Abstract
A new scaffold, azolylthioacetamide, was constructed and assayed against metallo-β-lactamases (MβLs). The obtained molecules specifically inhibited MβL ImiS, and 1c was found to be the most potent inhibitor, with a Ki = 1.2 μM using imipenem as substrate. Structure–activity relationships reveal that the aromatic carboxyl improves inhibitory activity of the inhibitors, but the aliphatic carboxyl does not. Compounds 1c–d and 1h–i showed the best antibacterial activities against E. coli BL21(DE3) cells producing CcrA or ImiS, resulting in 32- and 8-fold reduction in MIC values, respectively; 1c and 1f–j resulted in a reduction in MIC against P. aeruginosa. Docking studies revealed that 1a, 1c, and 1d fit tightly into the substrate binding site of CphA as a proxy for ImiS with the aromatic carboxylate forming interactions with Lys224, the Zn(II) ion, the backbone of Asn233, and hydrophobic portions of the inhibitors aligning with hydrophobic patches of the protein surface.
Keywords: Antibiotic resistance, metallo-β-lactamase, subclass B2, ImiS, inhibitor, azolylthioacetamide
The development of β-lactam antibiotics over the past 70 years has led to the availability of drugs to treat a wide range of Gram-positive and Gram-negative bacterial infections. However, overuse of antibiotics has resulted in a large number of bacteria that produce β-lactamases,1 and the resulting bacteria are resistant to the most commonly used antibiotics including penicillins, cephalosporins, and carbapenems. β-Lactamases are enzymes that inactivate β-lactam antibiotics by breaking the C–N bond of the β-lactam ring and render the drugs ineffective.2
There have been more than 1000 distinct β-lactamases identified, and these enzymes have been categorized into classes A to D, based on their amino acid sequence homologies.3 Class A, C, and D enzymes are collectively called serine β-lactamases. These enzymes use a common catalytic mechanism in which an active-site serine nucleophilically attacks the β-lactam carbonyl, ultimately leading to a cleaved β-lactam ring. Class B enzymes, also called metallo-β-lactamases (MβLs), utilize either 1 or 2 equiv of Zn(II) to catalyze the β-lactam hydrolysis.4 MβLs have been further subgrouped into subclasses B1 to B3, based on amino acid sequence homologies and Zn(II) content.3 MβLs belonging to the B1 and B3 subclasses can hydrolyze almost all known β-lactam antibiotics. In contrast, the B2 subclass enzymes have a narrow substrate profile including carbapenems, which have been called “last resort” antibiotics.5 There are no inhibitors of the MβLs available in the clinic to date.6
In a search for agents that could prevent the hydrolysis of antibiotics by MβLs, a large amount of effort has been made toward identifying novel inhibitors of these enzymes. However, the absence of a highly populated metastable covalent intermediate in the catalytic mechanism (as observed in serine β-lactamases) and the subtle but significant variations in MβLs’ active site architectures make it difficult to search for clinically useful inhibitors that act on all three subclasses of MβLs or even on all the enzymes within the same subclass.7 Toney et al. reported biphenyl tetrazoles and succinic acids to be effective inhibitors of the MβL IMP-1.8 Mercaptocarboxylate,9 3-subsutituted phthalic acid derivatives,10 and N-arylsulfonyl hydrazones11 also displayed inhibitory activities on IMP-1. A 2-picolinic acid-based inhibitor was found to inhibit the B2 subclass CphA.12 Also some broad-spectrum inhibitors of the MβLs have been reported, including mercaptophosphonate compounds, thiomandelic acid, and thiols.13
The MβL ImiS is a representative of the B2 subclass enzymes and exhibits maximal activity with only one Zn(II) bound.14 There has been a large amount of effort in structural, spectroscopic, mechanistic, steady-state kinetic, and inhibition studies of this enzyme.15 The crystal structures of several MβLs, including CcrA9 and L1,16 which are B1 and B3 subclass enzymes, respectively, have been solved. Although no structure of ImiS is available to date, crystal structures of the very closely related B2 enzyme CphA13 and the more distantly related Sfh-I17 have been reported. Spectroscopic studies indicate that the catalytic activity of ImiS requires that Zn(II) or Co(II) (in substituted ImiS) is bound by a cysteine and a histidine residue and the metal ion can adopt 4- or 5-fold coordination.18 Our studies showed that the reaction of ImiS with imipenem leads to a product-bound species, coordinated to Zn(II) via a carboxylate group;19 EPR studies revealed that the metal ion in Co(II)-ImiS is 4-coordinate.20 Site-directed mutagenesis studies indicated that Lys224 plays a catalytic role in ImiS, while the side chain of Asn233 does not play a role in binding or catalysis.21 Steady-state kinetic studies revealed that Co(II)-ImiS exhibited the catalytic constants kcat of 255 s–1 and Km of 99 μM when using imipenem as substrate.21 Inhibition studies revealed that N-heterocyclic dicarboxylic acid is a competitive inhibitor of ImiS with a Ki value of 7.1 μM.15
Azoles such as thiadiazoles, oxadiazoles, and triazoles22 have been found to possess broad-spectrum antimicrobial activity and MβL inhibition. The exploration of new heterocycles that have potency to multiple biological targets remains an intriguing scientific endeavor. Recently, we reported diaryl-substituted azolylthioacetamides as broad-spectrum inhibitors of MβLs.23 Our goal is to develop the specific or even broad-spectrum inhibitors of MβLs and to use these inhibitors as drug/inhibitor combinations to combat bacterial infections in which the bacteria produce a MβL. Toward this goal, novel azolylthioacetamides were synthesized and characterized (Figure 1). These compounds were tested as inhibitors against the purified MβLs CcrA, NDM-1, ImiS, and L1, which are representative enzymes belonging to the B1a, B1b, B2, and B3 subclasses of MβLs, respectively.24 Furthermore, antimicrobial activities of these inhibitors in combination with existing antibiotics against antibiotic resistant strains were evaluated.
Figure 1.
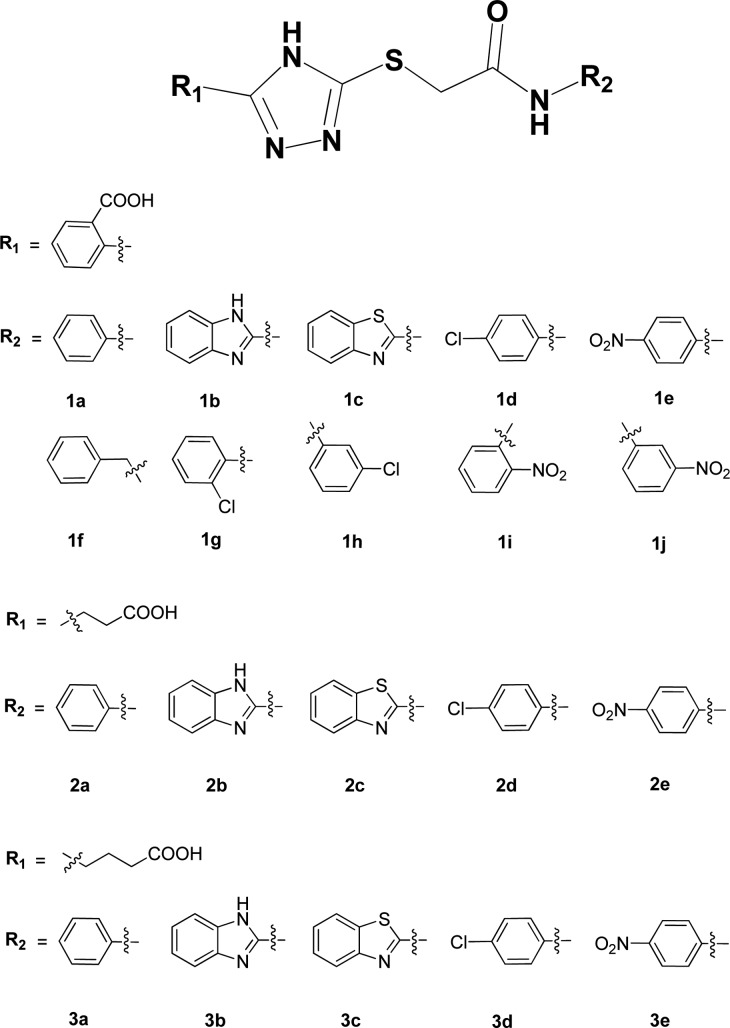
Structures of the synthesized azolylthioacetamides.
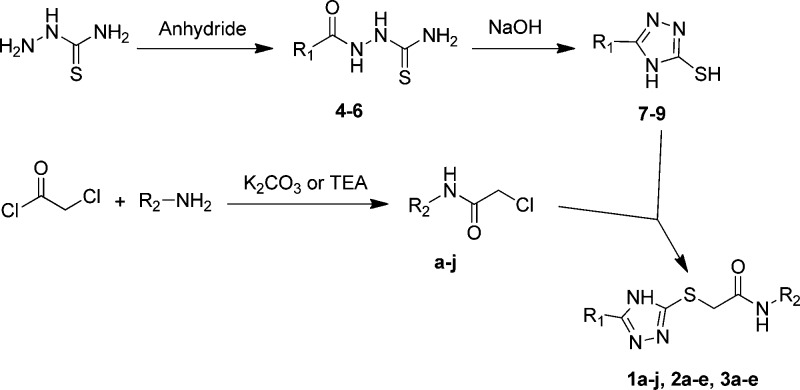
Twenty azolylthioacetamides (Figure 1) were synthesized as shown in Scheme 1. First, thiosemicarbazide was refluxed respectively with phthalic anhydride, succinic anhydride, or glutaric anhydride in acetonitrile to give substituted thiosemicarbazides (4–6). These intermediates were then refluxed in NaOH aqueous solution and acidified with dilute HCl to offer thiol triazoles (7–9).25 Anilines with various substituents were acylated with chloroacetyl chloride to get α-chloroacetamides (a–j). Finally, under alkaline conditions, the thiol triazoles and α-chloroacetamides were cross-linked by a nucleophilic substitution reaction to afford the target products azolylthioacetamides (1a–j, 2a–e, and 3a–e).26 All these azolylthioacetamides were characterized by 1H and 13C NMR and confirmed by MS.
Scheme 1. Synthetic Route of the Azolylthioacetamides.
To test whether these azolylthioacetamides were specific or even broad-spectrum inhibitors of the MβLs, the MβLs from subclasses B1a (CcrA), B1b (NDM-1), B2 (ImiS), and B3 (L1) were overexpressed and purified as previously described, respectively.27−30 The inhibition experiments of the azolylthioacetamides as inhibitors against MβLs were conducted on an Agilent UV8453 spectrometer. The initial reaction rates were determined in triplicate, and the average value was recorded. Kinetic studies were performed using cephalexin V as substrate of CcrA, NDM-1, and L1, and imipenem as substrate of ImiS. The substrate concentrations were varied between 24 and 140 μM, and inhibitor concentrations were varied between 0 and 10 μM. The mode of inhibition was determined by generating Lineweaver–Burk plots of the data, and the Ki values for the inhibitors were determined by fitting initial velocity versus substrate concentration at each inhibitor concentration. Hydrolysis of substrate was monitored at 262 and 300 nm, respectively.
The inhibition studies indicated that azolylthioacetamides had specific inhibition activity against ImiS, but no activity was observed against CcrA, NDM-1, and L1 at inhibitor concentrations of up to no more than 10 μM. The inhibitory constants (Ki) of azolylthioacetamides against ImiS are listed in Table 1. It is clear to be observed that 1a–e and 1g–j exhibited high inhibitory activities against ImiS with a Ki value range of 1.2–3.6 μM. Among these azolylthioacetamides, 1c showed the lowest Ki value of 1.2 μM. The fact that 1f did not inhibit ImiS indicates that an aromatic amine exhibits better inhibition efficiency than an aliphatic amine. However, the azolylthioacetamides 2a–b and 2d with an aliphatic carboxyl at R1 position showed little inhibitory activity against ImiS, exhibiting Ki > 2.5 mM, and 2c and 3b had no inhibitory activities, implying that the aliphatic carboxyl at R1 position weakens inhibition efficiency of the inhibitors.
Table 1. Inhibition Constants of Metallo-β-lactamase ImiS by Azolylthioacetamidesa.
| compd | Ki (μM) | compd | Ki (μM) |
|---|---|---|---|
| 1a | 1.8 ± 0.3 | 1i | 2.7 ± 0.3 |
| 1b | 3.6 ± 0.4 | 1j | 1.9 ± 0.2 |
| 1c | 1.2 ± 0.1 | 2a | >5000 |
| 1d | 1.4 ± 0.5 | 2b | >2500 |
| 1e | 2.2 ± 0.2 | 2c | |
| 1f | 2d | >5000 | |
| 1g | 2.3 ± 0.2 | 3b | |
| 1h | 2.4 ± 0.4 |
Ki values were determined using imipenem as substrate and ImiS as the enzyme in 50 mM Tris, pH 7.0, at 25 °C. The inhibitor concentrations were varied between 0 and 10 μM; empty cells showed no inhibition.
The above data of Ki values reveal a structure–activity relationship, which is that the aromatic carboxyl at the triazole ring (R1 position) improves the inhibitory activity of azolylthioacetamide against ImiS. It was of interest to explore the pontential role of an aromatic carboxyl at the R1 position. We will develop more azolylthioacetamides with various substituents for further structure–activity relationship studies.
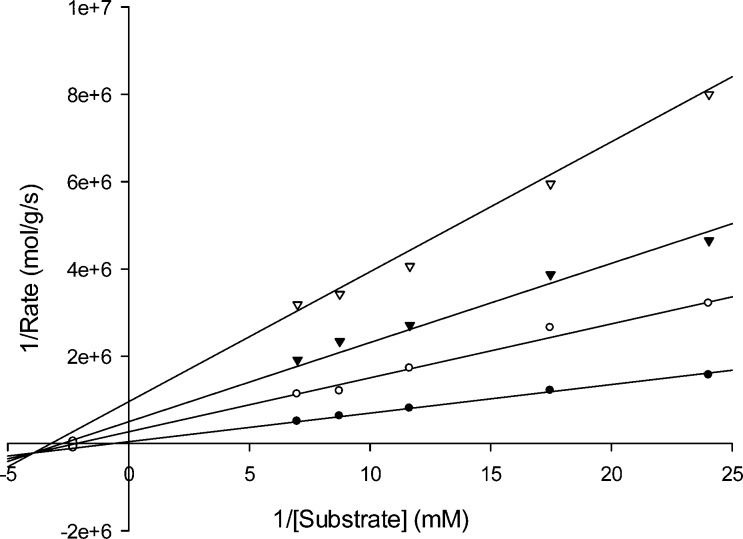
The Lineweaver–Burk plots with azolylthioacetamide inhibitors indicated that 1a–e and 1g–j are mixed inhibitors of ImiS. This inhibition mode is the same as that of the previously reported diaryl-substituted azolylthioacetamide derivatives.23 Inhibitor 1c was taken as an example to show its inhibition mode against ImiS in Figure 2.
Figure 2.
Lineweaver–Burk plots of inhibition of ImiS-catalyzed hydrolysis activity by azolylthioacetamide 1c. Imipenem was used as substrate of ImiS. Data points show inhibitor 1c concentrations of 0 (●), 2.5 (○), 5 (▼), and 10 (▽) μM.
The antibacterial activities of the azolylthioacetamides were investigated by determining the minimum inhibitory concentrations (MICs) of existing antibiotics in the presence and absence of 1a–j.31 Two clinical bacteria P. aeruginosa and K. pneumoniae and four bacterial strains of E. coli BL21(DE3) containing plasmids pMSZ02-CcrA, pET26b-NDM-1, pET26b-ImiS, and pET26b-L1 were used to assess these inhibitors (Table 2). While this strain of K. pneumoniae does not produce a metallo-β-lactamase, it is known to produce extended spectrum β-lactamases (ESBLs);32 therefore, the use of this strain in the MIC experiments allowed for us to evaluate the biological activity of the azolylthioacetamides toward other (serine) β-lactamases. P. aeruginosa is known to produce metallo-β-lactamase IMP-1;9 therefore, the use of this strain in the MIC experiments allowed for us to evaluate the biological activity of the azolylthioacetamide derivatives toward this clinically important strain.
Table 2. Antibacterial Activities (MICs in μg/mL) of β-Lactam Antibiotics in the Presence of the Azolylthioacetamides 1a–j at a Concentration of 16 μg/mL.
| azolylthioacetamides |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| antibiotic-resistant bacteria | inhibitor blank | 1a | 1b | 1c | 1d | 1e | 1f | 1g | 1h | 1i | 1j |
| E. coli-L1a | 8 | 8 | 8 | 8 | 8 | 16 | 16 | 64 | 64 | >128 | 8 |
| E. coli-NDM-1a | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| E. coli-CcrAa | 4 | 4 | 4 | <0.125 | <0.125 | 2 | 4 | 2 | 0.5 | 0.25 | 2 |
| E. coli-ImiSb | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 1 | 0.25 | <0.125 | <0.125 | 0.25 |
| P. aeruginosab | 0.25 | 0.25 | 0.25 | <0.125 | 0.25 | 0.25 | <0.125 | <0.125 | <0.125 | <0.125 | <0.125 |
| K. pneumoniaeb | 0.25 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
Antibiotic used: cefalexin V.
Antibiotic used: imipenem.
MIC data indicate that some of the inhibitors 1c–j increase the antimicrobial effect of antibiotics on Gram-negative P. aeruginosa and/or E. coli BL21(DE3) expressing ImiS or CcrA, and the largest effect was observed with E. coli BL21(DE3) expressing CcrA. However, these compounds showed no effect on K. pneumonia and on E. coli BL21(DE3) producing L1 and NDM-1 (Table 2). For K. pneumoniae this observation could be explained with decreased cell permeability but more likely with the compounds being ineffective against serine β-lactamases. Also for E. coli expressing L1 and NDM-1, permeability issues are unlikely because these cells are identical to those expressing CcrA and ImiS. More likely, it is a result of low binding affinity of these inhibitors as observed in the in vitro inhibition studies. It is not clear why the azolylthioacetamides 1e–i increase MIC values of existing antibiotics for E. coli containing the L1 plasmid and K. pneumoniae. This could be due to an allosteric effect of the compounds on L1 and the ESBL expressed in E. coli and K. pneumoniae, respectively, as has been suggested recently for the activation of BcII and ImiS by a β-phospholactam.36 Inclusion of compounds 1c–e and 1g–j resulted in 2–32-fold reduction in MIC for E. coli BL21(DE3) expressing CcrA, 1d–e and 1g–j resulted in 2–8-fold reduction in MIC for E. coli BL21(DE3) expressing ImiS, and 1c and 1f–j resulted in a 2-fold reduction in MIC for Gram-negative P. aeruginosa, but these three strains were not affected by 1a–b.
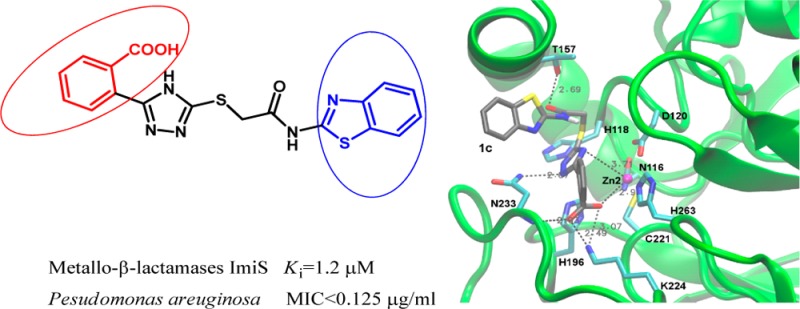
The compounds presented here differ from those of our previous study23 mostly in a negatively charged carboxylate group in R1. In some compounds that had an ortho-hydroxyl phenyl group at this position, the hydroxyl formed a hydrogen bond with Lys224, while the triazole coordinated the Zn(II) ion(s) in docking calculations.23 Introducing a carboxylate at the same position was expected to strengthen the interaction between this moiety and Lys224 through a salt bridge. The docking analysis procedure (see Supporting Information) resulted in 14, 13, and 10 conformations with average binding energies of −11.3, −11.8, and −11.7 kcal/mol for 1a, 1c, and 1d, respectively. These correspond to theoretical Kis in the micromolar range, in good agreement with the experimental Ki values (1.2–2.7 μM). The differences between clusters within 0.5 kcal/mol are not significant, consistent with the values that deviate by less than an order of magnitude. The conformations shown in Figure 3 are the highest ranked (lowest energy; between −12.8 and −11.6 kcal/mol) conformations of those clusters. As expected, these docking calculations reveal that in compounds 1a, 1c, and 1d, which exhibited the lowest Ki values with ImiS, the carboxylate forms a salt bridge with Lys224 of CphA, which serves as a proxy of ImiS. In addition, it interacts with Zn2 and the backbone amide of Asn233, thus tightly anchoring these compounds in the active site. Interestingly, the carboxylate is in an orientation perpendicular to the long axis of the binding site as well as to the phenyl ring it is attached to (Figure 3). The R2 thioacetamides, which are similar to the R group of penicillins and R1 of cephalosporins, are located in the area that is expected to be occupied by these antibiotic groups. R2 (phenyl in 1a, Figure 3B; benzylthiazolyl in 1c, Figure 3C; para-chlorophenyl in 1d, Figure 3D) binds in a hydrophobic area of the substrate binding site (Figure 3E), while either the carbonyl (Figure 3B,C) or amide (Figure 3D) of the thioacetamide interacts with Thr157. Somewhat unexpectedly, the triazole ring did not directly coordinate Zn2 edge-on, but rather interacted face-on with Zn2 at a distance of 3.4 to 3.7 Å, resembling a π electron–cation interaction. There are two possible explanations for this observation: (i) the tight binding of the carboxylphenyl ring to Lys224, Zn2, and Asn233 and resonance between the two rings does not allow the triazole ring to turn enough for an edge-on Zn2 coordination; (ii) there is also a hydrogen bond between the triazole ring and the Asn233 side chain, which could keep the triazole distant from Zn2. Figure 3E shows that compound 1c fits very tightly into the substrate binding site of CphA, taking advantage of hydrophobic interactions between R1 and R2 and protein residues as well as electrostatic interactions, most importantly the salt bridge between the carboxylate and Lys224 and those between the triazole and Zn2 as well as the Asn233 side chain. This image may also serve as a guide to further improve these molecules, perhaps by further exploring hydrophobic interactions between R1 and R2 and the protein.
Figure 3.
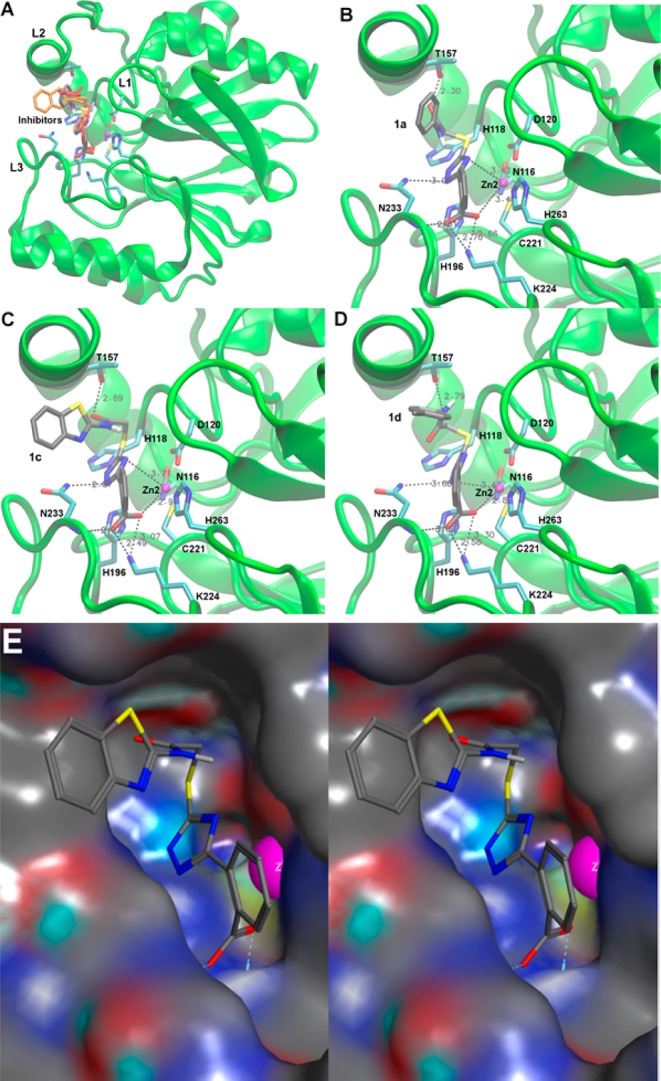
Low energy conformations of 1a, 1c, and 1d docked into the active site of CphA (PDB code 2QDS(13)). (A) Overview of the entire enzyme with loops L1, L2, and L3 (labeling according to Garau et al.33) flanking the active site. The enzyme backbone is shown as a cartoon in green and selected residues are shown as sticks colored by atom (C, cyan; N, blue; O, red; S, yellow) and labeled at Cα. The Zn(II) ion is shown as a magenta sphere. All three inhibitors are superimposed: 1a as sticks colored by atom (same colors as for protein residues except C in gray), 1c as orange sticks, and 1d as red sticks. (B–D) Detailed views of 1a, 1c, and 1d, respectively, with key residues displayed and interactions between the inhibitors and protein residues indicated by dashed lines. Panels A–D were generated with VMD.34 (E) Stereo view of 1c (same coloring and conformation as in panel C) with the enzyme represented as a surface with neutral areas in gray, positively charged areas in blue, and negatively charged areas in red. Zn2 is shown as a magenta sphere. The figure was generated with MOE.35
In addition to the tight fit of the compounds into the CphA binding site, we provide the following rationale for the preferential binding of the inhibitors studied here to ImiS (and CcrA in MIC assays), but not NDM-1 and L1. To that end, the different crystal structures used for docking here and previously23 as well as the chemical structures of the compounds were analyzed. The aromatic carboxylate in R1 of the 1a–j series of the compounds studied here may need a certain width of the binding site to be accommodated in the perpendicular orientation mentioned above, which is provided in CcrA and CphA, but not in NDM-1 and L1. In addition, Zn2 and the bound inhibitor are shifted toward the empty Zn1 binding site in CphA, which is occupied by a Zn(II) ion in CcrA, NDM-1, and L1. In CphA, the triazole ring is located at the position that corresponds to Zn1’s position in CcrA, NDM-1, and L1, which may explain why this favorable conformation cannot be accommodated in these enzymes.
In summary, 20 azolylthioacetamides 1a–j, 2a–e, and 3a–e were synthesized and characterized by 1H and 13C NMR and MS. Several azolylthioacetamides were tested as inhibitors of the MβLs CcrA, NDM-1, ImiS, and L1. Nine azolylthioacetamides inhibited ImiS, exhibiting Ki values of 1.2–3.6 μM using imipenem as substrate. All compounds tested did not inhibit CcrA, NDM-1, and L1 in vitro. Structure–activity relationship studies reveal that the aromatic carboxyl at the triazole ring of azolylthioacetamides improves inhibitory activity of the inhibitors against ImiS and that an aliphatic carboxyl does not. The azolylthioacetamides were tested for antibacterial activity by examining the MIC values for existing antibiotics in the presence/absence of these compounds. The inclusion of eight azolylthioacetamides resulted in lower MIC values when using E. coli BL21(DE3) cells expressing CcrA, ImiS, or P. aeruginosa. The best inhibitors of ImiS, compounds 1a, 1c, and 1d fit tightly into the substrate binding site of CphA as a proxy for ImiS with the aromatic carboxylate forming interactions with Lys224, Zn(II), and the backbone of Asn233, the triazole interacting with Zn(II) and the Asn233 side chain, and hydrophobic portions of the inhibitors occupying hydrophobic pockets. These studies demonstrate that the aromatic carboxyl-substituted azolylthioacetamides are good scaffolds for future inhibitors of the MβL ImiS and potentially other B2 subclass enzymes.
Supporting Information Available
Detailed synthesis procedure, NMR and ESI mass data for all target compounds, the methods for enzyme expression and purification, assay of inhibitory activity, determination of MIC, and docking study. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors.
This work was supported by grants 21272186 and 81361138018 (to K.W.Y.) from the National Natural Science Foundation of China.
The authors declare no competing financial interest.
Supplementary Material
References
- Kurosaki H.; Yamaguchi Y.; Higashi T.; Soga K.; Matsueda S.; Yumoto H.; Misumi S.; Yamagata Y.; Arakawa Y.; Goto M. Irreversible inhibition of metallo-beta-lactamase (IMP-1) by 3-(3-mercaptopropionylsulfanyl)propionic acid pentafluorophenyl ester. Angew. Chem., Int. Ed. 2005, 44, 3861–4. [DOI] [PubMed] [Google Scholar]
- Spencer J.; Walsh T. R. A new approach to the inhibition of metallo-beta-lactamases. Angew. Chem., Int. Ed. 2006, 45, 1022–6. [DOI] [PubMed] [Google Scholar]
- Bush K.; Jacoby G. A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toney J. H.; Moloughney J. G. Metallo-beta-lactamase inhibitors: promise for the future?. Curr. Opin. Investig. Drugs 2004, 5, 823–6. [PubMed] [Google Scholar]
- Papp-Wallace K. M.; Endimiani A.; Taracila M. A.; Bonomo R. A. Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawz S. M.; Bonomo R. A. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.; Ma S. Recent advances in the discovery of metallo-beta-lactamase inhibitors for beta-lactam antibiotic-resistant reversing agents. Curr. Drug Targets 2014, 15, 689–702. [DOI] [PubMed] [Google Scholar]
- Toney J. H.; Hammond G. G.; Fitzgerald P. M.; Sharma N.; Balkovec J. M.; Rouen G. P.; Olson S. H.; Hammond M. L.; Greenlee M. L.; Gao Y. D. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-beta-lactamase. J. Biol. Chem. 2001, 276, 31913–8. [DOI] [PubMed] [Google Scholar]
- Concha N. O.; Janson C. A.; Rowling P.; Pearson S.; Cheever C. A.; Clarke B. P.; Lewis C.; Galleni M.; Frere J. M.; Payne D. J.; Bateson J. H.; Abdel-Meguid S. S. Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 2000, 39, 4288–98. [DOI] [PubMed] [Google Scholar]
- Hiraiwa Y.; Morinaka A.; Fukushima T.; Kudo T. Metallo-beta-lactamase inhibitory activity of phthalic acid derivatives. Bioorg. Med. Chem. Lett. 2009, 19, 5162–5. [DOI] [PubMed] [Google Scholar]
- Siemann S.; Evanoff D. P.; Marrone L.; Clarke A. J.; Viswanatha T.; Dmitrienko G. I. N-arylsulfonyl hydrazones as inhibitors of IMP-1 metallo-beta-lactamase. Antimicrob. Agents Chemother. 2002, 46, 2450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfall L. E.; Garau G.; Lienard B. M.; Dideberg O.; Schofield C. J.; Frere J. M.; Galleni M. Competitive inhibitors of the CphA metallo-beta-lactamase from Aeromonas hydrophila. Antimicrob. Agents Chemother. 2007, 51, 2136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienard B. M.; Garau G.; Horsfall L.; Karsisiotis A. I.; Damblon C.; Lassaux P.; Papamicael C.; Roberts G. C.; Galleni M.; Dideberg O.; Frere J. M.; Schofield C. J. Structural basis for the broad-spectrum inhibition of metallo-beta-lactamases by thiols. Org. Biomol. Chem. 2008, 6, 2282–94. [DOI] [PubMed] [Google Scholar]
- Hernandez Valladares M.; Felici A.; Weber G.; Adolph H. W.; Zeppezauer M.; Rossolini G. M.; Amicosante G.; Frere J. M.; Galleni M. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-beta-lactamase activity and stability. Biochemistry 1997, 36, 11534–41. [DOI] [PubMed] [Google Scholar]
- Feng L.; Yang K. W.; Zhou L. S.; Xiao J. M.; Yang X.; Zhai L.; Zhang Y. L.; Crowder M. W. N-heterocyclic dicarboxylic acids: broad-spectrum inhibitors of metallo-beta-lactamases with co-antibacterial effect against antibiotic-resistant bacteria. Bioorg. Med. Chem. Lett. 2012, 22, 5185–9. [DOI] [PubMed] [Google Scholar]
- Ullah J. H.; Walsh T. R.; Taylor I. A.; Emery D. C.; Verma C. S.; Gamblin S. J.; Spencer J. The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution. J. Mol. Biol. 1998, 284, 125–36. [DOI] [PubMed] [Google Scholar]
- Fonseca F.; Bromley E. H. E.; Saavedra M. J.; Correia A.; Spencer J. Crystal Structure of Serratia fonticola Sfh-I: Activation of the Nucleophile in Mono-Zinc Metallo-β-Lactamases. J. Mol. Biol. 2011, 411, 951–9. [DOI] [PubMed] [Google Scholar]
- Sharma N.; Hu Z.; Crowder M. W.; Bennett B. Conformational changes in the metallo-beta-lactamase ImiS during the catalytic reaction: an EPR spectrokinetic study of Co(II)-spin label interactions. J. Am. Chem. Soc. 2008, 130, 8215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello A. L.; Sharma N. P.; Yang K. W.; Crowder M. W.; Tierney D. L. X-ray absorption spectroscopy of the zinc-binding sites in the class B2 metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry 2006, 45, 13650–8. [DOI] [PubMed] [Google Scholar]
- Sharma N. P.; Hajdin C.; Chandrasekar S.; Bennett B.; Yang K. W.; Crowder M. W. Mechanistic studies on the mononuclear ZnII-containing metallo-beta-lactamase ImiS from Aeromonas sobria. Biochemistry 2006, 45, 10729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford P. A.; Yang K. W.; Sharma N.; Bennett B.; Crowder M. W. Spectroscopic studies on cobalt(II)-substituted metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry 2005, 44, 5168–76. [DOI] [PubMed] [Google Scholar]
- Minond D.; Saldanha S. A.; Subramaniam P.; Spaargaren M.; Spicer T.; Fotsing J. R.; Weide T.; Fokin V. V.; Sharpless K. B.; Galleni M.; Bebrone C.; Lassaux P.; Hodder P. Inhibitors of VIM-2 by screening pharmacologically active and click-chemistry compound libraries. Bioorg. Med. Chem. 2009, 17, 5027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. L.; Yang K. W.; Zhou Y. J.; LaCuran A. E.; Oelschlaeger P.; Crowder M. W. Diaryl-Substituted Azolylthioacetamides: Inhibitor Discovery of New Delhi Metallo-beta-Lactamase-1 (NDM-1). ChemMedChem 2014, 9, 2445–8. [DOI] [PubMed] [Google Scholar]
- Bush K. The ABCD’s of beta-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–59. [DOI] [PubMed] [Google Scholar]
- Faridoon; Hussein W. M.; Vella P.; Islam N. U.; Ollis D. L.; Schenk G.; McGeary R. P. 3-mercapto-1,2,4-triazoles and N-acylated thiosemicarbazides as metallo-beta-lactamase inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 380–6. [DOI] [PubMed] [Google Scholar]
- Patel R. V.; Patel P. K.; Kumari P.; Rajani D. P.; Chikhalia K. H. Synthesis of benzimidazolyl-1,3,4-oxadiazol-2ylthio-N-phenyl (benzothiazolyl) acetamides as antibacterial, antifungal and antituberculosis agents. Eur. J. Med. Chem. 2012, 53, 41–51. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Benkovic S. J. Purification, characterization, and kinetic studies of a soluble Bacteroides fragilis metallo-beta-lactamase that provides multiple antibiotic resistance. J. Biol. Chem. 1998, 273, 22402–8. [DOI] [PubMed] [Google Scholar]
- Yang H.; Aitha M.; Hetrick A. M.; Richmond T. K.; Tierney D. L.; Crowder M. W. Mechanistic and spectroscopic studies of metallo-beta-lactamase NDM-1. Biochemistry 2012, 51, 3839–47. [DOI] [PubMed] [Google Scholar]
- Crawford P. A.; Sharma N.; Chandrasekar S.; Sigdel T.; Walsh T. R.; Spencer J.; Crowder M. W. Over-expression, purification, and characterization of metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Protein Expression Purif. 2004, 36, 272–9. [DOI] [PubMed] [Google Scholar]
- Crowder M. W.; Walsh T. R.; Banovic L.; Pettit M.; Spencer J. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 1998, 42, 921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlewska C.; Pancechowska K. D. Phosphorus. Sulfur Silicon Relat. Elem. 2006, 4, 737–744. [Google Scholar]
- Rasheed J. K.; Anderson G. J.; Yigit H.; Queenan A. M.; Domenech-Sanchez A.; Swenson J. M.; Biddle J. W.; Ferraro M. J.; Jacoby G. A.; Tenover F. C. Characterization of the extended-spectrum beta-lactamase reference strain, Klebsiella pneumoniae K6 (ATCC 700603), which produces the novel enzyme SHV-18. Antimicrob. Agents Chemother. 2000, 44, 2382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garau G.; Garcia-Saez I.; Bebrone C.; Anne C.; Mercuri P.; Galleni M.; Frere J. M.; Dideberg O. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 2004, 48, 2347–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–8. 27–8.. [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc.: Montreal, Canada, 2014. [Google Scholar]
- Yang K. W.; Feng L.; Yang S. K.; Aitha M.; LaCuran A. E.; Oelschlaeger P.; Crowder M. W. New b-phospholactam as a carbapenem transition state analog: Synthesis of a broad-spectrum inhibitor of metallo-b-lactamases. Bioorg. Med. Chem. Lett. 2013, 23, 5855–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.