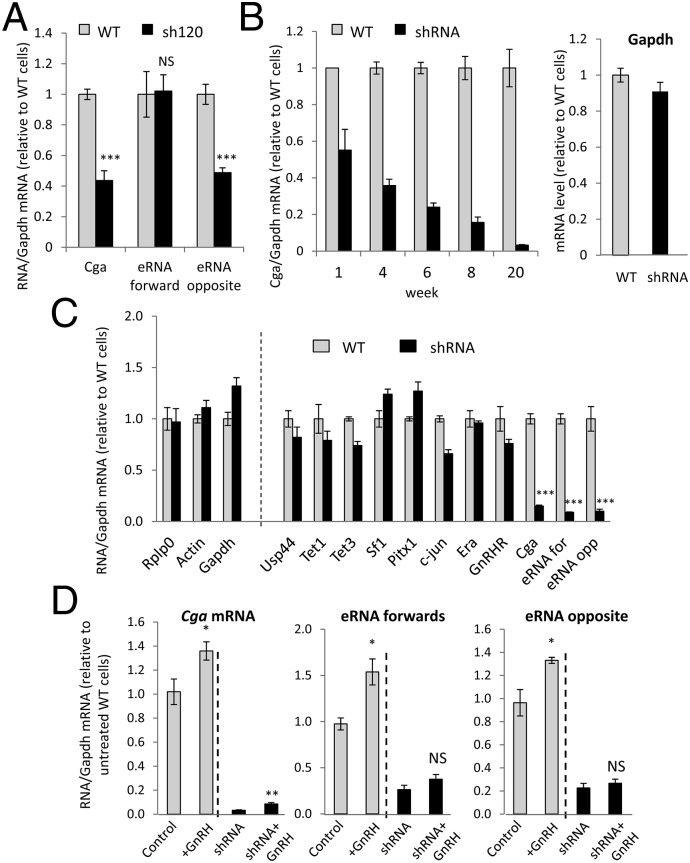
To determine whether these eRNAs have a function in Cga transcription, we attempted to knock down and/or abrogate their effects by stably transfecting various constructs encoding different shRNAs designed to target them. Most of the constructs led to a 60–80% drop in the level of the specific eRNA and to a 40–80% drop in Cga mRNA levels (Fig. S2). Notably, sequences targeting the eRNA-forward also appeared to alter eRNA-opposite levels, suggesting either nonspecific effects or possibly a functional dependence of its transcription on the eRNA-forward. Conversely, a sequence (sh632) targeting the 3′ end of the eRNA-opposite increased levels of the 5′ end of this eRNA and also of the eRNA-forward; this increase also could be caused by nonspecific targeting or might indicate an effect in stabilizing the eRNAs. In contrast, the effects of targeting the eRNA-opposite at its 5′ end (sh120) appeared to be specific (Fig. 2A) and, at least initially, did not affect the levels of the eRNA-forward, so these clones were chosen for further study.
Fig. 2.
Stable transfection of shRNA targeting the eRNA-opposite reduces Cga expression, and this effect intensifies and spreads. (A) αT3-1 cells were stably transfected with pSUPER-shRNA (sh120) targeting the eRNA-opposite. After selection, RNA levels were assessed by qPCR and are shown normalized to Gapdh, relative to levels in WT cells. Data are shown as mean ± SEM, n = 3–5; ***P < 0.001. (B) These shRNA clones then were analyzed over 20 wk, and Cga mRNA levels were assessed and are presented similarly; also shown are mRNA levels of Gapdh at 20 wk. n = 3–6. (C) At 14 wk, the expression levels of other genes were measured also. mRNA levels of Rplp0 (ribosomal protein, large, P0), Actin, and Gapdh are presented as a ratio to their levels in WT cells; all others were normalized to Rplp0. n = 2–5. ***P < 0.001, adjusted t-test comparing each RNA in shRNA cells with its level in WT cells. For all others, P > 0.05. (D) At 24 wk the GnRH responsiveness of the Cga mRNA and eRNAs was reevaluated in WT and shRNA cells, as in Fig. 1E, and values are presented relative to levels in untreated WT cells. Data are shown as mean ± SEM, n = 3. *P < 0.05; **P < 0.01; NS, P > 0.05, t-test compared with levels in treated and untreated cells for each cell line.
With prolonged culture of these shRNA clones, the repressive effect on Cga increased progressively, with mRNA levels reaching less than 5% those in WT cells at 20 wk, whereas Gapdh levels remained unaltered (Fig. 2B). We have not seen this progressive effect with similar long-term shRNA-targeted knockdown of other genes (e.g., Fig. S3). At 14 wk, the Cga mRNA and eRNA-opposite levels were reduced significantly in the shRNA cells, and so were those of the eRNA-forward, but the mRNA levels of various other genes were not similarly affected (Fig. 2C). Despite this major drop in basal Cga expression, the transcript still was elevated by GnRH treatment at 24 wk, although the eRNA levels no longer increased (Fig. 2D). Thus, with time, the effect of the shRNA on the eRNA-opposite and the Cga mRNA levels had intensified and also appeared to have spread along the chromatin to affect expression of the eRNA-forward as well.
H3K4me1, Histone H3 Lys9 Trimethylation, and the Distal CpG Island Are Unaffected by the eRNA Knockdown, but Association of the Chromatin Remodeling Factor CHD1 with the Promoter Is Reduced.
Having established that the repressive effect of the eRNA knockdown intensified and spread, we considered whether the eRNA might be involved in directing epigenetic modifications. We first looked at the level of H3K4me1 at the enhancer and found that it was no different in the shRNA cells than in WT cells at 8 wk after the knockdown (Fig. 3A). Such a lack of correlation between H3K4me1 levels and enhancer activity has been reported before (18, 19).
Fig. 3.
H3K4me1, H3K9me3, and the distal CpGI are unaffected by the eRNA knockdown, but the association of CHD1 with the promoter is reduced. ChIP for H3K4me1 (A), CHD1 (B), and H3K9me3 (C) was carried out in WT and eRNA-knockdown cells (after 8, 16, or 20 wk, respectively), using the sets of primers shown. The levels of IP DNA are presented relative to the levels in input samples. Negative and positive controls were the upstream region of Lhβ (as in Fig. 1C) and the Tet1 enhancer in A; Crystallin and Gtpbp in B; and Gapdh and Major satellites in C. Data are shown as mean ± SEM, n = 2–6. All means are similar (P > 0.05) for WT and shRNA cells at the same genomic location, except for those marked by a single asterisk in B, in which P < 0.05. (D) The CpGI found immediately upstream of the enhancer (−6672 to −7175) was examined in WT and eRNA-knockdown cells at 25 wk by bisulfite conversion and sequencing of 12 cloned amplicons. Filled circles represent methylated/unconverted cytosines.
Next we examined the association of the chromatin remodeling factor, chromodomain-helicase-DNA-binding protein 1 (CHD1), with the enhancer and found that at 16 wk after eRNA knockdown its levels were reduced at the distal region of the enhancer, at its untranscribed central region, and just upstream of the Cga proximal promoter (Fig. 3B). CHD1 allows promoter clearance of RNA polymerase II (RNAPII); however, its levels vary, being higher on processively transcribed genes than on stalled genes; it is also enriched at the 5′ ends of transcribed regions and throughout the distal regions, where it blocks cryptic transcription by suppressing nucleosome turnover (20). These distinct functions of CHD1 could explain its differing levels, although there may be another active chromatin locus that accounts for its high levels at the distal end of the enhancer (Discussion). In any case, the ability of the shRNA to reduce CHD1 levels at the enhancer and promoter regions indicates that the eRNA is able to mediate protein recruitment or binding at both genomic locations.
Given the major decrease in expression levels of the Cga mRNA and the eRNAs that occurred over time, we next looked at H3K9me3 and DNA methylation, which are both marks of silenced genes. H3K9me3 was seen to be enriched at the 3′ ends of both transcribed regions of the enhancer in WT cells, but at 20 wk this modification appeared largely unaffected by the eRNA knockdown (Fig. 3C). Just upstream of the distal-most region encoding the eRNA opposite, at −7175 to −6672 bp, there is a CpG island (CpGI) which was analyzed in WT and long-term (25 wk) eRNA-knockdown cells for DNA methylation. Bisulfite conversion and sequencing of 12 cloned amplicons from each cell type revealed that this region is completely unmethylated in both cell types (Fig. 3D).
Interaction of the Distal Enhancer with the Proximal Promoter of the Cga Gene Is Dependent on the eRNA.
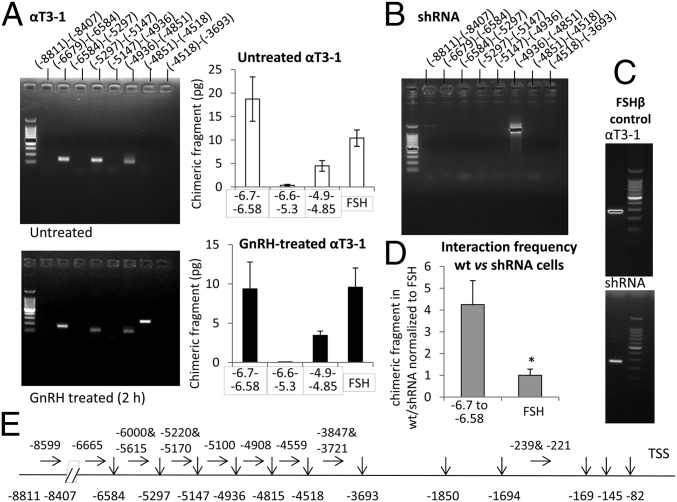
Given that the eRNA is required for transcription of the Cga and that its knockdown reduced the association of CHD1 with the proximal promoter, we went on to check whether the eRNA might be involved in the physical interaction between the enhancer and proximal promoter regions. We carried out chromatin confirmation capture (3C) assays in untreated WT cells, in cells treated with GnRH for 2 h, and in the eRNA-knockdown cells at 36 wk. In untreated WT cells, the proximal region was found in close physical proximity to three regions of the distal enhancer (−6.7 to −6.58; −5.3 to −5.15; and −4.9 to −4.85 kbp); these interactions and an additional region (−4.85 to −4.5 kbp) also were apparent in the GnRH-treated cells (Fig. 4A). Strikingly, however, these interactions were not seen in the eRNA-knockdown cells (Fig. 4B), although the interaction between a distal region and the proximal promoter of a control gene, follicle-stimulating hormone beta (Fshβ) still was observed (Fig. 4C). qPCR analysis confirmed the interaction of the distal enhancer regions (−6.7 to −6.58 and −4.9 to −4.85 kbp) with the proximal Cga promoter in both the treated and untreated cells and, with greater frequency, in WT and eRNA-knockdown cells after normalization to the relative levels of the Fshβ chimeric fragment (Fig. 4 A and D).
Fig. 4.
Interaction of the distal enhancer with the proximal promoter of the Cga gene is dependent on the eRNA. (A, Left) A 3C assay was carried out in untreated (Upper) or GnRH-treated (100 nM, 2 h) (Lower) αT3-1 cells. Chimeric fragments were detected using nested forward primers targeting −239 and −221 bp and sets of primers targeting the upstream regions as shown. The identity of all amplicons was confirmed by sequencing. (Right) qPCR measurement of some of these chimeric fragments in both 3C libraries; the fragment −6.6 to −5.3 kbp is the negative control, and an interacting fragment upstream of Fshβ is the positive control. Values are stated in picograms of DNA measured using standard curves from cloned fragments. Data are shown as mean ± SEM, n = 3–5. (B) 3C libraries also were prepared from eRNA-knockdown cells at 36 wk, and interactions were assessed similarly (the one visible fragment was sequenced and is an artifact). (C) Also shown is the Fshβ gene–control interaction in the same 3C libraries. (D) The frequency of the interaction of the distal fragment with the proximal Cga promoter in WT cells compared with that in the eRNA-knockdown cells was measured by qPCR and normalized to the relative levels of the Fshβ chimeric fragment in the same libraries. Data are shown as mean ± SEM, n = 3–4; *P < 0.05. (E) Dpn2 restriction sites (vertical arrows) and localization of PCR primers (horizontal arrows).
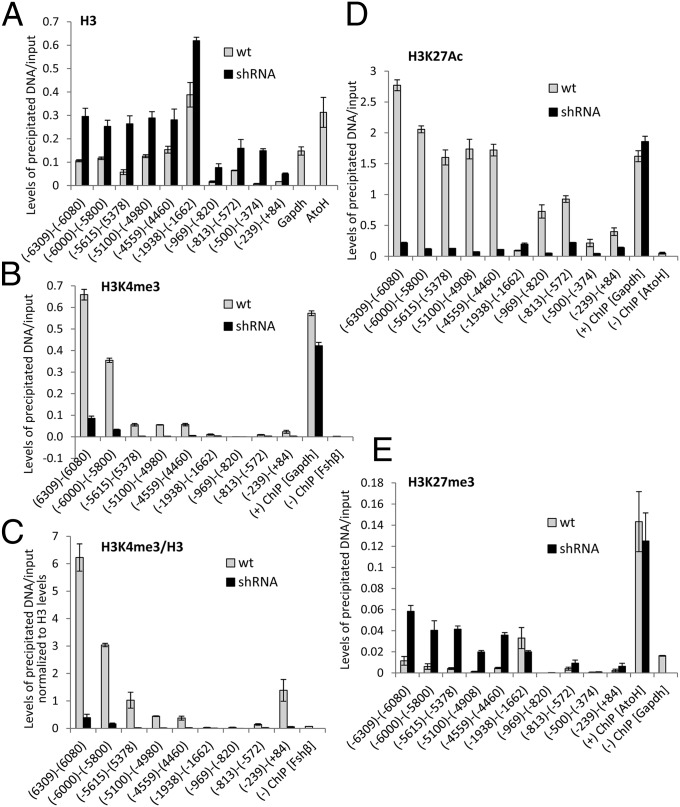
Knockdown of the eRNA Induces Major Changes in Histone H3 Lys 4 Trimethylation and at H3K27, Converting the Chromatin at both the Enhancer and the Proximal Promoter from an Active to a Repressed State.
To understand the impact of the loss of DNA looping on the Cga promoter, we examined the effect of long-term eRNA knockdown on additional histone modifications at both regions. We first saw that the eRNA knockdown (at 11 and 36–38 wk) led to an increase in H3 association throughout the enhancer, but the effect at the Cga promoter was even more pronounced (Fig. 5A and Fig. S4A). We next examined levels of histone H3 Lys 4 trimethylation (H3K4me3), which is characteristically bound at active promoters; at 36–38 wk we found very high levels of this modification at the distal end of the enhancer, but its levels were reduced dramatically at both enhancer and promoter regions in the eRNA-knockdown cells (Fig. 5 B and C). Such a reduction is not characteristic of other genes whose expression we have targeted through similar long-term stable transfection of shRNA (e.g., Fig. S5).
Fig. 5.
Knockdown of the eRNA induces major changes in H3K4me3 and at H3K27, converting the chromatin at both the enhancer and promoter from an active to a repressed state. ChIP for H3 (A and C), H3K4me3 (B and C), H3K27ac (D), and H3K27me3 (E) was carried out in WT and eRNA-knockdown cells at 36–38 wk. The immunoprecipitated DNA was measured by qPCR. Data are presented relative to input samples, as in Fig. 3, except in C, where modified histone levels are shown relative to total H3 levels at the same locus. Controls are the promoters of Gapdh, Atoh, and/or Fshβ, as marked. Data are shown as mean ± SEM, n = 2–4.
H3K27 is characteristically acetylated at active enhancers, and ChIP revealed that it is found at high levels throughout the enhancer region. Notably, in eRNA-knockdown cells this modification was reduced dramatically, at both the enhancer and promoter (Fig. 5D and Fig. S4B). Moreover, ChIP for the repressive H3K27me3 revealed that this modification was elevated throughout the enhancer region in the eRNA-knockdown cells but was barely detectable in the WT cells, except in one region where histone H3 Lys 27 acetylation (H3K27ac) was absent and histone H3 Lys 9 trimethylation (H3K9me3) also was found (Fig. 5E and Fig. S4C). These results indicate that the eRNA either facilitates or protects H3K27ac at the enhancer, without which it can be methylated to repress further the eRNA and thus also Cga expression.
Discussion
Our study indicates that eRNA transcribed from the Cga enhancer plays a crucial role in transcription of this gene. It facilitates the chromatin modifications that keep the proximal promoter, as well as the enhancer itself, in an active state, likely by promoting or stabilizing the interaction between these two regions. Our findings are in line with other recent studies showing a function for eRNAs (10–12, 21, 22), although we believe that ours is the first to show such an impact of the eRNA specifically on the chromatin at its target.
Characteristic of enhancer chromatin, the central region of the Cga enhancer contains high levels of H3K4me1 along with relatively low levels of H3K4me3 (23, 24). Only a single peak of H3K4me1 was observed; a single peak is less typical than a bimodal monomethylation peak but has been reported previously (5, 19, 23, 25). Notably, H3K4me3 was found at higher levels at the most distal end of the enhancer than at the Cga promoter, partly reflecting nucleosome depletion at this promoter and possibly also reflecting distinct patterns of polymerase pausing and transcriptional consistency (26). We have already reported that, although seemingly low, the level of H3K4me3 at the Cga promoter in these cells is greater than at the repressed gonadotropin β-subunit genes and appears to be significant in activating Cga expression (27). The major distal H3K4me3 peak also is seen in ENCODE data from non–Cga-expressing cell lines, spanning ∼1 kbp and peaking in the center of the CpGI. Moreover Cap analysis gene expression (CAGE) data, which do not include gonadotrope-specific events, indicate that the 5′ end of the CpGI is transcribed from the +strand (but is unlikely protein coding), whereas a hypothetical gene (AK039341) was reported for a more extensive region (−7069 to −3875 bp); this mRNA apparently was isolated from brain and spinal cord, although it is termed a “noncoding RNA” (28). However, we were unable to amplify fragments spanning the central region (−5.6 to −5.3 kbp) of the enhancer, detected only a single ∼1-kbp fragment in northern analysis using an eRNA-opposite probe, and could not amplify the eRNAs from poly(A) RNA. Moreover, RACE reactions showed clearly that the eRNAs are transcribed from different strands. Thus, we are confident that the Cga eRNA is distinct from these putative noncoding RNAs; however, there does appear to be a locus of active chromatin upstream of the enhancer that is not gonadotrope specific and likely is reflected in some of the chromatin marks observed. Therefore although we cannot rule out the possibility that some of the effects seen might be caused by changes in the expression of such a putative ncRNA, this possibility does not detract from the fact that the knockdown of the eRNA changes the chromatin structure in this entire region and also blocks Cga transcription.
Our study indicates that the eRNA facilitates DNA looping between the enhancer and the Cga promoter. eRNAs and other lncRNAs have been implicated in DNA looping and specifically in enhancer–promoter interactions, which were seen to correlate with the levels of eRNA and target gene expression (29). Knockdown of several estrogen receptor alpha (ERα)-activated eRNAs was seen to diminish the enhancer–promoter looping initiated by ERα, and it was proposed that estrogen alters these interactions via the eRNAs (10). However, another study on the same cell lines reported that the looping was unaffected by flavopiridol, which effectively blocked eRNA transcription (30). Also the androgen receptor (AR)-activated eRNA kallikrein-3e (KLK3e) mediates interaction of this enhancer with the target kallikrein-2 (KLK2) gene promoter (13). Such a role for eRNAs is supported by the finding that some eRNAs interact with components of the cohesion complex that regulate enhancer–promoter interactions in stem cells (10, 31). In addition, eRNAs and lncRNAs have been reported to interact with the Mediator complex, which also plays a role in looping, whereas the enhancer-like lncRNA HOTTIP interacts with WDR5, a component of Set1/MLL complexes that trimethylate H3K4me at gene promoters (21, 32).
Disruption of the enhancer–promoter interaction following eRNA knockdown appeared to have a major and progressive effect on the chromatin at both loci, leading to the repression of the Cga promoter and the entire distal region, even though only the eRNA-opposite was targeted. The DNA looping was only assessed at 36 wk, but the changes in K27ac, K27me3, and total H3 already were seen when first measured at 11 wk and were still present at 36–38 wk when H3K4me3 levels also were greatly reduced. H3K4me1, H3K9me3, and CpGI methylation appeared unchanged at 8, 20, and 25 wk, respectively, but were not examined at later time points. Notably, poised enhancers are activated in ES cells through H3K27ac, which replaces the methylation that likely had protected them from histone acetyl transferase (HAT) activity (18, 19, 24). It was suggested that the K27ac then prevents the subsequent spreading of K27me to active sites, which occurs rapidly after HAT inactivation (33). Acetylation of H3K27 at the Cga enhancer is thus likely a crucial step in keeping the enhancer active, possibly as a result of the interaction with modifying enzymes at the proximal promoter, which is lost after removal of the eRNA. Although the H3K27ac likely is catalyzed by locally bound HATs, which are commonly found at enhancers (23, 34), the eRNA-mediated interaction with the proximal promoter may be required for the stable binding of these HATs and/or to prevent access to histone deacetylases.
We have considered that the effects of the eRNA knockdown on the chromatin might be caused by the shRNA recruiting the RNAi machinery to this locus rather than by the lack of eRNA per se (35). However, we consider this possibility unlikely, because we do not see escalating repression following similar stable knockdown of other genes (e.g., Fig. S3), changes in H3K4me3 levels are not seen at 5′ targeted loci of other genes knocked down in this way (e.g., Fig. S5), and Argonaute (Ago) proteins do not appear to accumulate at the enhancer in the eRNA-knockdown cells (Fig. S6). Moreover mechanisms describing siRNA-induced heterochromatin implicate central roles for H3K9 and DNA methylation (36), and these modifications were not altered at a time when both the Cga mRNA and H3K27ac levels had already dropped dramatically, the chromatin was compacted, and H3K27me3 levels had increased.
Roles for eRNAs in modifying chromatin structure and/or the transcription initiation complex at the promoter of the target gene have been indicated, albeit to a lesser extent, in a number of recent studies. Knockdown of eRNAs in the myogenic gene regulatory network led to a decrease in chromatin accessibility at the target genes and reduced RNAPII occupancy (11), whereas ncRNA-a was shown to regulate the S10 kinase activity of the Mediator complex in HEK293 cells (21). In contrast, at hormone-inducible ERα-activated enhancers, eRNA depletion did not affect total H3K4me1, H3K27ac, or RNAPII levels (10, 30), although transfections were only transient and histone modifications at the target gene promoters were not reported. Notably, however, levels of RNAPII S5p were reduced at the AR-activated KLK2 gene promoter following KLK3e knockdown and dihydrotestosterone treatment (13).
The Cga enhancer was reported to facilitate basal and tissue-specific Cga expression, although its mechanisms of action were not elucidated (14, 15). Although there is some correlation between eRNA levels and the activity of the Cga proximal promoter, these two events are not tightly coupled, and the eRNA does not appear to mediate GnRH-induced Cga expression; however, in agreement with the functional studies, it is required for basal expression. The presence of Pitx1 at the enhancer and the effects of its partial knockdown suggest that this lineage-specific transcription factor may play a role in directing eRNA transcription, in line with the importance of Pitx1/2 in the development of the gonadotrope cell lineage (37, 38). Interestingly, we previously showed that Pitx1 can homodimerize and that, when bound to more than one response element on different parts of the Lhβ gene, it induces bending in the DNA (39). Other studies have reported that Pitx1 is associated with various enhancers of limb genes, being especially enriched on those carrying H3K27ac, and it was suggested that Pitx1 influences limb morphogenesis through activating hindlimb-specific enhancers (17). Such a role for lineage-specific factors could allow the regulation of enhancers during development and their ability to determine cell identity by providing control of gene expression in a tissue-specific manner (40).
The current study demonstrates the ability of a noncoding eRNA to alter the chromatin structure at its target gene, involving looping of the DNA, and in this case it promotes or protects the euchromatic state to allow basal gene expression. The eRNA in this study did not appear to mediate GnRH-induced Cga expression, and a picture is emerging of different classes of enhancers and eRNAs with distinct functions: Some, as in the current study, appear to determine cell-specific expression and cell lineage (e.g., 18, 40, 41), whereas others mediate the effects of specific stimuli such as hormones (e.g., 5, 10, 13, 30). Additional complexity has been suggested in which certain genes can use various enhancers through different stages of development and in different tissues, both to fine-tune levels of protein activity and to endow an element of tissue-specific control (18, 24, 40–42). Our findings, together with mounting evidence from other studies, suggest that eRNAs comprise a pivotal element in determining the chromatin architecture at their target gene, although they may use various modes of action under different conditions to modify both basal levels of expression and response to stimuli in distinct gene, and possibly cellular, contexts.
Experimental Procedures
For further details, see SI Experimental Procedures.
Cell Culture, Plasmid Constructs, and Transfections.
Murine gonadotrope-derived αT3-1 cells were cultured and transfected as described (43), and some were treated with GnRH (100 nM) or forskolin (10 μM; Sigma). HA- and FLAG-Pitx1 expression constructs have been reported previously (44). Pitx1 knockdown for 48 h used shRNA cloned into pSUPER-basic, and stable knockdown was achieved via pSR-GFP/neo plasmid (both from OligoEngine) expressing the shRNA (Table S1); clones were selected using G418 as in ref. 27.
Characterization of eRNAs and mRNA Levels by Real-Time PCR, RACE, and Northern Analysis.
Total RNA was extracted using TRIzol (Ambion) and was treated with DNase and reverse-transcribed with random hexamers (Applied Biosystems) for qPCR using gene-specific primers (Table S2 and Fig. S7), Absolute Blue SYBR-Green ROX Mix (Thermo Fisher), and the Illumina Eco Real-Time PCR as reported in ref. 43. Amplicon levels were quantitated relative to standard curves comprising cDNA or genomic DNA and were normalized to Gapdh mRNA. The 5′ and 3′ ends of the eRNAs were identified using nested gene-specific primers (Table S2) and SMARTer RACE cDNA amplification (Clontech) after poly(A) tailing and sequencing of the amplicons. Northern analysis was carried out according to established protocols, and the RNA was detected using the −6000 to −5630 bp sequence labeled with the PCR DIG Probe synthesis kit (Roche).
ChIP.
ChIP was carried out after formaldehyde cross-linking as described in ref. 43 and is detailed in SI Experimental Procedures. Immunoprecipitation and input levels were measured by real-time qPCR using standard curves comprised of sonicated genomic DNA, as above.
Chromatin Conformation Capture Assay.
The chromatin conformation capture (3C) assays were carried out as detailed in SI Experimental Procedures on serum-starved αT3-1 cells, some of which were exposed to GnRH for 2 h. All amplicons were verified by sequencing. Levels were quantitated according to standard curves using the same chimeric fragment that had been cloned, and averages were made after repeating (n-value) both rounds of the PCR.
Identification of Methylated DNA by Bisulfite Sequencing.
Bisulfite conversion was using the EZ-DNA methylation Gold kit (Zymo), and fragments were amplified and sequenced. Primers are listed in Table S2. All amplicons were cloned and sequenced.
Supplementary Material
Acknowledgments
We thank Pamela Mellon (University of California, San Diego) for the gift of the cell line. This work was supported by Israel Science Foundation Grant 840/12.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414841112/-/DCSupplemental.
References
- 1.Ong CT, Corces VG. Enhancers: Emerging roles in cell fate specification. EMBO Rep. 2012;13(5):423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch F, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18(8):956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12(11):R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10(9):1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 8.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14(2):103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 9.Natoli G, Andrau JC. Noncoding transcription at enhancers: General principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 10.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mousavi K, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51(5):606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melo CA, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh CL, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci USA. 2014;111(20):7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kendall SK, et al. Enhancer-mediated high level expression of mouse pituitary glycoprotein hormone α-subunit transgene in thyrotropes, gonadotropes, and developing pituitary gland. Mol Endocrinol. 1994;8(10):1420–1433. doi: 10.1210/mend.8.10.7531821. [DOI] [PubMed] [Google Scholar]
- 15.Brinkmeier ML, et al. Cell-specific expression of the mouse glycoprotein hormone alpha-subunit gene requires multiple interacting DNA elements in transgenic mice and cultured cells. Mol Endocrinol. 1998;12(5):622–633. doi: 10.1210/mend.12.5.0103. [DOI] [PubMed] [Google Scholar]
- 16.Tremblay JJ, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12(3):428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 17.Infante CR, Park S, Mihala AG, Kingsley DM, Menke DB. Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev Biol. 2013;374(1):234–244. doi: 10.1016/j.ydbio.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012 doi: 10.1038/ng.1064. 44(2):148–156. [DOI] [PubMed] [Google Scholar]
- 20.Skene PJ, Hernandez AE, Groudine M, Henikoff S. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife. 2014;3:e02042. doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 24.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21(8):1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benayoun BA, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158(3):673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wijerweera A, et al. Gonadotropin gene transcription is activated by menin-mediated effects on the chromatin. Biochim Biophys Acta. 2015;1849(3):328–341. doi: 10.1016/j.bbagrm.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Okazaki Y, et al. FANTOM Consortium RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 29.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23(8):1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53(1):49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 34.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13(9):793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg MS, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12(2):256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanctôt C, Gauthier Y, Drouin J. Pituitary homeobox 1 (Ptx1) is differentially expressed during pituitary development. Endocrinology. 1999;140(3):1416–1422. doi: 10.1210/endo.140.3.6549. [DOI] [PubMed] [Google Scholar]
- 38.Charles MA, et al. PITX genes are required for cell survival and Lhx3 activation. Mol Endocrinol. 2005;19(7):1893–1903. doi: 10.1210/me.2005-0052. [DOI] [PubMed] [Google Scholar]
- 39.Melamed P, Koh M, Preklathan P, Bei L, Hew C. Multiple mechanisms for Pitx-1 transactivation of a luteinizing hormone beta subunit gene. J Biol Chem. 2002;277(29):26200–26207. doi: 10.1074/jbc.M201605200. [DOI] [PubMed] [Google Scholar]
- 40.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155(7):1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14(6):838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nord AS, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155(7):1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pnueli L, Luo M, Wang S, Naor Z, Melamed P. Calcineurin mediates the gonadotropin-releasing hormone effect on expression of both subunits of the follicle-stimulating hormone through distinct mechanisms. Mol Cell Biol. 2011;31(24):5023–5036. doi: 10.1128/MCB.06083-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo Z, Wijeweera A, Oh Y, Liou YC, Melamed P. Pin1 facilitates the phosphorylation-dependent ubiquitination of SF-1 to regulate gonadotropin beta-subunit gene transcription. Mol Cell Biol. 2010;30(3):745–763. doi: 10.1128/MCB.00807-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.