Abstract
Although the pathogenesis of cardio-cerebrovascular disease (CCVD) is multifactorial, an increasing number of experimental and clinical studies have highlighted the importance of histone deacetylase (HDAC)-mediated epigenetic processes in the development of cardio-cerebrovascular injury. HDACs are a family of enzymes to balance the acetylation activities of histone acetyltransferases on chromatin remodeling and play essential roles in regulating gene transcription. To date, 18 mammalian HDACs are identified and grouped into four classes based on similarity to yeast orthologs. The zinc-dependent HDAC family currently consists of 11 members divided into three classes (class I, II, and IV) on the basis of structure, sequence homology, and domain organization. In comparison, class III HDACs (also known as the sirtuins) are composed of a family of NAD+-dependent protein-modifying enzymes related to the Sir2 gene. HDAC inhibitors are a group of compounds that block HDAC activities typically by binding to the zinc-containing catalytic domain of HDACs and have displayed anti-inflammatory and antifibrotic effects in the cardio-cerebrovascular system. In this review, we summarize the current knowledge about classifications, functions of HDACs and their roles and regulatory mechanisms in the cardio-cerebrovascular system. Pharmacological targeting of HDAC-mediated epigenetic processes may open new therapeutic avenues for the treatment of CCVD.
Keywords: Histone deacetylase, Epigenetic modification, Heart failure, Atherosclerosis, Stroke
1. Introduction
Cardio-cerebrovascular disease (CCVD) is a major leading cause of death in the world despite for great efforts being made to reduce the mortality.[1] Although the pathogenesis of CCVD is multifactorial, an increasing number of experimental and clinical studies have implicated that epigenetic modifications, such as DNA methylation, histone modification and microRNA, play vital roles in the development of cell injury in both cardiovascular and central nervous systems. Among them, the importance of histone deacetylase (HDAC)-mediated epigenetic processes in the development of cardio-cerebrovascular injury have been highlighted.[2] Recent studies have also demonstrated that HDAC inhibitors have anti-inflammatory and antifibrotic effects in the cardio-cerebrovascular system.[3],[4] In this review, we highlight the importance of HDACs on the regulation of the cardio-cerebrovascular function. A better understanding of the function of individual HDACs will provide unexpected opportunities to develop new therapies for CCVD by epigenetic modifications.
2. HDACs: classification and function
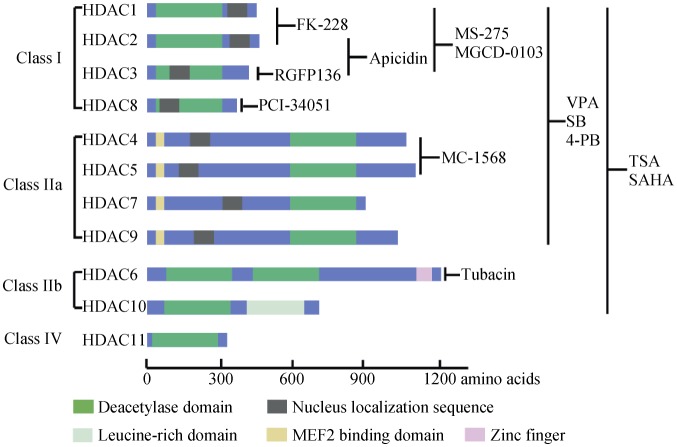
HDACs are a family of enzymes, which balance the acetylation activities of histone acetyltransferases (HAT) on the post-translational modifications of histone and a wide range of other non-histone substrates.[5],[6] To date, 18 mammalian HDACs are identified and grouped into four classes based on similarity to yeast orthologs. The Zn2+-dependent HDAC family currently consists of 11 members, which are divided into three classes (class I, II, and IV) on the basis of structure, sequence homology, and domain organization (Figure 1). Class I HDACs (HDAC 1, 2, 3 and 8) share sequence homology with the yeast protein reduced potassium dependency-3 (Rpd3), which are ubiquitously expressed and almost exclusively in the nucleus with the main gene transcriptional repression functions.[7] Class II (HDAC 4, 5, 6, 7, 9, and 10) are known to ‘shuttle’ between the cytoplasm and nucleus depending on the phosphorylation regulation. Class II HDACs are further divided into two subgroups, class IIa (HDAC 4, 5, 7, 9) and IIb (HDAC 6, 10) according to the number of catalytic domains. Class IIa HDACs share common molecular structures with other HDAC members with only one enzymatic domain, while class IIb are somehow atypical, such as containing two HDAC domains within a single molecule, but the second C-terminal domain has no catalytic function in HDCA10.[8] HDAC11 is the sole member of class IV localized in the nucleus, which has a catalytic domain in the N-terminal region. HDAC11 has been demonstrated to regulate the balance between immune activation and immune tolerance.[9] In comparison, class III HDACs (also known as the sirtuins, silent information regulator, SIRT 1–7) comprise a family of NAD+-dependent protein-modifying enzymes related to the yeast Sir2 gene.[10]. Sirtuins are localized mainly in nucleus (SIRT 1, 6, 7) cytoplasm (SIRT2) or mitochondria (SIRT 3, 4, 5)[11] and also with nucleocytoplasmic traffic under certain conditions (SIRT 1, 2).[12],[13] Sirtuins possess the activities in lysine deacetylation, adenosinediphospho (ADP)-ribosylation, and/ or deacylation.[10]
Figure 1. The classification, molecular structure and main inhibitors of Zn2+-dependent HDACs.
HDAC: histone deacetylase; MEF: myocyte enhancer factor; 4-PB: sodium-4-phenylbutyrate; SAHA: suberoylanilide hydroxamic acid; SB: sodium butyrate; TSA: trichostatin; VPA: valproic acid.
3. HDAC inhibitors: classification and selectivity
HDAC inhibitors are a group of compounds that block the activities of HDACs typically by binding to the zinc-containing catalytic domain of HDACs, which are originally a new class of anticancer drugs by therapeutically inducing cell cycle arrest, differentiation, apoptosis, as well as reducing proliferation, angiogenesis, migration, and cell resistance to chemotherapy.[14] Recent studies have also highlighted the important therapeutic potentials of HDAC inhibitors in non-cancer-related diseases, including neurological diseases, immune disorders, renal fibrosis and cardiovascular disease.[15] According to their chemical structure, HDAC inhibitors are classified into four classes, including hydroxamic acids, short chain fatty acids, cyclic peptides, and benzamides. Hydroxamic acids combine with the zinc ion at the catalytic site of HDACs, therefore these compounds exert nonspecific HDAC-inhibition activity affecting all classes of HDACs. Trichostatin (TSA) was the first one of the hydroxamic acids to be characterized as a potent HDAC inhibitor,[16] which was used as the core chemical structure for the design and synthesis of some new compounds of this class. Among them, suberoylanilide hydroxamic acid (SAHA, also known as vorinostat) is the first HDAC inhibitor approved by the U.S. Food and Drug Administration (USFDA) for the treatment of human cutaneous T-cell lymphoma (CTCL) in 2006.[17],[18] Short chain fatty acids, such as sodium butyrate (SB), sodium-4-phenylbutyrate (4-PB) and valproic acid (VPA), selectively inhibit class I and IIa HDACs and have less potent inhibitory effects compared with hydroxamic acids, but they can easily cross the blood-brain-barrier due to their smaller molecular weight. The best representative of this class is VPA, originally used as an antiepileptic drug, which has been tested for the treatment of other diseases, such as Parkinson's, Alzheimer's, Huntington's diseases and stroke.[6] Cyclic peptides are one group of Zn2+-dependent HDAC inhibitors with relative high selectivity. The most important member of this class is romidepsin (depsipeptide, FK-228) approved by the USFDA for marketing in the treatment of cancer in 2009 due to its potency to arrest cell growth.[19] Benzamides are considered as a kind of HDAC isoform-selective inhibitors with a long half-life which are currently in clinical trials for cancer treatment.
Although HDAC inhibitors have been used in experimental or clinical trials for the treatments of diverse diseases, the currently available HDAC inhibitors are mostly non-selective and inhibit multiple HDAC proteins (broad-spectrum HDAC inhibitors, or pan-HDAC inhibitors). Moreover, the effects of HDAC inhibitors are often evaluated by examining the alterations on bulk histone acetylation or the therapeutic results in a given experimental model and some clinical trials. Therefore, the use of broad–spectrum HDAC inhibitors could be problematic. With the increased knowledge of individual HDAC functions and key advances in the structural biology of various HDACs, reliable molecular homology models as well as suitable biological assays have provided new tools for the discovery of HDAC isoform-selective derivatives. For example: MS-275, an HDAC inhibitor belonging to the 2-aminophenyl benzamides class, has a higher affinity for HDAC 1, 2 and 3.[20] A cyclic peptide named Apicidin has more potent inhibition of HDAC 2 and 3 while no effects on HDAC1 and class II HDACs;[21] but FK-228, another cyclic peptide exhibits more potency on HDAC1, and 2.[22] Based on the analysis of HDAC8 crystal structure, selective HDAC inhibitors such as PCI-34051 targeting this isoform have recently been developed.[23]–[25] HDAC6 with a unique physiological function and structure has become a hot issue recently because of its involvement in tumorigenesis development and metastasis through α-tubulin, heat shock protein (HSP) 90 and ubiquitin-protein. Therefore, recently selective inhibitors of HDAC6, such as Tubacin, have been developed.[26] The general information of HDACs and the commonly used HDAC inhibitors are summarized in Table 1 and Figure 1. In this review, we mainly discuss the pathogenesis role of HDACs and their signaling pathways in CCVD.
Table 1. The HDAC family: classification, localization, substrates and functions.
| Class | Member | Localization | Substrate | Function |
| I | HDAC1 | Nucleus | Histones, MEF2, E2F1, p130, c-JUN, MKP1, p53, AMPK, p21,YY1, NF-κB, NOS, c-MYC, SMAD, pRb, SP1/SP3, TSHZ3, Myo-D, GATA, PCNA | Cell cycle progression, proliferation, differentiation, development, cancer. |
| HDAC2 | Nucleus | Histones, NF-κB, SRF, GATA, p53, PKC-δ, pRb, Ku70, CIITA, HSP70 | Cell proliferation, development, synaptic plasticity, cardiac hypertrophy | |
| HDAC3 | Nucleus/cytoplasm | Histones, MKP1, E2F1, NOS, MR, NKX2.5, p300, MCP, PCAF, NF-κB, pRb, YY1, HSP70, PPAR-γ | Cell proliferation, cell cycle control, development | |
| HDAC8 | Nucleus | Histones, HSP70, SMC3, cohesion | Smooth muscle differentiation and contractility, | |
| Iia | HDAC4 | Nucleus/cytoplasm | (Partners)Histones, MEF2, MyoD, p21, NFAT, Runx2, SRF, MRJ, p53, STAT1, FOXO, CAMTA2,HP1,14-3-3, | Cellular hypertrophy suppression, bone development, neuron survival and development, |
| HDAC5 | Nucleus/cytoplasm | (Partners)Histones, MEF2, MyoD, YY1, SRF, NKX2.5, CAMTA2, Runx2, FGF2, FOXO, HP1, 14-3-3 | Cellular hypertrophy suppression, bone development, axonal regeneration | |
| HDAC7 | Nucleus/cytoplasm | (Partners)Histones, MEF2, MyoD, MMP10, HIF2α, Nur77, 14-3-3, CAMTA2 | Cell survival, vascular development,immunomodulation | |
| HDAC9 | Nucleus/cytoplasm | (Partners)Histones, MEF2, MyoD, FOXP3, 14-3-3, CAMTA2 | Neuron development, synaptic plasticity,Immunomodulation | |
| IIb | HDAC6 | Cytoplasm | α-tubulin, HSP90, Prx1, Prx2 | Cytoskeletal dynamics, cell motility, aggresome formation, autophagy |
| HDAC10 | Nucleus/cytoplasm | pRb | Cell cycle | |
| III | SIRT1 | Mainly nucleus | Histone, MEF2, NF-κB, FOXO, α-MyHC, MyoD, IRF9, PCAF, TAFI68, p300, NOS, p53, PGC1α, BCL, HMG-B1, Prx, Egr-1 | Cellular survival, ageing, energy metabolism, inflammation |
| SIRT2 | Mainly cytoplasm | Histone, α-tubulin, FOXO, RIP1, HSP70 | Microtubule stability | |
| SIRT3 | Cytoplasm (Mitochondria) | Ku70, ACS2, MDH, GDH, Complex I, ICDH2, SOD2 | Energy metabolism | |
| SIRT4 | Cytoplasm | GDH | Energy metabolism, | |
| SIRT5 | Cytoplasm (Mitochondria) | HMG-B1, Cyt c, CPS I, Prx | Urea cycle, apoptosis | |
| SIRT6 | Nucleus | Histone (H3) | Telomeric DNA regulation | |
| SIRT7 | Nucleus | Histone (H3), p53 | Apoptosis | |
| IV | HDAC11 | Nucleus/cytoplasm | Histones,CDT1, HDAC6 | Immunomodulation |
ACS: Acetyl-CoA synthetase; AMPK: AMP-Activated Protein Kinase; Arg2: arginase 2; BCL: B-cell lymphoma; CAMTA: calmodulin binding transcription activator; CDT: chromatin licensing and DNA replication factor; CPS I: carbamoyl phosphate synthetase I; CIITA: class II transactivato; Cyt c: cytochrome c; E2F: E2 factor; FGF: Fibroblast growth factor; FOXO: Forkhead box class O; FOXP3: forkhead box P3; GATA: GATA binding factor; GDH: glutamate dehydrogenase; HDAC: histone deacetylase; HIF: hypoxia-inducible factor; HMG-B: high mobility group box; HP1: Heterochromatin protein 1; HSP: Heat shock protein; ICDH: isocitrate dehydrogenase; IRF: interferon regulatory factor; MCP: monocyte chemoattractant protein; MDH: malate dehydrogenase; MEF: myocyte enhancer factor; MKP: mitogen-activated protein kinase phosphatase; MMP: Matrix metalloproteinase; MR: mineralocorticoid receptor; MRJ: mammalian relative of DnaJ; MyHC: myosin heavy chain; NAD: nicotinamide adenine dinucleotide; NFAT: nuclear factor of activated T cells; NF-κB: nuclear factor-kappa B; NKX2.5: NK2 homeobox 5; NOS: nitric oxide synthase; Nur77: nerve growth factor-induced gene B; pCAF: p300/CbP-associated factor; PCNA: proliferating cell nuclear antigen; PGC1α: peroxisome proliferatoractivated receptor-γ coactivator 1α; PKC-δ: Protein kinase C δ; PPAR-γ: peroxisome proliferatoractivated receptor-γ; pRb: retinoblastoma protein; Prx: peroxiredoxin; RIP: receptor-interacting protein; Runx: runt-related transcription factor; SIRT: silent information regulator; SMC3: Structural maintenance of chromosomes 3; SOD: superoxide dismutase; SP: specificity protein; SRF: serum response factor; STAT: signal transducers and activators of transcription factor; TAFI: TATA box-binding protein-associated factor I; TSHZ: teashirt zinc finger homeobox; YY1: yin-yang 1.
4. HDACs and heart failure
Heart failure (HF) is a common, costly, and potentially fatal health problem worldwide, which results from structural or functional cardiovascular disorders that cause inadequate systemic perfusion. At the cellular levels, heart failure is associated with both myocyte hypertrophy, apoptosis or even death and myocardial or interstitial fibrosis. Experimental studies have indicated HDACs are involved in the pathogenesis of heart failure.
Cardiac hypertrophy is a form of remodeling in response to the request for higher workload from peripheral tissue, which is considered as a physiological compensatory mechanism in the initial process. However, prolonged cardiac hypertrophy caused by persistent stimuli leads to pathological changes,[27] characterized by diastolic dysfunction and massive interstitial fibrosis, and may result in irreversible heart failure.[28] Studies have indicated that HDAC inhibitors protect against cardiac hypertrophy in animal models,[29]–[31] and an increasing number of studies have also identified the different roles of individual HDACs in mediating cardiac hypertrophy. Class I HDACs have demonstrated to act as pro-hypertrophic activators in the heart. Recent studies indicate that HDAC1 contributes to cardiac differentiation and early embryonic development, which is involved in hypertrophy under pathological conditions.[32],[33] Numerous studies have focused on the role of HDAC2 in cardiac hypertrophy. HDAC2 deficiency prevents the re-expression of fetal genes and attenuates cardiac hypertrophy in hearts exposed to hypertrophic stimuli. Consistently, HDAC2 transgenic mice have augmented hypertrophy.[34] Mechanically, Eom, et al.[35] demonstrate that casein kinase-2α1 (CK-2α1) induces hypertrophic response by phosphorylation of HDAC2 S394, which is essential for its enzymatic activation in the heart. Another study emphasizes HSP70/HDAC2 as a novel mechanism regulating hypertrophy.[36] Although global deletion of HDAC8 in mice leads to perinatal lethality due to skull instability,[37] a recent study demonstrates that HDAC inhibition suppresses cardiac hypertrophy and fibrosis in DOCA-salt hypertensive rats via regulation of HDAC6/HDAC8 enzyme activity.[38] However, some studies report different results, among class I HDACs, HDAC3 induces cardiac myocyte proliferation rather than hypertrophy,[39] and cardiac specific deletion of HDAC3 results in severe cardiac hypertrophy,[40] indicating that HDAC3 is a negative regulator of hypertrophy.
Unlike most of class I HDACs, animal studies have indicated that class II HDACs, in particular class IIa, negatively regulate heart hypertrophy.[41] Either HDAC4 or HDAC5 knockout mice develop cardiac hypertrophy. In addition, HDAC5/9 double deletion results in even greater degree of cardiac hypertrophy than lacking one of them.[42],[43] Mechanically, class IIa (HDAC 4, 5, 7, 9) exhibit inhibitory effects on the transcriptional activity of myocyte enhancer factor-2 (MEF2), a key regulator of cardiac hypertrophy, by binding to a conserved DNA domain,[44] resulting in repression of downstream target genes. Apart from MEF2, other hypertrophic transcription factors, such as nuclear factor of activated T cells (NFAT),[45] GATA binding factor 4 (GATA 4),[46] serum response factor (SRF),[47] and calmodulin binding transcription activator 2 (CAMTA2),[48] are also mediated by class IIa HDACs.
The functional role of class IIb HDACs in the heart was unknown until recent studies found that the catalytic activity of HDAC6 was consistently increased in stressed myocardium, but not in a model of physiologic hypertrophy, indicating the contribution of HDAC6 to heart failure.[49] Further studies have shown that inhibition of HDAC6 protects against angiotensin II-induced cardiac and skeletal muscle remodeling.[38]
Sirtuins exhibit protective effects in the heart. SIRT1 protects against cardiac apoptosis and hypertrophy in response to aging and stress,[50] while suppression of endogenous SIRT1 activity with a small molecule inhibitor results in exaggerated apoptosis.[51] SIRT3,[52] SIRT6[53] and SIRT7[54] also exhibit beneficial effects in cardiac myocytes. However, opposite results regarding the role of SIRT1 were recently reported. Partially insufficiency of SIRT1 is proved to be cardiac protective,[55] while high levels of SIRT1 promote the development of cardiac hypertrophy and heart failure.[56]
Besides cardiac hypertrophy, cardiac fibrosis is regarded as another important feature of heart failure.[57] Recent studies have shown that HDAC inhibitors attenuate cardiac fibrosis, which is associated with the inhibition of cardiac fibroblast proliferation and differentiation from immature cardiac fibroblasts to collagen-producing myofibroblasts,[58] and suppression of the epithelial-to-mesenchymal transition (EMT) and endothelial-to-mesenchymal transition (Endo-MT)[59],[60]. These results indicate the importance of HDAC-mediated epigenetic processes in the pathogenesis of cardiac fibrosis.[61]
5. HDACs and atherosclerosis
Atherosclerosis is the most common underlying cause of cardiovascular diseases, such as myocardial infarction, hypertension and stroke, and is a slowly progressing disease in which plaques are formed in large and mid-sized arteries. The process of atherogenesis consists of several major pathophysiologic events including endothelial dysfunction, macrophage-derived chronic inflammation, and vascular smooth muscle cell (VSMC)-initiated neointimal remodeling.[62] Findeisen, et al.[63] find that inhibition of HDACs by TSA successfully prevents neointima formation after injury in cultured VSMC and carotid artery rings through suppressing the transcriptional activity of kruppel like factor 4 (KLF4), which in turn up-regulates the expression of p21, a potent negative regulator of cell cycle,[64] indicating that HDAC inhibition protects against atherosclerosis. However, some studies also report the pro-atherogenic effects of TSA by increasing the proliferation of VSMC or activating the Akt signaling pathway.[65] Considering the multiple functions of the HDAC family and contradictory results from HDAC inhibitors used in current studies, it is essential to identify the role of individual HDACs in the initiation and progression of atherosclerosis. Recent studies have indicated a potential protective role of HDAC2 in atherosclerosis. HDAC2 inhibits the transcriptional activity of class II transactivator (CIITA), a key mediator in immune response and the restructuring of extracellular matrix (ECM) in smooth muscle cells and macrophages in a deacetylation-dependent manner.[66] Moreover, a recent study also provides direct evidence showing that overexpression or activation of HDAC2 represents a novel therapy for endothelial dysfunction and atherosclerosis by deacetylation of endothelial arginase 2 (Arg2), a critical target in atherogenesis which controls endothelial nitric oxide, proliferation, fibrosis, and inflammation.[67] Similar to HDCA2, HDAC3 is proved to play a beneficial role in endothelial survival and atherosclerosis development.[68] Up-regulation of HDAC3 by disturbed flow is essential for endothelial survival under oxidative stress via activation of Akt phosphorylation, regulation of EMT and Galectin-9 mediated inflammatory responses or interaction with unspliced X-box-binding protein 1 (XBP 1).[69]–[71] However, Jung, et al.[72] demonstrate that HDAC3 can antagonize aspirin stimulated endothelial production of NO by deacetylation of aspirin-acetylated endothelial nitric oxide synthase (eNOS) on the lysine residue, so as to weaken low-dose aspirin-induced vasoprotection, which accounts, at least partly, for the phenomenon of aspirin resistance in the prevention and treatment of cardiovascular disease. In addition, a very recent study has described another previously unrecognized role of HDAC3 in regulating the atherosclerotic phenotype of macrophages. Using conditional knockout mice, it is found that myeloid HDAC3 deficiency induces a stable plaque phenotype in atherosclerotic lesions via promoting plaque collagen production and deposition, inducing macrophages turning to an anti-inflammatory and pro-fibrotic phenotype.[73]
Except for class I HDACs, recent studies have also demonstrated that class II HDACs are associated with atherosclerosis. By microarray analysis targeted cardiovascular single nucleotide polymorphism (SNP), Matthew, et al.[74] have identified that among the 2100 mapping genes of 50,000 SNPs from multiple ethnic individuals, HDAC4 (rs3791398) runs to the most robust one associated with carotid intima-media thickness, a validated indicators for clinical assessment of subclinical atherosclerosis, strongly indicating the participation of HDAC4 in atherogenesis. Recent studies further find that several stress factors, such as fluid shear and angiotensin II, may induce HDAC5 phosphorylation and nuclear export in SMCs or endothelial cells, and therefore, activate the transcriptional factor MEF2, which in turn enhances the expressions of some laminar flow atheroprotective genes, such as KLF2 and eNOS.[75],[76]. Margariti, et al.[77] demonstrate that overexpression of HDAC7 can suppress endothelial cell proliferation through preventing β-catenin nuclear translocation and down regulating T cell factor-1 (TCF-1)/Id2 and cyclin D1 expression, which results in G1 phase elongation, providing a novel insight into the HDAC7-mediated signal pathways in endothelial growth. Further studies report that HDAC7 promotes toll-like receptor 4 (TLR4)-dependent pro-inflammatory gene expression in macrophages.[78]
Recently, a SNP on chromosome 7p21.1 encoding HDAC9 has been identified to be associated with large vessel ischemic stroke, thus revealing a likely linkage between HDAC9 and other CCVD including atherosclerosis.[79] In both cerebral and systemic arteries, HDAC9 mRNA expressions are up-regulated in either carotid plaques, aortic plaques or femoral plaques compared with controls, which is in consistent with the genetic variant of HDAC9 associated with ischemic stroke, indicating the promoting role of HDAC9 on the pathogenesis of atherosclerosis.[80] Mechanically, some studies report that HDAC9 can repress cholesterol efflux and alternatively activate macrophages in the development of atherosclerosis.[81] These results suggest that targeting of HDAC9 might be a promising therapeutic approach for the prevention of atherosclerosis.
Moreover, studies have demonstrated the role of class III HDACs in atherosclerosis. SIRT1 exhibits protective effects in the vascular system by multiple signaling pathways, including directly deacetylation of some target proteins associated with atherosclerosis, such as eNOS, RelA/p65 and Egr-1 which in turn exhibit either endothelial function improvement, or anti-inflammatory functions to prevent atherogenesis.[82]–[84] In addition, the anti-atherosclerotic effects of SIRT1 are also due to some indirect process apart from deacetylation. SIRT1 can regulate the activity of liver X-receptor (LXR) to promote reverse cholesterol transport, or repress coagulation factor expression.[85],[86] These studies indicate that activation of SIRT1 is a possible therapeutic strategy for atherosclerosis. Although SIRT3 activity and NAD levels are proved to be vital in livers of obese animals,[87] SIRT3 deletion does not affect atherosclerosis in low-density lipoprotein receptor-deficient (LDLR−/−) mice, because gene deficiency of SIRT3 neither alters the vulnerability of atherosclerotic plaque, nor promotes the development of atherosclerotic lesions.[88] A very recent study indicates that circulating levels of SIRT4 might be a potential marker of oxidative metabolism, which is directly associated with the risk of endothelial dysfunction, early atherosclerosis, and coronary artery disease.[89] SIRT6 is involved in the regulation of cholesterol homeostasis and insulin signaling.[90] Taken together, these data strongly indicate specific HDAC isoforms exhibit different roles in the pathogenesis of atherosclerosis. Therefore, it is necessary to further explore the individual role of HDACs in atherosclerosis.
6. HDACs and stroke
Stroke is one of the most common pathologies and the second leading cause of death in the world. The large majority (60% to 80%) of strokes are ischemic, which result from an occlusion of a major cerebral artery by a thrombus, or embolism. In this review, we mainly focus on ischemic stroke. Despite numerous preclinical experiments, as well as clinical trials for exploring promising treatment strategies, currently recombinant tissue plasminogen activator (rtPA) remains to be the unique application for ischemic stroke with the approval of the USFDA and in many other countries. However, the narrow therapeutic window and other disadvantages limit its clinical effectiveness.[91] Given the better understanding of the pathogenesis of ischemic stroke, it is increasingly recognized that an effective treatment must be multi-targeted to provide substantial therapeutic benefit. Excitingly, an increasing number of studies have observed that HDAC inhibitors exhibit neuroprotective properties in various neurological and neurodegenerative diseases, including Huntington's disease (HD), Alzheimer's disease (AD), Parkinson's disease (PD) and stroke.[92] The most commonly studied HDAC inhibitors in experimental ischemic stroke are small molecular compounds with high blood brain barrier permeability, such as TSA, SAHA, VPA, SB and 4-PB. Either pretreatment or post injury administration with these compounds has been proved to reduce infarct volume in experimental animal stroke models, including permanent or transient middle cerebral artery occlusion (MCAO) with or without reperfusion.[93] Multiple mechanisms are involved in their neuroprotective effects, including inhibition of ischemia-induced H3 hypoacetylation, iNOS and cyclooxygenase-2 (COX2)-mediated inflammation, down-regulation of p53 gene expression and caspase-3 activation, and increases in HSP 70 and B-cell lymphoma 2 (Bcl-2) expression.[94]–[96] However, it should be noted that individual HDACs serve distinct functions within the central nervous system, ND it is not surprising that sometimes the use of currently available global HDAC inhibitors results in promoting neuronal death, rather than beneficial effects under certain circumstances.[93] In this review, we highlight the independent contribution of isoform-specific HDACs in the pathogenesis of ischemic stroke.
Class I HDACs are ubiquitously expressed in both central nervous system and peripheral tissues. Accumulating evidence shows that both HDAC1 and HDAC2 are involved in the process of neuron development and differentiation.[97]–[99] During the course of oxygen glucose deprivation induced neuronal ischemia, HDAC1 is recruited to form co-repressor complexes with neuron-restrictive silencer factor, a transcriptional repressor, which consequently antagonizes the cAMP-response element binding protein on cocaine- and amphetamine-regulated transcript activation, leading to augmented cell death, while HDAC2 is excluded in this process.[100] However, HDAC2-mediated histone deacetylation exerts a negative control on the neuronal differentiation of neural stem cells by regulating the interaction of neuronal nitric oxide synthase with postsynaptic density protein 95 (PSD-95) in neurons under ischemic conditions.[101] Although HDAC3 has been reported to be neuro-protective by transcriptional repression of the gene expression of E2 factor-1, a well-characterized neuronal apoptosis-inducing factor,[102] with mounting evidence to its detrimental role in stroke and other neurodegenerative disorders. Bates, et al.[103] report that HAD-3, the C. elegans homologue of HDAC3 can enhance neuronal toxicity in an HD animal model. Accordingly, the expression level of HDAC3 is also substantial increased at the early phase of MCAO induced ischemic stroke in mice.[104] Moreover, a very recent study has also provided evidence that HDAC3-mediated epigenetic regulation is closely associated with sodium-induced arterial stiffness, an independent predictor of stroke.[105]
In contrast to class I HDACs, the class IIa HDACs have tissue-specific expression properties. HDAC4 and HDAC5 are two major class IIa members expressed in neurons. HDAC4 can promote neuronal survival through suppressing cyclin-dependent kinase 1 (CDK1), or by increasing hypoxia-inducible factor-1alpha (HIF-1α) stabilization.[106] HDAC5 is another critical regulator by improving neuronal functions or promoting neuronal regeneration after injury.[107]–[109] In our recent studies, we find that HDAC4 and HDAC5 expressions are significantly decreased in experimental stoke, which is regulated by NADPH oxidase activity. We further note evidence showing that HDAC4 and HDAC5 can enhance cell viability via inhibition of high-mobility group box 1 (HMGB1), a central mediator of tissue damage after acute injury.[110] The identification of genetic variant in HDAC9 associated with ischemic stroke has aroused broad concern throughout the world.[79] In an experimental stroke model, we have observed that cerebral ischemia induces a significant increase of HDAC9 expression,[110] which contributes to deacetylation of autophagic related proteins (unpublished data).
Among the Zn2+-dependent HDACs, HDAC6 is unusual because of its two deacetylase domains. In the central nervous system, HDAC6 is expressed in most brain cells, but is particularly abundant in cerebellar Purkinje cells, with the main function in microtubule regulation by mediating α-tubulin acetylation.[111] HDAC6 has been shown to facilitate the clearance of accumulated Huntington protein by autophagic degradation in Huntington's disease.[112] Further studies have supported the beneficial role of HDAC6 because it may rescue polyQ-induced neurodegeneration by a ubiquitin-proteasome system dependent pathway in addition to autophagy.[113] Notably, despite the neuroprotective role in neurodegenerative diseases, inhibition of HDAC6 may increase vesicle transport of brain derived neurotrophic factor by promoting tubulin acetylation in HD.[114] Similar results have been obtained in experimental stroke showing that specific inhibition of HDAC6 improves the stroke outcome via boosting regulatory T cells, one of the key endogenous modulators of post-ischemic neuroinflammation,[115] indicating the involvement of HDAC6 in stroke and other neuroinflammatory disorders.
Although there is accumulating evidence to support the beneficial roles of SIRTs in ischemic stroke,[116],[117] the individual role of specific class III HDAC member requires further investigation. It has been reported that pretreatment with resveratrol, a SIRT1 activator, protects against cerebral ischemia.[118] The mechanism is probably attributed to regulating the acetylation levels of some molecules that are involved in the pathogenesis of stroke, including eNOS,[119] interferon regulatory factor 9 (IRF9),[120] p53 and NF-κB.[121] Conversely, SIRT2 appears to play a detrimental role in stroke. Recent studies have revealed that SIRT2 is required for programmed cell death by deacetylation of receptor-interacting protein 1, which in turn modulates RIP1-RIP3 complex formation, and thereby promotes tumor necrosis factor-α stimulated necrosis.[122]
7. Conclusions and perspectives
In this review, we summarize the current findings of HDACs in cardio-cerebrovascular system and outline several important molecular mechanisms of HDACs on the regulation of myocardial, vascular and neuronal functions. In addition, we also provide evidence showing the therapeutic effects of HDAC inhibitors for CCVD. However, a number of critical questions remain to be resolved. Currently, available HDAC inhibitors are pan-inhibitors and inhibit multiple HDAC proteins. Whether therapeutic efficacy can be enhanced by specifically targeting the disease relevant HDAC, or a subclass, while reducing or eliminating the side effects? What are the precise biochemical targets of HDACs that regulate these processes? Therefore, further studies are needed to continue deciphering the role of individual HDACs in the cardio-cerebrovascular system under different physiological and pathological situations. We anticipate that the exciting new developments regarding the isoform-specific HDAC inhibitors for optimized therapy in the near future.
Acknowledgments
This study was supported by grants from the National 973 Basic Research Program of China (2012CB517700); the National Nature Science Foundation of China (81470958, 81400730 and 81170772); Foundation of Program for New Century Excellent Talents in University (NCET-11-0311) to Yi F; Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, IRT13028); the Special Financial Grant from the China Postdoctoral Science Foundation (2014T70660); the China Postdoctoral Science Foundation (2012M521345) and the Shandong Province Post-doctoral Innovation Foundation (201202009).
References
- 1.Hunter DJ, Reddy KS. Noncommunicable diseases. N Engl J Med. 2013;369:1336–1343. doi: 10.1056/NEJMra1109345. [DOI] [PubMed] [Google Scholar]
- 2.Xie M, Hill JA. HDAC-dependent ventricular remodeling. Trends Cardiovasc Med. 2013;23(6):229–235. doi: 10.1016/j.tcm.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auzzas L, Larsson A, Matera R, et al. Non-natural macrocyclic inhibitors of histone deacetylases: design, synthesis, and activity. J Med Chem. 2010;53:8387–8399. doi: 10.1021/jm101092u. [DOI] [PubMed] [Google Scholar]
- 4.Tao H, Shi KH, Yang JJ, et al. Histone deacetylases in cardiac fibrosis: current perspectives for therapy. Cell Signal. 2014;26:521–527. doi: 10.1016/j.cellsig.2013.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Mai A, Massa S, Rotili D, et al. Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev. 2005;25:261–309. doi: 10.1002/med.20024. [DOI] [PubMed] [Google Scholar]
- 6.Chuang DM, Leng Y, Marinova Z, et al. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 8.Fischer DD, Cai R, Bhatia U, et al. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J Biol Chem. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- 9.Villagra A, Cheng F, Wang HW, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michishita E, Park JY, Burneskis JM, et al. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanno M, Sakamoto J, Miura T, et al. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 13.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks PA, Richon VM, Breslow R, et al. Histone deacetylase inhibitors as new cancer drugs. Curr Opin Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 15.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida M, Kijima M, Akita M, et al. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 17.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 18.Codd R, Braich N, Liu J, et al. Zn(II)-dependent histone deacetylase inhibitors: suberoylanilide hydroxamic acid and trichostatin A. Int J Biochem Cell Biol. 2009;41:736–739. doi: 10.1016/j.biocel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 19.Campas-Moya C. Romidepsin for the treatment of cutaneous T-cell lymphoma. Drugs Today (Barc) 2009;45:787–795. doi: 10.1358/dot.2009.45.11.1437052. [DOI] [PubMed] [Google Scholar]
- 20.Hess-Stumpp H, Bracker TU, Henderson D, et al. MS-275, a potent orally available inhibitor of histone deacetylases--the development of an anticancer agent. Int J Biochem Cell Biol. 2007;39:1388–1405. doi: 10.1016/j.biocel.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Khan N, Jeffers M, Kumar S, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 22.Itoh Y, Suzuki T, Miyata N. Isoform-selective histone deacetylase inhibitors. Curr Pharm Des. 2008;14:529–544. doi: 10.2174/138161208783885335. [DOI] [PubMed] [Google Scholar]
- 23.Vannini A, Volpari C, Filocamo G, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krennhrubec K, Marshall BL, Hedglin M, et al. Design and evaluation of ‘Linkerless’ hydroxamic acids as selective HDAC8 inhibitors. Bioorg Med Chem Lett. 2007;17:2874–2878. doi: 10.1016/j.bmcl.2007.02.064. [DOI] [PubMed] [Google Scholar]
- 25.Balasubramanian S, Ramos J, Luo W, et al. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 26.Haggarty SJ, Koeller KM, Wong JC, et al. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 28.Bernardo BC, Weeks KL, Pretorius L, et al. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol Ther. 2010;128:191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Kee HJ, Sohn IS, Nam KI, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 30.Kong Y, Tannous P, Lu G, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antos CL, McKinsey TA, Dreitz M, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278:28930–28937. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 32.Lagger G, O'Carroll D, Rembold M, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Li T, Liu Y, et al. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim Biophys Acta. 2009;1793:300–311. doi: 10.1016/j.bbamcr.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Trivedi CM, Luo Y, Yin Z, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 35.Eom GH, Cho YK, Ko JH, et al. Casein kinase-2alpha1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011;123:2392–2403. doi: 10.1161/CIRCULATIONAHA.110.003665. [DOI] [PubMed] [Google Scholar]
- 36.Kee HJ, Eom GH, Joung H, et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ Res. 2008;103:1259–1269. doi: 10.1161/01.RES.0000338570.27156.84. [DOI] [PubMed] [Google Scholar]
- 37.Haberland M, Mokalled MH, Montgomery RL, et al. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kee HJ, Bae EH, Park S, et al. HDAC inhibition suppresses cardiac hypertrophy and fibrosis in DOCA-salt hypertensive rats via regulation of HDAC6/HDAC8 enzyme activity. Kidney Blood Press Res. 2013;37:229–239. doi: 10.1159/000350148. [DOI] [PubMed] [Google Scholar]
- 39.Trivedi CM, Lu MM, Wang Q, et al. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008;283:26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montgomery RL, Potthoff MJ, Haberland M, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang CL, McKinsey TA, Chang S, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vega RB, Matsuda K, Oh J, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 43.Chang S, McKinsey TA, Zhang CL, et al. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han A, He J, Wu Y, et al. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J Mol Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 45.Dai YS, Xu J, Molkentin JD. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol Cell Biol. 2005;25:9936–9948. doi: 10.1128/MCB.25.22.9936-9948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morin S, Charron F, Robitaille L, et al. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 2000;19:2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis FJ, Gupta M, Camoretti-Mercado B, et al. Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J Biol Chem. 2003;278:20047–20058. doi: 10.1074/jbc.M209998200. [DOI] [PubMed] [Google Scholar]
- 48.Song K, Backs J, McAnally J, et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 49.Lemon DD, Horn TR, Cavasin MA, et al. Cardiac HDAC6 catalytic activity is induced in response to chronic hypertension. J Mol Cell Cardiol. 2011;51:41–50. doi: 10.1016/j.yjmcc.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alcendor RR, Gao S, Zhai P, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 51.Alcendor RR, Kirshenbaum LA, Imai S, et al. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 52.Sundaresan NR, Gupta M, Kim G, et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundaresan NR, Vasudevan P, Zhong L, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med. 2012;18:1643–1650. doi: 10.1038/nm.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vakhrusheva O, Smolka C, Gajawada P, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 55.Sundaresan NR, Pillai VB, Wolfgeher D, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 56.Oka S, Alcendor R, Zhai P, et al. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abonnenc M, Nabeebaccus AA, Mayr U, et al. Extracellular matrix secretion by cardiac fibroblasts: role of microRNA-29b and microRNA-30c. Circ Res. 2013;113:1138–1147. doi: 10.1161/CIRCRESAHA.113.302400. [DOI] [PubMed] [Google Scholar]
- 58.Guo W, Shan B, Klingsberg RC, et al. Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol. 2009;297:L864–L870. doi: 10.1152/ajplung.00128.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barter MJ, Pybus L, Litherland GJ, et al. HDAC-mediated control of ERK- and PI3K-dependent TGF-beta-induced extracellular matrix-regulating genes. Matrix Biol. 2010;29:602–612. doi: 10.1016/j.matbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Peinado H, Ballestar E, Esteller M, et al. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuetze KB, McKinsey TA, Long CS. Targeting cardiac fibroblasts to treat fibrosis of the heart: focus on HDACs. J Mol Cell Cardiol. 2014;70:100–107. doi: 10.1016/j.yjmcc.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:812–819. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Findeisen HM, Gizard F, Zhao Y, et al. Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arterioscler Thromb Vasc Biol. 2011;31:851–860. doi: 10.1161/ATVBAHA.110.221952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kee HJ, Kwon JS, Shin S, et al. Trichostatin A prevents neointimal hyperplasia via activation of Kruppel like factor 4. Vascul Pharmacol. 2011;55:127–134. doi: 10.1016/j.vph.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Song S, Kang SW, Choi C. Trichostatin A enhances proliferation and migration of vascular smooth muscle cells by downregulating thioredoxin 1. Cardiovasc Res. 2010;85:241–249. doi: 10.1093/cvr/cvp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kong X, Fang M, Li P, et al. HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells. J Mol Cell Cardiol. 2009;46:292–299. doi: 10.1016/j.yjmcc.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 67.Pandey D, Sikka G, Bergman Y, et al. Transcriptional regulation of endothelial arginase 2 by histone deacetylase 2. Arterioscler Thromb Vasc Biol. 2014;34:1556–1566. doi: 10.1161/ATVBAHA.114.303685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zampetaki A, Zeng L, Margariti A, et al. Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation. 2010;121:132–142. doi: 10.1161/CIRCULATIONAHA.109.890491. [DOI] [PubMed] [Google Scholar]
- 69.Zeng L, Wang G, Ummarino D, et al. Histone deacetylase 3 unconventional splicing mediates endothelial-to-mesenchymal transition through transforming growth factor beta2. J Biol Chem. 2013;288:31853–31866. doi: 10.1074/jbc.M113.463745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alam S, Li H, Margariti A, et al. Galectin-9 protein expression in endothelial cells is positively regulated by histone deacetylase 3. J Biol Chem. 2011;286:44211–44217. doi: 10.1074/jbc.M111.242289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin D, Li Y, Yang J, et al. Unspliced X-box-binding Protein 1 (XBP1) Protects Endothelial Cells from Oxidative Stress through Interaction with Histone Deacetylase 3. J Biol Chem. 2014;289:30625–30634. doi: 10.1074/jbc.M114.571984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung SB, Kim CS, Naqvi A, et al. Histone deacetylase 3 antagonizes aspirin-stimulated endothelial nitric oxide production by reversing aspirin-induced lysine acetylation of endothelial nitric oxide synthase. Circ Res. 2010;107:877–887. doi: 10.1161/CIRCRESAHA.110.222968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoeksema MA, Gijbels MJ, Van den Bossche J, et al. Targeting macrophage Histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol Med. 2014;6:1124–1132. doi: 10.15252/emmm.201404170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lanktree MB, Hegele RA, Yusuf S, et al. Multi-ethnic genetic association study of carotid intima-media thickness using a targeted cardiovascular SNP microarray. Stroke. 2009;40:3173–3179. doi: 10.1161/STROKEAHA.109.556563. [DOI] [PubMed] [Google Scholar]
- 75.Wang W, Ha CH, Jhun BS, et al. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood. 2010;115:2971–2979. doi: 10.1182/blood-2009-05-224824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu X, Ha CH, Wong C, et al. Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler Thromb Vasc Biol. 2007;27:2355–2362. doi: 10.1161/ATVBAHA.107.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Margariti A, Zampetaki A, Xiao Q, et al. Histone deacetylase 7 controls endothelial cell growth through modulation of beta-catenin. Circ Res. 2010;106:1202–1211. doi: 10.1161/CIRCRESAHA.109.213165. [DOI] [PubMed] [Google Scholar]
- 78.Shakespear MR, Hohenhaus DM, Kelly GM, et al. Histone deacetylase 7 promotes Toll-like receptor 4-dependent proinflammatory gene expression in macrophages. J Biol Chem. 2013;288:25362–25374. doi: 10.1074/jbc.M113.496281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.International Stroke Genetics C. Wellcome Trust Case Control C. Bellenguez C, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Markus HS, Makela KM, Bevan S, et al. Evidence HDAC9 genetic variant associated with ischemic stroke increases risk via promoting carotid atherosclerosis. Stroke. 2013;44:1220–1225. doi: 10.1161/STROKEAHA.111.000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao Q, Rong S, Repa JJ, et al. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arterioscler Thromb Vasc Biol. 2014;34:1871–1879. doi: 10.1161/ATVBAHA.114.303393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mattagajasingh I, Kim CS, Naqvi A, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci USA. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vedantham S, Thiagarajan D, Ananthakrishnan R, et al. Aldose reductase drives hyperacetylation of Egr-1 in hyperglycemia and consequent upregulation of proinflammatory and prothrombotic signals. Diabetes. 2014;63:761–774. doi: 10.2337/db13-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X, Zhang S, Blander G, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 86.Breitenstein A, Stein S, Holy EW, et al. Sirt1 inhibition promotes in vivo arterial thrombosis and tissue factor expression in stimulated cells. Cardiovasc Res. 2011;89:464–472. doi: 10.1093/cvr/cvq339. [DOI] [PubMed] [Google Scholar]
- 87.Choudhury M, Jonscher KR, Friedman JE. Reduced mitochondrial function in obesity-associated fatty liver: SIRT3 takes on the fat. Aging (Albany NY) 2011;3:175–178. doi: 10.18632/aging.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winnik S, Gaul DS, Preitner F, et al. Deletion of Sirt3 does not affect atherosclerosis but accelerates weight gain and impairs rapid metabolic adaptation in LDL receptor knockout mice: implications for cardiovascular risk factor development. Basic Res Cardiol. 2014;109:399. doi: 10.1007/s00395-013-0399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tarantino G, Finelli C, Scopacasa F, et al. Circulating levels of sirtuin 4, a potential marker of oxidative metabolism, related to coronary artery disease in obese patients suffering from NAFLD, with normal or slightly increased liver enzymes. Oxid Med Cell Longev. 2014;2014:920676. doi: 10.1155/2014/920676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kleindorfer D, Lindsell CJ, Brass L, et al. National US estimates of recombinant tissue plasminogen activator use: ICD-9 codes substantially underestimate. Stroke. 2008;39:924–928. doi: 10.1161/STROKEAHA.107.490375. [DOI] [PubMed] [Google Scholar]
- 92.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 93.Langley B, Brochier C, Rivieccio MA. Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke. 2009;40:2899–2905. doi: 10.1161/STROKEAHA.108.540229. [DOI] [PubMed] [Google Scholar]
- 94.Kim HJ, Rowe M, Ren M, et al. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: multiple mechanisms of action. J Pharmacol Exp Ther. 2007;321:892–901. doi: 10.1124/jpet.107.120188. [DOI] [PubMed] [Google Scholar]
- 95.Faraco G, Pancani T, Formentini L, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 96.Ren M, Leng Y, Jeong M, et al. Valproic acid reduces brain damage induced by transient focal cerebral ischemia in rats: potential roles of histone deacetylase inhibition and heat shock protein induction. J Neurochem. 2004;89:1358–1367. doi: 10.1111/j.1471-4159.2004.02406.x. [DOI] [PubMed] [Google Scholar]
- 97.Cunliffe VT. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development. 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- 98.Ballas N, Battaglioli E, Atouf F, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 99.Bai S, Ghoshal K, Datta J, et al. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol Cell Biol. 2005;25:751–766. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Zhang J, Wang S, Yuan L, et al. Neuron-restrictive silencer factor (NRSF) represses cocaine- and amphetamine-regulated transcript (CART) transcription and antagonizes cAMP-response element-binding protein signaling through a dual NRSE mechanism. J Biol Chem. 2012;287:42574–42587. doi: 10.1074/jbc.M112.376590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luo CX, Lin YH, Qian XD, et al. Interaction of nNOS with PSD-95 negatively controls regenerative repair after stroke. J Neurosci. 2014;34:13535–13548. doi: 10.1523/JNEUROSCI.1305-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Panteleeva I, Rouaux C, Larmet Y, et al. HDAC-3 participates in the repression of e2f-dependent gene transcription in primary differentiated neurons. Ann N Y Acad Sci. 2004;1030:656–660. doi: 10.1196/annals.1329.076. [DOI] [PubMed] [Google Scholar]
- 103.Bates EA, Victor M, Jones AK, et al. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26(10):2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen YT, Zang XF, Pan J, et al. Expression patterns of histone deacetylases in experimental stroke and potential targets for neuroprotection. Clin Exp Pharmacol Physiol. 2012;39:751–758. doi: 10.1111/j.1440-1681.2012.05729.x. [DOI] [PubMed] [Google Scholar]
- 105.Herrera VL, Decano JL, Giordano N, et al. Aortic and carotid arterial stiffness and epigenetic regulator gene expression changes precede blood pressure rise in stroke-prone Dahl salt-sensitive hypertensive rats. PLoS One. 2014;9:e107888. doi: 10.1371/journal.pone.0107888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Majdzadeh N, Wang L, Morrison BE, et al. HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev Neurobiol. 2008;68:1076–1092. doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsankova NM, Berton O, Renthal W, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 108.Cho Y, Sloutsky R, Naegle KM, et al. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell. 2013;155:894–908. doi: 10.1016/j.cell.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cho Y, Cavalli V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. EMBO J. 2012;31:3063–3078. doi: 10.1038/emboj.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.He M, Zhang B, Wei X, et al. HDAC4/5-HMGB1 signalling mediated by NADPH oxidase activity contributes to cerebral ischaemia/reperfusion injury. J Cell Mol Med. 2013;17:531–542. doi: 10.1111/jcmm.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 112.Iwata A, Riley BE, Johnston JA, et al. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 113.Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 114.Dompierre JP, Godin JD, Charrin BC, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liesz A, Zhou W, Na SY, et al. Boosting regulatory T cells limits neuroinflammation in permanent cortical stroke. J Neurosci. 2013;33:17350–17362. doi: 10.1523/JNEUROSCI.4901-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mokudai T, Ayoub IA, Sakakibara Y, et al. Delayed treatment with nicotinamide (Vitamin B(3)) improves neurological outcome and reduces infarct volume after transient focal cerebral ischemia in Wistar rats. Stroke. 2000;31:1679–1685. doi: 10.1161/01.str.31.7.1679. [DOI] [PubMed] [Google Scholar]
- 117.Chen TY, Lin MH, Lee WT, et al. Nicotinamide inhibits nuclear factor-kappa B translocation after transient focal cerebral ischemia. Crit Care Med. 2012;40:532–537. doi: 10.1097/CCM.0b013e31822f0b08. [DOI] [PubMed] [Google Scholar]
- 118.Raval AP, Dave KR, Perez-Pinzon MA. Resveratrol mimics ischemic preconditioning in the brain. J Cereb Blood Flow Metab. 2006;26:1141–1147. doi: 10.1038/sj.jcbfm.9600262. [DOI] [PubMed] [Google Scholar]
- 119.Hattori Y, Okamoto Y, Maki T, et al. Silent information regulator 2 homolog 1 counters cerebral hypoperfusion injury by deacetylating endothelial nitric oxide synthase. Stroke. 2014;45:3403–3411. doi: 10.1161/STROKEAHA.114.006265. [DOI] [PubMed] [Google Scholar]
- 120.Chen HZ, Guo S, Li ZZ, et al. A critical role for interferon regulatory factor 9 in cerebral ischemic stroke. J Neurosci. 2014;34:11897–11912. doi: 10.1523/JNEUROSCI.1545-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hernandez-Jimenez M, Hurtado O, Cuartero MI, et al. Silent information regulator 1 protects the brain against cerebral ischemic damage. Stroke. 2013;44:2333–2337. doi: 10.1161/STROKEAHA.113.001715. [DOI] [PubMed] [Google Scholar]
- 122.Narayan N, Lee IH, Borenstein R, et al. The NAD-dependent deacetylase SIRT2 is required for programmed necrosis. Nature. 2012;492:199–204. doi: 10.1038/nature11700. [DOI] [PubMed] [Google Scholar]