Abstract
Recently, autism-related research has focused on the identification of various genes and disturbed pathways causing the genetically heterogeneous group of autism spectrum disorders (ASD). The list of autism-related genes has significantly increased due to better awareness with advances in genetic technology and expanding searchable genomic databases. We compiled a master list of known and clinically relevant autism spectrum disorder genes identified with supporting evidence from peer-reviewed medical literature sources by searching key words related to autism and genetics and from authoritative autism-related public access websites, such as the Simons Foundation Autism Research Institute autism genomic database dedicated to gene discovery and characterization. Our list consists of 792 genes arranged in alphabetical order in tabular form with gene symbols placed on high-resolution human chromosome ideograms, thereby enabling clinical and laboratory geneticists and genetic counsellors to access convenient visual images of the location and distribution of ASD genes. Meaningful correlations of the observed phenotype in patients with suspected/confirmed ASD gene(s) at the chromosome region or breakpoint band site can be made to inform diagnosis and gene-based personalized care and provide genetic counselling for families.
Keywords: high-resolution chromosome ideograms, autism, genetic evidence, autism spectrum disorders (ASD), ASD genes
1. Introduction
Classical autism or autistic disorder is common, with developmental difficulties noted by three years of age. It belongs to a group of heterogeneous conditions known as autism spectrum disorders (ASDs) with significant impairments in verbal and non-verbal communication and social interactions with restricted repetitive behaviors, specifically in movements and interests [1,2,3]. Other symptoms include lack of eye contact or focus, sleep disturbances and tactile defensiveness beginning at an early age. Several validated rating scales are used at a young age to help establish the diagnosis, including the autism diagnostic observation schedule (ADOS) and the autism diagnostic interview-revised (ADI-R) supported by pertinent medical history and clinical findings [4,5,6]. ASD affects about 1% of children in the general U.S. population with a 4:1 male to female ratio, usually without congenital anomalies or growth retardation [7,8].
Autism was first used as a term by Kanner in 1943 when describing a group of children lacking the ability to establish interpersonal contact and communication [9]. About one-fourth of children with autism are diagnosed by 2–3 years of age and show regression of skills in about 30% of cases. About 60% of ASD subjects show intellectual disabilities at a young age [10,11]. When comparing the prevalence of health disorders involving the central nervous system, autism ranks higher than epilepsy (6.5 cases per 1000), brain paralysis or dementia (2.5 cases/1000 for each) and Parkinson disease (two cases per 1000); genetic factors are related to many of these disorders [12,13]. Autism also occurs more commonly than congenital malformations in the general population, but dysmorphic findings are present in about 25% of children with autism. Microcephaly is seen in about 10% of cases, but macrocephaly is documented with larger frontal and smaller occipital lobes in about 20% of children with autism. Those with autism and extreme macrocephaly are at a greater risk to have PTEN tumor suppressor gene mutations [14], while another autism-related gene (CHD8) can also lead to macrocephaly and autism [15].
Autism is due to a wide range of genetic abnormalities, as well as non-genetic causes, including the environment, environmental and gene interaction (epigenetics) and metabolic disturbances (e.g., mitochondrial dysfunction), with the recurrence risk dependent on the family history and presence or absence of dysmorphic features. Candidate genes for ASD are identified by different means, including cytogenetic abnormalities (i.e., translocations at chromosome breakpoints or deletions (e.g., the 22q11.2 deletion) indicating the location or loss of specific genes) in individuals with ASD along with overlapping linkage and functional data related to the clinical presentation, with certain chromosome regions identified by genetic linkage using DNA markers that co-inherit with the specific phenotype [16,17]. A representative example for such an occurrence is the proto-oncogene (MET) involved in pathways related to neuronal development [18] and found to be linked to the chromosome 7q31 band, where this gene is located. Decreased activity of the gene promoter was recognized when specific single nucleotide polymorphisms (SNPs) were present in this region by linkage studies. However, genetic linkage studies have received only limited success in the study of the genetics of autism. On the other hand, chromosomal microarray analysis using DNA probes disturbed across the genome can be used to detect chromosomal abnormalities at >100-times smaller than seen in high-resolution chromosome studies. Microarray studies have also become the first tier of genetic testing for this patient population and are recommended for all ASD patients [19]. Greater than 20% of studied patients with microarray analysis are found to have submicroscopic deletions or duplications in the genome containing genes that play a role in causing autism [20,21]. Identification of causative mutations is important to guide treatment selection and to manage medical co-morbidities, such as risks for seizures, developmental regression or for cancer (e.g., the PTEN gene).
Routine cytogenetic studies have shown abnormalities of chromosomes 2, 3, 4, 5, 7, 8, 11, 13, 15, 16, 17, 19, 22 and X, including deletions, duplications, translocations and inversions involving specific chromosome regions where known or candidate genes for ASD are located [22]. These studies further support the role of genetic factors in the causation of this common neurodevelopment disorder. Specifically, cytogenetic abnormalities involving the 15q11–q13 region are found in at least 1% of individuals with ASD and include CYFIP1, GABRB3 and UBE3A genes in this chromosome region [23] and most recently the 15q11.2 BP1-BP2 microdeletion (Burnside-Butler) syndrome [24]. DNA copy number changes have also shown recurrent small deletions or duplications of the chromosome 16p11.2 band using microarray analysis [25,26] and the chromosome 15q13.2–q13.3 region [27], whereas copy number changes are noted throughout the genome in individuals with ASD, indicating the presence of multiple candidate genes on every human chromosome. These copy number changes are more often of the deletion type.
For idiopathic or non-syndromic autism, the empirical risk for siblings to be similarly affected is between 2% and 8% with an average of 4% [28]. In multiplex families having two or more affected children with autism, the recurrence risk may be as high as 25%, but generally ranges from 13% [29] to 19% [30] if due to single-gene disturbances as the cause, a major focus of this illustrative review. Advances in genetic technology beyond linkage or cytogenetic analysis of affected families with ASD or other complex disorders have led to genome-wide association studies (GWAS) involving hundreds of affected and control individuals by analyzing the distribution and clustering of hundreds and thousands of SNPs that have successfully been searched for candidate genes. The first GWAS for ASD was undertaken by Lauritsen et al. in 2006 [31] using 600 DNA markers in an isolated population of affected individuals from the Faroe Islands. They found an association of the chromosome 3p25.3 band, and later, other investigators studied more subjects with larger collections of genotyped markers and found several chromosome bands and regions ascertained when specific SNPs were over-represented in the ASD subjects, including 5p14.1, 5p15 and 16p13–p21 [32,33,34,35,36,37]. The studies implicated several gene families, including the cadherin family, encoding proteins for neuronal cell adhesion, while other genes (e.g., SEMA5A) were implicated in axonal guidance with lower gene expression levels in brain specimens from individuals with ASD [33], reviewed by Holt and Monaco [17]. Since that time, several additional studies searching for clinically relevant and known genes for ASD have identified a new collection of ASD genes [38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53].
The ability to identify an increased number of SNPs with advanced genetic platforms and extensive approaches using bioinformatics have led to improved access and a more thorough analysis. This has led to comparing genotyping data from GWAS and DNA copy number variants (CNVs) with the identification of structural genetic defects, such as submicroscopic deletions or duplications of the genome, which was not possible a few years ago. Separate studies using array comparative genomic hybridization or microarray analysis to investigate those individuals with ASD continue to yield useful data in identifying candidate genes for ASD in affected individuals [20,21,54]. The yield for microarray analysis is reported to be approximately 20% for identifying deletions or duplications at sites where known or candidate ASD genes are present. The use of more advanced technology, such as next-generation sequencing (whole genome or exome) will yield additional valuable information on the location and description of lesions of genes contributing to ASD with increasing evidence for specific and recurring mutations of single genes involved with neurodevelopment and function, leading to potential therapeutic discoveries and interventions.
Autism is frequent in single-gene conditions, such as fragile X syndrome, tuberous sclerosis, Rett syndrome or neurofibromatosis, but single-gene disorders as a whole account for less than 20% of all cases; therefore, most individuals with ASD are non-syndromic. The heritability of ASD, which takes into consideration the extent of genetic factors contributing to autism, is estimated to be as high as 90% [55]; hence the relevance and continued importance of investigating the role of genetics in the causation of ASD and expanded diagnostic testing to inform and guide treatment for individuals with identifiable genetic disturbances.
A current list of clinically relevant and known candidate genes for ASD is needed for diagnostic testing and genetic counselling purposes in the clinical setting. Historically, a previous list of known or candidate genes showing an association with ASD was reported in 2011 by Holt and Monaco [17] with the placement of 175 genes on chromosome ideograms. A much greater number of validated genes are now recognized as playing a pivotal role in ASD, warranting an updated, revised summary. We will utilize high-resolution chromosome ideograms (850 band level) to plot the location of genes now recognized by searching the literature and website information as playing a documented role in ASD. In tabular form, we will list the individual gene symbol, expanded name or description and chromosome location.
2. Results and Discussion
The diagnostic approach for an individual with ASD should include a clinical genetics evaluation with interviews of parents and health caregivers for the collection and overview of historical problems, a three-generation family pedigree, recording of developmental milestones and description of atypical behaviors along with medical and surgical procedures and a current list of medications and ongoing treatments. Laboratory tests should include lead, thyroid function, lactate and pyruvate levels in order to assess metabolic and mitochondrial functions that may be impacted by an underlying genetic disturbance along with cholesterol and urine collection for organic acid levels. Brain imaging and electroencephalogram patterns should be reviewed, if available. In addition, the ADI-R and ADOS instruments are used to test the diagnosis of ASD.
To further increase the diagnostic yield in individuals with ASD presenting for genetic service, Schaefer et al. [19] proposed and utilized a three-tier approach to include a genetic work-up by a clinical geneticist with expertise in dysmorphology to identify known syndromes with or without dysmorphic features (e.g., birth marks), growth anomalies (e.g., microcephaly, macrocephaly and short stature), viral titers (e.g., rubella) and metabolic screening (urine for organic acids and mucopolysaccharides, plasma lactate and amino acid levels). DNA testing for fragile X syndrome and Rett syndrome in females and males is also available, along with chromosomal and DNA microarrays to examine structural DNA lesions in those with a sporadic form of autism and the use of SNP arrays to examine for regions of homozygosity or uniparental disomy, whereby both members of a chromosome pair come from one parent [56]. Exome sequencing is now available particularly to those affected subjects with a positive family history of autism (multiplex families), if other diagnostic tests are uninformative. PTEN gene mutation screening would be indicated in those patients with extreme macrocephaly (head size > 2 SD) [14], if not previously done, and a review of brain MRI results. Serum and urine uric acid levels and assays for adenylate succinase deficiency should be done to include biochemical genetic studies and mitochondrial genome screening and function [57] if the above testing protocols are not diagnostic. Up to one in five children with ASD show findings of mitochondrial dysfunction [57], and a detailed genetic work-up will significantly increase the yield for the diagnosis of ASD, leading to a better understanding of causation, treatment and more accurate genetic counselling for those presenting for genetic services [20,21,54].
Advances made in genetic technology and bioinformatics have led to vastly improved genetic testing options for application in the clinical setting in patients presenting for genetic services [54]. Significant discoveries have been made with the recognition of genetic defects in the causation of ASD using microarray technology and, now, next generation sequencing. This technology has flourished with a combination of DNA probes used for both copy number variation and SNPs being required to identify segmental deletions and duplications in the genome and regions of homozygosity for the determination of identical by descent for the calculation of inbreeding coefficients or consanguinity status along with uniparental disomy of individual chromosomes [56].
Next generation exome DNA sequencing and RNA sequencing allows for discoveries of disease-causing genes and regulatory sequences required for normal function. Identifying and characterizing molecular signatures for novel or disturbed gene or exon expression and disease-specific profiles and patterns with expression heat maps have led to the recognition of interconnected disturbed gene pathways in many diseases, including a growing body of genetic evidence for autism and other psychiatric or aberrant behavioral disorders [54].
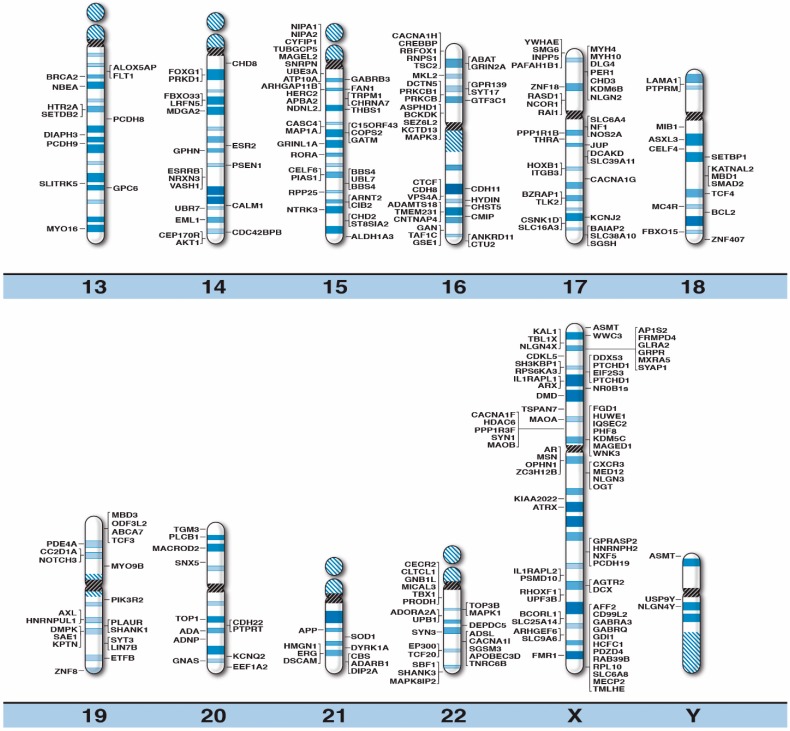
The position for each known or candidate gene for ASD susceptibility is plotted on high-resolution chromosome ideograms (850 band level), as shown in Figure 1 below. We have included gene symbols and expanded names along with the chromosome band location in Table 1 for the 792 genes recognized as playing a role in ASD.
Figure 1.
High-resolution human chromosome ideograms (850 band level) with the ASD gene symbol placed at the chromosomal band location. The centromere area, highlighted in black, separates the upper short “p” arm and lower long “q” arm for each chromosome. The gene symbols are arranged in alphabetical order with the expanded name and chromosome band position listed in Table 1.
Table 1.
Recognized genes for autism spectrum disorders (ASD) and their chromosome locations.
| Gene Symbol | Gene Name | Location |
|---|---|---|
| ABAT | 4-aminobutyrate aminotransferase | 16p13.2 |
| ABCA7 | ATP-binding cassette, sub-family A (ABC1), member 7 | 19p13.3 |
| ABI1 | Abl-interactor 1 | 10p12.1 |
| ABI2 | Abl-interactor 2 | 2q33.2 |
| ABL1 | C-Abl oncogene 1, non-receptor tyrosine kinase | 9q34.12 |
| ACY1 | Aminoacylase 1 | 3p21.2 |
| ADA | Adenosine deaminase | 20q13.12 |
| ADAMTS18 | A disintegrin-like and metalloproteinase with thrombospondin type 1 motif, 18 | 16q23.1 |
| ADARB1 | Adenosine deaminase, RNA-specific, B1 | 21q22.3 |
| ADCY5 | Adenylate cyclase 5 | 3q21.1 |
| ADK | Adenosine kinase | 10q22.2 |
| ADNP | Activity-dependent neuroprotector homeobox | 20q13.13 |
| ADORA2A | Adenosine A2A receptor | 22q11.23 |
| ADORA3 | Adenosine A3 receptor | 1p13.2 |
| ADRB2 | Adrenergic, β 2 receptor | 5q32 |
| ADSL | Adenylosuccinate lyase | 22q13.1 |
| AFF2 | AF4/fragile X mental retardation 2 (FMR2) family, member 2 | Xq28 |
| AFF4 | AF4/fragile X mental retardation 2 (FMR2) family, member 4 | 5q31.1 |
| AGBL4 | ATP/GTP binding protein-like 4 | 1p33 |
| AGMO | Alkylglycerol monooxygenase | 7p21.1 |
| AGTR2 | Angiotensin II receptor, type 2 | Xq23 |
| AHI1 | Abelson helper integration site 1 | 6q23.3 |
| AHRR | Aryl hydrocarbon receptor repressor | 5p15.33 |
| AKT1 | v-Akt murine thymoma viral oncogene homolog 1 | 14q32.33 |
| ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 15q26.3 |
| ALDH5A1 | Aldehyde dehydrogenase 5 family, member A1 | 6p22.3 |
| ALOX5AP | Arachidonate 5-lipoxygenase-activating protein | 13q12.3 |
| AMPD1 | Adenosine monophosphate deaminase 1 | 1p13.2 |
| AMT | Aminomethyltransferase | 3p21.31 |
| ANK2 | Ankyrin 2 | 4q25 |
| ANK3 | Ankyrin 3 | 10q21.2 |
| ANKRD11 | Ankyrin repeat domain 11 | 16q24.3 |
| ANXA1 | Annexin A1 | 9q21.13 |
| AP1S2 | Adaptor-related protein complex 1, sigma 2 subunit | Xp22.2 |
| APBA2 | Amyloid β precursor protein-binding, family A, member 2 | 15q13.1 |
| APC | Adenomatosis polyposis coli | 5q22.2 |
| APH1A | APH1A γ secretase subunit | 1q21.2 |
| APOBEC3D | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D | 22q13.1 |
| APP | Amyloid β precursor protein | 21q21.3 |
| AR | Androgen receptor | Xq12 |
| ARHGAP11B | Rho GTPase activating protein 11B | 15q13.2 |
| ARHGAP15 | Rho GTPase activating protein 15 | 2q22.2 |
| ARHGAP24 | Rho GTPase activating protein 24 | 4q22.1 |
| ARHGEF6 | RAC/CDC42 guanine nucleotide exchange factor (GEF) 6 | Xq26.3 |
| ARID1B | AT rich interactive domain 1B (SWI1-like) | 6q25.3 |
| ARID5A | AT rich interactive domain 5A (MRF1-like) | 2q11.2 |
| ARL6IP6 | ADP-ribosylation-like factor 6 interacting protein 6 | 2q23.3 |
| ARNT2 | Aryl-hydrocarbon receptor nuclear translocator 2 | 15q25.1 |
| ARX | Aristaless related homeobox | Xp21.3 |
| ASH1L | Ash1 (absent, small, or homeotic)-like (Drosophila) | 1q22 |
| ASMT | Acetylserotonin O-methyltransferase, X-chromosomal | Xp22.33 |
| ASMT | Acetylserotonin O-methyltransferase, Y-chromosomal | Yp11.32 |
| ASPHD1 | Aspartate β-hydroxylase domain containing 1 | 16p11.2 |
| ASPM | Asp (abnormal spindle) homolog, microcephaly associated | 1q31.3 |
| ASS1 | Argininosuccinate synthetase | 9q34.1 |
| ASTN2 | Astrotactin 2 | 9q33.1 |
| ASXL3 | Additional sex combs-like 3 | 18q12.1 |
| ATG7 | Autophagy related 7 | 3p25.3 |
| ATP10A | ATPase, Class V, type 10A | 15q11.2 |
| ATP2B2 | ATPase, Ca++ transporting, plasma membrane 2 | 3p25.3 |
| ATRNL1 | Attractin-like 1 | 10q25.3 |
| ATRX | α thalassemia/mental retardation syndrome X-linked | Xq21.1 |
| ATXN7 | Ataxin 7 | 3p14.1 |
| AUTS2 | Autism susceptibility candidate 2 | 7q11.22 |
| AVPR1A | Arginine vasopressin receptor 1A | 12q14.2 |
| AXL | AXL receptor tyrosine kinase | 19q13.2 |
| BAIAP2 | BAI1-associated protein 2 | 17q25.3 |
| BBS4 | Bardet-Biedl syndrome 4 | 15q24.1 |
| BCKDK | Branched chain ketoacid dehydrogenase kinase | 16p11.2 |
| BCL11A | B-Cell CLL/lymphoma 11A (zinc finger protein) | 2p16.1 |
| BCL2 | B-cell CLL/lymphoma 2 | 18q21.33 |
| BCORL1 | Bc16 co-repressor-like 1 | Xq26.1 |
| BDNF | Brain-derived neurotrophic factor | 11p14.1 |
| BIN1 | Bridging integrator 1 | 2q14.3 |
| BIRC6 | Baculoviral IAP repeat containing 6 | 2p22.3 |
| BRAF | v-Raf murine sarcoma viral oncogene homolog B | 7q34 |
| BRCA2 | Breast cancer 2, early onset | 13q13.1 |
| BTAF1 | RNA polymerase II, B-TFIID transcription factor-associated, 170 kDa (Mot1 homolog, S. cerevisiae) | 10q23.32 |
| BZRAP1 | Benzodiazepine receptor (peripheral) associated protein 1 | 17q23.2 |
| C11ORF30 | Chromosome 11 open reading frame 30 | 11q13.5 |
| C12ORF57 | Chromosome 12 open reading frame 57 | 12p13.31 |
| C15ORF43 | Chromosome 15 open reading frame 43 | 15q21.1 |
| C3ORF58 | Chromosome 3 open reading frame 58 | 3q24 |
| C4B | Complement component 4B | 6p21.33 |
| CA6 | Carbonic anhydrase VI | 1p36.2 |
| CACNA1B | Calcium channel, voltage-dependent, N type, α 1B subunit | 9q34.3 |
| CACNA1C | Calcium channel, voltage-dependent, L type, α 1C subunit | 12p13.33 |
| CACNA1D | Calcium channel, voltage-dependent, L type, α 1D subunit | 3p14.3 |
| CACNA1F | Calcium channel, voltage-dependent, α 1F subunit | Xp11.23 |
| CACNA1G | Calcium channel, voltage-dependent, T type, α 1G subunit | 17q21.33 |
| CACNA1H | Calcium channel, voltage-dependent, α 1H subunit | 16p13.3 |
| CACNA1I | Calcium channel, voltage-dependent, T type, α 1I subunit | 22q13.1 |
| CACNA2D3 | Calcium channel, voltage-dependent, α 2/δ subunit 3 | 3p21.1 |
| CACNB2 | Calcium channel, voltage-dependent, β 2 subunit | 10p12.33 |
| CADM1 | Cell adhesion molecule 1 | 11q23.3 |
| CADPS2 | Ca++-dependent activator protein for secretion 2 | 7q31.32 |
| CALM1 | Calmodulin 1 (phosphorylase kinase, δ) | 14q32.11 |
| CAMK4 | Calcium/calmodulin-dependent protein kinase | 5q22.1 |
| CAMSAP2 | Calmodulin regulated spectrin-associated protein family, member 2 | 1q32.1 |
| CAMTA1 | Calmodulin binding transcription activator 1 | 1p36.31 |
| CAPRIN1 | Cell cycle associated protein 1 | 11p13 |
| CASC4 | Cancer susceptibility candidate 4 | 15q15.3 |
| CBS | Cystathionine β-synthase | 21q22.3 |
| CCAR2 | Cell cycle and apoptosis regulator 2 | 8p21.3 |
| CC2D1A | Coiled-coil and C2 domain-containing 1A | 19p13.12 |
| CCDC19 | Coiled-coil domain-containing protein 19 | 1q23.2 |
| CCDC64 | Coiled-coil domain-containing 64 | 12q24.23 |
| CD38 | CD38 molecule | 4p15.32 |
| CD44 | CD44 molecule | 11p13 |
| CD163L1 | CD163 molecule-like 1 | 12p13.31 |
| CD99L2 | CD99 molecule-like 2 | Xq28 |
| CDC42BPB | CDC42 binding protein kinase β (DMPK-like) | 14q32.32 |
| CDH10 | Cadherin 10, type 2 | 5p14.2 |
| CDH22 | Cadherin-like 22 | 20q13.1 |
| CDH8 | Cadherin 8, type 2 | 16q22.1 |
| CDH9 | Cadherin 9, type 2 | 5p14.1 |
| CDH11 | Cadherin 11, type 2 | 16q21 |
| CDKL5 | Cyclin-dependent kinase-like 5 | Xp22.13 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | 12p13.1 |
| CECR2 | Cat eye syndrome chromosome region, candidate 2 | 22q11.21 |
| CELF4 | CUGBP, Elav-like family, member 4 | 18q12.2 |
| CELF6 | CUGBP, Elav-like family, member 6 | 15q23 |
| CENTG2 | Centaurin γ-2 | 2q37.2 |
| CEP170R | Centrosomal protein 170B | 14q32.33 |
| CEP290 | Centrosomal protein 290 kDa | 12q21.32 |
| CEP41 | Centrosomal protein 41 kDa | 7q32.2 |
| CHD1 | Chromodomain helicase DNA binding protein 1 | 5q21.1 |
| CHD2 | Chromodomain helicase DNA binding protein 2 | 15q26.1 |
| CHD3 | Chromodomain helicase DNA binding protein 3 | 17p13.1 |
| CHD7 | Chromodomain helicase DNA binding protein 7 | 8q12.2 |
| CHD8 | Chromodomain helicase DNA binding protein 8 | 14q11.2 |
| CHRM3 | Cholinergic receptor, muscarinic 3 | 1q43 |
| CHRNA7 | Cholinergic receptor, neuronal nicotinic, α 7 | 15q13.3 |
| CHRNB3 | Cholinergic receptor, neuronal nicotinic, β 3 | 8p11.21 |
| CHST5 | Carbohydrate sulfotransferase 5 | 16q22.3 |
| CIB2 | Calcium and integrin binding family member 2 | 15q25.1 |
| CKAP5 | Cytoskeleton associated protein 5 | 11p11.2 |
| CLCNKB | Chloride channel voltage-sensitive kidney, B | 1p36.13 |
| CLSTN3 | Calsyntenin 3 | 12p13.31 |
| CLTCL1 | Clathrin, heavy chain-like 1 | 22q11.21 |
| CMIP | c-MAF inducing protein | 16q23.2 |
| CNR1 | Cannabinoid receptor 1 | 6q15 |
| CNR2 | Cannabinoid receptor 2 | 1p36.11 |
| CNTN3 | Contactin 3 | 3p12.3 |
| CNTN4 | Contactin 4 | 3p26.3 |
| CNTN5 | Contactin 5 | 11q22.1 |
| CNTN6 | Contactin 6 | 3p26.3 |
| CNTNAP2 | Contactin associated protein-like 2 | 7q35 |
| CNTNAP3 | Contactin associated protein-like 3 | 9p13.1 |
| CNTNAP4 | Contactin associated protein-like 4 | 16q23.1 |
| CNTNAP5 | Contactin associated protein-like 5 | 2q14.3 |
| COL7A1 | Collagen, type VII, α 1 | 3p21.31 |
| COPS2 | Thyroid hormone receptor interactor 15 | 15q21.1 |
| CREBBP | CREB binding protein | 16p13.3 |
| CSMD1 | Cytoskeleton associated protein 5 | 11p11.2 |
| CSNK1D | Casein kinase 1, δ | 17q25 |
| CSTF2T | Cleavage stimulation factor, 3' pre-RNA, subunit 2, 64 kDa, tau | 10q21.1 |
| CTCF | CCCTC-binding factor | 16q22.1 |
| CTNNA3 | Catenin (cadherin-associated protein), α 3 | 10q21.3 |
| CTNNB1 | Catenin (cadherin-associated protein), β 1, 88 kDa | 3p22.1 |
| CTSB | Cathepsin B | 8p23.1 |
| CTTNBP2 | Cortactin binding protein 2 | 7q31.31 |
| CTU2 | Cytosolic thiouridylase subunit 2 homolog (S. pombe) | 16q24.3 |
| CUEDC2 | CUE domain containing 2 | 10q24.32 |
| CUL5 | Cullin 5 | 11q22.3 |
| CUL3 | Cullin 3 | 2q36.2 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 3p22.2 |
| CXCR3 | Chemokine, CXC motif, receptor 3 | Xq13.1 |
| CYFIP1 | Cytoplasmic FMRP interacting protein 1 | 15q11.2 |
| CYP11B1 | Cytochrome P450, subfamily XIB, polypeptide 1 | 8q24.3 |
| DAB1 | Disabled homolog 1 | 1p32.2 |
| DAG1 | Dystroglycan 1 (dystrophin-associated glycoprotein 1) | 3p21.31 |
| DAGLA | Diacylglycerol lipase, α | 11q12.2 |
| DAPK1 | Death-associated protein kinase 1 | 9q21.33 |
| DAPP1 | Dual adaptor of phosphotyrosine and 3-phosphoinositides 1 | 4q23 |
| DCAF13 | DDB1 and CUL4 associated factor 13 | 8q22.3 |
| DCAKD | Dephospho-CoA kinase domain-containing protein | 17q21.31 |
| DCTN5 | Dynactin 5 | 16p12.2 |
| DCUN1D1 | DCN1, domain containing protein 1 | 3q27.1 |
| DCX | Doublecortin | Xq23 |
| DDC | DOPA decarboxylase | 7p12.1 |
| DDX11 | DEAD (Asp-Glu-Ala-Asp)/H box 11 | 12p11.21 |
| DDX53 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 53 | Xp22.11 |
| DEAF1 | DEAF1 transcription factor | 11p15.5 |
| DEPDC5 | DEP domain containing 3 protein 5 | 22q12.2 |
| DHCR7 | 7-dehydrocholesterol reductase | 11q13.4 |
| DHX9 | DEAH (Asp-Glu-Ala-His) box helicase 9 | 1q25.3 |
| DIAPH3 | Diaphanous, Drosophila, homolog 3 | 13q21.2 |
| DIP2A | DIP2 disco-interacting protein 2 homolog A (Drosophila) | 21q22.3 |
| DISC1 | Disrupted in schizophrenia 1 | 1q42.2 |
| DLG4 | Discs, large, Drosophila, homolog 4 | 17p13.1 |
| DLGAP2 | Discs, large- associated protein 2 | 8p23.3 |
| DLGAP3 | Discs, large- associated protein 3 | 1p34.3 |
| DLL1 | δ-like 1 (Drosophila) | 6q27 |
| DLX1 | Distal-less homeobox 1 | 2q31.1 |
| DLX2 | Distal-less homeobox 2 | 2q31.1 |
| DLX6 | Distal-less homeobox 6 | 7q21.3 |
| DMD | Dystrophin | Xp21.1 |
| DMPK | Dystrophia myotonica-protein kinase | 19q13.32 |
| DNAJC19 | DNAJ Hsp40 homolog, subfamily C, member 19 | 3q26.33 |
| DNER | δ- and notch-like epidermal growth factor-related receptor | 2q36.3 |
| DNM1L | Dynamin 1-like | 12p11.21 |
| DNMT3A | DNA (cytosine-5)-methyltransferase 3 α | 2p23.3 |
| DOCK4 | Dedicator of cytokinesis 4 | 7q31.1 |
| DOCK10 | Dedicator of cytokinesis 10 | 2q36.2 |
| DOLK | Dolichol kinase | 9q34.1 |
| DPP10 | Dipeptidyl peptidase 10 | 2q14.1 |
| DPP6 | Dipeptidyl peptidase 6 | 7q36.2 |
| DPYD | Dihydropyrimidine dehydrogenase | 1p21.3 |
| DRD1 | Dopamine receptor D1 | 5q35.2 |
| DRD2 | Dopamine receptor D2 | 11q23.2 |
| DRD3 | Dopamine receptor D3 | 3q13.31 |
| DSCAM | Down syndrome cell adhesion molecule | 21q22.2 |
| DST | Dystonin | 6p12.1 |
| DUSP22 | Dual specificity phosphatase 22 | 6p25.3 |
| DYDC1 | DPY30 domain containing 1 | 10q23.1 |
| DYDC2 | DPY30 domain containing 2 | 10q23.1 |
| DYRK1A | Dual-specificity tyrosine-phosphorylation-regulated kinase 1A | 21q22.13 |
| EEF1A2 | Eukaryotic translation elongation factor 1 α 2 | 20q13.33 |
| EFR3A | EFR3 homolog A (S. cerevisiae) | 8q24.22 |
| EGR2 | Early growth response 2 | 10q21.3 |
| EHMT1 | Euchromatic histone methyltransferase 1 | 9q34.3 |
| EIF2S3 | Eukaryotic translation initiation factor 2, subunit 3 γ | Xp22.11 |
| EIF4E | Eukaryotic translation initiation factor 4E | 4q23 |
| EIF4EBP2 | Eukaryotic translation initiation factor 4E binding protein 2 | 10q22.1 |
| EML1 | Echinoderm microtubule associated protein like 1 | 14q32.2 |
| EN2 | Engrailed 2 | 7q36.3 |
| EP300 | E1A binding protein p300 | 22q13.2 |
| EP400 | E1A binding protein p400 | 12q24.33 |
| EPC2 | Enhancer of polycomb, Drosophila homolog of 2 | 2q23.1 |
| EPHA6 | Ephrin receptor A6 | 3q11.2 |
| EPHB2 | Ephrin receptor B2 | 1p36.12 |
| EPHB6 | Ephrin receptor B6 | 7q34 |
| EPS8 | Epidermal growth factor receptor pathway substrate 8 | 12p12.3 |
| ERBB4 | v-ERB-A avian erythroblastic leukemia viral oncogene homolog 4 | 2q34 |
| ERG | v-ETS avian erythroblastosis virus E26 oncogene homolog | 21q22.2 |
| ESR1 | Estrogen receptor 1 | 6q25.1 |
| ESR2 | Estrogen receptor 2 | 14q23.2 |
| ESRRB | Estrogen-related receptor β | 14q24.3 |
| ETFB | Electron-transfer-flavoprotein, β polypeptide | 19q13.41 |
| ETV1 | Ets variant 1 | 7p21.2 |
| EXOC6B | Exocyst complex component 6B | 2p13.2 |
| EXT1 | Exostosin 1 | 8q24.11 |
| F13A1 | Factor XIII, A1 subunit | 6p25.1 |
| FABP3 | Fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 1p35.2 |
| FABP5 | Fatty acid binding protein 5 | 8q21.13 |
| FABP7 | Fatty acid binding protein 7 | 6q22.31 |
| FAM135B | Family with sequence similarity 135, member B | 8q24.23 |
| FAN1 | FANCD2/FANCI-associated nuclease 1 | 15q13.2 |
| FAT1 | FAT tumor suppressor, Drosophila homolog of, 1 | 4q35.2 |
| FAT3 | FAT tumor suppressor, Drosophila homolog of , 3 | 11q14.3 |
| FBXO15 | F-box protein 15 | 18q22.3 |
| FBXO33 | F-box protein 33 | 14q21.1 |
| FBXO40 | F-box protein 40 | 3q13.33 |
| FBXW7 | F-box and WD repeat domain containing 7, E3 ubiquitin protein | 4q31.3 |
| FER | FPS/FES related tyrosine kinase | 5q21.3 |
| FEZF2 | FEZ family zinc finger 2 | 3p14.2 |
| FGA | Fibrinogen, A α polypeptide | 4q31.3 |
| FGD1 | FYVE, Rho GEF and PH domain containing 1 | Xp11.22 |
| FGFBP3 | Fibroblast growth factor binding protein 3 | 10q23.32 |
| FHIT | Fragile histidine triad | 3p14.2 |
| FLT1 | c-FMS-related tyrosine kinase 1 | 13q12.3 |
| FMR1 | Fragile X mental retardation 1 (FMR1) | Xq27.3 |
| FOLH1 | Folate hydrolase 1 | 11p11.2 |
| FOXG1 | Forkhead box G1 | 14q12 |
| FOXP1 | Forkhead box P1 | 3p13 |
| FOXP2 | Forkhead box P2 | 7q31.1 |
| FRK | FYN-related kinase | 6q22.1 |
| FRMPD4 | FERM and PDZ domain containing protein 4 | Xp22.2 |
| GABRA1 | γ-aminobutyric acid A receptor, α 1 | 5q34 |
| GABRA3 | γ-aminobutyric acid receptor, α 3 | Xq28 |
| GABRA4 | γ-aminobutyric acid receptor, α 4 | 4p12 |
| GABRB1 | γ-aminobutyric acid receptor, β 1 | 4p12 |
| GABRB3 | γ-aminobutyric acid receptor, β 3 | 15q12 |
| GABRQ | γ-aminobutyric acid receptor, θ | Xq28 |
| GAD1 | Glutamate decarboxylase 1 (brain, 67 kDa) | 2q31.1 |
| GALNT13 | UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyl-transferase 13 | 2q23.3 |
| GALNT14 | UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyl-transferase 14 | 2p23.1 |
| GAN | Gigaxonin | 16q24.1 |
| GAP43 | Growth associated protein 43 | 3q13.31 |
| GAS2 | Growth arrest-specific 2 | 11p14.3 |
| GATM | Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | 15q21.1 |
| GDI1 | GDP dissociation inhibitor 1 | Xq28 |
| GIGYF1 | GRB10 interacting GYF protein 1 | 7q22.1 |
| GLO1 | Glyoxalase I | 6p21.2 |
| GLRA2 | Glycine receptor, α 2 subunit | Xp22.2 |
| GNA14 | Guanine nucleotide-binding protein, α 14 | 9q21.2 |
| GNAS | Guanine nucleotide-binding protein, α-stimulating activity polypeptide I complex locus | 20q13.32 |
| GNB1L | Guanine nucleotide-binding protein, β 1-like | 22q11.21 |
| GPC6 | Glypican 6 | 13q31.3 |
| GPD2 | Glycerol-3-phosphate dehydrogenase 2 | 2q24.1 |
| GPHN | Gephyrin | 14q23.3 |
| GPR139 | G protein-coupled receptor 139 | 16p12.3 |
| GPR37 | G protein-coupled receptor 37 | 7q31.33 |
| GPRASP2 | G protein-coupled receptor associated sorting protein 2 | Xq22.1 |
| GPX1 | Glutathione peroxidase 1 | 3p21.31 |
| GRID1 | Glutamate receptor, ionotropic, δ 1 | 10q23.2 |
| GRID2 | Glutamate receptor, ionotropic, δ 2 | 4q22.1 |
| GRIK2 | Glutamate receptor, ionotropic, kainate 2 | 6q16.3 |
| GRIN1 | Glutamate receptor, ionotropic, N-methyl d-aspartate 1 | 9q34.3 |
| GRIN2A | Glutamate receptor, ionotropic, N-methyl d-aspartate 2A | 16p13.2 |
| GRIN2B | Glutamate receptor, ionotropic, N-methyl d-aspartate 2B | 12p13.1 |
| GRINL1A | GRINL1A complex locus 1 | 15q21.3 |
| GRIP1 | Glutamate receptor interacting protein 1 | 12q14.3 |
| GRM1 | Glutamate receptor, metabotropic 1 | 6q24.3 |
| GRM4 | Glutamate receptor, metabotropic 4 | 6p21.31 |
| GRM5 | Glutamate receptor, metabotropic 5 | 11q14.3 |
| GRM8 | Glutamate receptor, metabotropic 8 | 7q31.33 |
| GRPR | Gastrin-releasing peptide receptor | Xp22.2 |
| GSE1 | Gse1 coiled-coil protein | 16q24.1 |
| GSK3B | Glycogen synthase kinase 3 β | 3q13.33 |
| GSN | Gelsolin | 9q33.2 |
| GSTM1 | Glutathione S-transferase M1 | 1p13.3 |
| GTF2I | General transcription factor III | 7q11.23 |
| GTF2IRD1 | GTF2I repeat domain containing 1 | 7q11.23 |
| GTF3C1 | General transcription factor IIIC, polypeptide 1, α | 16p12.1 |
| GUCY1A2 | Guanylate cyclase 1, soluble, α 2 | 11q22.3 |
| HCAR1 | Hydroxycarboxylic acid receptor 1/G protein-coupled receptor 81 | 12q24.31 |
| HCFC1 | Host cell factor C1 | Xq28 |
| HCN1 | Hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | 5p12 |
| HDAC4 | Histone deacetylase 4 | 2q37.3 |
| HDAC6 | Histone deacetylase 6 | Xp11.23 |
| HDAC9 | Histone deacetylase 9 | 7p21.1 |
| HDLBP | High density lipoprotein binding protein | 2q37.3 |
| HEPACAM | Hepatic and glial cell adhesion molecule | 11q24.2 |
| HERC2 | HECT domain and RCC1-like domain 2 | 15q13.1 |
| HLA-A | Major histocompatibility complex, class I, A | 6p22.1 |
| HLA-DRB1 | Major histocompatibility complex, class II, DR β 1 | 6p21.32 |
| HMGN1 | High mobility group nucleosome binding domain 1 | 21q22.2 |
| HNRNPF | Heterogeneous nuclear ribonucleoprotein F | 10q11.21 |
| HNRNPH2 | Heterogeneous nuclear ribonucleoprotein H2 | Xq22.1 |
| HNRNPUL1 | Heterogeneous nuclear ribonucleoprotein U-like 1 | 19q13.2 |
| HOMER1 | Homer, Drosophila, homolog 1 of 1 | 5q14.1 |
| HOXA1 | Homeobox A1 | 7p15.3 |
| HOXB1 | Homeobox B1 | 17q21.32 |
| HRAS | v-HA-RAS Harvey rat sarcoma viral oncogene homolog | 11p15.5 |
| HS3ST5 | Heparan sulfate 3-O-sulfotransferase 5 | 6q22.31 |
| HSD11B1 | 11-β-hydroxysteroid dehydrogenase type 1 | 1q32.2 |
| HSPA4 | Heat shock 70 kDa protein 4 | 5q31.1 |
| HTR1B | 5-hydroxytryptamine receptor 1B | 6q14.1 |
| HTR2A | 5-hydroxytryptamine receptor 2A | 13q14.2 |
| HTR3A | 5-hydroxytryptamine receptor 3A | 11q23.2 |
| HTR3C | 5-hydroxytryptamine receptor 3, family member C | 3q27.1 |
| HTR7 | 5-hydroxytryptamine receptor 7 | 10q23.31 |
| HUWE1 | HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase | Xp11.22 |
| HYDIN | Hydrocephalus-inducing, mouse, homolog of | 16q22.2 |
| ICA1 | Islet cell autoantigen 1 | 7p21.3 |
| IL1R2 | Interleukin 1 receptor, type II | 2q11.2 |
| IL1RAPL1 | Interleukin 1 receptor accessory protein-like 1 | Xp21.3 |
| IL1RAPL2 | Interleukin 1 receptor accessory protein-like 2 | Xq22.3 |
| IMMP2L | Inner mitochondrial membrane peptidase, subunit 2, S. cerevisiae, homolog of | 7q31.1 |
| IMPDH2 | Inosine-5-prime monophosphate dehydrogenase 2 | 3p21.31 |
| INADL | Inactivation no after-potential D-like | 1p31.3 |
| INPP1 | Inositol polyphosphate-1-phosphatase | 2q32.2 |
| INPP5 | Inositol polyphosphate-5-phosphatase | 17p13.3 |
| IQSEC2 | IQ motif and Sec7 domain 2 | Xp11.22 |
| ITGA4 | Integrin, α 4 | 2q31.3 |
| ITGB3 | Integrin, β 3 | 17q21.32 |
| ITGB7 | Integrin, β 7 | 12q13.13 |
| ITK | IL20 inducible t-cell kinase | 5q33.3 |
| JARID2 | Jumonji, AT rich interactive domain 2 | 6p22.3 |
| JMJD1C | Jumonji domain containing 1C | 10q21.3 |
| JUP | Junction plakoglobin | 17q21.2 |
| KAL1 | Kallmann syndrome interval 1 | Xp22.31 |
| KANK1 | KN motif and ankyrin repeat domains 1 | 9p24.3 |
| KATNAL2 | Katanin p60 subunit A-like 2 | 18q21.1 |
| KCND2 | Potassium voltage-gated channel, Shal-related subfamily, member 2 | 7q31.31 |
| KCNJ2 | Potassium inwardly-rectifying channel, subfamily J, member 2 | 17q24.3 |
| KCNJ10 | Potassium inwardly-rectifying channel, subfamily J, member 10 | 1q23.2 |
| KCNMA1 | Potassium large conductance calcium-activated channel, subfamily M, α member 1 | 10q22.3 |
| KCNQ2 | Potassium voltage-gated channel, KQT-like subfamily, member 2 | 20q13.3 |
| KCNQ3 | Potassium voltage-gated channel, KQT-like subfamily, member 3 | 8q24.22 |
| KCNT1 | Potassium channel, subfamily T, member 1 | 9q34.3 |
| KCTD13 | Potassium channel tetramerization domain containing protein 13 | 16p11.2 |
| KDM5A | Lysine (K)-specific demethylase 5A | 12p13.33 |
| KDM5B | Lysine (K)-specific demethylase 5B | 1q32.1 |
| KDM5C | Lysine (K)-specific demethylase 5C | Xp11.22 |
| KDM6B | Lysine (K)-specific demethylase 6B | 17p13.1 |
| KHDRBS2 | KH domain containing, RNA binding, signal transduction associated protein 2 | 6q11.1 |
| KIAA1217 | Sickle tail protein homolog | 10p12.31 |
| KIAA1586 | KIAA1586 | 6p12.1 |
| KIAA2022 | KIAA2022 | Xq13.3 |
| KIF5C | Kinesin family member 5C | 2q23.1 |
| KIRREL3 | Kin of IRRE like 3 | 11q24.2 |
| KIT | v-KIT Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 4q12 |
| KLC2 | Kinesin light chain 2 | 11q13.2 |
| KMO | Kynurenine 3-monooxygenase | 1q43 |
| KMT2A | Lysine (K)-specific methyltransferase 2A | 11q23.3 |
| KMT2C | Lysine (K)-specific methyltransferase 2C | 7q36.1 |
| KMT2E | Lysine (K)-specific methyltransferase 2E | 7q22.3 |
| KPTN | Kaptin (actin binding protein) | 19q13.32 |
| LAMA1 | Laminin, α 1 | 18p11.23 |
| LAMB1 | Laminin, β 1 | 7q31.1 |
| LAMC3 | Laminin, γ 3 | 9q34.1 |
| LEP | Leptin | 7q32.1 |
| LIN7B | Lin-7 homolog B (C. elegans) | 19q13.33 |
| LMNA | Lamin A/C | 1q22 |
| LMX1B | LIM homeobox transcription factor 1, β | 9q33.3 |
| LRFN5 | Leucine-rich repeats and fibronectin type III domain containing 5 | 14q21.1 |
| LRGUK | Leucine-rich repeats and guanylate kinase domain containing | 7q33 |
| LRP2 | Low density lipoprotein receptor-related protein 2 | 2q31.1 |
| LRPPRC | Leucine-rich PPR motif containing protein | 2p21 |
| LRRC1 | Leucine-rich repeat-containing protein 1 | 6p12.1 |
| LRRC4 | Leucine-rich repeat-containing protein 4 | 7q32.1 |
| LRRC7 | Leucine-rich repeat-containing protein 7 | 1p31.1 |
| LZTS2 | Leucine zipper, putative tumor suppressor 2 | 10q24.31 |
| MACROD2 | Macro domain containing 2 | 20p12.1 |
| MAGED1 | Melanoma antigen family D, 1 | Xp11.22 |
| MAGEL2 | MAGE-like 2 | 15q11.2 |
| MAOA | Monoamine oxidase A | Xp11.3 |
| MAOB | Monoamine oxidase B | Xp11.23 |
| MAP1A | Microtubule-associated protein 1A | 15q15.3 |
| MAP2 | Microtubule-associated protein (MAP) 2 | 2q34 |
| MAP4 | Microtubule-associated protein (MAP) 4 | 3p21.31 |
| MAPK1 | Mitogen-activated protein kinase 1 | 22q11.22 |
| MAPK3 | Mitogen-activated protein kinase 3 | 16p11.2 |
| MAPK8IP2 | Mitogen-activated protein kinase 8 interacting protein 2 | 22q13.33 |
| MARK1 | MAP/microtubule affinity-regulating kinase 1 | 1q41 |
| MBD1 | Methyl-CpG binding domain protein 1 | 18q21.1 |
| MBD3 | Methyl-CpG binding domain protein 3 | 19p13.3 |
| MBD4 | Methyl-CpG binding domain protein 4 | 3q21.3 |
| MBD5 | Methyl-CpG binding domain protein 5 | 2q23.1 |
| MBD6 | Methyl-CpG binding domain protein 6 | 12q13.2 |
| MC4R | Melanocortin 4 receptor | 18q21.32 |
| MCC | Mutated in colorectal cancers | 5q22.2 |
| MCPH1 | Microcephalin 1 | 8p23.1 |
| MDGA2 | Mephrin, A5 antigen, protein tyrosine phosphatase mu (MAM) domain containing glycosylphosphatidylinositol anchor 2 | 14q21.3 |
| MDM2 | MDM2 oncogene, E3 ubiquitin protein ligase | 12q15 |
| MECP2 | Methyl CpG binding protein 2 | Xq28 |
| MED12 | Mediator complex subunit 12 | Xq13.1 |
| MED13L | Mediator complex subunit 13-like | 12q24.21 |
| MEF2C | MADS box transcription myocyte enhancer factor 2, polypeptide C | 5q14.3 |
| MET | Met proto-oncogene | 7q31.2 |
| MIB1 | Mind bomb E3 ubiquitin protein ligase 1 | 18q11.2 |
| MICAL3 | Microtubule-associated monooxygenase, calponin and lim domains-containing, 3 | 22q11.21 |
| MICALCL | MICAL C-terminus-like protein | 11p15.3 |
| MKL2 | Myocardin-like 2 | 16p13.12 |
| MOV10 | Moloney leukemia virus 10, mouse, homolog of | 1p13.2 |
| MSN | Moesin | Xq12 |
| MSNP1AS | Moesin pseudogene 1 antisense | 5p14.1 |
| MSR1 | Macrophage scavenger receptor | 8p22 |
| MTF1 | Metal-regulatory transcription factor 1 | 1p34.3 |
| MTHFR | 5-10-methylene-tetrahydrofolate reductase | 1p36.22 |
| MTR | 5-methyltetrahydrofolate-homocysteine S-methyltransferase | 1q43 |
| MTX2 | Metaxin 2 | 2q31.1 |
| MXRA5 | Matrix-remodelling associated 5 | Xp22.2 |
| MYH4 | Myosin, heavy chain 4, skeletal muscle | 17p13.1 |
| MYH10 | Myosin, heavy chain 10, non-muscle | 17p13.1 |
| MYO16 | Myosin XVI | 13q33.3 |
| MYO1A | Myosin IA | 12q13.3 |
| MYO9B | Myosin IXB | 19p13.11 |
| MYT1L | Myelin transcription factor 1-like | 2p25.3 |
| NAA15 | N(α)-acetyltransferase 15, NatA auxiliary subunit | 4q31.1 |
| NASP | Nuclear autoantigenic sperm protein (histone-binding) | 1p34.1 |
| NAV1 | Neuron navigator 1 | 1q32.1 |
| NBEA | Neurobeachin | 13q13.3 |
| NCKAP1 | NCK-associated protein 1 | 2q32.1 |
| NCKAP5 | NCK-associated protein 5 | 2q21.2 |
| NCKAP5L | NCK-associated protein 5-like | 12q13.12 |
| NCOR1 | Nuclear receptor corepressor 1 | 17p11.2 |
| NDNL2 | Necdin-like gene 2 | 15q13.1 |
| NDUFA5 | NADH-ubiquinone oxidoreductase 1 α subcomplex, 5 | 7q31.32 |
| NEFL | Neurofilament protein, light polypeptide | 8p21.2 |
| NELL1 | NEL-like 1 | 11p15.1 |
| NF1 | Neurofibromin 1 | 17q11.2 |
| NFIA | Nuclear factor I/A | 1p31.3 |
| NIPA1 | Non imprinted gene in Prader-Willi/Angelman syndrome chromosomal region 1 | 15q11.2 |
| NIPA2 | Non imprinted gene in Prader-Willi/Angelman syndrome chromosomal region 2 | 15q11.2 |
| NIPBL | Nipped-B-like | 5p13.2 |
| NLGN1 | Neuroligin 1 | 3q26.31 |
| NLGN2 | Neuroligin 2 | 17p13.1 |
| NLGN3 | Neuroligin 3 | Xq13.1 |
| NLGN4X | Neuroligin 4, X-linked | Xp22.31 |
| NLGN4Y | Neuroligin 4, Y-linked | Yq11.221 |
| NOS1AP | Nitric oxide synthase 1 (neuronal) adaptor protein | 1q23.3 |
| NOS2A | Nitric oxide synthase 2A | 17q11.2 |
| NOTCH3 | Notch 3 | 19p13.12 |
| NPAS2 | Neuronal PAS domain protein 2 | 2q11.2 |
| NR0B1 | Nuclear receptor subfamily 0, group B, member 1 | Xp21.2 |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 | 4q31.23 |
| NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | 12q13.13 |
| NRCAM | Neuronal cell adhesion molecule | 7q31.1 |
| NRG1 | Neuregulin 1 | 8p12 |
| NRP2 | Neuropilin 2 | 2q33.3 |
| NRXN1 | Neurexin I | 2p16.3 |
| NRXN2 | Neurexin II | 11q13.1 |
| NRXN3 | Neurexin III | 14q24.3 |
| NSD1 | Nuclear receptor-binding Sa-var, enhancer of zeste, and trithorax domain protein 1 | 5q35.3 |
| NTNG1 | Netrin G1 | 1p13.3 |
| NTRK1 | Neurotrophic tyrosine kinase, receptor, type 1 | 1q23.1 |
| NTRK3 | Neurotrophic tyrosine kinase, receptor, type 3 | 15q25.3 |
| NXF5 | Nuclear RNA export factor 5 | Xq22.1 |
| NXPH1 | Neurexophilin 1 | 7p21.3 |
| ODF3L2 | Outer dense fiber of sperm tails 3-like 2 | 19p13.3 |
| OGT | O-linked N-acetylglucosamine transferase | Xq13.1 |
| OPHN1 | Oligophrenin 1 | Xq12 |
| OPRM1 | Opioid receptor, mu 1 | 6q25.2 |
| OR1C1 | Olfactory receptor, family 1, subfamily C, member 1 | 1q44 |
| OTX1 | Orthodenticle Drosophila, homolog of | 2p15 |
| OXTR | Oxytocin receptor | 3p25.3 |
| P2RX4 | Purinergic receptor P2X, ligand-gated ion channel, 4 | 12q24.31 |
| PAFAH1B1 | Platelet-activating factor acetylhydrolase 1B, regulatory subunit 1 | 17p13.3 |
| PAH | Phenylalanine hydroxylase | 12q23.2 |
| PARD3B | PAR-3 family cell polarity regulator β | 2q33.3 |
| PARK2 | Parkin | 6q26 |
| PAX5 | Paired box 5 | 9p13.2 |
| PBRM1 | Polybromo 1 | 3p21.1 |
| PCDH10 | Protocadherin 10 | 4q28.3 |
| PCDH15 | Protocadherin 15 | 10q21.1 |
| PCDH19 | Protocadherin 19 | Xq22.1 |
| PCDH8 | Protocadherin 8 | 13q14.3 |
| PCDH9 | Protocadherin 9 | 13q21.32 |
| PCDHA1 | Protocadherin α 1 | 5q31.3 |
| PCDHA10 | Protocadherin α 10 | 5q31.3 |
| PCDHA11 | Protocadherin α 11 | 5q31.3 |
| PCDHA12 | Protocadherin α 12 | 5q31.3 |
| PCDHA13 | Protocadherin α 13 | 5q31.3 |
| PCDHA2 | Protocadherin α 2 | 5q31.3 |
| PCDHA3 | Protocadherin α 3 | 5q31.3 |
| PCDHA4 | Protocadherin α 4 | 5q31.3 |
| PCDHA5 | Protocadherin α 5 | 5q31.3 |
| PCDHA6 | Protocadherin α 6 | 5q31.3 |
| PCDHA7 | Protocadherin α 7 | 5q31.3 |
| PCDHA8 | Protocadherin α 8 | 5q31.3 |
| PCDHA9 | Protocadherin α 9 | 5q31.3 |
| PCDHAC1 | Protocadherin α subfamily C, member 1 | 5q31.3 |
| PCDHAC2 | Protocadherin α subfamily C, member 2 | 5q31.3 |
| PCDHGA11 | Protocadherin γ subfamily A, member 11 | 5q31.3 |
| PDE1C | Phosphodiesterase 1C | 7p14.3 |
| PDE4A | Phosphodiesterase 4A, cAMP-specific | 19p13.2 |
| PDE4B | Phosphodiesterase 4B, cAMP-specific | 1p31.3 |
| PDZD4 | PDZ domain containing 4 | Xq28 |
| PECR | Peroxisomal trans-2-enoyl-CoA reductase | 2q35 |
| PER1 | Period, Drosophila, homolog of | 17p13.1 |
| PEX7 | Peroxisomal biogenesis factor 7 | 6q23.3 |
| PGD | Phosphogluconate dehydrogenase | 1p36.22 |
| PHF2 | PHD finger protein 2 | 9q22.31 |
| PHF8 | PHD finger protein 8 | Xp11.22 |
| PIAS1 | Protein inhibitor of activated STAT, 1 | 15q23 |
| PIK3CG | Phosphatidylinositol-3-kinase, catalytic, γ | 7q22.3 |
| PIK3R2 | Phosphatidylinositol-3-kinase, regulatory subunit 2 | 19q13.11 |
| PINX1 | PIN2 interacting protein 1 | 8p23.1 |
| PITX1 | Paired-like homeodomain transcription factor 1 | 5q31.1 |
| PLAUR | Plasminogen activator receptor, urokinase-type | 19q13.31 |
| PLCB1 | Phospholipase C, β 1 | 20p12.3 |
| PLCD1 | Phospholipase C, δ 1 | 3p22.2 |
| PLN | Phospholamban | 6q22.31 |
| PLXNA4 | Plexin A4 | 7q32.3 |
| POGZ | POGO transposable element with ZNF domain | 1q21.3 |
| POLR2L | Polymerase (RNA) II (DNA directed) polypeptide L, 7.6 kDa | 11p15.5 |
| POMGNT1 | Protein O-mannose β-1, 2-N-acetylglucosaminyl-transferase | 1p34.1 |
| PON1 | Paraoxonase 1 | 7q21.3 |
| POT1 | Protection of telomeres 1 | 7q31.33 |
| PPFIA1 | Protein tyrosine phosphatase, receptor type, F polypeptide, interacting protein, α 1 | 11q13.3 |
| PPP1CB | Protein phosphatase 1, catalytic subunit, β isozyme | 2p23.2 |
| PPP1R1B | Protein phosphatase 1, regulatory (inhibitor) subunit 1B | 17q12 |
| PPP1R3F | Protein phosphatase 1, regulatory (inhibitor) subunit 3F | Xp11.23 |
| PRODH | Proline dehydrogenase (oxidase) 1 | 22q11.21 |
| PRICKLE1 | Prickle, Drosophila, homolog of, 1 | 12q12 |
| PRICKLE2 | Prickle, Drosophila, homolog of, 2 | 3p14.1 |
| PRKCB | Protein kinase C, β | 16p12.2 |
| PRKCB1 | Protein kinase C, β-1 | 16p12.2 |
| PRKD1 | Protein kinase D1 | 14q12 |
| PRDX1 | Peroxiredoxin 1 | 1p34.1 |
| PRSS38 | Protease, serine, 38 | 1q42.13 |
| PRUNE2 | Prune, Drosophila, homolog of, 2 | 9q21.2 |
| PSD3 | Pleckstrin and Sec7 domains-containing protein 3 | 8p22 |
| PSEN1 | Presenilin 1 | 14q24.2 |
| PSMD10 | Proteasome 26S subunit, non-ATPase, 10 | Xq22.3 |
| PTCHD1 | Patched domain containing protein 1 | Xp22.11 |
| PTEN | Phosphatase and tensin homolog | 10q23.31 |
| PTGER3 | Prostaglandin E receptor 3, EP3 subtype | 1p31.1 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1q31.1 |
| PTPN11 | Protein tyrosine phosphatase, non-receptor type 11 | 12q24.13 |
| PTPRB | Protein tyrosine phosphatase, receptor type, B | 12q15 |
| PTPRC | Protein tyrosine phosphatase, receptor type, C | 1q31.3 |
| PTPRM | Protein tyrosine phosphatase, receptor type, M | 18p11.23 |
| PTPRT | Protein tyrosine phosphatase, receptor type, T | 20q13.11 |
| PXDN | Peroxidasin, Drosophila homolog of | 2p25.3 |
| RAB11FIP5 | RAB11 family-interacting protein 5 | 2p13.2 |
| RAB19 | RAB19, member RAS oncogene family | 7q34 |
| RAB39B | RAS-associated protein RAB39B | Xq28 |
| RAI1 | Retinoic acid induced gene 1 | 17p11.2 |
| RAPGEF4 | Rap guanine nucleotide exchange factor | 2q31.1 |
| RASD1 | RAS protein, dexamethasone-induced, 1 | 17p11.2 |
| RASSF1 | RAS association (ralGDS/AF-6) domain family member 1 | 3p21.31 |
| RASSF5 | RAS association domain family protein 5 | 1q32.1 |
| RB1CC1 | RB1-inducible coiled-coil 1 | 8q11.23 |
| RBFOX1 | RNA binding protein FOX-1, C. elegans, homolog of, 1 | 16p13.3 |
| RBM8A | RNA binding motif protein 8A | 1q21.1 |
| RBMS3 | RNA binding motif protein, single stranded interacting, 3 | 3p24.1 |
| REEP3 | Receptor expression-enhancing protein 3 | 10q21.3 |
| RELN | Reelin | 7q22.1 |
| RERE | RE-repeats encoding gene | 1p36.23 |
| RFWD2 | Ring finger and WD repeat domains-containing protein 2 | 1q25.2 |
| RGS7 | Regulator of G protein signaling 7 | 1q43 |
| RHOXF1 | RHOX homeobox family, member 1 | Xq24 |
| RIC8A | RIC8 guanine nucleotide exchange factor A | 11p15.5 |
| RIMS1 | Regulating synaptic membrane exocytosis 1 | 6q13 |
| RIMS3 | Protein regulating synaptic membrane exocytosis 3 | 1p34.2 |
| RNPS1 | RNA binding protein S1 | 16p13.3 |
| ROBO1 | Roundabout, Drosophila, homolog of, 1 | 3p12.2 |
| ROBO2 | Roundabout, Drosophila, homolog of, 2 | 3p12.3 |
| RORA | RAR-related orphan receptor A | 15q22.2 |
| RPL10 | Ribosomal protein L10 | Xq28 |
| RPP25 | Ribonuclease P/MRP 25 kDa subunit | 15q24.2 |
| RPS6KA1 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 1 | 1p36.11 |
| RPS6KA2 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | 6q27 |
| RPS6KA3 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | Xp22.12 |
| RUVBL1 | RuvB-E. coli, homolog-like 1 | 3q21.3 |
| SAE1 | SUMO1 activating enzyme, subunit 1 | 19q13.32 |
| SATB2 | Special AT-rich sequence-binding protein 2 | 2q33.1 |
| SBF1 | SET binding factor 1 | 22q13.33 |
| SCFD2 | Sec1 family domain containing 2 | 4q12 |
| SCN1A | Sodium channel, neuronal, type I, α subunit | 2q24.3 |
| SCN2A | Sodium channel, voltage-gated, type II, α subunit | 2q24.3 |
| SCN7A | Sodium channel, voltage-gated, type VII, α subunit | 2q24.3 |
| SCN8A | Sodium channel, voltage-gated, type VIII, α subunit | 12q13.13 |
| SDC2 | Syndecan 2 | 8q22.1 |
| SDK1 | Sidekick cell adhesion molecule 1 | 7p22.2 |
| SEMA3F | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F | 3p21.31 |
| SEMA5A | Semaphorin 5A | 5p15.31 |
| SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 7q22.1 |
| SETBP1 | SET binding protein 1 | 18q12.3 |
| SETD2 | SET domain containing protein 2 | 3p21.31 |
| SETD5 | SET domain containing protein 5 | 3p25.3 |
| SETDB1 | SET domain, bifurcated, 1 | 1q21.3 |
| SETDB2 | SET domain, bifurcated, 2 | 13q14.2 |
| SEZ6L2 | Seizure related 6 homolog (mouse)-like 2 | 16p11.2 |
| SF1 | Splicing factor 1 | 11q13.1 |
| SFPQ | Splicing factor proline/glutamine-rich | 1p34.3 |
| SFTPD | Surfactant, pulmonary-associated protein D | 10q22.3 |
| SGSH | N-sulfoglucosamine sulfohydrolase | 17q25.3 |
| SGSM3 | Small G protein signaling modulator 3 | 22q13.1 |
| SH3KBP1 | SH3-domain kinase binding protein 1 | Xp22.12 |
| SHANK1 | SH3 and multiple ankyrin repeat domains 1 | 19q13.3 |
| SHANK2 | SH3 and multiple ankyrin repeat domains 2 | 11q13.4 |
| SHANK3 | SH3 and multiple ankyrin repeat domains 3 | 22q13.33 |
| SLC16A3 | Solute carrier family 16 (monocarboxylic acid transporter), member 3 | 17q25 |
| SLC16A7 | Solute carrier family 16 (monocarboxylic acid transporter), member 7 | 12q14.1 |
| SLC1A1 | Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter), member 1 | 9p24.2 |
| SLC22A15 | Solute carrier family 22, (organic cation transporter), member 15 | 1p13.1 |
| SLC24A2 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | 9p22.1 |
| SLC25A12 | Solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | 2q31.1 |
| SLC25A14 | Solute carrier family 25 (mitochondrial carrier, brain), member 14 | Xq26.1 |
| SLC25A24 | Solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 24 | 1p13.3 |
| SLC25A27 | Solute carrier family 25, member 27 | 6p12.3 |
| SLC29A4 | Solute carrier family 29 (equilibrative nucleoside transporter), member 4 | 7p22.1 |
| SLC30A5 | Solute carrier family 30 (zinc transporter), member 5 | 5q13.1 |
| SLC35A3 | Solute carrier family 35 (UDP-N-acetylglucosamine transporter), member 3 | 1p21.2 |
| SLC38A10 | Solute carrier family 38, member 10 | 17q25.3 |
| SLC39A11 | Solute carrier family 39 (metal ion transporter), member 11 | 17q21.31 |
| SLC4A10 | Solute carrier family 4 (sodium bicarbonate transporter-like), member 10 | 2q24.2 |
| SLC6A1 | Solute carrier family 6 (neurotransmitter transporter), member 1 | 3p25.3 |
| SLC6A3 | Solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | 5p15.33 |
| SLC6A4 | Solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 17q11.2 |
| SLC6A8 | Solute carrier family 6 (neurotransmitter transporter, creatine), member 8 | Xq28 |
| SLC9A6 | Solute carrier family 9 (sodium/hydrogen exchanger), member 6 | Xq26.3 |
| SLC9A9 | Solute carrier family 9 (sodium/hydrogen exchanger), member 9 | 3q24 |
| SLCO1B1 | Solute carrier organic anion transporter family, member 1B1 | 12p12.2 |
| SLCO1B3 | Solute carrier organic anion transporter family, member 1B3 | 12p12.2 |
| SLIT3 | Slit, Drosophila, homolog of, 3 | 5q35.1 |
| SLITRK5 | SLIT and NTRK-like family, member 5 | 13q31.2 |
| SLK | STE20-like kinase | 10q24.33 |
| SMAD2 | SMAD family member 2 | 18q21.1 |
| SMARCC2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily C, member 2 | 12q13.2 |
| SMG6 | SMG 6, C. elegans, homolog of | 17p13.3 |
| SND1 | EBNA2 coactivator p100 | 7q32.1 |
| SNRPN | Small nuclear ribonucleoprotein polypeptide N | 15q11.2 |
| SNTG2 | Syntrophin, γ 2 | 2p25.3 |
| SNX19 | Sorting nexin 19 | 11q25 |
| SNX5 | Sorting nexin 5 | 20p11.23 |
| SOD1 | Superoxide dismutase 1, soluble | 21q22.11 |
| SOS1 | Son of sevenless (SOS), Drosophila, homolog 1 | 2p22.1 |
| SOX5 | SRY (sex determining region Y)-box 5 | 12p12.1 |
| SOX7 | SRY (sex determining region Y)-box 7 | 8p23.1 |
| SPAST | Spastin | 2p22.3 |
| SRD5A2 | Steroid-5-α-reductase, 2 | 2p23.1 |
| ST7 | Suppressor of tumorigenicity 7 | 7q31.2 |
| ST8SIA2 | ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 2 | 15q26.1 |
| STK39 | Serine/threonine protein kinase 39 | 2q24.3 |
| STX6 | Syntaxin 6 | 1q25.3 |
| STX1A | Syntaxin 1A | 7q11.23 |
| STXBP1 | Syntaxin-binding protein 1 | 9q34.1 |
| STXBP5 | Syntaxin-binding protein 5 | 6q24.3 |
| STXBP5L | Syntaxin-binding protein 5-like | 3q13.33 |
| SUCLG2 | Succinate-CoA ligase, GDP-forming, β subunit | 3p14.1 |
| SUV420H1 | Suppressor of variegation 4–20, Drosophila, homolog of, 1 | 11q13.2 |
| SYAP1 | Synapse associated protein 1 | Xp22.2 |
| SYN1 | Synapsin 1 | Xp11.23 |
| SYN2 | Synapsin II | 3p25.2 |
| SYN3 | Synapsin III | 22q12.3 |
| SYNE1 | Spectrin repeat containing nuclear envelope 1 | 6q25.2 |
| SYNGAP1 | Synaptic RAS-GTPase-activating protein 1 | 6p21.32 |
| SYT17 | Synaptotagmin XVII | 16p12.3 |
| SYT3 | Synaptotagmin III | 19q13.33 |
| TAF1C | TATA box-binding protein-associated factor 1C | 16q24.1 |
| TAF1L | TATA box-binding protein-associated factor 1-like | 9p21.1 |
| TAS2R1 | Taste receptor, type 2, member 1 | 5p15.31 |
| TBC1D30 | TBC1 domain family, member 30 | 12q14.3 |
| TBC1D5 | TBC1 domain family, member 5 | 3p24.3 |
| TBC1D7 | TBC1 domain family, member 7 | 6p24 |
| TBL1X | Transducin-β-like 1, X-linked | Xp22.31 |
| TBL1XR1 | Transducin-β-like 1 receptor 1 | 3q26.32 |
| TBR1 | T-box, brain, 1 | 2q24.2 |
| TBX1 | T-box 1 | 22q11.21 |
| TCF3 | Transcription factor 3 | 19p13.3 |
| TCF4 | Transcription factor 4 | 18q21.2 |
| TCF20 | Transcription factor 20 (AR1) | 22q13.2 |
| TCF7L2 | Transcription factor 7-like 2 (t-cell specific, HMG-box) | 10q25.2 |
| TDO2 | Tryptophan 2,3-dioxygenase | 4q32.1 |
| TGM3 | Transglutaminase 3 | 20p13 |
| TH | Tyrosine hydroxylase | 11p15.5 |
| THBS1 | Thrombospondin 1 | 15q14 |
| THRA | Thyroid hormone receptor, α-1 | 17q21.1 |
| TLK2 | Tousled-like kinase 2 | 17q23.2 |
| TLX1 | T-cell leukemia homeobox 1 | 10q24.31 |
| TM4SF20 | Transmembrane 4 L6 family, member 20 | 2q36.3 |
| TMEM231 | Transmembrane protein 231 | 16q23.1 |
| TMLHE | Epsilon-trimethyllysine hydroxylase | Xq28 |
| TNIP2 | TNFAIP3 interacting protein 2 | 4p16.3 |
| TNRC6B | Trinucleotide repeat containing 6B | 22q13.1 |
| TOMM20 | MAS20P, S. cerevisiae, homolog of | 1q42.3 |
| TOP1 | Topoisomerase, DNA, I | 20q12 |
| TOP3B | Topoisomerase, DNA, III, β | 22q11.22 |
| TOPBP1 | Topoisomerase (DNA) II-binding protein 1 | 3q22.1 |
| TOPORS | Topoisomerase I-binding, arginine/serine-rich, E3 ubiquitin protein ligase | 9p21.1 |
| TPH2 | Tryptophan hydroxylase 2 | 12q21.1 |
| TPO | Thyroid peroxidase | 2p25.3 |
| TRIM33 | Tripartite motif containing protein 33 | 1p13.2 |
| TRIO | Trio Rho guanine nucleotide exchange factor | 5p15.2 |
| TRIP12 | Thyroid hormone receptor interactor 12 | 2q36.3 |
| TRPC6 | Transient receptor potential cation channel, subfamily C, member 6 | 11q22.1 |
| TRPM1 | Transient receptor potential cation channel, subfamily M, member 1 | 15q13.3 |
| TSC1 | Tuberous sclerosis 1 | 9q34.1 |
| TSC2 | Tuberous sclerosis 2 | 16p13.3 |
| TSN | Translin | 2q14.3 |
| TSPAN7 | Tetraspanin 7 | Xp11.4 |
| TTI2 | TELO2-interacting protein 2 | 8p12 |
| TTN | Titin | 2q31.2 |
| TUBA1A | Tubulin, α-1A | 12q13.12 |
| TUBGCP5 | Tubulin-γ complex-associated protein 5 | 15q11.2 |
| TYR | Tyrosinase | 11q14.3 |
| UBE1L2 | Ubiquitin-activating enzyme, E1-like 2 | 4q13.2 |
| UBE2H | Ubiquitin-conjugating enzyme E2H | 7q32.2 |
| UBE3A | Ubiquitin protein ligase E3A | 15q11.2 |
| UBE3B | Ubiquitin protein ligase E3B | 12q24.11 |
| UBE3C | Ubiquitin protein ligase E3C | 7q36.3 |
| UBL7 | Ubiquitin-like 7 | 15q24.1 |
| UBR5 | Ubiquitin protein ligase E3 component N-recognin 5 | 8q22.3 |
| UBR7 | Ubiquitin protein ligase E3 component N-recognin 7 | 14q32.12 |
| UIMC1 | Ubiquitin interaction motif containing 1 | 5q35.2 |
| UPB1 | Ureidopropionase, β 1 | 22q11.23 |
| UPF2 | UPF2, yeast, homolog of | 10p14 |
| UPF3B | UPF3, yeast, homolog of, B | Xq24 |
| USP54 | Ubiquitin specific peptidase 54 | 10q22.2 |
| USP9Y | Ubiquitin specific protease 9, Y-chromosome | Yq11.21 |
| VASH1 | Vasohibin 1 | 14q24.3 |
| VCP | Valosin containing protein | 9p13.3 |
| VIL1 | Villin 1 | 2q35 |
| VIP | Vasoactive intestinal peptide (VIP) | 6q25.2 |
| VPS13B | Vacuolar protein sorting 13, yeast, homolog of, B | 8q22.2 |
| VPS4A | Vacuolar protein sorting 4 homolog A (S. cerevisiae) | 16q22.1 |
| WAC | WW domain containing adaptor with coiled-coil | 10p12.1 |
| WDFY3 | WD repeat and FYVE domain containing 3 | 4q21.23 |
| WHSC1 | Wolf-Hirschhorn syndrome candidate 1 | 4p16.3 |
| WNK3 | Protein kinase lysine deficient 3 | Xp11.22 |
| WNT1 | Wingless-type MMTV integration site family, member 1 | 12q13.12 |
| WNT2 | Wingless-type MMTV integration site family, member 2 | 7q31.2 |
| WWC3 | WWC family member 3 | Xp22.32 |
| XIRP1 | Cardiomyopathy-associated protein 1 | 3p22.2 |
| XPC | Xeroderma pigmentosum complementation group C | 3p25.1 |
| XPO1 | Exportin 1 | 2p15 |
| XPO5 | Exportin 5 | 6p21.1 |
| YEATS2 | YEATS domain containing 2 | 3q27.1 |
| YTHDC2 | YTH domain containing 2 | 5q22.2 |
| YWHAE | Tyrosine 3-monooxygenase, tryptophan 5-monooxygenase activation protein, epsilon isoform | 17p13.3 |
| ZBTB16 | Zinc finger- and BTB domain-containing protein 16 | 11q23.1 |
| ZBTB20 | Zinc finger- and BTB domain-containing protein 20 | 3q13.31 |
| ZC3H12B | Zinc finger CCCH domain-containing protein 12B | Xq12 |
| ZFPL1 | Zinc finger protein-like 1 | 11q13.1 |
| ZMYND11 | Zinc finger, MYND-type containing 11 | 10p15.3 |
| ZNF18 | Zinc finger protein 18 | 17p12 |
| ZNF365 | Zinc finger protein 365 | 10q21.2 |
| ZNF385B | Zinc finger protein 385B | 2q31.3 |
| ZNF407 | Zinc finger protein 407 | 18q23 |
| ZNF517 | Zinc finger protein 517 | 8q24.3 |
| ZNF8 | Zinc finger protein 8 | 19q13.43 |
| ZNF713 | Zinc finger protein 713 | 7p11.2 |
| ZNF804A | Zinc finger protein 804A | 2q32.1 |
| ZNF827 | Zinc finger protein 827 | 4q31.22 |
| ZSWIM5 | Zinc finger, SWIM-type containing 5 | 1p34.1 |
3. Experimental Section
We used computer-based internet websites and PubMed (https://www.ncbi.nlm.nih.gov/pubmed) to search key words for genetics and autism. This included the integrated catalogue of human genetic studies related to autism found at the Simons Foundation Autism Research Initiative (SFARI) website (https://gene.sfari.org), which currently lists 667 genes reported as of 25 February 2015. This public access initiative is an ongoing curated collection of clinically proven ASD genes supported by clinical and autism experts, medical geneticists and laboratory specialists in the study of autism. This site includes gene description and evidence of support for causation with cited literature reports. We examined peer-reviewed articles found in the medical literature following our search for genetic evidence (i.e., gene variants, mutations or disturbed gene function) and the involvement of genetics playing a role in autism. Sources included whole-genome sequencing of ASD families randomly selected with at least one unaffected sibling [40] or gene expression profiles in ASD [39] along with other informative websites (e.g., Online Mendelian Inheritance in Man, www.OMIM.org). We then compiled the list of genes from these major sources for a total of 792 genes, whereby at least one mechanism was involved for each gene that could lead to ASD, a heterogeneous condition involving many genes; as our report is focused on the compilation of ASD genes from peer-reviewed research articles and authoritative computer website genomic databases for autism and not necessarily related to causal relationships between the individual gene and ASD. Those genes recognized, to date, as playing a role in ASD susceptibility and causation generally appear to impact chromatin remodeling, metabolism, mRNA translation, cell adhesion and synaptic function [39].
SFARI is a publicly available manually curated web-based searchable site of human genes with links to ASD and includes genes in catalogue form based on five categories—genetic association, syndromic, rare single-gene variant and functional and multi-genetic copy number variation—supported by cited research publications for each. Additional literature sources in our study consisted of both primary research articles and reviews summarizing genetic evidence. Many of the listed genes were identified in multiple research studies and widely reported in literature reviews, data repositories and/or computer genomic-based websites for autism (e.g., SFARI). A large number of genes showed a varied relationship to autism and neurodevelopment, but the mass of the literature surveyed limits the reliability of our relative strength estimates for the ASD and gene associations. The gene would be included if cited and recognized in peer-reviewed publications (e.g., PubMed) with supportive genetic evidence (e.g., genetic linkage, GWAS, functional gene expression patterns, informative SNPs, CNVs or identified gene mutations). Other supporting genetic evidence can be found at Simons Foundation Autism Research Initiative (SFARI) at https://sfari.org/sfari-initiatives/simons-simplex-collection, the National Institutes of Health (NIH) at https://www.ncbi.nlm.nih.gov/gap, the Online Inheritance in Man (OMIM) at www.omim.org or Genecards at https://www.genecards.org.
4. Conclusions
Readily available tissue sources, such as peripheral blood, established lymphoblastoid cell lines and saliva, hold promise for more advances in ASD by enabling the identification of new genes and a better understanding of the causation and disease mechanisms to further stimulate research with the hope to discover new treatment modalities impacted by the recognition of known disease-causing or candidate genes for ASD. We illustrated the master list of clinically relevant and known ASD genes in our summary by plotting individual genes on high-resolution chromosome ideograms and generated a tabular form to increase the awareness required for genetic testing and counselling purposes for family members presenting for genetic services. Creating a master list of genes related to ASD is a complicated process; new genes are continually identified, but not all genes are equally important or certain to be causative. Additional research is needed to further investigate the causal relationships between the specific gene and ASD. The authors encourage the use of this collection of known and clinically relevant candidate genes for ASD in their evaluation of patients and families presenting for genetic testing options and for accurate genetic counselling.
Acknowledgments
We thank Carla Meister for expert preparation of the manuscript and Lorie Gavulic for excellent artistic design and preparation of chromosome ideograms. We acknowledge support from National Institute of Child Health and Human Development (NICHD) HD02528.
Author Contributions
Merlin G. Butler conceived of the study, reviewed data from ASD gene literature reports and wrote the manuscript; Syed K. Rafi obtained and reviewed articles pertaining to ASD genes and summarized the master gene list; and Ann M. Manzardo contributed to gene data review and interpretation, contributed to the content of the manuscript and reviewed the literature.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. 4th ed. American Psychiatric Association; Washington, DC, USA: 2000. [Google Scholar]
- 2.Johnson C.P., Myers S.M., American Academy of Pediatrics Counsel on Children with Disablities Identifiction and evaluation of children with autism spectrum disorders. Pediatrics. 2007;120:1183–1215. doi: 10.1542/peds.2007-2361. [DOI] [PubMed] [Google Scholar]
- 3.Hughes J.R. Update on autism: A review of 1300 reports published in 2008. Epilepsy Behav. 2009;16:569–589. doi: 10.1016/j.yebeh.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Lord C., Risi S., Lambrecht L., Cook E.H., Jr., Leventhal B.L., DiLavore P.C., Pickles A., Rutter M. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000;30:205–223. doi: 10.1023/A:1005592401947. [DOI] [PubMed] [Google Scholar]
- 5.Le Couteur A., Lord C., Rutter M. Autism Diagnostic Interview-Reviewed (ADI-R) Western Psychological Services; Los Angeles, CA, USA: 2003. [Google Scholar]
- 6.Constantino J.N., Davis S.A., Todd R.D., Schindler M.K., Gross M.M., Brophy S.L., Metzger L.M., Shoushtari C.S., Splinter R., Reich W. Validation of a brief quantitative measure of autistic traits: Comparison of the social responsiveness scale with the autism diagnostic interview-revised. J. Autism Dev. Disord. 2003;33:427–433. doi: 10.1023/A:1025014929212. [DOI] [PubMed] [Google Scholar]
- 7.Rice C. Prevalence of autism spectrum disorders-autism and developmental disabilities monitoring network, United States, 2006. MMWR Surveill. Summ. 2009;58:3–8. [PubMed] [Google Scholar]
- 8.Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J. Clin. Psychiatry. 2005;66(Suppl. 10):3–8. [PubMed] [Google Scholar]
- 9.Kanner L. Autistic psychopathy in childhood. Nerv. Child. 1943;2:217–250. [Google Scholar]
- 10.Rapin I. Autistic regression and disintegrative disorder: How important the role of epilepsy? Semin. Pediatr. Neurol. 1995;2:278–285. doi: 10.1016/S1071-9091(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 11.Geschwind D.H., Levitt P. Autism spectrum disorders: Developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Kurtzke J. Neuroepidemiology. In: Bradley W., Daroff R., Fenichel G., Marsden C., editors. Neurology in Clinical Practice. Butterworth-Heinemann; Stoneham, MA, USA: 1991. pp. 545–560. [Google Scholar]
- 13.Gadia C.A., Tuchman R., Rotta N.T. Autism and pervasive developmental disorders. J. Pediatr. 2004;80:S83–S94. doi: 10.2223/1172. [DOI] [PubMed] [Google Scholar]
- 14.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prontera P., Ottaviani V., Toccaceli D., Rogaia D., Ardisia C., Romani R., Stangoni G., Pierini A., Donti E. Recurrent approximately 100 kb microdeletion in the chromosomal region 14q11.2, involving CHD8 gene, is associated with autism and macrocephaly. Am. J. Med. Genet. A. 2014;164:3137–3141. doi: 10.1002/ajmg.a.36741. [DOI] [PubMed] [Google Scholar]
- 16.Benvenuto A., Moavero R., Alessandrelli R., Manzi B., Curatolo P. Syndromic autism: Causes and pathogenetic pathways. World J. Pediatr. 2009;5:169–176. doi: 10.1007/s12519-009-0033-2. [DOI] [PubMed] [Google Scholar]
- 17.Holt R., Monaco A.P. Links between genetics and pathophysiology in the autism spectrum disorders. EMBO Mol. Med. 2011;3:438–450. doi: 10.1002/emmm.201100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell D.B., Sutcliffe J.S., Ebert P.J., Militerni R., Bravaccio C., Trillo S., Elia M., Schneider C., Melmed R., Sacco R., et al. A genetic variant that disrupts MET transcription is associated with autism. Proc. Natl. Acad. Sci. USA. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer G.B., Starr L., Pickering D., Skar G., Dehaai K., Sanger W.G. Array comparative genomic hybridization findings in a cohort referred for an autism evaluation. J. Child Neurol. 2010;25:1498–1503. doi: 10.1177/0883073810370479. [DOI] [PubMed] [Google Scholar]
- 20.Roberts J.L., Hovanes K., Dasouki M., Manzardo A.M., Butler M.G. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene. 2014;535:70–78. doi: 10.1016/j.gene.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler M.G., Usrey K., Roberts J.L., Schroeder S.R., Manzardo A.M. Clinical presentation and microarray analysis of Peruvian children with atypical development and/or aberrant behavior. Genet. Res. Int. 2014;2014:408516. doi: 10.1155/2014/408516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miles J.H. Autism spectrum disorders—A genetics review. Genet. Med. 2011;13:278–294. doi: 10.1097/GIM.0b013e3181ff67ba. [DOI] [PubMed] [Google Scholar]
- 23.Liu X., Takumi T. Genomic and genetic aspects of autism spectrum disorder. Biochem. Biophys. Res. Commun. 2014;452:244–253. doi: 10.1016/j.bbrc.2014.08.108. [DOI] [PubMed] [Google Scholar]
- 24.Cox D.M., Butler M.G. The 15q11.2 BP1-BP2 microdeletion syndrome: A review. Int. J. Mol. Sci. 2015;16:4068–4082. doi: 10.3390/ijms16024068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hempel M., Rivera Brugues N., Wagenstaller J., Lederer G., Weitensteiner A., Seidel H., Meitinger T., Strom T.M. Microdeletion syndrome 16p11.2-p12.2: Clinical and molecular characterization. Am. J. Med. Genet. A. 2009;149:2106–2112. doi: 10.1002/ajmg.a.33042. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez B.A., Roberts W., Chung B., Weksberg R., Meyn S., Szatmari P., Joseph-George A.M., Mackay S., Whitten K., Noble B., et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J. Med. Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 27.Miller D.T., Shen Y., Weiss L.A., Korn J., Anselm I., Bridgemohan C., Cox G.F., Dickinson H., Gentile J., Harris D.J., et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritvo E.R., Jorde L.B., Mason-Brothers A., Freeman B.J., Pingree C., Jones M.B., McMahon W.M., Petersen P.B., Jenson W.R., Mo A. The UCLA-university of Utah epidemiologic survey of autism: Recurrence risk estimates and genetic counseling. Am. J. Psychiatry. 1989;146:1032–1036. doi: 10.1176/ajp.146.8.1032. [DOI] [PubMed] [Google Scholar]
- 29.Sandin S., Lichtenstein P., Kuja-Halkola R., Larsson H., Hultman C.M., Reichenberg A. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauritsen M.B., Als T.D., Dahl H.A., Flint T.J., Wang A.G., Vang M., Kruse T.A., Ewald H., Mors O. A genome-wide search for alleles and haplotypes associated with autism and related pervasive developmental disorders on the Faroe islands. Mol. Psychiatry. 2006;11:37–46. doi: 10.1038/sj.mp.4001754. [DOI] [PubMed] [Google Scholar]
- 32.Ma D., Salyakina D., Jaworski J.M., Konidari I., Whitehead P.L., Andersen A.N., Hoffman J.D., Slifer S.H., Hedges D.J., Cukier H.N., et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann. Hum. Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss L.A., Arking D.E., Gene Discovery Project of Johns Hopkins & the Autism Consortium. Daly M.J., Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anney R., Klei L., Pinto D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., Sykes N., Pagnamenta A.T., et al. A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K., Zhang H., Ma D., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M., et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagnamenta A.T., Khan H., Walker S., Gerrelli D., Wing K., Bonaglia M.C., Giorda R., Berney T., Mani E., Molteni M., et al. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J. Med. Genet. 2011;48:48–54. doi: 10.1136/jmg.2010.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klei L., Sanders S.J., Murtha M.T., Hus V., Lowe J.K., Willsey A.J., Moreno-De-Luca D., Yu T.W., Fombonne E., Geschwind D., et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell M.G., Kohane I.S., Kong S.W. Pathway-based outlier method reveals heterogeneous genomic structure of autism in blood transcriptome. BMC Med. Genomics. 2013;6:34. doi: 10.1186/1755-8794-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Y.H., Yuen R.K., Jin X., Wang M., Chen N., Wu X., Ju J., Mei J., Shi Y., He M., et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huguet G., Ey E., Bourgeron T. The genetic landscapes of autism spectrum disorders. Ann. Rev. Genomics Hum. Genet. 2013;14:191–213. doi: 10.1146/annurev-genom-091212-153431. [DOI] [PubMed] [Google Scholar]
- 42.Correia C., Oliveira G., Vicente A.M. Protein interaction networks reveal novel autism risk genes within gwas statistical noise. PLoS One. 2014;9:e112399. doi: 10.1371/journal.pone.0112399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merikangas A.K., Segurado R., Heron E.A., Anney R.J., Paterson A.D., Cook E.H., Pinto D., Scherer S.W., Szatmari P., Gill M., et al. The phenotypic manifestations of rare genic CNVs in autism spectrum disorder. Mol. Psychiatry. 2014 doi: 10.1038/mp.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E., et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldani A.A.S., Downs S.R., Widjaja F., Lawton B., Hendren R.L. Biomarkers in autism. Front. Psychiatry. 2014;5:100. doi: 10.3389/fpsyt.2014.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ch’ng C., Kwok W., Rogic S., Pavlidis P. Meta-analysis of gene expression in autism spectrum disorder. 2015 doi: 10.1002/aur.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uddin M., Tammimies K., Pellecchia G., Alipanahi B., Hu P., Wang Z., Pinto D., Lau L., Nalpathamkalam T., Marshall C.R., et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat. Genet. 2014;46:742–747. doi: 10.1038/ng.2980. [DOI] [PubMed] [Google Scholar]
- 48.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadley D., Wu Z.L., Kao C., Kini A., Mohamed-Hadley A., Thomas K., Vazquez L., Qiu H., Mentch F., Pellegrino R., et al. The impact of the metabotropic glutamate receptor and other gene family interaction networks on autism. Nat. Commun. 2014 doi: 10.1038/ncomms5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butler M.G., Rafi S.K., Hossain W., Stephan D.A., Manzardo A.M. Whole exome sequencing in females with autism implicates novel and candidate genes. Int. J. Mol. Sci. 2015;16:1312–1335. doi: 10.3390/ijms16011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen R.K., Thiruvahindrapuram B., Merico D., Walker S., Tammimies K., Hoang N., Chrysler C., Nalpathamkalam T., Pellecchia G., Liu Y., et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 52.Willsey J.A., State M.W. Autism spectrum disorders: From genes to neurobiology. Curr. Opin. Neurobiol. 2015;30:92–99. doi: 10.1016/j.conb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hormozdiari F., Penn O., Borenstein E., Eichler E.E. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2015;25:142–154. doi: 10.1101/gr.178855.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler M.G., Youngs E.L., Roberts J.L., Hellings J.A. Assessment and treatment in autism spectrum disorders: A focus on genetics and psychiatry. Autism Res. Treat. 2012 doi: 10.1155/2012/242537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herman G.E., Henninger N., Ratliff-Schaub K., Pastore M., Fitzgerald S., McBride K.L. Genetic testing in autism: How much is enough? Genet. Med. 2007;9:268–274. doi: 10.1097/GIM.0b013e31804d683b. [DOI] [PubMed] [Google Scholar]
- 56.Butler M.G. Prader-willi syndrome: Obesity due to genomic imprinting. Curr. Genomics. 2011;12:204–215. doi: 10.2174/138920211795677877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dhillon S., Hellings J.A., Butler M.G. Genetics and mitochondrial abnormalities in autism spectrum disorders: A review. Curr. Genomics. 2011;12:322–332. doi: 10.2174/138920211796429745. [DOI] [PMC free article] [PubMed] [Google Scholar]