Abstract
Rubinstein-Taybi syndrome (RSTS) is a rare condition with a prevalence of 1 in 125,000–720,000 births and characterized by clinical features that include facial, dental, and limb dysmorphology and growth retardation. Most cases of RSTS occur sporadically and are caused by de novo mutations. Cytogenetic or molecular abnormalities are detected in only 55% of RSTS cases. Previous genetic studies have yielded inconsistent results due to the variety of methods used for genetic analysis. The purpose of this study was to use whole exome sequencing (WES) to evaluate the genetic causes of RSTS in a young girl presenting with an Autism phenotype. We used the Autism diagnostic observation schedule (ADOS) and Autism diagnostic interview revised (ADI-R) to confirm her diagnosis of Autism. In addition, various questionnaires were used to evaluate other psychiatric features. We used WES to analyze the DNA sequences of the patient and her parents and to search for de novo variants. The patient showed all the typical features of Autism, WES revealed a de novo frameshift mutation in CREBBP and de novo sequence variants in TNC and IGFALS genes. Mutations in the CREBBP gene have been extensively reported in RSTS patients, while potential missense mutations in TNC and IGFALS genes have not previously been associated with RSTS. The TNC and IGFALS genes are involved in central nervous system development and growth. It is possible for patients with RSTS to have additional de novo variants that could account for previously unexplained phenotypes.
Keywords: Rubinstein-Taybi syndrome (RSTS), Autism spectrum disorder (ASD), de novo variants, Tenascin C (TNC) gene, insulin-like growth factor-binding protein, acid labile subunit (IGFALS) gene, CREB (cAMP response element binding protein) binding protein (CREBBP) gene
1. Introduction
Rubinstein-Taybi syndrome (RSTS, OMIM 180849, 613684), also known as the broad thumb-hallux syndrome, is a rare condition with a prevalence of 1 in 125,000–720,000 births [1,2]. The syndrome is characterized by a group of well-defined clinical features including characteristic facial, dental, and limb dysmorphology, and growth retardation [3]. RSTS can also affect multiple internal organs including the heart, kidney, eyes, ears, and skin [4]. Patients are known to have an increased risk of developing non-cancerous and cancerous tumors, leukemia, and lymphoma [5]. Mental retardation and learning difficulties are other typical findings of RSTS. The average IQ of the patients is 35–50, although cognitive functioning outside this range has also been documented [6].
The diagnosis of RSTS is essentially a clinical diagnosis based on the features described above [7]. Most RSTS cases are sporadic and are caused by de novo mutations [8]. Cytogenetic or molecular abnormalities can be detected in only 55% of patients with RSTS [9]. Missense/non-sense mutations in genes encoding the CREB-binding protein (CREBBP) or the E1A-binding protein (EP300) are the most common, although micro-deletions in chromosome 16p13.3 and other rare mutations have also been reported as genetic causes of this condition [8,10,11,12,13,14,15]. Nevertheless, genetic analyses for identifying mutations in RSTS have often yielded inconsistent results, probably because of inadequacies in the methods used for the analyses, and many mutations remain unconfirmed. Previous studies examining genetic mutations in RSTS have mainly used fluorescence in situ hybridization (FISH) [16,17] or comparative genomic hybridization (CGH) array analyses [14]. Whole genome or exome sequencing (WES) methods have rarely been used. The purpose of this study was to use WES to evaluate the genetic causes of RSTS in a young girl presenting with RSTS and Autism spectrum disorder (ASD).
2. Results
2.1. Physical Characteristics
The typical dysmorphic features of RSTS were observed in the patient’s face and mouth: Microcephaly (head circumference of 32 cm (approximately 3rd to 5th percentile) at birth and 47.8 cm (below the 3rd percentile) at her current age (6.75 years), highly arched eyebrows, long eyelashes, downward-slanting palpebral fissures, a broad nasal bridge, a beaked nose with the nasal septum extending well below the alae, a pouting lower lip, mild micrognathia, and dental crowding.
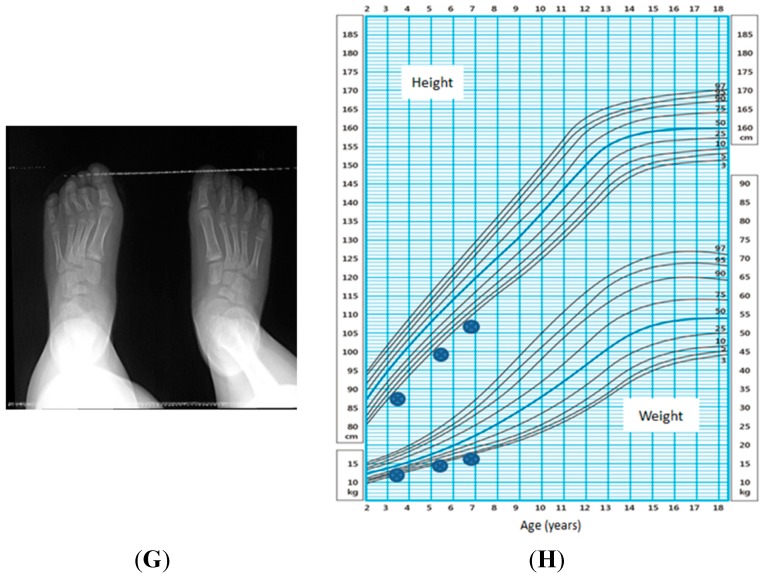
The patient had big toes that were short and broad, and mild clinodactyly of the fifth digits of both hands. The second, third, and fourth digits of the right hand had short distal phalanges, and the third and fourth digits of the left hand had broad distal phalanges. She also had a curly toe on the fourth digit of both feet and hallux valgus in the right foot. X-ray radiographs showed bilateral pes varus deformities and bilateral big-toe soft tissue thickening. There were abnormal accessory physes on the side of the distal phalanx, as well as other abnormalities in the physes of the big toes of both feet. Hand X-ray radiographs showed delayed carpal bone ossification in both hands and prominent thickening of the soft tissue of all fingers.
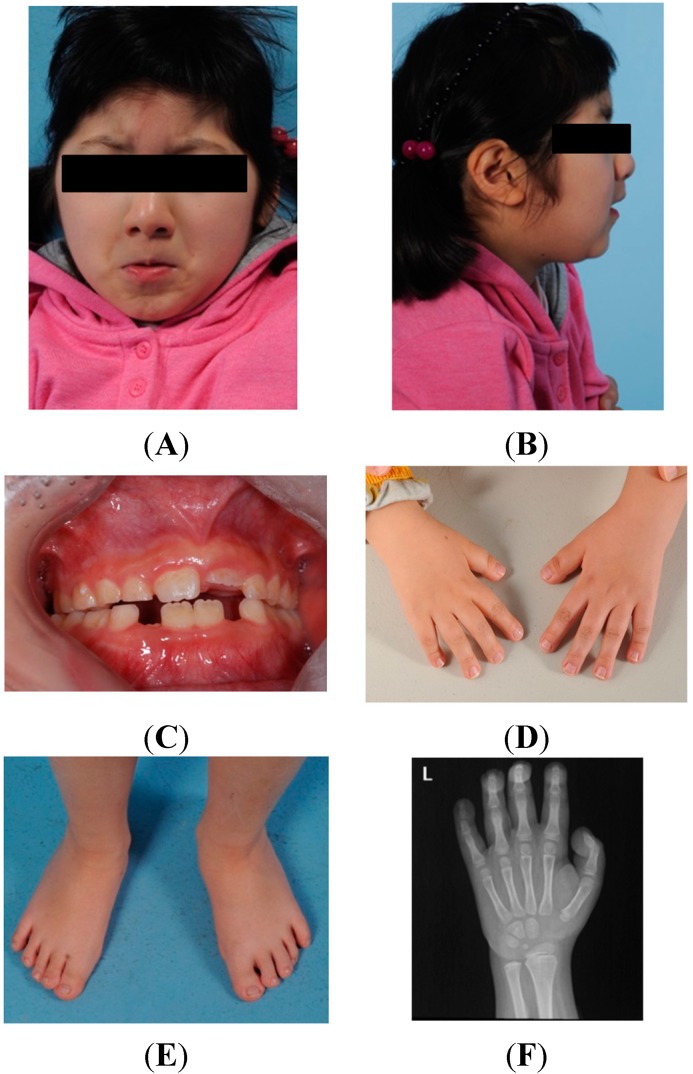
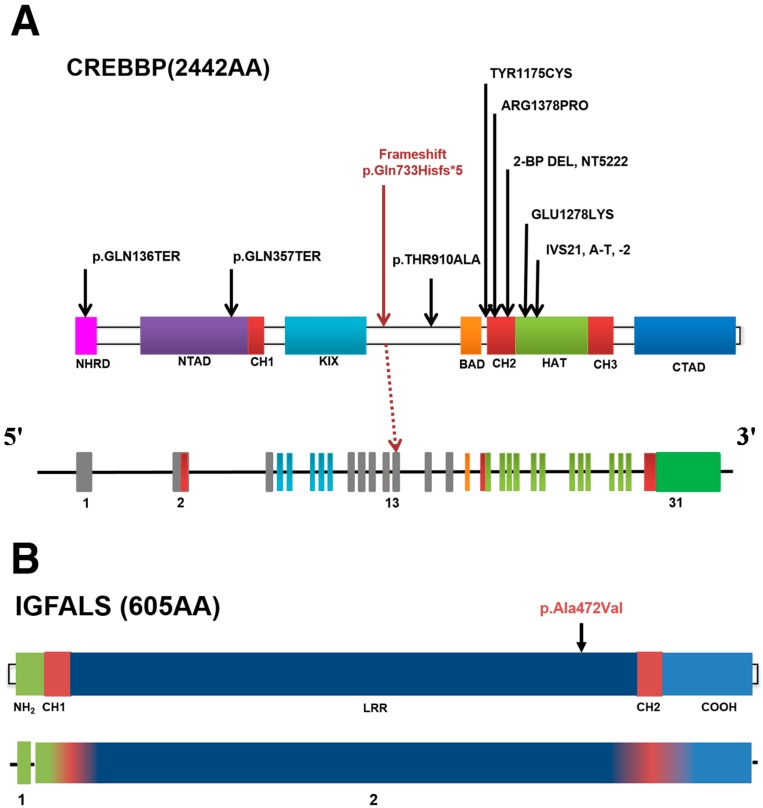
Marked growth retardation and poor weight gain were prominent in this patient from birth until her latest evaluation. Her height was 106.3 cm (below the 3rd percentile) and her weight was 17.2 kg (3rd percentile). A videofluoroscopic swallowing study revealed difficulty in swallowing. Furthermore, serum prealbumin and zinc levels were low. Other physical findings in the patient included anomalies of the eye (congenital cataract and nasolacrimal duct obstruction) and skin (pilomatrixoma on the left periauricular area, hypertrichosis on the forehead, anterior chest, and back, as well as nevus depigmentosus on the right calf). Gross hematuria had previously been found at five years of age, although a kidney ultrasound examination showed no abnormal findings. Results of cardiac evaluation were within normal limits. The characteristic physical features and growth curve of the patient are shown in Figure 1.
Figure 1.
Physical characteristics of the proband. Patient’s eyes are covered for reasons of confidentiality. Typical facial dysmorphic features can be observed in (A,B), including highly arched eyebrows, a broad nasal bridge, beaked nose with the nasal septum extending well below the alae, a pouting lower lip, and mild micrognathia. Dental crowding is shown in (C); The hand and feet abnormalities are shown in (D,E), showing short and broad big toes and thumbs, as well as mild clinodactyly on the 5th digit of both hands. The 2nd, 3rd, 4th digits of the right hand had short distal phalanges and the 3rd, 4th digits of the left hand had broad distal phalanges. The X-ray of the left hand shows delayed carpal bone ossification and prominent thickening of the soft tissue of all fingers (F); The X-ray of the feet shows bilateral pes varus deformities and abnormalities in the physes of big toe metatarsal bones (G); Growth curve (H) shows marked growth retardation in the proband (blue dot) superimposed on the normal growth curve of Korean girls of age 2 to 18 (black line). The growth curve shows the height and weight changes of the patient over time. These values never exceeded the third percentile since birth. Data for normal growth curve came from the Korea Center for Disease Control and Prevention.
2.2. Neuropsychiatric and Behavioral Features
The patient showed pronounced developmental delay with no meaningful vocabulary. She also showed moderate mental retardation with a full-scale intelligence quotient (IQ) score of 37 on the Korean Wechsler Preschool and Primary Scale for Intelligence (K-WPPSI) and a score of 45 on the Leiter International Performance Scale. Her adaptive functioning level was equivalent to a child of approximately 3–5 years of age. She had shown the typical behavioral characteristics of Autism from early infancy, including abnormalities in communication and social interaction, along with repetitive behavior and restricted interests. Her scores surpassed the diagnostic thresholds on all categories of both the ADOS and ADI-R. She presented the distinctive mannerism of repetitively waving her hand whenever emotionally disturbed. Scores for other questionnaires also supported the diagnosis of Autism including a score of 15 on the social communication questionnaire (SCQ) and a score of 113 on the social component of the social responsiveness scale (SRS). Further behavioral characteristics included marked hyperactivity, short attention span, withdrawal, and poor motor coordination. Frequent snoring was reported, although the possibility of obstructive sleep apnea was deemed low. Brain magnetic resonance imaging (MRI) showed mild pachygyria. Electroencephalogram (EEG) showed intermittent diffuse high-amplitude semirhythmic 1–1.5-Hz delta activity and one episode of diffuse 1.5-Hz rhythmic, synchronous bifrontal δ/ζ activity lasting up to 20 s.
2.3. Genetic Variants
Cytogenetic analyses showed normal karyotypes in the proband and both parents. Using in-depth statistics, we obtained a mean read-depth coverage of 120× over the targeted exome. Each exome contained more than 120,000 total variant calls. An average of 111 million individual paired-end reads of 100 bp were aligned to the human reference genome. More than 94% of targeted exon regions were covered with at least 10× depth, which is sufficient for variant discovery.
We found three de novo variants after filtering for all common variants in the dbSNP (except clinically associated variants) and an in-house control database comprising normal healthy individuals. We performed a visual inspection of read alignments using the Integrative Genomics Viewer (IGV) browser, and then removed “Benign” and “Likely Benign” variants after classification. A summary of the detected variants is described in Table S1.
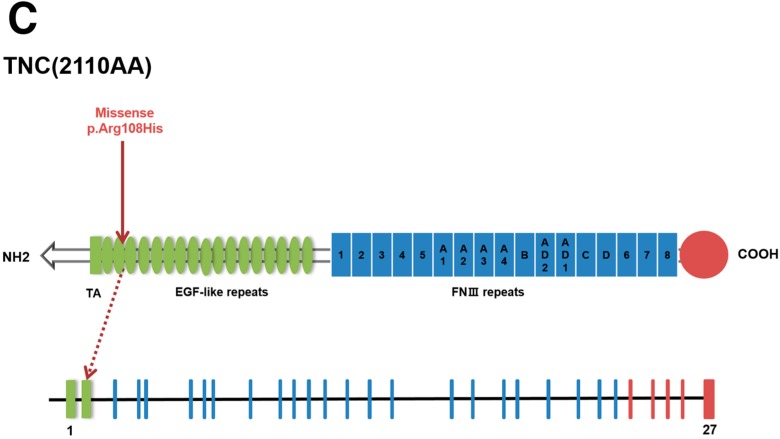
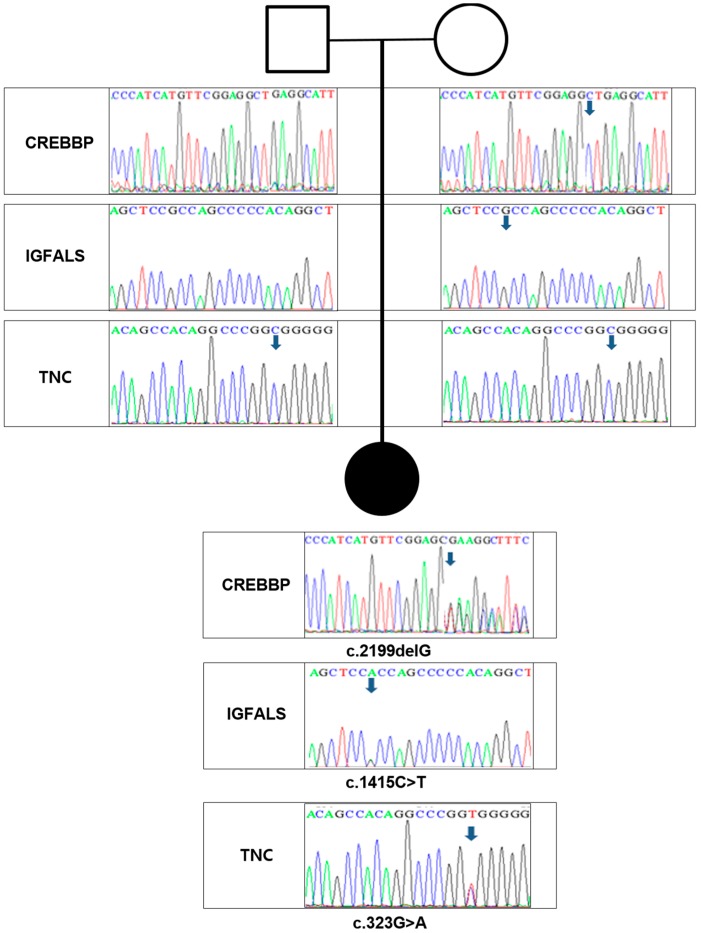
Three de novo variants were validated by Sanger sequencing: a frameshift mutation in CREBBP (c.2199delG), and missense mutations in TNC (c.323G>A) and IGFALS (c.1415C>T). The results of family sequencing analyses are shown in Figure 2 and the properties of the de novo variants are summarized in Table 1 and Figure 2. The genetic variants of the proband and both parents are shown in Figure 3.
Figure 2.
Schematic of de novo variants of CREBBP, IGFALS, and TNC in RSTS. (A) Genomic alterations and schematic drawing of proteins showing the predicted effects of selected missense mutations identified in CREBBP. The p.Thy1174Cys and p.Thr910Ala substitutions predicted by missense mutations do not affect known functional domains and are associated with a mild phenotype compared with the classic phenotype expected for mutations at other sites. The frameshift mutation p.Gln733Hisfs*5 is predicted to interrupt all of the following functional domains: BAD, CH2, HAT, CH3, and CTAD; (B) The IGFALS mutation site affects the LRR domain and (C) the TNC mutation one of the EGF-like repeats. The de novo variants are shown in red. CREBBP: CREB (cAMP response element-binding protein) binding protein; IGFALS: Insulin-like growth factor)-binding protein, acid labile subunit; TNC: Tenascin C; Protein domain names: (A) NHRD: Nuclear receptor-binding and receptor-interacting domain; NTAD: Amino-terminal transactivation domain; CH1: Cys/His-rich region; CREB/KIX: CREB-binding domain; BROMO/BRD: Bromo domain; HAT: Histone acetyltransferase domain; CH2: Cys/His-rich region 2; CH3: Cys/His-rich region 3; CTAD: C-terminal transactivation domain; (B) NH2: N-terminal; CH1: Cys-rich region 1; LRR: Leucine rich repeats; CH2: Cys-rich region 2; COOH: C-terminal; (C) TA: N-terminal tenascin assembly domain; EGF-like repeats: Epidermal growth factor-like repeats; FNIII repeats: Fibronectin type III-like repeats; FN Globe: C-terminal fibrinogen globe
Table 1.
The three de novo variants validated by Sanger Sequencing.
| Gene | TNC | IGFALS | CREBBP |
|---|---|---|---|
| Chromosome | chr9 | chr16 | chr16 |
| Position | 117852975 | 1841118 | 3823901 |
| Reference Allele | C | G | C |
| Alternative Allele | T | A | – |
| Mutation Type | Missense | Missense | Frameshift |
| Sequence Variant | c.323G>A | c.1415C>T | c.2199delG |
| Protein Variant | p.Arg108His | p.Ala472Val | p.Gln733Hisfs*5 |
| phyloP Score | 2.369 | 0.598 | 1.492 |
| GERP Score | 1200.8 | 2536.4 | 476.2 |
| SIFT Score | 0 | 0.30 | – |
| Mutation Assessor | 1.87 | 0.995 | – |
| Classification 1 | Likely pathogenic 2 | Variants of unknown significance | Pathogenic |
1 Variant Classification is based on the recommendations of Ambry Genetics, but pathogenicity of de novo variants in TNC and IGFALS are not certain; 2 TNC c.323G>A is reported in dbSNP as rs151119387, but MAF/MinorAlleleCount = 0.0002/1. Overall MAF in 1000 genomes, ESP6500, ExAc database is less than 0.0003. TNC: Tenascin C; IGFALS: Insulin-like growth factor-binding protein, acid labile subunit; CREBBP: CREB (cAMP response element-binding protein) binding protein; phyloP: Phylogenetic p-values; GERP: Genomic evolutionary rate profiling; SIFT: Sorting intolerant from tolerant.
Figure 3.
Sequences of CREBBP, IGFALS, and TNC for the family trio. A downward arrow indicates the mutated residue and nucleotides are shown in colored, single-letter codes. The black circle indicates the proband.
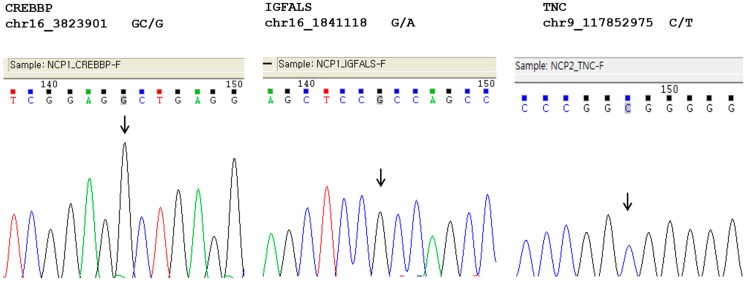
None of the observed sequence changes were observed in normal Korean subjects. The validation results from the normal population are shown in Figure 4.
Figure 4.
Validation results for de novo variants in CREBBP, IGFALS, and TNC in 80 normal Korean subjects. The downward arrow indicates the residue of de novo variants.
3. Discussion
We have shown that de novo variants in the CREBBP, TNC, and IGFALS genes are potential causes for RSTS by analyzing the whole genome of a female patient with RSTS. In WES analyses, we observed de novo variants in the CREBBP, TNC, and IGFALS genes. The girl showed typical morphological findings of RSTS, including abnormal features of the face, fingers, and toes, as well as growth retardation. Major characteristics of this patient were severe developmental/behavioral problems together with ASD with typical features that fulfilled the diagnostic criteria in multiple domains including communication, social interaction, specialized interests, and repetitive behavior.
Developmental/behavioral problems are common in patients with RSTS, with the most typical symptoms being moderate to severe mental retardation and language impairment [8,18]. There are no definitive reports regarding the specific frequency of psychiatric disorders in RSTS due to its low prevalence. In terms of autistic behavior, only a few reports have described this specific psychopathology associated with RSTS. In one relatively large-scale study exploring the socio-behavioral characteristics of RSTS, this syndrome was associated with behavioral problems such as a short attention span, motor stereotypies, and poor coordination [19]. These can also be part of the behavioral phenotype of ASD. The studies that have reported Autism-like phenotypes in patients with RSTS describe milder and atypical forms of ASD (i.e., pervasive developmental disorder not otherwise specified; PDD NOS) in only a small proportion (8%) of the subjects [20,21,22,23].
Mutations in the CREBBP gene are one of the most commonly reported genetic etiologies of RSTS (Table S2). CREBBP mutations are found in 50%–60% of RSTS cases, whereas EP300 (OMIM 602700) mutations are found in 5% of the cases [7,8]. RSTS is assumed to be a genetically heterogeneous disorder which is caused by mutations in the CREBBP gene as well as in other genes in up to 50% of cases [2]. The CREBBP mutations are heterogeneous, as 160 different mutations have been identified (Leiden Open Variation Database v.2.0 Build 35) [24]. The CREBBP gene is ubiquitously expressed and is involved in the transcriptional co-activation of many transcription factors [25]. CREBBP is also involved in multiple signaling pathways and in other cellular functions such as DNA repair, cell growth, differentiation, apoptosis, and tumor suppression [26]. The CREBBP protein plays an important role in regulating cell growth and division and is essential for normal fetal development. If one copy of CREBBP is deleted or mutated, cells make only half the normal amount of CREBBP protein. This reduction in the amount of this protein is thought to disrupt normal development before and after birth. The CREBBP mutation is known to be associated with more severe symptomatology than the EP300 mutations [24]. Based on previous studies, the severe cognitive symptomatology of the subject of this study, including the ASD phenotype, could be attributed to the CREBBP mutation.
We can infer that the de novo sequence changes of the TNC and IGFALS genes, featuring as missense mutations observed in our study could potentially explain any additional phenotypes in patients with RSTS, but experimental validation is required to establish disease causation. Tenascin C (TNC), encoded by TNC, is a glycoprotein expressed in the extracellular matrix of various tissues during development, disease, or injury, as well as in restricted areas of the central nervous system involved in neurogenesis [27]. TNC expression changes during development until adulthood. In the developing central nervous system, it is involved in regulating the proliferation of both oligodendrocyte precursor cells and astrocytes [28]. TNC expression by radial glia precedes the onset of gliogenesis, and is thought to drive astrocyte differentiation. In the adult brain, TNC expression is downregulated in all areas except the hypothalamus and areas that maintain neurogenesis into adulthood [28]. Male-specific association signals have been found in the intronic region of the TNC gene [29]. A missense mutation c.5317G>A (p.V1773M) in exon 19 of the TNC gene was recently reported as a causative gene for nonsyndromic hearing loss in family-based exome sequencing and linkage analyses [30]. Based on the fact that pronounced-to-profound bilateral hearing loss or deafness is diagnosed more commonly in ASD than in the general population, the causality of deafness and ASD might have a partial overlap [31]. This study suggests that mutations in the TNC gene could contribute to sensory impairment in ASD, including hearing loss. The patient with RSTS in this study showed slight hearing problems (data now provided), but it is still not clear which specific aspect of the patient’s phenotype can be attributed to the missense mutations in TNC.
To date, the presence of a genetic mutation in CREBBP has been linked with the behavioral phenotype in RSTS. For example, poor coordination is more prevalent in patients with RSTS and in subjects with genetic abnormalities in CREBBP than in those who have no abnormalities in this gene [19]. Severe cognitive impairments and autistic features have been reported in patients with large deletions in CREBBP [32]. This can be explained by the involvement of CREB-dependent gene transcription in long-term memory formation through signaling pathways that include multiple important kinases (protein kinase A, mitogen-activated protein kinase, and calcium/calmodulin-dependent kinase IV (CaMKIV)) in the nervous systems of many species [2]. The mental retardation and cognitive phenotype in patients with RSTS may be derived from impairments to the function of CREB-binding protein [2,13]. The role of the CREBBP mutation in long-term memory formation is supported by experiments showing that memory impairments in a CREBBP+/– mutant mouse model were ameliorated by phosphodiesterase 4 inhibitors [33]. Our observations suggest that de novo variants can have an additive effect and produce more severe and defined neurological/cognitive aberrations in subjects with RSTS.
The acid-labile subunit of insulin-like growth factor-binding protein (IGFALS) encoded by the IGFALS gene, is a serum protein that binds to IGFs. The IGFALS protein interacts with growth hormones to increase their half-life as well as their vascular localization [34]. IGFALS-dependent growth changes related to sex-specific body characteristics and bone size have been reported previously [35,36]. Defects in this gene cause acid-labile subunit deficiency which manifests as delayed and slow development during puberty [37]. The growth retardation in our patient, which has been present from birth until her current age, might be enhanced by the variants in the IGFALS gene.
One limitation of our study is that the analysis was restricted to only a single case. Validation of our results was also performed using relatively small samples from the general population. The precise function of the mutations and variants needs to be validated in future studies.
In conclusion, using WES, we observed de novo variants in the CREBBP, TNC, and IGFALS genes in a female patient with RSTS. To our knowledge, this is the first report suggesting that variants in the TNC and IGFALS genes can produce additional phenotypic effects. We assume that de novo variants in the TNC gene are related to the neurological phenotype and those in the IGFALS gene are related to growth retardation.
4. Methods
4.1. Ethics Statement
This study was approved by the Institutional Review Board (IRB) of the Seoul National University Bundang Hospital (Seongnam, Gyeonggi, Korea). Written informed consent was obtained from both parents of the patient.
4.2. Subject
The patient, a girl aged 6 years and 8 months, was examined in the child psychiatry clinic of the Seoul National University Bundang Hospital. She was thoroughly evaluated to assess problems related to delayed development and dysmorphology. A genetic assessment to diagnose RSTS, a possibility indicated during a previous diagnosis process, was also performed. The patient was the only child of a healthy non-consanguineous couple. Both biological parents were healthy with no clinically significant medical, developmental, or neuropsychological illnesses.
4.3. Clinical Evaluation
Thorough physical and laboratory evaluations were performed to confirm the diagnosis of RSTS and obtain detailed clinical and behavioral features. Physical, dental, ophthalmologic, and dermatologic examinations were performed. Chest and limb/digit X-ray radiography, urine and blood tests, a videofluoroscopic swallowing study, kidney ultrasonography, echocardiography, and electrocardiography were all performed as part of the laboratory analyses. Intelligence and adaptive functioning were assessed using the Korean version of the Wechsler preschool and primary scale of intelligence (K-WPPSI), the Leiter international performance scale [6,38], and the Vineland Adaptive Behavior scale (VABS) [39,40]. Two board-certified child psychiatrists evaluated the autistic symptoms. The Autism diagnostic observation schedule (ADOS), Module I [41,42], and the Autism diagnostic interview-revised (ADI-R) [43,44] were used, as well as the social communication questionnaire (SCQ) and social responsiveness scale (SRS) [45,46]. We measured the patients sleep quality using the Pittsburgh sleep quality index (PSQI) [47] and the “STOP” questionnaire (snoring, tiredness during daytime, observed apnea, high blood pressure) [48]. Brain magnetic resonance imaging (MRI) and electroencephalography (EEG) were performed to elucidate potential structural and functional abnormalities in the brain.
4.4. Genetic Analyses Using Bioinformatics Tools
Blood samples were drawn from the patient and both biological parents. Genomic DNA was extracted using the DNeasy Blood & Tissue kit (QIAGEN, Valencia, CA, USA). Raw reads in FASTQ format from exome sequencing were aligned to the hg19 reference genome from the UCSC genome browser using the Burrows–Wheeler Aligner (BWA) [49] with default parameters. Aligned reads were processed and polymerase chain reaction (PCR) duplicates were removed with SAMtools [50]. Single-nucleotide variants (SNVs) and insertions/deletions (indels) were identified using the Genome Analysis Toolkit (GATK) [51]. Regions near short indels were realigned using the IndelRealigner function in GATK according to default parameters. SNVs and indels affecting coding sequences or splicing sites were annotated by an in-house custom-made annotation system. All genomic changes were filtered against the Single Nucleotide Polymorphism Database (dbSNP; build 141) and the in-house control database comprising 54 Korean individuals.
We classified the de novo variants using the recommendations of Ambry Genetics (Scheme for AD and XD Mendelian disorders) into five categories: Benign, VLB (Variant, Likely benign), VUS (Variant, Unknown significance), VLP (Variant, Likely pathogenic), and Pathogenic mutation. Variants classified as VUS, VLP, and Pathogenic mutation were also confirmed using an IGV browser. To evaluate the mutations in the general population, we also applied Sanger sequencing for these mutations to 80 normal subjects belonging to the Korean ethnic group.
5. Conclusions
RSTS is a rare disorder characterized by facial, dental, and limb dysmorphology, growth retardation, and developmental disorders including intellectual disability and Autism spectrum disorder. Although many de novo mutations have been proposed as the genetic cause of RSTS, genetic analyses to identify mutations have often yielded inconsistent results due to inadequacies in the analysis methods, and many mutations are yet to be confirmed. The objective of this study was to investigate the genetic background of a young girl with RSTS presenting with an Autism phenotype, using whole exome sequencing (WES). We observed de novo variants in the CREBBP, TNC, and IGFALS genes. As far as the authors know, this is the first report to suggest that the TNC and IGFALS genes might contribute to clinical signs that are uncommon in RSTS patients.
Acknowledgments
A summary of this article was previously presented at the Annual Meeting of the American Society of Human Genetics, 2013, in Boston, MA, USA.
This work was supported by the Korea Healthcare Technology R&D project, Ministry of Health & Welfare, Republic of Korea [Grant Number A120029], Korean NRF grants (2011-0030049), and a grant from KRIBB Research Initiative Program.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/03/5697/s1.
Author Contributions
Hee Jeong Yoo and Seong-Hwan Rho recruited subjects. Hee Jeong Yoo and In Hyang Kim performed psychiatric and behavioral assessment of the subject. Namshin Kim, Hee Jeong Yoo and Ki Young Lee conceived and designed the experiments. Soon Ae Kim and Byung Yoon Choi performed the experiments. Kyung Kim and Namshin Kim analyzed the data. Kyung Kim, Hee Jeong Yoo, In Hyang Kim, Jong-Eun Park, and Namshin Kim wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hennekam R.C., Stevens C.A., van de Kamp J.J. Etiology and recurrence risk in Rubinstein-Taybi syndrome. Am. J. Med. Genet. Suppl. 1990;6:56–64. doi: 10.1002/ajmg.1320370610. [DOI] [PubMed] [Google Scholar]
- 2.Hallam T.M., Bourtchouladze R. Rubinstein-Taybi syndrome: Molecular findings and therapeutic approaches to improve cognitive dysfunction. Cell. Mol. Life Sci. 2006;63:1725–1735. doi: 10.1007/s00018-005-5555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinstein J.H., Taybi H. Broad thumbs and toes and facial abnormalities. A possible mental retardation syndrome. Am. J. Dis. Child. 1963;105:588–608. doi: 10.1001/archpedi.1963.02080040590010. [DOI] [PubMed] [Google Scholar]
- 4.Wiley S., Swayne S., Rubinstein J.H., Lanphear N.E., Stevens C.A. Rubinstein-Taybi syndrome medical guidelines. Am. J. Med. Genet. Part A. 2003;119A:101–110. doi: 10.1002/ajmg.a.10009. [DOI] [PubMed] [Google Scholar]
- 5.Miller R.W., Rubinstein J.H. Tumors in Rubinstein-Taybi syndrome. Am. J. Med. Genet. 1995;56:112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- 6.Hennekam R.C., Baselier A.C., Beyaert E., Bos A., Blok J.B., Jansma H.B., Thorbecke-Nilsen V.V., Veerman H. Psychological and speech studies in Rubinstein-Taybi syndrome. Am. J. Mental Retard. 1992;96:645–660. [PubMed] [Google Scholar]
- 7.Hennekam R.C. Rubinstein-Taybi syndrome. Eur. J. Hum. Genet. 2006;14:981–985. doi: 10.1038/sj.ejhg.5201594. [DOI] [PubMed] [Google Scholar]
- 8.Roelfsema J.H., White S.J., Ariyurek Y., Bartholdi D., Niedrist D., Papadia F., Bacino C.A., den Dunnen J.T., van Ommen G.J., Breuning M.H., et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: Mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartsch O., Schmidt S., Richter M., Morlot S., Seemanova E., Wiebe G., Rasi S. DNA sequencing of CREBBP demonstrates mutations in 56% of patients with Rubinstein-Taybi syndrome (RSTS) and in another patient with incomplete RSTS. Hum. Genet. 2005;117:485–493. doi: 10.1007/s00439-005-1331-y. [DOI] [PubMed] [Google Scholar]
- 10.Bentivegna A., Milani D., Gervasini C., Castronovo P., Mottadelli F., Manzini S., Colapietro P., Giordano L., Atzeri F., Divizia M.T. Rubinstein-Taybi syndrome: Spectrum of CREBBP mutations in Italian patients. BMC Med. Genet. 2006;7:77. doi: 10.1186/1471-2350-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negri G., Milani D., Colapietro P., Forzano F., Monica M.D., Rusconi D., Consonni L., Caffi L.G., Finelli P., Scarano G., et al. Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin. Genet. 2015;87:148–154. doi: 10.1111/cge.12348. [DOI] [PubMed] [Google Scholar]
- 12.Spena S., Milani D., Rusconi D., Negri G., Colapietro P., Elcioglu N., Bedeschi F., Pilotta A., Spaccini L., Ficcadenti A. Insights into genotype–phenotype correlations from CREBBP point mutation screening in a cohort of 46 Rubinstein-Taybi syndrome patients. Clin. Genet. 2014 doi: 10.1111/cge.12537. [DOI] [PubMed] [Google Scholar]
- 13.Petrij F., Giles R.H., Dauwerse H.G., Saris J.J., Hennekam R.C., Masuno M., Tommerup N., van Ommen G.J., Goodman R.H., Peters D.J., et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 14.Tsai A.C.-H., Dossett C.J., Walton C.S., Cramer A.E., Eng P.A., Nowakowska B.A., Pursley A.N., Stankiewicz P., Wiszniewska J., Cheung S.W. Exon deletions of the EP300 and CREBBP genes in two children with Rubinstein-Taybi syndrome detected by aCGH. Eur. J. Hum. Genet. 2011;19:43–49. doi: 10.1038/ejhg.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S.R., Kim H.J., Kim Y.J., Kwon J.Y., Kim J.W., Kim S.H. Cryptic microdeletion of the CREBBP gene from t(1;16) (p36.2;p13.3) as a novel genetic defect causing Rubinstein-Taybi syndrome. Ann. Clin. Lab. Sci. 2013;43:450–456. [PubMed] [Google Scholar]
- 16.Blough R.I., Petrij F., Dauwerse J.G., Milatovich-Cherry A., Weiss L., Saal H.M., Rubinstein J.H. Variation in microdeletions of the cyclic AMP-responsive element-binding protein gene at chromosome band 16p13. 3 in the Rubinstein-Taybi syndrome. Am. J. Med. Genet. 2000;90:29–34. doi: 10.1002/(SICI)1096-8628(20000103)90:1<29::AID-AJMG6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Gervasini C., Castronovo P., Bentivegna A., Mottadelli F., Faravelli F., Giovannucci-Uzielli M.L., Pessagno A., Lucci-Cordisco E., Pinto A.M., Salviati L. High frequency of mosaic CREBBP deletions in Rubinstein–Taybi syndrome patients and mapping of somatic and germ-line breakpoints. Genomics. 2007;90:567–573. doi: 10.1016/j.ygeno.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Stevens C.A., Hennekam R.C., Blackburn B.L. Growth in the Rubinstein-Taybi syndrome. Am. J. Med. Genet. Suppl. 1990;6:51–55. doi: 10.1002/ajmg.1320370609. [DOI] [PubMed] [Google Scholar]
- 19.Galera C., Taupiac E., Fraisse S., Naudion S., Toussaint E., Rooryck-Thambo C., Delrue M.A., Arveiler B., Lacombe D., Bouvard M.P. Socio-behavioral characteristics of children with Rubinstein-Taybi syndrome. J. Autism Dev. Disord. 2009;39:1252–1260. doi: 10.1007/s10803-009-0733-4. [DOI] [PubMed] [Google Scholar]
- 20.Calì F., Failla P., Chiavetta V., Ragalmuto A., Ruggeri G., Schinocca P., Schepis C., Romano V., Romano C. Multiplex ligation-dependent probe amplification detection of an unknown large deletion of the CREB-binding protein gene in a patient with Rubinstein-Taybi syndrome. CEP. 2013;14025:220. doi: 10.4238/2013.January.7.2. [DOI] [PubMed] [Google Scholar]
- 21.Hellings J.A., Hossain S., Martin J.K., Baratang R.R. Psychopathology, GABA, and the Rubinstein-Taybi syndrome: A review and case study. Am. J. Med. Genet. 2002;114:190–195. doi: 10.1002/ajmg.10156. [DOI] [PubMed] [Google Scholar]
- 22.Levitas A.S., Reid C.S. Rubinstein-Taybi syndrome and psychiatric disorders. J. Intellect. Disabil. Res. 1998;42:284–292. doi: 10.1046/j.1365-2788.1998.00136.x. [DOI] [PubMed] [Google Scholar]
- 23.Waite J., Moss J., Beck S.R., Richards C., Nelson L., Arron K., Burbidge C., Berg K., Oliver C. Repetitive behavior in Rubinstein-Taybi syndrome: Parallels with Autism spectrum phenomenology. J. Autism Dev. Disord. 2014 doi: 10.1007/s10803-014-2283-7. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Atalaya J.P., Valor L.M., Barco A. Epigenetic factors in intellectual disability: The Rubinstein-Taybi syndrome as a paradigm of neurodevelopmental disorder with epigenetic origin. Prog. Mol. Biol. Transl. Sci. 2014;128:139. doi: 10.1016/B978-0-12-800977-2.00006-1. [DOI] [PubMed] [Google Scholar]
- 25.Goodman R.H., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 26.Marzuillo P., Grandone A., Coppola R., Cozzolino D., Festa A., Messa F., Luongo C., del Giudice E.M., Perrone L. Novel cAMP binding protein-BP (CREBBP) mutation in a girl with Rubinstein-Taybi syndrome, GH deficiency, Arnold Chiari malformation and pituitary hypoplasia. BMC Med. Genet. 2013;14:28. doi: 10.1186/1471-2350-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nies D.E., Hemesath T.J., Kim J.-H., Gulcher J.R., Stefansson K. The complete cDNA sequence of human hexabrachion (Tenascin). A multidomain protein containing unique epidermal growth factor repeats. J. Biol. Chem. 1991;266:2818–2823. [PubMed] [Google Scholar]
- 28.Wiese S., Karus M., Faissner A. Astrocytes as a source for extracellular matrix molecules and cytokines. Front. Pharmacol. 2012;3:120. doi: 10.3389/fphar.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang S.C., Pauls D.L., Lange C., Sasanfar R., Santangelo S.L. Sex-specific association of a common variant of the XG gene with Autism spectrum disorders. Am. J. Med. Genet. Part B. 2013;162:742–750. doi: 10.1002/ajmg.b.32165. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., Zhao F., Zong L., Zhang P., Guan L., Zhang J., Wang D., Wang J., Chai W., Lan L., et al. Exome sequencing and linkage analysis identified tenascin-C (TNC) as a novel causative gene in nonsyndromic hearing loss. PLoS One. 2013;8:e69549. doi: 10.1371/journal.pone.0069549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenhall U., Nordin V., Sandstrom M., Ahlsen G., Gillberg C. Autism and hearing loss. J. Autism Dev. Disord. 1999;29:349–357. doi: 10.1023/A:1023022709710. [DOI] [PubMed] [Google Scholar]
- 32.Schorry E., Keddache M., Lanphear N., Rubinstein J., Srodulski S., Fletcher D., Blough-Pfau R., Grabowski G. Genotype–phenotype correlations in Rubinstein-Taybi syndrome. Am. J. Med. Genet. Part A. 2008;146:2512–2519. doi: 10.1002/ajmg.a.32424. [DOI] [PubMed] [Google Scholar]
- 33.Bourtchouladze R., Lidge R., Catapano R., Stanley J., Gossweiler S., Romashko D., Scott R., Tully T. A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. USA. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leong S.R., Baxter R.C., Camerato T., Dai J., Wood W.I. Structure and functional expression of the acid-labile subunit of the insulin-like growth factor-binding protein complex. Mol. Endocrinol. 1992;6:870–876. doi: 10.1210/mend.6.6.1379671. [DOI] [PubMed] [Google Scholar]
- 35.Courtland H.W., DeMambro V., Maynard J., Sun H., Elis S., Rosen C., Yakar S. Sex-specific regulation of body size and bone slenderness by the acid labile subunit. J. Bone Miner. Res. 2010;25:2059–2068. doi: 10.1002/jbmr.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domene H., Bengolea S., Jasper H., Boisclair Y. Acid-labile subunit deficiency: Phenotypic similarities and differences between human and mouse. J. Endocrinol. Investig. 2004;28:43–46. [PubMed] [Google Scholar]
- 37.Domené H.M., Scaglia P.A., Lteif A., Mahmud F.H., Kirmani S., Frystyk J., Bedecarrás P., Gutiérrez M., Jasper H.G. Phenotypic effects of null and haploinsufficiency of acid-labile subunit in a family with two novel IGFALS gene mutations. J. Clin. Endocrinol. Metab. 2007;92:4444–4450. doi: 10.1210/jc.2007-1152. [DOI] [PubMed] [Google Scholar]
- 38.Park H.W., Kwak K.J., Park G.B. Korean Wechsler Preschool and Primary Scale of Intelligence. Special Education Publishing; Seoul, Korean: 2002. [Google Scholar]
- 39.Sparrow S., Balla D., Cichetti D. Vineland Adaptive Behavior Scales. American Guidance Services; Circle Pines, MN, USA: 1984. [Google Scholar]
- 40.Aylward G. Practitioner’s Guide to Developmental and Psychological Testing. Springer; New York, NY, USA: 1994. [Google Scholar]
- 41.Lord C., Rutter M., DiLavore P.D., Risi S. Autism Diagnostic Observation Schedule. Western Psychological Services; Los Angeles, CA, USA: 2001. [Google Scholar]
- 42.Lord C., Rutter M., Goode S., Heemsbergen J., Jordan H., Mawhood L., Schopler E. Autism diagnostic observation schedule: A standardized observation of communicative and social behavior. J. Autism Dev. Disord. 1989;19:185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- 43.Le Couteur A., Lord C., Rutter M. The Autism Diagnostic Interview-Revised (ADI-R) Western Psychological Services; Los Angeles, CA, USA: 2003. [Google Scholar]
- 44.Lord C., Rutter M., Le Couteur A. Autism diagnostic interview-revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 45.Constantino J., Gruber C.P. Social Responsiveness Scale (SRS) Western Psychological Services; Los Angeles, CA, USA: 2005. [Google Scholar]
- 46.Rutter M., Bailey A., Lord C. Social Communication Questionnaire (SCQ) Western Psychological Services; Los Angeles, CA, USA: 2003. [Google Scholar]
- 47.Buysse D.J., Reynolds C.F., III, Monk T.H., Berman S.R., Kupfer D.J. The Pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989;28:193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- 48.Chung F., Yegneswaran B., Liao P., Chung S.A., Vairavanathan S., Islam S., Khajehdehi A., Shapiro C.M. Stop questionnaire: A tool to screen patients for obstructive sleep apnea. Anesthesiology. 2008;108:812–821. doi: 10.1097/ALN.0b013e31816d83e4. [DOI] [PubMed] [Google Scholar]
- 49.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murata T., Kurokawa R., Krones A., Tatsumi K., Ishii M., Taki T., Masuno M., Ohashi H., Yanagisawa M., Rosenfeld M,G., et al. Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 2001;10:1071–1076. doi: 10.1093/hmg/10.10.1071. [DOI] [PubMed] [Google Scholar]
- 53.Bartsch O., Locher K., Meinecke P., Kress W., Seemanova E., Wagner A., Ostermann K., Rodel G. Molecular studies in 10 cases of Rubinstein-Taybi syndrome, including a mild variant showing a missense mutation in codon 1175 of CREBBP. J. Med. Genet. 2002;39:496–501. doi: 10.1136/jmg.39.7.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalkhoven E., Roelfsema J.H., Teunissen H., den Boer A., Ariyurek Y., Zantema A., Breuning M.H., Hennekam R.C., Peters D.J. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 2003;12:441–450. doi: 10.1093/hmg/ddg039. [DOI] [PubMed] [Google Scholar]
- 55.Coupry I., Roudaut C., Stef M., Delrue M.A., Marche M., Burgelin I., Taine L., Cruaud C., Lacombe D., Arveiler B. Molecular analysis of the CBP gene in 60 patients with Rubinstein-Taybi syndrome. J. Med. Genet. 2002;39:415–421. doi: 10.1136/jmg.39.6.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 57.Udaka T., Samejima H., Kosaki R., Kurosawa K., Okamoto N., Mizuno S., Makita Y., Numabe H., Toral J.F., Takahashi T., et al. Comprehensive screening of CREB-binding protein gene mutations among patients with Rubinstein-Taybi syndrome using denaturing high-performance liquid chromatography. Congenit. Anom. (Kyoto) 2005;45:125–131. doi: 10.1111/j.1741-4520.2005.00081.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.