Abstract
There is a pressing need for more-efficient trial designs for biomarker-stratified clinical trials. We suggest a new approach to trial design that links novel treatment evaluation with the concurrent evaluation of a biomarker within a confirmatory phase II/III trial setting. We describe a new protocol using this approach in advanced colorectal cancer called FOCUS4. The protocol will ultimately answer three research questions for a number of treatments and biomarkers: (1) After a period of first-line chemotherapy, do targeted novel therapies provide signals of activity in different biomarker-defined populations? (2) If so, do these definitively improve outcomes? (3) Is evidence of activity restricted to the biomarker-defined groups? The protocol randomizes novel agents against placebo concurrently across a number of different biomarker-defined population-enriched cohorts: BRAF mutation; activated AKT pathway: PI3K mutation/absolute PTEN loss tumors; KRAS and NRAS mutations; and wild type at all the mentioned genes. Within each biomarker-defined population, the trial uses a multistaged approach with flexibility to adapt in response to planned interim analyses for lack of activity. FOCUS4 is the first test of a protocol that assigns all patients with metastatic colorectal cancer to one of a number of parallel population-enriched, biomarker-stratified randomized trials. Using this approach allows questions regarding efficacy and safety of multiple novel therapies to be answered in a relatively quick and efficient manner, while also allowing for the assessment of biomarkers to help target treatment.
INTRODUCTION
The concept of one treatment for all patients with a particular disease is increasingly outdated in oncology, and correspondingly, new approaches to trial designs are needed. Trial designs for evaluating biomarkers and treatment response have been well described by others,1-7 and a brief summary is presented here.
Much of the discussion of biomarker-based trial design focuses on evaluating new biomarkers that may help predict response to a treatment. Typically, in these situations, the treatment is already known to have some activity or efficacy, and emphasis is placed on whether the biomarker identifies those who do or do not gain benefit from the treatment. These can be either retrospective analyses of completed trials or prospectively planned studies.
Retrospective analyses of existing trial data remain a suitable approach for identifying potential predictive biomarkers. For example, KRAS was identified retrospectively as a predictor of the effectiveness of the epidermal growth factor receptor (EGFR) inhibitors panitumumab and cetuximab in advanced colorectal cancer.8 However, when considering such retrospective approaches, a prospectively defined analysis plan must be used, sufficient numbers in the biomarker subgroups are required to ensure adequate statistical power, and a high proportion of patients must have been assessed for the biomarker of interest, reducing the risk of potential selection bias.2,7
Alternatively, prospectively planned, so-called population-enriched designs define the eligible population by the presence of the biomarker and test the experimental agent in that population only. For example, HER2 amplification (present in approximately 15% of patients with breast cancer) and a translocation or inversion event involving the anaplastic lymphoma kinase (ALK) gene locus (which occurs in 4% of those with non–small-cell lung cancer) have been used prospectively to enrich the population for evaluation of trastuzumab and crizotinib, respectively. This design requires prior validation of the biomarker and ideally the use of a licensed in vitro diagnostic test for eligibility. However, the design is inefficient, because typically a large population of patients has to consent, be screened, and have their tumor tested to identify a minority biomarker-positive subgroup. This design also presupposes certainty of the link between the biomarker and treatment, which is often only possible at a late stage of development of both the biomarker and the treatment. It is, by design, not able to assess any effect of the agent in the biomarker-negative group.
Prospective designs have also been proposed, which aim to evaluate both a new treatment and a biomarker within one trial and are often referred to as having a biomarker-stratified design or marker-by–treatment interaction design. The designs that have been proposed are inefficient because of the need to size the trial either on the difference between the effect of the treatment in biomarker-positive and -negative patients (ie, interaction) or on the effect in all patients. Determining the size of the trial based on an interaction requires an extremely large number of patients, and the effect in all patients is likely to be modest because of an anticipated small or even negative effect in biomarker-negative patients. Another inefficiency of all these trial designs is that they typically evaluate only a single treatment or biomarker, which is wasteful. The treatment may prove ineffective, and/or the biomarker may not predict the outcome of treatment.
There are newer trial designs, such as those used in the I-SPY1/2 (Investigation of Serial Studies to Predict Your Therapeutic Response With Imaging and Molecular Analysis)9-12 and BATTLE (Biomarker-Integrated Approaches of Targeted Therapy for Lung Cancer Elimination) trials,4,13 which aim to evaluate multiple treatments and multiple biomarkers simultaneously in a purely exploratory way. The focus in these designs is earlier in the drug development process (ie, late phase I/early phase IIa). These trials are potentially efficient in terms of cost and time,14 using fewer patients and shorter-term treatment outcomes, although others have argued this is not necessarily always the case.15 They are able to incorporate relevant prior information and include adaptive randomisation, based on the accruing data, which relies on complex modeling and randomization schemes, which have been well discussed by others.16 Although no assumptions are required at the outset on the relationship between biomarker and treatment response, extra care is required to control the type I error rate in these types of studies,17,18 and they do not easily extend to the phase IIb or III setting. There are large numbers of new treatments emerging that need to be evaluated, first in a phase II setting and then, if appropriate, in a phase III setting. Most of these new treatments have been developed with a single target or few targets in mind and thus have putative biomarkers, often with preliminary clinical evidence, which might identify a group that may benefit most from that treatment. We consider this situation to be an important halfway house between the confirmatory biomarker designs discussed earlier and the more recent exploratory trial designs.
We propose an efficient approach for such a situation, exploiting the concept of a population-enriched design with the aim of evaluating multiple treatments and multiple biomarkers, thereby including most patients with a given type of cancer in the trial, irrespective of their biomarker categorization. Each treatment is evaluated first in the cohort of patients for whom the biomarker is hypothesized to be predictive of response. Then, if appropriate, we will subsequently test the hypothesis of the predictive ability of the biomarker by evaluating the agent in the biomarker-negative patients. Importantly, we do not assume that a treatment works only in biomarker-positive patients, although our design, like others, does assume that a treatment that does not demonstrate efficacy in the selected/enriched cohort need not be further tested in a biomarker-negative patient population. This more efficient sequential testing approach has been recently discussed and compared with other existing trial designs.19 Furthermore, by randomly assigning each comparison against placebo within the biomarker-defined groups, we can overcome the potential bias of the prognostic effects of certain biomarker-defined groups. Finally, the trial needs to be adaptive to be able to discontinue random assignment to treatments that do not seem to be sufficiently active, introduce both new biomarkers and treatments when warranted, and refine biomarkers as information from within or outside the trial emerges.
Our approach follows seven key principles outlined in Table 1. These are presented along with FOCUS4, the first trial using this approach. First-line chemotherapy for colorectal cancer was felt to be an appropriate setting for a trial employing such a design, for reasons explained in the Appendix (online only).
Table 1.
Seven Key Principles of FOCUS4 Trial Design
| Principle |
|---|
|
DESIGN PRINCIPLES
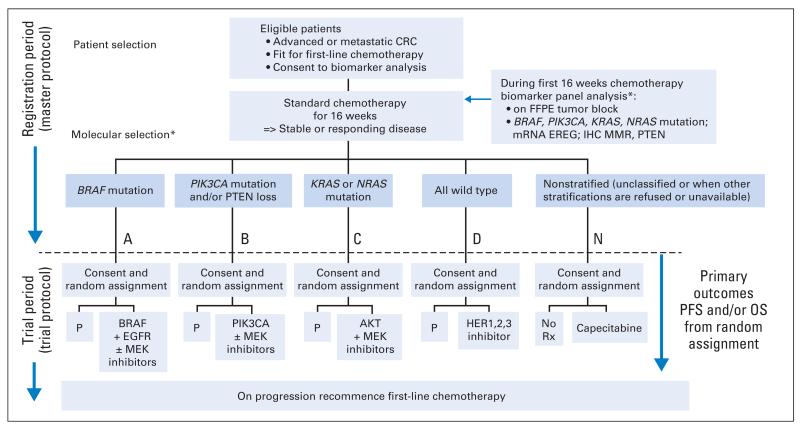
There are seven key principles in the FOCUS4 design. FOCUS4 uses some of the methods of the multiarm, multistage20-22 randomized trial design. After registration and biomarker assessment during a planned 16 weeks of standard first-line chemotherapy, patients are stratified into one of four biologically defined cohorts (A to D; Fig 1). If stable or responding disease is confirmed at the end of 16 weeks, patients are then enrolled onto the corresponding randomized trial of novel targeted agent(s) or, for travel, logistic, or technical reasons, to the one conventional chemotherapy maintenance trial (FOCUS4-N).
Fig 1.
Trial schema for FOCUS4. (*) The molecular cohorts are arranged in a hierarchy from left to right. For example, a patient with both a PIK3CA mutation and a KRAS mutation will be classified into the PIK3CA mutation cohort. CRC, colorectal cancer; EGFR, epidermal growth factor receptor; EREG, epiregulin; FFPE, formalin fixed, paraffin embedded; HER, human epidermal growth factor receptor; IHC, immunohistochemistry; MMR, mismatch repair; OS, overall survival; P, placebo; PFS, progression-free survival; Rx, treatment.
Key Principle One
Evaluate multiple treatments and biomarkers in the same protocol, including as many patients as possible with a given disease, with separate clinical questions for as many marker-defined subgroups as are supported by current evidence.
Incorporating multiple treatments across multiple population-enriched biomarker-defined trials fits easily into conventional clinical practice patterns, in which most patients with one type of cancer (by conventional criteria) are generally referred and/or managed in a common manner and with similar clinical protocols. In colorectal cancer, a single approach for all has now evolved into two clinical pathways and chemotherapy approaches: one for patients with KRAS wild-type tumors, for whom EGFR-targeted monoclonal antibodies may be planned, and one for those with KRAS-mutated tumors. As further (appropriate) segregation of treatment approaches occurs, managing separately coordinated clinical research efforts, which often involve different pharmaceutical collaborators and different research teams, will become progressively more unwieldy and inefficient. Our approach filters all fit and consenting patients into one overarching clinical trial program and is therefore inclusive and consequently more attractive to patients. This design also offers clear efficiency gains in both cost and time compared with running multiple individual trials to evaluate different treatments under separate protocols. It increases the likelihood that the investment in cost and effort of setting up such trials will lead to discovery of (at least one) effective treatment and will allow us to stop further development of ineffective treatments in this disease setting.
Further efficiency is inherent in biomarker analysis being set up to include all diagnostic tests for the differing subgroups. So far as is scientifically feasible, an inclusive trial allows the maximum number of patients to participate and maximizes the potential to recruit rare subtypes. It allows for maximum flexibility in refinement of the biomarker cohort definitions in response to developing clinical data from both within and outside of the trial and provides administrative and organizational efficiencies.
Key Principle Two
In initial stages, assess each treatment in the presumptive biomarker-enriched subset (thus exploiting the putative link between biomarkers and novel treatments with corresponding mechanisms of action) but without assuming in the design that this association will be confirmed in later stages.
In oncology, even when novel agents are found to be active, the expected biomarker selection may not apply.23 A key strength of the FOCUS4 protocol is that we neither assume that any encouraging outcome results are limited to the biomarker selection, nor expend numbers of biomarker-negative patients until we have a positive signal from the initial staged analyses (stages 1 to 2). Thus, we restrict entry in the earlier phases of the evaluation of a novel treatment to those patients thought most likely to respond. Once the significance level associated with activity of the experimental treatment falls to less than a given value, we have the option to open a similar efficacy evaluation among those patients who do not show the positivity of the biomarker in their tumor (ie, off-target effect), using the same type of lack-of-activity assessments, to refute or confirm activity in this complementary population of patients. This approach builds on the putative link between the biomarker and drug efficacy but does not presume that this is certainly the case.
Key Principle Three
Use randomized evidence with a control group for each biomarker/treatment cohort evaluation (eliminating confounding resulting from prognostic biomarker effects).
Constructing the protocol as a set of parallel randomized comparisons assures that the measured or unmeasured prognostic effects of different biomarkers do not confound the assessment of treatment efficacy, meaning that we can ascribe any benefits to the new treatment and not the potential prognostic effect of the marker. The concern about prognostic differences in relation to biomarker expression is not just theoretic; there are recognized clinical patterns associated with predictive biomarkers in a number of cancers, such as EGFR mutations and the EML4-ALK fusion protein in non–small-cell lung cancer,24,25 human epidermal growth factor receptor–positive and triple-negative breast cancers,26-29 and markers of mismatch repair (MMR) in colorectal cancer.30,31
Key Principle Four
Ensure rapid evaluation of each new treatment, which involves (a) incorporating the flexibility of phase II and III components into each trial and (b) targeting a reasonably large treatment effect, with discontinuation of random assignment to treatments that are unpromising or overwhelmingly effective as early and reliably as possible.
For the individual trials within the protocol, a larger effect size can be targeted than might be chosen in a more traditional trial. This can be done for two reasons. First, enrichment of the population means that we may expect a somewhat larger effect, even if enrichment excludes only a proportion of those who do not benefit. Second, there are currently a large number of potential treatments available for evaluation, and thus, it is reasonable to seek a larger treatment effect than if only a limited number of new treatments were available for testing.
A key aspect of the FOCUS4 protocol is the flexibility to have both phase II and III components for each randomized comparison, with the potential to move seamlessly from phase II to III (Table 2). The aim of the phase III component is to determine if, in the interval after standard first-line chemotherapy, the proposed novel agents improve progression-free survival (PFS), and potentially (in some of the larger cohorts) overall survival (OS), compared with placebo within the biomarker-defined populations. Use of PFS and OS may be particularly important when agents with various, quite different mechanisms of action are being tested. This is in preference to other earlier outcomes of treatment response (eg, disease response by RECIST criteria), which may not reliably translate into longer-term outcomes of importance to the patient. For each agent-versus-placebo comparison, FOCUS4 employs a maximum of four stages, with the relevant primary end point given in parentheses (Tables 2 and 3):
Safety and screening for sufficient activity (PFS)—stage 1
Screening for sufficient activity (PFS)—stage 2
Efficacy (PFS)—stage 3
Efficacy (for those cohorts with sufficient patients; OS)—stage 4
Table 2.
Sample Size Estimates for FOCUS4 Trial
| Maximum No. of Events Required |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Molecular Cohort | Random Allocation Ratio | Phase | Outcome | Stage | Target HR | Total | Control Arm | Estimated Cumulative Analysis Time (months) | Maximum No. of Patients Required |
| BRAF mutation (trial A) | 2:1 | II | PFS | 1 | 0.5 | 41 | 16 | 20.4 | 61 |
| PFS | 2 | 0.5 | 76 | 28 | 32.5 | 97 | |||
| III | PFS | 3 | 0.5 | 118 | 42 | 46.5 | 139 | ||
| OS | 4 (potential) | 0.65 | 217 | 79 | 100.4 | 301 | |||
| PIK3CA mutation and/or PTEN loss (trial B) | 2:1 | II | PFS | 1 | 0.65 | 107 | 40 | 17.0 | 170 |
| PFS | 2 | 0.65 | 197 | 71 | 26.5 | 264 | |||
| III | PFS | 3 | 0.65 | 303 | 107 | 37.2 | 373 | ||
| OS | 4 (potential) | 0.7 | 289 | 109 | 54.6 | 546 | |||
| KRAS or NRAS mutation (trial C) | 2:1 | II | PFS | 1 | 0.65 | 109 | 41 | 16.1 | 177 |
| PFS | 2 | 0.65 | 198 | 72 | 22.8 | 273 | |||
| III | PFS | 3 | 0.65 | 302 | 107 | 31.4 | 378 | ||
| OS | 4 (potential) | 0.7 | 287 | 109 | 50.6 | 574 | |||
| All wild type (trial D) | 2:1 | II | PFS | 1 | 0.65 | 109 | 41 | 20.0 | 180 |
| PFS | 2 | 0.65 | 198 | 72 | 30.6 | 275 | |||
| III | PFS | 3 | 0.65 | 301 | 107 | 42.3 | 381 | ||
| OS | 4 (potential) | 0.7 | 289 | 109 | 60.8 | 547 | |||
Abbreviations: HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
Table 3.
Operating Characteristics for Generic Trial Design in FOCUS4-D: All Wild-Type Cohort
| Phase II |
Phase III |
|||
|---|---|---|---|---|
| Characteristic | Stage 1: Safety and LSA | Stage 2: LSA | Stage 3: Efficacy for PFS | Stage 4: Efficacy for OS (potential) |
| Outcome | PFS | PFS | PFS | OS |
| One-sided α | 0.30 | 0.10 | 0.025 | 0.025 |
| Power (overall power maintained at 80%) | 0.95 | 0.95 | 0.95 | 0.85 |
| Target HR | 0.65 | 0.65 | 0.65 | 0.70 |
| Critical HR | 0.91 | 0.83 | 0.79 | 0.80 |
| Time required, months | 20 | 11 | 12 | 19 |
| Cumulative time, months | 20 | 31 | 42 | 61 |
| Cumulative events required | ||||
| Control arm | 41 | 72 | 107 | 109 |
| Total | 109 | 198 | 301 | 289 |
| Total expected cumulative random assignments | 180 | 275 | 381 | 547 |
Abbreviations: HR, hazard ratio; LSA, lack of sufficient activity; OS, overall survival; PFS, progression-free survival.
Stages 1 and 2 together can be considered analogous to a traditional phase II trial, whereas continuing into stages 3 and 4 can be considered functionally to complete a traditional phase III trial. Such designs can be adapted for different end points at different stages and use different decision criteria for moving from one stage to another.32
For all therapies that pass activity screening in stage 2, a number of paths are possible. If no major changes are to be made to the research arm (eg, in biomarker selection criteria or agent dosing), a seamless move to stage 3 is possible, with outcome data on all patients entered during stages 1 and 2 being used in stages 3 and 4.
Alternatively, if a major change to a research arm is to be made, the trial can still continue to the phase III component (stages 3 and 4) but with outcome data from only newly entered patients from that time forward contributing to the final (phase III) analysis. However, there is considerable efficiency even in this situation. This so-called new phase III trial does not require a new protocol and can be initiated by amendment and activated rapidly at all the sites already participating in the single FOCUS4 protocol.
A final possibility is that for reasons external to the trial itself, the sponsor or funder may decide not to support continuation to the phase III stages. This might even happen in the face of a positive outcome in the activity screening stages (eg, if testing of the agent in other settings has already been planned). Note that such an outcome would still be appropriately viewed as useful, if it served to stimulate or facilitate additional trials. In such a situation, other novel agent(s) could then be tested in the relevant cohort of FOCUS4.
Table 2 lists the design characteristics, required number of patients and events, analysis timings, and target effect sizes for each individual trial in the FOCUS4 program, assuming that each trial continues to its phase III component, with no major changes after the phase II component. All sample size calculations were performed using the nstage program in STATA software (version 12.0; STATA, College Station, TX).21,33 More-detailed operational characteristics, including analysis timings for a particular trial, are listed in Table 3. For each of the trials within FOCUS4, the overall power is maintained at 80%, allowing for multiple interim data looks, with a maximum 5% two-sided overall significance level (type I error rate). To maintain 80% power overall (for each trial), the power of each trial for the primary analysis varies from 85% to 95%, depending on the number and timing of the interim looks. Each biomarker-defined trial is considered separately in terms of the effect size (hazard ratio) to be detected, to suit issues relevant to specific biomarker cohorts or agents. An important distinction between stages 2 (lack of sufficient activity) and 3 (efficacy) is the difference in type I error, set higher for the stage 2 interim PFS analysis (one-sided 10%), compared with the stage 3 main PFS efficacy analysis (one-sided 2.5%).
Data for each of the trials will be reviewed by the independent data monitoring committee at each interim analysis. The committee can also advise early closure of a trial in the event of overwhelming evidence of efficacy, using a significance level of .001 as a guideline on the phase III efficacy outcome measure. This will be considered at approximately the halfway point in terms of accrued number of events for each biomarker-defined trial. Adopting this approach allows for preservation of the overall type I error rate at the end of the trial.34,35
Key Principle Five
Allow the possibility of refining any biomarkers through the course of the trial, either from internal data or more typically from data emerging from outside the trial.
Biomarker definitions are constantly evolving and being refined. Although, for example, KRAS mutation was a multiply confirmed predictive marker for use of anti-EGFR antibody therapy, it subsequently became clear that not all KRAS mutations had identical effects, with some perhaps (eg, KRAS G13D) not even carrying the same negative predictive value.36 Evolving data now suggest that expression of EGFR ligands such as epiregulin and amphiregulin may additionally modulate response to this class of agents.37 Other alterations elsewhere in the RASRAF-MEK-ERK and interacting pathways (eg, phosphoinositide-3 kinase [PI3K] –AKT–mammalian target of rapamycin [mTOR]) are almost certainly also of considerable importance.
An example of biomarker refinement would be the introduction of a new platform for mutation assessment. Developments in molecular diagnostics are occurring at such a pace that older technologies are continuously being outperformed in terms of cost and sample requirements by newer technologies. Such refinements could be introduced into a continuing program such as FOCUS4, but with close attention to quality assurance and a preplanned parallel evaluation using both platforms for a period of time to ensure comparability of assessment. If there proves a need to revise one or another biomarker assay during the first two (signal-seeking) analysis stages, patients assigned on the basis of the earlier assay would likely have to be excluded from the definitive third- and fourth-stage analyses and their possible use toward registration. The third-stage sample size might need to be increased, but the overall time delay introduced in the stage 3 analysis would be minimal (months), because the trial would already be activated and recruiting from a large number of sites.
Key Principle Six
Allow the possibility of introducing a new biomarker and treatment pairing into the overall trial program when evidence warrants.
The flexibility inherent in the design means there are opportunities to further adapt the trial to accommodate additional findings, typically from other research outside of the trial. For instance, it is well known that there is a cohort of patients with MMR deficiency within colorectal cancer. In earlier disease, these patients amount to 15% of the total, but they have an improved prognosis, and series among metastatic patients reveal only approximately 4% prevalence of MMR deficiency. Currently, there is no convincing case for testing a specific class of agent in this cohort, but if such evidence were to emerge within the course of the study, we would be able to amend the protocol to open up a new biomarker/treatment cohort by identifying the MMR-deficient cohort and testing the appropriate novel agent(s) versus placebo in this group. The introduction of such a new biomarker-defined cohort during the trial would remove such patients from the other cohorts, but as we have discussed regarding changes in assay methodology, the introduction of a new cohort would not in any way compromise the study design, requiring only sample size adjustments.
Key Principle Seven
Investigate new treatments in the earliest and most likely responsive settings that are clinically feasible.
With many agents being developed and recognition that drug development is a lengthy and expensive process, it is critical that we seek strong positive signals as early as possible in testing. When new agents are tested against all comers and late in the natural history of the disease, it is usually difficult to determine whether observed modest improvements are likely to hold up with further testing, especially in earlier stages of disease.
In the FOCUS4 protocol, we propose a quadruple selection process of patients for each trial, which improves the chance of identifying clinical benefit from novel agent(s). First, we exclude patients with aggressive disease as manifest in a raised baseline platelet count.38 Second, we include only those patients with stable or responding disease during 16 weeks of first-line systemic therapy. This therefore specifically selects responding patients, in comparison with most study designs, which select resistant patients to evaluate novel agents. Third, by testing the novel agents first in the molecular cohort in which theoretically they should have the greatest benefit, we maximize the likelihood of success. Finally, new agents are used on their own or in novel-novel combinations, after standard treatment, thereby avoiding unpredictable negative pharmacologic or toxicity interactions with conventional chemotherapy, which has been seen repeatedly in colorectal cancer chemotherapy.39-42 Consequently, randomization occurs in a treatment window of opportunity or treatment break, which is a clinically reasonable and safe strategy on the basis of randomized data from the COIN (Continuous or Intermittent) trial.38 Although this strategy is somewhat unusual in colorectal cancer, there are many settings in the management of other tumors in which periods of observation of patients off treatment are standard and could be used for such window-of-opportunity trials. In FOCUS4, the setting has the advantage of allowing relatively new agents to be tested in patients before the onset of chemotherapy resistance and yet well before comprehensive data would become available with regard to combined administration along with chemotherapy.
DISCUSSION
The FOCUS4 protocol introduces a new paradigm in clinical research of solid tumors, evaluating a number of treatments and biomarkers rapidly and in an adaptive way. The umbrella protocol presents a structure in which there is characterization of patients on the basis of the presence of specific mutations or expression markers, which define a number of biologic cohorts. After an initial period of disease control with standard chemotherapy, eligible and consenting patients within each of these biologically defined cohorts are given a chemotherapy break and are randomly assigned between a control group and one or more new targeted treatments for which there is a priori evidence suggesting this biologic cohort is more likely to benefit from the new treatment than others.
As a component of a large national study that can potentially include practically all patients, each individual randomized comparison can adapt efficiently to refinement of biomarker and signature data. The structure will enable rapid accrual, even in rare subtypes such as those with BRAF mutation, who comprise only 8% of the metastatic colorectal cancer population. This trial will further build on lessons learned from the Medical Research Council–funded FOCUS3 feasibility trial, which tested this general approach in a cohort of 240 patients.43
FOCUS4 has been provided here as a current real example of our proposed design. However, we believe the key principles underpinning this design are transferable to other cancer settings and indeed to other diseases.
Supplementary Material
Table A1. Biomarker-Defined Cohorts and Novel Agent Interventions for FOCUS4
Acknowledgments
FOCUS4 is jointly funded by Cancer Research UK and Medical Research Council/National Institute of Health Research Efficacy and Mechanism Evaluation Programme. Work on the design was supported by core funding from each academic institution.
Footnotes
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST Although all authors completed the disclosure declaration, the following author(s) and/or an author’s immediate family member(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO’s conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors. Employment or Leadership Position: None Consultant or Advisory Role: Richard Wilson, Roche (C), Hospira (C), sanofi-aventis (C), Merck Serono (C) Stock Ownership: None Honoraria: Timothy Maughan, Merck Serono; Richard Wilson, Roche, Hospira, sanofi-aventis, Merck Serono Research Funding: None Expert Testimony: None Patents: None Other Remuneration: None
Contributor Information
Richard Kaplan, Medical Research Council Clinical Trials Unit, London.
Timothy Maughan, University of Oxford, Oxford.
Angela Crook, Medical Research Council Clinical Trials Unit, London.
David Fisher, Medical Research Council Clinical Trials Unit, London.
Richard Wilson, Queen’s University Belfast, Belfast, United Kingdom..
Louise Brown, Medical Research Council Clinical Trials Unit, London.
Mahesh Parmar, Medical Research Council Clinical Trials Unit, London.
REFERENCES
- 1.Sargent DJ, Conley BA, Allegra C, et al. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23:2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 2.Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: Theoretical considerations and practical challenges. J Clin Oncol. 2009;27:4027–4034. doi: 10.1200/JCO.2009.22.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: Design issues. J Natl Cancer Inst. 2010;102:152–160. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JJ, Xuemin G, Suyu L. Bayesian adaptive randomization designs for targeted agent development. Clin Trials. 2010;7:584–596. doi: 10.1177/1740774510373120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandrekar SJ, Sargent DJ. Predictive bio-marker validation in practice: Lessons from real trials. Clin Trials. 2010;7:567–573. doi: 10.1177/1740774510368574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai TL, Lavori PW, Shih MC, et al. Clinical trial designs for testing biomarker-based personalized therapies. Clin Trials. 2012;9:141–154. doi: 10.1177/1740774512437252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 9.Barker AD, Sigman CC, Kelloff GJ, et al. I-SPY 2: An adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 10.Esserman LJ, Berry DA, Cheang MC, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: Results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res Treat. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Ning J, Li Y, et al. Using short-term response information to facilitate adaptive randomization for survival clinical trials. Stat Med. 2009;28:1680–1689. doi: 10.1002/sim.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C, Buxton MB, Moore D, et al. Locally advanced breast cancers are more likely to present as Interval Cancers: Results from the I-SPY 1 TRIAL (CALGB 150007/150012, ACRIN 6657, InterSPORE Trial) Breast Cancer Res Treat. 2012;132:871–879. doi: 10.1007/s10549-011-1670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Liu S, Kim ES, et al. Bayesian adaptive design for targeted therapy development in lung cancer: A step toward personalized medicine. Clin Trials. 2008;5:181–193. doi: 10.1177/1740774508091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledford H. Clinical drug tests adapted for speed. Nature. 2010;464:1258. doi: 10.1038/4641258a. [DOI] [PubMed] [Google Scholar]
- 15.Freidlin B, Korn EL. Biomarker-adaptive clinical trial designs. Pharmacogenomics. 2010;11:1679–1682. doi: 10.2217/pgs.10.153. [DOI] [PubMed] [Google Scholar]
- 16.Korn EL, Freidlin B. Outcome: Adaptive randomization—Is it useful? J Clin Oncol. 2011;29:771–776. doi: 10.1200/JCO.2010.31.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Time to adapt. Nature. 2010;464:1245–1246. doi: 10.1038/4641245b. [No authors listed] [DOI] [PubMed] [Google Scholar]
- 18.McShane LM, Hunsberger S, Adjei AA. Effective incorporation of biomarkers into phase II trials. Clin Cancer Res. 2009;15:1898–1905. doi: 10.1158/1078-0432.CCR-08-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freidlin B, Sun Z, Gray R, et al. Phase III clinical trials that integrate treatment and biomarker evaluation. J Clin Oncol. 2013;31:3158–3161. doi: 10.1200/JCO.2012.48.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parmar MK, Barthel FM, Sydes M, et al. Speeding up the evaluation of new agents in cancer. J Natl Cancer Inst. 2008;100:1204–1214. doi: 10.1093/jnci/djn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Royston P, Barthel FM, Parmar MK, et al. Designs for clinical trials with time-to-event outcomes based on stopping guidelines for lack of benefit. Trials. 2011;12:81. doi: 10.1186/1745-6215-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royston P, Parmar MK, Qian W. Novel designs for multi-arm clinical trials with survival outcomes with an application in ovarian cancer. Stat Med. 2003;22:2239–2256. doi: 10.1002/sim.1430. [DOI] [PubMed] [Google Scholar]
- 23.Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 24.Kwak EL, Bang YJ, Camidge DR, et al. Ana-plastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu SG, Kuo YW, Chang YL, et al. EML4-ALK translocation predicts better outcome in lung adenocarcinoma patients with wild-type EGFR. J Thorac Oncol. 2012;7:98–104. doi: 10.1097/JTO.0b013e3182370e30. [DOI] [PubMed] [Google Scholar]
- 26.Bayraktar S, Glück S. Molecularly targeted therapies for metastatic triple-negative breast cancer. Breast Cancer Res Treat. 2013;138:21–35. doi: 10.1007/s10549-013-2421-5. [DOI] [PubMed] [Google Scholar]
- 27.Boyle P. Triple-negative breast cancer: Epidemiological considerations and recommendations. Ann Oncol. 2012;23(suppl 6):vi7–vi12. doi: 10.1093/annonc/mds187. [DOI] [PubMed] [Google Scholar]
- 28.Cortesi L, De Matteis E, Cirilli C, et al. Outcome evaluation in pre-trastuzumab era between different breast cancer phenotypes: A population-based study on Italian women. Tumori. 2012;98:743–750. doi: 10.1177/030089161209800611. [DOI] [PubMed] [Google Scholar]
- 29.Dreyer G, Vandorpe T, Smeets A, et al. Triple negative breast cancer: Clinical characteristics in the different histological subtypes. Breast. 2013;22:761–766. doi: 10.1016/j.breast.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Brixen LM, Bernstein IT, Bülow S, et al. The survival of patients with stage III colon cancer is improved in HNPCC compared with sporadic cases: A Danish registry based study. Colorectal Dis. 2013;15:816–823. doi: 10.1111/codi.12150. [DOI] [PubMed] [Google Scholar]
- 31.Laghi L, Malesci A. Microsatellite instability and therapeutic consequences in colorectal cancer. Dig Dis. 2012;30:304–309. doi: 10.1159/000337003. [DOI] [PubMed] [Google Scholar]
- 32.Korn EL, Freidlin B, Abrams JS, et al. Design issues in randomized phase II/III trials. J Clin Oncol. 2012;30:667–671. doi: 10.1200/JCO.2011.38.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barthel FM, Royston P, Parmar MK. A menu-driven facility for sample size calculation in novel multiarm, multistage randomized controlled trials with a time-to-event outcome. Stata J. 2009;9:505–523. [Google Scholar]
- 34.Haybittle JL. Repeated assessment of results in clinical trials of cancer treatment. Br J Radiol. 1971;44:793–797. doi: 10.1259/0007-1285-44-526-793. [DOI] [PubMed] [Google Scholar]
- 35.Peto R, Pike MC, Armitage P, et al. Design and analysis of randomized clinical trials requiring prolonged observation of each patient: I. Introduction and design. Br J Cancer. 1976;34:585–612. doi: 10.1038/bjc.1976.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 37.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 38.Maughan TS, Adams RA, Smith CG, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: Results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hecht JR, Mitchell E, Chidiac T, et al. A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J Clin Oncol. 2009;27:672–680. doi: 10.1200/JCO.2008.19.8135. [DOI] [PubMed] [Google Scholar]
- 40.Tol J, Koopman M, Cats A, et al. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360:563–572. doi: 10.1056/NEJMoa0808268. [DOI] [PubMed] [Google Scholar]
- 41.Adams RA, Meade AM, Madi A, et al. Toxicity associated with combination oxaliplatin plus fluoropyrimidine with or without cetuximab in the MRC COIN trial experience. Br J Cancer. 2009;100:251–258. doi: 10.1038/sj.bjc.6604877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madi A, Fisher D, Wilson RH, et al. Oxaliplatin/capecitabine vs oxaliplatin/infusional 5-FU in advanced colorectal cancer: The MRC COIN trial. Br J Cancer. 2012;107:1037–1043. doi: 10.1038/bjc.2012.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maughan TS, Wilson RH, Williams GT, et al. Developing a biomarker-stratified trial design in advanced colorectal cancer: The MRC FOCUS 3 feasibility study. J Clin Oncol. 2011;29(suppl) abstr TPS165. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A1. Biomarker-Defined Cohorts and Novel Agent Interventions for FOCUS4