Abstract
pH is a ubiquitous regulator of biological activity, including protein-folding, protein-protein interactions and enzymatic activity. Existing constant pH molecular dynamics (CPHMD) models that were developed to address questions related to the pH-dependent properties of proteins are largely based on implicit solvent models. However, implicit solvent models are known to underestimate the desolvation energy of buried charged residues, increasing the error associated with predictions that involve internal ionizable residue that are important in processes like hydrogen transport and electron transfer. Furthermore, discrete water and ions cannot be modeled in implicit solvent, which are important in systems like membrane proteins and ion channels. We report on an explicit solvent constant pH molecular dynamics framework based on multi-site λ-dynamics (CPHMDMSλD). In the CPHMDMSλD framework, we performed seamless alchemical transitions between protonation and tautomeric states using multi-site λ-dynamics, and designed novel biasing potentials to ensure that the physical end-states are predominantly sampled. We show that explicit solvent CPHMDMSλD simulations model realistic pH-dependent properties of proteins such as the Hen-Egg White Lysozyme (HEWL), binding domain of 2-oxoglutarate dehydrogenase (BBL) and N-terminal domain of ribosomal L9 (NTL9), and the pKa predictions are in excellent agreement with experimental values, with a RMSE ranging from 0.72 to 0.84 pKa units. With the recent development of the explicit solvent CPHMDMSλD framework for nucleic acids, accurate modeling of pH-dependent properties of both major class of biomolecules – proteins and nucleic acids is now possible.
Keywords: CPHMD, pKa values, protein electrostatics, λ-dynamics, pH, molecular dynamics, protein dynamics, explicit solvent
1. INTRODUCTION
pH is one of the critical regulators of biological activity. Enzymatic activity is optimized within a narrow pH range,1 often requiring the participation or presence of ionizable residues such as aspartic acid, glutamic acid and/or histidine in the active site,2 and accurate measurement of their pKa values is crucial in understanding the catalytic mechanism.3–5 In recent years, the role of pH regulation in nucleic acid systems has been acknowledged,6,7 where parallels to proteins can be drawn, such as the catalytic activity of ribozymes (ribonucleic acid enzyme),8–11 demonstrating the ubiquity of pH regulation in biological processes. Apart from its influence on catalytic activity, pH regulation has been observed in numerous other processes including protein folding,12–15 protein-protein interactions,16 protein-substrate binding,17,18 translational recoding,19 and aberrant pH regulation has even been implicated in cancer-related physiology.20 As such, specific examples of pH-dependent properties encompass a wide variety of systems, such as the catalytic mechanism of dihydrofolate reductase,21 proton gradient driven ATP synthesis,22 activity of the U6 intramolecular stem-loop of the spliceosome complex23 and the influenza virus haemagglutinin.24
While the pKa values of amino acid monomers have been known for decades, the microenvironment around the residue located in a protein environment may alter its pKa value. Thus, the ability to measure the microscopic pKa of a site-specific residue is invaluable in identifying key titrating residues and understanding the mechanism of these pH-dependent biological processes. Using staphylococcal nuclease and its mutants as a model system, Garcia-Moreno and co-workers undertook a series of comprehensive investigations into the effect that the local microenvironment has on the perturbation of the pKa values of protein residues.25–28 Their effort has cumulated in a joint collaboration between experimentalist and theoreticians, where the current state-of-the-art computational methods for predicting protein pKa values were evaluated against experimentally measured pKa values.29,30
One major physics-based approach that has emerged over the years to treat electrostatics in proteins and nucleic acids is the Poisson-Boltzmann (PB) equation methodology, which has achieved reasonable success in predicting protein pKa values.31 A key limitation of initial PB methods was the lack of conformational flexibility, although this has been partially addressed using approaches like tuning the effective protein dielectric constant32 and including representations of multiple conformations.33,34 The need for conformational flexibility led to the development of the other major physics-based approach in computational pKa predictions, which is based on molecular dynamics (MD) simulation. Warshel and co-workers were the first to demonstrate the use of free energy calculations to calculate the pKa values of protein residues.35–38 Subsequent developments in the MD community have sought to couple the protonation state of the titrating residue with the dynamics of the protein itself. Such pH-coupled simulations, which have been termed constant pH molecular dynamics (CPHMD), are uniquely suited to model realistic pH-dependent responses, even in systems where there is limited experimental data because no a priori information on the identity of key titrating residues and their protonation state is required.
The CPHMD methodology has been implemented using two distinct approaches, which vary in the manner in which the titration coordinates are treated – either discretely or continuously.39 In the discrete CPHMD variant, the MD sampling of atomic coordinates is combined with the Monte Carlo (MC) sampling of protonation states. At regular intervals during a typical MD simulation, a MC step is performed to determine the change of the protonation state. Discrete CPHMD was first reported by Bürgi et. al.,40 which was computationally expensive at that time and suffered from convergence issues, owing to the fact that it was performed in explicit solvent and used the more expensive thermodynamic integration approach to calculate the energies used in the MC evaluation step. Baptista and co-workers reported a similar discrete CPHMD implementation but used the Poisson-Boltzmann finite-difference method to calculate the energies used in the MC evaluation step.41–43 With the advances in implicit solvation models around this time,44,45 and the initial convergence issues reported for explicit solvent CPHMD,40 subsequent developments in discrete CPHMD by Dlugosz and Antosiewicz,46,47 and Mongan et. al.,48 were implemented using a Generalized-Born (GB) implicit solvent model. More recent improvements in the discrete CPHMD community have been focused on achieving better sampling by enhanced sampling techniques, such as Accelerated Molecular Dynamics by Williams et. al.49 and replica exchange strategies by Roitberg and co-workers.50–52 Others in the field, namely Warshel and co-workers have focused on developing a more physically realistic form of CPHMD,53 using time-dependent MC sampling of the proton transfer process,54 and the empirical valence bond (EVB) framework to simulate proton transfer between solute and solvent.55
By contrast, in the continuous CPHMD variant, which were first reported by Baptista et. al.56 and Borjesson et. al.,57 titration coordinates can be treated as mixed states. In the continuous CPHMD variant developed by Brooks and co-workers, the titration coordinate represents an instantaneous microstate, and it is propagated continuously between the protonated and unprotonated states using the λ dynamics approach.58–60 Continuous CPHMD allows one to avoid sudden jumps in potential energy that occur after a successful MC move in the discrete CPHMD variant, and potentially avoids artifacts that may be caused by the MC moves in titration coordinates. Additionally, continuous CPHMD facilitates coupled proton moves, which would need to be engineered as specific move types in the MC-based variant. Continuous CPHMD was originally implemented in implicit solvent,61 improved to account for proton tautomerism,62 and it provided the first demonstration of using enhanced sampling strategies to accelerate sampling and convergence in CPHMD simulations.63 The effectiveness of continuous CPHMD has been demonstrated on numerous pH-dependent systems, including of protein folding,64,65 aggregation of Alzheimer's beta-amyloid peptides,66 pH-triggered chaperon activity of HdeA dimers,67 electrostatic effects on protein stability,68 self-assembly of spider silk proteins,69 and RNA silencing in the carnation italian ringspot virus.70 Other investigators in the field have also seen a number of successes using discrete CPHMD simulations.71–73
While the move to implicit solvent CPHMD has obvious advantages in sampling and convergence, a number of unresolved issues have emerged over the years. It has been reported that the generalized Born implicit solvent model underestimates the desolvation of buried charge-charge interactions,63 causing a systematic overstabilization of the ionized form74 and consequently increasing the error of predicted pKa values. In addition, these models are known to cause structural compaction which may distort the overall structure,68,75 introducing another source of error to the pKa calculations. Furthermore, in systems such as ion channels76–78 and some transmembrane proteins,79 where the microscopic interactions of discrete ions and water with the protein are important, the use of an explicit solvent representation of the solvent environment is desirable. Thus, recent developments in the continuous CPHMD community have been focused on re-introducing explicit solvent into the CPHMD framework. Wallace and Shen were the first to report a hybrid solvent CPHMD model, and showed that using an explicit solvent representation of the protein’s conformational dynamics can reduce the errors introduced by the GB implicit model.75 Around the same time, the first “pure” explicit solvent CPHMD was reported by Grubmüller and co-workers, with a proof of concept demonstration for model amino acid compounds.80 Brooks and co-workers subsequently reported another “pure” explicit solvent constant pH MD simulation, termed as CPHMDMSλD as it is based on the newer multi-site λ-dynamics (MSλD) framework.81,82 CPHMDMSλD was developed initially for investigating pH-dependent behavior of nucleic acid systems and it has been validated on both model nucleotides83 and larger RNA systems.84 The initial challenges associated with convergence in explicit solvent were noted by practitioners in the field,80,83 and have been addressed to some extent using enhanced sampling strategies.75,85 More recent developments in enhanced sampling methods, such as Orthogonal Space Random Walk by Yang and co-workers have demonstrated that accurate pKa calculations for buried protein residues in explicit solvent simulations can be achieved,86 indicating that practical challenges first encountered by Bürgi et. al. over a decade ago will be resolved eventually.
In this article, we will adopt the explicit solvent CPHMDMSλD framework and extend its application to include proteins. We apply the multi-site λ-dynamics (MSλD) algorithm to seamlessly perform alchemical reactions between protonation and tautomeric states, and develop a novel biasing potential to ensure that the physical end-states are adequately sampled. The quality of the CPHMDMSλD model for proteins will be demonstrated by its ability to reproduce the pKa values of model compounds, simulate coupled pH-dependent behavior of dipeptides, and to accurately reproduce experimental pKa values of proteins, such as the hen egg-white lysozyme (HEWL), the binding domain of 2-oxoglutarate dehydrogenase (BBL) and the N-terminal of ribosomal L9 protein (NTL9).
2. THEORY
2.1 Constant pH Molecular Dynamics Simulations in Explicit Solvent
We briefly review the theory behind constant pH molecular dynamics (CPHMD). In CPHMD, the protonation state of the titrating residue is described by a continuous variable, λ. The dynamics of λ is described according to multi-site λ-dynamics (MSλD),81,82 a formalism that couples the dynamics of λ to the dynamics of the protein system. The simulation is under the influence of a hybrid Hamiltonian and its potential energy is described by:
| (1) |
where Nsites is the total number of titrating residues, X represents the coordinates of the environment atoms (i.e., the parts of the protein that are not titrating), and xα,1 and xα,2 represent the coordinates of atoms in residue α that are associated with the protonated and unprotonated states, respectively. λ serves as a scaling factor that is associated with each titrating residue α and its value describes the physically relevant protonated (λα,1 = 1) and unprotonated (λα,2 = 1) states. Details about the theoretical and methodological treatment of λ dynamics can be found in the following references.81,82
CPHMD simulations are calibrated on model compounds (i.e., amino acids) to reproduce the external pH environment. Modeling of the external pH is achieved by introducing a fixed biasing potential parameter () to the unprotonated state, which results in the biased potential energy function:
| (2) |
The free energy of protonation (ΔGprotonation) is used to calibrate the biasing potential () that simulates the effect of an external pH environment. By setting the value of to ΔGprotonation, approximately equal populations of protonated and unprotonated states are sampled in the simulation. Under this condition, the external pH environment is equal to the pKa value of the model compound. To change the pH of the simulation, can be adjusted by the following equation:
| (3) |
where pH is the external pH of the simulation and pKa is the experimental pKa of the model compound. The fixed biasing potential is pre-calculated and its value, corresponding to the specified external pH, is universally applied to all residues of the same type regardless of the protein environment it is in. In explicit solvent CPHMD simulations, when the titration coordinates are allowed to propagate dynamically, the two end points that correspond to physical protonation states may not be sufficiently sampled to yield converged estimates of the pKa shifts. To ameliorate this issue, the inclusion of an extra variable biasing potential (Fvar) is introduced, which can be adjusted to tune the sampling efficiency of titration coordinates and the fraction of physical protonation states:
| (4) |
Thus, in the CPHMD treatment, titratable groups in proteins may be viewed as model compounds that are perturbed by the introduction of the protein environment. Further details on the implementation and calibration of explicit solvent CPHMDMSλD can be obtained from the following reference.83
2.2 pH Replica Exchange Sampling Protocol
The potential for slow convergence of protonation state sampling in CPHMD simulations has been well documented, and is exacerbated for residues with conformationally-coupled pKa values, where they undergo a local conformation change that causes them to sample different electrostatic environments yielding distinct microscopic pKa values.49,84,87 Early work by Brooks and co-workers on protein CPHMD simulations has demonstrated that the introduction of a temperature replica exchange (T-REX) protocol can significantly accelerate sampling to address such issues.63 However, using T-REX in explicit solvent MD simulations typically incurs a large computational expense, for example, a moderate sized protein of ~100 residues (40k atoms when solvated) requires at least 20 replicas to achieve reasonable exchange rates between adjacent temperature replicas, and when simulating CPHMD across a reasonable pH range (e.g., pH 5 to 9), the total number of replicas required increases to ~100. Therefore, in this paper, we used a pH replica exchange (pH-REX) sampling strategy instead, and the pH-REX sampling protocol implemented in our work is based on the work of Wallace and Shen,75 where simulations performed at various pH conditions are exchanged based on the following Metropolis criterion:
| (5) |
where β is 1/kbT, the first two terms, UpH({λi};pH') and UpH ({λ i'};pH) are the pH-biasing potential energies for the two adjacent replicas after the exchange, and the next two terms, UpH({λi};pH') and UpH ({λ i'};pH') are the corresponding energies for the respective replicas before the exchange.
3. METHODS
3.1 Generating Input Structures
Input structures of the peptide compounds were generated from the CHARMM topology files using the IC facility in CHARMM.88 The input structure for the protein hen egg-white lysozyme (HEWL), the 45-residue binding domain of 2-oxoglutarate dehydrogenase multienzyme complex (BBL) and the 56-residue N-terminal domain of ribosomal L9 protein (NTL9) were generated from the PDB file (accession codes: 2LZT, 1W4H, 1CQU respectively). Hydrogen atoms were added using the HBUILD facility in CHARMM. Model compounds (single amino acids), test compounds (dipeptide sequences) and the proteins were solvated in a cubic box of explicit TIP3P water89 using the convpdb.pl tool from the MMTSB toolset.90 For each protein, the system was first neutralized, before an appropriate number of Na+ and Cl− counterions was added to match the experimental ionic strength of 100mM NaCl. All systems were capped at the N-terminus and C-terminus using CHARMM’s ACE and CT2 patches. Additional patches were constructed to represent the protonated forms of Asp, Glu, His and Lys. All of the associated bonds, angles and dihedrals were explicitly defined in the patch. Each titratable residue was simulated as a hybrid model that explicitly included atomic components of both the protonated and unprotonated forms. The CHARMM parameters for the partial charges of aspartic acid, glutamic acid and lysine used in this study were reported previously by Lee et. al.61 Partial charges for the three protonation states of histidine were obtained without modification from the HSP, HSE and HSD residues as reported in the CHARM22 all-atom force field for proteins.91
3.2 Simulation Details
MD simulations were performed within the CHARMM macromolecular modeling program (version c36a6) using the CHARMM22 all-atom force field for proteins91 and TIP3P water.89 The SHAKE algorithm92 was used to constrain the hydrogen-heavy atom bond lengths. The Leapfrog Verlet integrator was used with an integration time step of 2 fs. A non-bonded cutoff of 12 Å was used with an electrostatic force shifting function and a van der Waals switching function. While group-based 8 Å cutoffs investigated in the 1990s were notoriously poor in reproducing accurate dynamics of biomolecules relative to the Ewald summation technique,93,94 modern atom-based cutoff schemes with sufficiently long cutoff distances (12 Å),95 such as those employed in this study, has been shown to be comparable to the Ewald summation technique in modeling the dynamics of both proteins96 and nucleic acids.97 Titration was performed in the multi-site λ-dynamics framework (MSλD)81,82 within the BLOCK facility, using the λNexp functional form for λ (FNEX) with a coefficient of 5.5.81,82 The titratable fragment included the protonation site and adjacent atoms whose partial charge differed according to the protonation state. The environment atoms were defined as all atoms that were not included in the titratable fragments. Linear scaling by λ was applied to all energy terms except bond, angle and dihedral terms, which were treated at full strength regardless of λ value to retain physically reasonable geometries. Each θα was assigned a fictitious mass of 12 amu•Å2 and λ values were saved every 10 steps. The temperature was maintained at 298K by coupling to a Langevin heatbath using a frictional coefficient of 10ps−1. After an initial minimization, the system was heated for 100 ps and equilibrated for 100ps (for amino acid compounds and dipeptides) or 400 ps (for HEWL, BBL and NTL9). This was followed by a production run of 3 ns (for amino acid compounds, dipeptides and NTL9), 5 ns (for BBL) or 20 ns (for HEWL). CPHMDMSλD simulations were performed across the pH range, with integer value pH spacing, as indicated in the titration curves (see Results and Discussion), from pH 1 to 7 for Asp and Glu, pH 4 to 9 for His, pH 7 to 12 for Lys, pH 0 to 8 for HEWL, and pH 0 to 5 for BBL and NTL9. In the pH-REX simulations, exchange attempts were made at every 1 ps. All CPHMDMSλD simulations were performed in triplicate.
3.3 Calculation of pKa value
The populations of unprotonated (Nunprot) and protonated (Nprot) states are defined as the total number of times in the trajectory where conditions λα,1 > 0.8 and λα,2 > 0.8 are satisfied respectively. They are used in the calculation of the fraction of physical states, which is the ratio of Nunprot and Nprot states over all states (which include intermediate λ values). The unprotonated fraction (Sunprot) is calculated for each pH window:
| (6) |
Sunprot values computed across the entire pH range, were then fitted to a generalized version of the Henderson-Hasselbalch (HH) formula98 to obtain a single pKa value:
| (7) |
To illustrate the effect of coupled titrating residues, CPHMDMSλD simulations on several dipeptides (see Supporting Information for discussion) were also performed. For these dipeptide simulations, the protonation state statistics for a specific residue may not be associated with the titrating residue, because the symmetry of the system may render the environment around each titrating residue to be similar. Therefore, the pKa calculation has to be performed using a modified version of eqn. 7, where the combined Sunprot ratio for all i residues is fitted to the following equation:
| (8) |
In this study, the reported pKa value and its uncertainty correspond to the mean and standard deviation calculated from 3 sets of independent runs.
4. RESULTS & DISCUSSION
4.1 Optimization of Model Potential Parameters for 2-State Titrations
As with the previous implementation of CPHMDMSλD for nucleic acids,83 we used the free energy of deprotonation as the fixed biasing potential (Ffixed) in our simulation. The free energy of deprotonation was calculated for each isolated model compound embedded in explicit solvent using traditional λ-dynamics at zero ionic strength. In order to facilitate transitions between the two protonation states, we optimized the force constant (kbias) on the variable biasing potential (Fvar) that was applied to each model compound, and targeted to achieve a maximal value of the transition rate in λ-space (i.e., titration coordinate sampling), while maintaining a high fraction of physical ligands. The optimized parameters for the model potentials are reported in Table I. Calculation of the sampling statistics (see Table S1) indicates that the fraction of physical states was maintained at ~70% and transitions in λ-space were ~50 transitions/ns. The sampling properties of our model amino acids are comparable to previous work performed on model nucleosides in explicit solvent.83
Table I.
Parameters for the Model Potential for 2-state Titrations
| Residue | ΔGprotonation (kcal/mol) | Fvar (kcal/mol) | Ref pKa |
|---|---|---|---|
| kbias | |||
| Asp | 43.71 | 34.00 | 4.00 |
| Glu | 46.00 | 34.25 | 4.40 |
| His-δ | −3.58 | 26.00 | 7.00 |
| His-ε | −12.26 | 26.00 | 6.60 |
| Lys | −23.02 | 29.50 | 10.40 |
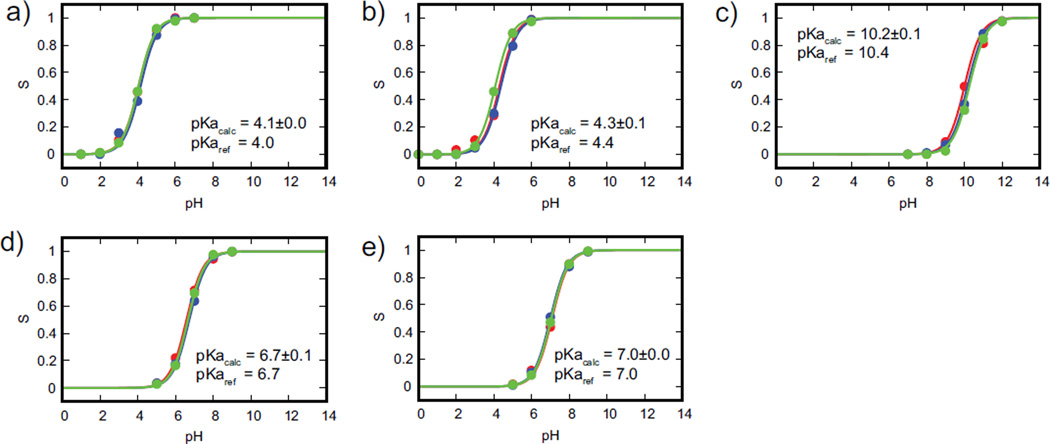
Next, we performed a 2-state titration simulation, where only two titrating states (protonated and unprotonated) were simulated, and tautomers for each protonation state were not explicitly modeled. The titration curves for our model compounds are illustrated in Figure 1. The calculated pKa of aspartic and glutamic acid for a two-state titration (i.e., without proton tautomerism) was 4.1 and 4.3 pKa respectively, which is within ±0.1 pKa units from their experimental pKa values of 4.0 and 4.4 respectively.99 For the two-state titration of histidine, where either Nδ or Nε was titrated, the pKa values obtained were 6.7 and 7.0 respectively,100 which is identical to their experimental pKa values. Finally, the calculated pKa of lysine was 10.2, which is in close agreement with the experimental pKa value of 10.4.99 The excellent agreement between our model compounds calculated pKa values and their experimental values indicate that the sampling of titration coordinates in our CPHMDMSλD simulations was sufficient to yield accurate results.
Figure 1.
Titration curve of model compounds: (a) aspartic acid, (b) glutamic acid, (c) lysine, (d) (d) histidine-δ and (e) histidine-ε. Calculated pKa values of model compounds are in excellent agreement with experimental pKa values. Colors represent the results from the triplicate runs.
4.2 Optimization of Model Potential Parameters for 3-State Titrations
The original form of the Fvar potential assumed the existence of only two states. When accounting for proton tautomerism and thus three states, the original form was not suitable because it frequently sampled an intermediate state of the two tautomers. This intermediate state is typically characterized by λα,1 ≈ 0, λα,2 ≈ 0.5 and λα,3 ≈ 0.5, which corresponds to a half proton on both the Nδ and Nε protonation sites (using His as an example), and this represents an unphysical state whose sampling should be minimized. The existence of the intermediate state can be rationalized by considering that the free energy barrier for conversion between the two protonation states would be larger than the conversion between the two tautomers, as in the former process there is a change in the net charge of the system and a greater reorganization of the distribution of partial charges. The combined functional form of the original Fvar potential that uses the same 0.8 cutoff in the definition of physical protonation states (see Methods section) as expressed in eqn. 9, where λα,1, λα,2 and λα,3 denote the alchemical scaling factors associated with each of the 3 states for some residue α, does not account for the uneven barrier height of the different alchemical reactions.
| (9) |
To avoid the intermediate tautomeric states, we modified the existing Fvar potential by including additional cross terms (k2 expressions) to account for uneven barrier heights, and a final term (k3 expressions) was added to ensure that the relative free energy of the end-states were not altered. The resulting functional form as outlined in eqn. 10 results in a more versatile biasing potential that is suited to address the asymmetry of the potential energy surface associated with changes in both protonation and tautomeric states.
| (10) |
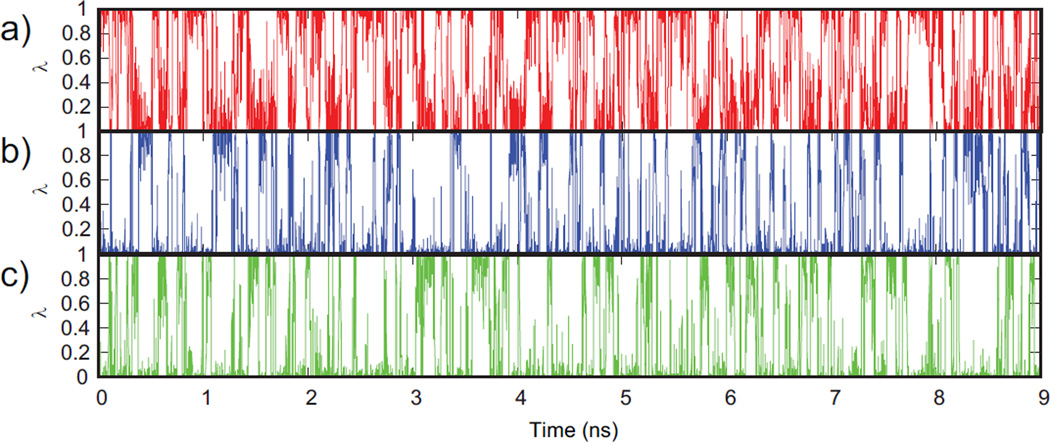
An iterative grid search strategy was used in testing various combinations of the force constants (k1, k2, k3), and the optimal combination is reported in Table II. As illustrated in Figure 2, which shows the time-evolution of λ, all 3 end states for the model compounds were well sampled. Calculation of the sampling statistics as summarized in Table III indicates that the fraction of physical states was maintained above 70%, confirming that the modified Fvar potential does not trap λ in an unphysical intermediate state. The transitions in λ-space were ~50 transitions/ns, which is comparable to the statistics obtained from 2-state titrations of the model compounds.
Table II.
Parameters for the Model Potential for 3-state Titrations
| Residue | ΔGprotonation (kcal/mol) |
Fvar (kcal/mol) | Ref pKa | ||
|---|---|---|---|---|---|
| k1 | k2 | k3 | |||
| Asp-T | 43.30 | −16.5 | 18.5 | −18.5 | 4.00 |
| Glu-T | 45.59 | −16.0 | 18.5 | −18.5 | 4.40 |
| His-T | −3.58/−12.26 | 8.0 | 6.0 | −6.0 | 6.45 |
Figure 2.
Titration coordinate transitions of aspartic acid at pH 4 for (a) unprotonated state, (b) protonated tautomer #1 and (c) protonated tautomer #2 shows that the physical end states are well sampled.
Table III.
Sampling characteristics of 3-state titration simulations performed at the pH closest to the model compound’s pKa value. Using pH-REX greatly accelerates sampling of titration coordinates with minimal loss in the fraction of physical states (FPS).
| Residue | pH | Normal MD | pH-REX | ||
|---|---|---|---|---|---|
| FPS | Transition(ns−1) | FPS | Transition(ns−1) | ||
| Asp-T | 4 | 0.78 ± 0.01 | 50 ± 1 | 0.78 ± 0.01 | 294 ± 16 |
| Glu-T | 4 | 0.76 ± 0.01 | 46 ± 7 | 0.75 ± 0.00 | 322 ± 21 |
| His-T | 7 | 0.81 ± 0.02 | 60 ± 6 | 0.81 ± 0.01 | 298 ± 12 |
While the sampling efficiency in λ-space of model compounds allows us to reproduce the pKa values of the model compounds, the transition rate is nevertheless limited to ~50 transitions/ns. In our previous evaluation of explicit solvent CPHMDMSλD simulations of larger nucleic acid structures, slower pKa convergence was observed,84 and it is likely that protein systems will encounter similar issues as well. The sampling of titration and spatial coordinates can be accelerated using a pH-REX sampling strategy.85 Therefore, we have applied pH-REX sampling, and as illustrated in Table III, it resulted in a 6-fold improvement in λ-space sampling of model compounds with effectively no loss in the fraction of physical states. As pH-REX sampling confers significant improvement over straightforward MD simulations and requires negligible overhead in terms of computational cost, the results presented in the subsequent sections are obtained from pH-REX CPHMDMSλD simulations unless specified otherwise.
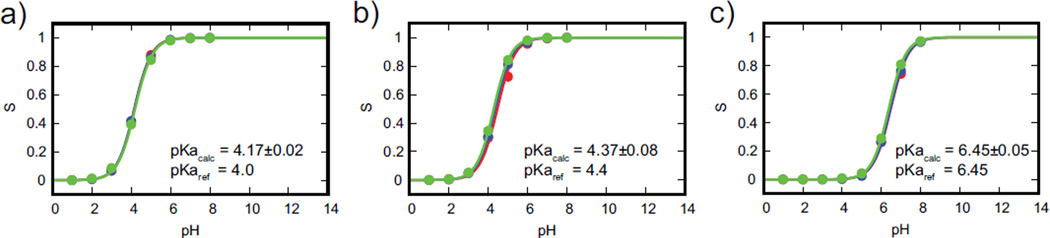
We performed a 3-state titration on the model compounds, where alchemical transformations across different protonation states and across different tautomers of the same protonation state were explicitly modeled. The tautomeric titrations of aspartic and glutamic yielded a pKa of 4.4 and 4.8 respectively, which matches well with the macroscopic pKa of 4.35 and 4.70 when the double degeneracy of the protonated states is taken into account.62 However, since the experimental pKa measured does not distinguish between the tautomeric forms, we recalibrated the fixed biasing potential in our CPHMDMSλD simulations to reproduce the experimentally measured macroscopic pKa values. This was achieved by reducing the biasing potential at pH=pKa by kbTln(2) = 0.41 kcal/mol, which accounts for the degeneracy of the tautomeric protonated states. Our approach is different from that of Khandogin and co-workers,62 where a post-correction factor of 0.3 pKa units was applied to tautomeric residues. However, the net result in both approaches is the same, in the sense that the final pKa value calculated accounts for tautomer degeneracy. The titration curves for our model compounds with proton tautomerism are illustrated in Figure 3. The calculated pKa of aspartic and glutamic acid was 4.17 and 4.37 respectively, which is good agreement with experimental pKa values. For histidine tautomeric titration, no re-calibration was performed because the pKa measured by experiments were microscopic pKa associated with the titration at the Nε and Nδ sites, and the fixed biasing potential applied to each tautomer was identical to those used in the 2-state titration setup. Our calculated pKa for the histidine tautomer was 6.45, which is identical to the expected macroscopic pKa value of 6.45.62
Figure 3.
Titration curve of model compounds with proton tautomerism: (a) aspartic acid, (b) glutamic acid and (c) histidine. Calculated pKa values of model compounds show excellent agreement with experimental pKa values. Colors represent the results from the triplicate runs.
4.3 pKa Calculations of Proteins: Hen-Egg White Lysozyme (HEWL)
The HEWL protein is a well-studied protein system that contains the 3 most common titrating residues (Asp, Glu, His) with microscopic pKa values for each residue that have been measured in a number of experimental studies.101–106 It is perhaps the closest thing to a “universal benchmark” system that has been evaluated by numerous CPHMD implementations over the years.43,48,49,51,63,75,107 To the best of our knowledge, all existing “pure” explicit solvent CPHMD simulations reported in the literature have only been demonstrated on small peptide compounds80 and simple organic molecules.108 We performed a 20 ns pH-REX CPHMDMSλD simulation of HEWL, which is the first example of explicit solvent CPHMD simulation on a full protein to be reported.
pKa calculations over 5 ns interval segments of our pH-REX CPHMDMSλD trajectory show that good convergence is achieved within 20 ns (Table S3). The difference in pKa values across our triplicate runs is small, typically between 0.2 to 0.3 pKa units, demonstrating that our results are robust and reproducible. The accuracy of our calculated pKa values are then compared to experimental measurements from consensus NMR titrations.106 As summarized in Table IV and Figure S3, the calculated pKa values are in good agreement with experiment, with a root-mean-square-error (RMSE) of 0.85 pKa units and an average unsigned error (AUE) of 0.68 pKa units. Nielson and co-workers previously estimated that experimental pKa values reported in the literature on average may vary by 0.5 pKa units depending on the experimental method and/or protocol used to make the measurements.106 This suggests that the accuracy of our pH-REX CPHMDMSλD simulations are approaching the uncertainty of experimental pKa values. Next, we identified the residues that had errors in their calculated pKa values, which we defined as having more than 1.0 pKa unit difference between calculated and experimental values. Asp-119 was underpredicted by −1.9 pKa units, which suggests that the unprotonated state is overstabilized in our simulations. Analysis of its microenvironment indicates that persistent hydrogen bond interactions between the carboxylic oxygens of Asp-119 and the amide backbone hydrogen of Gln-121 and Ala-122 were present even in a low pH environment (Figure S4), which accounts for the extra stabilization of the unprotonated state of Asp-119. Similar underprediction of Asp pKa values has been documented in other CPHMD work, where salt bridge interactions were responsible.49 When non-salt-bridge configurations were sampled, it resulted in more accurate pKa results.49 This suggests that the apparent error in the Asp-119 pKa value could be a sampling issue, and more extensive sampling or more aggressive sampling methods may be required when dealing with residues that are “locked” to their initial conformation by strong interactions like hydrogen bonds or salt bridges.
Table IV.
pKa values of HEWL calculated using implicit and hybrid solvent pH-REX CPHMD simulations as reported by Wallace and Shen,75 compared to pKa values calculated using explicit solvent pH-REX CPHMDMSλD simulations in this work. Calculated pKa values with error greater than 1.0 pKa unit relative to experimental values based on consensus NMR titrations106 are identified in red.
| Residue | Exp pKa | Implicit CPHMD | Hybrid CPHMD | Explicit CPHMDMSλD | |||
|---|---|---|---|---|---|---|---|
| pKa | Error | pKa | Error | pKa | Error | ||
| GLU-7 | 2.6 ± 0.2 | 2.6 ± 0.1 | 0.0 | 2.7 ± 0.0 | 0.1 | 2.7 ± 0.1 | 0.1 |
| HIS-15 | 5.5 ± 0.2 | 5.3 ± 0.5 | −0.2 | 6.6 ± 0.1 | 1.1 | 6.0 ± 0.2 | 0.5 |
| ASP-18 | 2.8 ± 0.3 | 2.9 ± 0.0 | 0.1 | 3.1 ± 0.1 | 0.3 | 2.1 ± 0.2 | −0.7 |
| GLU-35 | 6.1 ± 0.4 | 4.4 ± 0.2 | −1.8 | 7.2 ± 0.2 | 1.1 | 7.0 ± 0.3 | 0.9 |
| ASP-48 | 1.4 ± 0.2 | 2.8 ± 0.2 | 1.4 | 1.6 ± 0.5 | 0.2 | 1.3 ± 0.0 | −0.1 |
| ASP-52 | 3.6 ± 0.3 | 4.6 ± 0.0 | 1.0 | 2.9 ± 0.1 | −0.7 | 4.5 ± 0.3 | 0.9 |
| ASP-66 | 1.2 ± 0.2 | 1.2 ± 0.4 | −0.1 | 1.5 ± 0.6 | 0.3 | 1.5 ± 0.1 | 0.3 |
| ASP-87 | 2.2 ± 0.1 | 2.0 ± 0.1 | −0.2 | 1.5 ± 0.4 | −0.7 | 1.3 ± 0.0 | −0.9 |
| ASP-101 | 4.5 ± 0.1 | 3.3 ± 0.3 | −1.2 | 3.0 ± 0.1 | −1.5 | 5.1 ± 0.5 | 0.6 |
| ASP-119 | 3.5 ± 0.3 | 2.5 ± 0.1 | −1.1 | 2.9 ± 0.1 | −0.7 | 1.6 ± 0.0 | −1.9 |
| RMSE | 0.94 | 0.80 | 0.84 | ||||
| AUE | 0.70 | 0.66 | 0.68 | ||||
We compared the performance of explicit solvent pH-REX CPHMDMSλD simulations to CPHMD models implemented in other solvation models. A number of CPHMD variations have been implemented in AMBER48 and GROMACS.43 However, they will not be included our analysis as deconvoluting the effects originating from force field differences to those arising from solvation model differences is not straightforward. Instead, we will focus our analysis on CPHMD variations implemented in CHARMM. The original CPHMD in CHARMM was implemented with a GB implicit solvent model,61 and we have used the HEWL pKa values reported by Wallace and Shen for comparison.75 Since that work was reported using a pH-REX sampling strategy, we have also eliminated the effects of using different sampling strategies. At the time of writing, there is no “pure” explicit solvent CPHMD based on the CHARMM force field that has been tested on the HEWL protein. However, a close comparison can be made with Shen’s hybrid solvent CPHMD model.75 The key methodological difference between explicit and hybrid solvent models is that the evaluation of free energies of deprotonation and the forces on the fictitious λ particles that govern the titration coordinates are calculated using a GB implicit solvent model in Shen’s hybrid solvent CPHMD model, whereas in our explicit solvent CPHMDMSλD model there is no use of the GB implicit solvent model in any part of the calculation. Unfortunately, the use of such hybrid sampling means there is no clear Hamiltonian for this system and correspondence to results from any specific statistical mechanical cannot be demonstrated. Lastly, the sampling of titration coordinates in implicit solvent is typically ~2000 transitions/ns,51 which is an order of magnitude higher than those obtained in our explicit solvent simulations. Therefore, to compensate for the differential sampling speed associated with different solvent models, we compared the results of our 20 ns pH-REX CPHMDMSλD trajectories to the previously reported 2 ns pH-REX trajectories that uses the implicit and hybrid CPHMD model.
As summarized in Table IV, in terms of overall pKa predictive performance, our explicit solvent CPHMDMSλD results had a RMSE error of 0.84 pKa units. This is an improvement from the results obtained using implicit solvent CPHMD (RMSE = 0.94), and our model performance is close to that of the hybrid solvent CPHMD (RMSE = 0.80). A similar trend was also noted using alternative error metrics, such as the average unsigned error (AUE). We then identified the number of residues that had errors of more than 1.0 pKa unit relative to experimental values. Our explicit solvent CPHMDMSλD model had only 1 such residue (i.e., Asp-119) compared to the implicit and hybrid solvent CPHMD models which had 5 and 3 residues respectively. Notable improvements in moving from a hybrid solvent to a “pure” explicit solvent model can be observed in His-15, where the overestimation of its pKa value is reduced from 1.1 to 0.5 pKa units. Similarly, the hybrid solvent CPHMD model incorrectly predicted the direction of pKa shift for residue Asp-101, whereas the explicit solvent CPHMDMSλD model not only predicted the right direction of pKa shift, but the magnitude of error was also smaller (−1.5 vs +0.6). Our findings suggest that when corrected for differences in titrating coordinates sampling, the explicit solvent CPHMDMSλD model produces more accurate pKa predictions than the original implicit solvent CPHMD.
4.4 pKa Calculations of Proteins: Other Proteins, BBL and NTL9
Lastly, to demonstrate that the pKa calculations obtained from the CPHMDMSλD framework for proteins is not specific to HEWL protein, we performed pKa calculations on two additional proteins, the BBL and NTL9 protein. Given that we have only investigated a single His residue in a protein environment, for BBL we only titrated the two His residues. NTL9 has no His residues, and the Glu and Asp residues that have experimental pKa measurements were titrated. As summarized in Table V and Figure S5 the calculated pKa values have are reasonably accurate (RMSE = 0.72, AUE = 0.59).109,110 From the experimental data, most of the residues titrate close to the pKa of their reference compounds, but two residues had more than a 1.0 pH unit shift. For His-166 of BBL, the residue is buried and its experimental pKa is 5.4. For Asp-8 of NTL9, its experimental pKa of 3.0 can be traced to the salt bridge interactions it forms with the amide backbone of adjacent residues. Our calculated pKa values demonstrate a similar downward shift, although in both cases the extent of the shift tends to be overestimated. We suggest that this overestimation may be due to the lack of sampling stemming from the shorter 3 to 5 ns simulations performed for these systems. In other proteins like staphylococcal nuclease, residues with shifted pKa values are known to undergo local conformational changes,26,28 and sampling these states will be required to improve the accuracy of pKa calculations. Together with our observations for Asp-119 in HEWL, our work suggests that while short pH-REX CPHMDMSλD simulations are capable of reproducing experimental pKa values of most protein residues, accurate reproduction of highly shifted pKa values (e.g., buried charged residues) or those involving salt-bridge or similarly strong interactions remains a challenge that may be better addressed with more aggressive conformational sampling techniques.
Table V.
pKa values of BBL and NTL9 calculated using explicit solvent pH-REX CPHMDMSλD simulations in this work. Calculated pKa values with error greater than 1.0 pKa unit relative to experimental values109,110 based are identified in red.
| Residue | Explicit CPHMDMSλD | ||
|---|---|---|---|
| Exp pKa | pKa | Error | |
| BBL | |||
| HIS-142 | 6.5 | 6.6 ± 0.1 | 0.1 |
| HIS-166 | 5.4 | 4.8 ± 0.0 | −0.6 |
| NTL9 | |||
| ASP-8 | 3.0 | 1.5 ± 0.1 | −1.5 |
| GLU-17 | 3.6 | 4.0 ± 0.5 | 0.4 |
| ASP-23 | 3.1 | 3.7 ± 0.2 | 0.6 |
| GLU-38 | 4.0 | 3.9 ± 0.2 | −0.1 |
| GLU-48 | 4.2 | 3.4 ± 0.3 | −0.8 |
| GLU-54 | 4.2 | 3.6 ± 0.2 | −0.6 |
| RMSE | 0.72 | ||
| AUE | 0.59 | ||
5. CONCLUSION
In conclusion, we have demonstrated the use of the constant pH molecular dynamics framework based on multi-site λ-dynamics (CPHMDMSλD) to simulate realistic pH-dependent properties of proteins. In the CPHMDMSλD framework, we performed seamless alchemical transitions between protonation and tautomeric states using multi-site λ-dynamics, and designed a novel biasing potential to ensure that only the physical end-states are predominantly sampled. Then, we applied explicit solvent CPHMDMSλD simulations to the proteins HEWL, BBL and NTL9, which are the first examples of a “pure” explicit solvent CPHMD on full protein systems to be reported. Our pKa calculations for HEWL protein are in excellent agreement with experimental values, with a RMSE of 0.84 pKa units, and this is close to the uncertainty of 0.50 pKa units associated with experimental measurements. Our pKa calculations on the other model protein systems, BBL and NTL9 also provide similarly good agreement with experiments. In addition, comparison with implicit solvent CPHMD shows that explicit solvent CPHMDMSλD produces results that are more accurate, reducing the number of residues with large errors in their pKa predictions from 5 to 1. With the development of explicit solvent CPHMDMSλD for proteins, it will finally allow us to confidently address questions related to pH-dependent properties of membrane proteins and ion channels, where discrete representation of ions and water is important. Coupled with the explicit solvent CPHMDMSλD framework for nucleic acids, accurate modeling of pH-dependent properties for all major classes of biomolecules – proteins, nucleic acids and even protein-nucleic acid complexes is now a reality.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (GM037554 and GM057513). Additionally, we acknowledge grant support for BH from the Interdisciplinary Opportunities in Biophysics REU at the University of Michigan (PHY-1004618). We thank Dr. Afra Panahi for critically reviewing the manuscript and providing helpful comments.
REFERENCES
- 1.Warshel A. Calculations of Enzymatic-Reactions - Calculations of Pka, Proton-Transfer Reactions, and General Acid Catalysis Reactions in Enzymes. Biochemistry. 1981;20:3167–3177. doi: 10.1021/bi00514a028. [DOI] [PubMed] [Google Scholar]
- 2.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen JE, Mccammon JA. Calculating pKa values in enzyme active sites. Protein Sci. 2003;12:1894–1901. doi: 10.1110/ps.03114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demchuk E, Genick UK, Woo TT, Getzoff ED, Bashford D. Protonation states and pH titration in the photocycle of photoactive yellow protein. Biochemistry. 2000;39:1100–1113. doi: 10.1021/bi991513p. [DOI] [PubMed] [Google Scholar]
- 5.Dillet V, Dyson HJ, Bashford D. Calculations of electrostatic interactions and pKas in the active site of Escherichia coli thioredoxin. Biochemistry. 1998;37:10298–10306. doi: 10.1021/bi980333x. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox JL, Ahluwalia AK, Bevilacqua PC. Charged Nucleobases and Their Potential for RNA Catalysis. Acc Chem Res. 2011;44:1270–1279. doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnamurthy R. Role of pK(a) of Nucleobases in the Origins of Chemical Evolution. Acc Chem Res. 2012;45:2035–2044. doi: 10.1021/ar200262x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih IH, Been MD. Involvement of a Cytosine Side Chain in Proton Transfer in the Rate- determining Step of Ribozyme Self-cleavage. Proc Natl Acad Sci U S A. 2001;98:1489–1494. doi: 10.1073/pnas.98.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A Conformational Switch Controls Hepatitis Delta Virus Ribozyme Catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 10.Ravindranathan S, Butcher SE, Feigon J. Adenine Protonation in Domain B of the Hairpin Ribozyme. Biochemistry. 2000;39:16026–16032. doi: 10.1021/bi001976r. [DOI] [PubMed] [Google Scholar]
- 11.Ryder SP, Oyelere AK, Padilla JL, Klostermeier D, Millar DP, Strobel SA. Investigation of Adenosine Base Ionization in the Hairpin Ribozyme by Nucleotide Analog Interference Mapping. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 12.Bierzynski A, Kim PS, Baldwin RL. A Salt Bridge Stabilizes the Helix Formed by Isolated C-Peptide of Rnase-A. Proc Natl Acad Sci U S A. 1982;79:2470–2474. doi: 10.1073/pnas.79.8.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoemaker KR, Kim PS, Brems DN, Marqusee S, York EJ, Chaiken IM, Stewart JM, Baldwin RL. Nature of the Charged-Group Effect on the Stability of the C-Peptide Helix. Proc Natl Acad Sci U S A. 1985;82:2349–2353. doi: 10.1073/pnas.82.8.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer M, Van Vlijmen HWT, Karplus M. Electrostatic contributions to molecular free energies in solution. Adv Protein Chem. 1998;51:1–57. doi: 10.1016/s0065-3233(08)60650-6. [DOI] [PubMed] [Google Scholar]
- 15.Kelly JW. Alternative conformations of amyloidogenic proteins govern their behavior. Curr Opin Struct Biol. 1996;6:11–17. doi: 10.1016/s0959-440x(96)80089-3. [DOI] [PubMed] [Google Scholar]
- 16.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 17.Warshel A. Electrostatic Basis of Structure-Function Correlation in Proteins. Acc Chem Res. 1981;14:284–290. [Google Scholar]
- 18.Hunenberger PH, Helms V, Narayana N, Taylor SS, McCammon JA. Determinants of ligand binding to cAMP-dependent protein kinase. Biochemistry. 1999;38:2358–2366. doi: 10.1021/bi982064g. [DOI] [PubMed] [Google Scholar]
- 19.Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D'Souza VM. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. 2011;480 doi: 10.1038/nature10657. 561-U193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: A Perfect Storm for Cancer Progression. Nat Rev Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 21.Howell EE, Villafranca JE, Warren MS, Oatley SJ, Kraut J. Functional-Role of Aspartic Acid-27 in Dihydrofolate-Reductase Revealed by Mutagenesis. Science. 1986;231:1123–1128. doi: 10.1126/science.3511529. [DOI] [PubMed] [Google Scholar]
- 22.Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402:263–268. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- 23.Reiter NJ, Blad H, Abildgaard F, Butcher SE. Dynamics in the U6 RNA Intramolecular Stem-loop: A Base Flipping Conformational Change. Biochemistry. 2004;43:13739–13747. doi: 10.1021/bi048815y. [DOI] [PubMed] [Google Scholar]
- 24.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of Influenza Hemagglutinin at the Ph of Membrane-Fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 25.Harms MJ, Schlessman JL, Sue GR, Garcia-Moreno B. Arginine residues at internal positions in a protein are always charged. Proc Natl Acad Sci U S A. 2011;108:18954–18959. doi: 10.1073/pnas.1104808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci U S A. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pey AL, Rodriguez-Larrea D, Gavira JA, Garcia-Moreno B, Sanchez-Ruiz JM. Modulation of buried ionizable groups in proteins with engineered surface charge. J Am Chem Soc. 2010;132:1218–1219. doi: 10.1021/ja909298v. [DOI] [PubMed] [Google Scholar]
- 28.Isom DG, Cannon BR, Castaneda CA, Robinson A, Garcia-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc Natl Acad Sci U S A. 2008;105:17784–17788. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen JE, Gunner MR, Garcia-Moreno BE. The pKa Cooperative: a collaborative effort to advance structure-based calculations of pKa values and electrostatic effects in proteins. Proteins: Struct, Funct, Bioinf. 2011;79:3249–3259. doi: 10.1002/prot.23194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexov E, Mehler EL, Baker N, Baptista AM, Huang Y, Milletti F, Nielsen JE, Farrell D, Carstensen T, Olsson MH, Shen JK, Warwicker J, Williams S, Word JM. Progress in the prediction of pKa values in proteins. Proteins: Struct, Funct, Bioinf. 2011;79:3260–3275. doi: 10.1002/prot.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bashford D. Macroscopic electrostatic models for protonation states in proteins. Front Biosci. 2004;9:1082–1099. doi: 10.2741/1187. [DOI] [PubMed] [Google Scholar]
- 32.Antosiewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 33.You TJ, Bashford D. Conformation and hydrogen ion titration of proteins: a continuum electrostatic model with conformational flexibility. Biophys J. 1995;69:1721–1733. doi: 10.1016/S0006-3495(95)80042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell ST, Warshel A. Calculations of electrostatic energies in proteins. The energetics of ionized groups in bovine pancreatic trypsin inhibitor. J Mol Biol. 1985;185:389–404. doi: 10.1016/0022-2836(85)90411-5. [DOI] [PubMed] [Google Scholar]
- 36.Lee FS, Chu ZT, Warshel A. Microscopic and Semimicroscopic Calculations of Electrostatic Energies in Proteins by the Polaris and Enzymix Programs. J Comput Chem. 1993;14:161–185. [Google Scholar]
- 37.Warshel A, Sussman F, King G. Free-Energy of Charges in Solvated Proteins - Microscopic Calculations Using a Reversible Charging Process. Biochemistry. 1986;25:8368–8372. doi: 10.1021/bi00374a006. [DOI] [PubMed] [Google Scholar]
- 38.Sham YY, Chu ZT, Warshel A. Consistent calculations of pKa's of ionizable residues in proteins: semi-microscopic and microscopic approaches. J Phys Chem B. 1997;101:4458–4472. [Google Scholar]
- 39.Mongan J, Case DA. Biomolecular simulations at constant pH. Curr Opin Struct Biol. 2005;15:157–163. doi: 10.1016/j.sbi.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Burgi R, Kollman PA, van Gunsteren WF. Simulating proteins at constant pH: an approach combining molecular dynamics and Monte Carlo simulation. Proteins: Struct, Funct, Bioinf. 2002;47:469–480. doi: 10.1002/prot.10046. [DOI] [PubMed] [Google Scholar]
- 41.Baptista AM, Teixeira VH, Soares CM. Constant-pH Molecular Dynamics Using Stochastic Titration. J Chem Phys. 2002;117:4184–4200. [Google Scholar]
- 42.Machuqueiro M, Baptista AM. Constant-pH molecular dynamics with ionic strength effects: protonation-conformation coupling in decalysine. J Phys Chem B. 2006;110:2927–2933. doi: 10.1021/jp056456q. [DOI] [PubMed] [Google Scholar]
- 43.Baptista AM, Machuqueiro M. Acidic range titration of HEWL using a constant-pH molecular dynamics method. Proteins: Struct, Funct, Bioinf. 2008;72:289–298. doi: 10.1002/prot.21923. [DOI] [PubMed] [Google Scholar]
- 44.Im WP, Lee MS, Brooks CL., III Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 45.Chen JH, Im WP, Brooks CL., III Balancing solvation and intramolecular interactions: Toward a consistent generalized born force field. J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dlugosz M, Antosiewicz JM. Constant-pH molecular dynamics simulations: a test case of succinic acid. Chem Phys. 2004;302:161–170. [Google Scholar]
- 47.Dlugosz M, Antosiewicz JM, Robertson AD. Constant-pH molecular dynamics study of protonation-structure relationship in a heptapeptide derived from ovomucoid third domain. Phys Rev E. 2004;69:021915. doi: 10.1103/PhysRevE.69.021915. [DOI] [PubMed] [Google Scholar]
- 48.Mongan J, Case DA, McCammon JA. Constant pH Molecular Dynamics in Generalized Born Implicit Solvent. J Comput Chem. 2004;25:2038–2048. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 49.Williams SL, de Oliveira CA, McCammon JA. Coupling Constant pH Molecular Dynamics with Accelerated Molecular Dynamics. J Chem Theory Comput. 2010;6:560–568. doi: 10.1021/ct9005294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng Y, Roitberg AE. Constant pH replica exchange molecular dynamics in biomolecules using a discrete protonation model. J Chem Theory Comput. 2010;6:1401–1412. doi: 10.1021/ct900676b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swails JM, Roitberg AE. Enhancing Conformation and Protonation State Sampling of Hen Egg White Lysozyme Using pH Replica Exchange Molecular Dynamics. J Chem Theory Comput. 2012;8:4393–4404. doi: 10.1021/ct300512h. [DOI] [PubMed] [Google Scholar]
- 52.Sabri Dashti D, Meng Y, Roitberg AE. pH-Replica Exchange Molecular Dynamics in Proteins Using a Discrete Protonation Method. J Phys Chem B. 2012;116:8805–8811. doi: 10.1021/jp303385x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Messer BM, Roca M, Chu ZT, Vicatos S, Kilshtain AV, Warshel A. Multiscale Simulations of Protein Landscapes: Using Coarse-grained Models as Reference Potentials to Full Explicit Models. Proteins: Struct, Funct, Bioinf. 2010;78:1212–1227. doi: 10.1002/prot.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsson MHM, Warshel A. Monte Carlo simulations of proton pumps: On the working principles of the biological valve that controls proton pumping in cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103:6500–6505. doi: 10.1073/pnas.0510860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aaqvist J, Warshel A. Simulation of enzyme reactions using valence bond force fields and other hybrid quantum/classical approaches. Chem Rev. 1993;93:2523–2544. [Google Scholar]
- 56.Baptista AM, Martel PJ, Petersen SB. Simulation of protein conformational freedom as a function of pH: constant-pH molecular dynamics using implicit titration. Proteins: Struct, Funct, Bioinf. 1997;27:523. [PubMed] [Google Scholar]
- 57.Borjesson U, Hunenberger PH. Explicit-solvent molecular dynamics simulation at constant pH: Methodology and application to small amines. J Chem Phys. 2001;114:9706–9719. [Google Scholar]
- 58.Kong X, Brooks CL., III Lambda-Dynamics-A New Approach to Free-Energy Calculations. J Chem Phys. 1996;105:2414–2423. [Google Scholar]
- 59.Knight JL, Brooks CL., III Lambda-dynamics free energy simulation methods. J Comput Chem. 2009;30:1692–1700. doi: 10.1002/jcc.21295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Z, Brooks CL, III, Kong X. Efficient and flexible algorithm for free energy calculations using the λ-dynamics approach. J Phys Chem B. 1998;102:2032–2036. [Google Scholar]
- 61.Lee MS, Salsbury FR, Brooks CL., III Constant-pH Molecular Dynamics Using Continuous Titration Coordinates. Proteins: Struct, Funct, Bioinf. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 62.Khandogin J, Brooks CL., III Constant pH Molecular Dynamics with Proton Tautomerism. Biophys J. 2005;89:141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khandogin J, Brooks CL., III Toward the accurate first-principles prediction of ionization equilibria in proteins. Biochemistry. 2006;45:9363–9373. doi: 10.1021/bi060706r. [DOI] [PubMed] [Google Scholar]
- 64.Khandogin J, Chen J, Brooks CL., III Exploring Atomistic Details of pH-dependent Peptide Folding. Proc Natl Acad Sci U S A. 2006;103:18546–18550. doi: 10.1073/pnas.0605216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khandogin J, Raleigh DP, Brooks CL., III Folding Intermediate in the Villin Headpiece Domain Arises From Disruption of a N-terminal Hydrogen-bonded Network. J Am Chem Soc. 2007;129:3056–3057. doi: 10.1021/ja0688880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khandogin J, Brooks CL., III Linking Folding with Aggregation in Alzheimer's Beta-amyloid Peptides. Proc Natl Acad Sci U S A. 2007;104:16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang BW, Brunetti L, Brooks CL., III Probing pH-dependent Dissociation of HdeA Dimers. J Am Chem Soc. 2011;133:19393–19398. doi: 10.1021/ja2060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen JK. Uncovering specific electrostatic interactions in the denatured states of proteins. Biophys J. 2010;99:924–932. doi: 10.1016/j.bpj.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallace JA, Shen JK. Unraveling A Trap-and-Trigger Mechanism in the pH-Sensitive Self- Assembly of Spider Silk Proteins. J Phys Chem Lett. 2012;3:658–662. doi: 10.1021/jz2016846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law SM, Zhang BW, Brooks CL., III pH-sensitive residues in the p19 RNA silencing suppressor protein from carnation Italian ringspot virus affect siRNA binding stability. Protein Sci. 2013;22:595–604. doi: 10.1002/pro.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dlugosz M, Antosiewicz JM. Effects of solute-solvent proton exchange on polypeptide chain dynamics: a constant-pH molecular dynamics study. J Phys Chem B. 2005;109:13777–13784. doi: 10.1021/jp0505779. [DOI] [PubMed] [Google Scholar]
- 72.Machuqueiro M, Baptista AM. The pH-dependent Conformational States of Kyotorphin: A Constant-pH Molecular Dynamics Study. Biophys J. 2007;92:1836–1845. doi: 10.1529/biophysj.106.092445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campos SR, Machuqueiro M, Baptista AM. Constant-pH molecular dynamics simulations reveal a beta-rich form of the human prion protein. J Phys Chem B. 2010;114:12692–12700. doi: 10.1021/jp104753t. [DOI] [PubMed] [Google Scholar]
- 74.Arthur EJ, Yesselman JD, Brooks CL., III Predicting extreme pK(a) shifts in staphylococcal nuclease mutants with constant pH molecular dynamics. Proteins: Struct, Funct, Bioinf. 2011;79:3276–3286. doi: 10.1002/prot.23195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallace JA, Shen JK. Continuous Constant pH Molecular Dynamics in Explicit Solvent with pH-Based Replica Exchange. J Chem Theory Comput. 2011;7:2617–2629. doi: 10.1021/ct200146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang WZ, Chu XP, Li MH, Seeds J, Simon RP, Xiong ZG. Modulation of acid-sensing ion channel currents, acid-induced increase of intracellular Ca2+, and acidosis-mediated neuronal injury by intracellular pH. J Biol Chem. 2006;281:29369–29378. doi: 10.1074/jbc.M605122200. [DOI] [PubMed] [Google Scholar]
- 77.Hesselager M, Timmermann DB, Ahring PK. pH dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem. 2004;279:11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- 78.Berdiev BK, Mapstone TB, Markert JM, Gillespie GY, Lockhart J, Fuller CM, Benos DJ. pH alterations"reset" Ca2+ sensitivity of brain Na+ channel 2, a degenerin/epithelial Na+ ion channel, in planar lipid bilayers. J Biol Chem. 2001;276:38755–38761. doi: 10.1074/jbc.M107266200. [DOI] [PubMed] [Google Scholar]
- 79.Damaghi M, Bippes C, Koster S, Yildiz O, Mari SA, Kuhlbrandt W, Muller DJ. pH-Dependent Interactions Guide the Folding and Gate the Transmembrane Pore of the beta-Barrel Membrane Protein OmpG. J Mol Biol. 2010;397:878–882. doi: 10.1016/j.jmb.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 80.Donnini S, Tegeler F, Groenhof G, Grubmuller H. Constant pH Molecular Dynamics in Explicit Solvent with lambda-Dynamics. J Chem Theory Comput. 2011;7:1962–1978. doi: 10.1021/ct200061r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knight JL, Brooks CL., III Applying Efficient Implicit Non-geometric Constraints in Alchemical Free Energy Simulations. J Comput Chem. 2011;32:3423–3432. doi: 10.1002/jcc.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Knight JL, Brooks CL., III Multisite λ Dynamics for Simulated Structure–Activity Relationship Studies. J Chem Theory Comput. 2011;7:2728–2739. doi: 10.1021/ct200444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goh GB, Knight JL, Brooks CL., III Constant pH Molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent. J Chem Theory Comput. 2012;8:36–46. doi: 10.1021/ct2006314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goh GB, Knight JL, Brooks CL., III pH-dependent Dynamics of Complex RNA Macromolecules. J Chem Theory Comput. 2013;9:935–943. doi: 10.1021/ct300942z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goh GB, Knight JL, Brooks CL., III Toward Accurate Prediction of the Protonation Equilibrium of Nucleic Acids. J Phys Chem Lett. 2013;4:760–766. doi: 10.1021/jz400078d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng L, Chen M, Yang W. Random Walk in Orthogonal Space to Achieve Efficient Free-energy Simulation of Complex Systems. Proc Natl Acad Sci U S A. 2008;105:20227–20232. doi: 10.1073/pnas.0810631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi CY, Wallace JA, Shen JK. Thermodynamic Coupling of Protonation and Conformational Equilibria in Proteins: Theory and Simulation. Biophys J. 2012;102:1590–1597. doi: 10.1016/j.bpj.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 90.Feig M, Karanicolas J, Brooks CL., III MMTSB Tool Set: Enhanced Sampling and Multiscale Modeling Methods for Applications in Structural Biology. J Mol Graphics Modell. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 91.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 92.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 93.Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA. Molecular-Dynamics Simulations on Solvated Biomolecular Systems - the Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, Rna, and Proteins. J Am Chem Soc. 1995;117:4193–4194. [Google Scholar]
- 94.Schreiber H, Steinhauser O. Cutoff Size Does Strongly Influence Molecular-Dynamics Results on Solvated Polypeptides. Biochemistry. 1992;31:5856–5860. doi: 10.1021/bi00140a022. [DOI] [PubMed] [Google Scholar]
- 95.Steinbach PJ, Brooks BR. New Spherical-Cutoff Methods for Long-Range Forces in Macromolecular Simulation. J Comput Chem. 1994;15:667–683. [Google Scholar]
- 96.Beck DAC, Armen RS, Daggett V. Cutoff Size Need Not Strongly Influence Molecular Dynamics Results for Solvated Polypeptides. Biochemistry. 2005;44:609–616. doi: 10.1021/bi0486381. [DOI] [PubMed] [Google Scholar]
- 97.Norberg J, Nilsson L. On the truncation of long-range electrostatic interactions in DNA. Biophys J. 2000;79:1537–1553. doi: 10.1016/S0006-3495(00)76405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Onufriev A, Case DA, Ullmann GM. A novel view of pH titration in biomolecules. Biochemistry. 2001;40:3413–3419. doi: 10.1021/bi002740q. [DOI] [PubMed] [Google Scholar]
- 99.Nozaki Y, Tanford C. Examination of Titration Behavior. Methods Enzymol. 1967;11:715–734. [Google Scholar]
- 100.Bashford D, Case DA, Dalvit C, Tennant L, Wright PE. Electrostatic Calculations of Side-Chain Pk(a) Values in Myoglobin and Comparison with Nmr Data for Histidines. Biochemistry. 1993;32:8045–8056. doi: 10.1021/bi00082a027. [DOI] [PubMed] [Google Scholar]
- 101.Parsons SM, Raftery MA. Ionization behavior of the catalytic carboxyls of lysozyme. Effects of ionic strength. Biochemistry. 1972;11:1623–1629. doi: 10.1021/bi00759a013. [DOI] [PubMed] [Google Scholar]
- 102.Kuramitsu S, Ikeda K, Hamaguchi K, Fujio H, Amano T. Ionization constants of Glu 35 and Asp 52 in hen, turkey, and human lysozymes. J Biochem. 1974;76:671–683. [PubMed] [Google Scholar]
- 103.Kuramitsu S, Ikeda K, Hamaguchi K. Participation of the catalytic carboxyls, Asp 52 and Glu 35, and Asp 101 in the binding of substrate analogues to hen lysozyme. J Biochem. 1975;77:291–301. doi: 10.1093/oxfordjournals.jbchem.a130726. [DOI] [PubMed] [Google Scholar]
- 104.Takahashi T, Nakamura H, Wada A. Electrostatic forces in two lysozymes: calculations and measurements of histidine pKa values. Biopolymers. 1992;32:897–909. doi: 10.1002/bip.360320802. [DOI] [PubMed] [Google Scholar]
- 105.Bartik K, Redfield C, Dobson CM. Measurement of the Individual Pk(a) Values of Acidic Residues of Hen and Turkey Lysozymes by 2-Dimensional H-1-Nmr. Biophys J. 1994;66:1180–1184. doi: 10.1016/S0006-3495(94)80900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Webb H, Tynan-Connolly BM, Lee GM, Farrell D, O'Meara F, Sondergaard CR, Teilum K, Hewage C, McIntosh LP, Nielsen JE. Remeasuring HEWL pK(a) values by NMR spectroscopy: methods, analysis, accuracy, and implications for theoretical pK(a) calculations. Proteins: Struct, Funct, Bioinf. 2011;79:685–702. doi: 10.1002/prot.22886. [DOI] [PubMed] [Google Scholar]
- 107.Machuqueiro M, Baptista AM. Is the prediction of pKa values by constant-pH molecular dynamics being hindered by inherited problems? Proteins: Struct, Funct, Bioinf. 2011;79:3437–3447. doi: 10.1002/prot.23115. [DOI] [PubMed] [Google Scholar]
- 108.Wallace JA, Shen JK. Charge-leveling and proper treatment of long-range electrostatics in all-atom molecular dynamics at constant pH. J Chem Phys. 2012;137:184105. doi: 10.1063/1.4766352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arbely E, Rutherford TJ, Sharpe TD, Ferguson N, Fersht AR. Downhill versus Barrier-Limited Folding of BBL 1: Energetic and Structural Perturbation Effects upon Protonation of a Histidine of Unusually Low pK(a) J Mol Biol. 2009;387:986–992. doi: 10.1016/j.jmb.2008.12.055. [DOI] [PubMed] [Google Scholar]
- 110.Kuhlman B, Luisi DL, Young P, Raleigh DP. pK(a) values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry. 1999;38:4896–4903. doi: 10.1021/bi982931h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.