Abstract
Recent studies suggest that single genome amplification (SGA) as compared to standard bulk PCR and virus stocks from 293T transfection versus short term passage in peripheral blood mononuclear cells (PBMC) yield a less biased representation of HIV-1 envelope characteristics. In 9 different subjects, genetic diversity, divergence, and population structure was not significantly different among SGA or bulk PCR amplified envelope V1–V3 segments. In these subjects, 293T transfection derived virus stocks with SGA or bulk PCR amplified envelopes had similar infectivity, replication kinetics, co-receptor usage, and neutralization susceptibility. While PBMC passage as compared to the 293T derived virus stocks had similar co-receptor usage, PBMC viruses were less neutralization susceptible to some specific antibodies. Our results suggest that the method of envelope sequence amplification, either SGA or bulk PCR, does not have a significant impact on the genotypic and phenotypic properties of the virus envelope quasispecies.
Keywords: HIV-1, envelope, single genome amplification, diversity, replication, neutralization
1. INTRODUCTION
Genotypic and phenotypic properties of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein have important implications for understanding disease pathogenesis and developing prevention strategies. Interrogation of the HIV-1 envelope properties, however, is complicated because an infected subject often harbors a large number of different HIV-1 envelopes, termed quasispecies (Coffin, 1995; Gaschen et al., 2002). An amplification reaction starting with multiple templates can be used to sample the diverse envelopes present within the virus quasispecies. In addition, combining the product from multiple independent PCRs can be used to overcome resampling bias (Liu et al., 1996). This bulk PCR amplification strategy, however, has been supplanted in more recent studies with single genome amplification (SGA) or limiting dilution PCR (Keele et al., 2008; Sagar et al., 2004; Salazar-Gonzalez et al., 2008). As named, SGA starts with a presumed single template; this strategy avoids polymerase induced recombination artifacts observed in PCRs with multiple genomes as the starting template. In comparison to bulk PCR, however, SGA is both labor intensive and expensive. In addition, a large number of independent SGAs are required to achieve the same sampling depth as that from multiple independent bulk PCRs. Previous studies that compared the SGA and bulk PCR methodology found no significant differences in the amplified sequence genotypic characteristics (Buzon et al., 2014; Jordan et al., 2010). These comparisons, however, have been in relatively short or genetically constant regions which may limit detection of differences among SGA versus bulk PCR amplified sequences. Furthermore, no study has compared phenotypic differences among envelope products generated either through SGA or bulk PCR. Thus, it remains unclear if envelope sequences generated from either SGA or bulk PCR will yield similar genotypic and phenotypic characteristics.
Once envelopes are amplified, they are routinely incorporated into a heterologous HIV-1 backbone to examine their phenotypic properties within a viral context. Viruses with the envelopes of interest are often generated from transfections or peripheral blood mononuclear cell (PBMC) passages. While transfection derived virus stocks are relatively easy to generate, they yield viruses that have non-physiologic envelope content and different types of glycosylation commonly observed in patient derived virions (Binley et al., 2010; Nawaz et al., 2011; Raska et al., 2010). On the other hand, while PBMC derived viruses are more physiologically relevant, passaging viruses in PBMCs may favor the replication of specific variants, which changes the proportion and affects the virus quasispecies phenotype (Voronin et al., 2007). Thus, it is possible that transfection and PBMC passaged virus stocks have differences in the phenotype of interest.
In this study, we compared both genotypic and phenotypic characteristics among envelope products isolated from 9 subjects that were amplified using both SGA and bulk PCR. We also examined the effect of short term PBMC passage of 293T derived virus stocks on co-receptor usage and neutralization profile.
2. MATERIAL AND METHODS
2.1 Study subjects
Envelopes were isolated from 9 different HIV-1 infected subjects. Three subjects were from the AIDS Linked to the IntraVenous Experience (ALIVE) cohort, which follows HIV-1 uninfected and HIV-1 infected injection drug users in Baltimore, Maryland through semiannual visits (Etemad et al., 2014; Vlahov et al., 1998). Samples from these 3 subjects were obtained prior to HIV-1 seroconversion. The remaining 6 samples were from subjects with known HIV-1 seropositivity with infection of undetermined duration. All subjects were not on antiretroviral therapy. The study was deemed as not human subject research by the Boston University human subjects review board.
2.2 Envelope amplification and virus stocks
HIV-1 RNA was isolated from 100 ul of the sample, and RNA was reverse transcribed to generate cDNA using previously described primers and conditions (Salazar-Gonzalez et al., 2008). Both bulk PCR and SGA used the same primers and amplification conditions as previously described (Salazar-Gonzalez et al., 2008). Bulk PCRs were done with a minimum of 1 ul of cDNA as the starting template. For the SGAs, cDNA was diluted to ensure that a maximum of 30 from a total of 96 independent PCRs yielded a positive reaction. Sequence of the SGA amplicons was examined directly after ExoSap IT (Affymetrix) treatment. Bulk PCRs were cloned as detailed below and sequences from individual clones were picked for sequence examination. All SGA and 4 independent bulk PCRs were combined to generate a SGA and bulk PCR amplified envelope library respectively. SGA and bulk PCR envelope pools were inserted into linearized pCMV-NL4-3-PBS→LTRΔGp160 plasmid using yeast gap-repair homologous recombination as previously described (Chatziandreou et al., 2012; Dudley et al., 2009). Clone pools were transfected into HEK293T cells to yield 293T transfection virus stocks. The 293T virus stock from bulk envelope pools was passaged in PBMC for 4 days to generate PBMC virus stock. The number of infectious particles (IP) was estimated on TZM-bl cells as previously described (Kimpton and Emerman, 1992; Sagar et al., 2006). The p24gag content was estimated using commercially available ELISA kit (Perkin Elmer). V1–V3 SGA and bulk derived sequences were used to construct a maximum likelihood phylogenetic (ML) tree. For each subject, ML phylogenies were generated using Paup with parameters from FindModel best fit evolutionary model as described previously (Sagar et al., 2009). The ML trees were used to estimate a MRCA. Within each subject, SGA and bulk PCR sequences were grouped, and the average of pairwise distances was used to estimate genetic diversity within a group. Group sequence divergence was estimated as the average distance from the MRCA to a node. Within each subject, population structure among SGA and bulk sequences was examined using a web based non-parametric panmixia test http://wwwabi.snv.jussieu.fr/~achaz/hudsontest.html (Achaz et al., 2004). This test compares intra-group average pairwise genetic distances among user specified groups or among sequences randomly allocated to 2 different groups. For each subject, sequences were randomly allocated to different groups 10,000 different times to generate a probability that the observed as compared to the random population structure was significantly different.
2.3 Replication kinetics
PBMCs were isolated from HIV-1 negative blood donation volunteer’s buffy coats using Ficoll Hypaque density centrifugation. Primary human CD4+ T cells were positively isolated from the PBMCs using antibody conjugated magnetic beads (Stem Cell Technologies) according to manufacturer’s instructions. CD4+ T cells were activated with 2% phytohaemagglutinin (PHA) and 20 U/ml recombinant human IL-2 (r-IL-2) for 2 days. CD4+ T cells from 3 different blood donation volunteers were combined to assess replication kinetics. Around 2×106 CD4+ T cells were exposed to 1,000 infectious particles in the presence of 20 ug/ml diethylaminoethyl (DEAE) Dextran. After two hours, cultures were washed a minimum of three times to remove unbound virus. Infectious virus concentration was estimated by infecting 1 × 104 TZM-bl cells with 4 to 8 serial two-fold dilutions of supernatant culture starting at 50 ul (Etemad et al., 2014; Pena-Cruz et al., 2013). All infections were done in triplicate in a 96 well format. Two days post-infection, TZM-bls were examined for beta-galactosidase production using Galacto-Light Plus System (Applied Biosystems). Virus stock dilutions in the non-linear range of the TZM-bl assay were discarded. A linear interpolated curve of the relative light units (RLUs) versus supernatant dilution was used to estimate RLU/ul. The AUC was generated from the plot of RLU/ul versus days post infection (Pena-Cruz et al., 2013). Replication kinetics and infectivity but not the genotypic characteristics or other phenotypic properties have been described for 6 of 9 subjects in our previous work (Etemad et al., 2014).
2.4 Co-receptor usage
Co-receptor usage was determined on TZM-bl cells in the presence or absence of CCR5 inhibitor, TAK779, or CXCR4 antagonist, AMD3100. Each virus infection was done in triplicate in a 96 well format under 4 different conditions: 1) without any inhibitor; 2) with 800nM TAK779; 3) with 800nM AMD3100, 3) and with both TAK779 and AMD3100 at 800nM. Two days after virus exposure, RLU values from each well were log transformed. As a first test, a virus was deemed as both infectious and using no other co-receptor, other than CCR5 or CXCR4, if the RLUs in the presence of no inhibitor as compared to the wells with both inhibitors was greater than 0.4 log10 and significantly different (p < 0.05, t-test). No subsequent tests were done if a virus failed this first test. A virus was deemed as exclusively CCR5 tropic if the RLU in the presence of no inhibitor as compared to the wells with TAK779 was greater than 0.4 log10 and significantly different (p < 0.05, t-test). A virus was deemed as exclusively CXCR4 tropic if the RLU in the presence of no inhibitor as compared to the wells with AMD3100 was greater than 0.4 log10 and significantly different (p < 0.05, t-test). A virus stock was deemed dual-mixed if it had a positive result in both the CCR5 and CXCR4 usage tests. In every assay, reference viruses with known co-receptor usage, such as NL4-3, YU-2, and 89.6, were used as controls.
2.5 Neutralization sensitivity
TZM-bl, TAK779, AMD3100, and HIV-1 IgG were obtained through Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (Bjorndal et al., 1997; Wang et al., 1999; Wei et al., 2002). VRC01, PGT121, and 10E8 antibodies were generated by the transfection of 293F cells with immunoglobulin heavy and light chain plasmids containing the appropriate variable loop domains as described previously (Huang et al., 2012; Sagar et al., 2012; Walker et al., 2011; Wu et al., 2010). Infection of TZM-bl cells in the absence and presence of two-fold serial dilution of the antibody was used to estimate the 50% inhibitory concentration (IC50) as previously described (Etemad et al., 2009). All reported IC50s are mean estimates from a minimum of 2 independent assays.
2.6 Statistical analysis
Estimations from SGA and bulk PCR methods were compared using Bland Altman plots and by analyzing summary statistics of their differences. Different estimates were also compared using Spearman correlation coefficients. The neutralization IC50s from 293T virus stocks with either SGA or bulk PCR, and PBMC passaged virus were compared among each other using the matched pairs Wilcoxon rank sum test. All p-values were based on a two-sided test.
3. RESULTS
3.1 Subjects
We chose 9 convenience samples to examine if envelope amplification using either SGA or bulk PCR influenced its genotypic and phenotypic properties. Three of the 9 samples were collected prior to HIV-1 seroconversion, and the remaining 6 samples were from subjects known to be HIV-1 seropositive with infection of unknown duration. No subject was on antiretroviral therapy. As expected the virus levels in the individuals sampled prior to seroconversion (median HIV-1 copies per ml 1,031,929, range 499,713 – 1,289,549) was significantly higher compared to those that were known seropositive (median HIV-1 copies per ml 227,198, range 126,346 – 416,487, p = 0.02) (Table 1).
Table 1.
Subject demographics and envelope V1–V3 genotypic characteristics
| Virus | Stage1 | Plasma virus level (copies /ml) |
# SGA Seq2 |
# Bulk seq2 |
Panmixia Prob. (P) 3 |
Diversity SGA4 |
Diversity Bulk4 |
Divergence SGA5 |
Divergence Bulk5 |
|---|---|---|---|---|---|---|---|---|---|
| A33 | Pre | 1,031,929 | 12 | 12 | 0.48 | 0.0069 | 0.0119 | 0.0264 | 0.0280 |
| A41 | Pre | 1,289,549 | 12 | 12 | 0.91 | 0.0011 | 0.0005 | 0.0019 | 0.0019 |
| A43 | Pre | 499,713 | 12 | 12 | 1 | 0.0005 | 0.0005 | 0.0003 | 0.0003 |
| C | Post | 416,487 | 10 | 11 | 0.24 | 0.0455 | 0.0377 | 0.0355 | 0.0310 |
| D | Post | 152,902 | 12 | 12 | 0.06 | 0.0037 | 0.0089 | 0.0024 | 0.0061 |
| E | Post | 133,850 | 12 | 19 | 0.0002 | 0.0027 | 0.0025 | 0.0017 | 0.0030 |
| F | Post | 326,389 | 8 | 13 | 0.06 | 0.0276 | 0.0235 | 0.0186 | 0.0323 |
| G | Post | 126,346 | 17 | 20 | 0.003 | 0.0122 | 0.0091 | 0.0139 | 0.0138 |
| H | Post | 301,494 | 10 | 20 | 0.22 | 0.0344 | 0.0388 | 0.0452 | 0.0384 |
: Pre: pre-seroconversion, Post: post-seroconversion
: Number of V1–V3 sequences analyzed
: Panmixia test for envelope sequence population structure
: Average genetic distance between any 2 envelope sequences
: Average genetic distance between an envelope sequence and the predicted most recent common ancestor (MRCA).
3.2 Genotypic characteristics
For each sample, full-length envelopes were amplified using both bulk PCR or SGA with the same primer set and cycling conditions (Salazar-Gonzalez et al., 2008). To examine genotypic differences among bulk PCR versus SGA envelopes, we compared the most diverse portion of the HIV-1 envelope, namely the variable loop 1 to variable loop 3 segment (V1 – V3) (Gaschen et al., 2002). A median of 12 (range 8 – 17) SGA and a median of 12 (range 11 – 20) bulk amplified envelope V1–V3 segments were sequenced from each sample (Table 1). Phylogenetic analysis revealed that each individual’s bulk and SGA envelope V1–V3 sequences clustered together (Fig. 1). This suggests that there was no evidence of contamination or super-infection in this small envelope segment at this relatively limited level of sampling. The phylogenetic analysis further revealed that although SGA and bulk PCR cloning often yielded different unique envelope V1–V3 variants, sequences acquired for both methodologies were intermingled. A panmixia test was used to detect differences among SGA and bulk PCR population structure (Achaz et al., 2004; Hudson, Boos, and Kaplan, 1992). Panmixia probability (P) of 1 denotes identical populations while a value less than 1 indicates different populations (Table 1). Four of the 9 subjects (D, E, F, and G) had relatively low P values which are potentially driven by a cluster of sequences selectively found with the bulk PCR methodology (Fig.1). Values for P below or at 0.003 has previously been deemed as significant population structure difference because it accounts for sampling error and multiple nucleotide comparisons (Achaz et al., 2004). Thus, 7 of the 9 subjects had no significant population structure difference among SGA versus bulk PCR sequences. Importantly, Bland Altman plots revealed that the difference in estimated genetic diversity (mean 0.00013, range −0.0052 – 0.0078, standard deviation (SD) 0.0046) and divergence (mean −0.00099, range −0.014 – 0.0068, SD 0.0057) between SGA versus bulk amplified V1–V3 sequences did not appear substantially different, and overall, differences were very small relative to the standard deviation of the differences (Table 1) (Fig. 2). Furthermore, SGA and bulk PCR estimations of genetic diversity (ρ = 0.96) and divergence (ρ = 0.93) were highly correlated among the different subjects. The phylogenetic tree also showed that samples A33, C, E, F, and H potentially harbored recombinant V1–V3 sequences (Fig. 1). Because these potential recombinant sequences were observed with both the SGA and bulk PCR strategy, it is difficult to know whether any or all of these variants are present in the sample or represent a PCR artifact. In aggregate, our results suggest that by sampling a median of 12 sequences, bulk PCR cloning yields similar gross envelope genotypic characteristics as that obtained by examining a median of 12 SGA variants.
Figure 1. Bulk PCR and SGA derived envelope sequences cluster together.
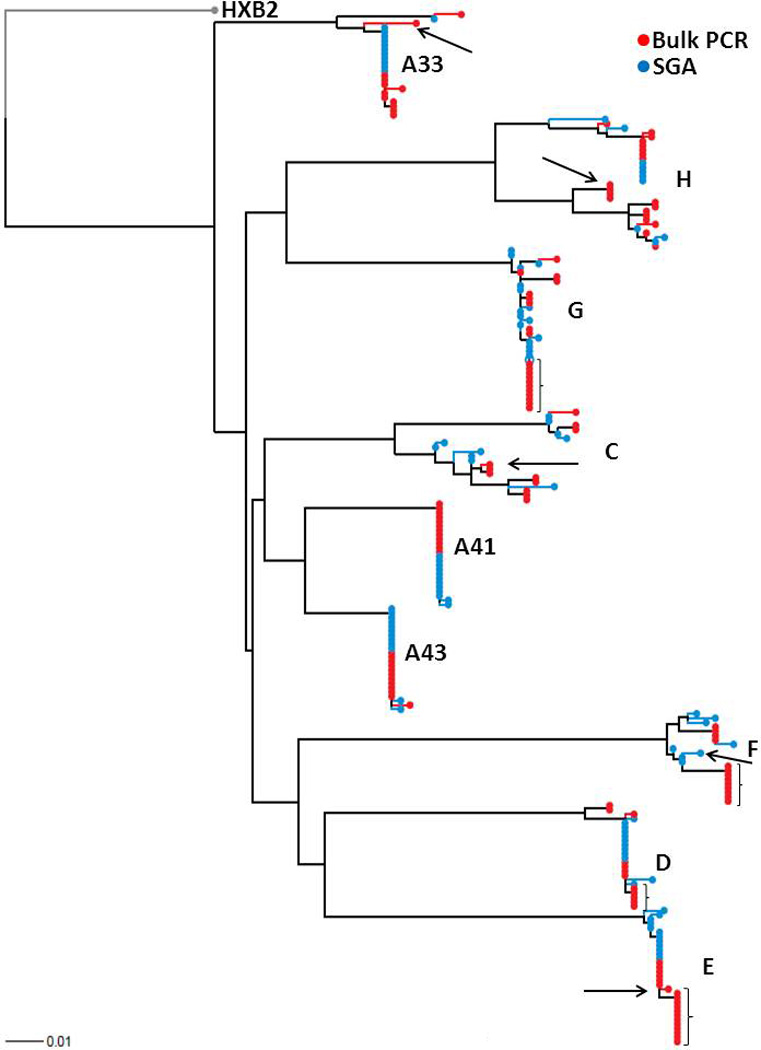
Envelope V1–V3 sequences derived either using bulk PCR (red) and SGA (blue) were aligned with reference sequence HXB2 from the Los Alamos database using Clustal X. Paup was used to generate the maximum likelihood tree using parameters from FindModel best fit evolutionary model. Subject IDs and HXB2 reference outgroup node are noted. Arrows point to potential recombinant sequences within specific subjects. Brackets denote cluster of sequences isolated using the bulk PCR methodology but not SGA. The tree was generated using a web tool at http://www.hiv.lanl.gov/content/sequence/RAINBOWTREE/rainbowtree.html.
Figure 2. Genetic diversity and divergence are similar among envelope V1–V3 sequences derived using either SGA or bulk PCR strategies.
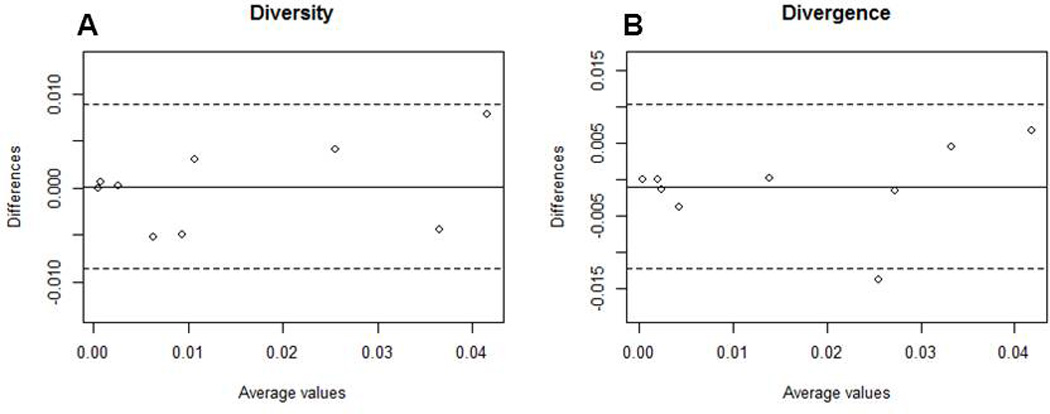
Bland Altman plots of genetic diversity (A) and divergence (B) among sequences obtained from SGA compared to bulk PCR. The solid line shows the mean difference among the 2 methodologies and the dotted lines delineate values at 2 standard deviations of the difference. The mean difference in diversity and divergence was 0.00013 and −0.00099 respectively. The standard deviation in diversity and divergence was 0.0046 and 0.0057 respectively. The x-axis shows the average and the y-axis shows the difference of the value obtained by the 2 methodologies.
3.3 Infectivity and replication kinetics of SGA versus bulk PCR envelope virus stocks
Because bulk PCR cloning as compared to SGA showed envelope variants in different proportions although the population structure was the same in the majority of subjects, we wished to investigate if envelopes generated using these 2 different methods yielded viruses with different phenotypic characteristics. For each sample, we generated independent virus stocks incorporating envelopes amplified using either SGA or bulk PCR. A median of 18 (range 9 – 32) different SGA full-length envelope amplicons were combined and 4 independent bulk PCRs were pooled to generate HIV-1 recombinant clones (Table 2). All SGA derived clones and a median of 106 (range 13 – 258) clones generated from bulk PCR pools were combined to make virus stocks. Virus stocks were generated by transfecting 293T cells with either SGA or bulk PCR envelope recombinant clone pools.
Table 2.
Number of SGAs and clones pooled to generate virus stocks and subsequent phenotypic characteristics
| Virus | # of SGA1 |
# of bulk2 |
Inf – Bulk (p24/IP)2 |
Inf – SGA (p24/IP) 2 |
AUC – Bulk3 |
AUC- SGA3 |
293T SGA4 |
293T Bulk4 |
PBMC Bulk4 |
|---|---|---|---|---|---|---|---|---|---|
| A33 | 32 | 258 | 0.06 | 0.03 | 102815 | 115225 | R5 | R5 | R5 |
| A41 | 18 | 133 | 0.13 | 0.14 | 1361 | 569.2 | R5 | R5 | R5 |
| A43 | 22 | 13 | 0.02 | 0.02 | 44018 | 40350 | R5 | R5 | R5 |
| C | 12 | 51 | 0.03 | 0.02 | 152257 | 95753 | DM | DM | DM |
| D | 16 | 13 | 0.02 | 0.06 | 120278 | 75390 | R5 | R5 | R5 |
| E | 30 | 106 | 0.02 | 0.12 | 126612 | 80576 | R5 | R5 | R5 |
| F | 9 | 167 | 0.08 | 0.01 | 132121 | 137911 | R5 | R5 | R5 |
| G | 31 | 108 | 0.03 | 0.01 | 42317 | 89949 | R5 | R5 | R5 |
| H | 13 | 81 | 0.01 | 0.01 | 149969 | 160434 | R5 | R5 | R5 |
: Number of SGA derived sequences pooled to generate clones; all clones were combined to make the virus stocks with SGA envelopes
: Number of bulk derived clones pooled to generate virus stocks with bulk envelopes
: Inf: Infectivity (p24gag/ul per infectious particle (IP)/ul)
: AUC: area under the replication curve
: Co-receptor tropism: R5 denotes ability to use CCR5 only. DM indicates dual-mixed with the capacity to utilize both CCR5 and CXCR4.
Infectivity and replication kinetic differences among virus stocks incorporating either SGA or bulk PCR envelopes in 3 further subjects (C, D, and H) was combined with previously published data on 6 individuals (Etemad et al., 2014). Infectivity was defined as the pg p24gag content per ul relative to infectious particles (IP) per ul. SGA versus bulk PCR envelope 293T derived virus stock infectivity difference was not significantly different from 0 (mean 0.0022, range −0.070 – 0.10, SD 0.047) (Table 2) (Fig. 3A). SGA and bulk PCR infectivity, however, failed to show a strong correlation (ρ= 0.18). Discarding infectivity data from 1 outlier subject (E), however, showed that the infectivity in the remaining subjects was more strongly correlated (ρ= 0.46).
Figure 3. Virus stocks with either SGA or bulk PCR derived envelopes have similar infectivity and replication kinetics.
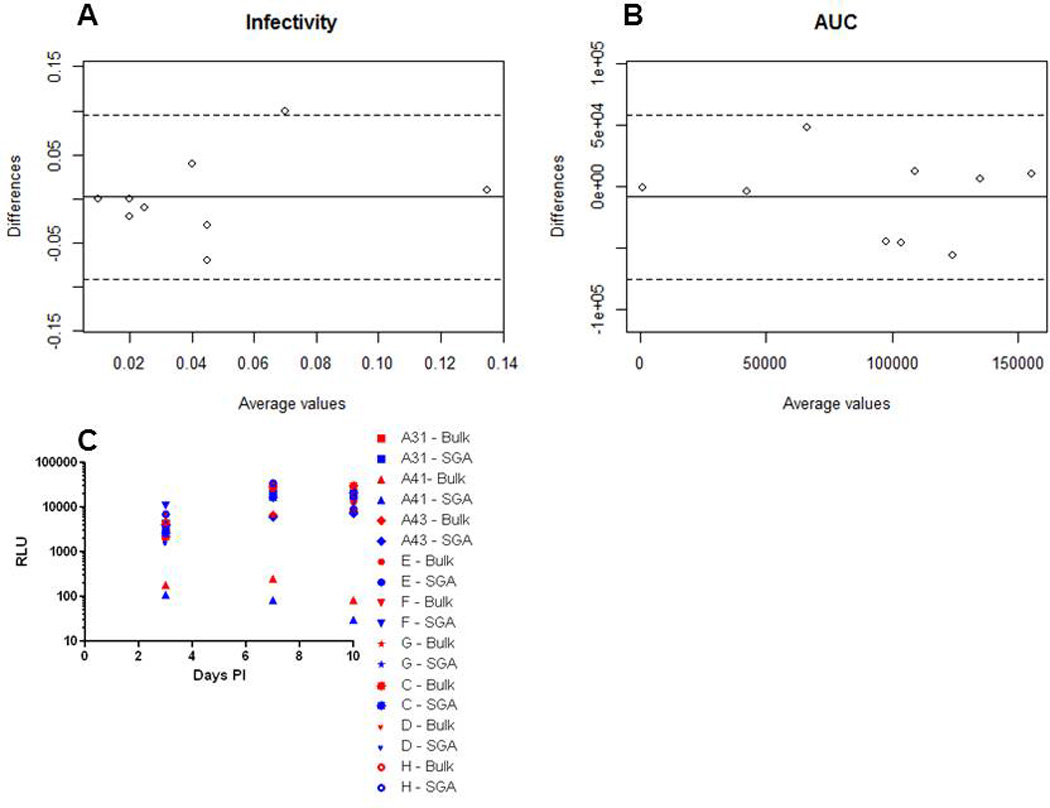
Bland Altman plots of infectivity (A) and replication kinetics (B) among virus stocks with pool of either SGA or bulk PCR envelopes. The solid line shows the mean difference among the 2 methodologies and the dotted lines delineate values at 2 standard deviations of the difference. Graph C shows the relative light units (RLU) per ul (y-axis) generated from infecting TZM-bl cells with virus stocks collected at different days post – infection (x-axis). Replication kinetics was quantified by measuring the area under the curve (AUC) for a plot of RLU per ul of virus stock versus days post-infection on TZM-bl cells at days 3, 7 and 10 post-infection of CD4+ T cells. The mean difference in infectivity and AUC was 0.0022 and −8,399 respectively. The standard deviation in infectivity and AUC was 0.047 and 34,044 respectively. The x-axis shows the average and the y-axis shows the difference of the value obtained by the 2 methodologies.
Replication kinetics was examined by measuring infectious virus in the culture supernatant of CD4+ T cells at multiple times after virus exposure. For these assays, the target cells consisted of CD4+ T cells combined from three different blood donation volunteers to alleviate concerns about host variability. We found that envelopes conferring high replication replicated well and envelopes that led to low replication replicated less well regardless of whether the virus stock contained SGA or bulk PCR amplified envelopes (Table 2) (Fig. 3C). To quantify differences among SGA versus bulk PCR amplified envelopes, we estimated the area under the replication curve (AUC). SGA versus bulk PCR envelope 293T derived virus stock replication AUC difference was not significantly different from 0 (mean −8,399, range −56,500 – 47,630, SD 34,044) (Fig. 3B). In addition, AUC from viruses with bulk amplified envelopes was highly correlated (p = 0.01) to the AUC obtained from HIV-1 with SGA derived envelopes (ρ= 0.68). Thus, the method used to amplify the HIV-1 envelopes does not have significant impact on the replication kinetics of the subsequent virus stock.
3.4 Co-receptor usage
To enhance virus titers and generate more physiologically relevant viruses, 293T derived viruses are often passaged in PBMC cultures. Because virus passage can select for specific virus strains, it is possible this methodology may change virus phenotype (Voronin et al., 2007). To examine this possibility, we passaged 293T virus stocks containing bulk amplified envelopes in PBMC cultures for a maximum of 7 days. We compared the phenotype of 293T virus stocks with SGA or bulk PCR amplified envelopes and PBMC derived viruses. First, we examined co-receptor usage. One subject’s, C, 293T and PBMC derived viruses was classified as dual-mixed (DM) because it demonstrated the ability to infect target cells using either CCR5 or CXCR4 (Table 2). On the other hand, virus stocks from all the other remaining subjects were not able to infect cells in the absence of CCR5, and thus they were classified as CCR5 tropic only (R5). Importantly, in all subjects, the 293T virus stocks incorporating either SGA or bulk amplified envelopes and the PBMC viruses demonstrated the same co-receptor phenotype. Thus, the method of envelope amplification, either SGA or bulk PCR, and virus stock generation, either 293T transfection or PBMC passage, did not alter the co-receptor tropism of the envelope quasispecies.
3.5 Neutralization sensitivity
We further compared the neutralization sensitivity of 293T virus stocks containing either SGA or bulk PCR amplified envelopes and PBMC passaged viruses. Neutralization sensitivity was examined against broadly neutralizing antibodies (bnAbs) that target the CD4 binding site, VRC01, the envelope V3 loop, PGT121, and the trans-membrane proximal external region (MPER), 10E8 (Huang et al., 2012; Walker et al., 2011; Wu et al., 2010) and polyclonal HIV IgG. Neutralization profiles were used to cluster the different virus stocks (Fig. 4) (Table 3). In all 9 different subjects, the 293T virus stocks with either SGA or bulk PCR derived envelopes clustered together based on their neutralization profile. The IC50s for the 293T derived virus stocks with either SGA or bulk PCR envelopes were highly correlated both when IC50s values that exceeded the maximum tested antibody concentration was set as 5 (ρ = 0.90) or when this data was censored (ρ = 0.63).
Figure 4. Virus stocks with either SGA or bulk PCR derived envelopes have similar neutralization profile.
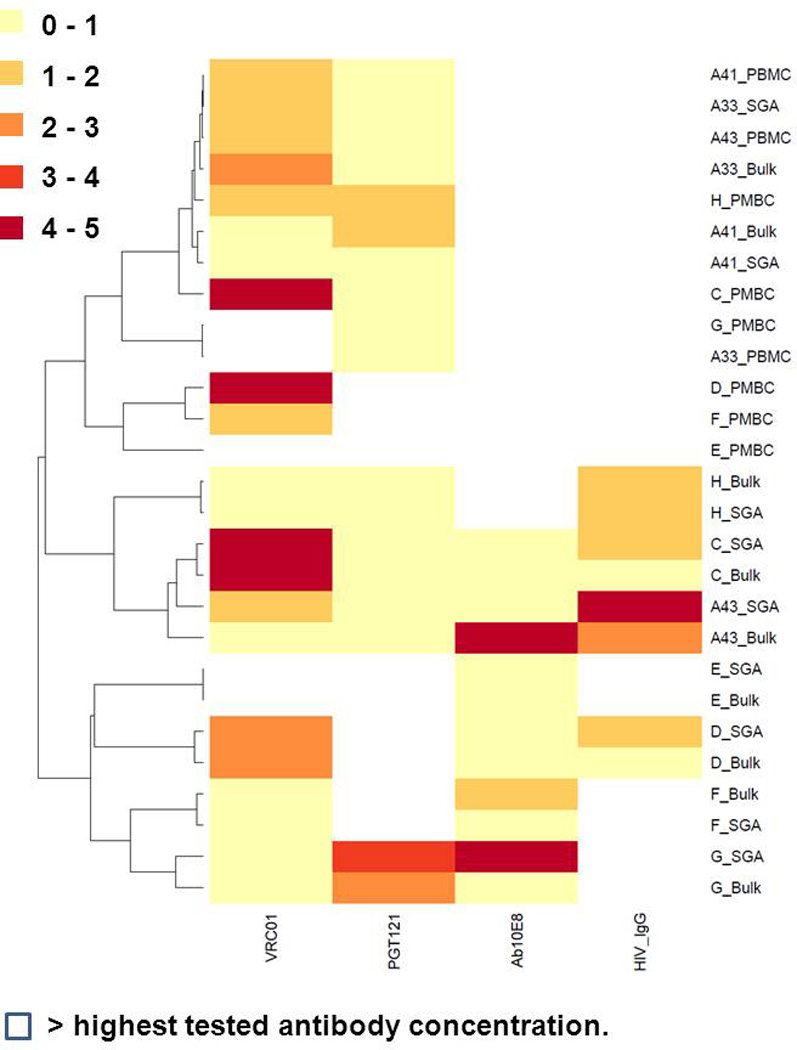
The heatmap shows neutralization IC50s to VRC01, PGT121, 10E8, and HIV IgG among different virus stocks. The different colors used to represent the range of IC50s are displayed on the left. All IC50s represent mean of 2 independent experiments. The IC50s for VRC01, PGT121, and 10E8 are in ug/ml. The HIV IgG IC50s are ×102 ug/ml. For the heatmap, virus stocks that failed to be neutralized by more than 50% at the highest tested antibody concentration (5 ug/ml for VRC01, PGT121, and 10E8 and 500ug/ml for HIV IgG) were deemed above the threshold. The dendogram on the right shows the clustering of the different virus stocks based on the neutralization profile. Hierarchical clustering was used to group the different virus stocks. Heatmaps were generated using web based tools at http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap
Table 3.
Neutralization IC50s of different virus stocks to different antibodies.
| Virus | 293T Bulk VRC01 |
293T SGA VRC01 |
PBMC VRC01 |
293T Bulk 10E8 |
293T SGA 10E8 |
PBMC 10E8 |
293T Bulk PGT121 |
293T SGA PGT121 |
PBMC PGT121 |
293T Bulk HIVIgG |
293T SGA HIVIgG |
PBMC HIVIgG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A33 | 2.25 | 1.92 | >5 | 1.71 | 1.21 | >5 | 0.01 | 0.06 | 0.01 | >500 | >500 | >500 |
| A41 | 0.93 | 0.73 | 0.74 | 1.81 | 1.25 | >5 | 1.25 | 0.82 | 0.47 | >500 | >500 | >500 |
| A43 | 0.63 | 1.73 | 1.74 | 4.43 | 0.02 | >5 | 0.02 | 0.02 | 0.01 | 274.63 | 490.01 | >500 |
| C | 4.26 | 4.91 | 4.93 | 0.03 | 0.06 | >5 | 0.00005 | 0.00008 | 0.02 | 95.95 | 184.07 | >500 |
| D | 2.73 | 2.43 | 4.24 | 0.75 | 0.78 | >5 | >5 | >5 | >5 | 47.97 | 196.98 | >500 |
| E | >5 | >5 | >5 | 0.21 | 0.36 | >5 | >5 | >5 | >5 | >500 | >500 | >500 |
| F | 3.33 | 2.66 | 3.47 | 0.95 | 0.26 | >5 | >5 | >5 | >5 | >500 | >500 | >500 |
| G | 0.09 | 0.001 | >5 | 0.07 | 0.62 | >5 | >5 | >5 | >5 | >500 | >500 | >500 |
| H | 0.51 | 0.96 | 1.95 | >5 | >5 | >5 | >5 | >5 | >5 | 126.75 | 129.79 | >500 |
All IC50s are expressed in ug/ml
In all subjects except 2 (A33 and A41), the PBMC virus stocks did not cluster with their respective 293T generated viruses (Fig. 4). The PBMC virus stocks failed to cluster with their corresponding 293T derived viruses because in general they exhibited less neutralization susceptibility. For instance, all PBMC virus stocks were not neutralized at the highest tested concentration of 10E8 (5 ug/ml) and HIV IgG (500 ug/ml). PBMC virus stocks were significantly more neutralization resistant compared to the corresponding 293T virus with either bulk PCR (p = 0.0006) or SGA envelopes (p = 0.0008) when the IC50s values above the maximum tested antibody concentration was set as 5. If IC50s beyond the maximum threshold were censored in the analysis, the IC50s for the 293T virus stocks with bulk PCR envelopes were significantly correlated to the corresponding PBMC viruses (n = 10, ρ = 0.92) (Fig. 5). This suggests that, in general, PBMC passage increases neutralization resistance in an antibody dependent manner. In specific virus antibody combinations, however, PBMC passage as compared to 293T transfection virus stocks do not have significantly different neutralization susceptibility.
Figure 5. The IC50s for 293T derived and PBMC passage virus stocks are correlated.
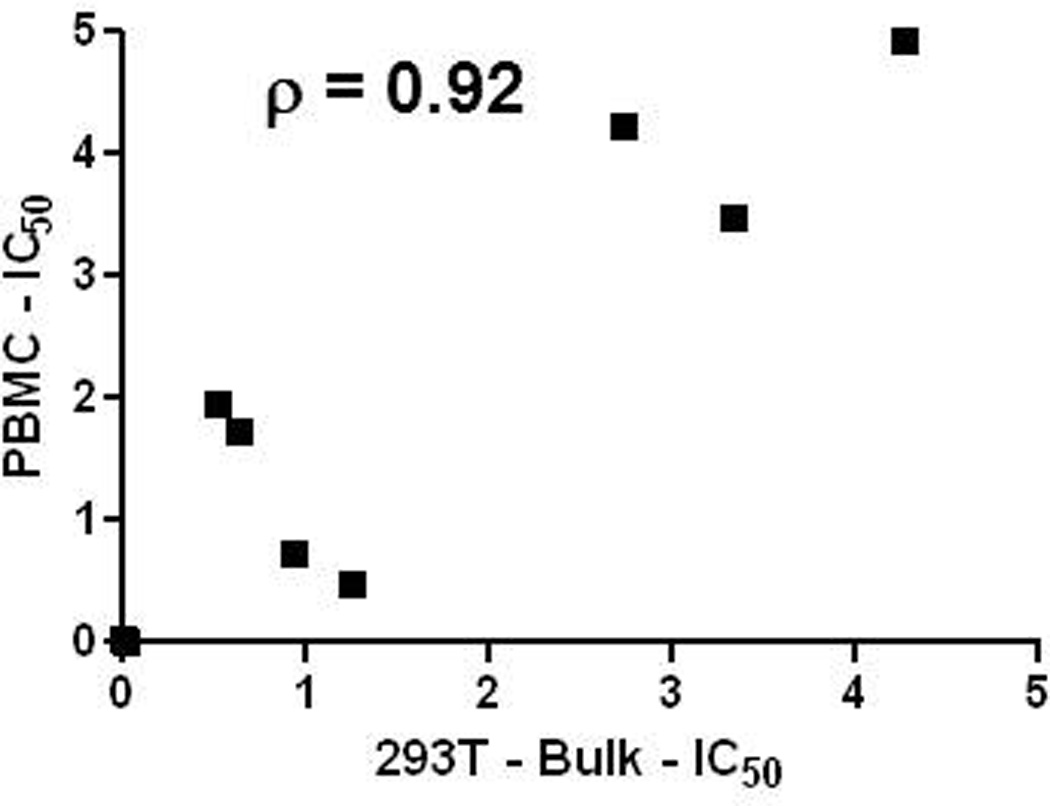
Graph shows IC50s for 293T virus stocks with bulk envelope PCRs (x – axis) compared to short term PBMC passage virus (y-axis). All virus antibody combinations that yielded an IC50 above the maximum tested antibody concentration were not included in this analysis. Graph also shows the estimated correlation coefficient for a best fit linear regression curve.
4. DISCUSSION
Characterizing the HIV-1 envelope quasispecies is important for understanding virus pathogenesis and devising prevention strategies. As compared to the standard bulk PCR cloning, newer studies have employed SGA or limiting dilution PCR to generate HIV-1 envelope amplicons (Sagar et al., 2004; Salazar-Gonzalez et al., 2008). These newer studies that use SGA have questioned the validity of the results from studies that used standard bulk PCR strategy because bulk PCR can introduce polymerase induced recombination and resampling bias. In this study, we show that SGA and bulk PCR derived envelope segments had similar genetic diversity, divergence, and population structure. Viruses with SGA or bulk PCR amplified envelopes have similar infectivity, replication kinetics, co-receptor usage, and neutralization sensitivity. In addition, PBMC passage, which is often used to enhance virus stock titers and generate more physiological viruses, does not change co-receptor usage as compared to 293T virus stocks. On the other hand, PBMC as compared to 293T derived viruses were less neutralization susceptible although this was antibody and virus strain dependent. Our studies suggest that the method of envelope isolation, either SGA or bulk PCR, have minimal impact on the genotypic and phenotypic properties of the virus quasispecies.
Similar to previous studies, we found that SGA and bulk PCR amplification strategies yield envelopes with no significant difference in genetic diversity, divergence, or population structure (Buzon et al., 2014; Jordan et al., 2010). These previous investigations, however, examined either relatively short segments (around 100 base-pairs of the envelope V3 loop) (Buzon et al., 2014) or relatively conserved portions (pro-pol gene) (Jordan et al., 2010) of the HIV-1 genome. In addition, the envelope V3 loop is relatively conserved compared to other variable portions of the HIV-1 envelope (Andrabi et al., 2012; Gaschen et al., 2002; Lin et al., 2012). In contrast to these studies, we examined approximately 700 bp of the envelope V1–V3 segment, which is the most diverse part of the HIV-1 genome (Gaschen et al., 2002). Examination of relatively long highly polymorphic sequences will be better at identifying differences among the two amplification methods. In addition, we studied subjects both prior to seroconversion who had extremely limited viral diversity and individuals sampled during the chronic phase of infection with more extensive number of variants (Sagar, 2010). Both amplification methods yielded similar envelope genotypic characteristics after sampling an average 12 sequences. In conjunction with the previous studies (Buzon et al., 2014; Jordan et al., 2010), we conclude that SGA and bulk PCR yield similar genotypic characteristics regardless of the genetic diversity in the viral segment of interest.
To our knowledge, no study has examined phenotypic differences among virus stocks with either bulk PCR or SGA derived envelopes. One would hypothesize that the envelope phenotypes should be not be different because SGA and bulk PCR yielded similar sequences. In our studies, however, virus stocks incorporated a much larger number of envelope products compared to the number of sequences examined for the genotypic analysis. If the differences in the proportion of viral variants among SGA versus bulk PCR cannot be observed by examining an average of 12 sequences, incorporating a larger number of envelope products in the virus stocks will potentially identify a significant frequency bias. We found no significant difference in infectivity, replication kinetics, co-receptor usage, and neutralization sensitivity among 293T transfection generated virus stocks with either SGA or bulk PCR derived envelopes. While, it is quite possible that individual envelopes isolated using SGA or bulk PCR may differ in the phenotype of interest, there is no evidence that the characteristics of the quasispecies differ. Indeed, viruses incorporating a pool of either SGA or bulk PCR envelopes may have similar phenotypes because the property of interest may be primarily determined by a small number of variants that are preferentially amplified using either methodology. Thus, pooling around 18 independent SGA envelope amplicons will yield virus quasispecies with similar phenotypic characteristics as pooling amplicons from 4 bulk PCRs.
We also compared co-receptor usage and neutralization sensitivity among virus stocks generated by 293T transfections and short term PBMC passage of 293T virus supernatants. It is possible that PBMC cultures may alter the virus phenotype because in-vitro passage potentially selects for specific variants. We found that virus stocks from 293T stocks had the same co-receptor usage as compared to viruses from PBMC passage. This conclusion, however, is limited because we only had 1 subject in which we isolated envelopes that could use both the CCR5 and CXCR4 co-receptor. In this 1 subject, short term PBMC passage did not select for or against viruses with the ability to use a specific receptor.
In contrast to co-receptor usage, we found that PBMC generated virus stocks had lower neutralization susceptibility as compared to 293T transfection derived viruses. Our results corroborate findings from previous publications that also found that PBMC generated viruses as compared to 293T transfection stocks have decreased sensitivity to neutralizing antibodies (Louder et al., 2005; Mann et al., 2009; Provine et al., 2012). Similar to these studies, we also found that neutralization susceptibility differences between 293T transfection virus stocks and PBMC passaged virus varied between different antibodies and were virus specific. In contrast to these previous studies, we examined neutralization susceptibility of virus quasispecies as opposed to individual variants. Short term PBMC passage of both viral quasispecies and individual variants have decreased neutralization susceptibility compared to the 293T transfection virus. This suggests that the changes likely do not occur because of the enrichment of specific variants. Indeed, in a previous study, we have shown that short duration PBMC passage virus stocks contain similar level of envelope genotypic diversity as evident in bulk PCR clones, and there was no evidence of selection sweep (Pena-Cruz et al., 2013). Rather differences in neutralization sensitivity are potentially related to changes in envelope content or types and density of glycosylation among the viruses from the 2 different producer cells.
5. CONCLUSIONS
In summary, we found that the method of envelope amplification, either SGA or bulk PCR, did not have a significant impact on the genotype or phenotype of the quasispecies. Thus, pooling 4 independent bulk PCRs and subsequently sampling around 12 sequences should yield similar genotypic and phenotypic characteristics as the SGA methodology. Therefore, the more expensive and labor intensive SGA methodology does not necessarily have any superiority over the standard bulk PCR strategy as long as the goal is to examine characteristics of the quasispecies rather than an individual variant. Furthermore, while one can compare neutralization sensitivities among virus stocks that contain either SGA or bulk amplified envelopes, one cannot compare relative neutralization differences among viruses generated from different producer cells.
ACKNOWLEDGEMENTS
We thank all the subjects who have contributed samples for these studies. This study was supported by NIH grants AI077473 (MS) and AI102774 (MS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achaz G, Palmer S, Kearney M, Maldarelli F, Mellors JW, Coffin JM, Wakeley J. A robust measure of HIV-1 population turnover within chronically infected individuals. Molecular biology and evolution. 2004;21:1902–1912. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- Andrabi R, Kumar R, Bala M, Nair A, Ss P, Kushwaha V, Luthra K. Envelope diversity, characteristics of V3 region and predicted co-receptor usage of human immunodeficiency viruses infecting north Indians. J Microbiol. 2012;50:869–873. doi: 10.1007/s12275-012-2136-z. [DOI] [PubMed] [Google Scholar]
- Binley JM, Ban YE, Crooks ET, Eggink D, Osawa K, Schief WR, Sanders RW. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. Journal of virology. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. Journal of virology. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nature medicine. 2014;20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziandreou N, Arauz AB, Freitas I, Nyein PH, Fenton G, Mehta SH, Kirk GD, Sagar M. Sensitivity changes over the course of infection increases the likelihood of resistance against fusion but not CCR5 receptor blockers. AIDS research and human retroviruses. 2012;28:1584–1593. doi: 10.1089/aid.2011.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science (New York, N.Y. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Dudley DM, Gao Y, Nelson KN, Henry KR, Nankya I, Gibson RM, Arts EJ. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques. 2009;46:458–467. doi: 10.2144/000113119. [DOI] [PubMed] [Google Scholar]
- Etemad B, Fellows A, Kwambana B, Kamat A, Feng Y, Lee S, Sagar M. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. Journal of virology. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad B, Gonzalez OA, White L, Laeyendecker O, Kirk GD, Mehta S, Sagar M. Characterization of HIV-1 envelopes in acutely and chronically infected injection drug users. Retrovirology. 2014;11:106. doi: 10.1186/s12977-014-0106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. Diversity considerations in HIV-1 vaccine selection. Science (New York, N.Y. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Molecular biology and evolution. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Jordan MR, Kearney M, Palmer S, Shao W, Maldarelli F, Coakley EP, Chappey C, Wanke C, Coffin JM. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. Journal of virological methods. 2010;168:114–120. doi: 10.1016/j.jviromet.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. Journal of virology. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NH, Becerril C, Giguel F, Novitsky V, Moyo S, Makhema J, Essex M, Lockman S, Kuritzkes DR, Sagar M. Env sequence determinants in CXCR4-using human immunodeficiency virus type-1 subtype C. Virology. 2012;433:296–307. doi: 10.1016/j.virol.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. HIV quasispecies and resampling. Science (New York, N.Y. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Louder MK, Sambor A, Chertova E, Hunte T, Barrett S, Ojong F, Sanders-Buell E, Zolla-Pazner S, McCutchan FE, Roser JD, Gabuzda D, Lifson JD, Mascola JR. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology. 2005;339:226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Mann AM, Rusert P, Berlinger L, Kuster H, Gunthard HF, Trkola A. HIV sensitivity to neutralization is determined by target and virus producer cell properties. AIDS (London, England) 2009;23:1659–1667. doi: 10.1097/QAD.0b013e32832e9408. [DOI] [PubMed] [Google Scholar]
- Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, Ogundare O, Pascuccio M, Patel N, Wei D, Fauci AS, Arthos J. The genotype of early-transmitting HIV gp120s promotes alpha (4) beta(7)-reactivity, revealing alpha (4) beta(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS pathogens. 2011;7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Cruz V, Etemad B, Chatziandreou N, Nyein PH, Stock S, Reynolds SJ, Laeyendecker O, Serwadda D, Gray RH, Lee S, Quinn TC, Sagar M. HIV-1 envelope replication and α4β7 utilization among newly infected subjects and their corresponding heterosexual partners. Retrovirology. 2013;10:162. doi: 10.1186/1742-4690-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine NM, Cortez V, Chohan V, Overbaugh J. The neutralization sensitivity of viruses representing human immunodeficiency virus type 1 variants of diverse subtypes from early in infection is dependent on producer cell, as well as characteristics of the specific antibody and envelope variant. Virology. 2012;427:25–33. doi: 10.1016/j.virol.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M, Takahashi K, Czernekova L, Zachova K, Hall S, Moldoveanu Z, Elliott MC, Wilson L, Brown R, Jancova D, Barnes S, Vrbkova J, Tomana M, Smith PD, Mestecky J, Renfrow MB, Novak J. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M. HIV-1 transmission biology: selection and characteristics of infecting viruses. The Journal of infectious diseases. 2010;202(Suppl 2):S289–S296. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Akiyama H, Etemad B, Ramirez N, Freitas I, Gummuluru S. Transmembrane Domain Membrane Proximal External Region but Not Surface Unit-Directed Broadly Neutralizing HIV-1 Antibodies Can Restrict Dendritic Cell-Mediated HIV-1 Trans-infection. The Journal of infectious diseases. 2012;205:1248–1257. doi: 10.1093/infdis/jis183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Kirkegaard E, Long EM, Celum C, Buchbinder S, Daar ES, Overbaugh J. Human immunodeficiency virus type 1 (HIV-1) diversity at time of infection is not restricted to certain risk groups or specific HIV-1 subtypes. Journal of virology. 2004;78:7279–7283. doi: 10.1128/JVI.78.13.7279-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer MJ, Gray RH, Serwadda D, Sewankambo NK, Shepherd JC, Toma J, Huang W, Quinn TC. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. The Journal of infectious diseases. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. HIV-1 V1–V2 envelope loop sequences expand and add glycosylation sites over the course of infection and these modifications affect antibody neutralization sensitivity. Journal of virology. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. Journal of virology. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahov D, Graham N, Hoover D, Flynn C, Bartlett JG, Margolick JB, Lyles CM, Nelson KE, Smith D, Holmberg S, Farzadegan H. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA. 1998;279:35–40. doi: 10.1001/jama.279.1.35. [DOI] [PubMed] [Google Scholar]
- Voronin Y, Chohan B, Emerman M, Overbaugh J. Primary isolates of human immunodeficiency virus type 1 are usually dominated by the major variants found in blood. Journal of virology. 2007;81:10232–10241. doi: 10.1128/JVI.01035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Sawyer LS, Murthy KK, Fang X, Walfield AM, Ye J, Wang JJ, Chen PD, Li ML, Salas MT, Shen M, Gauduin MC, Boyle RW, Koup RA, Montefiori DC, Mascola JR, Koff WC, Hanson CV. Postexposure immunoprophylaxis of primary isolates by an antibody to HIV receptor complex. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10367–10372. doi: 10.1073/pnas.96.18.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrobial agents and chemotherapy. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (New York, N.Y. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]


