Abstract
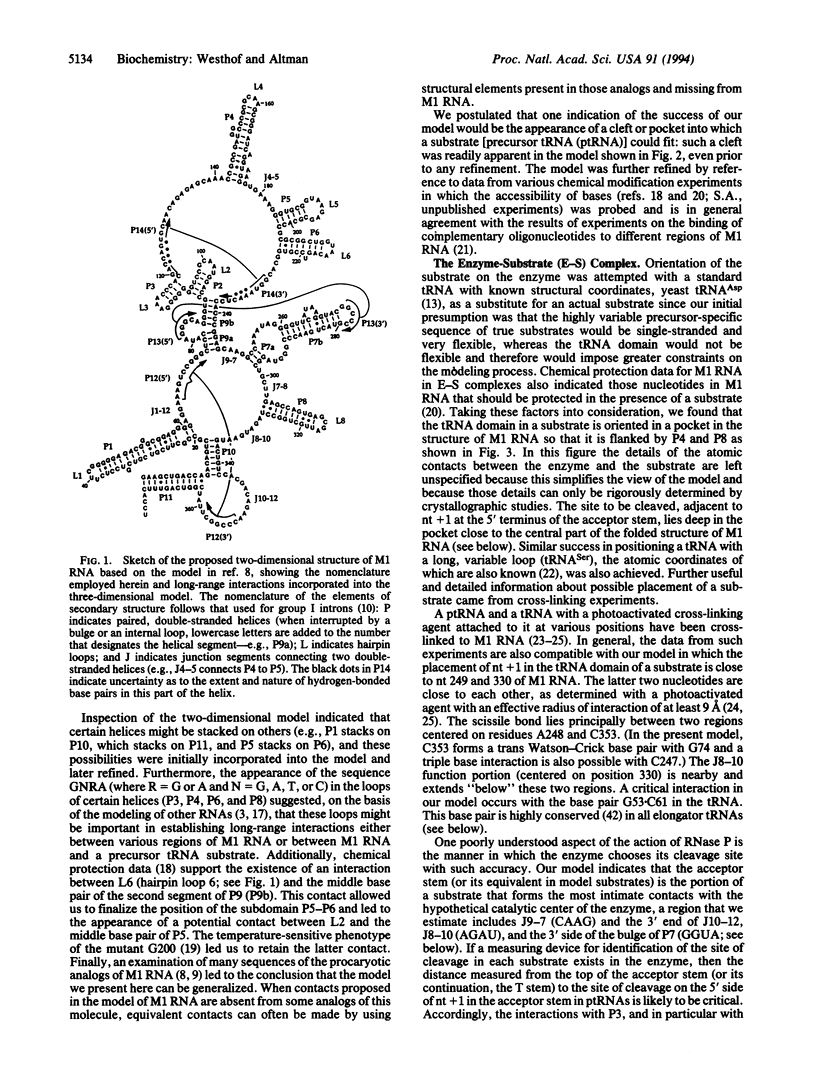
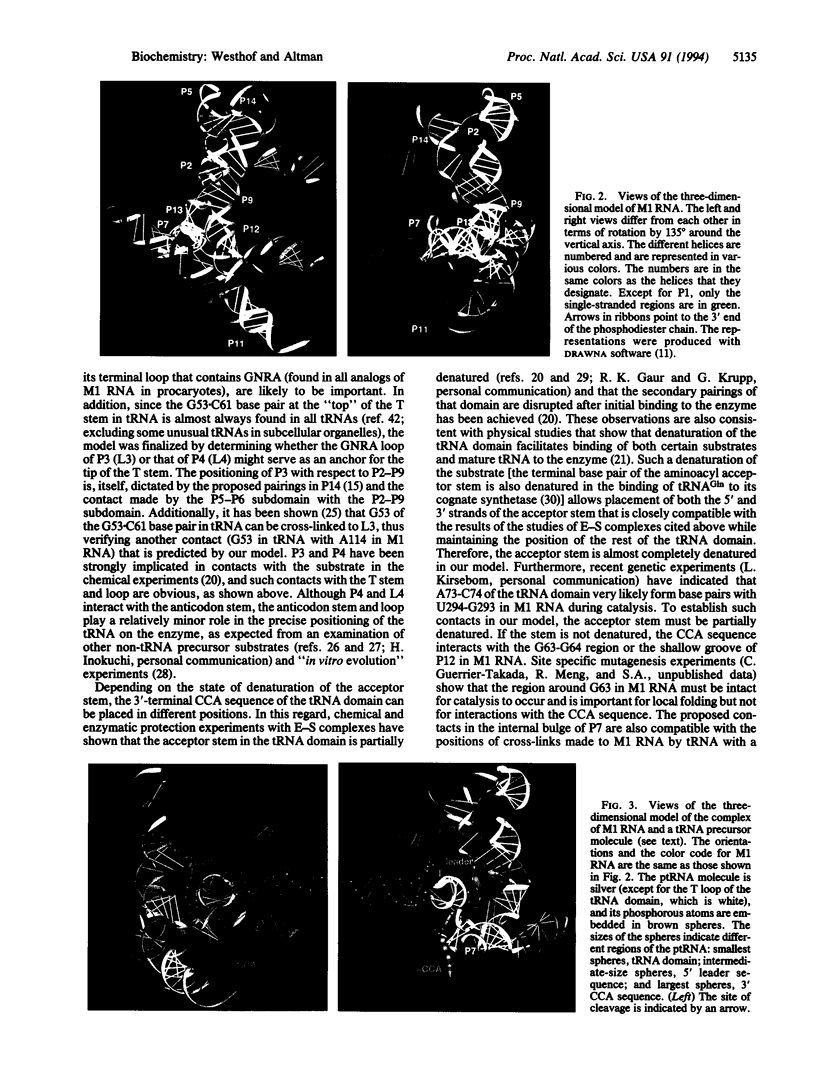
A three-dimensional model of M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli, was constructed with the aid of a computer. The modeling process took into account data from chemical and enzymatic protection experiments, phylogenetic analysis, studies of the activities of mutants, and the kinetics of reactions catalyzed by the binding of substrate to M1 RNA. The model provides a plausible picture of the binding to M1 RNA of the tRNA domain of a precursor tRNA substrate. The scissile bond and adjacent segments of the aminoacyl acceptor stem of a precursor tRNA substrate can fit into a cleft that leads to the phylogenetically conserved, central part of the structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Altman S., Wesolowski D., Puranam R. S. Nucleotide sequences of the RNA subunit of RNase P from several mammals. Genomics. 1993 Nov;18(2):418–422. doi: 10.1006/geno.1993.1488. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Pace N. R. Ribonuclease P RNA and protein subunits from bacteria. Nucleic Acids Res. 1992 Apr 11;20(7):1451–1456. doi: 10.1093/nar/20.7.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel C., Romby P., Westhof E., Ehresmann C., Ehresmann B. Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J Mol Biol. 1991 Sep 5;221(1):293–308. doi: 10.1016/0022-2836(91)80220-o. [DOI] [PubMed] [Google Scholar]
- Burgin A. B., Pace N. R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990 Dec;9(12):4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Belfort M., Cech T. R., Davies R. W., Schweyen R. J., Shub D. A., Szostak J. W., Tabak H. F. Structural conventions for group I introns. Nucleic Acids Res. 1987 Sep 25;15(18):7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr S. C., Brown J. W., Pace N. R. The varieties of ribonuclease P. Trends Biochem Sci. 1992 May;17(5):178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Westhof E., Giegé R., Moras D. Solution structure of a tRNA with a large variable region: yeast tRNASer. J Mol Biol. 1989 Apr 20;206(4):707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- Dumas P., Moras D., Florentz C., Giegé R., Verlaan P., Van Belkum A., Pleij C. W. 3-D graphics modelling of the tRNA-like 3'-end of turnip yellow mosaic virus RNA: structural and functional implications. J Biomol Struct Dyn. 1987 Apr;4(5):707–728. doi: 10.1080/07391102.1987.10507674. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. A physical assay for and kinetic analysis of the interactions between M1 RNA and tRNA precursor substrates. Biochemistry. 1993 Jul 20;32(28):7152–7161. doi: 10.1021/bi00079a012. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Reconstitution of enzymatic activity from fragments of M1 RNA. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1266–1270. doi: 10.1073/pnas.89.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C., Haydock K., Allen L., Altman S. Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli. Biochemistry. 1986 Apr 8;25(7):1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Lumelsky N., Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989 Dec 22;246(4937):1578–1584. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Michel F., Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994 Mar 11;236(5):1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Westhof E., Michel F. Monitoring of the cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3' terminal intron components. J Mol Biol. 1993 Nov 20;234(2):331–346. doi: 10.1006/jmbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. EMBO J. 1990 Jun;9(6):1929–1937. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov S., Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. Identification of a region within M1 RNA of Escherichia coli RNase P important for the location of the cleavage site on a wild-type tRNA precursor. J Mol Biol. 1993 Jun 5;231(3):594–604. doi: 10.1006/jmbi.1993.1312. [DOI] [PubMed] [Google Scholar]
- Knap A. K., Wesolowski D., Altman S. Protection from chemical modification of nucleotides in complexes of M1 RNA, the catalytic subunit of RNase P from E coli, and tRNA precursors. Biochimie. 1990 Nov;72(11):779–790. doi: 10.1016/0300-9084(90)90187-l. [DOI] [PubMed] [Google Scholar]
- Lumelsky N., Altman S. Selection and characterization of randomly produced mutants in the gene coding for M1 RNA. J Mol Biol. 1988 Aug 5;202(3):443–454. doi: 10.1016/0022-2836(88)90277-x. [DOI] [PubMed] [Google Scholar]
- Mans R. M., Guerrier-Takada C., Altman S., Pleij C. W. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990 Jun 25;18(12):3479–3487. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Nolan J. M., Burke D. H., Pace N. R. Circularly permuted tRNAs as specific photoaffinity probes of ribonuclease P RNA structure. Science. 1993 Aug 6;261(5122):762–765. doi: 10.1126/science.7688143. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Altman S. Important 2'-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J Mol Biol. 1992 Jul 20;226(2):399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Shiraishi H., Shimura Y. Functional domains of the RNA component of ribonuclease P revealed by chemical probing of mutant RNAs. EMBO J. 1988 Dec 1;7(12):3817–3821. doi: 10.1002/j.1460-2075.1988.tb03266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Steitz J. A. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Tallsjö A., Svärd S. G., Kufel J., Kirsebom L. A. A novel tertiary interaction in M1 RNA, the catalytic subunit of Escherichia coli RNase P. Nucleic Acids Res. 1993 Aug 25;21(17):3927–3933. doi: 10.1093/nar/21.17.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Altman S. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease P. Science. 1994 Mar 4;263(5151):1269–1273. doi: 10.1126/science.8122108. [DOI] [PubMed] [Google Scholar]