Abstract
Heart is the first organ formed during organogenesis. The fetal heart undergoes several structural and functional modifications to form the four-chambered mammalian heart. The adult heart shows different adaptations during compensatory and decompensatory heart failure. However, one common adaptation in the pathological heart is fetal reprogramming, where the adult heart expresses several genes and miRNAs which are active in the fetal stage. The fetal reprogramming in the failing heart raises several questions, such as whether the switch of adult to fetal genetic programming is an adaptive response to cope with adverse remodeling of the heart, does the expression of fetal genes protect the heart during compensatory and/or decompensatory heart failure, does repressing the fetal gene in the failing heart is protective to the heart? To answer these questions, we need to understand the expression of genes and miRNAs that are reprogrammed in the failing heart. In view of this, we provided an overview of differentially expressed genes and miRNAs, and their regulation in this review. Further, we elaborated novel strategies for a plausible future therapy of cardiovascular diseases.
Keywords: heart failure, miRNA, fetal reprogramming, therapy, remodeling
1. Introduction
Heart failure is a leading cause of morbidity and mortality across the world [1]. There are several molecular changes in the myocardium, which lead to structural and functional remodeling in the failing heart [2,3]. Several co-morbidities such as hypertension, diabetes, and coronary artery disease promote myocardial apoptosis and ventricular dysfunction leading to heart failure [2,4–6]. One of the common features of the failing heart is fetal reprogramming, which is re-activation of fetal miRNAs [7], fetal genes such as A-, and B-types of natriuretic peptides, β-myosin heavy chain (β-MHC) [8], α-skeletal actin [9], and fetal type cardiac ion channels such as hyperpolarization activated cyclic nucleotide-gated channel and T-type Ca2+ channel [10], and suppression of several adult cardiac genes such as sarco-endoplastic reticulum Ca2+ ATPase [11] and α-myosin heavy chain [12]. The upregulation of fetal genes is often used as a marker of pathological remodeling. For example, induction of β-MHC is a marker of cardiac hypertrophy [9]. However, the role of genetic reprogramming in cardiac adaptation is poorly understood. Here, we reviewed the developmental stages of cardiogenesis leading to formation of a four-chambered heart, genetics of congenital heart failure, fetal reprogramming of genes and miRNAs and differential regulation of miRNAs in the failing heart, and novel therapeutic strategies for treating heart failure.
2. Cardiogenesis to form a four-chambered heart
2.1 The developmental stages of the heart
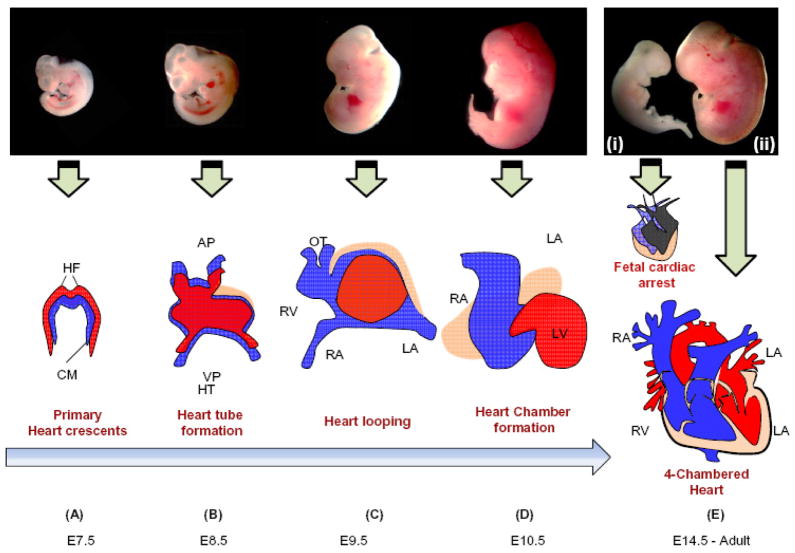
Heart is formed from mesodermal cells of the primitive streak during early gastrulation [13]. Cardiac progenitor cells present in the anterior mesoderm form cardiac crescent (the primary heart field) at embryonic day 7.5. The pharyngeal mesoderm, which is located medial and anterior to the primary heart field, gives rise to the secondary heart field. The primary heart field later (at day 8) migrates to the midline and forms the heart tube [14]. The expansion of primary and secondary heart fields give rise to primitive atria and ventricles [14–17]. The mid–portion of the heart tube becomes twisted to form the ventricular loop, which later forms left and right ventricles. On day 9, truncus arteriosus, endocardial cushions and future valves of the heart are formed. The ventricular surface is enclosed by the epicardial cells on day 10 except the truncus arteriosus, which becomes ensheathed by epicardial cells on day 11. The sub-epicardial connective tissue and capillary containing blood cells are observed on the 11th day. By day 15, septum in atria and ventricles, and valve are formed. The left ventricle and left and right atria are formed by primary heart field, whereas the right ventricle, the outflow tract, and the left and right atria are formed by the secondary heart field [14]. Different steps of development of the heart are elaborated elsewhere [18–24]. The timeline and formation of the four-chambered mouse heart from the primary heart crescents is shown in Figure 1.
Figure 1. Schematic presentation of embryonic cardiac developmental stages, growth and formation of four chambered heart.
A. Formation of primary heart crescent at ~E7.5. B. Development of heart tube at ~E8.5 from the cardiac crescent cells. C. Formation of primitive atria and ventricles at ~E9.5 by looping of heart tube. D. Heart chamber formation at ~E10.5 from primitive chambers. E (i) Defective cardiac development leads to embryonic heart failure and in-utero death by cardiac arrest at ~E14.5. E (ii) Formation of the four-chambered heart at ~E14.5. The successive developmental stages during mouse embryogenesis are shown in top panel with representative pictures. Abbreviations: HF, Heart Fields; CM, Cardiogenic Mesoderm; AP, Arterial Pole; OT, Outflow Tract; HT, Heart Tube; VP, Venous Pole; RV, Right Ventricle; LV, Left Ventricle; RA, Right Atrium; LA, Left Atrium; PA, Pulmonary Artery; A; Aorta.
2.2 Differentiation of fetal cardiac cell types
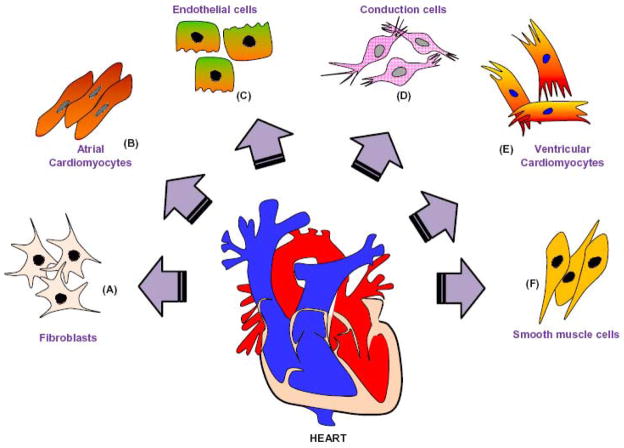
Embryonic stem cells differentiate into a primitive tubular heart that forms the adult heart. The lineage specific precursor cells differentiate into different cells that form myocardium, vascular endothelium, fibroblasts and smooth muscle cells. These cells contribute to contractility, conduction system and vasculature of the adult heart [25,26]. An example of different types of cells in the adult heart is shown in Figure 2. Specific genes are differentially expressed in a spatial and temporal manner during the development of the heart. These genes are comprehensively reviewed by Thomas Brand [27].
Figure 2. Schematic presentation of cardiac cell types.
The adult heart consists of several cell types, which maintain structural, mechanical, electrical and functional integrity of the heart. A. Fibroblasts contribute in the formation of extracellular matrix, which provides mechanical support to the heart. B. Atrial cardiomyocytes contribute to contractility of atrium. C. Endothelial cells form the inner lining of cardiac blood vessels. D. Conduction cells generate electrical impulses for cardiac contractility. E. Ventricular cardiomyocytes are involved in contractility of the ventricles. F. Smooth muscle cells render support to the coronary arteries and vasculatures.
3. Pathophysiology of embryonic hearts
3.1 Fetal heart failure
In the fetal heart, progenitor cells differentiate into specific lineages to form specific types of cells. The differentiation of cells into a particular lineage is a tightly regulated process, which depends on the microenvironment and the signaling cues in the extracellular matrix [26,27]. Deregulation of regulatory genes involved in lineage determination, differentiation, or cardiac development causes fetal heart failure, and may lead to embryonic lethality. Impaired contractility and myocardial compliance in response to hemodynamic stress increases the incidence of fetal heart failure [28–31]. The phenotype of fetal cardiac arrest in mouse is shown in Figure 1.
3.2 Congenital heart disease
Congenital heart disease (CHD) is the manifestation of structural abnormalities of the heart such as valve defect, hole inside wall of the heart, atrial and ventricular septal defects, and stenosis before birth [30,32]. CHD leads to heart failure or death and is caused due to genetic defects, chromosomal abnormalities, excessive use of alcohol during pregnancy, and/or maternal viral infection [32]. Mutations in the heart muscle specific MYH6 (α-myosin heavy chain) and several proteins that interact with MYH6 such as GATA4 that forms complex with the TBX5 (T-box gene 5) lead to heart failure [33]. Mutation of NKX2-5, another protein that interacts with MYH6 is associated with ventricular and atrial septal defects [24]. NKX2-5 is also associated with the electrical conduction and impulse distribution in the heart. Defects in the electrical conductance in the heart causes arrhythmia, which is detrimental to the heart [29]. TBX5 is implicated in the Holt-Oram Syndrome, which includes abnormalities in neuromuscular electrical conduction and defects of the heart and upper limb. Another TBX gene, the TBX1 causes DiGeorge Syndrome, which has symptoms and features including defects of the cardiac outflow tract and Tetralogy of Fallot (TOF). In TOF, the ventricular septal defect (VSD) causes mixing of oxygenated and deoxygenated blood in the left ventricle. Further, due to defects in the pulmonary valve, there is a preference of mixed blood to flow through the aorta [31]. The causes, diagnosis, symptoms and presently available treatments of CHD are elaborated in a recent review by Sun et al [32].
4. Functionally identified fetal genes that are re-activated in the failing heart
4.1 Transcriptional regulation of fetal contractile gene expression: β-myosin heavy chain
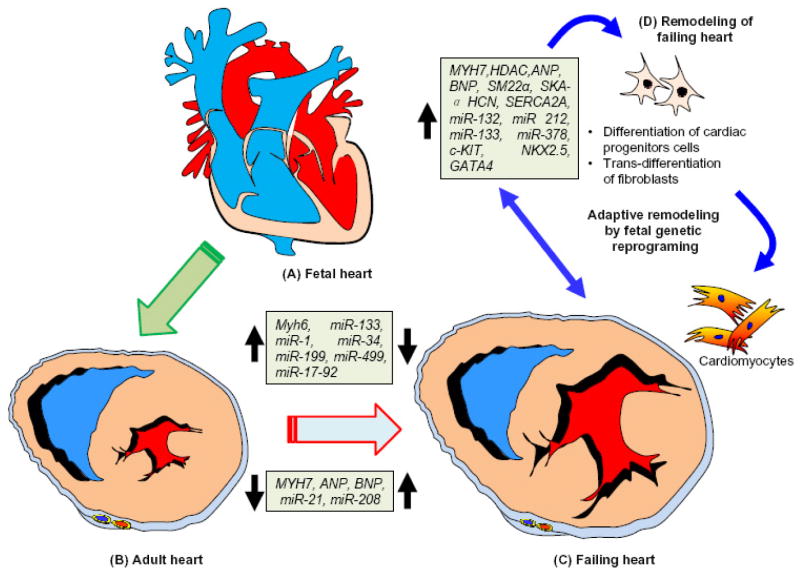
Several fetal genes are re-activated in the failing heart (Figure 3). For example, isoform switch in the expression of sarcomeric proteins after the birth. Similarly, in the ventricle, β-myosin heavy chain (MHC) is the predominant isoform in the fetal heart, which is switched to α-MHC after birth [34]. The change in the MHC isoform appears to be an important process via which the heart adapts its mechanical performance and efficiency to the postnatal circulation. During cardiac hypertrophy and heart failure, the expression of β-MHC gene is induced together with several other fetal cardiac genes. In the embryo, brahma-related gene-1 (Brg1) promotes myocytes proliferation by maintaining Bmp10 (bone morphogenic protein-10) and suppressing p57 (a cyclin dependent kinase inhibitor 1C) [28]. Brg1 preserves fetal cardiac development and differentiation by interacting with HDAC (histone deacetylase) and PARP (poly ADP ribose polymerase) to repress α-MHC and activate β-MHC. In adults, Brg1 (Smarca4) is turned off in cardiomyocytes, but is re-activated by cardiac stress and forms a complex with its fetal partners HDAC and PARP to induce a pathological α-MHC to β-MHC shift. Preventing Brg1 re-expression decreases hypertrophy and reverses the switching of isoforms of MHC in failing hearts. Brg1 is activated in patients with hypertrophic cardiomyopathy and its expression level is positively correlated with severity of the heart failure [35–38]. Brg1 is essential for embryonic cardiomyocytes. The cross-talk among the three chromatin-modifying factors Brg1, HDAC and PARP are regulated by epigenetic mechanisms and they control the expression of developmental and pathological genes [39].
Figure 3. Schematic presentation of cardiac genetic reprogramming in the fetal, adult, and failing heart.
A. The four-chambered fetal heart. The genes and miRNAs involved in normal development of the fetal heart are highlighted in the box. B. Representation of transverse sections of the adult heart with normal ventricular myocardium. Genes and miRNAs that are upregulated and downregulated during heart failure are highlighted in the box. C. Representative transverse sections of the failing heart with abnormal ventricular myocardium and hypertrophy. Genes and miRNAs that are upregulated and downregulated are highlighted in the box. D. Adaptive remodeling in the failing heart through re-activation of the fetal genetic programing to generate new cardiac cells through differentiation of cardiac progenitor cells and trans-differentiation of fibroblasts into cardiomyocytes can be induced for the therapy of heart failure.
4.2 Transcriptional regulation of ANP gene expression
Studies using transgenic mice carrying a 500-bp or 2.4-kbp segment of the 5′ flanking region (5′-FR) of the human atrial natriuretic peptide (ANP) gene, or a 638-bp or 3-kbp 5′-FR segment of the rat ANP gene, and fused to a reporter gene have demonstrated that these regions are sufficient to confer cardiac-restricted gene expression [40]. Moreover, the ventricular activities of these 5′-FR segments are down regulated after birth even though the atrial activity remains high [41].
4.3 Transcriptional regulation of BNP gene expression
The 5′-FR of the B-type natriuretic peptide (BNP) gene has also been extensively studied in order to understand the regulatory mechanisms that control its cardiac-specific and inducible expression. A study using transgenic mice carrying a 1.8-kbp or 400-bp segment of the 5′-FR of the human BNP gene coupled to a luciferase gene (−1818hBNPluc and −400hBNPluc, respectively) demonstrates that the proximal region of the human BNP promoter is sufficient to activate ventricle-specific expression [42]. In addition, BNP mRNA has an AT-rich region in its 3′-untranslated region (UTR), which makes the transcript unstable. This could be important for post-transcriptional regulation of BNP. The BNP mRNA has a shorter half-life than ANP mRNA [43].
4.4 Fetal ion channel genes: HCN channels
Hyperpolarization-activated cyclic nucleotide gated (HCN) channels are comprised of 1–4 ion channels and they generate local currents in the heart. In the adult heart, these channels are expressed predominantly in the conduction system, especially in the SA (sinoatrial) node, where HCN4 is the key isoform that controls cardiac rhythmicity [44]. HCN channels are widely expressed in ventricular myocytes where HCN2 is the dominant isoform. The levels of HCN channel expression in the adult ventricular myocardium are much lower than that in the conduction system. During development, HCN channels are profusely expressed in the fetal ventricular myocardium, but their ventricular expression progressively declines after birth [45].
4.5 Fetal reprogramming of skeletal α-actin
Skeletal α-actin (SkA) is a principal component of adult skeletal muscle thin filaments. It is also a prominent actin isoform in the fetal heart [46,47]. After birth, ventricular expression of SkA declines and the gene is suppressed in the normal adult ventricular myocardium. In the hypertrophied and failing hearts, SkA is re-activated [48].
4.6 Transcriptional regulation of SM22α
SM22α (also named transgelin1) is a member of the calponin family and is specifically expressed in mature smooth muscle cells. During embryogenesis, SM22α is transiently expressed in the cardiac and skeletal muscle lineages, but later its expression is restricted to smooth muscle [49]. The 445-bp SM22α promoter, which contains SRF-binding sequences (CArG), a SBE (a Smad-binding site), and a TCE (TGF-β control element) is sufficient to direct the expression of a linked reporter gene in cardiac and skeletal muscle during cardiac development. In the adult heart, SM22α gene is re-expressed under pathological condition in both mice and humans [50].
4.7 Transcriptional regulation of T-type Ca2+ channel
During fetal development, T-type Ca2+ channels are abundantly expressed in embryonic ventricle, but their expression is reduced in the adult ventricle [10]. However, T-type Ca2+ channels are re-expressed in the ventricle in the failing hearts and contribute to the pathological cardiac remodeling leading to arrhythmogenesis and systolic dysfunction [51].
5. Differential regulation of non-coding RNAs in adult heart failure
5.1 miRNAs in the adult heart failure
MiRNAs are a novel class of tiny non-coding, conserved, regulatory RNAs [52,53] that modulate gene expression post-transcriptionally [54], and have emerged as a therapeutic target for cardiovascular disease [55–57]. Similar to gene reprogramming, the fetal miRNAs are re-expressed in the failing heart and contribute to the genetic changes in the failing heart [7]. Several miRNAs are involved in different types of heart failure. For example, miR-208 and Mef2 regulate the decompensation of right ventricular function in pulmonary hypertension [58], miR-212/132 family regulates cardiac hypertrophy and autophagy [59], miR-133a is involved in mitigation of cardiac hypertrophy [60] and fibrosis [61] and emerged as cardioprotective miRNA [62], and miR-17-92 cluster regulates cardiac ischemia-reperfusion injury [63].
5.2 MiRNAs in pathological remodeling
Differential expressions of specific miRNAs are associated with heart failure [55,56,64–66]. For example, downregulation of miR-1 and miR-133 causes cardiac hypertrophy [60]. Chromosomal deletion of miR-133 prevents the compensatory left ventricular hypertrophy in mice after transaortic constriction surgery, resulting in dilated cardiomyopathy [67]. Conversely, upregulation of miR-132 and miR-212 contributes to heart failure. Genetic deletion of miR-132 and miR-212 protects the heart from the pressure overload–induced ventricular hypertrophy [59]. MiR-378 attenuates hypertrophic growth by suppressing the MAPK (mitogen-activated protein kinase) pathway [68]. MiR-21 increases the ability of fibroblast survival and progression of the cardiac fibrosis by inducing ECM remodeling and turnover [69]. The global miRNA profile in the failing heart resembles partly with those of fetal hearts, which indicates these miRNAs play a pivotal role in adaption of the heart in stress condition by re-activating the fetal genetic program. For example, in the adult heart, fast and rapid contracting isoform α-MHC is activated and slow contracting β-MHC, which is active in the fetal heart is suppressed. However, in the failing heart α-MHC is inhibited and β-MHC is activated to cope with the stress condition and maintain the contractility of the heart in efficient manner. It is reported that the heart undergoes hypertrophy, fibrosis, and reduced contractility to respond to the stress condition [70]. The intron region of α-MHC encode miR-208, which contributes to cardiomyocytes fibrosis, hypertrophy, and regulates the expression of β-MHC in response to stress [70].
5.3 Circulating miRNAs as biomarkers of heart failure
MiRNAs are transcribed in the tissue and are encapsulated into vesicles such as exosome before being released into the blood stream, where they remain stable. The plasma level of miRNAs are emerged as a biomarker for heart failure [71]. Recently, it is demonstrated that acute and chronic exercise changes the miRNA profile in the plasma [72]. Similarly, the levels of specific miRNAs change during reduced and preserved left ventricular ejection fraction [73]. These findings suggest that circulating level of miRNAs can be used as a biomarker and promising therapeutic target for heart disease [55,64]. However, how the level of circulatory miRNAs change in the fetal versus adult heart is yet to be explored.
5.4 Long non-coding (lnc) RNAs in regulation of heart failure
LncRNAs are newly emerged regulatory non-coding RNAs, which are defined as transcripts of greater than 200 nucleotides without protein coding function [74]. On the basis of molecular mechanisms, they are classified into four categories: 1) Signal, 2) Decoy, 3) Guide, and 4) Scaffold [74]. The signal lncRNAs regulate transcriptional silencing of several genes by recruiting chromatin–modifying machinery and interacting with chromatin. In response to specific spatial and temporal stimuli, they initiate transcriptional silencing [75,76]. The decoy lncRNAs are involved in removing a specific protein by binding to it. They alter the transcriptome by changing the pattern of alternative splicing [77]. The guide lncRNAs regulate the localization of ribonucleoproteins to specific chromatin targets. The change in localization of ribonucleoproteins causes change in gene expression of the neighboring genes [78]. The scaffold lncRNAs regulates transcriptional activation and repression by binding to different proteins with its multiple domains [79]. Although the role of lncRNAs is beginning to unfold in cardiovascular pathophysiology [80–85], its role in cardiac development and in the fetal heart is an open avenue for investigation.
6. Novel therapeutic strategies
Despite gigantic strides made towards treating heart disease, it is the number one cause of mortality in the world, which warrants novel therapeutic strategies to ameliorate heart failure. The recent emergence of non-coding RNAs, the miRNAs and lncRNAs in the regulation of pathological remodeling and heart failure suggest that they have potential to be a novel therapeutic target for heart disease. It is also necessary to understand the role of different genes and specifically those genes, which change their expression in the failing heart. In this regard, the genes and miRNAs that express in the fetal heart and reactivate in the failing heart are promising targets because these genes inherently and physiologically important for the cardiac adaptation in the stress condition. Understanding the specific role of these genes and their regulation may provide a novel therapeutic strategy to inhibit progression of adverse remodeling and thereby mitigate cardiac dysfunction. Recent studies show that fibroblast can be trans-differentiated into cardiomyocytes with the help of specific transcription factors and miR-133a [86,87].
Reprogramming of non-myocytes is also emerged as novel strategy for cardiac repair [88,89]. These findings suggest that regulating specific gene/transcription factors and or miRNA has potential to turn the devil (fibroblasts, because in the failing heart accumulation of fibroblasts leads to fibrosis) into god (cardiomyocytes, which are less in number due to increased apoptosis or are hypertrophied due to excessive workload in the failing heart). Further, it is plausible to ameliorate cardiac dysfunction in the failing heart by manipulating the above regulators. These are novel therapeutic strategies to find a better cure for heart disease.
Acknowledgments
This work is supported in part by NIH grants: HL-113281 and HL-116205 to Paras K. Mishra.
We would like to acknowledge Bryan T. Hackfort, a postdoctoral fellow in our laboratory for editing the final version of the manuscript.
Footnotes
Conflict of interest: No conflicts declared.
References
- 1.Hata J, Kiyohara Y. Epidemiology of stroke and coronary artery disease in Asia. Circ J. 2013;77:1923–1932. doi: 10.1253/circj.cj-13-0786. [DOI] [PubMed] [Google Scholar]
- 2.Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Myocyte death, growth, and regeneration in cardiac hypertrophy and failure. Circ Res. 2003;92:139–150. doi: 10.1161/01.res.0000053618.86362.df. [DOI] [PubMed] [Google Scholar]
- 3.Sarkar S, Chawla-Sarkar M, Young D, Nishiyama K, Rayborn ME, Hollyfield JG, et al. Myocardial cell death and regeneration during progression of cardiac hypertrophy to heart failure. J Biol Chem. 2004;279:52630–52642. doi: 10.1074/jbc.M402037200. [DOI] [PubMed] [Google Scholar]
- 4.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 5.Sutendra G, Dromparis P, Paulin R, Zervopoulos S, Haromy A, Nagendran J, et al. A metabolic remodeling in right ventricular hypertrophy is associated with decreased angiogenesis and a transition from a compensated to a decompensated state in pulmonary hypertension. J Mol Med (Berl) 2013;91:1315–1327. doi: 10.1007/s00109-013-1059-4. [DOI] [PubMed] [Google Scholar]
- 6.Yokota H, Imai Y, Tsuboko Y, Tokumaru AM, Fujimoto H, Harada K. Nocturnal Blood Pressure Pattern Affects Left Ventricular Remodeling and Late Gadolinium Enhancement in Patients with Hypertension and Left Ventricular Hypertrophy. PLoS One. 2013;8:e67825. doi: 10.1371/journal.pone.0067825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 8.Barry SP, Davidson SM, Townsend PA. Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol. 2008;40:2023–2039. doi: 10.1016/j.biocel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Black FM, Packer SE, Parker TG, Michael LH, Roberts R, Schwartz RJ, et al. The vascular smooth muscle alpha-actin gene is reactivated during cardiac hypertrophy provoked by load. J Clin Invest. 1991;88:1581–1588. doi: 10.1172/JCI115470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita H, Kuwahara K, Takano M, Arai Y, Kuwabara Y, Yasuno S, et al. T-type Ca2+ channel blockade prevents sudden death in mice with heart failure. Circulation. 2009;120:743–752. doi: 10.1161/CIRCULATIONAHA.109.857011. [DOI] [PubMed] [Google Scholar]
- 11.Currie S, Smith GL. Enhanced phosphorylation of phospholamban and downregulation of sarco/endoplasmic reticulum Ca2+ ATPase type 2 (SERCA 2) in cardiac sarcoplasmic reticulum from rabbits with heart failure. Cardiovasc Res. 1999;41:135–146. doi: 10.1016/s0008-6363(98)00241-7. [DOI] [PubMed] [Google Scholar]
- 12.Carniel E, Taylor MR, Sinagra G, Di LA, Ku L, Fain PR, et al. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54–59. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
- 13.Tam PP, Parameswaran M, Kinder SJ, Weinberger RP. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development. 1997;124:1631–1642. doi: 10.1242/dev.124.9.1631. [DOI] [PubMed] [Google Scholar]
- 14.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Martinez V, Schoenwolf GC. Primitive-streak origin of the cardiovascular system in avian embryos. Dev Biol. 1993;159:706–719. doi: 10.1006/dbio.1993.1276. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki HR, Solursh M, Baldwin HS. Relationship between fibronectin expression during gastrulation and heart formation in the rat embryo. Dev Dyn. 1995;204:259–277. doi: 10.1002/aja.1002040305. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, Baldini A. Genetic pathways to mammalian heart development: Recent progress from manipulation of the mouse genome. Semin Cell Dev Biol. 2007;18:77–83. doi: 10.1016/j.semcdb.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viragh S, Challice CE. Origin and differentiation of cardiac muscle cells in the mouse. J Ultrastruct Res. 1973;42:1–24. doi: 10.1016/s0022-5320(73)80002-4. [DOI] [PubMed] [Google Scholar]
- 19.Fishman MC, Stainier DY. Cardiovascular development. Prospects for a genetic approach. Circ Res. 1994;74:757–763. doi: 10.1161/01.res.74.5.757. [DOI] [PubMed] [Google Scholar]
- 20.Anderson RH, Webb S, Brown NA, Lamers W, Moorman A. Development of the heart: (3) formation of the ventricular outflow tracts, arterial valves, and intrapericardial arterial trunks. Heart. 2003;89:1110–1118. doi: 10.1136/heart.89.9.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson RH, Webb S, Brown NA, Lamers W, Moorman A. Development of the heart: (2) Septation of the atriums and ventricles. Heart. 2003;89:949–958. doi: 10.1136/heart.89.8.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moorman A, Webb S, Brown NA, Lamers W, Anderson RH. Development of the heart: (1) formation of the cardiac chambers and arterial trunks. Heart. 2003;89:806–814. doi: 10.1136/heart.89.7.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moorman AF, Lamers WH. A molecular approach towards the understanding of early heart development: an emerging synthesis. Symp Soc Exp Biol. 1992;46:285–300. [PubMed] [Google Scholar]
- 24.Moorman AF, Christoffels VM. Development of the cardiac conduction system: a matter of chamber development. Novartis Found Symp. 2003;250:25–34. [PubMed] [Google Scholar]
- 25.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 26.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 27.Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 28.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornberger LK, Sahn DJ. Rhythm abnormalities of the fetus. Heart. 2007;93:1294–1300. doi: 10.1136/hrt.2005.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huhta JC. Fetal congestive heart failure. Semin Fetal Neonatal Med. 2005;10:542–552. doi: 10.1016/j.siny.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Sun R, Liu M, Lu L, Zheng Y, Zhang P. Congenital Heart Disease: Causes, Diagnosis, Symptoms, and Treatments. Cell Biochem Biophys. 2015 doi: 10.1007/s12013-015-0551-6. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, et al. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol. 2009;29:2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circ Res. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Yao W, Irwin MG, Wang T, Wang S, Zhang L, et al. Adiponectin Ameliorates Hyperglycemia-Induced Cardiac Hypertrophy and Dysfunction by Concomitantly Activating Nrf2 and Brg1. Free Radic Biol Med. 2015 doi: 10.1016/j.freeradbiomed.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Lei S, Liu Y, Gao X, Irwin MG, Xia ZY, et al. Antioxidant N-acetylcysteine attenuates the reduction of Brg1 protein expression in the myocardium of type 1 diabetic rats. J Diabetes Res. 2013;2013:716219. doi: 10.1155/2013/716219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikimi T, Kuwahara K, Nakao K. Current biochemistry, molecular biology, and clinical relevance of natriuretic peptides. J Cardiol. 2011;57:131–140. doi: 10.1016/j.jjcc.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Horsthuis T, Houweling AC, Habets PE, de Lange FJ, el AH, Clout DE, et al. Distinct regulation of developmental and heart disease-induced atrial natriuretic factor expression by two separate distal sequences. Circ Res. 2008;102:849–859. doi: 10.1161/CIRCRESAHA.107.170571. [DOI] [PubMed] [Google Scholar]
- 42.Lanfear DE. Genetic variation in the natriuretic peptide system and heart failure. Heart Fail Rev. 2010;15:219–228. doi: 10.1007/s10741-008-9113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonow RO. New insights into the cardiac natriuretic peptides. Circulation. 1996;93:1946–1950. doi: 10.1161/01.cir.93.11.1946. [DOI] [PubMed] [Google Scholar]
- 44.Heckert RA, Saif LJ, Myers GW. Mucosal and systemic isotype-specific antibody responses to bovine coronavirus structural proteins in naturally infected dairy calves. Am J Vet Res. 1991;52:852–857. [PubMed] [Google Scholar]
- 45.Schweizer PA, Yampolsky P, Malik R, Thomas D, Zehelein J, Katus HA, et al. Transcription profiling of HCN-channel isotypes throughout mouse cardiac development. Basic Res Cardiol. 2009;104:621–629. doi: 10.1007/s00395-009-0031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boheler KR, Carrier L, de la Bastie D, Allen PD, Komajda M, Mercadier JJ, et al. Skeletal actin mRNA increases in the human heart during ontogenic development and is the major isoform of control and failing adult hearts. J Clin Invest. 1991;88:323–330. doi: 10.1172/JCI115295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wlodarski K, Kobus M, Luczak M. Orthotopic bone induction at sites of Moloney murine sarcoma virus inoculation in mice. Nature. 1979;281:386–387. doi: 10.1038/281386a0. [DOI] [PubMed] [Google Scholar]
- 48.Driesen RB, Verheyen FK, Debie W, Blaauw E, Babiker FA, Cornelussen RN, et al. Re-expression of alpha skeletal actin as a marker for dedifferentiation in cardiac pathologies. J Cell Mol Med. 2009;13:896–908. doi: 10.1111/j.1582-4934.2008.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abeyratne UR, Kinouchi Y, Oki H, Okada J, Shichijo F, Matsumoto K. Artificial neural networks for source localization in the human brain. Brain Topogr. 1991;4:3–21. doi: 10.1007/BF01129661. [DOI] [PubMed] [Google Scholar]
- 50.Huang J, Elicker J, Bowens N, Liu X, Cheng L, Cappola TP, et al. Myocardin regulates BMP10 expression and is required for heart development. J Clin Invest. 2012;122:3678–3691. doi: 10.1172/JCI63635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ono K, Iijima T. Cardiac T-type Ca(2+) channels in the heart. J Mol Cell Cardiol. 2010;48:65–70. doi: 10.1016/j.yjmcc.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 52.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 53.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 54.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 55.Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013;113:676–689. doi: 10.1161/CIRCRESAHA.113.300226. [DOI] [PubMed] [Google Scholar]
- 56.Mishra PK, Tyagi N, Kumar M, Tyagi SC. MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med. 2009;13:778–789. doi: 10.1111/j.1582-4934.2009.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klimczak D, Paczek L, Jazdzewski K, Kuch M. MicroRNAs: powerful regulators and potential diagnostic tools in cardiovascular disease. Kardiol Pol. 2015;73:1–6. doi: 10.5603/KP.a2014.0210. [DOI] [PubMed] [Google Scholar]
- 58.Paulin R, Sutendra G, Gurtu V, Dromparis P, Haromy A, Provencher S, et al. A miR-208-Mef2 axis drives the decompensation of right ventricular function in pulmonary hypertension. Circ Res. 2015;116:56–69. doi: 10.1161/CIRCRESAHA.115.303910. [DOI] [PubMed] [Google Scholar]
- 59.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 61.Matkovich SJ, Hu Y, Eschenbacher WH, Dorn LE, Dorn GW. Direct and indirect involvement of microRNA-499 in clinical and experimental cardiomyopathy. Circ Res. 2012;111:521–531. doi: 10.1161/CIRCRESAHA.112.265736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishra PK. Is miR-133a a promising therapeutic target for heart failure. J Diabetes & Metabolism. 2014;5:e118. [Google Scholar]
- 63.Zhou M, Cai J, Tang Y, Zhao Q. MiR-17-92 cluster is a novel regulatory gene of cardiac ischemic/reperfusion injury. Med Hypotheses. 2013;81:108–110. doi: 10.1016/j.mehy.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 64.Condorelli G, Latronico MV, Cavarretta E. microRNAs in cardiovascular diseases: current knowledge and the road ahead. J Am Coll Cardiol. 2014;63:2177–2187. doi: 10.1016/j.jacc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 65.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thum T. MicroRNA therapeutics in cardiovascular medicine. EMBO Mol Med. 2012;4:3–14. doi: 10.1002/emmm.201100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 2013;9:e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. 2013;127:2097–2106. doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]
- 69.Dong S, Ma W, Hao B, Hu F, Yan L, Yan X, et al. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int J Clin Exp Pathol. 2014;7:565–574. [PMC free article] [PubMed] [Google Scholar]
- 70.van RE, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 71.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 72.Xu T, Liu Q, Yao J, Dai Y, Wang H, Xiao J. Circulating microRNAs in response to exercise. Scand J Med Sci Sports. 2015 doi: 10.1111/sms.12421. [DOI] [PubMed] [Google Scholar]
- 73.Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015 doi: 10.1002/ejhf.223. [DOI] [PubMed] [Google Scholar]
- 74.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- 76.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman SC, Dorn GW. Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci U S A. 2014;111:12264–12269. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J. 2015;36:353–368. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piccoli MT, Gupta SK, Thum T. Noncoding RNAs as regulators of cardiomyocyte proliferation and death. J Mol Cell Cardiol. 2015 doi: 10.1016/j.yjmcc.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 83.Thum T, Condorelli G. Long Noncoding RNAs and MicroRNAs in Cardiovascular Pathophysiology. Circ Res. 2015;116:751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 84.Uchida S, Dimmeler S. Long Noncoding RNAs in Cardiovascular Diseases. Circ Res. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 85.Wu C, Arora P. Long Noncoding Mhrt RNA: Molecular Crowbar Unravel Insights into Heart Failure Treatment. Circ Cardiovasc Genet. 2015;8:213–215. doi: 10.1161/CIRCGENETICS.115.001019. [DOI] [PubMed] [Google Scholar]
- 86.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palpant NJ, Murry CE. Regenerative medicine: Reprogramming the injured heart. Nature. 2012;485:585–586. doi: 10.1038/485585a. [DOI] [PubMed] [Google Scholar]
- 89.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]