Abstract
The Human Variome Project (HVP) is a global effort to collect and curate all human genetic variation affecting health. Mutations of mitochondrial DNA (mtDNA) are an important cause of neurogenetic disease in humans; however, identification of the pathogenic mutations responsible can be problematic. In this article, we provide explanations as to why and suggest how such difficulties might be overcome. We put forward a case in support of a new Locus Specific Mutation Database (LSDB) implemented using the Leiden Open-source Variation Database (LOVD) system that will not only list primary mutations, but also present the evidence supporting their role in disease. Critically, we feel that this new database should have the capacity to store information on the observed phenotypes alongside the genetic variation, thereby facilitating our understanding of the complex and variable presentation of mtDNA disease. LOVD supports fast queries of both seen and hidden data and allows storage of sequence variants from high-throughput sequence analysis. The LOVD platform will allow construction of a secure mtDNA database; one that can fully utilize currently available data, as well as that being generated by high-throughput sequencing, to link genotype with phenotype enhancing our understanding of mitochondrial disease, with a view to providing better prognostic information.
Keywords: MtDNA, LOVD, mutation, human variome project, haplogroup
Mitochondria, Mitochondrial DNA, and Disease
Mitochondria are found in all nucleated cells and are the main producers of adenosine triphosphate (ATP) by oxidative phosphorylation (OXPHOS); they also have a role in other critical cellular processes, including signaling and cell death. Mitochondrial DNA is a circular molecule, which in humans is approximately 16.6 kb in length (Fig. 1). One of the hallmark features of mtDNA is that it is present in multiple copies within the cell, with several mtDNA molecules being packaged into a nucleoid structure (mtDNA along with associated proteins). The coding content of mtDNA is modest—13 essential polypeptides of the OXPHOS system and the necessary RNA machinery (2 rRNAs and 22 tRNAs) for their translation in the organelle [Anderson et al., 1981]. The majority of the protein subunits that make up the OXPHOS complexes, along with those required for mtDNA maintenance, assembly, and gene expression are nuclear-encoded and are imported into mitochondria from the cytosol.
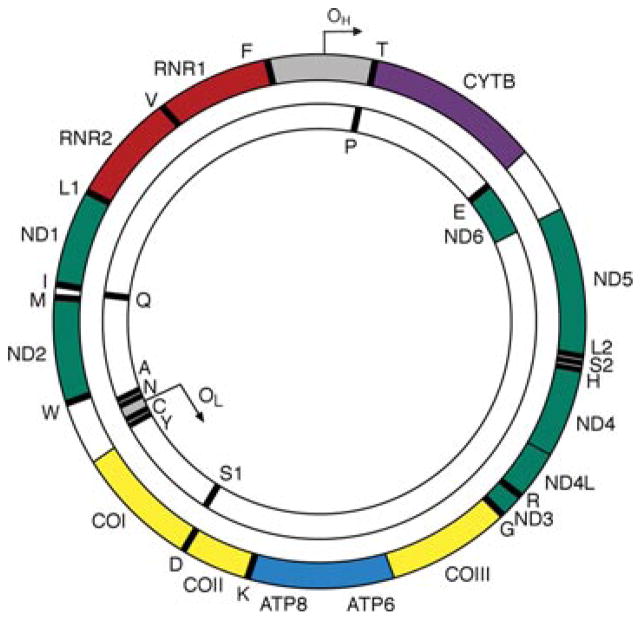
Figure 1.
A schematic of the 16.6 kb circular, double-stranded human mitochondrial DNA molecule. The outer circle represents the heavy (H) strand of mtDNA and the inner circle the light (L) strand. OH and OL are the origins heavy-strand and light strand and replication. The gray region represents the mitochondrial control region also known as the D-loop. Genes encoding subunits of respiratory chain complex I are shown in green, the MTCYB gene of complex III in purple, the three catalytic subunits of complex IV in yellow and MTATP6 and MTATP8 of complex V in blue. The two ribosomal RNAs are shown in red and the 22 transfer RNAs represented as black bars and denoted by their single-letter abbreviations.
A number of important differences exist between mtDNA and nuclear DNA, many of which are relevant to mtDNA disease expression. First, in most animals including humans, mtDNA is inherited maternally and as a result, mitochondrial lineages are uniparental. As such, only mothers are at risk of transmitting mtDNA disease. Due to strict maternal inheritance and the resulting lack of biparental recombination [Elson et al., 2001] the evolution of mtDNA is characterized by the emergence of distinct lineages called mitochondrial haplotypes or haplogroups. The classification of all human mtDNA sequences into one of a now large number of well-defined haplogroups is possible [Torroni et al., 1996; van Oven and Kayser, 2009]. Haplogroups are defined by a single block of characteristic nucleotide polymorphisms (SNPs) (commonly known as the “sequence motif”) upon which new polymorphisms occur that give rise to new subhaplogroups. It has been suggested that some of the frequent mtDNA variants might alter susceptibility to a number of common complex diseases.
As mentioned, another major difference between the mitochondrial chromosome and those found in the nucleus is that there are multiple copies of mtDNA in a cell; hundreds or thousands depending on cell type. When all mtDNA copies in a cell are identical, the population of mtDNA molecules is described as homoplasmic [Monnat and Loeb, 1985]. When there are two or more populations of nonidentical mtDNA present in a cell, the situation is referred to as heteroplasmy (literally “heterogeneity of the cytoplasm”). Next generation sequencing is revealing heteroplasmy to be a common rather than exceptional occurrence [He et al., 2010; Li et al., 2010], which has implications for our understanding of mutation rates, inheritance, and disease.
The first disease linked to mtDNA mutations was chronic progressive external ophthalmoplegia (CPEO) due to large-scale rearrangements of mtDNA [Holt et al., 1988]. This was quickly followed by the demonstration that Leber hereditary optic neuropathy (LHON) was linked to a mtDNA point-mutation (m.11778G>A) in a subunit of complex I [Wallace et al., 1988]. Unusually, this and other LHON mutations may be seen as either a homoplasmic or heteroplasmic change in affected individuals. Subsequently, over 250 point-mutations and rearrangements of mtDNA have been demonstrated to be able to cause clinically manifesting disease in patients. MtDNA disease is frequently seen in conjunction with heteroplasmy with the vast majority of mtDNA mutations being functionally recessive, a biochemical defect only being evident when a threshold level of mutation is exceeded; this threshold is generally greater than 50% of the cellular mtDNA. This threshold level varies for each mutation and differs between tissues, possibly as a consequence of the variable energy demands placed upon individual tissues [Tuppen et al., 2009]. Our focus in this article is the inherited pathogenic mtDNA mutations causing clinical manifestations in patients. We put forward the case for the implementation of a LOVD database, which will allow the investigation of genotype–phenotype correlations in this complex group of disease.
The prevalence of mtDNA disease has been notoriously difficult to estimate due to clinical heterogeneity of mitochondrial disorders and multitude causative mutations. Estimates carried out in the North East of England have found as many as 9 in 100,000 people have clinically manifesting mtDNA disease, making mtDNA disease one of the most common inherited causes of neuromuscular disorders [Schaefer et al., 2008]. As a first step to estimating the unseen and underlying levels of mtDNA mutations in the population, Elliott and colleagues utilized a multiplex primer extension assay to determine the frequency of ten common mtDNA point mutations in the North Cumbrian (UK) Birth Cohort, and found that at least 1 in 200 healthy humans harbours a low level of a pathogenic mtDNA mutation, albeit at levels far below that required to cause disease [Elliott et al., 2008]. However, the presence of even low levels of mtDNA mutation in a female means they could potentially have children that manifest mtDNA disease due to the segregation of mtDNA during the germline bottleneck [Cree et al., 2008]. Making inherited mtDNA disease potentially of clinical importance to a wider section of the population than other inherited disorders in this disease category with a typical autosomal dominant or recessive pattern of inheritance.
The clinical diagnosis of mitochondrial disease caused by mutations in mtDNA is far from straightforward and is often complicated by a diverse, variably penetrant clinical phenotype that may be associated with several different genotypes. Each of these genotypes may, in turn, have multiple phenotypic associations. For instance, the m.3243A>G mutation is commonly seen in patients with maternally inherited diabetes and deafness (MIDD), CPEO, and mitochondrial encephalomyopathy lactic acidosis and stroke-like episodes (MELAS); but each of these phenotypes is independently associated with several other mtDNA mutations. When further considerations such as multisystem involvement and variable age of onset are taken into account, it can be appreciated how the diagnosis may be missed and why the estimates of disease prevalence referred to above are probably rather conservative. Disease caused by mutations in mtDNA can be largely confined to a single organ, or even specific cells within that organ (e.g., retinal ganglion cells in LHON), or involve many different tissues—typically, muscle, brain, and heart—but many other organs can also be affected. Indeed clinical diagnosis is dependent upon having a high index of suspicion for mtDNA disease and recognizing unusual conjunctions of affected organs, for example, cochlea and pancreas. Table 1 gives a summary of the diversity and variability of clinical phenotypes present in patients as a result of mtDNA mutations. Natural history studies using validated rating scales [Achouitar et al., 2011; Phoenix et al., 2006; Schaefer et al., 2006] are in progress and initiatives in several countries to develop mitochondrial disease patient cohorts will make an enormous contribution to our understanding of disease progression and facilitate clinical trials when suitable therapies or interventions are available. There has been substantial progress in understanding the genetic basis of mitochondrial disease, with important implications for the diagnosis, investigation, and multidisciplinary management of affected patients [McFarland et al., 2010].
Table 1.
Tissues Affected and Symptoms Seen in Mitochondrial Patients
| Tissue | Symptom |
|---|---|
| Skeletal muscle | Exercise intolerance and fatigue, hypotonia, myopathy, chronic progressive external ophthalmoplegia, ptosis |
| Peripheral nervous system | Neuropathy, dorsal root ganglionopathy |
| Central nervous system | Seizures, movement disorders (Parkinsonism, dystonia, myoclonus, ataxia), leukodystrophy, encephalopathy, spasticity, stroke-like episodes, learning difficulties and psychomotor retardation |
| Neuro-ophthalmic | Cataracts, retinopathy and retinitis pigmentosa, optic atrophy cortical visual impairment |
| Neuroauditory | Sensorineural deafness (cochlear and auditory nerve) |
| Neuropsychiatric | Dementia, depression |
| Heart | Cardiac arrhythmia, cardiomyopathy (hypertrophic and dilated) |
| Liver | Hepatic dysfunction and failure |
| Kidney | Proximal renal tubulopathy, nephrotic syndrome |
| Endocrine | Diabetes, exocrine pancreatic insufficiency, hypothyroidism, hypoparathyroidism, Addison’s disease, adrenal insufficiency, infertility (primary and secondary gonadal failure), multiple endocrine disturbance, short stature |
| Hematological | Sideroblastic anemia, pancytopenia |
| Metabolic | Organic acidopathy, lactic acidosis |
| Gastrointestinal | Cyclical vomiting, gastro-oesophageal reflux, pseudo-obstruction, constipation, diarrhea |
This table illustrates the wide variety of tissues involved in diseases associated with mtDNA mutations and the wide range of symptoms that may present. Disease presentation and future course of disease cannot always be predicted by knowing the primary mutation alone. Building a database that is able to link phenotypes, primary mutation, and other variants is likely to be critical to advancing the prognosis that can be provided to patients.
Assessment and Reporting of mtDNA Variation Seen in Patients with Mitochondrial Disease
It is presently not uncommon for specialist diagnostic laboratories to sequence the complete mtDNA of patients suspected of manifesting mtDNA disease. The advent of cheaper and quicker sequencing technologies is likely to make this even more frequent. Indeed, frequent complete mtDNA sequencing will almost certainly extend beyond nations that are currently offering extensive clinical genetics services [Smuts et al., 2010; van der Walt et al., 2012]. When conducted, complete mtDNA sequencing may reveal a known pathogenic variant, but in some cases no previously confirmed pathogenic mutation is identified, though one or more novel (and potentially pathogenic) changes may be observed. Deciding if one of these changes is genuinely causing disease is of the utmost importance regarding the accuracy of genetic counseling that will be provided to the patient (and their family) and upon which future reproductive choices may be made.
Confusion regarding which mitochondrial variants cause disease is exacerbated by the lack of an agreed “burden of proof” that must be met prior to scientific publication of specific mtDNA variations in association with disease. The absence of critical functional tests from many published reports, coupled with a failure to display the evidence upon which decisions regarding pathogenicity are reached on databases further compounds this confusion [Yarham et al., 2011]. Also important in this regard are the poor application of human phylogenetic knowledge and the historical idiosyncrasies of mtDNA nomenclature, which have not followed the HGVS (Human Genome Variation Society) guidelines. This has made it difficult for some investigators to search the literature to identify previous confirmation of mutation status. Finally, insufficient curation of frequently used databases causes inaccuracies to be propagated. This latter point may well reflect the lack of adequate funding and resources for the current databases. The seemingly ever-increasing quantities of sequence data, of variable quality, has now exceeded the coping capabilities of small, unconnected groups of dedicated individuals.
Prime among the causes of confusion in the classification of mtDNA changes as pathogenic is a distinct lack of agreement about what should be required to class a change seen in a patient as being causal of the disease. In their seminal article, Dimauro and Schon described the canonical pathogenic criteria for confirming mtDNA point mutations as disease causing; that the mutation must be heteroplasmic; mutated mtDNA should segregate with a biochemical defect in clinically relevant tissues and likewise with disease severity in maternally related family members; that the variant should be at an evolutionarily conserved site and absent from an ethnically matched control population [Dimauro and Schon, 2001]. These criteria provided a robust, early framework for separation of disease causing mutations from polymorphisms. The criteria focused on some of the unique features of mtDNA genetics that provide powerful methods to determine pathogenicity. The existence of heteroplasmy in association with disease, for example, allows not only the segregation of a mutation within a family to be studied, but also in cells within tissues, providing the basis for functional assays critical to correctly identifying disease causing variants. The importance of understanding population variation is also central to the criteria.
Some pathogenic mtDNA mutations identified since have, however, failed to meet the canonical criteria. For instance, an increasing number of pathogenic mtDNA mutations are homoplasmic [McFarland et al., 2002; Perli et al., 2012; Prezant et al., 1993] and, therefore, cannot by definition be heteroplasmic or demonstrate a clear segregation with biochemical deficiency in tissues. Some mtDNA pathogenetic mutations are not evolutionarily well conserved therefore conservation alone is insufficient to determine if a change is pathogenic or a polymorphism [McFarland et al., 2004b; Tuppen et al., 2008].
A paradigmatic case of a frame shift change that appears to have all the typical characteristics of a pathogenic mutation is represented by the insertion m.3308+C that occurs in parallel with the transversion m.3308T>A. A pathogenic role for m.3308+C has been suggested [Simon et al. 2001]. It seems that the negative effect of this frameshift insertion, which would produce a truncation of most of the ND1 subunit, finds a mechanism of compensation [Achilli et al., 2008] when occurring together with m.3308T>A. Thus, while m.3308T>A eliminates the original methionine codon (AUA), a second methionine codon (AUG) is present at the third codon of the peptide (and therefore leaving aside the deleterious effect of m.3308+C). The final result is an mRNA that codes for a slightly truncated protein that only lacks two amino acids when compared to the most common variation of the peptide. This shorter protein does not seem to have a functional deficit, as the combined motif m.3308T>A plus m.3308+C represents the motif of a common Native America haplogroup, named A2i. In addition, the transversion m.3308T>A occurs as a diagnostic variant of several other haplogroup backgrounds, including the Sub-Saharan L1b; and, therefore, this can be considered to be a quite common polymorphism in several human population groups. As such it is highly unlikely that theses variants are the cause of clinical manifestations of mitochondrial disease. However, an effect of such population variants in a common or complex disease cannot be discarded, such a role in susceptibility or outcome of disease, would have to be investigated by an association study. Such an association study would evaluate haplogroup frequencies in both patients and controls attempting to demonstrate that the genotype and phenotype were significantly associated using a statistical methodology.
There have been a number of studies re-examining how best to determine if a mtDNA change has a causal role in a disease with these and other complications in mind for mt-tRNAs genes [McFarland et al., 2004a] and for the protein encoding genes of complex I [Mitchell et al., 2006]. These publications set out a score of pathogenicity for mtDNA mutations, which include several parameters such as presence of heteroplasmy, segregation within the family, biochemical defects, interspecies amino acid conservation, and functional studies. The weighted criteria for mt-tRNA’s mutations was refined recently given the increase in the number of available reported pathogenic changes [Yarham et al., 2011].
Insufficient knowledge (or application of what is known) about mitochondrial phylogeny appears to have been a major contributor to the misattribution of pathogenicity to haplogroup defining variants, as highlighted by Bandelt and colleagues [Bandelt et al., 2005, 2007]. It is agreed that if a variant defines a whole major or minor subhaplogroup then it would be very unlikely that the variant would be the primary disease causing mutation of an inherited mitochondrial syndrome. Such changes may of course play a role in disease though a “common variant, common disease” hypothesis, or indeed modulate the penetrance of a primary mutation. Phylogeographic analysis of mtDNA haplogroup F2 in China was critical in revealing m.12338T>C in the initiation codon of the MTND5 gene not to be pathogenic [Kong et al., 2004]. Given the consequences of incomplete understanding of the phylogeny in the study of disease there is a strong case for researchers in the areas of evolutionary biology, population genetics, and genealogy to contribute to a database on the variation seen on human mtDNA [Bandelt et al., 2008; Kong et al., 2006]. Another persistent difficulty has been the presence of mitochondrial pseudogenes from the nuclear genome, which have been mistakenly amplified in the past [Yao et al., 2008]. An additional complication has been generated by the previously used nomenclature to describe mtDNA variants, which did not conform to the HGVS guidelines. Mitochondrial point mutations have been described in a number of different formats, for example, the m.3243A>G point mutation which is the recommended HGVS nomenclature has been described variously as A3243G, 3243A>G, and 3243G. This can complicate PubMed and other searches made to determine if a variant has been previously detected. This unconventional nomenclature hampers literature searches to confirm prior reporting of identified mtDNA variants.
The first complete human genome to be sequenced was human mtDNA [Anderson et al., 1981], sometimes referred to as the “Cambridge Reference Sequence.” Given there were a number of errors in this original sequence, Howell and colleagues reanalyzed the original DNA source to update the consensus sequence [Andrews et al., 1999], now known as the revised Cambridge reference sequence (rCRS; GenBank accession number NC_012920.1). In the rCRS, the m.3107del is retained being represented by an N to enable historical nucleotide numbering to be maintained. This can cause problems with annotation, for example, the GeneChip Human Mitochondrial sequencing array from Affymetrix does not account for the m.3107del error. Hence substantial efforts have to be made to follow the HGVS guidelines. There have been many cases where a mtDNA variant has been asserted to be novel when in fact this is not the case [Bandelt et al., 2006, 2009]. Novel should refer to mtDNA mutations or polymorphisms that have not been observed or described in the scientific literature; although novelty has been misused to refer to changes that are particularly rare or “de novo” that is mutations not observed in maternal tissues [Bandelt et al., 2009]. The belief that a change is novel has often been taken as strong and in some instances sufficient evidence for that change to be viewed as causal of disease [Brandon et al., 2005]. By utilizing a number of different online databases and Internet resources it is possible to identify most changes in mtDNA that have been previously reported [Bandelt et al., 2009]. However, this has not always proven to be straightforward, especially for investigators new to the field. A central reference database for disease causing mutations, where evidence for their pathogenicity can be presented along with the variant would allow efficient evaluation of the probable role of a change in disease. There are a number of mtDNA databases and tools available to the community, and we summarize the major ones in Table 2. The most frequently updated of these reference points is the MITOMAP database (http://www.mitomap.org/MITOMAP), which is updated on a weekly basis having undergone major rebuilds to include information on phylogenetic context [Ruiz-Pesini et al., 2007a] and to incorporate analytical tools such as Mitomaster [Zaragoza et al., 2011]. A number of the other frequently referenced databases have not been updated in many years; mtDB http://www.mtdb.igp.uu.se/ was last updated on March 1, 2007, and mtSNP http://mtsnp.tmig.or.jp/mtsnp/index_e.shtml on January 25, 2006. Such a fossilization process is not uncommon when a database has been built and run by a single group. The member of staff that built the database might move on and with them goes the key knowledge and enthusiasm to maintain the resource. The fact that a number of the fossil databases are still well used is a testament to their user-friendly nature and need for the resource. They can, however, represent a data interpretation trap for the unwary. As an example, mtDB is often used as a population variation database; however; it contains many sequences from patients with mitochondrial disease. Ideally, any new database should not be the effort of a single group but should draw together experience and resources from interested research groupings across the globe to avoid the duplication of effort and database fossilization seen in the past.
Table 2.
Summary of the Online mtDNA Variation Databases
| Name | URL | Description | Updated | Sequences available | mtDNA region covered | Evidence of variant pathogenicity |
|---|---|---|---|---|---|---|
| GenBank (NCBI) | http://www.ncbi.nlm.nih.gov/nuccore | An annotated collection of all publicly available DNA sequences | Daily | 9,659 | Complete | – |
| GOBASE | http://gobase.bcm.umontreal.ca/ | Taxonomically broad organelle genome database | 31/08/2010 | 1,898 sequences from 1283 species* | Complete | – |
| HMPD | http://bioinfo.nist.gov/hmpd/ | Human mitochondrial protein database | 01/02/2007 | 643** | Complete | – |
| HmtDB | http://www.hmtdb.uniba.it | Database of human mitochondrial genome sequences annotated with population and variability data | 19/06/2011 | 7,825 complete genomes and 868 coding region sequences | Complete | Individual’s complete mitochondrial genome sequencing data with references |
| HvrBase++ | http://www.hvrbase.org/ | A phylogenetic database for primate species | 01/01/2006 | 1,376 | Complete | – |
| Mamit-tRNA | http://mamit-trna.u-strasbg.fr/ | A compilation of mammalian mitochondrial tRNA genes | 24/10/2007 | 3,064 tRNA gene sequences from 150 genomes* | tRNAs | Mutation references |
| MitoLSDB | http://mitolsdb.igib.res.in/ | Database of mitochondrial variants | 19/10/2011 | – | – | – |
| MITOMAP | www.mitomap.org | A compendium of polymorphisms and mutations of the human mitochondrial DNA | weekly | – | – | Classification system and mutation references |
| MitoTool | http://www.mitotool.org/ | A bioinformatics platform for handling human mtDNA sequence data | 10/09/2011 | 9,328 | Complete | Description of amino acid change and mutation references |
| MitoVariome | http://variome.kobic.re.kr/MitoVariome/ | A variome database of human mitochondrial DNA sequences | 08/07/2009 | 5,344 | Complete | – |
| mtDB | http://www.mtdb.igp.uu.se/ | Database of human mitochondrial genomes and SNPs | 01/03/2007 | 1,865 complete sequences and 839 coding region sequences | Complete | Description of resulting amino acid change |
| mtDNAmanager | http://mtmanager.yonsei.ac.kr/index.php | Database of mtDNA control region sequences | 03/10/2011 | 9,294 | Control Region | – |
| mtSNP | http://mtsnp.tmig.or.jp/mtsnp | Human mitochondrial genome SNP database | 25/01/2006 | 672 | Complete | – |
| OGRe | http://drake.physics.mcmaster.ca/ogre | Database of complete mitochondrial metazoan genome sequences | 2006 | 1,244* | Complete | – |
| tRNAdb 2009 | http://trnadb.bioinf.uni-leipzig.de/ | Compilation of tRNA sequences and sequences of tRNA genes | 2009 | 12,099 tRNA gene sequences from 587 species* | tRNAs | – |
This table displays relevant details for many of the currently available and freely accessible mtDNA variant databases. These details include the database name, an active URL address, a description of the database, the last known date the database was updated, how many sequences are available (*—not all human sequences; **—cannot access sequence data), the mtDNA region covered by the database, and a description of any evidence of variant pathogenicity presented by the database. Resources primarily considered tools were omitted. None of the current databases allow phenotypes to be sorted to allow the links to be built between genotypes and phenotypes.
Future Prospects with the LOVD Database Platform
Many of the current problems with gathering and using mtDNA variation data seem to stem from uncoordinated approaches to the problem within the field. The International society for Gastrointestinal Hereditary Tumours (InSiGHT), is an excellent example of what might be achieved when the efforts of a community are pooled and directed [Peltomäki and Vasen, 2004]. In 2003, the The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) and the Leeds Castle Polyposis Group (LCPG), the two major groups with databases in this area of disease, merged into a new group, InSiGHT. The merger allowed an unification effort in the collection, curation and study of DNA mismatch repair gene mutations in a single database [Ou et al., 2008]. The InSiGHT group use the Leiden Open Variation Database (LOVD) to store and organize the curated data. LOVD is a well-supported piece of open access software that allows creation of Web-based locus-specific databases (LSDBs) by providing a tool-kit for users. The use of LOVD as a LSDBs has been growing rapidly [Fokkema et al., 2001]. LOVD’s gene centered approach with a modular design coupled with human mtDNA’s nonrecombining nature, means it can be considered as a single locus. As such, a single LOVD database would be suitable to store all mtDNA genes. Using a LOVD database with all the variants from the rCRS may help unravel the modifying genetic backgrounds and explain the variable phenotype/penetrance seen in mitochondrial disorders.
It will also be essential to build databases for the important nuclear genes linked to mitochondrial disease mtDNA disease. The process is well underway with the creation of a locus-specific database for mutations in GDAP1 which makes use of LOVD’s facilities for the analysis of genotype–phenotype correlations in Charcot–Marie–Tooth disease [Cassereau et al., 2011]. The LOVD platform has recently released an updated version, which has encouraged many curators to move their database or static PDF and HTML files into a LOVD format, for example, OPA1 database http://lbbma.univ-angers.fr/lbbma.php?id=22&type=m (Vincent Procaccio et al., 2012, personal communication).
MITOMAP [Brandon et al., 2005; Kogelnik et al., 1996], the most well-known, widely used, and frequently updated of the available mtDNA databases, has evolved and grown over the years. It is in many senses a locus specific database, but also provides many other tools and widely used resources. It is, and has been for many years, the “go to” point of reference for information in the mtDNA field [Ruiz-Pesini et al., 2007b; Zaragoza et al., 2011]. The LOVD database system is, however, able to offer additional features. Having excellent database security LOVD was designed to withstand Web-based system attacks [Fokkema et al., 2011] and as such, is suitable for the storage of data that cannot be made available to all members of the public in addition to open access data.
To truly study the impact of variants on human health, we need to link variation to the clinical features of the patient. The entry of any such data even in an anonymized fashion presents important confidentiality considerations. LOVD is not only secure but also offers a variety of search options. One option, “Search through hidden entries,” can be enabled (by the database curators) to allow queries of restricted data. Any such searches do not return the data records, but the number of records matching the query and allows for the possibility of contacting the data contributor. This potentially allows clinicians with patients having rare genotype–phenotype combinations to make contact. Also, to understand the subtleties of mtDNA diseases and how mtDNA background might affect penetrance and disease course, it is essential that not only the suspected pathogenic variant is added to the database, but all variants from the reference sequence. This could also be implemented in a flexible fashion on a LOVD database. Given these data storage and search options, a LOVD database dedicated to mtDNA would make a valuable companion to MITOMAP.
mtDNA Variation in Common Disease
The discussion so far has been focused on single pathogenic point mutations causing clinical manifestations of disease in patients. This might be the “tip of the iceberg” when considering the contribution of mtDNA changes to disease. It has been suggested that some of the population mtDNA variants might alter susceptibility to a number of more common diseases [Coskun et al., 2011] or alter the course taken by a disease [Achilli et al., 2011]. Mitochondrial defects have also been implicated in ageing and susceptibility to cancer [Wallace, 1999]. Such data has traditionally been collected in the form of haplogroup association studies with almost all studies analyzing haplogroup variation in association with a complex trait having used a contingency table analysis. A common problem is that such association studies often lack power due to small sample sizes [Samuels et al., 2006]. There have been a few higher resolution case-control studies where the whole of the mtDNA was sequenced for all the samples used in the study [Elson et al., 2006]. In the light of next generation sequencing (NGS), high-resolution studies looking at all SNPs might become more common. Given the goal of the HVP to collect and curate all human genetic variation affecting human health, it would be essential to find a way to include data from these studies in a LOVD database dedicated to the role of mtDNA variation in human disease. Inherited mtDNA polymorphisms may play a role in the aetiology of complex disease in at least two ways. First, a disease might be significantly associated with one mtDNA haplogroup, suggesting one or more haplogroup defining polymorphisms modify risk of the disease. This hypothesis is akin to the “common-disease, common-variant” hypothesis. Second, it is possible that the cumulative effects of multiple phenotypically subtle mtDNA mutations are risk factors for disease, the “mtDNA mutational load hypothesis” [Elson et al., 2006]. Considering the wider view on the possible role of mtDNA variants in disease underscores the need for investigators to enter all variants from the rCRS in order that we are able to unravel the role of mtDNA variants in the aetiology of disease. The ability of a LOVD database to link genotypes with phenotypes will also be essential in this category of study with information on geographical location of subject and tissue sampled being important data to store alongside the variants. We hope that this new database will allow questions of such statistical associations to be addressed. LOVD’s modular design provides a great deal of flexibility which will be needed to cope with the specific interests of the users studying patient-disease (mtDNA mutations) and population-disease and the role of population variants (mtDNA polymorphisms).
A significant amount of research has also suggested that mtDNA somatic mutations that have undergone clonal expansion play an important role in aging [Loeb et al., 2005], cancer [Brandon et al., 2006], and neurodegeneration [Coskun et al., 2004]. These studies are a very active area of research and one that is controversial. A LOVD database could allow input of data from studies of somatic mtDNA variants using specific fields to keep the somatic study data distinct from that of inherited mutations, a similar solution was implemented in MITOMAP.
It is anticipated that each of these three fields of study; inherited disease, association with population variants, and somatic mutations would have separate modules within the one database, which would allow searches to be performed from a common interface.
Conclusions
Gathering data on all areas in which the mtDNA variome contributes to human health outcomes in a single database will require careful consideration and a flexible well-supported platform. It is, however, clear that many challenges must be met if we are to make the step changes in understanding made possible by the vast amounts of data that new sequencing technologies are allowing us to gather.
In summary, the field of neurogenetics is extensive and the confluence of many experts will be needed in systematic attempts to collect and annotate as much of the relevant data as possible to build high-quality databases of genetic variation and its significance. The issues related to information and data collection for different neurogenetic disorders should be worked out in a coordinated manner if the best possible level of integration is sought. Funding support will be a key issue to provide the necessary continuity and to guarantee the delivery of open-access, high-quality, and up-to-date information. Involvement of the wider scientific community as a whole, including patient advocates, will be crucial and is highly encouraged. To this end, we invite all members of the neurogenetics community worldwide to join this effort, which we believe will be for a widespread benefit of patients suffering from neurological disorders.
Acknowledgments
Contract grant sponsor: Research Council UK RCUK Academic fellowship (to JLE); Department of Health’s NIHR Biomedical Research Centre (to MGS); UK NHS Specialised Services “Rare Mitochondrial Disorders of Adults and Children” Diagnostic Service (to MS and RDSP); Fondation pour la Recherche Médicale,” “Union Nationale des Aveugles et Déficients Visuels, and “Association contre les Maladies Mitochondriales (to VP); Medical Research Council, UK (JWY); Ministerio de Ciencia e Innovación (SAF2011-26983 [to AS]); MRC (grant number G0800674 [to RDSP]); Australian National Health and Medical Research Council (Principal Research Fellowship [to DRT]); National Institutes of Health (grants NS21328, AG24373, DK73671, NS41850 [to DCW and ML]); Simons Foundation (grant #205844 [to DCW and ML]); Wellcome Trust (to RWT); Center support (074454/Z/04/Z); Medical Research Council, UK (G1000848 [to RWT and RM]).
References
- Achilli A, Olivieri A, Pala M, Hooshiar Kashani B, Carossa V, Perego UA, Gandini F, Santoro A, Battaglia V, Grugni V, et al. Mitochondrial DNA backgrounds might modulate diabetes complications rather than T2DM as a whole. PLoS. 2011;6:e21029. doi: 10.1371/journal.pone.0021029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achouitar S, Mohamed M, Gardeitchik T, Wortmann SB, Sykut-Cegielska J, Ensenauer R, de Baulny HO, Õunap K, Martinelli D, de Vries M, et al. Nijmegen paediatric CDG rating scale: a novel tool to assess disease progression. J Inherit Metab Dis. 2011;34:923–927. doi: 10.1007/s10545-011-9325-5. Epub 2011 May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, Debruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Achilli A, Kong QP, Salas A, Lutz-Bonengel S, Sun C, Zhang YP, Torroni A, Yao YG. Low “penetrance” of phylogenetic knowledge in mitochondrial disease studies. Biochem Biophys Res Commun. 2005;333:122–130. doi: 10.1016/j.bbrc.2005.04.055. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Salas A, Bravi CM. What is a ‘novel’ mtDNA mutation–and does ‘novelty’ really matter? J Hum Genet. 2006;51:1073–1082. doi: 10.1007/s10038-006-0066-5. Epub 2006 Oct 5. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Salas A, Taylor RW, Yao YG. Exaggerated status of “novel” and “pathogenic” mtDNA sequence variants due to inadequate database searches. Hum Mutat. 2009;30:191–6. doi: 10.1002/humu.20846. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Yao YG, Salas A. The search of ‘novel’ mtDNA mutations in hypertrophic cardiomyopathy: MITOMAPping as a risk factor. Int J Cardiol. 2008;126:439–42. doi: 10.1016/j.ijcard.2007.02.049. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Yao YG, Salas A, Kivisild T, Bravi CM. High penetrance of sequencing errors and interpretative shortcomings in mtDNA sequence analysis of LHON patients. Biochem Biophys Res Commun. 2007;352:283–91. doi: 10.1016/j.bbrc.2006.10.131. [DOI] [PubMed] [Google Scholar]
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database - 2004 update. Nucleic Acids Research. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassereau J, Chevrollier A, Bonneau D, Verny C, Procaccio V, Reynier P, Ferre M. A locus-specific database for mutations in GDAP1 allows analysis of genotype-phenotype correlations in Charcot–Marie–Tooth diseases type 4A and 2K. Orphanet. 2011;6:87. doi: 10.1186/1750-1172-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun P, Wyrembak J, Schriner S, Chen HW, Marciniack C, Laferla F, Wallace DC. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2012;1820:553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, Lopes S, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HHM, Chinnery PF. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–254. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- Dimauro S, Schon EA. Mitochondrial DNA mutations in human disease. A J Med Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson JL, Andrews RM, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Analysis of European mtDNAs for recombination. Am J Hum Genet. 2001;68:145–153. doi: 10.1086/316938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson JL, Herrnstadt C, Preston G, Thal L, Morris CM, Edwardson JA, Beal MF, Turnbull DM, Howell N. Does the mitochondrial genome play a role in the etiology of Alzheimer’s disease? Hum Genet. 2006;119:241–54. doi: 10.1007/s00439-005-0123-8. Epub 2006 Jan 12. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2001;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- He YP, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–U175. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morganhughes JA. Deletions of muscle mitochondrialdna in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: A human mitochondrial genome database. Nucleic Acids Res. 1996;24:177–179. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–86. doi: 10.1093/hmg/ddl130. Epub 2006 May 19. [DOI] [PubMed] [Google Scholar]
- Kong QP, Yao YG, Sun C, Zhu CL, Zhong L, Wang CY, Cai WW, Xu XM, Xu AL, Zhang YP. Phylogeographic analysis of mitochondrial DNA haplogroup F2 in China reveals T12338C in the initiation codon of the ND5 gene not to be pathogenic. J Hum Genet. 2004;49:414–23. doi: 10.1007/s10038-004-0170-3. [DOI] [PubMed] [Google Scholar]
- Li M, Schönberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland R, Clark KM, Morris AAM, Taylor RW, Macphail S, Lightowlers RN, Turn-bull DM. Multiple neonatal deaths due to a homoplasmic mitochondrial DNA mutation. Nat Genet. 2002;30:145–146. doi: 10.1038/ng819. [DOI] [PubMed] [Google Scholar]
- McFarland R, Elson JL, Taylor RW, Howell N, Turnbull DM. Assigning pathogenicity to mitochondrial tRNA mutations: when ‘definitely maybe’ is not good enough. Trends Genet. 2004a;20:591–596. doi: 10.1016/j.tig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- McFarland R, Taylor RW, Elson JL, Lightowlers RN, Turnbull DM, Howell N. Proving pathogenicity: when evolution is not enough. Am J Med Genet A. 2004b;131:107–108. doi: 10.1002/ajmg.a.30318. author reply 109–110. [DOI] [PubMed] [Google Scholar]
- McFarland R, Taylor RW, Turnbull DM. A neurological perspective on mitochondrial disease. Lancet Neurol. 2010;9:829–840. doi: 10.1016/S1474-4422(10)70116-2. [DOI] [PubMed] [Google Scholar]
- Mitchell AL, Elson JL, Howell N, Taylor RW, Turnbull DM. Sequence variation in mitochondrial complex I genes: mutation or polymorphism? J Med Genet. 2006;43:175–179. doi: 10.1136/jmg.2005.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnat RJ, Loeb LA. Nucleotide-sequence preservation of human mitochondrialdna. Proc Natl Acad Sci USA. 1985;82:2895–2899. doi: 10.1073/pnas.82.9.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Niessen RC, Vonk J, Westers H, Hofstra RM, Sijmons RH. A database to support the interpretation of human mismatch repair gene variants. Hum Mutat. 2008;29:1337–1341. doi: 10.1002/humu.20907. [DOI] [PubMed] [Google Scholar]
- Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perli E, Giordano C, Tuppen HA, Montopoli M, Montanari A, Orlandi M, Pisano A, Catanzaro D, Caparrotta L, Musumeci B, et al. Isoleucyl-tRNA synthetase levels modulate the penetrance of a homoplasmic m.4277T>C mitochondrial tRNAIle mutation causing hypertrophic cardiomyopathy. Hum Mol Genet. 2012;21:85–100. doi: 10.1093/hmg/ddr440. [DOI] [PubMed] [Google Scholar]
- Phoenix C, Schaefer AM, Elson JL, Morava E, Bugiani M, Uziel G, Smeitink JA, Turn-bull DM, McFarland R. A scale to monitor progression and treatment of mitochondrial disease in children. Neuromusc Disorders. 2006;16:814–820. doi: 10.1016/j.nmd.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu XD, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et al. Mitochondrial ribosomalrna mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007b;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DC, Carothers AD, Horton R, Chinnery PF. The power to detect disease associations with mitochondrial DNA haplogroups. Am J Hum Genet. 2006;78:713–720. doi: 10.1086/502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, Chinnery PF, Turnbull DM. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, Phoenix C, Elson JL, McFarland R, Chinnery PF, Turnbull DM. Mitochondrial disease in adults: a scale to monitor progression and treatment. Neurology. 2006;66:1932–1934. doi: 10.1212/01.wnl.0000219759.72195.41. [DOI] [PubMed] [Google Scholar]
- Smuts I, Louw R, du Toit H, Klopper B, Mienie L, van der Westhuizen F. An overview of a cohort of South African patients with mitochondrial disorders. 2010. [DOI] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC. Classification of European mtDNAs from an analysis three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2009;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Tuppen HA, Fattori F, Carrozzo R, Zeviani M, DiMauro S, Seneca S, Martindale JE, Olpin SE, Treacy EP, McFarland R, et al. Further pitfalls in the diagnosis of mtDNA mutations: homoplasmic mt-tRNA mutations. J Med Genet. 2008;45:55–61. doi: 10.1136/jmg.2007.051185. [DOI] [PubMed] [Google Scholar]
- van der Walt EM, Smuts I, Taylor RW, Elson JL, Turnbull DM, Louw R, van der Westhuizen FH. Characterization of mtDNA variation in a cohort of South African paediatric patients with mitochondrial disease. Eur J Hum Genet. 2012;20:650–656. doi: 10.1038/ejhg.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AMS, Elsas LJ, Nikoske-lainen EK. Mitochondrialdna mutation associated with lebers hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Yao YG, Kong QP, Salas A, Bandelt HJ. Pseudomitochondrial genome haunts disease studies. J Med Genet. 2008;45:769–772. doi: 10.1136/jmg.2008.059782. [DOI] [PubMed] [Google Scholar]
- Yarham JW, Al-Dosary M, Blakely EL, Alston CL, Taylor RW, Elson JL, McFarland R. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat. 2011;32:1319–1325. doi: 10.1002/humu.21575. [DOI] [PubMed] [Google Scholar]
- Zaragoza MV, Brandon MC, Diegoli M, Arbustini E, Wallace DC. Mitochondrial cardiomyopathies: how to identify candidate pathogenic mutations by mitochondrial DNA sequencing, MITOMASTER and phylogeny. Eur J Hum Genet. 2011;19:200–207. doi: 10.1038/ejhg.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]