Summary
During embryogenesis and tissue maintenance and repair in an adult organism, a myriad of stem cells are regulated by their surrounding extracellular matrix (ECM) enriched with tissue/organ-specific nanoscale topographical cues to adopt different fates and functions. Attributed to their capability of self-renewal and differentiation into most types of somatic cells, stem cells also hold tremendous promise for regenerative medicine and drug screening. However, a major challenge remains as to achieve fate control of stem cells in vitro with high specificity and yield. Recent exciting advances in nanotechnology and materials science have enabled versatile, robust, and large-scale stem cell engineering in vitro through developments of synthetic nanotopographical surfaces mimicking topological features of stem cell niches. In addition to generating new insights for stem cell biology and embryonic development, this effort opens up unlimited opportunities for innovations in stem cell-based applications. This review is therefore to provide a summary of recent progress along this research direction, with perspectives focusing on emerging methods for generating nanotopographical surfaces and their applications in stem cell research. Furthermore, we provide a review of classical as well as emerging cellular mechano-sensing and -transduction mechanisms underlying stem cell nanotopography sensitivity and also give some hypotheses in regard to how a multitude of signaling events in cellular mechanotransduction may converge and be integrated into core pathways controlling stem cell fate in response to extracellular nanotopography.
Keywords: Biomaterials, Nanotopography, Stem cell, Tissue engineering and regenerative medicine, Mechanobiology
Introduction
Rapid advances in stem cell studies have unveiled the great potential of stem cells as promising solutions for regenerative medicine, disease modeling, developmental biology studies, and drug screening [1–6]. Stem cells, including adult stem cells [7], embryonic stem cells (ESCs) [8], and induced pluripotent stem cells (iPSCs) [9], share the ability of self-renewal and differentiation into specific cell lineages, providing invaluable cell sources for various biomedical and biological applications [6]. The self-renewal capability of stem cells enables rapid cell expansion without losing cell stemness, critical for generating enough cell quantities for large-scale cell-based applications. Directing stem cell differentiation into defined lineages with tissue-specific mature functions is also important for treating tissue-specific degenerative diseases (such as neurological, hepatic, hematopoietic, and diabetic diseases [1, 4, 6]) and in vitro disease modeling and drug screening. Among different stem cells, adult stem cells possess limited, tissue-specific regenerative potential and thus can only differentiate into a few lineages [7]. In contrast, pluripotent stem cells (PSCs), including both ESCs and iPSCs, possess the potential of differentiating into all three germ layers, i.e. endoderm, mesoderm, and ectoderm, and subsequently into any type of somatic cells [1, 10]. Although together, both adult and pluripotent stem cells can provide virtually unlimited cell sources for in vitro and in vivo cell-based applications, a major technical hurdle remains as to achieve large-scale, high-efficiency cell expansion as well as directed differentiation into cell lineages of mature functions with high specificity and yield.
In the physiological stem cell niche, stem cells are constantly challenged by both soluble cues and insoluble, physical stimuli dynamically regulated in the local extracellular matrix (ECM) [11, 12]. The stem cell-ECM interface is composed of structural units of nanometer length scales, which in turn regulate stem cell fate along with other physical factors [13–16]. Specifically, in vivo ECM is enriched with hierarchical fibers and fibrils consisting of filamentous proteins such as collagen, elastin, fibronectin, vitronectin, and laminin, presenting adhesive ligands on a structured landscape with spatial organizations and characteristic dimensions of a few to hundreds of nanometers [17]. The helical surface topographical periodicity of individual ECM fibrils (e.g., collagen I) is also a physical cue that can dictate stem cell behaviors [18]. In direct contact with the ECM, the cell membrane is enriched with adhesive molecules and protrusive structures with characteristic nanometer length scales. For instance, integrin, the transmembrane receptor directly linking ECM ligands to intracellular adaptor proteins and the actin cytoskeleton (CSK), has a dimension of 20 – 50 nm [13, 19]. Nanoscale filopodia (“nanopodia”), cell membrane protrusions containing bundled actin filaments, also have been shown in cellular probing of extracellular nanotopographical features [20]. Cellular sensing of extracellular nanotopographical cues through nanoscale architecture and dynamics of cell-ECM adhesions initiates downstream intracellular mechanotransductive events, resulting in a multitude of nanotopography-sensitive cellular behaviors, including cell adhesion, morphology, proliferation, gene expression, self-renewal, and differentiation [16, 21–27]. Owing to its potent role in regulating stem cell fate, extracellular nanotopography recently attracts much attention from bioengineers and materials scientists in an effort to achieve stem cell fate control using synthetic nanotopographical surfaces generated from different novel nanofabrication technologies and material synthesis methods [17, 25].
A few key molecular players have emerged in accompany with several principal mechanotransductive pathways for regulating stem cell nanotopography sensitivity [19, 28]. Specifically, existing evidence has suggested the involvement of integrin-mediated adhesion signaling [29], CSK contractility (tension) and integrity [30, 31], and nuclear mechanics [32, 33] in mechanosensing and mechanotransduction of extracellular nanotopographical cues [19]. However, it remains unclear how these different mechanotransductive cellular machineries function or collaborate differently in different stem cell systems. Moreover, it remains elusive how mechanoregulation at multiple levels (genetic and epigenetic, transcriptional, and post-transcriptional including microRNA) and time scales are integrated into a core regulatory network to control stem cell nanotopography sensitivity. Future exploration of nanotopography-sensitive pathways will help improve rational designs of functional biomaterials for enhancing their performance in stem cell-based applications.
A major goal of this review is to offer a summary of recent progress on the new trend of engineering synthetic nanotopographical surfaces for controlling stem cell fate. We first provide a review of state of the art nanofabrication methods for generating functional nanotopographical surfaces for stem cell studies, with a focus on those applicable for large-scale stem cell culture. We then highlight applications of nanotopographical surfaces in recent investigations for the control of stem cell fate. We discuss a few important intracellular mechanotransductive mechanisms that have been implicated in cellular responses to extracellular nanotopography. We also provide some speculations as regard to how these mechanotransductive events may converge with classical signal transduction pathways to control stem cell fate. We conclude by offering some perspectives on future research directions and opportunities for leveraging stem cell nanotopography sensitivity for engineering stem cell fate and function.
Fabrication of nanotopographical surfaces
Various nanoengineering tools and synthesis methods have been successfully developed and utilized to generate nanotopographical surfaces or scaffolds for in vitro stem cell research. Based on their fabrication principles, these techniques can be classified into four different groups: lithographic patterning, pattern transfer, surface roughening, and material synthesis (Fig. 1, Table 1–2). Lithographic patterning and pattern transfer are two top-down approaches that utilize predefined patterns to create nanotopographical features on two-dimensional planar surfaces. Surface roughening and material synthesis, on the contrary, directly generate nanostructures on material surfaces from the bottom up using chemical or physical means. Together, these methods present a wide spectrum of fabrication tools capable of generating nanotopographical features of a wide range of sizes and geometries, and even hierarchical (micro-)nanotopographical surfaces. To successfully utilize stem cell-nanotopography interactions for stem cell applications, it is important to understand and appreciate advantages and limitations of each of available nanoengineering tools and synthesis methods for generating extracellular nanotopography in terms of fabrication cost, throughput, materials, controllability of feature shape, size and accuracy (Table 1).
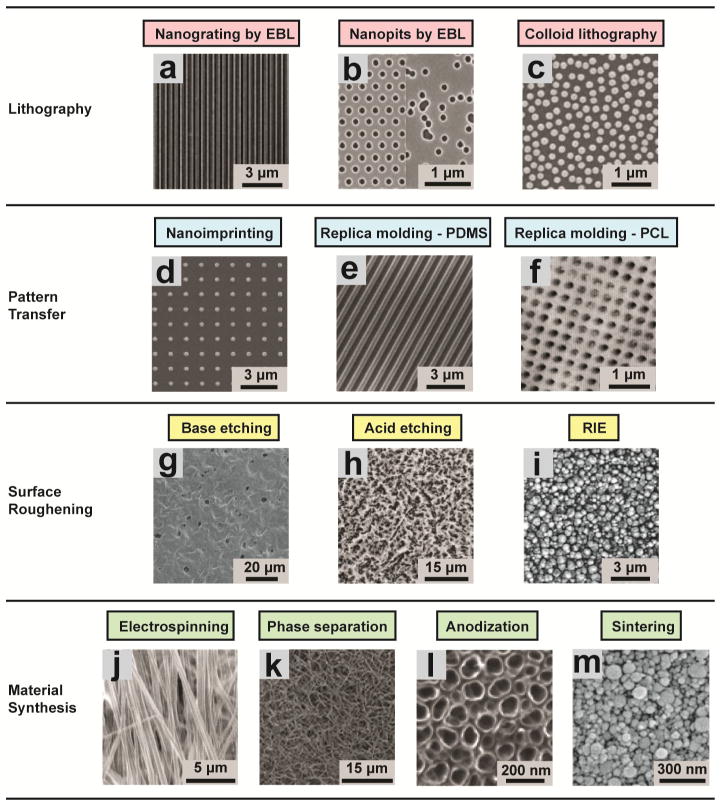
Figure 1. Fabrication of nanotopographic surfaces.
Lithographic patterning. (a) A nanogrooved silicon substrates with 70 nm wide ridge and 400 nm pitch fabricated by EBL [36]. Reproduced with permission [36]. Copyright 2003, Biologists Ltd. (b) Regular (left) and random (right) arrays of 120-nm-diameter, 100-nm-deep nanopits on silicon substrates fabricated by EBL [37]. Reproduced with permission [37]. Copyright 2007, Nature Publishing Group. (c) A nanotopographic substrate fabricated by the self-assembly of 110-nm-diameter nanoparticles [54]. Reproduced with permission [54]. Copyright 2003, IEEE. Pattern transfer. (d) Nanostructured polyurethane acrylate (PUA) surface with a patterned array of nanopillars fabricated by nanoimprinting from a silicon master. The diameter of the pillars was 300 nm and the gap between the pillars was 900 nm [68]. Reproduced with permission [68]. Copyright 2013, American Chemical Society. (e) PDMS nanograting produced by replica molding from a PMMA master [69]. Reproduced with permission [69]. Copyright 2005, Elsevier. (f) AFM scan of a PCL surface with nanopits produced by replica molding from a pillared quartz master [38]. Reproduced with permission [38]. Copyright 2002, IEEE. Surface roughening. (g) Nanostructured PCL with feature dimensions of 50–100 nm fabricated by NaOH etching [77]. Reproduced with permission [77]. Copyright 2003, Wiley Periodicals, Inc. (h) Nanoroughened Ti surface (surface roughness Ra = 0.87 ± 0.03 μm) fabricated by acid etching in hydrochloric acid/sulfuric acid [78]. Reproduced with permission [78]. Copyright 1998, John Wiley & Sons, Inc. (i) Glass surfaces with surface roughness of 100 nm fabricated by RIE [80]. Reproduced with permission [80]. Copyright 2013, American Chemical Society. Material synthesis. (j) Aligned nanofibrous hydroxybutyl chitosan (HBC) scaffolds fabricated by electrospinning [104]. Reproduced with permission [104]. Copyright 2007, WILEY-VCH Verlag GmbH & Co. (k) Nanofibrous PLLA matrix with an average fiber diameter of 148 ± 21 nm and a porosity of 92.9% fabricated by phase separation [114]. Reproduced with permission [114]. Copyright 2009, Mary Ann Liebert, Inc. (l) Self-aligned TiO2 nanotubes with a diameter of 100 nm generated by anodizing Ti sheets under a potential of 20 V [139]. Reproduced with permission [139]. Copyright 2009, National Academy of Sciences of the USA. (m) Nanostructured alumina substrates with 24-nm grain-like structures fabricated by sintering [146]. Reproduced with permission [146]. Copyright 2008, Wiley Periodicals, Inc.
Table 1.
Comparison of methods for generating nanotopography.
| Fabrication Technique | Cost | Throughput | Controllability of feature shape | Controllability of feature size | Material compatibility |
|---|---|---|---|---|---|
| Photolithography | ** | *** | *** | * | *** |
| E-beam lithography | *** | * | *** | *** | * |
| Colloid lithography | ** | *** | * | * | * |
| Nanoimprinting | *** | *** | *** | *** | ** |
| Replica molding | * | *** | *** | *** | ** |
| Chemical etching | * | ** | * | ** | * |
| RIE | ** | *** | * | *** | * |
| Electrospinning | * | ** | ** | * | *** |
| Phase separation | * | ** | ** | ** | ** |
| Anodization | * | ** | * | ** | * |
| Sintering | * | ** | * | ** | ** |
High,
Medium,
Low
Table 2.
Summary of various nanotopographic methods for stem cell studies.
| Fabrication Technique | Topography | Feature Size | Material | Cell Type | Major Result | Reference |
|---|---|---|---|---|---|---|
| Lithography & replica-molding | Grating | 600 nm | PDMS | Endothelial progenitor cells | Supports in vitro Capillary Tube Formation | [34] |
| Lithography & replica-molding | Grating | 600 nm | PDMS | Human ESC | Alters ESC morphology and proliferation | [49] |
| E-beam lithography | Nanopit Array | 120 nm | Silicon | Human MSC | Control of human MSC differentiation using nanoscale symmetry and disorder | [37, 277] |
| Colloid lithography | Island | 120–600 nm | Silica | Mouse ESC | Maintains mouse ESC self-renewal | [57] |
| RIE | Random roughness | 1–150 nm | Glass | Human ESC | Regulates ESC adhesion, proliferation, self-renewal and differentiation | [80] |
| Electrospinning | Random nanofibers | 200–400 nm | PCL | Human & rat MSC | Promoted osteogenesis of MSCs | [95, 96] |
| Electrospinning | Random nanofibers | 500–700 nm | PCL | Human MSC | Support in vitro chondrogenesis of MSCs | [97] |
| Electrospinning | Random & aligned nanofibers | 250–930 nm | PCL | Rat NSC | Aligned nanofibers promote neuronal differentiation | [98] |
| Electrospinning | Random nanofibers | 280 nm | PCL | Human ESC | Support ESC colony formation and maintain stemness. | [99] |
| Electrospinning | Random & aligned nanofibers | 250 nm | PCL | Mouse ESC | Aligned nanofibers promote neuronal differentiation and neurite outgrowth | [100] |
| Electrospinning | Random nanofiber | 283–1452 nm | PES | Rat NSC | Fiber diameter regulate differentiation and proliferation of rat NSCs | [101] |
| Electrospinning | Nanofiber | 529 ± 114 nm | PES | Human HSC | Nanofibrous surface supports HSC expansion | [102] |
| Electrospinning | Nanofiber | 180 nm | Polyamide | Mouse ESC | Promote the proliferation and self-renewal of mouse ESCs | [94, 157] |
| Electrospinning | Random and aligned nanofibers | 150–1150 nm | PLLA | Mouse NSC | Aligned nanofibers enhance NSC differentiation | [87, 88] |
| Electrospinning | Random and aligned nanofibers | 500 nm | CNT/PLLA | Mouse ESC | Aligned nanofibers enhance NSC differentiation | [89] |
| Electrospinning | Random and aligned nanofibers | 196–253 nm | P(LLA-CL) | Mouse NSC | Promote NSC proliferation and differentiation | [90] |
| Electrospinning | Random nanofibers | 230–620 nm | P(LLA-CL) | Mouse MSC | Promote neuronal differentiation | [91] |
| Electrospinning | Random nanofibers | 250–300 nm | PLGA | Mouse ESC | Affects ESC fate | [92] |
| Electrospinning | Random nanofibers | 50–300 nm/2–4 μm | Fibrin/PLGA | Human MSC | Promotes cardiomyogenesis | [148] |
| Electrospinning | Random nanofiber | Not available | PMGI | Mouse ESC | Maintain mouse ESC culture under feeder-free conditions | [103] |
| Electrospinning | Aligned nanofiber | 436 nm | HBC | Human MSC | Induces cell alignment and myogenesis | [104] |
| Electrospinning | Random and aligned nanofibers | 369–554 nm | PHBV/HA | Rat MSC | Random nanofibers promote in vitro proliferation and early osteogenesis | [105] |
| Electrospinning | Random nanofibers | Not available | HA/CTS | Mouse MSC | Enhances osteogenic differentiation | [106] |
| Phase separation | Random nanofiber | 50–500 nm | PLLA | Mouse ESC | Promotes osteogenic differentiation | [114] |
| Phase separation | worm-like and dot-like patterns | 160 nm | polystyrene-blockpoly(2-vinylpyridine) | Human MSC | Influences MSC morphology, proliferation | [127] |
| Anodization | Nanotube | 30–100 nm | TiO2 | human MSC | Regulates human MSC differentiation toward osteoblast lineage | [139] |
| Anodization | Nanotube | 15–100 nm | TiO2 | Rat MSC | Influences adhesion, morphology proliferation and osteogenesis | [14] |
| Anodization | Nanopillars | 15–100 nm | Titania | Human MSC | Feature height affects human MSC adhesion, morphology and differentiation | [140] |
| Sintering | Grain-like structure | 24–1500 nm | Alumina, titania, and hydroxyapatite | Human MSC | Affects human MSC adhesion and proliferation | [146] |
| Graft | Random nanofibers | Not available | poly(acrylic acid) /CNT | human ESC | Direct and promote human ESC derived EBs differentiation into neuron cells | [166] |
| Nano-imprinting | Grooves and pillars | 300–1500 nm | PUA | Human NSC | Nanopatterns influence morphology, alignment, focal adhesion, and neuronal differentiation | [68] |
| Nano-imprinting | Ridge/groove | 350 nm | PUA | Human ESC | Induces neuronal differentiation | [168] |
| Nano-imprinting and soft lithography | Grating | 350–1000 nm | PDMS | Human MSC | Induce differentiation of human MSCs into neuronal lineage | [75] |
| Nano-imprinting | Nanopit | 50 nm | polycarbonate | Human ESC | Influences human ESC differentiation | [162] |
Lithographic patterning
A variety of lithographic patterning methods, including photolithography [34], electron beam lithography [35–38], and colloidal lithography [39–44], have been successfully applied to generate extracellular nanotopography of different size ranges and spatial organizations on planar 2D surfaces following pre-defined patterns (Table 2).
Photolithography
Developed from semiconductor microfabrication, photolithography, or optical lithography, is the most popular technique for surface patterning at micron and sub-micron scales. In photolithography, defined geometric patterns are transferred from a photomask to a light-sensitive organic material (photoresist) coated on a planar substrate via ultraviolet (UV) light exposure. After photolithographic patterning of positive / negative photoresist, exposed / protected regions of the photoresist can be removed with the protected / exposed regions remaining on the substrate, serving as a lithographic mask faithfully inheriting pre-defined patterns from the photomask and transferring it to the substrate with subsequent etching processes [45, 46]. The finest resolvable dimension (resolution) of photolithography is limited by UV light wavelength as well as the ability of reduction lens to capture enough diffraction orders from illuminated photomask [45]. State of the art photolithography using deep UV light from excimer lasers with wavelengths of 248 and 193 nm allows fabrications of nanoscale structures with a minimum feature size down to 50 nm [45–48]. Due to the expense and limited accessibility of photolithography instruments for sub-100 nm fabrication, application of photolithography for fabrication of nanotopographical features has been limited to a length scale of hundreds of nanometers (Table 1). It should be noted, however, that the capability of photolithography in fabricating large-area, arbitrarily designed sub-micron topographical features has rendered it the most popular surface patterning technique for biomedical applications. As an example, nanoscale gratings have been fabricated using photolithography on silicon substrates for studying nanotopographic sensing by human ESCs and endothelial progenitor cells [34, 49].
Electron beam lithography
To overcome resolution of photolithography limited by UV wavelength, other sources of illumination have been exploited for lithography, such as electron beams and x-rays. As a powerful nanofabrication method, electron beam lithography (EBL) [50] has been developed for creating well-defined patterns with feature sizes < 10 nm, owing to a reduced wavelength of electron waves. Instead of using photomasks and UV light, in EBL a focused electron beam is utilized to selectively expose an electron-sensitive organic resist (such as poly(methyl methacrylate), or PMMA) following a pre-defined scanning path. Fabrication of periodic line patterns with a line width of 5 – 7 nm has been successfully demonstrated using EBL [35]. Owing to its versatility in generating nanoscale features with dimensions comparable to cell adhesion structures, EBL has been commonly utilized for generating nanotopographical surfaces to regulate cell-ECM adhesive interactions on both silicon and fused silica surfaces (Fig. 1a, b).
Although EBL can achieve precise surface patterning with a sub-100 nm resolution, given its “direct writing” nature, its throughput is extremely low, limiting its application to low-volume productions of nanotopographical surfaces with a limited surface area. The high cost of EBL machine has also limited its access for pilot stem cell studies where a large number of nanotopographical surfaces with different geometrical patterns are desired for high-content screening of nanotopography that is optimal for specific stem cell fate regulation.
Colloidal lithography
Both photolithography and EBL are top-down lithography-based approaches. As a totally different bottom-up approach, colloid lithography has been developed to generate surface topological features down to a nanometer scale with high-throughput, relatively simple fabrication strategies and reasonable cost [38, 53–56]. In colloidal lithography, self-assembled nanoparticle crystal structures with a short-range intrinsic order are created on planar surfaces to serve as masks for subsequent etching processes (Fig. 1c) [39–42]. Colloidal lithography does not possess the accuracy and on-demand control over spatial patterns it can generate, as compared to photolithography and EBL. To generate nanotopography with different spatial patterns, colloidal lithography can utilize different crystal structures of self-assembled colloidal masks and alter incidence angle of plasma that etches the underlying surface [39–42].
With proper surface functionalization, self-assembled colloidal monolayers can be directly used as nanotopographical substrates for cell assays [54, 57, 58]. In addition, colloid lithography has been applied to generate nanoscale topographical features on curvilinear surfaces of microscale particles and scaffolds to create hierarchical topographical biomaterials that are difficult to obtain using conventional methods [39–41, 56]. Combining nanoscale patterned surfaces generated by photolithography and EBL with colloid self-assembly, it is also possible to achieve regular patterns of nanotopography with curvilinear local geometries [59]. Instead of serving as an etching mask, self-assembled colloidal nanoparticle layers can also act as a deposition mask to generate nanotopography with a negative pattern of the colloidal mask [60–62].
Pattern transfer
Pattern transfer utilizes pre-existing nanotopographical features on a rigid mold to transfer such features using molding processes to other materials with high-efficiency and high-fidelity. Here we discuss two types of pattern transfer techniques, nanoimprinting and replica molding, that have been commonly used for generating nanotopographical surfaces for stem cell studies.
Nanoimprinting
Nanoimprinting utilizes hot embossing of thermoplastics to achieve nanoscale surface patterning [63–65]. In nanoimprinting, a thin layer of thermoplastic polymer is spread or spin-coated onto a planar, featureless substrate. A hard template (“mold”) containing prefabricated nanotopographical features is then brought into a direct contact with the polymer layer (“imprinting”), and they are pressed together under pressure. When heated up above the glass transition temperature of the polymer, the polymer on the substrate melts and conforms to the pattern of the template. The polymer layer forms an inverse replica of the template after cooling and separation from the template (“demolding”) (Fig. 1d). In nanoimprinting, adhesion between the polymer and the template is carefully controlled to allow proper release of polymeric substrates.
A key advantage of nanoimprinting is its ability to achieve high resolution nanofabrication (down to 14 nm [66]) over a large surface area using a relatively simple procedure; however, nanoimprinting requires specialized equipments to orchestrate its different processing steps [67]. To date, synthetic nanotopographical surfaces have been created by nanoimprinting using thermoplastic polymers such as PMMA [69] and polyurethane acrylate (PUA) [68] and more recently with platinum-based bulk metallic glass alloy (Pt-BMG), an inorganic material susceptible to thermoplastic forming [70].
Replica molding
While nanoimprinting uses thermoplastic materials, replica molding is a method of replicating structures into an elastomeric polymer material that hardens after baking under elevated temperature. Conceptually, replica molding belongs to a larger class of methods called soft lithography that was developed complementary to traditional photolithography [71, 72]. Polydimethylsiloxane (PDMS) is the most frequently used elastomeric polymer material for replica molding owing to its excellent physical property and biocompatibility. The procedure of replica molding is straightforward and easy for implementation in a regular research laboratory, rendering this method ideal for rapid prototyping.
In replica molding, PDMS precursor in a liquid state is poured directly onto a mold with surface topography inverse to that is desired. Upon heating, PDMS cures, resulting in a hardened PDMS layer that can be readily released from the mold. The final PDMS layer presents a replica that retains the size and geometry of the original topography on the mold with high fidelity (Fig. 1e). As typical elastomers including PDMS can be readily deformed by surface tension, they are not suitable for molding to generate nanoscale topographical features with high fidelity. Instead, “hard” thermally curable polymers such as h-PDMS [73] and PUA [74] have been utilized for replica molding when desired topographical features have critical dimensions of less than 500 nm. To study cellular responses to extracellular nanotopography, replica molding has been applied to generate nanoscale gratings on PDMS using either silicon [34] or nanoimprinted PMMA molds [69, 75]. Other than PDMS, polycaprolactone (PCL), a biodegradable polyester, has also been used as a elastomeric polymer for replica molding to transfer nanopit structures with a diameter of 50 nm from a silicon mold fabricated by EBL [38] (Fig. 1f).
Surface roughening
Even without external templates or masks, a uniform physical or chemical etching environment can generate random nanotopographic features on certain material surfaces through “surface roughening” processes [76–80]. Although it is intrinsically difficult for surface roughening to achieve nanotopographic features with accurate feature patterns and sizes, they are effective for generating nanotopographic surfaces of large surface areas owing to its simple methodology and compatibility with bulk processing. To date, surface roughening has been utilized for generating random nanotopographic features using either chemical etching or physical reactive ion etching (RIE) processes.
Chemical etching
As its name suggests, chemical etching relies on chemical reactions between etchants and substrate surfaces to be processed to remove materials from the surfaces. Due to material inhomogeneities at the atomic scale or nanoscale within the substrate, etchants locally react and remove materials from the surface in an anisotropic fashion at different rates, resulting in an uneven landscape with surface undulations (surface roughness) down to a nanometer scale (Fig. 1g, h). Surface roughening during chemical etching depends on experimental conditions such as etchant composition and concentration and etching time and temperature [76, 81]. It is known, for example, that etching of silicon surfaces with KOH can result in nanoscale surface roughness, the level of which increases with etching time but decreases with increasing reaction temperature [76]. Importantly, chemical etching can also be applied to some biocompatible materials commonly used in tissue engineering. For example, etching using acids or bases can create nanotopography on surfaces of biocompatible polymer materials such as poly (lactide-co-glycolide) (PLGA), poly-ether-urethane (PU), and polycaprolactone (PCL) [77]. Acid etching can also generate nanotopography on titanium (Ti) surfaces, a common metallic material used for implant [78].
Reactive ion etching
Besides chemical etching, nanotopographical surfaces can also be generated on silica-based glass surfaces with RIE, a well-established semiconductor microfabrication technique. During RIE etching on glass surfaces, bombardment by reactive ion species generated using SF6 and C4F8 gases disrupts un-reactive glass substrate and causes damage such as dangling bonds and dislocations, resulting in the glass surface reactive toward etchant species. Importantly, since small concentrations of impurities such as Al, K and Na exist in silica glass, these impurities result in accumulations of less volatile species (such as AlF3, KF, NaF, etc.) on glass surface during the RIE process [82, 83]. Some of these less-volatile compounds are then backscattered onto glass surface and form randomly distributed small clusters that can shield glass surface from bombardment and reaction with reactive ions. These compound clusters effectively generate the so-called “micro masking” effect that can randomly shadow glass surface and thus result in nanoscale roughening of glass surface during the RIE process (Fig. 1i) [82, 83]. In practice, nanotopography of glass surface can be controlled by adjusting the RIE process duration. Compared with chemical etching, RIE usually has a lower etching rate and thereby allows a more precise control over nanotopography generated on substrate surfaces. In addition, RIE is compatible with other semiconductor microfabrication techniques such as photolithography, and as such one can generate patterned nanorough islands on a flat glass surface combing RIE with photolithography [80].
Material synthesis
Different material synthesis methods developed from tissue engineering and other research fields, such as electrospinning, phase separation, anodization, and sintering, can also be utilized for fabricating nanotopographical biomaterials useful for stem cell studies.
Electrospinning
Electrospinning is a long-existing polymer processing technique which was rediscovered in the early 1990s for its applications in the field of tissue engineering owing to its capability of generating nanofibrous constructs from a broad range of polymers in a simple setting [84–86]. Since electrospinning does not require the use of coagulation chemistry or high temperature to produce solid threads from solution, this process is suited to production of fibers with large and complex molecules.
In electrospinning, a high voltage in the range of kilovolts is applied to a pendent droplet of polymer solution, and the body of the solution becomes charged. Electrostatic repulsion in the droplet counteracts surface tension, resulting in stretching of the droplet. When the voltage passes a certain threshold, force balance between electrostatic repulsion and surface tension breaks, and a liquid jet erupts from the droplet surface. The liquid jet undergoes further stretching due to electrostatic force and solvent evaporation, reducing fiber diameter to as small as a few nanometers. Arrangements of electrospun nanofibers can be conveniently controlled from completely random to unidirectionally aligned, producing a wide span of extracellular nanotopographical textures (Fig. 1j) [87–92].
A significant advantage of electrospinning for studying stem cell-nanotopography interactions is its capability to generate three-dimensional nanofibrous architectures that can properly mimic the structure and organization of in vivo ECM [93]. Its compatibility with many types of polymers and feasibility for multiplexed functionalization has rendered electrospinning a popular method for different tissue engineering applications. For example, both random and aligned nanofibrillar matrices have been fabricated using electrospinning of polymers such as polyamide [94], PCL [95–100], polyethersulfone (PES) [101, 102], poly(L-lactic acid) (PLLA) [87–91], PLGA [92], polymethylglutari-mide (PMGI) [103], hydroxybutyl chitosan (HBC) [104], poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) [105], and hydroxyapatite/chitosan (HA/CTS) [106]. Fiber functionalization can be achieved by simply mixing adhesive ECM proteins such as collagen and gelatin into polymer solution before electrospinning [90, 91]. Multiplexed functionalization of electrospun nanofibrous constructs can be accomplished by tailored grafting of polymer chains. Patel et al., for example, have developed a multifunctional PLLA nanofiber biomaterial presenting both laminin and basic fibroblast growth factor (bFGF) in addition to the intrinsic nanotopography, via grafting through heparin and di-amino-poly(ethylene glycol) (di-NH2-PEG) as linkers [107]. Electrospinning has also been applied to fabricate nanofibrous structures composed of silk fibroin [108], showing its compatibility with native proteins.
Phase separation
Thermally induced phase separation has long been utilized for fabrication of nanoporous scaffolds for tissue engineering applications [109–112]. Phase separation occurs when the concentration of polymer or polymer blends exceeds its solubility in solvent. It is convenient to control the mixture temperature to initiate phase separation in polymer solution and subsequent generations of micro- and nanoscale polymer-rich and polymer-lean phase domains within the mixture [112]. After removal of solvent, polymer-rich phase domains solidify and form polymeric foam that appears as nanotopographical scaffolds (Fig. 1k). By varying the types of polymers and solvents, polymer concentration, and phase separation temperature, different nanotopographic morphologies and structures can be created from phase separation. For example, to recapitulate the fibrous architecture of type I collagen, phase separation has been utilized for fabricating synthetic biodegradable PLLA nanofibrous matrices with fiber diameters ranging from 50 – 500 nm [113, 114]. Other than nanofibrous matrices, phase separation has also been used for generation of nanotopography such as nanoscale islands and pits using blends of poly(p-bromostyrene) and poly(deuteriostyrene) in conjunction with a spin-coating method [115].
Phase separation can also be induced at room temperature to fabricate nanoscale islands of polymers based on a bulk demixing process for a binary polymer mixture, such as the polystyrene (PS) and poly(4-bromostyrene) (PBrS) mixture in a spin-cast thin film [116–119]. Dimension and density of nanotopographical features generated by polymer demixing depend on mixture composition and component concentration. Using demixing of a PS/PBrS mixture, Dalby et al. have successfully fabricated arrays of circular islands with a diameter of 10 – 100 nm to study cellular responses to nanotopography [117]. Nanotopography patterns generated by polymer demixing can also be regulated by pre-defined interfaces at the polymer mixture-substrate boundary. In an early study, Sprenger et al. have developed a hierarchical nanotopographical surface by applying ternary polymer demixing onto a pre-patterned substrate prepared by microcontact printing [120]. Specifically, a pre-defined microscale pattern of octadecylthiol was generated on a gold surface using microcontact printing to exclude poly(2-vinylpyridine) (PVP) phase of the PS/PMMA/PVP mixture onto polar, bare gold regions, where they retain a non-polar PS phase, resulting in a wall-like PMMA phase of 200 – 300 nm thickness in between the microscale patterns.
Block copolymer lithography is another phase separation-based nanofabrication method with unique advantages of low cost and rapid casting [121–124]. Due to phase separation driven by interactions between different segments of block copolymers, block copolymer lithography can generate a rich set of domain-like patterns with a characteristic dimension ranging from micro- to nanoscale [122]. Interestingly, block copolymer lithography was named after “lithography” because the patterns obtained via block copolymer self-assembly was first used as lithography masks [121]. In recent years, block copolymer lithography has been directly applied to generate nanotopographical surfaces for cell assays. Maclaine et al., for example, have used poly(styrene-block-poly-2-vinylpyridine) (PS-b-P2VP) block copolymer micelles to generate nanoscale islands of 20 nm in height and 150 nm in in-plane periodicity [125]. Using a novel PS-PDMS diblock copolymer, Salaun et al. have achieved fabrication of PDMS nanopillars of 10 nm in height and 20 nm in width [126]. Interestingly, by using PS-b-P2VP and PS-b-P4VP diblock copolymers, Khor et al. have generated two distinct patterns of nanotopography, namely, “dot-like” (“salt-and-pepper”) and “work-like” (“labrynth”) patterns, respectively [127]. In addition, emergent patterns generated in functionalized block copolymer thin films can also be used to spatially define nanoscale arrays of gold nanoparticles [124, 128, 129] and proteins [130], thus expanding the diversity of nanotopographical cues for cell assays.
Anodization
Anodization is an electrochemical process commonly used for increasing thickness of the oxide layer on metal or alloy surfaces [131–138]. During anodization, metal is immersed in an electrolytic solution and connected as the anode (the positive electrode) of an electrical circuit. A current passing directly through the electrolytic solution releases hydrogen at the cathode (the negative electrode) and oxygen at the surface of the metal anode, building up an oxide layer of a few to tens of nanometers thick. Due to heterogeneous local reaction rates at the metal surface, the electrochemical reaction in anodization can generate nano- and microscale textures on the metal surface.
Anodization has been successfully used for fabrication of nanotopographic surfaces for stem cell studies (Fig. 1l). For example, Park et al. [14] have applied controlled anodization of Ti in a fluoride-containing electrolyte to generate highly ordered layers of titania (TiO2) nanotube structures for promoting osteogenic differentiation of MSCs [14, 139]. Similarly, Sjöström et al. have fabricated TiO2 nanopillars with pillar heights of 15 – 100 nm on Ti surfaces using anodization through a porous alumina mask [140].
Other than additive nanofabrication, anodization has also been applied in fabrication of nanoporous substrates via electrochemical etching. In a recent study, Clements et al. have developed a gradient nanoporous silicon (pSi) biomaterial for rat MSC culture [141]. Using an asymmetric electrode setup, a gradient of anodic etching generated a corresponding gradient of nanoscale pores in a Si wafer, with pore sizes gradually decreasing from 920 nm to 20 nm as the distance to the electrode increased. Surface functionalization using electrografting further generated an orthogonal gradient of RGD peptides and thus enabled study of synergistic effect of nanotopography and biochemical gradients on MSC behaviors [141].
Sintering
Sintering is a widely used method for creating objects from metal and ceramic powders [142–145]. In sintering, powdered materials are heated to a temperature below the material’s melting point but high enough for diffusion of atoms, causing fusion of different powder particles into one solid piece. Although sintering is mostly used in bulk processing, it can also create granular surface patterns with critical dimensions in nanometer size scales (Fig. 1m) when sintering temperature and duration are carefully controlled. Dulgar-Tulloch et al., for example, have fabricated alumina, titania, and hydroxyapatite surfaces with grain-like nanotopography of critical dimensions ranging from 24 nm to 1,500 nm for MSC culture [146].
Nanotopography controls stem cell fate
In vivo, stem cells interact with and interrogate their surroundings at the micrometer and/or nanometer length scale. Plenty of evidence exists to suggest that nanotopographic signals from the local stem cell niche instruct behaviors of stem cells. Here we provide illustrative examples using bioengineering and nanofabrication approaches to control nanotopographic features of the local stem cell niche and, where evidence suggests, regulate stem cell fate through synergistic regulations of stem cell shape [16, 22, 23], CSK tension [30, 31], integrin-mediated adhesion signaling [29], and nuclear mechanics [32, 33].
Mesenchymal stem cells
Mesenchymal stem cells (MSCs) are multipotent cells that can give rise to cells of the stromal lineage, namely, osteoblastic (bone) [14, 18, 37, 95, 96, 105, 106, 139, 140], adipogenic (fat) [23, 147], myoblastic (muscle) [104, 148], chondrogenic (cartilage) [97], and fibroblastic (connective tissue) [37] lineages. In recent studies, abundant evidence has been obtained to support that MSC fate can be influenced by the size, symmetry, and regularity of nanotopographical features.
It has been commonly observed that both rat and human MSCs on nanofiber scaffolds show significantly enhanced osteogenic differentiation compared to conventional tissue culture plates [14, 95, 96, 105, 106, 139]. In addition, Park et al. [14] observed that 15 nm diameter TiO2 nanotubes promoted osteogenic differentiation of rat MSCs (Fig. 2a). However, when the tube diameter increased to 50 nm or above, osteogenic differentiation of rat MSCs was significantly diminished. More recently, Oh et al. [139] demonstrated that TiO2 nanotube arrays directed human MSC differentiation toward the osteoblast lineage even in the absence of soluble inductive factors. In this study, among arrays of 30, 50, 70, and 100 nm diameter TiO2 nanotubes fabricated on Ti substrates, the one composed of 100 nm diameter nanotubes showed the highest potential for promoting human MSC osteogenic differentiation.
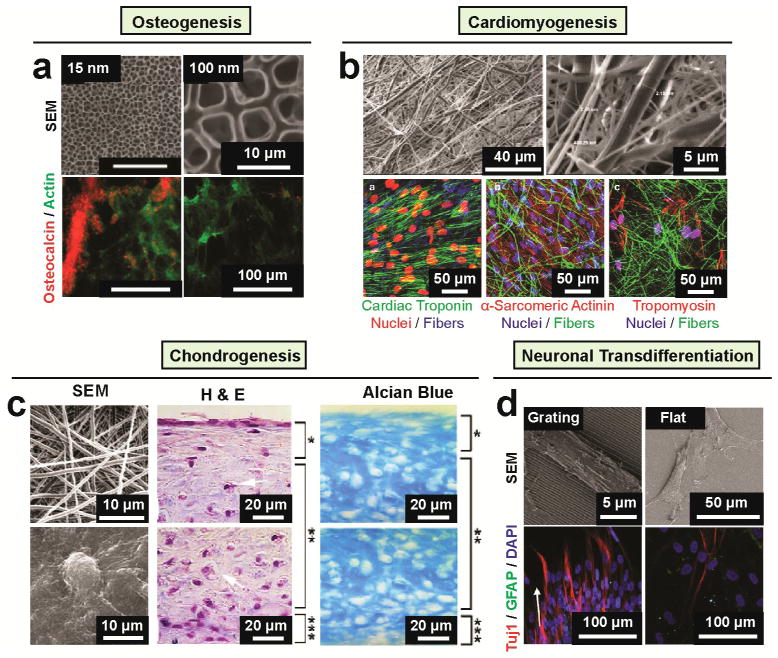
Figure 2. Nanotopography regulates MSC lineage.
(a) TiO2 nanotube diameter directs MSC osteogenic differentiation [14]. The top row presents SEM images of highly ordered TiO2 nanotubes of small (15 nm; left) and large (100 nm; right) pore sizes. The bottom row presents immunofluorescence images of cells with osteocalcin staining in red and F-actin staining is in green on 15 and 100 nm nanotubes after 2 weeks in culture in osteoblast differentiation medium. Osteogenic differentiation occurred 15 nm nanotubes as seen by osteocalcin staining but rarely detectable on 100 nm nanotubes. Reproduced with permission [14]. Copyright 2007, American Chemical Society. (b) Using of fibrin nanofiber and PLGA microfiber composite scaffold for human MSC differentiation toward myocardial lineage [148]. The top row presents SEM micrographs of PLGA–fibrin electrospun membrane at different magnification. The bottom row presents confocal microscopy images of MSCs grown on PLGA–fibrin composite fibers after 14 days of induction of cardiac differentiation expressing of cardiac specific markers including cardiac troponin, α-sarcomeric actinin, tropomyosin as indicated. Reproduced with permission [148]. Copyright 2013, Mary Ann Liebert, Inc. (c) Nanofibrous scaffold for human MSC chondrogenesis [97]. The left column shows SEM images of nanofibrous surface (top) produced by the electrospinning process showing random orientation of ultra-fine fibers with an diameter ranging from 500 to 900 nm and MSCs (bottom) with round, ECM-embedded chondrocyte-like cells on the surface after 21 days culture in the presence of TGF-β1. The middle and right columns present the histological analysis of human MSC cultured on nanofibrous surface in a chondrogenic medium supplemented with TGF-β1 for 21 days. Sections from the upper and lower portions of the three-dimensional constructs were stained with H&E (middle column) and Alcian blue (right column). H&E staining showed flat fibroblast-like cells on the top zone (bracket, *), round chondrocyte-like cells embedded in lacunae (arrows) in the middle zone (bracket, **), and small, flat cells at the bottom zone (bracket, ***). Alcian blue staining showed the presence of sulfated proteoglycan-rich ECM in the construct. Reproduced with permission [97]. Copyright 2005, Elsevier. (d) Nanograting surfaces induce neuroal transdifferetiation of MSCs [75]. SEM (top) and immunofluorescence (bottom) images of human MSCs cultured on nanograting and unpatterned PDMS as indicated. In the immunofluorescence images, cells were differentiated for 14 days in the presence of retinoic acid and were stained with neuronal marker Tuj1in red and GFAP in green. Reproduced with permission [75]. Copyright 2007, Elsevier.
Notably, while recent studies have shown enhanced ECM proteins adsorption on nanotopographical surfaces [149, 150], direct effect of such ECM protein adsorption may be secondary compared to the effect of nanotopography sensitivity on stem cell fate regulation. Supporting this view, Oh et al. [139] proposed that instead of regulating stem cell behaviors through adsorbing more proteins onto surface, instead, nanotopographic surfaces resulted in distinct sizes and spacing of aggregated ECM protein on nanotopographic surfaces, which in turn induced changes in integrin-mediated focal adhesion (FA) formation and stem cell function. Similar results were also obtained by Chen et al. [80] for hESCs on glass surfaces with varied nanoroughness.
In addition to in-plane nanotopographical features, the third dimension, i.e., the nanoscale height of topological features, can also play a potent role in regulating stem cell fate. Using 15, 55, and 100 nm high Ti nanopillars, Sjöström et al. [140] showed that Ti nanopillars with the smallest (15 nm) height resulted in the most significant bone matrix nodule formation, suggesting an increased level of osteogenic differentiation of human skeletal stem cells. A different observation, however, was reported recently by Zouani et al., wherein the authors developed nanotopographical features with similar in-plane dimensions yet different depths using UV-mediated photodegradation of polyethylene terephthalate (PET) [151]. Zouani et al. observed that a greater nanotopographical depth (100 nm) promoted osteogenic differentiation of human MSCs, whereas much less osteogenic differentiation was observed for surfaces with shallower (10 nm depth) features. The inconsistent observations about different nanotopographical feature sizes favorable for osteogenic differentiation support cell-type specific response to nanotopography. Interestingly, recent studies have also shown that MSC fate can be influenced by symmetry and regularity of nanotopographical patterns. Specifically, nanoscale disorder of a nanopit array significantly promoted osteogenesis of human MSCs even in the absence of soluble inductive factors, whereas highly ordered nanotopographical surfaces produced much limited cellular adhesion or osteogenic differentiation [37]. Using silica nanohelices grafted on glass surfaces, Das et al. [18] also demonstrated that the periodicity of chiral extracellular topography directed commitment of human MSCs toward the osteoblast lineage.
In addition to osteogenesis, other specific cell lineage commitments, including cardiomyogenesis [148], myogenesis [104] and chondrogenesis [97], have also been explored using nanotopographical biomaterials. For example, synthetic biodegradable PCL nanofibrous scaffolds were utilized for enhancing in vitro chondrogenesis of MSCs compared to the established cell pellet culture method (Fig. 2c) [97]. In addition to chondrogenesis, it was demonstrated that nanofibrous topography regulated the actin CSK and nuclear shape of MSCs and induced MSC differentiation into the myogenic/myocardial lineages even without soluble inductive factors [104, 148].
Nanotopographic cues have also been demonstrated in recent studies for inducing neural transdifferentiation of MSCs. A recent report by Woodbury et al. [152], for example, has shown that MSCs seeded on nanotopographical surfaces can transdifferentiate into neural cells when cultured in a neural induction medium. Interestingly, another recent study by Yim et al. [75] has shown that grating-like nanotopography alone can induce neural transdifferentiation of MSCs (Fig. 2d). Gene expression and immunostaining assays demonstrated significant up-regulation of neuronal markers such as microtubule-associated protein 2 (MAP2) and β-tubulin III (Tuj1) for MSCs cultured on nanograting surfaces compared to unpatterned or micropatterned controls. Yim et al. [75] further proposed that nanotopography-induced CSK rearrangement and nuclei elongation in MSCs might play an important role for neural transdifferentiation of MSCs. Interestingly, Yim et al. [75] also reported that even though nanograting surfaces in conjunction with soluble inductive factors such as retinoic acid could enhance neuronal marker expression in MSCs to the maximal extent, nanotopography showed a stronger independent effect on inducing neural transdifferentiation compared to retinoic acid alone on unpatterned surfaces. Similarly, Prabhakaran et al. [91] utilized nanofibrous surfaces in conjunction with suitable inductive factors for neuronal transdifferentiation of human MSCs in vitro.
Neural stem cells
Neural stem cells (NSCs) are multipotent stem cells intrinsically capable of self-renewal and differentiation into different neural lineages, offering promising applications for NSC-based cell therapies to treat neurodegenerative diseases and traumatic injuries [153]. However, clinical applications of NSC-based cell therapies are hindered due to a lack of efficient methods for large-scale expansion as well as controlled differentiation of NSCs.
In recent studies, nanotopographical substrates, especially nanofibrous scaffolds, have been widely applied for regulating NSC differentiation for neural tissue engineering applications [68, 87, 88, 90, 98, 101, 108]. NSC differentiation has been shown dependent on both nanofiber size and alignment. For example, Yang et al. [87] showed an enhanced NSC differentiation on aligned PLLA nanofibers compared to PLLA microfibers. In other studies, nanofiber scaffolds with well-aligned structural features were demonstrated for enhanced proliferation and differentiation of NSCs compared to randomly aligned nanofiber scaffolds [90, 98].
Importantly, nanotopographical surfaces not only promote NSC neural differentiation, but also selectively enhance other lineage specifications of NSCs. For example, Christopherson et al. [101] demonstrated that on nanofibrous matrix composed of 283 nm diameter fibers, rat NSCs could spread unhindered, adopt a cell morphology suggesting glial lineages, and preferentially differentiate into oligodendrocytes; whereas cells seeded on matrix composed of larger fibers with diameters of 749 nm and 1452 nm were confined by individual fibers and preferentially differentiated into neural lineages (Fig. 3). More recently, by coating electrospun PCL nanofibers with graphene oxide (GO), Shah et al. [154] developed a hybrid nanotopographical biomaterial that preferentially guided NSC differentiation into oligodendrocytes. Together, the nanofibrous materials have been widely demonstrated for functional regulation of NSC neural differentiation including differentiation efficiency and lineage specifications. Notably, the nanotopographical materials (especially those with nanofibrous forms) may not only improve NSC differentiation, but also offer favorable microenvironment for neural tissue engineering [155, 156]. With careful design and engineering of material features at nanoscale, nanofibrous materials can potentially serve as a unique matrix platform for neural regeneration throughout the process of stem cell neural differentiation, neural culture, and transplantation.
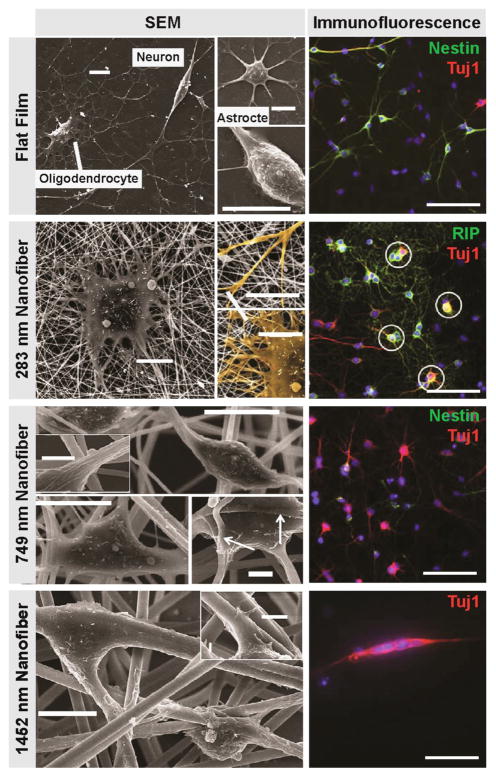
Figure 3. Nanotopography regulates NSC lineage[101].
SEM (left) and immunofluorescence (right) images of rat NSCs cultured on various substrates (laminin-coated PES films (flat film), 283-nm, 749-nm, 1452-nm nanofibers) in the presence of 1 mM retinoic acid and 1% fetal bovine serum for 5 days. For the immunofluorescence images, the cells are stained with RIP (oligodendrocyte marker), Tuj-1 (neuronal marker) or nestin (neural progenitor marker) as indicted. Scale bars for SEM images are 10 μm, and for inserts are 2 μm. The arrows in the SEM images indicate cell attachment to nanofibers. Scale bar for all immunofluorescence images with are 100 μm. Circled cells on 283-nm fiber mesh are cells stained double positive for RIP and Tuj1. Reproduced with permission [101]. Copyright 2009, Elsevier.
Pluripotent stem cells
Pluripotent stem cells including both ESCs and iPSCs, possess the ability of differentiating into any specialized cell type of the human body. The pluripotent nature of ESCs and iPSCs opens unprecedented opportunities for potential stem cell-based regenerative therapies for various degenerative diseases and the development of drug discovery platforms. Yet, the current PSC research for regenerative medicine, although has attracted much enthusiasm, is still in a nascent state largely due to a lack of effective technologies and functional biomaterials for large scale maintaining and expansion of PSCs as well as accurate control of lineage specification of PSCs. Recent advances in nanotechnology and materials science have raised a widespread hope of developing promising nanoengineered biomaterials for PSC-based regenerative medicine, among which nanotopographical matrixes mimicking the in vivo stem cell niche have shown exceptional potentials of regulation of stem cell function and fate.
Stemness maintenance and self-renewal
One of the major challenges in PSC-based cell engineering and regenerative medicine is to achieve large-scale, long-term PSC expansion without losing pluripotency. Although the standard protocol maintaining PSCs on feeder cells is successful and widely adopted, such feeder based culture protocol is labor-intensive, prone to contaminations from feeder cells, and difficult for high-throughput automation. Excitingly, recent developments of nanotopographical biomaterials have brought a few novel, feeder-free methods for PSC culture and thus pose promising routes for future large-scale PSC engineering (Fig. 4).
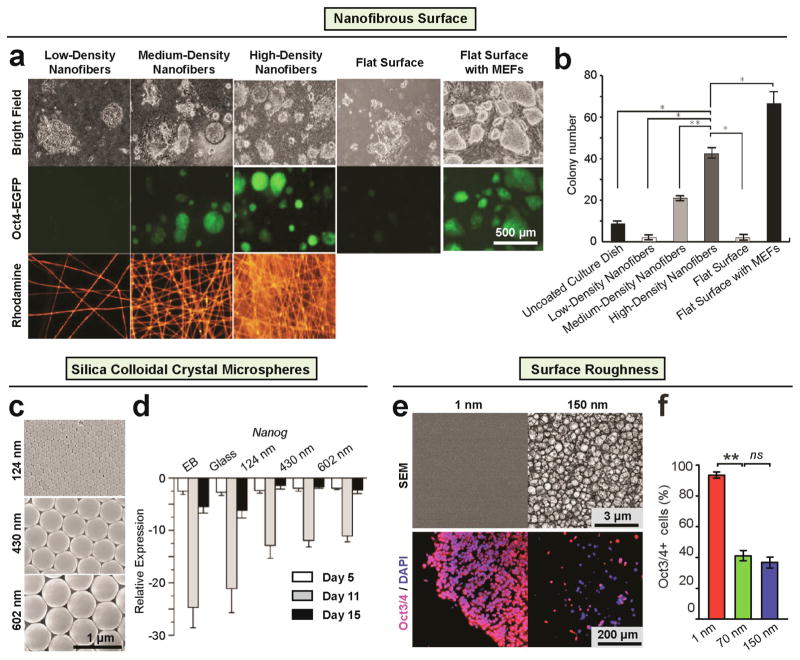
Figure 4. Nanotopography regulates ESC self-renewal.
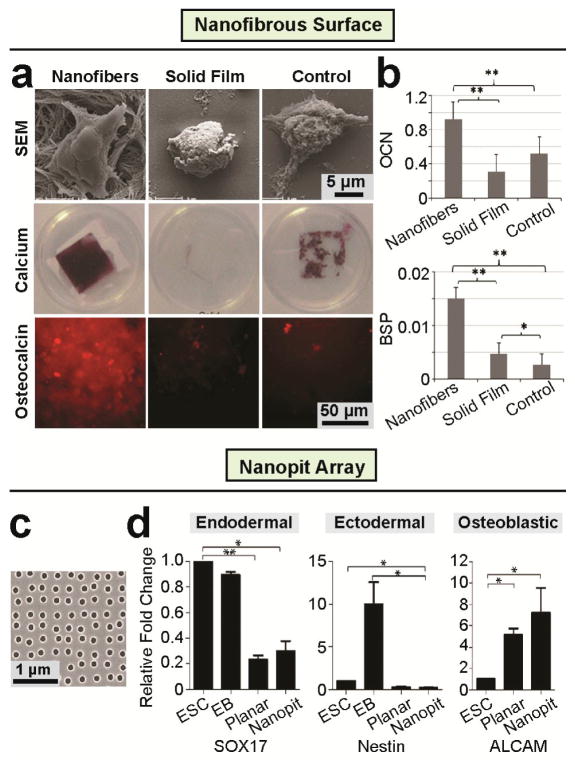
(a–b) Polymethylglutarimide (PMGI) nanofibrous surfaces serve as a cellular scaffold for maintaining self-renewal of mouse ESCs without MEFs [103]. (a) Bright field (top) and fluorescence images (middle) of R1-Oct4-EGFP mouse ESCs cultured on nanofibers at three different densities (i.e., low, medium and high) and controls. Bottom panels show the density of PMGI nanofibers doped with Rhodamine 6G for visualization. (b) The number of colonies within 5 mm2 after culturing for 3 days on various substrates. Data are presented as mean ± SD of three independent experiments. (*p < 0.001, **p < 0.01, n>3). Reproduced with permission [103]. Copyright 2012 Springer. (c–d) Nanotopographical surface formed from silica colloidal crystal microspheres (SCC) maintain the expression of mouse ESC self-renewal in comparison to flat glass [57]. (c) SEM images of SCC substrates with particles of 124 ± 4 nm, 430 ± 4 nm, and 602 ± 12 nm diameter, respectively, showing their ordered face-centered cubic packing. Quantitative PCR showing (d) Nanog, a stem cell marker was less down-regulated on the SCC substrates than on glass cover slips; N= 3. Reproduced with permission [57]. (e–f) Smooth vitronectin-coated glass surfaces supported cell adhesion, rapid cell proliferation, and long-term self-renewal of human ESCs without using mouse MEF feeder cells [80]. (e) Representative SEM images of glass surfaces (top) and immunofluorescence images of human ESCs (bottom) plated on glass surfaces with their root-mean-square (rms) nanoroughness Rq indicated. In the immunofluorescence images, the cells were co-stained for Oct3/4 (red) and nuclei (DAPI; blue). (f) Percentage of Oct3/4+ human ESCs on the glass substrates with different levels of nanoroughness as indicated, after culture for 7 days. Error bars represent (standard error of the mean (SE, n = 3). ns (> 0.05) and ** (p < 0.01) (Student’s t-test). Adapted from [80].
Recently, Nur-E-Kamal et al. [157] reported an electrospun nanofibrillar substrate composed of polyamide nanofibers to promote proliferation and self-renewal of mouse ESCs via the Rac and phosphoinositide 3-kinase (PI3K) signaling. Similar results were obtained by Liu et al. [103], where high density electrospun polymethylglutarimide (PMGI) nanofibers were used to achieve sustained feeder-free maintenances of mouse ESCs (Fig. 4a, b). In another related study, feeder-free mouse ESC maintenance was achieved using a topographical surface formed with silica colloidal crystal (SCC) spheres to generate highly ordered topographical features of 120 – 600 nm in diameter [57] (Fig. 4c, d).
In addition to mouse ESCs, it is also challenging to achieve long-term culture of human ESCs while maintaining pluripotency. It has recently been reported that the current feeder based human ESC culture method can be significantly improved by introducing collagen or gelatin coated electrospun PCL nanofibrous scaffolds [99]. It should be noted, however, that a significant difference exists between human and mouse ESCs for their culture environment requirements for self-renewal and clonal growth [8]. For example, human ESCs do not require leukemia inhibitory factor (LIF) for self-renewal, which is however needed for mouse ESCs [8]. Such difference has also been observed in the context of mechanosensitive behaviors of human and mouse ESCs. For instance, although it was found that both intrinsic (cell) and extrinsic (substrate) softness facilitated maintenance of mouse ESC pluripotency [158, 159], soft cell culture surfaces promoted spontaneous differentiation rather than self-renewal of human ESCs [24, 160, 161]. In another recent study, Chen et al. reported that nanorough substrates with topographical features of 100 nm high promoted spontaneous differentiation of human ESCs, whereas flat, featureless coverslips were supportive for stemness maintenance [80] (Fig. 4e, f). A similar observation was reported by the Dalby group, wherein disordered nanopit arrays made of polycarbonates were observed to suppress self-renewal and promote early osteogenesis of human ESCs [162]. However, one conflicting observation has been reported by Kong et al., wherein fibronectin-coated nanotopographical surfaces, rather than flat controls, supported stemness maintenance of human ESCs, and switching nanotopography features from a hexagonal pattern to a honeycomb one further improved such an effect [60].
Directed differentiation
Nanotopography cues have also been shown important for directed differentiation of ESCs toward neural cells [89, 100, 163–168], osteoblasts [92, 114, 162, 169, 170], muscle cells [171, 172], hematopoietic cells [173, 174], adipocytes [175], and chondrocytes [176]. Here for illustrations, we only discuss how nanotopographical biomaterials have been utilized for improving directed neurogenesis and osteogenesis from ESCs. For ESC differentiation toward other cell lineages, the readers are referred to references cited above.
Neural lineage
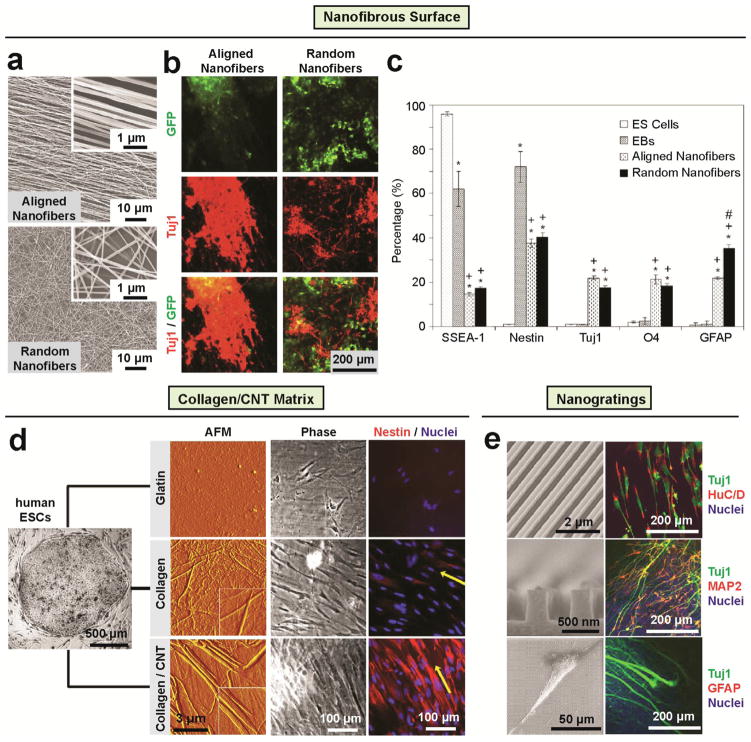
Nanotopographical materials have been widely demonstrated for promoting neural lineage commitments of PSCs. In a study by Xie et al. [100], uniaxially aligned nanofiber substrates were used to induce mouse ESC differentiation into different neural lineages, such as neurons, oligodendrocytes, and astrocytes (Fig. 5a–c). Aligned nanofibers enhanced not only neural differentiation efficiency but also neurite outgrowth along nanofibers, critical for neuronal network development. Carbon nanotubes (CNT) were also recently utilized for promoting ESC neurogenic differentiation. For example, CNT/PLLA composite scaffolds were applied to enhance neural differentiation of mouse ESCs even in the absence of soluble inductive factors [89]. Similarly, human ESCs directly seeded onto type I collagen-coated CNT matrix showed enhanced ectodermal lineage commitment as well as cell alignment along collagen/carbon nanotube fibrils [167] (Fig. 5d). Thin film scaffolds composed of biocompatible polymer poly(acrylic acid) or silk grafted with CNTs were also shown to promote human ESC-derived embryoid body (EB) differentiation into neural cells with heightened cell viability [23, 166]. In addition to nanofibrous materials and CNT composite scaffolds with random nanotopological features, well controlled nano-scale surface patterns such as nanoscale ridge/groove arrays also showed efficiently induced differentiation of human ESCs into neural lineages without any soluble inductive factor (Fig. 5e) [168]. Interestingly, human ESCs cultured on such nanoscale ridge/grooves arrays preferentially differentiated into neurons but not glial cells, such as astrocytes. Such selective suppression of glial cell formation is desirable for potential applications in therapies for spinal cord injury, as astrocytes are known to contribute to glial scar formation, creating barriers to axons in the central nerve system (CNS) [177].
Figure 5. Nanotopography regulates ESCs toward neural lineage.
(a–c) The differentiation of mouse ESCs on electrospun nanofibrous surfaces into neural lineages [100]. (a) SEM images of aligned (top) and randomly oriented (bottom) PCL nanofibers prepared by electrospinning. (b) Representative immunofluorescence images of differentiated mouse ESCs on aligned (left) and random (right) PCL nanofibers for 14 days. The mouse ESCs expressed GFP in green color (top) and stained in red color with neuron marker Tuj1 (middle), the bottom images show and superimposed image of the top and middle images in the same region. (c) Cell phenotype analysis of mouse EBs cultured on PCL nanofibrous scaffolds for 14 days. The markers examined were SSEA-1 (stage specific embryonic antigen, for undifferentiated mouse ES cells), nestin (for neural precursors), Tuj1 (β-tubulin III, for neurons), O4 (for oligodendrocytes), and GFAP (glia fibrillary acidic protein, for astrocytes). # indicates p < 0.05 for markers compared with embryonic stem cells. + indicates p < 0.05 for markers compared with embryoid bodies. * Indicates p < 0.05 for markers compared with random PCL fibers. Reproduced with permission [100]. Copyright 2009, Elsevier. (d) Direct human ESC neural differentiation on collagen/CNT matrix [167]. AFM characterization of the surface structures, cell morphology, and nestin expression of human ESCs after cultured in the medium of spontaneous differentiation for three days on gelatin (top), collagen (middle) and collagen/CNT (bottom) matrices. Inset Image size in the AFM images: inset 2×2 μm2. The yellow arrows in the staining images indicate the coarse alignments of the cells. Reproduced with permission [167]. Copyright 2009, Elsevier. (e) Direct differentiation of human ESCs into selective neurons on nanoscale ridge/groove pattern arrays [168]. The left column are representative SEM images of a bird’s eyes view (left top) and a cross-section (left middle) of 350-nm ridge/groove pattern arrays (spacing of 350 nm, height of 500 nm) and a SEM image showing human ESCs on the 350-nm nanoscale ridge/groove pattern arrays (left bottom). The right column are representative immunofluorescence images of human ESCs stained with nuclei, neural and glial markers as indicated after cultured for 10 days on the 350-nm ridge/groove pattern arrays. Differentiated hESCs stained positively for HuC/D (human neuronal protein: RNA-binding protein) and MAP2 (mature neuronal marker: microtubule-associated protein 2), but were not for GFAP (intermediate filament proteins of mature astrocytes: glial fibrillary acidic protein). This suggests that hESCs differentiated into mature neurons without differentiation into a glial lineage such as astrocytes. Reproduced with permission [168]. Copyright 2010, Elsevier.
Osteogenic lineage
Another important direction is the osteogenesis of PSC for bone repair and regeneration. Enhanced osteogenic differentiation of mouse ESCs has been observed on nanofibrous matrices (Fig. 6a, b) as indicated by heightened expression of osteogenic markers [114, 170]. In another related study, Massumi et al. [92] showed that in synergy with other scaffold properties such as Matrigel coating, both the nanoroughness and alignment of PLGA nanofibrous scaffolds played a significant role in directing differentiation of mouse ESCs toward mesoderm lineages. Following their work showing the effect of symmetry of nanotopographical cues on osteogenic differentiation of MSCs [37], Dalby and colleagues recently extended their polycarbonate nanotopographical substrates for regulation of human ESC differentiation [162] (Fig. 6c, d). Human ESCs cultured on nanopit arrays exhibited enhanced expression of mesenchymal or stromal markers, resulting in improved production of osteogenic progenitors. Gene expression analysis confirmed that nanopit arrays did not induce endodermal or ectodermal lineage commitment. Interestingly, EB-based differentiations of human ESCs toward mesodermal lineages was less responsive to nanotopographical cues [162], suggesting efficient cell-substrate interactions critical for mediating stem cell nanotopography sensitivity.
Figure 6. Nanotopography regulates ESCs toward osteogenic lineage.
(a–b) Enhancing osteogenic differentiation of mouse ESCs by nanofibers matrix [114]. (a) SEM micrographs (top) of mouse ESCs after 12 hrs under differentiation conditions, calcium staining (middle) and immunofluorescence staining (bottom) images of late bone differentiation (Osteocalcin) marker expression after 26 days under osteogenic differentiation conditions on nanofibrous matrix, solid films, and gelatin-coated tissue culture plastic (Control). Quantitative PCR of osteogenic markers (b) osteocalcin (top) and bone sialoprotein (BSP; bottom) RNAs isolated from cells on nanofibrous matrix, solid films, and gelatin-coated tissue culture plastic (Control) after 26 days of culture under osteogenic differentiation conditions. Reproduced with permission [114]. Copyright 2009, Mary Ann Liebert, Inc. (c–d) Nanopit surfaces augment mesenchymal differentiation of Human ESCs [162]. (c) SEM image of polycarbonate nanopit substrate. Nanopits of 120 nm diameter and 100 nm depth are arranged in a near square geometry with centre-centre spacing 300 nm ± 50 nm. (d) Expression of endodermal (SOX17), ectodermal (Nestin) and osteogenic progenitor (ALCAM) markers was assessed by qPCR for self-renewing human ESCs, EBs and human ESCs seeded onto planar or nanopit substrates in basal media for 14 days (n = 6). Nanopit substrates do not direct endodermal or ectodermal differentiation, but significantly enhanced osteoblastic differentiation. * indicates p < 0.05, ** indicates p < 0.01. Reproduced with permission [162]. Copyright 2013, Wiley-VCH (Germany).
Seeding mouse ESCs on PLLA nanofibrous matrices resulted in upregulated β1 integrin expression, consistent with the important role of β1 integrins in ESC osteogenesis and mesodermal lineage commitment [178]. In another study, Chen et al. showed that nanotopography significantly affected the local molecular arrangement and formation of integrin-mediated FA that might in turn regulate the spatial organization of myosin II activity, CSK contractility and E-cadherin mediated intercellular adhesion of hESCs [80]. It suggests that the nanoscale topography in the stem cell niche influences the molecular organization of integrins and triggers integrin-mediated FA signaling to elicit downstream biochemical signals important for the regulation of gene expression and stem cell fate. Although similar effects were also observed for other biophysical factors like matrix rigidity [24, 161], nanotopography in the stem cell microenvironment might serve as a more direct cue to affect integrin conformation and clustering and thus organizations of adaptor and signaling proteins in FAs due to their similarity in size with integrins [13].
Molecular insights for stem cell nanotopography sensitivity
Much effort has been directed to study behaviors of stem cells in response to nanotopographic cues. However, molecular mechanisms underlying stem cell nanotopography sensitivity remain elusive. Some major questions remain unanswered including detailed molecular mechanisms relaying extracellular nanotopography to intracellular signaling pathways and regulatory networks controlling stem cell fate. Hereby, we review some prominent cellular mechanosensory machineries that have been implicated in stem cell nanotopography sensitivity. We discuss how intracellular signaling pathways downstream of these machineries relay extracellular nanotopography through cytoplasmic transducers, cytoskeletal integrators, and transcriptional actuators. Such an integrated mechanotransduction network may provide a roadmap for future studies to understand stem cell nanotopography sensitivity.
Adhesion-based mechanosensors and cytoplasmic transducers
Playing a pivotal role in cell-ECM adhesions, integrins are heterodimeric transmembrane proteins composed of an α and a β subunit. Currently, there are 24 known integrin heterodimers constituted by different combinations of 18 α and 8 β subunits [179–182]. Human ESCs express a broad range of integrins including α1, α2, α3, α5, α6, α7, α11, αV, αE,and β 1, β2, β3, β5, β6 integrins. Human iPSCs express mainly α5, α6, αV, β1, and β5 integrins, with variations among different iPSC lines [183–185]. There are abundant studies showing functional regulation of adult stem cells through integrin-mediated adhesion signaling [14, 37, 139, 186].
The first step adherent stem cells take to probe surrounding ECM is mediated by “inside-out” and “outside-in” bidirectional regulation of integrin-ligand binding [187–189]. Specifically, initial binding of integrin to adhesive molecules initiates clustering of intracellular adaptor proteins including talin, which in turn bind the cytoplasmic domain of integrin, resulting in a conformational change of integrin’s extracellular domain and its heightened affinity to ECM ligands. Firm binding of integrin to ECM ligand in turn activates integrin clustering through their intracellular domains and further recruits adaptor and signaling proteins to adhesion sites. Key molecular players in the early stage of integrin-mediated cell-ECM adhesion (also known as “focal adhesion”, or FA) include adaptor proteins talin and paxillin and tyrosine kinases such as focal adhesion kinase (FAK) and Src family kinases [190, 191].
Importantly, FA formation is sensitive to spatial arrangement and presentation of extracellular ECM ligands [192]. As the head of an integrin heterodimer is about 20 nm in diameter [13], a nanometer scale variation in substrate topography can directly affect integrin conformation and clustering and thus dynamic organization of adaptor and signaling proteins in FA. Results reported from different groups have suggested that successful integrin clustering requires a spacing between individual ECM ligands less than a threshold value between 50 – 70 nm, and a too sparse presentation of individual ligands inhibits integrin crosslinking and thus FA formation [124, 193–195]. Interestingly, simply clustering individual ECM ligands into oligomer nanopatterns without changing the overall ligand density still promotes FA formation and cell spreading, supporting the crucial role of nanoscale organization of integrins in FA in controlling cell behaviors [196]. In line with this observation, Huang et al. reported that a local disorder of ECM ligand arrangement could still promote integrin clustering when the average ligand spacing was greater than 70 nm., likely owing to the emergence of local sub-70 nm-threshold ligand spacing [197].
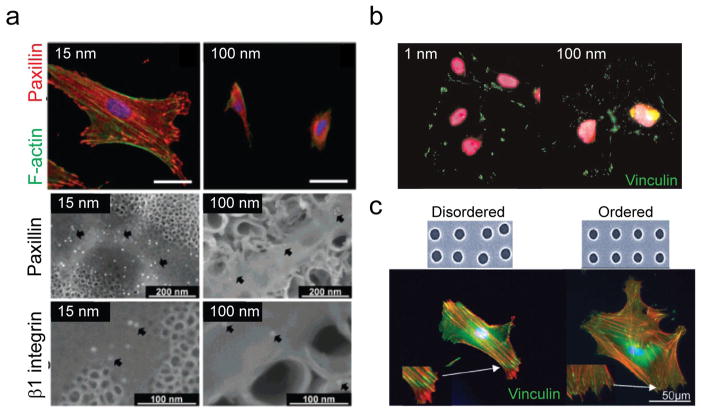
A few recent studies have directly highlighted the effect of nanotopography on ligand spacing/density and local order/disorder and thus integrin clustering and downstream stem cell behaviors. For example, Park et al. observed that the larger TiO2 nanotube diameter was (e.g., > 50 nm), the less occurrence of integrin clustering and FA formation was observed (Fig. 7a) [14]. In another study by Chen et al., it was also observed that FA formation was promoted in human ESCs cultured on nanotopographical surfaces with smaller (e.g., 1 nm), rather than greater (e.g., 100 nm), nanorough features (Fig. 7b) [80]. In a series of works published by Dalby and colleagues [186, 198], disordered arrangements of nanopits were observed to induce larger and more mature FAs with elongated morphology in human MSCs compared to ordered nanopit arrays (Fig. 7c). This observation again supports the mechanism proposed by Huang et al. [197] that local disorder of ECM presentation might promote the emergence of integrin activation and FA formation under a constant global average of ECM ligand density.
Figure 7. Sensitivity of integrin clustering and FA morphogenesis to nanotopography.
(a) (Top panel) Immunofluorescence images showing that TiO2 nanotube arrays of tube diameter 15 nm promoted the growth of large FAs and prominent F-actin stress fibers in MSCs; while larger tube diameter (100 nm) strongly inhibited FA maturation and resulted in diffusive actin staining. (Middle and bottom panels) SEM immunogold staining images showing that the density of FAs (middle) and β1 integrin clustering was much higher on nanotube arrays of 15 nm diameter TiO2 nanotubes. Reproduced with permission [14]. Copyright 2007, American Chemical Society. (b) Immunofluorescence images showing decreased FA sizes in human ESCs cultured on nanotopographical surfaces of 100 nm roughness features. Adapted with permission from [80]. Copyright 2012, American Society of Chemistry. (c) (Top panel) SEM images of nanopit arrays of disorder and ordered arrangements. (Bottom panel) Immunofluorescence images showing that larger and more elongated FAs formed on disordered nanopit arrays. Adapted from [186]. Copyright 2014, Wiley.
An important signaling axis downstream of integrin-mediated FA is the FAK-Src pathway [190, 199, 200]. Activated by integrin ligation, FAK is recruited to FA by binding integrin, which in turn activates FAK through autophosphorylation on Y397. Further phosphorylation of FAK is completed by binding of FAK to Src family proteins. Together, FAK and Src form a signaling complex that relays signals from integrin to control downstream Rho GTPase activities through, for instance, the FAK/p190RhoGEF/RhoA, FAK/p190RhoGAP/RhoA, and Grb2/SOS/Rac pathways [199]. Integrin-regulated FAK-Src signaling also mediates the MAPK pathways to regulate cell proliferation, apoptosis, and differentiation through the FAK/Ras/Raf/MEK/ERK, Fyn/Shc/Grb2/SOS/Ras, and p38 MAPK signaling [201–204].
Various studies have reported that nanotopographical substrates could promote FAK phosphorylation [205]. However, some other studies have suggested that maximal FAK phosphorylation and activation might occur at an intermediate nanotopographic level. For example, Park et al. reported that TiO2 nanotubes of 15 nm in diameter induced greater FAK phosphorylation compared to either smooth surfaces or TiO2 nanotube arrays of greater diameters [14]. Through FAK inhibition using small molecule drugs and siRNA, Teo et al. further showed that FAK was not only responsive to but also required for nanotopography sensitivity on 250 nm gratings and subsequent neurogenesis of human MSCs [205]. Importantly, constitutively activated FAK could rescue neurogenesis of human MSCs on smooth surfaces to an extent comparable to that on nanograting surfaces, further underscoring the critical role of FAK in stem cell nanotopography sensitivity. Given the dual, opposite effects of FAK on RhoA activity [199, 206, 207] (e.g., activation through p190RhoGEF [208, 209] or inactivation via p190RhoGAP [210, 211]), it remains an open question whether FAK phosphorylation induced by nanotopography could activate or inhibit RhoA activity.
Other signaling events downstream of integrin-mediated FAK-Src signaling, such as the FAK/MEK/ERK [14, 212], PI3K/Akt [213] and BMP and TGFβ/SMAD [214] pathways, which are important for cell proliferation, self-renewal, and differentiation [215–217], might also be involved in regulating nanotopography sensitivity of stem cells. For example, Park et al. observed maximal phosphorylation and activation of ERK in human MSCs on TiO2 nanotube arrays with a tube diameter of 15 nm [14]. Through whole proteome analysis, Kantawong et al. observed that disordered nanopit arrays elicited changes of protein expression involved in the ERK pathway for human MSCs [212]. Interestingly, Kim et al. reported that ERK in human MSCs was maximally activated on nanogroove substrates with an optimal groove width: spacing ratio (1:3 and 1:1), indicating a biphasic activating effect of nanotopography on ERK [218]. In another recent study, Yang et al. reported that disordered nanopit arrays promoted osteogenesis of human MSCs through co-localizing BMP receptors to integrins, upregulating expression of BMPs and BMP receptors, and thus enhancing transcriptional activity of SMAD1/5 and Runx2 [219]. Although it has not yet been demonstrated in nanotopography sensitivity per se, the Wnt/GSK3β signaling, which functions downstream of FAK/PI3K/Akt, was also implied by recent studies to be a potential regulator of differentiation, especially in human MSCs [220]. In light of these observations, a critical question to answer in future studies is how these different signaling pathways initiated by FA signaling and relayed by kinase cascades are interconnected and whether they converge on an integrative downstream signaling mechanism governing nanotopography sensitivity of stem cells.
Beside multifaceted FAK signaling, the RhoA/ROCK pathway originated from G-protein coupled receptor (GPCR) signaling [221–224] and downstream of integrin-mediated FA signaling has also been shown important for mechanotransduction in stem cells [223, 225]. When functioning, RhoA/ROCK signaling could feedback to mediate integrin-regulated adhesion signaling through its effect on the actin CSK contractility. Specifically, FAK activation downstream of integrin ligation activates Rho family GTPases, including RhoA, which in turn activates two effectors ROCK and mDia to regulate structural quality of filamentous actin CSK and contractile activity of non-muscle myosin II motor proteins, respectively [221]. Actin CSK contractility has been shown important for FA maturation and signaling, which in turn enhances RhoA/ROCK activation. Altogether, it implicates RhoA, ROCK, and CSK contractility as an interconnected, tripartite module responsible for nanotopography sensitivity. This hypothesis, in fact, is supported by a few recent studies. For example, McMurray et al. reported that ROCK activity and CSK contractility were required for sensing different arrangement orders of nanopit arrays by human MSCs, as their inhabitation by small molecule inhibitors Y27632 and blebbistatin, respectively, abolished nanotopography sensitivity of human MSCs [226]. Similarly, Teo et al. also showed that inhibition of either ROCK or CSK contractility was sufficient to inhibit nanotopography sensitivity of human MSCs on 250 nm nanogratings [227]. In addition, in a recent study by Chen et al., the authors demonstrated decreased CSK contractility on nanorough polymeric substrates, implying that stem cells might sense nanotopography through changes of CSK contractility [79]. However, it is still unclear whether such nanotopography-regulated CSK contractility is mediated directly through the RhoA/ROCK pathway.
Cytoskeletal integrators
Although mounting evidence has suggested involvements of integrin-mediated adhesion signaling and downstream effectors, such as FAK, ERK, RhoA/ROCK, and CSK contractility, in stem cell nanotopography sensitivity, a critical link between such diverse cytoplasmic signal transducers and downstream transcriptional regulators is stillmissing. Some important hints provided from mechanotransduction studies have supported the actin CSK and its integrity to serve as an integrator of the multitude of upstream signals relayed from extracellular mechanical cues including nanotopography (Fig. 8) [228]. Supporting this view, recent studies have demonstrated disorganized F-actin CSK and compromised CSK contractility in adherent cells cultured on nanorough substrates [80]. Herein we briefly review current understanding of the role of the actin CSK in mechanotransduction and speculate its possible involvement in stem cell nanotopography sensitivity through controlling downstream transcriptional activity of Hippo/YAP and MAL/SRF signaling. For detailed discussions about Hippo/YAP signaling, the readers can refer to excellent reviews published elsewhere [229–232].
Figure 8. Potential mechanotransduction mechanisms in cellular responsiveness to nanotopographical biomaterials.
(left) On pro-integrin clustering surfaces, which could be either smooth or composed of certain nanotopographical features, integrins could undergo free lateral recruitment and ligation with ECM proteins, and thus cluster and form mature, stable FAs together with the unhindered recruitment of FA adaptor proteins. This process initiates signals from plasma membrane, enhancing FAK/ERK signaling, as well as RhoA activity, which further promoting CSK contractility (through RhoA/ROCK) as well as stress fiber formation. CSK contractility, as an important mediator of mechanotransduction, could provide positive feedback to the FAs. In addition, CSK force could also induce phosphorylation of emerin, a nuclear envelope-located protein, and thus initiate nuclear mechanotransduction through enhanced nuclear actin polymerization and subsequent nuclear translocation of MAL, a transcription cofactor of SRF. In the meantime, prominent stress fibers formed in cells on smooth surfaces could promote the nuclear translocation of YAP/TAZ, a transcription co-factor of TEAD. Nuclear shuttling of SMAD 2/3 (R-SMAD), the transcription factor downstream of TGFβ signaling, is also controlled by YAP/TAZ translocation. (right) On surfaces containing anti-integrin clustering nanotopographical cues, although integrins could still freely diffuse laterally, the nanoscale surface features restrict the ligation of additional integrins to the ECM proteins, which further restrict successful recruitment and clustering of integrins and other FA proteins, resulting in smaller, less stable FAs. Such process disrupts the activation of RhoA and therefore limits the formation of stress fibers and might induce high cytoplasmic G-actin level, which inhibits the nuclear translocation of MAL and thus SRF signaling. Unlike nanotopographical surfaces that promote integrin clustering, anti-integrin clustering surfaces containing different types of nanoscale structures could not sustain FAK activation and downstream signaling. Last but not the least, compromised stress fibers and FAs on nanotopographical substrates could potentially intersect with Hippo/YAP pathway by enhancing YAP/TAZ phosphorylation via either Lats-dependent or –independent mechanisms, resulting in cytoplasmic retention of YAP/TAZ.
The Hippo/YAP pathway, which is important for organ size control and cancer [231, 233], has recently been identified as a downstream effector of the quality, dynamics as well as intrinsic contractility of the F-actin CSK [234–240]. Hippo/YAP signaling has also been shown important in controlling stem cell differentiation [240, 241]. Interestingly, experimental evidence has been accumulated implicating an emerging pattern of mechanical regulation of Hippo/YAP activity: extracellular mechanical cues (e.g., rigid substrate, external stretch, large cell size) resulting in prominent F-actin CSK formation promote high nuclear YAP activity, while those cues (e.g., soft substrate and small cell size) compromising the integrity of F-actin CSK lead to low YAP activity and dominant cytoplasmic YAP retention due to their association with the scaffold protein 14-3-3 [234, 235]. Such a role of the F-actin CSK as a potent mechanical signal integrator has been supported by recent studies in both 2D and 3D contexts on a myriad of cell behaviors and functions [235]. Furthermore, convergent signaling through the F-actin CSK to suppress YAP activity has also been shown involved in regulating mechanosensitive motor neuronal differentiation of human ESCs [24].
In addition to the Hippo/YAP pathway, the integrity and polymerization of actin CSK have also been implicated in regulating SRF (serum response factor) signaling [242, 243]. Specifically, enhanced polymerization of cytoplasmic actin monomers (G-actin) releases MAL, a transcription co-factor, from its association with G-actin, resulting in MAL nuclear translocation and thus elevated SRF signaling. Following this mechanism, large cell size with elevated cytoplasmic G-actin level has been found to inhibit SRF signaling and differentiation of epidermal stem cells [242].
Combining these observations with the recent finding of diffusive F-actin CSK in human ESCs cultured on nanotopographical substrates [80], it is tempting to speculate that nanotopography may either activate the actin CSK-dependent Hippo pathway and thus suppress nuclear YAP activity or inhibit nuclear translocation of MAL and thus SRF signaling (Fig. 8). Yet, how Hippo/YAP and/or MAL/SRF signaling are involved in stem cell nanotopography sensitivity has not been specifically examined so far. It is worth noting that although cellular sensing of nanotopography and other extracellular mechanical cues (such as substrate mechanics, geometric confinements, and surface electric charge, as reviewed by Higuchi et al. [244]) might share common signaling pathways, like those already been shown via RhoA/ROCK, FAK, and CSK contractility, a possibility exists that nanotopography sensitivity might elicit certain unique intracellular mechanisms. It calls for future careful examinations of the actin CSK and its integrity and their functional relationship with Hippo/YAP and MAL/SRF signaling in stem cell nanotopography sensitivity.
Nuclear mechanosensors and transcriptional actuators
Although deeply embedded within a cell, the nucleus is an emerging, active mechanosensor [33, 245]. As the most important mechanical component of the nucleus, the nuclear lamina is composed mainly of intermediate filament proteins lamin A and C, endowing a significant mechanical stiffness to the nucleus. Lamin A/C has been demonstrated critical for mechanotransduction and transcription regulation through signaling molecules such as MAL, emerin, and F-/G-actin [12, 33, 246–248]. Importantly, the nucleus of ESCs, which lacks lamin A/C, is extremely deformable, and emergence of lamin A/C in the nucleus has been proposed as an ESC differentiation marker [249].
Through its connection to the actin CSK via KASH and SUN domain proteins (together known as the LINC - linker of nucleoskeleton and cytoskeleton - complex) across the nuclear envelope as well as perinuclear adaptor proteins such as nesprin and plectin, the nuclear lamina is structurally connected to the mechanoregulatory network of actin filaments, microtubules, and intermediate filaments [33, 250, 251]. Thus, it is not surprising that the nucleus itself serves an important role in mechanoresponsive stem cell fate regulation. Herein, we briefly review nuclear mechanotransduction mechanisms involved in cellular sensing of mechanical cues such as substrate mechanics and external forces, in the hope of providing prospective mechanisms for future studies on the role of nuclear mechanotransduction in stem cell nanotopography sensitivity, which has so far remained a largely unexplored field.
Using a cysteine shot gun technique in combination with in vitro shear force application [252], Discher and colleagues identified a group of proteins, including lamin A, that could change conformation and expose cryptic hydrophobic domains under unfolding forces [253]. Recently, the same authors observed a positive scaling relation between nuclear lamin expression and in vivo ECM rigidity, suggesting a functional link between rigidity-dependent CSK contractility and force-dependent nuclear lamin expression and function [253]. Nuclear mechanical stiffness, as endowed mainly by lamin A/C expression, thus may scale with ECM rigidity, consistent with a recent study by Liu and colleagues using nanoindentation [254].
Recently, Poh et al. observed that an intact linkage between the actin CSK and nucleoskeleton was required for transmission of integrin-mediated external forces from the cytoplasm membrane to the Cajal bodies in the nucleus to directly dissociate coilin and SMN protein complexes [255]. Furthermore, transmission of CSK tension within actin stress fibers and microtubules has been found critical for mediating long-range force-sensing involving the nucleus [251, 256]. In addition, it has been reported that confining adherent cell shapes to high aspect ratio morphology results in a corresponding high aspect ratio of the nucleus and prominent chromatin condensation [257].
In another very recent study, the inner nucleus membrane-located protein, emerin, is demonstrated to be mechanosensitive and undergo force-dependent phosphorylation [258]. Using isolated nuclei, this study demonstrated nuclear stiffening in response to external cyclic pulling forces and its regulation by recruitment of lamin A/C through Src-mediated tyrosine-phosphorylated emerins under tension. Interestingly, such force-mediated emerin phosphorylation depended on both substrate rigidity and mechanical forces applied through integrin ligation.
So far, there are few studies of nuclear mechanotransduction in the context of stem cell nanotopography sensitivity. Although it has been reported that nanotopography can induce nucleus deformation [104, 259], detailed examinations of such nanotopography-dependent nuclear events have not yet been demonstrated until very recently. Specifically, Dalby and colleagues have reported mechanosensitive chromosome positioning in response to disordered nanopit arrays, wherein nanotopographic cue poses chromosome 1 closer to the nuclear membrane and increases its mean inter-territory distance during human MSC osteogenesis [260]. The authors have also observed that positions of gene regulation along the chromosome are sensitive to nanotopography [261], with nanotopography-enhanced gene regulation concentrated toward the telomeric end of the chromosome, where osteogenesis-related genes are clustered [260].
A few mechanosensitive nuclear actuators relaying extracellular mechanical signals to transcription activities in the nucleus have been identified recently. As a nuclear transcription co-factor, YAP/TAZ can bind transcription factors TEAD as well as co-translocate SMAD 2/3 (regulatory SMAD, R-SMAD) to the nucleus, thus relaying cytoplasmic mechanotransductive signals to transcriptional control machineries [24, 238, 262, 263]. Importantly, the nucleus protein emerin and nuclear localization of lamin A/C are both critical for MAL/SRF signaling in the nucleus [248, 267]. In particular, emerin promotes nuclear actin polymerization, which in turn prohibits MAL nuclear export and thus promotes transcriptional activity of MAL. In addition, emerin phosphorylation is correlated with both substrate rigidity and nuclear YAP localization and activity [258]. Mechanical tension through nesprin-1 on isolated nuclei can also induce RhoA activation in the nucleus, although the exact role of RhoA in transcriptional control remains unclear [258]. Together, it is foreseeable that mechanistic studies of nanotopography sensitivity of stem cells involving nuclear deformation, nuclear envelope mechanics, lamin A/C expression, nuclear lamina protein expression and phosphorylation, and nuclear transcription activation will help elucidate nuclear mechanosensitive players completing the signaling axis through which substrate nanotopography controls stem cell fate (Fig. 7).
Conclusion remarks and outlook
Stem cells are promising cell sources for tissue engineering, regenerative medicine, drug screening, and toxicity assays. The major challenge faced by stem cell biologists and bioengineers is the large-scale, long-term stem cell maintenance and high-specificity, high-yield, directed stem cell differentiation toward clinically relevant lineages with mature tissue-specific functions. Rapid advances in nanotechnology and materials science have leveraged regulation of stem cell fate and function via microenvironmental physical factors such as nanotopographical cues to enhance functional performances of synthetic nanoengineered biomaterials in stem cell engineering.
In this review, we have summarized state of the art nanofabrication methods for generating nanotopographical biomaterials and surfaces for regulating stem cell fate. Different synthetic nanotopographical biomaterials have been successfully developed to control behaviors and phenotypes of stem cells, such as adhesion, migration, and morphology, and eventually determine transcriptional activities and stem cell fate. By designing nanotopographical features mimicking in vivo stem cell niches, a diverse toolbox of nanotopographical biomaterials have enabled either long-term maintenance or directed lineage commitment of stem cells, producing promising solutions for stem cell-based applications. Although successful as a proof of concept, static nanotopographical biomaterials alone may not sufficiently dictate stem cell fate desired for different applications. Soluble biochemical cues, dynamic control and regulation of topographical features, as well as cell co-culture systems, have all been demonstrated to play in synergy with physical cues in regulating stem cell fate [11, 12, 89, 166, 167, 268, 269]. In the future, design and fabrication of nanotopographical biomaterials will need to be integrated with multifunctional biochemical modifications and even spatiotemporal patterning for simultaneously control of multiple aspects of in vitro stem cell microenvironment and fine tuning of specific terminal lineage commitment at a large scale.
Besides controlled stem cell differentiation, new strategies to support long-term stem cell self-renewal and large-scale expansion (maintaining an undifferentiated stem cell state without spontaneous differentiation) will be crucial to obtain sufficient cell source for stem cell-based regenerative therapies. While it is currently still challenging to design well-defined biomaterials and protocols promoting stem cell culture, nanotopographical materials have been shown great potential to form a favorable microenvironment for maintenance of stem cell potencies. Such approaches, alongside the potential of nanotopography to modulate stem cell functions and to mimic the stem cell niche, remain to be investigated in depth.
It is also worth noting that possible variance in nanotopography sensitivity might occur as a consequence of studies using different cell lines with distinct culture conditions and genetic origins and from different species. For example, rate and human MSCs have been shown distinct preferences for different nanotopographic feature sizes for optimal osteogenic differentiation [14, 139]. There are also a growing number of studies indicating differences between iPSCs and ESCs [270, 271]. As such, it is important that multiple cell lines from the same species are examined and compared in the same set of nanotopography studies. Especially for PSCs, iPSCs are always recommended to be investigated alongside ESCs.
Another important direction in stem cell research is induction of pluripotency or transdifferentiation of one cell type to another [272, 273]. Abundant evidence has shown potent regulation of adult stem cell function and signaling by nanotopography sensitivity, yet questions of whether and how nanotopography at the cell-substrate interface can be leveraged for improving cell reprogramming and transdifferentiation remain unexplored. Such studies will potentially provide novel approaches to improve efficiency of reprogramming and transdifferentiation processes and yield significant mechanistic insights related to mechanoregulation of development and diseases [9, 274–276].
To improve performance of nanotopography-driven stem cell fate regulation, it is imperative to understand the mechano-sensing and -transductive mechanisms underlying cellular responses to extracellular nanotopography. In this review, we have discussed a few key mechano-sensing and -transductive mechanisms implicated in regulation of stem cell fate via nanotopographical biomaterials. Based on current understanding of stem cell mechanobiology, we have also proposed a few speculations regarding how different mechanosensitive signaling events may eventually be integrated and converge onto several core mechanotransduction axes, which constitute the “chain of commands” from extracellular nanotopography to nuclear gene expression. Future studies of nanotopography-responsive stem cell behaviors may have to involve spatiotemporal dynamics and regulations of molecular mechanosensors, transducers, integrators, and actuators residing in different cellular compartments and organelles. Elucidating interconnections and cross-regulations between each component involved in mechanotransduction remain the central goal for the field to address in the future. Multidisciplinary approaches merging stem cell biology, materials science, biomedical engineering, and nanotechnology will be the most promising and powerful route toward such goal.
Supplementary Material
Highlights.
Synthetic nanotopographical surfaces for stem cell fate control.
Emerging nanotechnologies for generation of nanotopographical surfaces.
Cellular mechano-sensing and -transduction mechanisms for nanotopography sensing.
Acknowledgments
We acknowledge financial support from the National Science Foundation (CMMI 1129611 and CBET 1149401), the National Institutes of Health (R21 HL114011 and R21 EB017078), the American Heart Association (12SDG12180025), the UM Comprehensive Cancer Center Prostate SPORE Pilot Project (P50 CA069568), and the Michigan Institute for Clinical & Health Research (MICHR) Pilot Program (UL1 RR024986). W. Chen is supported in part by the American Heart Association Predoctoral Fellowship (13PRE16510018). Finally, we extend our apologies to all our colleagues in the field whose work we are unable to discuss formally because of space constraints.
Biographies
Dr. Weiqiang Chen has been an assistant professor of Mechanical and Aerospace Engineering at New York University since 2014. Dr. Chen received his Ph.D. in Mechanical Engineering from the University of Michigan in 2014. He was an American Heart Association Predoctoral Fellow at the University of Michigan from 2013 to 2014. Dr. Chen’s research interests focus on Bio-Microelectromechanical Systems (BioMEMS), Lab-on-a-Chip, biomaterials, mechanobiology and stem cell biology.

Yue Shao is a PhD candidate in the Department of Mechanical Engineering at the University of Michigan, Ann Arbor. He received his B.E. and M.E. degrees on engineering mechanics and solid mechanics from Tsinghua University, China, in 2008 and 2011, respectively. His current research interests include biomechanics of cellular mechanosensing and mechanotransduction of extracellular physical cues, stem cell mechanobiology, and micro/nanoengineering ex vivo cell microenvironment for regenerative medicine.

Xiang Li is a PhD candidate in the Department of Mechanical Engineering at the University of Michigan, Ann Arbor. He received his B.S. degree in theoretical and applied mechanics from Peking University in 2012. His current research interest is to develop novel microengineering tools for single cell studies and tissue engineering.

Dr. Gang Zhao has been a professor at Biomedical Engineering Research Center of University of Science and Technology of China (USTC) since April 2011. Dr. Zhao received his Ph.D. degree in power engineering and engineering thermophysics from USTC in June 2004. He was a postdoctor at USTC from July 2004 to September 2006, and a JSPS (Japan Society for Promotion of Science) research fellow from July 2008 to October 2010. His research interests include cryobiology, biomicro/nanofluidics and biomedical microsystems. He received the CAREER Award for Young Teachers (CAYT) of USTC in 2013.

Dr. Jianping Fu has been an assistant professor of Mechanical and Biomedical Engineering at the University of Michigan, Ann Arbor since 2009. Dr. Fu received his Ph.D. degree from the Massachusetts Institute of Technology in 2007. He was an American Heart Association Postdoctoral Fellow at the University of Pennsylvania from 2007 to 2009. Dr. Fu’s research focuses on mechanobiology, stem cell biology, and applying microfabrication technology to illuminate biological systems at both the molecular and cellular levels. Dr. Fu is the recipient of the American Heart Association Scientist Development Grant (2012) and the National Science Foundation CAREER Award (2012).

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daley GQ, Scadden DT. Cell. 2008;132:544–548. doi: 10.1016/j.cell.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Nishikawa SI, Jakt LM, Era T. Nat Rev Mol Cell Biol. 2007;8:502–507. doi: 10.1038/nrm2189. [DOI] [PubMed] [Google Scholar]
- 3.Jopling C, Boue S, Belmonte JCI. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 4.Singec I, Jandial R, Crain A, Nikkhah G, Snyder EY. Annu Rev Cell Dev Biol. 2007;58:313–328. doi: 10.1146/annurev.med.58.070605.115252. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien LE, Bilder D. Annu Rev Cell Dev Biol. 2013;29:107–136. doi: 10.1146/annurev-cellbio-101512-122319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadig RR. J Conserv Dent. 2009;12:131–138. doi: 10.4103/0972-0707.58329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young HE, Black AC. Anat Rec Part A. 2004;276A:75–102. doi: 10.1002/ar.a.10134. [DOI] [PubMed] [Google Scholar]
- 8.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Murry CE, Keller G. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Tenney RM, Discher DE. Curr Opin Cell Biol. 2009;21:630–635. doi: 10.1016/j.ceb.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. J Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi J, Petre BM, Walz T, Springer TA. Cell. 2002;110:599–511. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 14.Park J, Bauer S, von der Mark K, Schmuki P. Nano Lett. 2007;7:1686–1691. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 15.Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng PN, Traas J, Schugar R, Deasy BM, Badylak S, Buhring HJ, Giacobino JP, Lazzari L, Huard J, Peault B. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Kingham E, Oreffo RO. ACS Nano. 2013;7:1867–1881. doi: 10.1021/nn3037094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HN, Jiao A, Hwang NS, Kim MS, Kang DH, Kim DH, Suh KY. Adv Drug Deliver Rev. 2013;65:536–558. doi: 10.1016/j.addr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das RK, Zouani OF, Labrugere C, Oda R, Durrieu MC. ACS Nano. 2013;7:3351–3361. doi: 10.1021/nn4001325. [DOI] [PubMed] [Google Scholar]
- 19.Dalby M, Gadegaard N, Oreffo R. Nature Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 20.McNamara LE, Sjostrom T, Seunarine K, Meek RD, Su B, Dalby MJ. J Tissue Eng. 2014;5:2041731414536177. doi: 10.1177/2041731414536177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemischka IR, Moore KA. Nature. 2003;425:778–779. doi: 10.1038/425778a. [DOI] [PubMed] [Google Scholar]
- 22.Spradling A, Drummond-Barbosa D, Kai T. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, Chen CS, Fu J. Annu Rev Biophys. 2012;41:519–542. doi: 10.1146/annurev-biophys-042910-155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Nature Mater. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim DH, Provenzano PP, Smith CL, Levchenko A. J Cell Biol. 2012;197:351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu KC, Tseng CL, Wu CC, Kao FC, Tu YK, So EC, Wang YK. Sci Technol Adv Mater. 2013;14:054401. doi: 10.1088/1468-6996/14/5/054401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 29.Geiger B, Spatz JP, Bershadsky AD. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 30.DuFort CC, Paszek MJ, Weaver VM. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Tytell JD, Ingber DE. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 32.Buxboim A, Ivanovska I, Discher D. J Cell Sci. 2010;123:297–308. doi: 10.1242/jcs.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isermann P, Lammerding J. Curr Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettinger CJ, Zhang ZT, Gerecht S, Borenstein JT, Langer R. Adv Mater. 2008;20:99–103. doi: 10.1002/adma.200702487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Ahmed H. Appl Phys Lett. 1993;62:1499–1501. [Google Scholar]
- 36.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. J Cell Sci. 2003;116:1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. Nature Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 38.Gallagher J, McGhee K, Wilkinson C, Riehle M. IEEE Trans Nanobiosci. 2002;1:24–28. doi: 10.1109/tnb.2002.806918. [DOI] [PubMed] [Google Scholar]
- 39.Yang SM, Jang SG, Choi DG, Kim S, Yu HK. Small. 2006;2:458–475. doi: 10.1002/smll.200500390. [DOI] [PubMed] [Google Scholar]
- 40.Zhang G, Wang DY. Chem Asian J. 2009;4:236–245. doi: 10.1002/asia.200800298. [DOI] [PubMed] [Google Scholar]
- 41.Bhawalkar SP, Qian J, Heiber MC, Jia L. Langmuir. 2010;26:16662–16666. doi: 10.1021/la1035147. [DOI] [PubMed] [Google Scholar]
- 42.Iovan A, Fischer M, Lo Conte R, Korenivski V. Beilstein J Nanotechnol. 2012;3:884–892. doi: 10.3762/bjnano.3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalby MJ, Berry CC, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Exp Cell Res. 2004;295:387–394. doi: 10.1016/j.yexcr.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis AS. Biomaterials. 2004;25:5415–5422. doi: 10.1016/j.biomaterials.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 45.Jaeger RC. Introduction to microelectronic fabrication. 2. Prentice Hall; Upper Saddle River, N.J: 2002. [Google Scholar]
- 46.Jain K, Willson CG, Lin BJ. Electron Device Lett. 1982;3:53–55. [Google Scholar]
- 47.Jain K, Kerth RT. Appl Optics. 1984;23:648–650. doi: 10.1364/ao.23.000648. [DOI] [PubMed] [Google Scholar]
- 48.Wong AK-K. Resolution enhancement techniques in optical lithography. SPIE Press; Bellingham, Wash: 2001. [Google Scholar]
- 49.Gerecht S, Bettinger CJ, Zhang Z, Borenstein JT, Vunjak-Novakovic G, Langer R. Biomaterials. 2007;28:4068–4077. doi: 10.1016/j.biomaterials.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieu C, Carcenac F, Pepin A, Chen Y, Mejias M, Lebib A, Manin-Ferlazzo L, Couraud L, Launois H. Appl Surf Sci. 2000;164:111–117. [Google Scholar]
- 51.Unadkat HV, Hulsman M, Cornelissen K, Papenburg BJ, Truckenmuller RK, Post GF, Uetz M, Reinders MJT, Stamatialis D, van Blitterswijk CA, de Boer J. Proc Natl Acad Sci U S A. 2011;108:16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn EH, Kim Y, Kshitiz, An SS, Afzal J, Lee S, Kwak M, Suh KY, Kim DH, Levchenko A. Biomaterials. 2014;35:2401–2410. doi: 10.1016/j.biomaterials.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood M, Meredith D, Owen G. IEEE Trans Nanobiosci. 2002;1:133–140. doi: 10.1109/tnb.2003.809460. [DOI] [PubMed] [Google Scholar]
- 54.Andersson AS, Brink J, Lidberg U, Sutherland DS. IEEE Trans Nanobiosci. 2003;2:49–57. doi: 10.1109/tnb.2003.813934. [DOI] [PubMed] [Google Scholar]
- 55.Wood M, Wilkinson C, Curtis A. IEEE Trans Nanobiosci. 2006;5:20–31. doi: 10.1109/tnb.2005.864015. [DOI] [PubMed] [Google Scholar]
- 56.Wood MA. J R Soc Interface. 2007;4:1–17. doi: 10.1098/rsif.2006.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji L, LaPointe VL, Evans ND, Stevens MM. Eur Cell Mater. 2012;23:135–146. doi: 10.22203/ecm.v023a10. [DOI] [PubMed] [Google Scholar]
- 58.Solanki A, Shah S, Yin P, Lee K-B. Sci Rep. 2013;3:1553. doi: 10.1038/srep01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grego S, Jarvis TW, Stoner BR, Lewis JS. Langmuir. 2005;21:4971–4975. doi: 10.1021/la0468850. [DOI] [PubMed] [Google Scholar]
- 60.Kong Y, Tu C, Donovan P, Yee A. Acta Biomater. 2013;9:6369–6380. doi: 10.1016/j.actbio.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 61.Malmström J, Lovmand J, Kristensen S, Sundh M, Duch M, Sutherland D. Nano Lett. 2011;11:2264–2271. doi: 10.1021/nl200447q. [DOI] [PubMed] [Google Scholar]
- 62.Ng V, Lee YV, Chen BT, Adeyeye AO. Nanotechnology. 2002;13:554–558. [Google Scholar]
- 63.Chou SY, Krauss PR, Renstrom PJ. Science. 1996;272:85–87. [Google Scholar]
- 64.Guo LJ. J Phys D Appl Phys. 2004;37:R123–R141. [Google Scholar]
- 65.Schift H. J Vac Sci Technol B. 2008;26:458–480. [Google Scholar]
- 66.Austin MD, Ge HX, Wu W, Li MT, Yu ZN, Wasserman D, Lyon SA, Chou SY. Appl Phys Lett. 2004;84:5299–5301. [Google Scholar]
- 67.Tan H, Kong L, Li MT, Steere C, Koecher L. Emerging Lithographic Technologies Viii. 2004;5374:213–221. [Google Scholar]
- 68.Yang K, Jung K, Ko E, Kim J, Park KI, Kim J, Cho S-W. ACS Appl Mater Interfaces. 2013;5:10529–10540. doi: 10.1021/am402156f. [DOI] [PubMed] [Google Scholar]
- 69.Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Biomaterials. 2005;26:5405–5413. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Padmanabhan J, Kinser E, Stalter M, Duncan-Lewis C, Balestrini J, Sawyer A, Schroers J, Kyriakides T. ACS Nano. 2014;8:4366–4375. doi: 10.1021/nn501874q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xia Y, Whitesides GM. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 72.Qin D, Xia Y, Whitesides G. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 73.Odom TW, Love CJ, Wolfe DB, Paul KE, Whitesides GM. Langmuir. 2002;18:5314–5320. [Google Scholar]
- 74.Kim DH, Han K, Gupta K, Kwon KW, Suh KY, Levchenko A. Biomaterials. 2009;30:5433–5444. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yim EKF, Pang SW, Leong KW. Exp Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palik ED, Glembocki OJ, Heard I, Burno PS, Tenerz L. J Appl Phys. 1991;70:3291–3300. [Google Scholar]
- 77.Thapa A, Webster TJ, Haberstroh KM. J Biomed Mater Res A. 2003;67A:1374–1383. doi: 10.1002/jbm.a.20037. [DOI] [PubMed] [Google Scholar]
- 78.Boyan BD, Batzer R, Kieswetter K, Liu Y, Cochran DL, Szmuckler-Moncler S, Dean DD, Schwartz Z. J Biomed Mater Res. 1998;39:77–85. doi: 10.1002/(sici)1097-4636(199801)39:1<77::aid-jbm10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 79.Chen W, Sun Y, Fu J. Small. 2013;9:81–89. doi: 10.1002/smll.201201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen W, Villa-Diaz L, Sun Y, Weng S, Kim J, Lam R, Han L, Fan R, Krebsbach P, Fu J. ACS Nano. 2012;6:4094–4103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zubel I, Kramkowska M. Sensor Actuat A Phys. 2001;93:138–147. [Google Scholar]
- 82.Metwalli E, Pantano CG. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms. 2003;207:21–27. doi: 10.1016/s0168-583x(01)00693-0. [DOI] [PubMed] [Google Scholar]
- 83.Leech PW. Vacuum. 1999;55:191–196. [Google Scholar]
- 84.Bhardwaj N, Kundu SC. Biotechnol Adv. 2010;28:325–347. doi: 10.1016/j.biotechadv.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 85.Li D, Xia YN. Adv Mater. 2004;16:1151–1170. [Google Scholar]
- 86.Greiner A, Wendorff JH. Angew Chem Int Edit. 2007;46:5670–5703. doi: 10.1002/anie.200604646. [DOI] [PubMed] [Google Scholar]
- 87.Yang F, Murugan R, Wang S, Ramakrishna S. Biomaterials. 2005;26:2603–2610. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 88.He L, Liao S, Quan D, Ma K, Chan C, Ramakrishna S, Lu J. Acta Biomater. 2010;6:2960–2969. doi: 10.1016/j.actbio.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 89.Kabiri M, Soleimani M, Shabani I, Futrega K, Ghaemi N, Ahvaz HH, Elahi E, Doran MR. Biotechnol Lett. 2012;34:1357–1365. doi: 10.1007/s10529-012-0889-4. [DOI] [PubMed] [Google Scholar]
- 90.Kijenska E, Prabhakaran MP, Swieszkowski W, Kurzydlowski KJ, Ramakrishna S. J Biomed Mater Res Part B Appl Biomater. 2012;100:1093–1102. doi: 10.1002/jbm.b.32676. [DOI] [PubMed] [Google Scholar]
- 91.Prabhakaran MP, Venugopal JR, Ramakrishna S. Biomaterials. 2009;30:4996–5003. doi: 10.1016/j.biomaterials.2009.05.057. [DOI] [PubMed] [Google Scholar]
- 92.Massumi M, Abasi M, Babaloo H, Terraf P, Safi M, Saeed M, Barzin J, Zandi M, Soleimani M. Tissue Eng Part A. 2012;18:609–620. doi: 10.1089/ten.tea.2011.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Cell Tissue Res. 2000;299:39–46. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 94.Schindler M, Ahmed I, Kamal J, Nur-E-Kamal A, Grafe TH, Chung HY, Meiners S. Biomaterials. 2005;26:5624–5631. doi: 10.1016/j.biomaterials.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 95.Gandhimathi C, Venugopal J, Ravichandran R, Sundarrajan S, Suganya S, Ramakrishna S. Macromol Biosci. 2013;13:696–706. doi: 10.1002/mabi.201200435. [DOI] [PubMed] [Google Scholar]
- 96.Yoshimoto H, Shin YM, Terai H, Vacanti JP. Biomaterials. 2003;24:2077–2082. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 97.Li WJ, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ, Tuan RS. Biomaterials. 2005;26:599–609. doi: 10.1016/j.biomaterials.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 98.Lim SH, Liu XY, Song H, Yarema KJ, Mao HQ. Biomaterials. 2010;31:9031–9039. doi: 10.1016/j.biomaterials.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gauthaman K, Venugopal JR, Yee FC, Peh GS, Ramakrishna S, Bongso A. J Cell Mol Med. 2009;13:3475–3484. doi: 10.1111/j.1582-4934.2009.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie JW, Willerth SM, Li XR, Macewan MR, Rader A, Sakiyama-Elbert SE, Xia YN. Biomaterials. 2009;30:354–362. doi: 10.1016/j.biomaterials.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Christopherson GT, Song H, Mao HQ. Biomaterials. 2009;30:556–564. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 102.Chua KN, Chai C, Lee PC, Tang YN, Ramakrishna S, Leong KW, Mao HQ. Biomaterials. 2006;27:6043–6051. doi: 10.1016/j.biomaterials.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 103.Liu L, Yuan Q, Shi J, Li X, Jung D, Wang L, Yamauchi K, Nakatsuji N, Kamei K, Chen Y. Biotechnol Lett. 2012;34:1951–1957. doi: 10.1007/s10529-012-0973-9. [DOI] [PubMed] [Google Scholar]
- 104.Dang JM, Leong KW. Adv Mater. 2007;19:2775–2779. doi: 10.1002/adma.200602159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu LX, Zhang XF, Wang YY, Ortiz L, Mao X, Jiang ZL, Xiao ZD, Huang NP. ACS Appl Mater Interfaces. 2013;5:319–330. doi: 10.1021/am302146w. [DOI] [PubMed] [Google Scholar]
- 106.Peng H, Yin Z, Liu H, Chen X, Feng B, Yuan H, Su B, Ouyang H, Zhang Y. Nanotechnology. 2012;23:485102. doi: 10.1088/0957-4484/23/48/485102. [DOI] [PubMed] [Google Scholar]
- 107.Patel S, Kurpinski K, Quigley R, Gao H, Hsiao B, Poo M-M, Li S. Nano Lett. 2007;7:2122–2128. doi: 10.1021/nl071182z. [DOI] [PubMed] [Google Scholar]
- 108.Qu J, Wang D, Wang H, Dong Y, Zhang F, Zuo B, Zhang H. J Biomed Mater Res A. 2013;101:2667–2678. doi: 10.1002/jbm.a.34551. [DOI] [PubMed] [Google Scholar]
- 109.Nam YS, Park TG. J Biomed Mater Res. 1999;47:8–17. doi: 10.1002/(sici)1097-4636(199910)47:1<8::aid-jbm2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 110.Zhang RY, Ma PX. J Biomed Mater Res. 1999;44:446–455. doi: 10.1002/(sici)1097-4636(19990315)44:4<446::aid-jbm11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 111.Lee SH, Kim BS, Kim SH, Kang SW, Kim YH. Macromol Biosci. 2004;4:802–810. doi: 10.1002/mabi.200400021. [DOI] [PubMed] [Google Scholar]
- 112.Smith LA, Liu XH, Ma PX. Soft Matter. 2008;4:2144–2149. doi: 10.1039/b807088c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ma PX, Zhang R. J Biomed Mater Res. 1999;46:60–72. doi: 10.1002/(sici)1097-4636(199907)46:1<60::aid-jbm7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 114.Smith LA, Liu X, Hu J, Wang P, Ma PX. Tissue Eng Part A. 2009;15:1855–1864. doi: 10.1089/ten.tea.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Affrossman S, Henn G, ONeill SA, Pethrick RA, Stamm M. Macromolecules. 1996;29:5010–5016. [Google Scholar]
- 116.Walheim S, Boltau M, Mlynek J, Krausch G, Steiner U. Macromolecules. 1997;30:4995–5003. [Google Scholar]
- 117.Dalby MJ, Riehle MO, Johnstone HJH, Affrossman S, Curtis ASG. Tissue Eng. 2002;8:1099–1108. doi: 10.1089/107632702320934191. [DOI] [PubMed] [Google Scholar]
- 118.Dalby MJ, Giannaras D, Riehle MO, Gadegaard N, Affrossman S, Curtis AS. Biomaterials. 2004;25:77–83. doi: 10.1016/s0142-9612(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 119.Dalby MJ, Pasqui D, Affrossman S. IEEE Proc Nanobiotechnol. 2004;151:53–61. doi: 10.1049/ip-nbt:20040534. [DOI] [PubMed] [Google Scholar]
- 120.Sprenger M, Walheim S, Schafle C, Steiner U. Adv Mater. 2003;15:703–706. [Google Scholar]
- 121.Park M, Harrison C, Chaikin PM, Register RA, Adamson DH. Science. 1997;276:1401–1404. [Google Scholar]
- 122.Hawker CJ, Russell TP. MRS Bull. 2005;30:952–966. [Google Scholar]
- 123.Hamley IW. Nanotechnology. 2003;14:R39–R54. [Google Scholar]
- 124.Glass R, Möller M, Spatz J. Nanotechnology. 2003;14:1153. [Google Scholar]
- 125.Maclaine SE, Gadhari N, Pugin R, Meek RMD, Liley M, Dalby MJ. J Orthopaed Res. 2012;30:1190–1197. doi: 10.1002/jor.22076. [DOI] [PubMed] [Google Scholar]
- 126.Salaun M, Zelsmann M, Archambault S, Borah D, Kehagias N, Simao C, Lorret O, Shaw MT, Torres CMS, Morris MA. J Mater Chem C. 2013;1:3544–3550. [Google Scholar]
- 127.Khor HL, Kuan Y, Kukula H, Tamada K, Knoll W, Moeller M, Hutmacher DW. Biomacromolecules. 2007;8:1530–1540. doi: 10.1021/bm0611533. [DOI] [PubMed] [Google Scholar]
- 128.Haupt M, Miller S, Glass R, Arnold M, Sauer R, Thonke K, Moller M, Spatz JP. Adv Mater. 2003;15:829–831. [Google Scholar]
- 129.Ku SJ, Woo SA, Seo D, Song H, Kim JB. Eur Polym J. 2011;47:305–310. [Google Scholar]
- 130.Reynhout IC, Delaittre G, Kim HC, Nolte RJM, Cornelissen JJLM. J Mater Chem B. 2013;1:3026–3030. doi: 10.1039/c3tb20411a. [DOI] [PubMed] [Google Scholar]
- 131.Lee K, Mazare A, Schmuki P. Chem Rev. 2014;114:9385–9454. doi: 10.1021/cr500061m. [DOI] [PubMed] [Google Scholar]
- 132.Zhou XM, Nguyen NT, Ozkan S, Schmuki P. Electrochem Commun. 2014;46:157–162. [Google Scholar]
- 133.Yao C, Webster TJ. J Nanosci Nanotechno. 2006;6:2682–2692. doi: 10.1166/jnn.2006.447. [DOI] [PubMed] [Google Scholar]
- 134.Yao C, Perla V, McKenzie JL, Slamovich EB, Webster TJ. J Biomed Nanotechnol. 2005;1:68–73. [Google Scholar]
- 135.Oh SH, Finones RR, Daraio C, Chen LH, Jin SH. Biomaterials. 2005;26:4938–4943. doi: 10.1016/j.biomaterials.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 136.Ohgai T, Hoffer X, Gravier L, Ansermet JP. J Appl Electrochem. 2004;34:1007–1012. [Google Scholar]
- 137.Watanabe K, Sakairi M, Takahashi H, Hirai S, Yamaguchi S. J Electroanal Chem. 1999;473:250–255. [Google Scholar]
- 138.Tsuchiya H, Macak JM, Ghicov A, Tang YC, Fujimoto S, Niinomi M, Noda T, Schmuki P. Electrochim Acta. 2006;52:94–101. [Google Scholar]
- 139.Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, Jin S. Proc Natl Acad Sci U S A. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sjostrom T, Dalby MJ, Hart A, Tare R, Oreffo ROC, Su B. Acta Biomater. 2009;5:1433–1441. doi: 10.1016/j.actbio.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 141.Clements LR, Wang PY, Tsai WB, Thissen H, Voelcker NH. Lab Chip. 2012;12:1480–1486. doi: 10.1039/c2lc20732j. [DOI] [PubMed] [Google Scholar]
- 142.Jarcho M, Bolen CH, Thomas MB, Bobick J, Kay JF, Doremus RH. J Mater Sci. 1976;11:2027–2035. [Google Scholar]
- 143.Nishimura T, Mitomo M, Hirotsuru H, Kawahara M. J Mater Sci Lett. 1995;14:1046–1047. [Google Scholar]
- 144.Nygren M, Shen ZJ. Solid State Sci. 2003;5:125–131. [Google Scholar]
- 145.Rankin J, Sheldon BW. Materials Science and Engineering a-Structural Materials Properties Microstructure and Processing. 1995;204:48–53. [Google Scholar]
- 146.Dulgar-Tulloch AJ, Bizios R, Siegel RW. J Biomed Mater Res A. 2009;90A:586–594. doi: 10.1002/jbm.a.32116. [DOI] [PubMed] [Google Scholar]
- 147.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sreerekha PR, Menon D, Nair SV, Chennazhi KP. Tissue Eng Part A. 2013;19:849–859. doi: 10.1089/ten.TEA.2012.0374. [DOI] [PubMed] [Google Scholar]
- 149.Namgung S, Kim T, Baik KY, Lee M, Nam JM, Hong S. Small. 2011;7:56–61. doi: 10.1002/smll.201001513. [DOI] [PubMed] [Google Scholar]
- 150.Khang D, Kim SY, Liu-Snyder P, Palmore GTR, Durbin SM, Webster TJ. Biomaterials. 2007;28:4756–4768. doi: 10.1016/j.biomaterials.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 151.Zouani OF, Chanseau C, Brouillaud B, Bareille R, Deliane F, Foulc MP, Mehdi A, Durrieu MC. J Cell Sci. 2012;125:1217–1224. doi: 10.1242/jcs.093229. [DOI] [PubMed] [Google Scholar]
- 152.Woodbury D, Schwarz EJ, Prockop DJ, Black IB. J Neurosci Res. 2000;61:364–370. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 153.Gage FH, Temple S. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 154.Shreyas S, Perry TY, Thiers MU, Sy-Tsong Dean C, Letao Y, Ki-Bum L. Adv Mater. 2014. In Press. [Google Scholar]
- 155.Lee S, Leach MK, Redmond SA, Chong SYC, Mellon SH, Tuck SJ, Feng ZQ, Corey JM, Chan JR. Nat Methods. 2012;9:917. doi: 10.1038/nmeth.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gerardo-Nava J, Fuhrmann T, Klinkhammer K, Seiler N, Mey J, Klee D, Moller M, Dalton PD, Brook GA. Nanomedicine (Lond) 2009;4:11–30. doi: 10.2217/17435889.4.1.11. [DOI] [PubMed] [Google Scholar]
- 157.Nur EKA, Ahmed I, Kamal J, Schindler M, Meiners S. Stem Cells. 2006;24:426–433. doi: 10.1634/stemcells.2005-0170. [DOI] [PubMed] [Google Scholar]
- 158.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Nature Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chowdhury F, Li YZ, Poh YC, Yokohama-Tamaki T, Wang N, Tanaka TS. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Keung AJ, Asuri P, Kumar S, Schaffer DV. Integr Biol. 2012;4:1049–1058. doi: 10.1039/c2ib20083j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sun YB, Villa-Diaz LG, Lam RHW, Chen WQ, Krebsbach PH, Fu JP. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kingham E, White K, Gadegaard N, Dalby MJ, Oreffo ROC. Small. 2013;9:2140–2151. doi: 10.1002/smll.201202340. [DOI] [PubMed] [Google Scholar]
- 163.Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 164.Wichterle H, Lieberam I, Porter JA, Jessell TM. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 165.Brustle O, Jones KN, Learish RD, Karram K, Choudhary K, Wiestler OD, Duncan ID, McKay RD. Science. 1999;285:754–756. doi: 10.1126/science.285.5428.754. [DOI] [PubMed] [Google Scholar]
- 166.Chao TI, Xiang S, Chen CS, Chin WC, Nelson AJ, Wang C, Lu J. Biochem Bioph Res Co. 2009;384:426–430. doi: 10.1016/j.bbrc.2009.04.157. [DOI] [PubMed] [Google Scholar]
- 167.Sridharan I, Kim T, Wang R. Biochem Bioph Res Co. 2009;381:508–512. doi: 10.1016/j.bbrc.2009.02.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, Kim KS. Biomaterials. 2010;31:4360–4366. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 169.zur Nieden NI, Kempka G, Ahr HJ. Differentiation. 2003;71:18–27. doi: 10.1046/j.1432-0436.2003.700602.x. [DOI] [PubMed] [Google Scholar]
- 170.Garreta E, Genove E, Borros S, Semino CE. Tissue Eng. 2006;12:2215–2227. doi: 10.1089/ten.2006.12.2215. [DOI] [PubMed] [Google Scholar]
- 171.Yamashita J, Itoh H, Hirashima M, Ogawa M, Nishikawa S, Yurugi T, Naito M, Nakao K, Nishikawa S. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 172.Drab M, Haller H, Bychkov R, Erdmann B, Lindschau C, Haase H, Morano I, Luft FC, Wobus AM. FASEB J. 1997;11:905–915. doi: 10.1096/fasebj.11.11.9285489. [DOI] [PubMed] [Google Scholar]
- 173.Schmitt RM, Bruyns E, Snodgrass HR. Gene Dev. 1991;5:728–740. doi: 10.1101/gad.5.5.728. [DOI] [PubMed] [Google Scholar]
- 174.Wiles MV, Keller G. Development. 1991;111:259–267. doi: 10.1242/dev.111.2.259. [DOI] [PubMed] [Google Scholar]
- 175.Dani C, Smith AG, Dessolin S, Leroy P, Staccini L, Villageois P, Darimont C, Ailhaud G. J Cell Sci. 1997;110:1279–1285. doi: 10.1242/jcs.110.11.1279. [DOI] [PubMed] [Google Scholar]
- 176.Kramer J, Hegert C, Guan KM, Wobus AM, Muller PK, Rohwedel J. Mech Develop. 2000;92:193–205. doi: 10.1016/s0925-4773(99)00339-1. [DOI] [PubMed] [Google Scholar]
- 177.Chua KN, Chai C, Lee PC, Ramakrishna S, Leong KW, Mao HQ. Exp Hematol. 2007;35:771–781. doi: 10.1016/j.exphem.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Siebers MC, ter Brugge PJ, Walboomers XF, Jansen JA. Biomaterials. 2005;26:137–146. doi: 10.1016/j.biomaterials.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 179.Schwartz Martin, Ginsberg Mark. Nat Cell Biol. 2002;4:E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 180.Arnaout M, Goodman S, Xiong J-P. Curr Opin Cell Biol. 2007;19:495–507. doi: 10.1016/j.ceb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rossier O, Octeau V, Sibarita J-B, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albigès-Rizo C, Tampé R, Cognet L, Choquet D, Lounis B, Giannone G. Nat Cell Biol. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 182.Schiller H, Hermann M-R, Polleux J, Vignaud T, Zanivan S, Friedel C, Sun Z, Raducanu A, Gottschalk K-E, Théry M, Mann M, Fässler R. Nat Cell Biol. 2013;15:625–636. doi: 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- 183.Li L, Bennett SAL, Wang LS. Cell Adhes Migr. 2012;6:59–70. doi: 10.4161/cam.19583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Meng Y, Eshghi S, Li YJ, Schmidt R, Schaffer DV, Healy KE. FASEB J. 2010;24:1056–1065. doi: 10.1096/fj.08-126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rowland TJ, Miller LM, Blaschke AJ, Doss EL, Bonham AJ, Hikita ST, Johnson LV, Clegg DO. Stem Cells Dev. 2010;19:1231–1240. doi: 10.1089/scd.2009.0328. [DOI] [PubMed] [Google Scholar]
- 186.Tsimbouri P, Gadegaard N, Burgess K, White K, Reynolds P, Herzyk P, Oreffo R, Dalby MJ. J Cell Biochem. 2014;115:380–390. doi: 10.1002/jcb.24673. [DOI] [PubMed] [Google Scholar]
- 187.Hynes R. Cell. 2002 [Google Scholar]
- 188.Qin J, Vinogradova O, Plow E. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Askari JA, Buckley PA, Mould AP, Humphries MJ. J Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Geiger B, Spatz J, Bershadsky A. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 191.Zaidel-Bar R, Geiger B. J Cell Sci. 2010;123:1385–1388. doi: 10.1242/jcs.066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. J Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 193.Arnold M, Cavalcanti-Adam E, Glass R, Blümmel J, Eck W, Kantlehner M, Kessler H, Spatz J. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 194.Cavalcanti-Adam E, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz J. Biophys J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schvartzman M, Palma M, Sable J, Abramson J, Hu X, Sheetz M, Wind S. Nano Lett. 2011;11:1306–1312. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. J Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 197.Huang J, Grater S, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding J, Spatz J. Nano Lett. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dalby M, Gadegaard N, Tare R, Andar A, Riehle M, Herzyk P, Wilkinson C, Oreffo R. Nature Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 199.Mitra S, Hanson D, Schlaepfer D. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 200.Seong J, Tajik A, Sun J, Guan JL, Humphries MJ, Craig SE, Shekaran A, Garcia AJ, Lu SY, Lin MZ, Wang N, Wang YX. Proc Natl Acad Sci U S A. 2013;110:19372–19377. doi: 10.1073/pnas.1307405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schlaepfer DD, Hanks SK, Hunter T, Vandergeer P. Nature. 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 202.Hong SY, Jeon YM, Lee HJ, Kim JG, Baek JA, Lee JC. Mol Cell Biochem. 2010;335:263–272. doi: 10.1007/s11010-009-0276-1. [DOI] [PubMed] [Google Scholar]
- 203.Wary KK, Mariotti A, Zurzolo C, Giancotti FG. Cell. 1998;94:625–634. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
- 204.Aikawa R, Nagai T, Kudoh S, Zou YZ, Tanaka M, Tamura M, Akazawa H, Takano H, Nagai R, Komuro I. Hypertension. 2002;39:233–238. doi: 10.1161/hy0202.102699. [DOI] [PubMed] [Google Scholar]
- 205.Teo BKK, Wong ST, Lim CK, Kung TYS, Yap CH, Ramagopal Y, Romer LH, Yim EKF. ACS Nano. 2013;7:4785–4798. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- 206.Hsia DA, Mitra SK, Hauck CR, Streblow D, Nelson JA, Ilic D, Huang S, Li EG, Nemerow GR, Leng J, Spencer KSR, Cheresh DA, Schlaepfer DD. J Cell Biol. 2003;160:753–767. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Schlaepfer DD, Mitra SK, Ilic D. Bba-mol Cell Res. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 208.Zhai JB, Lin H, Nie ZY, Wu JH, Canete-Soler R, Schlaepfer WW, Schalepfer DD. J Biol Chem. 2003;278:24865–24873. doi: 10.1074/jbc.M302381200. [DOI] [PubMed] [Google Scholar]
- 209.Rico B, Beggs HE, Schahin-Reed D, Kimes N, Schmidt A, Reichardt LF. Nat Neurosci. 2004;7:1059–1069. doi: 10.1038/nn1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hildebrand JD, Taylor JM, Parsons JT. Mol Cell Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ren XD, Kiosses WB, Sieg DJ, Otey CA, Schlaepfer DD, Schwartz MA. J Cell Sci. 2000;113:3673–3678. doi: 10.1242/jcs.113.20.3673. [DOI] [PubMed] [Google Scholar]
- 212.Kantawong F, Burgess KEV, Jayawardena K, Hart A, Burchmore RJ, Gadegaard N, Oreffo ROC, Dalby MJ. Biomaterials. 2009;30:4723–4731. doi: 10.1016/j.biomaterials.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 213.Xia H, Nho RS, Kahm J, Kleidon J, Henke CA. J Biol Chem. 2004;279:33024–33034. doi: 10.1074/jbc.M313265200. [DOI] [PubMed] [Google Scholar]
- 214.Du J, Chen X, Liang X, Zhang G, Xu J, He L, Zhan Q, Feng X-Q, Chien S, Yang C. Proc Natl Acad Sci U S A. 2011;108:9466–9471. doi: 10.1073/pnas.1106467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, Wappler I, Peters H, Walter T, Stojkovic P, Evans J, Stojkovic M, Lako M. Hum Mol Genet. 2006;15:1894–1913. doi: 10.1093/hmg/ddl112. [DOI] [PubMed] [Google Scholar]
- 216.Chambers S, Fasano C, Papapetrou E, Tomishima M, Sadelain M, Studer L. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun YH, Menendez L, Kulik M, Dalton S. Cell Stem Cell. 2012;10:312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim J, Kim HN, Lim KT, Kim Y, Seonwoo H, Park SH, Lim HJ, Kim DH, Suh KY, Choung PH, Choung YH, Chung JH. Sci Rep. 2013;3 doi: 10.1038/srep03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yang J, McNamara L, Gadegaard N, Alakpa E, Burgess K, Meek R, Dalby M. ACS Nano. 2014;8:9941–9953. doi: 10.1021/nn504767g. [DOI] [PubMed] [Google Scholar]
- 220.Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 221.Burridge K, Wennerberg K. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 222.Mammoto A, Huang S, Moore K, Oh P, Ingber DE. J Biol Chem. 2004;279:26323–26330. doi: 10.1074/jbc.M402725200. [DOI] [PubMed] [Google Scholar]
- 223.Bhadriraju K, Yang M, Alom Ruiz S, Pirone D, Tan J, Chen C. Exp Cell Res. 2007;313:3616–3623. doi: 10.1016/j.yexcr.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. Nat Cell Biol. 2011;13:722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 226.McMurray R, Gadegaard N, Tsimbouri P, Burgess K, McNamara L, Tare R, Murawski K, Kingham E, Oreffo R, Dalby M. Nature Mater. 2011;10:637–644. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 227.Teo BK, Wong S, Lim C, Kung T, Yap C, Ramgopal Y, Romer L, Yim E. ACS Nano. 2013 doi: 10.1021/nn304966z. In Press. [DOI] [PubMed] [Google Scholar]
- 228.DuFort C, Paszek M, Weaver V. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Halder G, Dupont S, Piccolo S. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 230.Piccolo S, Dupont S, Cordenonsi M. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 231.Mo JS, Park HW, Guan KL. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Halder G, Camargo F. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Dong JX, Feldmann G, Huang JB, Wu S, Zhang NL, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan DJ. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 235.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 236.Wada KI, Itoga K, Okano T, Yonemura S, Sasaki H. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 237.Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, Janody F. Development. 2011;138:2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 238.Mo JS, Park HW, Guan KL. EMBO Rep. 2014 doi: 10.15252/embr.201438638. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sansores-Garcia L, Bossuyt W, Wada KI, Yonemura S, Tao CY, Sasaki H, Halder G. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yang C, Tibbitt MW, Basta L, Anseth KS. Nature Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sun YB, Aw Yong KM, Villa-Diaz LG, Zhang XL, Chen WQ, Philson R, Weng SN, Xu HX, Krebsbach PH, Fu JP. Nature Mater. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Connelly JT, Gautrot JE, Trappmann B, Tan DWM, Donati G, Huck WTS, Watt FM. Nat Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 243.Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. J Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Higuchi A, Ling Q-D, Chang Y, Hsu S-T, Umezawa A. Chem Rev. 2013;113:3297–3328. doi: 10.1021/cr300426x. [DOI] [PubMed] [Google Scholar]
- 245.Shivashankar GV. Annu Rev Biophys. 2011;40:361–378. doi: 10.1146/annurev-biophys-042910-155319. [DOI] [PubMed] [Google Scholar]
- 246.Lammerding J, Schulze C, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Circulation. 2003;108:289–289. [Google Scholar]
- 247.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. J Biol Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang N, Tytell J, Ingber D. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 252.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Swift J, Ivanovska I, Buxboim A, Harada T, Dingal P, Pinter J, Pajerowski J, Spinler K, Shin J-W, Tewari M, Rehfeldt F, Speicher D, Discher D. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Liu H, Wen J, Xiao Y, Liu J, Hopyan S, Radisic M, Simmons C, Sun Y. ACS Nano. 2014;8:3821–3828. doi: 10.1021/nn500553z. [DOI] [PubMed] [Google Scholar]
- 255.Poh YC, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N. Nat Commun. 2012;3 doi: 10.1038/ncomms1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Na S, Collin O, Chowdhury F, Tay B, Ouyang MX, Wang YX, Wang N. Proc Natl Acad Sci U S A. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Versaevel M, Grevesse T, Gabriele S. Nat Commun. 2012;3:671. doi: 10.1038/ncomms1668. [DOI] [PubMed] [Google Scholar]
- 258.Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Nat Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chalut KJ, Kulangara K, Giacomelli MG, Wax A, Leong KW. Soft Matter. 2010;6:1675–1681. doi: 10.1039/B921206J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tsimbouri PM, Murawski K, Hamilton G, Herzyk P, Oreffo ROC, Gadegaard N, Dalby MJ. Biomaterials. 2013;34:2177–2184. doi: 10.1016/j.biomaterials.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Dalby MJ, Gadegaard N, Herzyk P, Sutherland D, Agheli H, Wilkinson CDW, Curtis ASG. J Cell Biochem. 2007;102:1234–1244. doi: 10.1002/jcb.21354. [DOI] [PubMed] [Google Scholar]
- 262.Mammoto T, Mammoto A, Ingber DE. Annu Rev Cell Dev Biol. 2013;29:27–61. doi: 10.1146/annurev-cellbio-101512-122340. [DOI] [PubMed] [Google Scholar]
- 263.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 264.Piccolo S, Cordenonsi M, Dupont S. Clin Cancer Res. 2013;19:4925–4930. doi: 10.1158/1078-0432.CCR-12-3172. [DOI] [PubMed] [Google Scholar]
- 265.Halder G, Dupont S, Piccolo S. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 266.Mammoto T, Mammoto A, Ingber D. Annu Rev Cell Dev Biol. 2013;29:27–61. doi: 10.1146/annurev-cellbio-101512-122340. [DOI] [PubMed] [Google Scholar]
- 267.Baarlink C, Wang HC, Grosse R. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 268.Kim J, Kim HN, Lim KT, Kim Y, Pandey S, Garg P, Choung YH, Choung PH, Suh KY, Chung JH. Biomaterials. 2013;34:7257–7268. doi: 10.1016/j.biomaterials.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 269.Guvendiren M, Burdick JA. Adv Healthcare Mater. 2013;2:155–164. doi: 10.1002/adhm.201200105. [DOI] [PubMed] [Google Scholar]
- 270.Xi J, Khalil M, Shishechian N, Hannes T, Pfannkuche K, Liang H, Fatima A, Haustein M, Suhr F, Bloch W, Reppel M, Saric T, Wernig M, Janisch R, Brockmeier K, Hescheler J, Pillekamp F. FASEB J. 2010;24:2739–2751. doi: 10.1096/fj.09-145177. [DOI] [PubMed] [Google Scholar]
- 271.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Condic ML, Rao M. Stem Cells. 2008;26:2753–2758. doi: 10.1634/stemcells.2008-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Corti S, Nizzardo M, Simone C, Falcone M, Nardini M, Ronchi D, Donadoni C, Salani S, Riboldi G, Magri F, Menozzi G, Bonaglia C, Rizzo F, Bresolin N, Comi GP. Sci Transl Med. 2012;4:165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Amabile G, Meissner A. Trends Mol Med. 2009;15:59–68. doi: 10.1016/j.molmed.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 275.Lee H, Park J, Forget BG, Gaines P. Regen Med. 2009;4:759–769. doi: 10.2217/rme.09.46. [DOI] [PubMed] [Google Scholar]
- 276.Wernig M, Meissner A, Foreman R, Brambrink T, Ku MC, Hochedlinger K, Bernstein BE, Jaenisch R. Nature. 2007;448:318–U312. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 277.McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo RO, Dalby MJ. Nature Mater. 2011;10:637–644. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.