Abstract
The Adhesion family forms a large branch of the pharmacologically important superfamily of G protein–coupled receptors (GPCRs). As Adhesion GPCRs increasingly receive attention from a wide spectrum of biomedical fields, the Adhesion GPCR Consortium, together with the International Union of Basic and Clinical Pharmacology Committee on Receptor Nomenclature and Drug Classification, proposes a unified nomenclature for Adhesion GPCRs. The new names have ADGR as common dominator followed by a letter and a number to denote each subfamily and subtype, respectively. The new names, with old and alternative names within parentheses, are: ADGRA1 (GPR123), ADGRA2 (GPR124), ADGRA3 (GPR125), ADGRB1 (BAI1), ADGRB2 (BAI2), ADGRB3 (BAI3), ADGRC1 (CELSR1), ADGRC2 (CELSR2), ADGRC3 (CELSR3), ADGRD1 (GPR133), ADGRD2 (GPR144), ADGRE1 (EMR1, F4/80), ADGRE2 (EMR2), ADGRE3 (EMR3), ADGRE4 (EMR4), ADGRE5 (CD97), ADGRF1 (GPR110), ADGRF2 (GPR111), ADGRF3 (GPR113), ADGRF4 (GPR115), ADGRF5 (GPR116, Ig-Hepta), ADGRG1 (GPR56), ADGRG2 (GPR64, HE6), ADGRG3 (GPR97), ADGRG4 (GPR112), ADGRG5 (GPR114), ADGRG6 (GPR126), ADGRG7 (GPR128), ADGRL1 (latrophilin-1, CIRL-1, CL1), ADGRL2 (latrophilin-2, CIRL-2, CL2), ADGRL3 (latrophilin-3, CIRL-3, CL3), ADGRL4 (ELTD1, ETL), and ADGRV1 (VLGR1, GPR98). This review covers all major biologic aspects of Adhesion GPCRs, including evolutionary origins, interaction partners, signaling, expression, physiologic functions, and therapeutic potential.
I. Introduction
G protein–coupled receptors (GPCRs) consist of five main families in mammals, the largest being the Rhodopsin family, or class A, with about 284 members (plus about 380 olfactory receptors) in humans, followed by the Adhesion GPCR family with 33 members, and then the Glutamate family (class C), Secretin family (class B), and Frizzled family, with 22, 15, and 11 members, respectively (Civelli et al., 2013). Originally, it was suggested that the Adhesion GPCRs belong to class B (Baud et al., 1995; Hamann et al., 1995, 1996a), but evidence emerged that they are different from the Secretin receptors in many aspects, including their unique autocatalytic processing, their multitude of domains in the often long N termini, their evolutionary conservation, and their roles in cell–cell and cell–matrix adhesion. This is in contrast to the Secretin GPCRs, which are not autocatalytically processed and often mediate hormonal responses.
Different groups of researchers commonly studying the Adhesion GPCRs with epidermal growth factor (EGF) domains within their N termini started a series of workshops that was the foundation for the current larger Adhesion GPCR Consortium (http://www.adhesiongpcr.org/) and the biennial Adhesion GPCR Workshops (e.g., Arac et al., 2012a). The Adhesion GPCR Consortium has also worked to establish descriptions of the Adhesion GPCRs for the International Union of Basic and Clinical Pharmacology (IUPHAR)/British Society for Pharmacology (BPS) Guide to Pharmacology (http://www.guidetopharmacology.org/). Therein, general gene and protein information on all 33 human Adhesion GPCRs, including data on binding partners, transduction mechanisms, tissue distribution, functional assays, physiologic functions, relevant mutations, and involvement in pathophysiology, are provided. This review, written by members of the Adhesion GPCR Consortium, extends this effort toward a comprehensive description of the Adhesion GPCRs in relation to human health and disease, and their prospects as pharmacological interventions, which is a hallmark of the GPCR superfamily.
II. Recommended Nomenclature
Adhesion GPCR nomenclature has been highly diverse for many historical reasons. Initial names, like CELSR (cadherin EGF LAG seven-pass G-type receptor), EGF-TM7 (epidermal growth factor–seven-span transmembrane), BAI (brain-specific angiogenesis inhibitor), VLGR (very large GPCR), and others, were created by pioneers of this research field, but without harmonization with regard to nomenclature efforts. In collaboration with the Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC), about half of the Adhesion GPCR genes were assigned GPR# names (Fredriksson et al., 2002, 2003a), but these were regarded as temporary identifiers until more information could be elucidated about the protein products. Today, we know that the Adhesion GPCRs are a unique set of proteins that share fundamental structural properties. The research field has expanded, and the high use of genome-wide efforts (omics), including studies on genomics/genetics, expression, and epigenetics, calls for a naming system that clearly illustrates the relationship between these proteins/genes for a wide range of researchers. The IUPHAR Committee on Receptor Nomenclature and Drug Classification (NC-IUPHAR) has noted this high level of name diversity and ambiguity. Therefore, NC-IUPHAR took the initiative to develop a new nomenclature and contacted members of the Adhesion GPCR Consortium, who subsequently worked on an alternative naming system guided by HGNC. We aimed to give the Adhesion GPCRs a prefix that identifies any Adhesion GPCR homolog, independent of species or subfamily. Such a coherent and systematic naming system will help to name orthologs and other genetic variants in different species in the future. This is particularly important as Adhesion GPCRs are found in a wide range of species, where they have important biologic functions (see section VIII).
The prefix that was found to be the most appropriate and unique was ADGR, standing for Adhesion G protein–coupled receptor. Each subfamily was then assigned a letter to relate to previous names, such as L for the latrophilins, E for the EGF-TM7 receptors, C for the CELSRs, B for the BAIs, and V for VLGR, while the subfamilies with GPR# names have been given a letter in alphabetic order (A, D, F, G). These subfamilies, formerly indicated with Roman numerals I–IX, were defined on the basis of previous phylogenetic classifications according to the molecular signature of the 7TM regions and extracellular regions (Bjarnadóttir et al., 2004; Lagerström and Schiöth, 2008; Langenhan et al., 2013). The members within each subfamily received a number in an order similar to that of former names. This nomenclature (Fig. 1; Table 1) has been accepted by both HGNC and NC-IUPHAR, and both organizations encourage the use of this nomenclature in all literature and databases. We suggest that the new nomenclature be used together with the old names, for example “ADGRE5 (CD97)” or “ADGRL1 (latrophilin-1),” until the new names are fully established. It is important that these new names are mentioned in abstracts of scientific papers to ensure that researchers outside of the field can easily identify the relevant literature.
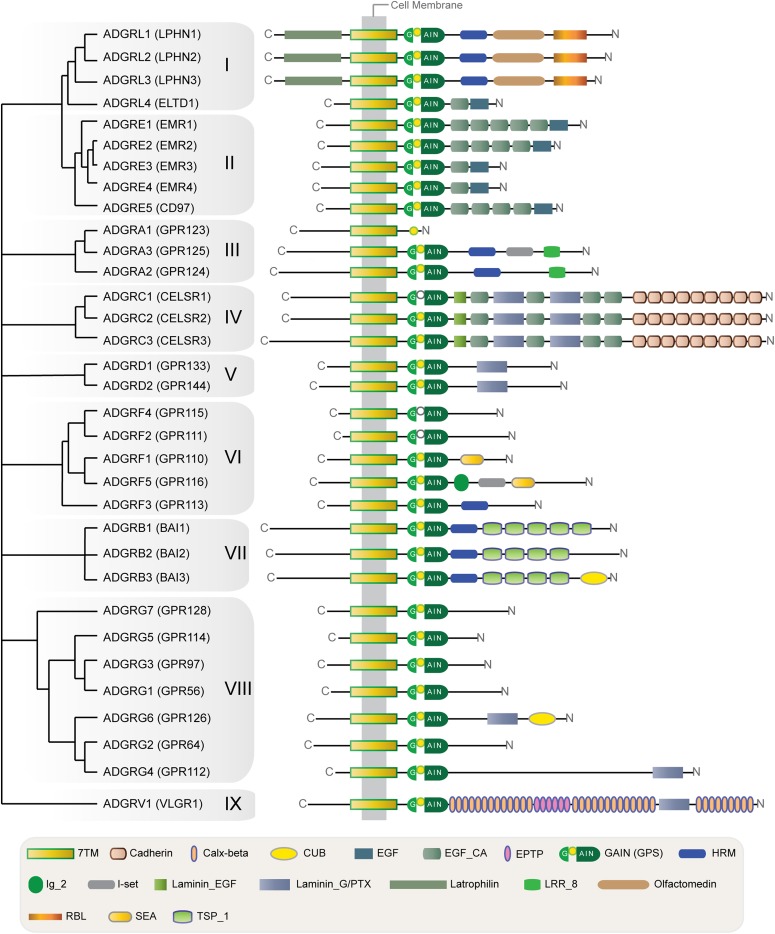
Fig. 1.
Representation of the phylogenetic relationships of Adhesion GPCRs and their N-terminal domain architecture. Adhesion GPCRs can be subdivided into nine distinct subfamilies with 33 homologs in the human genome on the basis of phylogenetic analysis of the conserved 7TM domain region. Each subfamily has a unique combination of N-terminal domains and varies widely in length of their N termini. The conserved sequence motif within the GPS, found within the GAIN domain of most Adhesion GPCRs, mediates autoproteolysis and subsequent attachment of the cleaved NTF and CTF fragments. All subfamilies share a GAIN and 7TM domain (Adhesion GPCR signature), whereas the depicted N-terminal functional domains are not present in every homolog of the subfamilies. ADGRA1 (GPR123) is exceptional as it is the only mammalian adhesion GPCR that does not contain a GAIN domain. Of note, improper GPS motifs that (may) preclude cleavage are found in more Adhesion GPCRs. Open GPS symbols indicate Adhesion GPCRs that do not undergo cleavage, according to experimental evidence. The tree topology shown on the left was obtained from consensus of previous phylogenetic studies of Adhesion GPCRs (Bjarnadóttir et al., 2004; Gloriam et al., 2007; Haitina et al., 2009). It should be noted that the hierarchy within some of these divergent subfamilies, such as VI (ADGRFs) and VIII (ADGRGs), may vary in a few nodes, depending on the methodology, dataset, and parameters used in each phylogenetic analysis. Calx, calnexin; CUB, Cs1 and Csr/Uegf/BMP1; EGF_CA, calcium-binding EGF; EPTP, epitempin; HRM, hormone receptor motif; I-set, immunoglobulin I-set domain; LRR, leucine-rich repeat; PTX, pentraxin; RBL, rhamnose-binding lectin; SEA, sperm protein, enterokinase, agrin module; TSP, thrombospondin.
TABLE 1.
Proposed new nomenclature for Adhesion GPCRs
| Subfamily | Receptor | |||
|---|---|---|---|---|
| Current Number |
Proposed New Name |
Current Gene Name |
Current Protein Name
(Alternative Names) |
Proposed New Name |
| I | L (Latrophilin) | LPHN1 | Latrophilin 1 (CIRL-1, CL1, LEC2) | ADGRL1 |
| LPHN2 | Latrophilin 2 (CIRL-2, CL2, LPHH1, LEC1) | ADGRL2 | ||
| LPHN3 | Latrophilin 3 (CIRL-3, CL3, LEC3) | ADGRL3 | ||
| ELTD1 | ELTD1 (ETL) | ADGRL4 | ||
| II | E (EGF-TM7) | EMR1 | EMR1 (F4/80) | ADGRE1 |
| EMR2 | EMR2 (CD312) | ADGRE2 | ||
| EMR3 | EMR3 | ADGRE3 | ||
| EMR4 | EMR4 (FIRE, GPR127) | ADGRE4 | ||
| CD97 | CD97 (BL-Ac(F2)) | ADGRE5 | ||
| III | A | GPR123 | GPR123 | ADGRA1 |
| GPR124 | GPR124 (TEM5) | ADGRA2 | ||
| GPR125 | GPR125 | ADGRA3 | ||
| IV | C (CELSR) | CELSR1 | CELSR1 | ADGRC1 |
| CELSR2 | CELSR2 (MEGF3) | ADGRC2 | ||
| CELSR3 | CELSR3 (MEGF2, Fm1, EGFL1) | ADGRC3 | ||
| V | D | GPR133 | GPR133 | ADGRD1 |
| GPR144 | GPR144 | ADGRD2 | ||
| VI | F | GPR110 | GPR110 | ADGRF1 |
| GPR111 | GPR111 | ADGRF2 | ||
| GPR113 | GPR113 | ADGRF3 | ||
| GPR115 | GPR115 | ADGRF4 | ||
| GPR116 | GPR116 (Ig-Hepta) | ADGRF5 | ||
| VII | B (BAI) | BAI1 | BAI1 | ADGRB1 |
| BAI2 | BAI2 | ADGRB2 | ||
| BAI3 | BAI3 | ADGRB3 | ||
| VIII | G | GPR56 | GPR56 (TM7XN1) | ADGRG1 |
| GPR64 | GPR64 (HE6) | ADGRG2 | ||
| GPR97 | GPR97 (Pb99) | ADGRG3 | ||
| GPR112 | GPR112 | ADGRG4 | ||
| GPR114 | GPR114 | ADGRG5 | ||
| GPR126 | GPR126 (VIGR, DREG) | ADGRG6 | ||
| GPR128 | GPR128 | ADGRG7 | ||
| IX | V | GPR98 | VLGR1 (GPR98, MASS1, USH2C, FEB4) | ADGRV1 |
BAI, brain-specific angiogenesis inhibitor; CD, cluster of differentiation; CELSR, cadherin EGF LAG seven-pass G-type receptor; CIRL, calcium-independent receptor of α-latrotoxin; CL, CIRL/latrophilin; DREG, developmentally regulated GPCR; EGFL, epidermal growth factor–like; EGF-TM7, epidermal growth factor–seven-span transmembrane; ELTD, EGF, latrophilin and seven transmembrane domain–containing protein; EMR, EGF-like molecule containing mucin-like hormone receptor; ETL, EGF-TM7-latrophilin–related protein; FEB, febrile seizures gene disease locus; FIRE, F4/80-like receptor; Fm, Flamingo; HE, human epididymal; LEC, lectomedin; LPHH, latrophilin homolog in humans; MASS, monogenic audiogenic seizure susceptibility; MEGF, multiple epidermal growth factor–like domains; TEM, tumor endothelial marker; VIGR, vascular inducible GPCR; VLGR, very large GPCR.
While the full version of the new names may be somewhat cumbersome to pronounce, for example at conferences, it is likely that the shorthand versions, such as A1, B3, F6, V1, etc., will be widely used during oral presentations when there is no doubt that it is the Adhesion GPCRs that are being addressed. Such shorthand versions may also be very useful for tables and figures where space is limited and clarification can be provided in the legends and notes.
III. Taxonomy and Evolutionary Origin
Adhesion GPCRs are of ancient origin and found in all vertebrates (Fredriksson and Schiöth, 2005), the closest relatives to the vertebrates (Kamesh et al., 2008; Nordstrom et al., 2008), primitive animals (Putnam et al., 2007; Nordstrom et al., 2011), the most ancient metazoan phyla (Srivastava et al., 2010), and also in unicellular metazoan relatives, such as Capsaspora owczarzaki and Monosiga brevicollis (Krishnan et al., 2012). Recently, Adhesion GPCRs with short extracellular regions have been identified in several basal fungi, indicating that the Adhesion family is likely to have evolved before the split of unikonts from the common ancestor of eukaryotes about 1275 million years ago (Krishnan et al., 2012). Adhesion GPCRs, like other GPCR families, were proposed to have evolved from a common ancestor with the ancient cAMP receptors (Nordstrom et al., 2011; Krishnan et al., 2012) typically found in Dictyostelium discoideum. Furthermore, analysis of metazoan GPCR data sets indicates that Adhesion GPCRs are ancestral to the Secretin family, as Secretin GPCRs probably diverged from a specific family of Adhesion GPCRs (Nordstrom et al., 2009). Intriguingly, Adhesion GPCR gene-mining studies revealed a vast array of N-terminal domains that includes both well known domains, like cadherin, EGF, and immunoglobulin, as well as domains that are unusual to Adhesion GPCRs, such as R (scavenger receptor cysteine-rich) and Kringle, that are possibly the evolutionary result of lineage-specific innovations (Nordstrom et al., 2008). Interestingly, the GPCR autoproteolysis–inducing (GAIN) domain is evolutionarily well conserved among Adhesion GPCRs and seems to be present from tetrahymena to mammals (Arac et al., 2012b; Prömel et al., 2013).
As described in the previous section, human Adhesion GPCRs are classified into nine distinct subfamilies (Bjarnadóttir et al., 2004). From an evolutionary perspective, potential homologs for genes belonging to subfamilies ADGRL (latrophilins), ADGRA, ADGRC (CELSRs), ADGRD, ADGRG, and ADGRV (GPR98) are present in most invertebrates, including ascidians (Kamesh et al., 2008), lancelets (Nordstrom et al., 2008), acorn worms (Krishnan et al., 2013), and cnidarians (Nordstrom et al., 2009), whereas the subfamilies ADGRE (EGF-TM7), ADGRF, and ADGRB (BAIs) are likely restricted to vertebrates (see Fig. 1) (Kwakkenbos et al., 2004, 2006; Yona et al., 2008b; Strotmann et al., 2011). However, there are also Adhesion GPCR genes in the genomes of choanoflagellates and fungi that cannot be classified according to the specific metazoan families (Krishnan et al., 2012). These likely represent ancestral versions of Adhesion GPCRs that acquired more specified functions during the course of metazoan multicellularity evolution. Several gene-mining studies have also delineated the early evolution and diversification of characteristic extracellular Adhesion GPCR motifs and domains, with as primary examples the emergence of the GPCR proteolysis site (GPS) and Calx_beta domains in the unicellular filasterean C. owczarzaki and the appearance of EGF_CA domains in free-living unicellular organisms, such as the choanoflagellate Salpingoeca rosetta (Krishnan et al., 2012).
Overall, Adhesion GPCRs have an ancient origin, and they are likely to be one of the evolutionary innovations that allowed cells to adhere to and communicate with one another during the evolution of metazoan multicellularity. At the same time, the evolution of ADGRE (EGF-TM7) subfamily members is a recent and ongoing process. Human ADGRE2 (EMR2) and ADGRE3 (EMR3) have no murine orthologs (Kwakkenbos et al., 2006), whereas human ADGRE4 (EMR4) is probably a pseudogene attributable to a single-nucleotide deletion acquired after divergence from the great apes (Hamann et al., 2003; Caminschi et al., 2006). Moreover, ADGRE2 (EMR2) arose in early placental mammals and has since coevolved with ADGRE5 (CD97) and ADGRE3 (EMR3) through regional gene conversion (Kwakkenbos et al., 2006). As a consequence, ADGRE2 (EMR2) is a chimeric molecule with a 7TM region highly similar to ADGRE3 (EMR3) and EGF-like domains almost identical to ADGRE5 (CD97) (Lin et al., 2000).
IV. Receptor Terminology
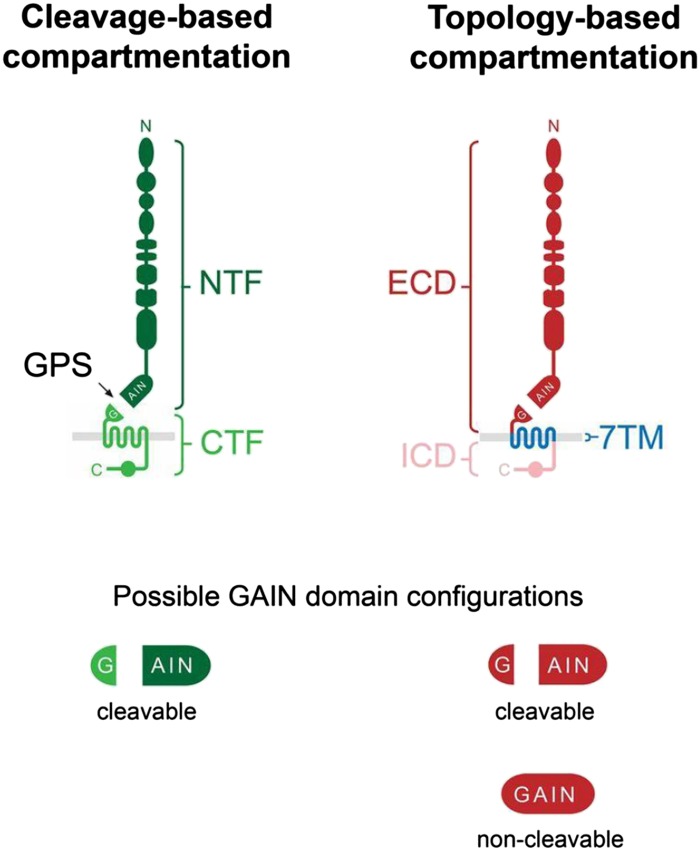
As introduced above, Adhesion GPCRs display a peculiar molecular architecture. Besides the canonical 7TM domain, which is shared by all GPCRs, Adhesion GPCRs are distinguished from other GPCR families in that they mostly possess large N and C termini (Bjarnadóttir et al., 2004) and a GAIN domain (Arac et al., 2012b). With reference to topological considerations regarding membrane-spanning proteins, Adhesion GPCRs can be divided into an extracellular domain (ECD), a 7TM domain, and an intracellular domain (ICD; Fig. 2) (Langenhan et al., 2013; Liebscher et al., 2013).
Fig. 2.
Cleavage- and topology-based compartmentation of Adhesion GPCR architecture. Most Adhesion GPCRs undergo autoproteolysis at the GPS within their GAIN domain, resulting in a two-partite structure containing the NTF and the CTF (left). Alternatively, the protein layout of all Adhesion GPCRs is marked by a three-partite structure consisting of an ECD, a 7TM domain, and an ICD.
The N-termini of most Adhesion GPCRs contain, in varying copy number, multiple types of protein domains that are associated with adhesive functions, which makes many Adhesion GPCRs exceptionally large membrane-bound proteins (Fig. 1). Moreover, the combinatorial complexity of these domains within individual Adhesion GPCRs generates the high structural (and probably also functional) diversity of the receptor family.
Most intriguingly, the GAIN domain is the single extracellular structural feature shared by all family members (Arac et al., 2012a) with only one exception, ADGRA1 (GPR123). Owing to the juxtamembranous position of the GAIN domain and its autoproteolytic function, Adhesion GPCR molecules can also be divided into an N-terminal fragment (NTF) and a C-terminal fragment (CTF; Fig. 2) (Langenhan et al., 2013; Liebscher et al., 2013). The NTF contains all extracellular protein domains and the larger part of the cleaved GAIN domain, whereas the CTF harbors a small part of the proteolysed GAIN domain, a linker region, the 7TM domain, and the entire ICD.
It is important to note that not all Adhesion GPCRs are predicted to be cleavable at the GAIN domain, because of the lack of a consensus catalytic triad sequence within their GPS, a prerequisite for autoproteolysis (see section V). These noncleavable Adhesion GPCRs include ADGRF2 (GPR111), ADGRF4 (GPR115) (Prömel et al., 2012b), ADGRC1 (CELSR1) (Formstone et al., 2010), whose lack of cleavage has been experimentally confirmed, and ADGRE1 (EMR1), ADGRA1 (GPR123), ADGRA2 (GPR124), ADGRA3 (GPR125), and ADGRC3 (CELSR3), which are predicted noncleavable. Therefore, some Adhesion GPCRs might be expressed as single-chain receptors (Prömel et al., 2012b; Langenhan et al., 2013). Notably, endogenous expression of several Adhesion GPCRs is notoriously low, and/or suitable specific antibodies for investigating cleavage are missing. Hence, it is often complicated to harvest Adhesion GPCR protein from primary sources in order to assess physiologic autocatalytic cleavage.
Given the complex molecular design of Adhesion GPCRs, it is important to precisely follow the topology- and cleavage-based receptor terminology depicted here and agreed upon by the Adhesion GPCR Consortium (see http://www.adhesiongpcr.org/) in all publications.
V. Autoproteolytic Processing
A defining feature of Adhesion GPCRs is the presence of a highly conserved cysteine-rich GPS (GPCR proteolysis site) of ∼50 amino acids, located immediately before the first TM helix (Stacey et al., 2000; Lin et al., 2010). The GPS motif is named due to a common proteolytic modification of Adhesion GPCRs (and polycystins), which cleaves the receptors into an extracellular NTF and a membrane-spanning/cytoplasmic CTF (Krasnoperov et al., 1997; Ponting et al., 1999). Thus, the majority of mature Adhesion GPCRs are cleaved and exist as a noncovalently-attached NTF-CTF complex. GPS proteolysis has been suggested to be critical for the maturation, stability, trafficking, and function of Adhesion GPCRs (Yona et al., 2008b). However, recent studies indicate that for some Adhesion GPCRs, the GPS motif itself, but not protein cleavage, is probably more important for receptor signaling and function (Prömel et al., 2012a; Langenhan et al., 2013).
Proteolysis at the GPS occurs between a conserved aliphatic residue (usually a leucine) and a threonine, serine, or cysteine (L↓T/S/C) (Stacey et al., 2000) (Fig. 3). Detailed analysis has revealed that proteolysis occurs as an autocatalytic intramolecular reaction (Lin et al., 2004). A plausible mechanism was suggested in which a series of nucleophilic attacks and subsequent generation and hydrolysis of an ester intermediate facilitates cleavage (Lin et al., 2004, 2010). The autoproteolytic reaction proceeds in the lumen of the endoplasmic reticulum during receptor biosynthesis and is absolutely dependent on the GPS motif; however, the GPS motif alone is not sufficient to mediate proteolysis (Chang et al., 2003b).
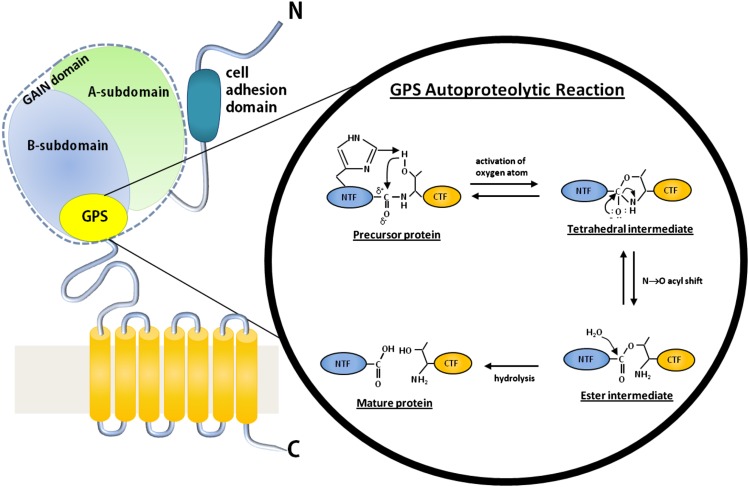
Fig. 3.
The GAIN domain and GPS autoproteolysis. Diagram of an Adhesion-GPCR. The GAIN domain, located at the C-terminal half of the NTF, consists of A-subdomain (green) and B-subdomain (light blue). The GPS motif, which is a part of subdomain B, is colored yellow. The proposed mechanism of the GPS autoproteolytic reaction is shown inside the black circle. A histidine or another general base withdraws a proton from the hydroxyl group of a serine or threonine at position +1. The resulting negatively charged oxygen makes a nucleophilic attack on the carbonyl group of the residue at position –1 (e.g., a leucine), yielding a tetrahedral intermediate and subsequently an ester intermediate. The resulting ester is then hydrolyzed to produce the NTF and CTF that form the final mature protein.
Recent structural analysis revealed that the GPS motif is actually a part of a much larger (∼320-residue) novel domain, termed the GAIN domain (Arac et al., 2012b). The crystal structures of the GAIN domains of ADGRL1 (latrophilin 1) and ADGRB3 (BAI3) revealed that the GPS motif comprises the last five β-strands of subdomain B, and that cleavage occurs between the last two β-strands (Fig. 3). Furthermore, the entire GAIN domain was necessary and, as opposed to the GPS motif, sufficient for autoproteolysis (Arac et al., 2012b). Of note, the GAIN domain need not consist of two subdomains, as illustrated by members of the ADGRE (EGF-TM7) subfamily, which undergo proteolysis despite the lack of a subdomain A (Kwakkenbos et al., 2002; Stacey et al., 2002; Matmati et al., 2007).
Apart from a putative role for biosynthesis of Adhesion GPCRs, autoproteolysis at the GPS motif within the GAIN domain and subsequent reassociation of the NTF and CTF (homogeneric heterodimerization) has also been suggested to be ligand dependent (Volynski et al., 2004). In addition, Adhesion GPCRs may display promiscuity in NTF and CTF pairings, such that the NTF of one individual Adhesion GPCR can form a dimer with the CTF of another family member (heterogeneric heterodimerization) (Davies et al., 2007; Silva et al., 2009; Huang et al., 2012). Although this mechanism might not hold true for all Adhesion GPCR homologs in all species (Prömel et al., 2012a), the implications for Adhesion GPCR signaling complexity and biologic impact are vast (Langenhan et al., 2013; Liebscher et al., 2013; Prömel et al., 2013).
VI. Extracellular Interaction Partners
Several Adhesion GPCR–interacting partners have been described over the years, leading to the notion that the receptors mainly interact with cellular and matricellular ligands, in contrast to other GPCR classes that mainly interact with small molecules or peptides as ligands (Langenhan et al., 2013) (Fig. 4). A further feature of the Adhesion GPCR binding profile seems to be a high promiscuity in ligand recognition, with one receptor binding to multiple partners. Lastly, the location of these partners is not always found on opposing cells or in the extracellular space (in trans: Hamann et al., 1996b; Park et al., 2007; Das et al., 2011; Silva et al., 2011); in some cases, receptor and ligand may reside on the surface of the same cell (in cis: Little et al., 2004; Nishimura et al., 2012; Prömel et al., 2012a).
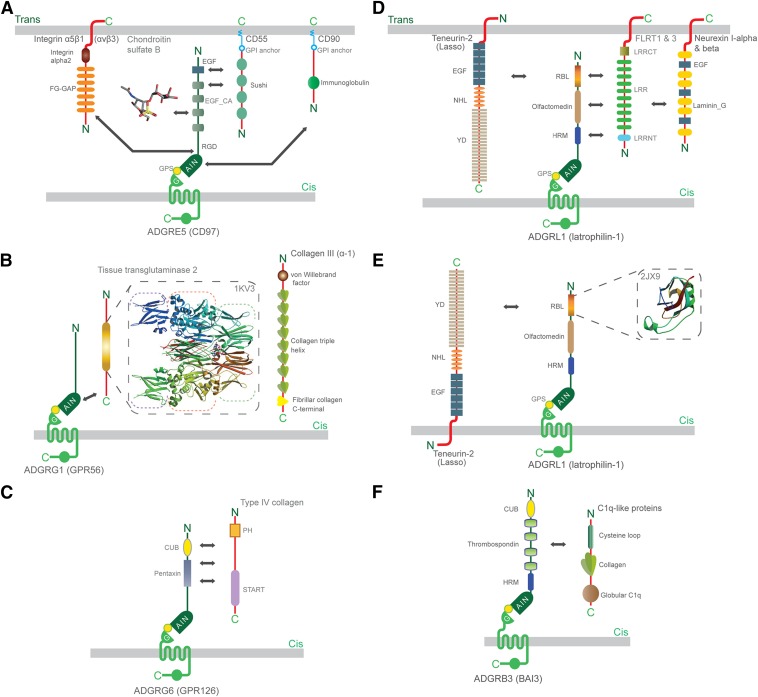
Fig. 4.
Representation of Adhesion GPCR interactions within the NTFs. As examples, interactions of the Adhesion GPCRs (A) ADGR5 (CD97), (B) ADGRG1 (GPR56), (C) ADGRG6 (GPR126), (D and E) ADGRL1 (latrophilin-1), and (F) ADGRB3 (BAI3) are depicted. The interactions of the binding partners with the receptors are shown in cis and trans configurations. Arrows indicate the protein domains—as far as these are known—that facilitate the interactions. The figure also shows some of the solved three-dimensional structures of protein domains, including RBL domain (PDB ID 2JX9) and tissue transglutaminase 2 (PDB ID 1KV3) obtained from the PDB database. The three-dimensional structure of chondroitin sulfate B (a glycosaminoglycan) has been obtained from the PubChem Compound Database (compound ID: 32756). CUB, Cs1 and Csr/Uegf/BMP1; EGF_CA, calcium-binding EGF; FG–GAP, phenylalanine-glycine–glycine-alanine-proline; GPI, glycosylphosphatidylinositol; HRM, hormone receptor motif; LRR, leucine-rich repeat; LRRCT, leucine-rich repeat C-terminal; LRRNT, leucine-rich repeat N-terminal; NHL, NCL-1, HT2A, and Lin-41; PH, Pleckstrin homology; RBL, rhamnose-binding lectin; RGD, arginine-glycine-aspartate; START, StAR-related lipid-transfer; YD, tyrosine-aspartate.
The first Adhesion GPCR ligand identified was CD55 (also known as decay-accelerating factor), which interacts with the EGF-like domains of ADGRE5 (CD97) (Hamann et al., 1996b). Later, chondroitin sulfate B (Stacey et al., 2003), α5β1 and αvβ3 integrins (Wang et al., 2005), and CD90 (Wandel et al., 2012) were shown to bind distinct sites within the NTF of ADGRE5 (CD97) (Fig. 4A). In vitro, multivalent probes loaded with the NTF of full-length CD97 can bind CD55 and chondroitin sulfate B in parallel (Kwakkenbos et al., 2005). In vivo, the two interactions are less likely to occur simultaneously, since affinity for CD55 and chondroitin sulfate B is fairly restricted to the distinct isoforms (Hamann et al., 1998; Lin et al., 2001). Chondroitin sulfate B also interacts with ADGRE2 (EMR2) (Stacey et al., 2003), which possesses an EGF-domain region very similar to ADGRE5 (CD97) (Lin et al., 2000). Similarly, ADGRG1 (GPR56) has different binding partners. Collagen III and tissue transglutaminase 2, a large calcium-dependent enzyme involved in cytoskeletal regulation, both interact with the NTF of GPR56 (Xu et al., 2006; Luo et al., 2011) (Fig. 4B). More recently, ADGRG6 (GPR126) was shown to bind collagen IV (Paavola et al., 2014) (Fig. 4C). Of note, the known binding partners of ADGRE5 (CD97), ADGRE2 (EMR2), ADGRG1 (GPR56), and ADGRG6 (GPR126) are all widely expressed by stromal cells and may facilitate the positioning of motile cells that express these Adhesion GPCRs.
ADGRLs (latrophilins) are well studied with regard to interacting partner binding, and recently, teneurin-2 (also known as Lasso) was suggested to be an endogenous binding partner for ADGRL1 (latrophilin-1) (Silva et al., 2011). In rats, teneurin-2 interacts strongly and specifically with ADGRL1 (latrophilin-1) in trans (Silva et al., 2011) (Fig. 4D), while work on the relationship between the respective Caenorhabditis elegans homologs TEN-1 and LAT-1 suggested that both molecules reside on the same membrane and thus engage in cis with each other (Prömel et al., 2012a) (Fig. 4E). Further, teneurin-2 binds very weakly to ADGRL2 (latrophilin-2) and does not bind to ADGRL3 (latrophilin-3) (Silva et al., 2011). However, ADGRL3 (latrophilin-3) does interact with teneurin-3 (O’Sullivan et al., 2012). In addition, all three mammalian latrophilins are known to interact with fibronectin leucine-rich transmembrane (FLRT) family members. The extracellular domains of ADGRL3 (latrophilin-3) and FLRT3 interact and promote glutamatergic synapse development (O’Sullivan et al., 2012). Both ADGRL1 (latrophilin-1) and ADGRL3 (latrophilin-3) interact with FLRT1 and FLRT3, whereas ADGRL2 (latrophilin-2) only interacts with FLRT3. Additionally, ADGRL1 (latrophilin-1) interacts with the presynaptically localized neurexin-1α, -1β, -2β, and -3β proteins forming a trans-synaptic adhesion complex (Boucard et al., 2012) (Fig. 4E). Interestingly, all three distinct interacting partners of the latrophilins—teneurins, FLRTs, and neurexins—are single-span transmembrane molecules that have roles in axon guidance, neuronal connectivity, and synapse formation (Lise and El-Husseini, 2006; Tucker et al., 2007; Bottos et al., 2011; O'Sullivan et al., 2012). This suggests that latrophilins have a general role in recognizing transmembranous molecules involved in neuronal biology and may function to signal during neuronal development mediated via interactions with these binding partners.
Members of subfamily ADGRB (BAIs) and ADGRF also have known interacting partners. In macrophages, the thrombospondin type 1 repeats of ADGRB1 (BAI1) bind phosphatidylserine, which enables both the recognition and subsequent internalization of apoptotic cells (Park et al., 2007). ADGRB1 (BAI1) also binds lipopolysaccharide of Gram-negative bacteria, resulting in Rac signaling and phagocytic uptake (Das et al., 2011). Moreover, ADGRB3 (BAI3) interacts with high affinity with C1q-like molecules and thereby might regulate synapse formation (Bolliger et al., 2011) (Fig. 4F). Finally, surfactant protein D was recently suggested to be a binding partner for ADGRF5 (GPR116) (Fukuzawa et al., 2013), adding collectins to the list of potential molecules that interact with Adhesion GPCRs. Thus, ADGRBs (BAIs) and ADGRF5 (GPR116) bind further molecule types adding to the structural diversity of Adhesion GPCR interaction partners.
Although several novel ligands have been identified recently, our knowledge of molecules that interact with of Adhesion GPCRs is far from complete, with about two-thirds of the family members remaining to be deorphanized and a fair chance of multiple interactions. Another question that warrants further investigation is whether single receptor molecules bind multiple partners simultaneously, both in trans and in cis.
VII. Signal Transduction
The lack of a biologic stimulus that induces receptor activity has been the major obstacle in elucidating the signal-transduction mechanisms of Adhesion GPCRs. As discussed above, numerous interaction partners have been identified, but until lately, none of these have been demonstrated to act as a pharmacological on/off signal. Only very recently was collagen IV shown to activate the signaling of ADGRG1 (Gpr126) (Paavola et al., 2014). Nevertheless, Adhesion GPCRs clearly belong to the GPCR superfamily, since their common 7TM core has similarities with the other well established GPCR families (Stacey et al., 2000; Lagerström and Schiöth, 2008). However, actual coupling and signaling via intracellular G proteins was not proven until recently.
A. G Protein–Mediated Intracellular Signaling
Unfortunately, there is no known sequence motif that clearly identifies whether a given protein interacts with G proteins. Interactions between ADGRL1 (latrophilin-1) and Gαo (Lelianova et al., 1997), as well as between ADGRG1 (GPR56) and Gαq/11 (Little et al., 2004), have been proposed on the basis of affinity chromatography and immunoprecipitation approaches. Additionally, teneurin-2 binding to ADGRL1 (latrophilin-1) induces Ca2+ signals (Silva et al., 2011), and an antibody against the extracellular domain of ADGRG1 (GPR56) activates the RhoA pathway (Iguchi et al., 2008). Furthermore, the binding of collagen III to ADGRG1 (GPR56) activates RhoA, which can be attenuated by dominant-negative Gα13, supporting the notion that ADGRG1 (GPR56) couples to Gα12/13 upon stimulation. Mice lacking either GPR56 or collagen III display a malformed cerebral cortex, characterized by overmigration of neurons beyond the pial basement membrane (Li et al., 2008; Luo et al., 2011; Jeong et al., 2012). A very similar phenotype was found in mice lacking Gα12/13 (Moers et al., 2008), further supporting a model in which Gα12/13 is downstream of ADGRG1 (GPR56) activation. Mutant phenotypic analysis of another Adhesion GPCR, ADGRG6 (GPR126), also supported G protein coupling for this receptor. Zebrafish (Danio rerio) adgrg6 (gpr126) mutants exhibit myelination defects; these phenotypes were reversible through forskolin-induced cAMP increase (Monk et al., 2009), suggestive of Gαs coupling. Recently, collagen IV was shown to stimulate cAMP signaling in cells expressing ADGRG6 (Gpr126) (Paavola et al., 2014). Additionally, in vitro analysis showed Gαs and Gαi coupling for ADGRG6 (GPR126) (Mogha et al., 2013) and Gαi coupling for ADGRV1 (VLGR1) (Hu et al., 2014).
More direct evidence of G protein coupling was obtained for ADGRD1 (GPR133). Here, an overexpression strategy was applied to measure the basal activity of this Adhesion GPCR. The basal activity of any GPCR results from an equilibrium between inactive and active conformations of receptors (Lefkowitz et al., 1993). Receptor overexpression does not change this equilibrium but increases the number of receptors in both conformations, hence reaching a threshold at which the active conformation can be detected owing to constitutive activation of signaling pathways. Using this approach, ADGRD1 (GPR133) showed a receptor concentration–dependent increase in cAMP levels, which is indicative of Gαs coupling (Bohnekamp and Schoneberg, 2011) and IP3 levels when using a Gαqi chimera, indicating Gαi coupling (Liebscher et al., 2013). Using a similar approach, coupling to Gαs, Gαq, Gαi/o, or Gα12/13 proteins was demonstrated for several other Adhesion GPCRs (ADGRE2 [EMR2], ADGRF1 [GPR110], ADGRF4 [GPR115], ADGRB1 [BAI1], ADGRG3 [GPR97], ADGRG5 [GPR114], ADGRG6 [GPR126]) (Gupte et al., 2012; Stephenson et al., 2013). In sum, multiple lines of direct and indirect evidence support the notion that most members of the Adhesion GPCR family can likely mediate signals by activation of G protein cascades.
B. G Protein–Independent Intracellular Signaling
Experimental evidence suggests that Adhesion GPCRs are also capable of activating non–G protein signaling cascades. This evidence comes mainly from interaction studies with intracellular proteins. For example, members of the ADGRB (BAI) subfamily present with a relatively long ICD, relative to most other Adhesion GPCRs, which is predisposed for such complex formation. Indeed, the ICD of ADGRB1 (BAI1) forms a complex with ELMO (engulfment and cell motility) and Dock180 (dedicator of cytokinesis) and mediates phagocytosis of apoptotic cells and Gram-negative bacteria as well as myoblast fusion via an ELMO/Dock180/Rac signaling module (Park et al., 2007; Das et al., 2011, 2014; Hochreiter-Hufford et al., 2013). Similarly, BAI3 promotes myoblast fusion through ELMO/DOCK1 (Hamoud et al., 2014). A recent study showed that ADGRB1 (BAI1) also interacts with Par3/Tiam1 and recruits these proteins to synaptic sites for spinogenesis and synaptogenesis (Duman et al., 2013). However, unlike in phagocytosis and myoblast fusion, the ADGRB1 (BAI1)-ELMO/DOCK180 interaction is dispensable for the role of ADGRB1 (BAI1) in synapse development. Further, ADGRF5 (GPR116) was recently shown to engage a Gαq-p63-RhoGEF-Rho GTPase pathway (Tang et al., 2013), and ADGRE3 (GPR97) signaling is involved in Rho kinase activation (Valtcheva et al., 2013). Other examples come from planar cell polarity (PCP) signaling during embryonic development. During avian neural tube closure, actomyosin-dependent planar-polarized contraction is mediated by ADGRC1 (CELSR1) via direct crosstalk to frizzled/dishevelled DAAM1 (disheveled-associated activator of morphogenesis 1) and PDZ-RhoGEF to upregulate Rho kinase (Nishimura et al., 2012). In the developing zebrafish gastrula, the intracellular C terminus of ADGRA3 (Gpr125) directly interacts with dishevelled, and together, ADGRA3 (Gpr125) and dishevelled recruit a subset of PCP components into membrane subdomains (Li et al., 2013).
G protein–independent signaling via recruitment of PDZ domain- and SH3 domain-containing proteins, calmodulin, and arrestin is also common in other GPCR families (Magalhaes et al., 2012). β-Arrestins were originally discovered to desensitize activated GPCRs. However, arrestins are now well established mediators of receptor endocytosis, ubiquitylation, and G protein–independent signaling (Shukla et al., 2011). It is therefore interesting that recruitment of β-arrestin and ubiquitination has been demonstrated for the Adhesion GPCRs ADGRG1 (GPR56), ADGRG3 (GPR97), ADGRB1 (BAI1), and ADGRB3 (BAI3) (Paavola et al., 2011; Stephenson et al., 2013; Southern et al., 2013). Taken together, recruitment of G protein–independent cascades significantly contributes to the signaling repertoire of Adhesion GPCRs and probably accounts for the exceptional signaling complexity and specificity of Adhesion GPCRs.
C. Modes of Signaling
Although our knowledge of signaling pathways employed by Adhesion GPCRs has significantly increased in recent years, the molecular mode(s) whereby Adhesion GPCRs switch between active and inactive states is (are) still enigmatic. Despite a lack of agonists, several hypotheses describing distinct aspects of Adhesion GPCR activation and signaling mechanisms have recently emerged (Langenhan et al., 2013). One main paradigm, which dominates discussions about Adhesion GPCR signaling, is autoproteolysis at the GPS. Because most Adhesion GPCRs undergo autoproteolysis, it is hypothesized that this cleavage modulates receptor activity. Indeed, receptor mutants without an NTF (mimicking the situation after GPS proteolysis and NTF release) have been found to display increased activity in studies on ADGRG1 (GPR56), ADGRB1 (BAI1), ADGRB2 (BAI2), and ADGRE5 (CD97) (Okajima et al., 2010; Paavola et al., 2011; Ward et al., 2011; Stephenson et al., 2013). How this is achieved in detail still remains elusive, but at least two activation models of intracellular signaling cascades have been suggested (Arac et al., 2012a; Paavola and Hall, 2012; Langenhan et al., 2013; Liebscher et al., 2013; Prömel et al., 2013): 1) A segment of the ECD functions as an inverse agonist of constitutive signaling by the 7TM, with this ECD-mediated inhibition being removed upon binding of a ligand, and 2) a segment of the ECD functions as a tethered agonist, which is exposed to the 7TM core following ligand binding. The second scenario might be more likely, since all other known GPCRs signal via an agonist-driven switch-on mechanism. Similar mechanisms may facilitate inhibitory functions of Adhesion GPCRs, e.g., the CTF of ADGRV1 (VLGR1) alone inhibits adenylate cyclase activity through Gαi coupling (Hu et al., 2014).
Evidence from various studies indicates that the GPS motif and the GAIN domain play vital roles in Adhesion GPCR function independent of receptor cleavage, potentially functioning as a hinge for receptor activity and signal transduction (Arac et al., 2012a). For example, human disease mutations in numerous Adhesion GPCRs map to the GAIN domain (Qian et al., 2002; Piao et al., 2004; Kan et al., 2010; Prömel et al., 2013). A frequently discussed hypothesis proposes the GAIN domain as a potential interaction interface for ligands or even for intramolecular interactions (Arac et al., 2012a). Identification of such tethered ligands is one key future goal for the Adhesion GPCR field.
Another general question regarding Adhesion GPCR signaling concerns whether the NTF and CTF of a given receptor have separate functions. It has been shown for the C. elegans ADGRL (latrophilin) homolog LAT-1 that the complete ECD, anchored to the membrane, mediates a physiologic function, fertility, independently from the CTF in vivo (Prömel et al., 2012a). Moreover, when the GPS of lat-1 is mutated such that the receptor is rendered entirely noncleavable, this mutant remains capable of mediating its two activities, the CTF-dependent mode (tissue polarity) and the CTF-independent mode (fertility). A further example is ADGRG6 (GPR126), which is required for heart and peripheral nervous system development. Notably, ectopic expression of the NTF of GPR126 in a zebrafish knockdown model rescued defective trabeculation in the heart but not the myelination phenotype in the peripheral nervous system, supporting a model in which the NTF of GPR126, in contrast to the CTF, plays an important role in heart development (Patra et al., 2013). The ECD also appears to be involved in homo- and heterodimerization of Adhesion GPCRs. For example, ADGRC1 (CELSR1) forms a homophilic trans interaction that is required for PCP signaling (Nishimura et al., 2012). Moreover, ADGRC2 (CELSR2), ADGRC3 (CELSR3), and ADGRG1 (GPR56) have also been reported to undergo homophilic trans interactions that influence receptor activity (Shima et al., 2007; Paavola et al., 2011).
These studies indicate that Adhesion GPCRs can signal via at least two different modes, cis (e.g., intracellular G protein signaling) or second cis/trans, where second cis refers to coreception in complex with a second transmembrane receptor and trans refers to interaction with extracellular partners. Importantly, the trans mode would be unique for Adhesion GPCRs among the other GPCR families. ADGRE5 (CD97) may serve as an example for the second cis interaction mode: in both prostate and thyroid cancer cell lines, ADGRE5 (CD97) amplifies [lysophosphatidic acid (LPA)–induced RhoA activation via interaction with LPA receptor 1 (Ward et al., 2011, 2012)]. These studies provide evidence for heterodimerization of an Adhesion GPCR with a canonical GPCR and the physiologic consequence of this association.
Finally, Adhesion GPCR NTFs, upon shedding, can also initiate noncell autonomous activities at distant locations. For example, soluble ADGRE5 (CD97) stimulates angiogenesis through binding to α5β1 and αvβ3 integrins (Wang et al., 2005). Soluble ADGRA2 (GPR124) has an exposed cryptic arginine-glycine-asparagine motif in the NTF that binds αvβ3 integrin and supports the survival of endothelial cells (Vallon and Essler, 2006), and ADGRB1 (BAI1) similarly possesses an NTF arginine-glycine-asparagine motif that inhibits angiogenesis by engaging with integrins (Koh et al., 2004).
There have been numerous reports of the shedding of a 120-kDa fragment from the NTF of ADGRB1 (BAI1) (termed vasculostatin) and the role it has in inhibiting angiogenesis (Koh et al., 2004; Kaur et al., 2005, 2009; Hardcastle et al., 2010; Klenotic et al., 2010). However, these results are hard to reconcile with the solved structure of the GAIN domain, which suggests that the cleaved NTF will remain tightly bound to the CTF (Arac et al., 2012a). An elegant explanation for this conundrum was the discovery of an additional furin- and matrix metalloproteinase-dependent cleavage location in the NTF of ADGRB1 (BAI1) creating vasculostatin-40 (Cork et al., 2012). This smaller 40-kDa fragment of ADGRB1 (BAI1), which contains a single thrombospondin repeat, is capable of angiogenesis activity and therefore can reconcile the functional and structural studies. Additional cleavage sites close to the GAIN domain may be a common theme observed in Adhesion GPCRs that is not yet fully appreciated, as ADGRL1 (latrophilin-1), ADGRB2 (BAI2), and ADGRF5 (GPR116) have all been reported to possess such sites (Fukuzawa and Hirose, 2006; Krasnoperov et al., 2009; Okajima et al., 2010). These findings indicate that that Adhesion GPCRs may easily have at least two separate functions: one is cell autonomous and another is not, increasing the number of pathways that each family member may be involved in.
VIII. Expression
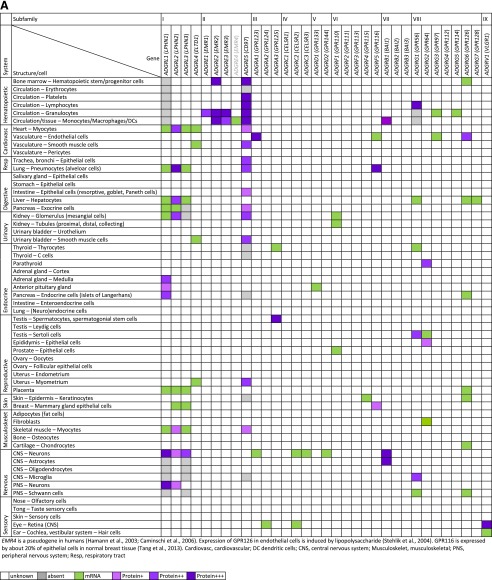
Adhesion GPCRs are present on almost all mammalian cells. Figure 5, A and B, provides a summary of the available data on the expression of Adhesion GPCRs in humans and rodents (mice and rats), obtained by transcriptional profiling and protein analysis techniques.
Fig. 5.
Expression of Adhesion GPCRs. Summary of available published data on the expression of Adhesion GPCRs in (A) humans and (B) mice and rats, obtained by transcriptional profiling and protein analysis techniques. For additional information and references, see the descriptions of individual Adhesion GPCRs at http://www.guidetopharmacology.org/.
Expression of some Adhesion GPCR subfamilies initially appeared confined to distinct cell types or organs, like the immune system or the central nervous system (CNS). For example, cells of the immune system prominently express ADGRE (EGF-TM7) subfamily receptors as well as the ADGRG subfamily members ADGRG1 (GPR56), ADGRG3 (GPR97), and ADGRG5 (GPR114). The presence of ADGRE (EGF-TM7) subfamily receptors 1–4 (EMR1–4) is a defining feature of myeloid leukocytes. The tightly restricted expression of murine ADGRE1 (EMR1 or F4/80) has allowed its use as an excellent marker of resident murine tissue macrophages for over 30 years (Austyn and Gordon, 1981; Gordon et al., 2011). Human ADGREs (EGF-TM7 receptors) are useful markers for granulocytes, with ADGRE1 (EMR1) found on eosinophils and ADGRE3 (EMR3) on mature polymorphogenic granulocytes (Hamann et al., 2007; Matmati et al., 2007; Legrand et al., 2014). In contrast, ADGRE5 (CD97) is not restricted to myeloid cells but also found on lymphoid, epithelial, muscle, and other cell types (Eichler et al., 1994; Jaspars et al., 2001; Aust et al., 2006; Veninga et al., 2008; Zyryanova et al., 2014).
Members of subfamilies ADGRL, ADGRC, and ADGRB (latrophilins, CELSRs, and BAIs) were first identified in the CNS (Hadjantonakis et al., 1997; Krasnoperov et al., 1997; Lelianova et al., 1997; Shiratsuchi et al., 1997; Shima et al., 2002; Stephenson et al., 2014). Later studies actually revealed that expression of these subfamilies is not restricted to the CNS, and in particular, human ADGRLs (latrophilins) and murine ADGRCs (CELSRs) have been identified in many cell types. Similarly, several isoforms of ADGRV1 (VLGR1) were reported to be widely expressed in a spatiotemporally regulated manner (reviewed in McMillan and White, 2010). This receptor shows a particularly strong expression in sensory cells of the eye and the inner ear, as well as in the brain (van Wijk et al., 2006; Maerker et al., 2008; Zallocchi et al., 2012; Shin et al., 2013).
Some Adhesion GPCRs have been reported to be expressed in only a few cell types or organs, including human ADGRE1 (EMR1) in eosinophils (Hamann et al., 2007; Legrand et al., 2014), and human and mouse ADGRF4 (GPR115) in the skin (Prömel et al., 2012b; Gerber et al., 2013). Many more Adhesion GPCRs are widely distributed, such as the above-discussed ADGRE5 (CD97) and the ADGRLs, ADGRCs, and ADGRBs (latrophilins, CELSRs, and BAIs). Likewise, ADGRG1 (GPR56), originally identified in melanoma cells, is also expressed by various progenitor cells and by cytotoxic lymphocytes (Zendman et al., 1999; Piao et al., 2004; Della Chiesa et al., 2010; Peng et al., 2011). Of note, extracellular interaction partners (see section V) have been ascribed mostly to Adhesion GPCRs that have a rather wide cellular distribution.
The presence of Adhesion GPCRs on stem and progenitor cells is an important theme and likely at the basis of their role in various developmental processes and in tumorigenesis (see section IX.C). For example, ADGRE5 (CD97) and ADGRG1 (GPR56) are expressed by hematopoietic stem and progenitor cells (van Pel et al., 2008a) (https://www.immgen.org/), ADGRG1 (GPR56) is found on neural stem cells (Bai et al., 2009), and ADGRA3 (GPR125) is a specific marker of adult spermatogonial progenitor cells, a source of multipotent stem cells (Seandel et al., 2007; Dym et al., 2009; Izadyar et al., 2011).
While the cellular distributions of some Adhesion GPCRs provide clues to their involvement in physiologic processes, a substantial number of receptors are still poorly characterized. For example, expression of ADGRCs (CELSRs) has been studied in detail only in the mouse and zebrafish.
Marked differences in the expression of some Adhesion GPCRs between human and mouse have been observed, stressing the importance of a comprehensive investigation of expression patterns in different species. For example, the characteristic presence of ADGRE1 (EMR1) on resident tissue macrophages is mouse-specific, while the specific presence of ADGRG1 (GPR56) on cytotoxic lymphocytes is a human trait (https://www.immgen.org/). Future interpretations of the biologic roles of Adhesion GPCRs on the basis of model organism studies need to consider these potential differences.
IX. Physiology and Disease
Cell biologic effects of Adhesion GPCRs have been studied in great detail using a wide range of genetic models, including invertebrate and non–mammalian vertebrate species, and in vitro assays. This extensive body of work is covered within section IX.A and delineates fundamental structure-function relationships of Adhesion GPCRs at the cellular level. These models have helped to sketch a preliminary picture of Adhesion GPCR impact on individual organs described in section IX.B, and is the basis to understand clinical implications of Adhesion GPCR signals as discussed in section IX.C. For a summary of the functions of Adhesion GPCRs in physiology and disease see Fig. 6.
Fig. 6.
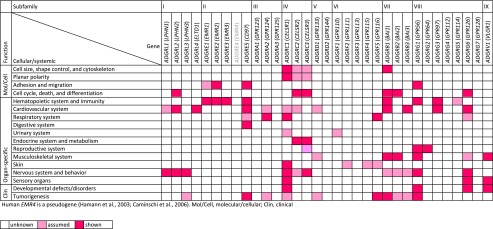
Functional involvement of Adhesion GPCRs. Summary of available published data on the function of Adhesion GPCRs, obtained in vitro and in vivo from humans and mouse models.
A. Molecular and Cellular Functions
1. Cell Size, Shape Control, and Cytoskeleton.
The ability of many Adhesion GPCRs to mediate regulation of cytoskeletal organization has long been appreciated (Oda et al., 1999; Gao et al., 1999; Usui et al., 1999; Li and Gao, 2003). The known developmental functions of the ADGRB (BAI) and ADGRC (Flamingo/Starry night/CELSRs; also designated as 7TM-cadherins by developmental biologists [Hulpiau and van Roy (2009)] subfamily members rely on actin- and microtubule-based processes (Antic et al., 2010; Hashimoto et al., 2010; Park and Ravichandran, 2010). Although cadherin repeats are present, for example in Flamingo and ADGRC3 (CELSR3) (Berger-Muller and Suzuki, 2011), evidence of classic cadherin-mediated signaling cascade activation has not yet been obtained.
Elegant studies have recently revealed that members of ADGRC (CELSR) and ADGRB (BAI) subfamilies facilitate changes in the size and shape of cell membranes via actin reorganization downstream of signaling by Rho/Rac guanine-nucleotide exchange factors (GEFs) to Rho, Rac, and Cdc42 (Stephenson et al., 2014). These small GTPases link membrane receptors to the assembly/disassembly of the cytoskeleton via cell signaling. Apart from the previously discussed functions, ADGRB1 (BAI1) additionally regulates the size and shape of actin-rich protrusions (spines) on the dendrites of cultured neurons (Duman et al., 2013) through the spatial restriction of the RacGEF Tiam1 and the cell polarity protein Par3 (Suzuki and Ohno, 2006). The observation that ADGRB1 (BAI1) also binds to integrins suggests an exciting potential link between ADGRB1 (BAI1), cell adhesion, and cell polarity.
Subfamily ADGRC (CELSR) receptors recruit RhoGEFs during PCP signaling (see next section) to control the size and shape of asymmetric epithelial membrane domains. In the insect tracheal system, anisotropic enrichment (asymmetric enrichment across opposing cell membranes) of components, including the Drosophila melanogaster Adhesion GPCR Flamingo, results in the associated planar polarity of RhoGEF2, leading to increased endocytosis of E-cadherin (Warrington et al., 2013). Rho signaling plays essential roles in endocytosis through cytoskeletal modulation (de Curtis and Meldolesi, 2012). Adherens junction turnover facilitates membrane shrinkage, which drives the cell rearrangements essential for tracheal tube morphogenesis. Asymmetric enrichment of adherens junction–associated ADGRC1 (CELSR1) and PDZ-RhoGEF was also observed within the ventral neural tube during avian neural tube closure (Nishimura et al., 2012). PDZ-RhoGEF forms part of a protein complex with frizzled/dishevelled/Daam1, which upregulates Rho kinase activity, resulting in the contraction of planar polarized actin-myosin cables and apical constriction of neuroepithelial cells. Thus, the vital inward bending of the neural plate is facilitated by actin-myosin–mediated shrinkage of neuroepithelial cell-apical membranes.
2. Planar Cell Polarity.
The Drosophila Adhesion GPCR Flamingo was originally identified because of its essential role in PCP signaling (Usui et al., 1999). PCP, which acts orthogonally to apico-basal polarity, was first recognized in insects (Lawrence, 1973; Lawrence and Shelton, 1975), but its effects can be visualized on the body surface of most animals by the orderly alignment of hairs on mammalian skin and insect cuticle (Seifert and Mlodzik, 2007; Devenport and Fuchs, 2008). Flamingo and its vertebrate homolog, ADGRC1 (CELSR1), function in a defined signaling pathway, called the core-PCP pathway, in which PCP information is locally propagated from one cell neighbor to another across epithelial/epidermal fields. First described for Flamingo (Shimada et al., 2001), the molecular signature of PCP signaling is the asymmetric distribution of pathway components. Studies in Drosophila have also revealed that asymmetry of core-PCP proteins is preceded by the uniform cell-surface localization of Flamingo close to apical adherens junctions, followed by recruitment of the 7TM receptor Frizzled and the multidomain protein Dishevelled (Strutt et al., 2002). This core group alone can transmit local PCP but does so less efficiently than protein complexes, which include the transmembrane protein Vang (Struhl et al., 2012). It is generally believed that ADGRC (CELSR) proteins form homodimeric molecular bridges between neighboring cells. ADGRCs (CELSRs) are therefore fundamental to local PCP transmission, and their asymmetric enrichment at both interfaces of opposing cell membranes is unique (Strutt et al., 2002; Devenport et al., 2011). Frizzled and Vang each enrich opposing cell membranes and are thought to interact in trans. It remains an open question whether the role of ADGRCs (CELSRs) in PCP signaling is instructive or merely one of stabilization of asymmetric protein complexes. Downstream of ADGRCs (CELSRs), small GTPase signaling elicits cytoskeleton alignment, cell junction turnover, and cell shape changes. Notably, mice mutant in gene homologs of Drosophila core-PCP components, including ADGRC (CELSR) homologs, exhibit severe developmental defects in multiple tissue and organ systems, including the neural tube and the ependyma, the latter causing abnormal cerebrospinal fluid circulation (Wansleeben and Meijlink, 2011; Boutin et al., 2014).
Similar in effect to ADGRCs (CELSRs), the ADGRL (latrophilin) subfamily homolog LAT-1 organizes cell division planes across the anterior-posterior axis of the C. elegans embryo, acting in parallel with noncanonical Wnt/Frizzled signaling (Langenhan et al., 2009). The molecular requirements for LAT-1 function in this process argue for a role(s) involving intercellular interactions coupled to cell signaling, as has been proposed for ADGRCs (Flamingo and CELSRs). Thus, it is tempting to speculate that these Adhesion GPCRs may fulfill similar functions in PCP transmission in different tissue contexts.
3. Cell Adhesion and Migration.
The long ECDs of most Adhesion GPCRs contain structural domains that are implicated in cell–cell and cell–matrix interactions, including EGF-like, thrombospondin, leucine-rich, lectin-like, immunoglobulin, and cadherin repeats (Fig. 1). Indeed, the term Adhesion was proposed for this family to reflect potential roles in cellular adhesion (Fredriksson et al., 2003b).
ADGRE5 (CD97) provides a model for Adhesion GPCR-mediated cell–cell and cell–matrix interactions. ADGRE5 (CD97) is expressed on almost all leukocytes, and the first identified binding partner of ADGRE5 (CD97), CD55, is found on stromal cells in addition to its presence on leukocytes (Hamann et al., 1996b). Interestingly, in the presence of arterial shear stress, the binding of leukocyte ADGRE5 (CD97) to stromal cell CD55 results in shedding of the NTF of ADGRE5 (CD97) and subsequent downregulation of the CTF (Karpus et al., 2013). This observation implies that the ADGRE5 (CD97)-CD55 interaction facilitates the engagement of adhesive contacts between ADGRE5 (CD97)-positive immune cells and CD55-positive stromal cells, possibly to enable the retention of leukocytes at specific tissue sites, while the shear stress–dependent downregulation of ADGRE5 (CD97) upon contact with CD55 prevents cell aggregation in the circulation.
ADGRG1 (GPR56) is also reported to mediate cell–matrix adhesion in developing neurons and hematopoietic stem cells. Loss of ADGRG1 (GPR56) resulted in decreased granule cell adhesion to laminin and fibronectin (Koirala et al., 2009). Moreover, knockdown of ADGRG1 (GPR56) decreased cellular adhesion of acute myeloid leukemia (AML) cells to fibronectin, laminin-1, and collagen III, corresponding to a significant decrease in the number of hematopoietic stem cells (HSCs) in the bone marrow of Adgrg1 (Gpr56) knockout mice and, conversely, an increase in HSC number in spleen, liver, and peripheral blood (Saito et al., 2013). These data suggest that ADGRG1 (GPR56) plays a role in the maintenance of HSCs and/or leukemia stem cells in the bone marrow niche.
Adhesion and migration are closely linked processes, especially in noncirculating cells. Several Adhesion GPCRs increase cell migration in vitro, probably via signaling to members of the Rho family of small GTPases (see section VII), which act to regulate cytoskeletal structure as well as cell or extracellular matrix contacts. Some examples include ADGRG3 (GPR97), regulating the migration of lymphatic endothelial cells to facilitate angiogenesis via RhoA and Cdc42 (Valtcheva et al., 2013); ADGRA2 (GPR124), mediating Cdc42-dependent directional migration of brain endothelial cells toward embryonic forebrain cell conditioned medium (Kuhnert et al., 2010); and, finally, ADGRE5 (CD97) overexpression, increasing single random tumor cell migration (Galle et al., 2006).
In most cases, the extracellular signals for Adhesion GPCR-regulated migration have not yet been identified; adhesive interactions, soluble molecules, such as LPA, or mechanical stress are likely candidates. Of note, an antibody directed to the ECD of ADGRE2 (EMR2) induced adhesion and CXCL12-directed migration of neutrophils in vitro (Yona et al., 2008a).
Finally, direct consequences on cell migration, such as guiding of cells in developmental processes (see section IX.C.1), have been shown for several Adhesion GPCRs in vivo. The C. elegans ADGRC (CELSR) homolog FMI-1 appears to be involved in the navigation and shaping of neuronal processes during the development of GABAergic neurons (Steimel et al., 2010; Najarro et al., 2012; Huarcaya Najarro and Ackley, 2013). The instructive effects by a pioneer axon on follower axon movements were shown to rely on the ECD but not the 7TM nor ICD of FMI-1, indicating that the adhesion and/or trans signaling events figure prominently in this function (Steimel et al., 2010). Moreover, ADGRG1 (GPR56) inhibits migration of neural progenitors through coordinated action with α3β1 integrin (Iguchi et al., 2008; Luo et al., 2011; Jeong et al., 2013).
4. Cell Cycle, Cell Death, and Differentiation.
Although there is no mechanistic data supporting a direct role for Adhesion GPCRs in controlling canonical cell cycle pathways, there is growing evidence that certain Adhesion GPCRs are important for the regulation of cell proliferation. In vitro, conditioned medium containing cleavage products of the ADGRB1 (BAI1) NTF suppresses proliferation of endothelial cells by blocking αvβ5 integrin (Koh et al., 2004; Kaur et al., 2005; Cork et al., 2012). In vivo, hippocampal neuron proliferation increases in Adgrb2 (Bai2) knockout mice relative to wild-type animals, although how ADGRB2 (BAI2) inhibits neurogenesis is unclear (Okajima et al., 2011). Similarly, conditional loss of Adgrg6 (Gpr126) in Schwann cells causes increased proliferation of this cell type (Mogha et al., 2013). Moreover, proliferation of neuronal progenitors is modulated by ADGRG1 (GPR56), as indicated by opposing effects of loss- and gain-of-function studies in mice (Bae et al., 2014).
In addition, Adhesion GPCRs are regulators of cell survival. Overexpression of ADGRB1 (BAI1) causes increased clearance of apoptotic human umbilical vein endothelial cells (HUVECs) (Koh et al., 2004). Recently, ADGRCs (CELSRs) were shown to potentially regulate the inhibition of apoptosis during early brain development in mice. Although levels of apoptosis are not significantly affected in Adgrc2 (Celsr2) and Adgrc3 (Celsr3) single-knockout mice, there is a considerable increase in apoptosis in medial rhombomeres 4/5 in Adgrc2;Adgrc3 (Celsr2;Celsr3) double-knockout embryos when compared with wild type or single-knockout embryos (Qu et al., 2010). The molecular basis of this observation has yet to be explored.
Furthermore, Adhesion GPCRs also regulate essential developmental transitions at both tissue and cellular levels. ADGRL2 (latrophilin-2) expression is required for normal epithelial-mesenchymal transition of endothelial cells in the atrioventricular canal of the heart during early chick development; however, it is unclear how ADGRL2 (latrophilin-2) controls this transition (Doyle et al., 2006). In zebrafish and mice, ADGRG6 (GPR126) signaling is required for Schwann cell differentiation, as elevation of cAMP mediated by ADGRG6 (GPR126) is necessary for the terminal differentiation of myelinating Schwann cells (Monk et al., 2009, 2011; Glenn and Talbot, 2013; Mogha et al., 2013). Hence, Adhesion GPCRs appear also to figure in the regulation of cell differentiation.
Finally, Adhesion GPCRs have recently been implicated in promoting cell–cell fusion. ADGRB1 (BAI1), ADGRB3 (BAI3), and ADGRG1 (GPR56) promote myoblast fusion to form mature multinucleated skeletal muscle fibers (Hochreiter-Hufford et al., 2013; Wu et al., 2013; Hamoud et al., 2014). Apoptotic myoblasts expose phosphatidylserine, which activates ADGRB1 (BAI1) signaling on healthy myoblasts to promote fusion with myoblast and myotubes.
B. Organ Systems
1. Hematopoietic System and Immunity.
Members of subfamily ADGRE (EGF-TM7), ADGRB (BAI), and ADGRG are prominently expressed by cells of the immune system. The ADGRE (EGF-TM7) receptors ADGRE1–4 (EMR1–4) and ADGRE5 (CD97) have been studied in vitro and in vivo using antibodies and genetic mouse models. ADGRE5 (CD97) has been implicated in innate and adaptive immune processes through its role in cellular migration (Leemans et al., 2004), proliferation (Capasso et al., 2006), and mobilization from the bone marrow (van Pel et al., 2008b). Many of these effects are presumed to be either adhesion- or signaling-dependent, as they are also observed in mice lacking the binding partner CD55. Addition of F4/80 [mouse ADGRE1 (EMR1)]-blocking antibodies to cell culture prevented natural killer cell–macrophage cellular contacts necessary for mediating a robust interferon-γ response toward the bacteria Listeria monocytogenes (Warschkau and Kiderlen, 1999). Moreover, in mouse models, ADGRE1 (EMR1) was essential for the induction of peripheral immune tolerance from eye-derived antigens (Lin et al., 2005). Elucidation of the function of other ADGRE (EGF-TM7) subfamily receptors is hampered by the absence of ADGRE2 (EMR2) and ADGRE3 (EMR3) orthologs in rodents. In vitro studies demonstrated a role for human ADGRE2 (EMR2) in enhancing neutrophil migration, degranulation, and cytokine secretion, as well as in suppressing lipopolysaccharide-induced neutrophil survival (Yona et al., 2008a; Huang et al., 2012).
Although not restricted to the hematopoietic lineage, BAI1 has been shown to bind phosphatidylserine and mediate the uptake of apoptotic cells by numerous cell types, including macrophages and microglia (Park et al., 2007; Elliott et al., 2010; Sokolowski et al., 2011; Mazaheri et al., 2014). In macrophages, it also acts act as a pattern-recognition receptor binding to lipopolysaccharide of Gram-negative bacteria resulting in Rac signaling and phagocytic uptake (Das et al., 2011). In zebrafish microglia, BAI1 controlled the formation of phagosomes around dying neurons and cargo transport (Mazaheri et al., 2014).
The presence of ADGRG1 (GPR56), ADGRG3 (GPR97), and ADGRG5 (GPR114) in leukocyte subsets (Della Chiesa et al., 2010; Peng et al., 2011) implies that these Adhesion GPCRs have immune-related functions that need to be explored. These receptors may also mediate hematopoietic stem cell repopulation and retention of myeloid cells within the bone marrow niche (Saito et al., 2013). In mice, the highest expression of Adgrg3 (Gpr97) mRNA was found in the bone marrow. Although a previous study failed to demonstrate defects in lymphocyte development in Adgrg3 (Gpr97) mutant mice (Sleckman et al., 2000), a more recent study showed altered follicular-versus-marginal zone B-lymphocyte fate decision in the spleen and decreased numbers of B220+ lymphocytes in the bone marrow in an independently raised Adgrg3 (Gpr97) mouse model (Wang et al., 2013a).
2. Cardiovascular System.
Several Adhesion GPCRs are associated with the cardiovascular system. ADGRL2 (latrophilin-2) is required for heart valve formation, acting as a component of the epithelial-mesenchymal transition in the atrioventricular canal (Doyle et al., 2006). ADGRL1 (latrophilin-1) expression has been detected during rat heart development, but its function is unclear (Ferrand et al., 1999). The Adgrg6 (Gpr126) knockout is lethal mice, with most dying in utero ∼E12. Mutant embryos show signs of myocardial wall thinning, hypotrabeculation, defective mitochondrial function (Patra et al., 2013), and circulatory failure (Waller-Evans et al., 2010).
ADGRE5 (CD97) and ADGRL4 (ELTD1) regulate angiogenesis. The NTF of human ADGRE5 (CD97) is a chemoattractant for migration and invasion of HUVECs in vitro. ADGRE5 (CD97) binds to αvβ3 and α5β1 integrins, and when overexpressed in tumor cells, soluble ADGRE5 (CD97)-NTF promotes angiogenesis after injection in mice (Wang et al., 2005). ADGRL4 (ELTD1) is expressed in endothelial cells of human renal and colorectal tumors and is required for proper HUVEC sprouting and vessel formation in zebrafish (Masiero et al., 2013). Adgrl4 (Eltd1) silencing in a mouse orthotopic ovarian cancer model markedly reduced tumor growth and metastatic dissemination, probably by reduction of microvessel density (Masiero et al., 2013).
The NTF of ADGRA2 (GPR124) is shed by cultured HUVECs, thus mediating cell survival by linking glycosaminoglycans of the extracellular matrix to integrin αVβ3 (Vallon and Essler, 2006). In vivo, ADGRA2 (GPR124) functions autonomously in CNS endothelial cells to regulate sprouting and migration. Adgra2 (Gpr124) knockout results in embryonic lethality owing to CNS-specific angiogenesis arrest and hemorrhage (Kuhnert et al., 2010; Anderson et al., 2011; Cullen et al., 2011), whereas vascular overexpression results in CNS-specific hyperproliferative vascular malformations, suggesting relevance for ADGRA2 (GPR124) in various CNS-related vascular pathologies.
ADGRG3 (GPR97) is the most highly and specifically expressed GPCR in mouse intestinal lymphatic endothelium (Valtcheva et al., 2013). It regulates migration of human lymphatic endothelial cells in vitro via the small GTPases RhoA and Cdc42 (Valtcheva et al., 2013). Expression studies in human gliomas and ischemia models in rats identified ADGRBs 1–3 (BAIs 1–3) as antiangiogenic (Nishimori et al., 1997; Kee et al., 2002, 2004), and BAI1 at least acts in a paracrine way through its NTF, called vasculostatin (Kaur et al., 2005; Cork et al., 2012). Overexpression of ADGRB1 (BAI1) (Duda et al., 2002; Kudo et al., 2007) or vasculostatin (Kaur et al., 2005, 2009) in mice drastically reduced tumor growth. Recent genome-wide association studies have also implicated Adhesion GPCRs in heart rate control [ADGRD1 (GPR133)] (Marroni et al., 2009), brain arteriovenous malformation [ADGRA2 (GPR124)] (Weinsheimer et al., 2012), stroke [ADGRC1 (CELSR1)] (Yamada et al., 2009; Gouveia et al., 2011), and myocardial infarction [ADGRC2 (CELSR2)] (Kathiresan et al., 2009; Qi et al., 2011).
3. Respiratory Tract.
Although understudied in terms of Adhesion GPCR biology, the respiratory system displays a wide array of Adhesion GPCR expression patterns and phenotypes. Expression profiling shows that several Adhesion GPCRs, including ADGRE5 (CD97), ADGRF5 (GPR116), and ADGRG6 (GPR126), are present at high levels in lung tissue (Abe et al., 1999; Veninga et al., 2008; Prömel et al., 2012b; Bridges et al., 2013; Fukuzawa et al., 2013; Yang et al., 2013). Moreover, ADGRC1 (CELSR1) expression is found in restricted spatial domains within the murine lung epithelium. In line with this observation, lung branching is perturbed in Crash [Adgrc1 (Celsr1) mutant] mice, which results in disrupted lung development and defects in lung architecture (Yates et al., 2010b). The effect is regulated at least in part via Rho kinase–mediated regulation of the cytoskeleton. Adgra2 (Gpr124)-deficient mouse embryos show reduced lung size (Anderson et al., 2011). To investigate lung hypoplasia, conditional knockout mice will be necessary, because Adgra2 (Gpr124) deficiency results in embryonic lethality caused by abnormal angiogenesis, as detailed above (Kuhnert et al., 2010; Anderson et al., 2011). ADGRF5 (GPR116) mRNA is highly enriched in fetal and adult lung (Bridges et al., 2013; Fukuzawa et al., 2013; Yang et al., 2013). Gene targeting in mice revealed a critical role of ADGRF5 (GPR116) in lung surfactant homeostasis (Bridges et al., 2013; Fukuzawa et al., 2013; Yang et al., 2013). Normally expressed by alveolar type II pneumocytes, ADGRF5 (GPR116) functions as a molecular sensor of alveolar surfactant pool sizes by regulating surfactant secretion. Adgrf5 (Gpr116) deficiency results in progressive accumulation of surfactant lipids and proteins in the alveolar space in young mice, causing labored breathing and decreasing lifespan. These mice can serve as a model to understand human pulmonary alveolar proteinosis, which is characterized by an accumulation of lung surfactant, a compound of phospholipids and proteins that regulates tension in the lung. Finally, single-nucleotide polymorphisms (SNPs) in ADGRG6 (GPR126) are associated with impaired pulmonary function in humans, measured as the ratio of forced expiratory volume in the first second/forced vital capacity (FEV1/FVC), in a meta-analysis of 28,890 participants (Hancock et al., 2010).
4. Gastrointestinal Tract.
ADGRE5 (CD97) is located in lateral cell contacts of human intestinal epithelial cells (Aust et al., 2013). Its overexpression in this cell type in transgenic mice attenuated experimental colitis, probably by strengthening adherens junctions (Becker et al., 2010). Furthermore, Adgre5 (Cd97) transgenic mice show a massive enlargement of the small intestine. This megaintestine phenotype develops after birth, before weaning, with a normal microscopic anatomy and provides a unique model for studying postnatal cylindrical intestinal growth (Aust et al., 2013).
5. Urinary System (Kidney, Urinary Bladder).
Only a handful of Adhesion GPCRs are expressed in the urinary system, and the functional consequences of their presence are unknown. ADGRF1 (GPR110) gene expression was localized in the papilla-ureter transition zone in humans, i.e., the region of the renal pelvis where the tip of the papilla and the ureter meet, yet no obvious defects were observed in Adgrf1 (Gpr110) knockout/LacZ knockin animals (Prömel et al., 2012b). Moreover, murine ADGRC1 (CELSR1) protein was noted in collecting duct stalks and S-shaped bodies, just below the nephrogenic zone, and in proximal tubule and podocyte epithelia in E18.5 kidneys (Yates et al., 2010a).
6. Endocrine System and Metabolism.
For a few Adhesion GPCRs, there is a clear indication of an essential function in endocrine organs and metabolism in general. In mice with adipose tissue–specific absence of Adgrf5 (Gpr116), a pronounced glucose intolerance and insulin resistance developed (Nie et al., 2012). Mice deficient for Adgrc2/3 (Celsr2/3) had severe defects in pancreatic beta cell differentiation, resulting in decreased glucose clearance (Cortijo et al., 2012). Finally, in humans, a genetic association was identified for ADGRC2 (CELSR2), for which SNPs correlate with decreased low-density lipoprotein levels (Kathiresan et al., 2008, 2009).
7. Reproductive Organs.
While key reproductive functions in both sexes are under the control of GPCR signaling, evidence is sparse to this point for roles of Adhesion GPCRs in the reproductive system. As yet, only ADGRG1 (GPR56) and ADGRG2 (GPR64), also named HE6 (human epididymis 6), both members of subfamily ADGRG, have been shown to be essential. ADGRG1 (GPR56) is expressed in peritubular myoid cells, Sertoli cells, and germ cells of the testis (Chen et al., 2010). ADGRG2 (GPR64) is mainly expressed in the nonciliated principal cells of the proximal excurrent ducts (Kirchhoff et al., 2008), which are primarily implicated in testicular fluid reabsorption and sperm concentration. Targeted disruption of either of the encoding genes led to severe subfertility or complete infertility in homozygous knockout males (Davies et al., 2004; Chen et al., 2010). Female reproductive functions, in contrast, were unaffected. Hormonal levels in either of the knockout males were not significantly different from those in wild-type males, suggesting that both receptors exert their functions immediately in a tissue or cell type of the male genital tract. Male subfertility in Adgrg1 (Gpr56) knockout males originated in defects in basal lamina formation during testis development, finally leading to seminiferous tubule disruption (Chen et al., 2010). Targeted disruption of Adgrg2 (Gpr64) led to sperm stasis and duct obstruction, resulting from dysregulation of fluid reabsorption (Davies et al., 2004). Consequences of loss-of-function mutations in human orthologs of these Adhesion GPCRs in male fertility remain unknown.
8. Skeletal Muscle and Bone.
Mounting evidence indicates pivotal functions for Adhesion GPCRs in the musculoskeletal system. Two laboratories recently made exciting discoveries regarding the functions of ADGRB1 (BAI1) and ADGRB3 (BAI3) in this system (Hochreiter-Hufford et al., 2013; Hamoud et al., 2014). Both proteins were reported to be expressed by myoblasts and to promote myoblast fusion into muscle fibers in vitro, but there appear to be differences regarding the in vivo context in which each protein functions, and these differences will be an interesting area for future study.
ADGRG1 (GPR56) was shown to be upregulated during early differentiation of human cultured myoblasts (Wu et al., 2013). In line with these findings, Adgrg1 (Gpr56)-deficient myoblasts showed decreased myoblast fusion in vitro, but Adgrg1 (Gpr56) knockout mice exhibited no overt skeletal muscle phenotype (Wu et al., 2013). Recently, ADGRE5 (CD97) was detected not only in the sarcolemma as other Adhesion GPCRs but also in the sarcoplasmatic reticulum of myocytes. Adgre5 (Cd97)-knockout mice showed a dilated sarcoplasmatic reticulum; yet, despite this severe ultrastructural alteration, the mice had no overt skeletal muscle phenotype (Zyryanova et al., 2014), which is similar to as Adgrb1 (Bai1)- and Adgrg1 (Gpr56)-deficient mice (Hochreiter-Hufford et al., 2013; Wu et al., 2013) and may indicate compensatory mechanisms.
Human height and weight, and thus body stature, are polygenic quantitative traits. Several studies support an association of the locus around ADGRD1 (GPR133) with human height (Tonjes et al., 2009; Kim et al., 2012; Kim et al., 2013). In parallel, Adgrd1 (Gpr133) has been identified as a positional candidate gene controlling body weight in mice in selection experiments for extreme body weight (Chan et al., 2012). ADGRG6 (GPR126) SNPs are also coupled to human skeletal frame size. The SNP rs6570507 has the potential to regulate transcriptional activity and is associated inversely with trunk length (Soranzo et al., 2009) and adolescent idiopathic scoliosis, the most common pediatric skeletal disease (Kou et al., 2013). These data suggest that alterations in ADGRG6 (GPR126) levels result in abnormal skeletal growth. Finally, ADGRV1 (VLGR1) is involved in bone turnover. An increased risk of fractures, a clinical sign of osteoporosis, is associated with SNPs in the 3′-flanking region of ADGRV1 (VLGR1) in postmenopausal women (Urano et al., 2012). Correspondingly, Adgrv1 (Vlgr1) null mice (Yagi et al., 2005) showed decreased femoral bone mineral density and trabeculae/cortical thickness resulting in increased fragility, which are probably caused by increased osteoclast activity (Urano et al., 2012).
9. Skin (Including Hair, Nails, and Mammary Gland).
Adhesion GPCRs can also be found in epidermal epithelia, where most expression studies have been performed in murine tissues. Besides ADGRC1 (CELSR1), ADGRF2 (GPR111), ADGRF4 (GPR115), and ADGRF5 (GPR116) are, among other tissues, also localized to epidermis in the mouse (Bjarnadóttir et al., 2004; Devenport and Fuchs, 2008; Prömel et al., 2012b). While ADGRC1 (CELSR1) has been identified as a key player in hair follicle polarization and PCP in mice (see section IX.A), the function of the other Adhesion GPCRs in the skin remains elusive. Screens for novel skin-associated genes (Gerber et al., 2013) as well as for glucocorticosteroid response genes (Wang et al., 2004) showed ADGRF4 (GPR115) localization to keratinocytes. The appearance of ADGRF2 (GPR111) and ADGRF4 (GPR115) in vertebrate genomes is coincident with terrestrial evolution, suggesting the hypothesis that these Adhesion GPCRs play a function in skin specialization that is essential for life on land (Prömel et al., 2012b). In mouse, both are expressed in stratified squamous epithelium with onset of expression very early during development of the epidermis in embryogenesis. ADGRF2 (GPR111) and ADGRF4 (GPR115) are highly homologous, suggesting a level of functional redundancy, which might be one reason why no phenotype in knockout mice lacking either gene has yet been detected (Prömel et al., 2012b).
10. Nervous System and Behavior.
Most Adhesion GPCRs are present in nervous tissues, and some are especially highly expressed in the brain (Haitina et al., 2008; Regard et al., 2008). While not critical for the survival of individual neurons per se, these Adhesion GPCRs seem to jointly control various high-level functions that are characteristic of the brain as an organ.
ADGRL1 (latrophilin-1) is involved in the control of spontaneous exocytosis in neurons: Upon binding an exogenous agonist, the α-latrotoxin of black widow spiders, it activates Gαq and phospholipase C, leading to the release of stored Ca2+ and massive exocytosis (Davletov et al., 1998; Ashton et al., 2001; Capogna et al., 2003; Volynski et al., 2004). Presynaptic ADGRL1 (latrophilin-1) strongly binds postsynaptic teneurin-2, and this interaction regulates presynaptic Ca2+ dynamics (Silva et al., 2011). Deletion of ADGRL1 (LPHN1), together with five other genes, leads to a neuropsychiatric syndromic disorder in humans (Bonaglia et al., 2010). Adgrl1 (Lphn1) knockout mice are viable, but demonstrate abnormal maternal behavior (Tobaben et al., 2002). ADGRL3 (latrophilin-3) interacts with FLRT3, and this affects the formation and regulation of excitatory synapses (O’Sullivan et al., 2012). SNPs in ADGRL3 (LPHN3) are a risk factor for attention deficit/hyperactivity disorder (Arcos-Burgos et al., 2010; Domene et al., 2011; Ribasés et al., 2011). Loss of Adgrl3 (Lphn3) function in mice and zebrafish disturbs dopaminergic brain circuits, leading to a hyperactive/impulsive phenotype, which can be rescued by anti–attention deficit/hyperactivity disorder drugs (Lange et al., 2012; Wallis et al., 2012).
ADGRC subfamily members (Flamingo/Starry night/CELSRs) influence neural tube closure, the subcellular distribution of ependymal cilia, axon guidance, dendritic morphogenesis, and neuronal migration (Steimel et al., 2010; Berger-Muller and Suzuki, 2011; Boutin et al., 2012; Chai et al., 2014). ADGRC1 (CELSR1) facilitates neural tube closure via core-PCP signaling (Nishimura et al., 2012). However, this process also requires ADGRC3 (CELSR2) and ADGRC3 (CELSR3), hinting at a complex functional network that may involve long-range PCP in conjunction with shorter-range interactions and/or cell adhesion. That ADGRCs (CELSRs) influence neuronal cell adhesion is apparent from studies of axon guidance in insects (Chen and Clandinin, 2008). In distinct organisms, ADGRCs (CELSRs) differentially interact, either genetically or directly, with various proteins, such as Frizzled, Gogo, Espinas, Vangl2, and RhoA, to control axon outgrowth and targeting, as well as dendritic self-avoidance (Berger-Muller and Suzuki, 2011; Matsubara et al., 2011; Hakeda and Suzuki, 2013; Huarcaya Najarro and Ackley, 2013; Chai et al., 2014; Qu et al., 2014). These studies give credence to the idea that the function of 7TM-cadherins could be defined by their binding partners in cis (Berger-Muller and Suzuki, 2011). Notably, in mammals, ADGRC2 (CELSR2) and ADGRC3 (CELSR3) regulate dendrite morphogenesis in opposite ways, but also interact homophilically to trigger intracellular Ca2+ release, suggesting an involvement of G protein signaling (Shima et al., 2007). Adgrc3 (Celsr3) mutant mice exhibit severe defects in neural circuit development within the CNS, similar to frizzled3 and vangl2 mouse mutants (Wang et al., 2002; Tissir et al., 2005; Shafer et al., 2011).
As detailed above, in the central nervous system ADGRB (BAI) proteins may contribute to synapse formation and/or function. In neurons, the ADGRB1 (BAI1) and ADGRB3 (BAI3) proteins are localized postsynaptically, and ADGRB1 (BAI1) expression levels can regulate synaptogenesis (Duman et al., 2013; Lanoue et al., 2013; Stephenson et al., 2013). This function may involve the interaction of ADGRB (BAI) proteins with secreted C1q-like proteins, which strongly bind ADGRB3 (BAI3) and regulate synapse density in vitro (Bolliger et al., 2011). SNPs in ADGRB3 (BAI3) are significantly associated with schizophrenia and addiction predisposition (Liu et al., 2006; DeRosse et al., 2008), and Adgrb2 (Bai2) mutant mice showed significant resistance to depression after repeated stress in the social defeat test (Okajima et al., 2011).
ADGRG1 (GPR56) is likely responsible for mediating the interaction between early-born neurons and the pial basement membrane (Singer et al., 2013). Its binding to collagen III in basement membrane activates the Gα12/13–RhoA pathway, inhibiting neuronal migration. Lack of ADGRG1 (GPR56) leads to breaches in basement membrane and neuronal ectopias during cerebral cortical development, resulting in bilateral frontoparietal polymicrogyria (BFPP) in humans and similar brain malformations in mice (Piao et al., 2004; Li et al., 2008; Luo et al., 2011). Recently, a role for ADGRG1 (GPR56) in gyral patterning and, potentially, neocortex evolution has been described (Bae et al., 2014). ADGRG6 (GPR126) signaling is required in Schwann cells to elevate cAMP and activate protein kinase A to initiate myelination (Monk et al., 2009; Glenn and Talbot, 2013; Mogha et al., 2013). In Adgrg6 (Gpr126) mouse and zebrafish mutants and conditional knockouts, Schwann cells are arrested in development and cannot generate a myelin sheath (Monk et al., 2009, 2011; Mogha et al., 2013).
Studies on the Frings mouse indicated that deficiency in Adgrv1 (Vlgr1) is associated with audiogenic seizures and epilepsy (Skradski et al., 2001). More recently the expression of Adgrv1 (Vlgr1) in oligodendrocytes has also been described, and myelin-associated glycoprotein levels were reduced in the Frings mutant, suggesting a potential role in regulating myelination for this Adhesion GPCR (Shin et al., 2013).
11. Sensory Organs.
Although some Adhesion GPCRs are strongly expressed in rodent sensory organs (Haitina et al., 2008; Regard et al., 2008), relatively little is known about their role in sensory function. However, ADGRF3 (GPR113) has been shown to be selectively expressed in a subset of taste receptor cells (LopezJimenez et al., 2005), whereas ADGRCs 1–3 (CELSRs 1–3) are expressed in the developing murine olfactory epithelium and cochlea (Shima et al., 2002). Adgrc1 (Celsr1) mouse mutants exhibit defects in PCP of the inner ear sensory epithelium, revealed by misorientation of the outer but not inner hair cells (Curtin et al., 2003). Furthermore, zebrafish mutant for adgrg6 (gpr126) have semicircular canal vestibular morphogenesis defects and thus vestibular dysfunction of the inner ear (Geng et al., 2013).
Mutations in the ADGRV1 (VLGR1) gene are associated with a severe sensory-neuronal disorder, the human Usher syndrome (USH) that affects vision and hearing (Weston et al., 2004). In the mouse inner ear, ADGRV1 (VLGR1) is essential for the correct organization of signal-uptaking hair bundles during the differentiation of the mechanosensitive hair cells. There, ADGRV1 (VLGR1) was identified as a component of the transient ankle-links, spanning neighboring stereocilia (McGee et al., 2006; Michalski et al., 2007; Yagi et al., 2007). These links stabilize the growing stereocilia during the differentiation of the hair bundle. In murine retinal photoreceptor cells, ADGRV1 (VLGR1) is also associated with adhesion fibers. The analysis of Adgrv1 (Vlgr1)-deficient mice revealed that ADGRV1 (VLGR1) is an integral component of fibrous links between the membranes of the apical inner segment and the connecting cilium at the base of the photosensitive outer segment (Maerker et al., 2008). In both cell types, ADGRV1 (VLGR1) is part of protein networks composed of other USH proteins (Reiners et al., 2005; van Wijk et al., 2006). In photoreceptor cells, ADGRV1 (VLGR1) is a component of a periciliary USH protein network, which is crucial for the transport of cargo from the inner to the outer segment (Maerker et al., 2008). In addition, ADGRV1 (VLGR1) is found at synapses of cochlear and retinal cells (Reiners et al., 2005; Specht et al., 2009; Zallocchi et al., 2012); however, its synaptic function remains to be elucidated.
C. Clinical Aspects
1. Developmental Defects.
The most intensively-studied human disorder associated with loss of an Adhesion GPCR is BFPP, which is caused by mutations in ADGRG1 (GPR56), as discussed above (Piao et al., 2004, 2005). BFPP is an autosomal recessive disorder (Piao et al., 2002, 2004, 2005; Chang et al., 2003a). Affected individuals present with moderate to severe intellectual disability, motor developmental delay, seizures, ataxia, and abnormal eye movement. Magnetic resonance images of BFPP brains reveal bilateral polymicrogyria with an anterior-to-posterior gradient, decreased white matter volume with associated bilateral signal changes, as well as brainstem and cerebellar hypoplasia. Histologic studies in a Adgrg1 (Gpr56) knockout mouse model and a human postmortem BFPP forebrain demonstrate that BFPP is a cobblestone-like cortical malformation, characterized by neuronal overmigration through a breached pial basement membrane (Li et al., 2008; Bahi-Buisson et al., 2010). Studies in Adgrg1 (Gpr56) knockout mice further demonstrated that ADGRG1 (GPR56) is essential for proper morphogenesis of the rostral cerebellum by its regulation of the external granule cell adhesion to the extracellular matrix proteins (Koirala et al., 2009). Collagen III (gene symbol: Col3a1) is the ligand of ADGRG1 (GPR56) in the developing brain (Luo et al., 2011), and mouse Col3a1 mutants phenocopy Adgrg1 (Gpr56) mutants in cortical malformation (Jeong et al., 2012).
As noted above, mutations in the ADGRV1 (VLGR1) gene cause human Usher syndrome type 2C (Weston et al., 2004). USH is the most common form of combined hereditary deaf-blindness (Wolfrum, 2012), and USH type 2 is characterized by congenital moderate-to-severe hearing impairment and onset of retinal dysfunction (retinitis pigmentosa) in the first or second decade of life. In contrast to USH type 1, the vestibular system is not affected. Vlgr1/del7TM mice lack the fibrous links between membranes of neighboring stereocilia of hair cells and in the periciliary region of photoreceptor cells (McGee et al., 2006; Michalski et al., 2007; Maerker et al., 2008). In the inner ear, the lack of ankle links results in disturbance of the organization of the hair bundles leading to profound deafness by 3 weeks of age, phenocopying the symptoms found in USH2C patients. In retinal photoreceptor cells, the loss of the homologous fibrous links associated with photoreceptor cilia causes only a mild age-related phenotype in mouse models (Michalski et al., 2007), indicating that ADGRV1 (VLGR1) has a more crucial role in human photoreceptor cells. Furthermore, Adgrv1 (Vlgr1) mutations in mice are associated with audiogenic seizures (Skradski et al., 2001) and defects in the human ortholog are possibly related to febrile seizures (Nakayama et al., 2002; Deprez et al., 2006). Interestingly, ADGRV1 (VLGR1) has also been shown to be associated with osteoporosis susceptibility (Urano et al., 2012), indicating an additional crucial role of the receptor in non-neuronal cell types.
In humans, LPHN1 gene deletion has been linked to mental retardation, language delay, hyperactivity, hearing impairment, and cranial malformation (Bonaglia et al., 2010). Members of the ADGRB (BAI) subfamily have also been associated with human disease. As described above, the ADGRB1 (BAI1) and ADGRB3 (BAI3) proteins are localized postsynaptically in neurons, and ADGRB1 (BAI1) expression levels have been shown to regulate synaptogenesis (Selimi et al., 2009; Duman et al., 2013; Stephenson et al., 2013). ADGRB (BAI) proteins may regulate synaptogenesis by interacting with the secreted C1q-like proteins, which were shown to be high-affinity binding partners for the previously orphaned ADGRB3 (BAI3) receptor and to regulate synapse density in vitro (Bolliger et al., 2011). Consistent with a function for the ADGRB (BAI) proteins at synapses, SNPs in ADGRB3 (BAI3) are significantly associated with schizophrenia and addiction predisposition in genome-wide association studies (Liu et al., 2006; DeRosse et al., 2008), and Adgrb2 (Bai2) mutant mice display antidepressant-like behavior (Okajima et al., 2011).
Loss-of-function mutations in ADGRCs 1–3 (CELSRs 1–3) are associated with several human disorders. Mutations in ADGRC1 (CELSR1) cause neural tube defects as well as caudal agenesis in humans (Allache et al., 2012; Robinson et al., 2012). Similarly, mice with missense mutations in Adgrc1 (Celsr1) show craniorachischisis, a severe neural tube defect (Curtin et al., 2003). Furthermore, improperly oriented hair cells in the inner ear and disorganized fur patterning have been reported (Curtin et al., 2003; Ravni et al., 2009). Adgrc2 (Celsr2) knockout and Adgrc2;Adgrc3 (Celsr2;Celsr3) double-knockout mice exhibit impaired development of epidymal cilia resulting in hydrocephalus resulting from defective cerebrospinal fluid dynamics (Tissir et al., 2010), whereas Adgrc3 (Celsr3) knockout mice have defects in the development of the anterior-posterior axon tract organization in the brainstem (Fenstermaker et al., 2010). All three ADGRC (CELSR) proteins are essential for hindbrain neuron migration (Qu et al., 2010).
2. Tumorigenesis.
Cancer genome sequencing has identified putative driver mutations in numerous Adhesion GPCR genes (Greenman et al., 2007; Wood et al., 2007; Jones et al., 2008; Kan et al., 2010; Kang et al., 2013; Weischenfeldt et al., 2013). Notably, missense mutations in ADGRL3 (LPHN3) and ADGRB3 (BAI3) in lung cancer have been predicted to result in loss-of-function of these receptors (Kan et al., 2010). This and the high “frequency of copy loss”–to–“frequency of copy gain” ratio are consistent with a role of these Adhesion GPCRs as tumor suppressors. However, none of the cancer-associated mutations in Adhesion GPCRs have been studied in detail, and thus their impact in cancer progression is not understood.
ADGRE5 (CD97) was the first Adhesion GPCR identified to be involved in cancer (Aust et al., 1997). Widely distributed in normal cells, ADGRE5 (CD97) is regularly induced, upregulated, or post-translationally modified in corresponding malignancies (Aust et al., 1997, 2002, 2006; Steinert et al., 2002; Wobus et al., 2004; Ward et al., 2012; Wu et al., 2012; Mirkowska et al., 2013; Safaee et al., 2013; Zyryanova et al., 2014). In thyroid cancer, ADGRE5 (CD97) expression levels correlate with dedifferentiation (Aust et al., 1997; Ward et al., 2012), and in colorectal cancer, this Adhesion GPCR is overexpressed at the invasion front (Steinert et al., 2002; Galle et al., 2006). Moreover, in gall bladder carcinoma and glioblastoma, ADGRE5 (CD97) expression is inversely related to overall survival (Wu et al., 2012; Safaee et al., 2013). Correspondingly, in mouse models of colorectal and gastric cancer, ADGRE5 (CD97) supported local tumor growth and promoted metastatic spread (Galle et al., 2006; Liu et al., 2012).
Further evidence for a role of ADGRE5 (CD97) in tumorigenesis comes from screening studies demonstrating that ADGRE5 (CD97) is a direct target of the tumor suppressor microRNA-126 in MDA-MB-231 cells (Lu et al., 2014). Gene expression studies and characterization of the leukemia cell surface proteome identified ADGRE5 (CD97) as a marker for minimal residual disease in ALL (acute lymphoblastic leukemia) (Coustan-Smith et al., 2011). ADGRE5 (CD97) expression also accounts for the most informative differences between normal and malignant cells in this leukemia (Mirkowska et al., 2013) and ADGRE5 (CD97) has been identified as a leukemic stem cell marker in acute myeloid leukemia (AML) (Bonardi et al., 2013).
The function of ADGRG1 (GPR56) in cancer seems to be cell- and/or context-specific. On one hand, expression levels of ADGRG1 (GPR56) were found to be inversely correlated with the metastatic potential of melanoma cell lines (Zendman et al., 1999; Xu et al., 2006), and re-expression of the receptor led to a reduction in melanoma growth and metastasis (Yang et al., 2011). On the other hand, ADGRG1 (GPR56) was found to be upregulated in various cancer types (Shashidhar et al., 2005; Kausar et al., 2011) and in tumor cell lines (Ke et al., 2007), compared with normal samples.
In melanoma, the regulatory mechanisms of ADGRG1 (GPR56) appear to involve protein kinase C (PKC)α activation, vascular endothelial growth factor secretion, and tumor angiogenesis (Yang et al., 2011). Overexpression of full-length ADGRG1 (GPR56) versus variants missing the NTF or those parts of the NTF that bind tissue transglutaminase 2 (Xu et al., 2006) had opposing downstream effects in tumor models (Yang et al., 2011). Recently, the mechanism by which ADGRG1 (GPR56) inhibits melanoma growth has been clarified: The tumor-promoting function of tissue transglutaminase 2 and its extracellular matrix-modifying activity is antagonized by ADGRG1 (GPR56) via receptor-mediated internalization and degradation (Yang et al., 2014).
Expression of ADGRF5 (GPR116) is associated with progression, metastasis, and poor prognosis in breast cancer patients (Tang et al., 2013). In breast cancer cell lines, ADGRF5 (GPR116) modulates motility and morphology via the Gαq–p63RhoGEF-Rho–GTPase pathway (Tang et al., 2013).
ADGRB1 (BAI1) expression is absent or downregulated in glioblastoma (Nishimori et al., 1997; Kaur et al., 2003), colorectal cancer (Fukushima et al., 1998), metastatic brain tumors derived from lung (Zohrabian et al., 2007), renal cell carcinoma (Izutsu et al., 2011), and astrocytoma (Wang et al., 2013b). Further, ADGRB1 (BAI1) expression levels are inversely correlated with tumor vascularization (Fukushima et al., 1998; Hatanaka et al., 2000; Duda et al., 2002; Kang et al., 2006; Kudo et al., 2007; Hardcastle et al., 2010), suggesting antiangiogenic functions. Changes in ADGRB1–3 (BAI1–3) expression levels in the experimental focal ischemia model in rodents further suggest a role for these proteins in neovascularization (Koh et al., 2001, Kee et al., 2002, 2004; Jeong et al., 2006). Indeed, proteolysis of ADGRB1 (BAI1) releases 120- or 40-kDa fragments from the NTF, named vasculostatin or vasculostatin-40, respectively, which inhibit migration of endothelial cells in vitro and angiogenesis and/or tumor growth in vivo (Koh et al., 2004; Kaur et al., 2005, 2009; Cork et al., 2012).
In summary, conventional clinical and biologic research as well as genomic and proteomic analyses highlight the importance of Adhesion GPCRs in tumorigenesis.
X. Perspectives on Pharmacological Opportunities
There are no marketed drugs known to act on Adhesion GPCRs, and we are not aware of any current clinical trials targeting these receptors with new drug compounds (Rask-Andersen et al., 2014). However, there is a growing interest in Adhesion GPCR pharmacology, and the structural features of the Adhesion GPCRs (i.e., having multiple extracellular epitopes) can be effectively targeted by antibodies. Numerous studies demonstrate a direct involvement of Adhesion GPCRs in immune reactions and tumor pathogenesis, providing perhaps the best current foundation for their therapeutic potential.
The selective expression of several Adhesion GPCR genes may provide opportunities for immune cell ablation strategies. A recent study showed that afucosylated monoclonal antibodies directed against ADGRE1 (EMR1) dramatically enhanced natural killer cell–mediated cytolysis of human eosinophils and induced rapid and sustained depletion of eosinophils in monkeys (Legrand et al., 2014), providing promise as treatment of eosinophilic disorders. Notably, monoclonal antibodies directed against different EGF domains of the related ADGRE5 (CD97) receptor have been shown to deplete neutrophils specifically under inflammatory conditions (Veninga et al., 2011), with ameliorating consequences in experimental inflammatory disease models (Hamann et al., 2010). These findings indicate that targeting subfamily ADGRE (EGF-TM7) members may have potency for the treatment of inflammatory conditions caused by polymorphonuclear granulocytes.
As summarized in section IX.C.2, Adhesion GPCRs are involved in several aspects of tumorigenesis. Targeting angiogenesis during solid tumor growth may provide potential (onco)therapeutic benefits. ADGRA2 (GPR124) was first described as tumor-endothelial marker 5 (TEM5) because it is present in endothelial cells of the tumor stroma but not of the corresponding normal colonic tissue (Carson-Walter et al., 2001). ADGRA2 (GPR124) is required for vascular endothelial growth factor–induced tumor angiogenesis in vivo (Wang et al., 2014). Moreover, in vitro knockdown of ADGRA2 (GPR124) overcomes resistance to gefitinib (Gao et al., 2014), an EGF receptor tyrosine kinase inhibitor that is a clinically effective treatment of non–small cell lung cancer. Thus, ADGRA2 (GPR124) might serve as a potential therapeutic target for overcoming non–small cell lung cancer gefitinib resistance.
Moreover, modulation of Adhesion GPCR functionality in tumors, either by specific antibodies or drugs, may be an effective approach for preventing tumor progression. Indeed, ectopic expression of ADGRE5 (CD97) was found in human thyroid carcinomas (Aust et al., 1997; Ward et al., 2012), whereas the corresponding normal thyrocytes were ADGRE5 (CD97)-negative. The levels of ADGRE5 (CD97) protein correlated strongly to tumor dedifferentiation, and targeting this Adhesion GPCR might plausibly provide therapeutic benefit. Similarly, ADGRF5 (GPR116) protein levels increased in breast cancer with progression through clinical stages and was highest in tumors with distant metastases (Davies et al., 2011; Tang et al., 2013). However, as described in section VIII, the tumor-associated Adhesion GPCRs ADGRE5 (CD97), ADGRF5 (GPR116), and ADGRG1 (GPR56) are widely expressed and not restricted to a specific cell type or tissue. Thus, their pharmacological targeting will require carefully designed preclinical safety studies and perhaps cell- and/or tissue-specific application of the therapeutic compounds.
There are other burgeoning fields in which Adhesion GPCRs may have a therapeutic role. As mentioned earlier, Adgrf5 (Gpr116) deficiency in mice has been shown to result in progressive accumulation of surfactant lipids and proteins in the alveolar space (Bridges et al., 2013; Fukuzawa et al., 2013; Yang et al., 2013), thus enabling a better understanding of human pulmonary alveolar proteinosis, which is characterized by an accumulation of lung surfactant. Thus, ADGRF5 (GPR116) may provide an opportunity to pharmacologically manipulate pathways that modulate surfactant pool sizes and to achieve therapeutic benefit.
While most of the focus in Adhesion GPCR drug discovery to this point has been related to targeting the receptors’ extracellular domains with antibodies, the potential modulation of these receptors with low-molecular-weight compounds is also an area of interest. Recently, the first high-resolution structures of the related Secretin GPCRs were published (Hollenstein et al., 2013) in complexes with small-molecule drugs. This development may prompt novel means of drug discovery for the Adhesion GPCRs as well. It has been shown that Adhesion GPCRs with a truncated NTF can be expressed at the cell surface, showing basal activity in cAMP assays (Bohnekamp and Schoneberg, 2011), and such truncated receptors could be used for small molecule screening. Beclomethasone dipropionate was recently identified to specifically induce G protein activation via binding to ADGRG3 (GPR97) (Gupte et al., 2012). The availability of low-molecular-weight substances would likely facilitate approaches to target the wide range of Adhesion GPCRs that are expressed in the brain, a tissue that cannot easily be accessed by peripherally administered antibodies owing to the presence of the blood-brain barrier. As mentioned earlier, numerous Adhesion GPCRs of clinical interest are found in the brain, with ADGRL3 (LPHN3) and ADGRB3 (BAI3) being two members of the family that have been associated with psychiatric disorders (Liu et al., 2006; DeRosse et al., 2008; Arcos-Burgos et al., 2010; Domene et al., 2011; Ribasés et al., 2011).
In sum, Adhesion GPCRs possess novel extracellular elements that can be targeted by antibodies serving as diagnostic and therapeutic tools, while their common features of “classic” GPCRs also have potential in the development of low-molecular-weight drugs. Moreover, the cell-specific expression of several Adhesion GPCRs may extend our diagnostic repertoire to identification of cell subpopulations, with the largest progress in this area so far being made for immune and tumor cells. Given the large number of important physiologic processes that Adhesion GPCRs have been shown to regulate, it seems likely that interest in the therapeutic targeting of this receptor family will continue to escalate in the years to come. To harvest the potential of pharmacological modulation of Adhesion GPCRs, the fundamental aspects of the biology of these receptors, including function, ligand interaction, and transmembrane and intracellular signal transduction, need to be studied and forged into quantitative assays to characterize the kinetics and dynamics of Adhesion GPCR activity.
Acknowledgments
This paper was a joint effort of the Adhesion-GPCR Consortium, NC-IUPHAR, and the DFG Research Unit FOR 2149. The authors thank numerous colleagues from these organizations for helpful discussions.
Abbreviations
- BFPP
bilateral frontoparietal polymicrogyria
- CNS
central nervous system
- CTF
C-terminal fragment
- ECD
extracellular domain
- EGF
epidermal growth factor
- FLRT
fibronectin leucine-rich transmembrane
- GAIN
GPCR autoproteolysis–inducing
- GPCR
G protein–coupled receptor
- GPS
GPCR proteolysis site
- HSC
hematopoietic stem cells
- HUVEC
human umbilical vein endothelial cells
- ICD
intracellular domain
- LPA
lysophosphatidic acid
- NTF
N-terminal fragment
- PCP
planar cell polarity
- SNP
single-nucleotide polymorphism
- TM
transmembrane
- USH
Usher syndrome
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Hamann, Aust, Araç, Engel, Formstone, Fredriksson, Hall, Harty, Kirchhoff, Knapp, Krishnan, Liebscher, Lin, Martinelli, Monk, Peeters, Piao, Prömel, Schöneberg, Schwartz, Singer, Stacey, Ushkaryov, Vallon, Wolfrum, Wright, Xu, Langenhan, Schiöth.
Footnotes
NC-IUPHAR is supported in part by Wellcome Trust Grant 099156/Z/12/Z.
References
- Abe J, Suzuki H, Notoya M, Yamamoto T, Hirose S. (1999) Ig-hepta, a novel member of the G protein-coupled hepta-helical receptor (GPCR) family that has immunoglobulin-like repeats in a long N-terminal extracellular domain and defines a new subfamily of GPCRs. J Biol Chem 274:19957–19964. [DOI] [PubMed] [Google Scholar]
- Allache R, De Marco P, Merello E, Capra V, Kibar Z. (2012) Role of the planar cell polarity gene CELSR1 in neural tube defects and caudal agenesis. Birth Defects Res A Clin Mol Teratol 94:176–181. [DOI] [PubMed] [Google Scholar]
- Anderson KD, Pan L, Yang XM, Hughes VC, Walls JR, Dominguez MG, Simmons MV, Burfeind P, Xue Y, Wei Y, et al. (2011) Angiogenic sprouting into neural tissue requires Gpr124, an orphan G protein-coupled receptor. Proc Natl Acad Sci USA 108:2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D, Stubbs JL, Suyama K, Kintner C, Scott MP, Axelrod JD. (2010) Planar cell polarity enables posterior localization of nodal cilia and left-right axis determination during mouse and Xenopus embryogenesis. PLoS ONE 5:e8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araç D, Aust G, Calebiro D, Engel FB, Formstone C, Goffinet A, Hamann J, Kittel RJ, Liebscher I, Lin HH, et al. (2012a) Dissecting signaling and functions of adhesion G protein-coupled receptors. Ann N Y Acad Sci 1276:1–25. [DOI] [PubMed] [Google Scholar]
- Araç D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Südhof TC, Brunger AT. (2012b) A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J 31:1364–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos-Burgos M, Jain M, Acosta MT, Shively S, Stanescu H, Wallis D, Domené S, Vélez JI, Karkera JD, Balog J, et al. (2010) A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry 15:1053–1066. [DOI] [PubMed] [Google Scholar]
- Ashton AC, Volynski KE, Lelianova VG, Orlova EV, Van Renterghem C, Canepari M, Seagar M, Ushkaryov YA. (2001) alpha-Latrotoxin, acting via two Ca2+-dependent pathways, triggers exocytosis of two pools of synaptic vesicles. J Biol Chem 276:44695–44703. [DOI] [PubMed] [Google Scholar]
- Aust G, Eichler W, Laue S, Lehmann I, Heldin NE, Lotz O, Scherbaum WA, Dralle H, Hoang-Vu C. (1997) CD97: a dedifferentiation marker in human thyroid carcinomas. Cancer Res 57:1798–1806. [PubMed] [Google Scholar]
- Aust G, Steinert M, Schütz A, Boltze C, Wahlbuhl M, Hamann J, Wobus M. (2002) CD97, but not its closely related EGF-TM7 family member EMR2, is expressed on gastric, pancreatic, and esophageal carcinomas. Am J Clin Pathol 118:699–707. [DOI] [PubMed] [Google Scholar]
- Aust G, Wandel E, Boltze C, Sittig D, Schütz A, Horn LC, Wobus M. (2006) Diversity of CD97 in smooth muscle cells. Cell Tissue Res 324:139–147. [DOI] [PubMed] [Google Scholar]
- Aust G, Kerner C, Gonsior S, Sittig D, Schneider H, Buske P, Scholz M, Dietrich N, Oldenburg S, Karpus ON, et al. (2013) Mice overexpressing CD97 in intestinal epithelial cells provide a unique model for mammalian postnatal intestinal cylindrical growth. Mol Biol Cell 24:2256–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austyn JM, Gordon S. (1981) F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol 11:805–815. [DOI] [PubMed] [Google Scholar]
- Bae BI, Tietjen I, Atabay KD, Evrony GD, Johnson MB, Asare E, Wang PP, Murayama AY, Im K, Lisgo SN, et al. (2014) Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science 343:764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahi-Buisson N, Poirier K, Boddaert N, Fallet-Bianco C, Specchio N, Bertini E, Caglayan O, Lascelles K, Elie C, Rambaud J, et al. (2010) GPR56-related bilateral frontoparietal polymicrogyria: further evidence for an overlap with the cobblestone complex. Brain 133:3194–3209. [DOI] [PubMed] [Google Scholar]
- Bai Y, Du L, Shen L, Zhang Y, Zhang L. (2009) GPR56 is highly expressed in neural stem cells but downregulated during differentiation. Neuroreport 20:918–922. [DOI] [PubMed] [Google Scholar]
- Baud V, Chissoe SL, Viegas-Péquignot E, Diriong S, N’Guyen VC, Roe BA, Lipinski M. (1995) EMR1, an unusual member in the family of hormone receptors with seven transmembrane segments. Genomics 26:334–344. [DOI] [PubMed] [Google Scholar]
- Becker S, Wandel E, Wobus M, Schneider R, Amasheh S, Sittig D, Kerner C, Naumann R, Hamann J, Aust G. (2010) Overexpression of CD97 in intestinal epithelial cells of transgenic mice attenuates colitis by strengthening adherens junctions. PLoS ONE 5:e8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Müller S, Suzuki T. (2011) Seven-pass transmembrane cadherins: roles and emerging mechanisms in axonal and dendritic patterning. Mol Neurobiol 44:313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnadóttir TK, Fredriksson R, Höglund PJ, Gloriam DE, Lagerström MC, Schiöth HB. (2004) The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics 84:23–33. [DOI] [PubMed] [Google Scholar]
- Bohnekamp J, Schöneberg T. (2011) Cell adhesion receptor GPR133 couples to Gs protein. J Biol Chem 286:41912–41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolliger MF, Martinelli DC, Südhof TC. (2011) The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci USA 108:2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Marelli S, Novara F, Commodaro S, Borgatti R, Minardo G, Memo L, Mangold E, Beri S, Zucca C, et al. (2010) Genotype-phenotype relationship in three cases with overlapping 19p13.12 microdeletions. Eur J Hum Genet 18:1302–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi F, Fusetti F, Deelen P, van Gosliga D, Vellenga E, Schuringa JJ. (2013) A proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Mol Cell Proteomics 12:626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottos A, Rissone A, Bussolino F, Arese M. (2011) Neurexins and neuroligins: synapses look out of the nervous system. Cell Mol Life Sci 68:2655–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Ko J, Südhof TC. (2012) High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem 287:9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin C, Goffinet AM, Tissir F. (2012) Celsr1-3 cadherins in PCP and brain development. Curr Top Dev Biol 101:161–183. [DOI] [PubMed] [Google Scholar]
- Boutin C, Labedan P, Dimidschstein J, Richard F, Cremer H, André P, Yang Y, Montcouquiol M, Goffinet AM, Tissir F. (2014) A dual role for planar cell polarity genes in ciliated cells. Proc Natl Acad Sci USA 111:E3129–E3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges JP, Ludwig MG, Mueller M, Kinzel B, Sato A, Xu Y, Whitsett JA, Ikegami M. (2013) Orphan G protein-coupled receptor GPR116 regulates pulmonary surfactant pool size. Am J Respir Cell Mol Biol 49:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I, Vandenabeele S, Sofi M, McKnight AJ, Ward N, Brodnicki TC, Toy T, Lahoud M, Maraskovsky E, Shortman K, et al. (2006) Gene structure and transcript analysis of the human and mouse EGF-TM7 molecule, FIRE. DNA Seq 17:8–14. [DOI] [PubMed] [Google Scholar]
- Capasso M, Durrant LG, Stacey M, Gordon S, Ramage J, Spendlove I. (2006) Costimulation via CD55 on human CD4+ T cells mediated by CD97. J Immunol 177:1070–1077. [DOI] [PubMed] [Google Scholar]
- Capogna M, Volynski KE, Emptage NJ, Ushkaryov YA. (2003) The alpha-latrotoxin mutant LTXN4C enhances spontaneous and evoked transmitter release in CA3 pyramidal neurons. J Neurosci 23:4044–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. (2001) Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res 61:6649–6655. [PubMed] [Google Scholar]
- Chai G, Zhou L, Manto M, Helmbacher F, Clotman F, Goffinet AM, Tissir F. (2014) Celsr3 is required in motor neurons to steer their axons in the hindlimb. Nat Neurosci 17:1171–1179. [DOI] [PubMed] [Google Scholar]
- Chan YF, Jones FC, McConnell E, Bryk J, Bünger L, Tautz D. (2012) Parallel selection mapping using artificially selected mice reveals body weight control loci. Curr Biol 22:794–800. [DOI] [PubMed] [Google Scholar]
- Chang BS, Piao X, Bodell A, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, et al. (2003a) Bilateral frontoparietal polymicrogyria: clinical and radiological features in 10 families with linkage to chromosome 16. Ann Neurol 53:596–606. [DOI] [PubMed] [Google Scholar]
- Chang GW, Stacey M, Kwakkenbos MJ, Hamann J, Gordon S, Lin HH. (2003b) Proteolytic cleavage of the EMR2 receptor requires both the extracellular stalk and the GPS motif. FEBS Lett 547:145–150. [DOI] [PubMed] [Google Scholar]
- Chen G, Yang L, Begum S, Xu L. (2010) GPR56 is essential for testis development and male fertility in mice. Dev Dyn 239:3358–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Clandinin TR. (2008) The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron 58:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelli O, Reinscheid RK, Zhang Y, Wang Z, Fredriksson R, Schiöth HB. (2013) G protein-coupled receptor deorphanizations. Annu Rev Pharmacol Toxicol 53:127–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork SM, Kaur B, Devi NS, Cooper L, Saltz JH, Sandberg EM, Kaluz S, Van Meir EG. (2012) A proprotein convertase/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Oncogene 31:5144–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo C, Gouzi M, Tissir F, Grapin-Botton A. (2012) Planar cell polarity controls pancreatic beta cell differentiation and glucose homeostasis. Cell Reports 2:1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustan-Smith E, Song G, Clark C, Key L, Liu P, Mehrpooya M, Stow P, Su X, Shurtleff S, Pui CH, et al. (2011) New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood 117:6267–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Elzarrad MK, Seaman S, Zudaire E, Stevens J, Yang MY, Li X, Chaudhary A, Xu L, Hilton MB, et al. (2011) GPR124, an orphan G protein-coupled receptor, is required for CNS-specific vascularization and establishment of the blood-brain barrier. Proc Natl Acad Sci USA 108:5759–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM, et al. (2003) Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol 13:1129–1133. [DOI] [PubMed] [Google Scholar]
- Das S, Owen KA, Ly KT, Park D, Black SG, Wilson JM, Sifri CD, Ravichandran KS, Ernst PB, Casanova JE. (2011) Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc Natl Acad Sci USA 108:2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Sarkar A, Ryan KA, Fox S, Berger AH, Juncadella IJ, Bimczok D, Smythies LE, Harris PR, Ravichandran KS, et al. (2014) Brain angiogenesis inhibitor 1 is expressed by gastric phagocytes during infection with Helicobacter pylori and mediates the recognition and engulfment of human apoptotic gastric epithelial cells. FASEB J 28:2214–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Baumann C, Kirchhoff C, Ivell R, Nubbemeyer R, Habenicht UF, Theuring F, Gottwald U. (2004) Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol Cell Biol 24:8642–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JQ, Chang GW, Yona S, Gordon S, Stacey M, Lin HH. (2007) The role of receptor oligomerization in modulating the expression and function of leukocyte adhesion-G protein-coupled receptors. J Biol Chem 282:27343–27353. [DOI] [PubMed] [Google Scholar]
- Davies JQ, Lin HH, Stacey M, Yona S, Chang GW, Gordon S, Hamann J, Campo L, Han C, Chan P, et al. (2011) Leukocyte adhesion-GPCR EMR2 is aberrantly expressed in human breast carcinomas and is associated with patient survival. Oncol Rep 25:619–627. [DOI] [PubMed] [Google Scholar]
- Davletov BA, Meunier FA, Ashton AC, Matsushita H, Hirst WD, Lelianova VG, Wilkin GP, Dolly JO, Ushkaryov YA. (1998) Vesicle exocytosis stimulated by alpha-latrotoxin is mediated by latrophilin and requires both external and stored Ca2+. EMBO J 17:3909–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Curtis I, Meldolesi J. (2012) Cell surface dynamics - how Rho GTPases orchestrate the interplay between the plasma membrane and the cortical cytoskeleton. J Cell Sci 125:4435–4444. [DOI] [PubMed] [Google Scholar]
- Della Chiesa M, Falco M, Parolini S, Bellora F, Petretto A, Romeo E, Balsamo M, Gambarotti M, Scordamaglia F, Tabellini G, et al. (2010) GPR56 as a novel marker identifying the CD56dull CD16+ NK cell subset both in blood stream and in inflamed peripheral tissues. Int Immunol 22:91–100. [DOI] [PubMed] [Google Scholar]
- Deprez L, Claes LR, Claeys KG, Audenaert D, Van Dyck T, Goossens D, Van Paesschen W, Del-Favero J, Van Broeckhoven C, De Jonghe P. (2006) Genome-wide linkage of febrile seizures and epilepsy to the FEB4 locus at 5q14.3-q23.1 and no MASS1 mutation. Hum Genet 118:618–625. [DOI] [PubMed] [Google Scholar]
- DeRosse P, Lencz T, Burdick KE, Siris SG, Kane JM, Malhotra AK. (2008) The genetics of symptom-based phenotypes: toward a molecular classification of schizophrenia. Schizophr Bull 34:1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D, Fuchs E. (2008) Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol 10:1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport D, Oristian D, Heller E, Fuchs E. (2011) Mitotic internalization of planar cell polarity proteins preserves tissue polarity. Nat Cell Biol 13:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domené S, Stanescu H, Wallis D, Tinloy B, Pineda DE, Kleta R, Arcos-Burgos M, Roessler E, Muenke M. (2011) Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. Am J Med Genet B Neuropsychiatr Genet 156B:11–18. [DOI] [PubMed] [Google Scholar]
- Doyle SE, Scholz MJ, Greer KA, Hubbard AD, Darnell DK, Antin PB, Klewer SE, Runyan RB. (2006) Latrophilin-2 is a novel component of the epithelial-mesenchymal transition within the atrioventricular canal of the embryonic chicken heart. Dev Dyn 235:3213–3221. [DOI] [PubMed] [Google Scholar]
- Duda DG, Sunamura M, Lozonschi L, Yokoyama T, Yatsuoka T, Motoi F, Horii A, Tani K, Asano S, Nakamura Y, et al. (2002) Overexpression of the p53-inducible brain-specific angiogenesis inhibitor 1 suppresses efficiently tumour angiogenesis. Br J Cancer 86:490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman JG, Tzeng CP, Tu YK, Munjal T, Schwechter B, Ho TS, Tolias KF. (2013) The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J Neurosci 33:6964–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dym M, He Z, Jiang J, Pant D, Kokkinaki M. (2009) Spermatogonial stem cells: unlimited potential. Reprod Fertil Dev 21:15–21. [DOI] [PubMed] [Google Scholar]
- Eichler W, Aust G, Hamann D. (1994) Characterization of an early activation-dependent antigen on lymphocytes defined by the monoclonal antibody BL-Ac(F2). Scand J Immunol 39:111–115. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Zheng S, Park D, Woodson RI, Reardon MA, Juncadella IJ, Kinchen JM, Zhang J, Lysiak JJ, Ravichandran KS. (2010) Unexpected requirement for ELMO1 in clearance of apoptotic germ cells in vivo. Nature 467:333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenstermaker AG, Prasad AA, Bechara A, Adolfs Y, Tissir F, Goffinet A, Zou Y, Pasterkamp RJ. (2010) Wnt/planar cell polarity signaling controls the anterior-posterior organization of monoaminergic axons in the brainstem. J Neurosci 30:16053–16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrand N, Pessah M, Frayon S, Marais J, Garel JM. (1999) Olfactory receptors, Golf alpha and adenylyl cyclase mRNA expressions in the rat heart during ontogenic development. J Mol Cell Cardiol 31:1137–1142. [DOI] [PubMed] [Google Scholar]
- Formstone CJ, Moxon C, Murdoch J, Little P, Mason I. (2010) Basal enrichment within neuroepithelia suggests novel function(s) for Celsr1 protein. Mol Cell Neurosci 44:210–222. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Gloriam DE, Höglund PJ, Lagerström MC, Schiöth HB. (2003a) There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem Biophys Res Commun 301:725–734. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Höglund PJ, Schiöth HB. (2002) Novel human G protein-coupled receptors with long N-terminals containing GPS domains and Ser/Thr-rich regions. FEBS Lett 531:407–414. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. (2003b) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63:1256–1272. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Schiöth HB. (2005) The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol 67:1414–1425. [DOI] [PubMed] [Google Scholar]
- Fukushima Y, Oshika Y, Tsuchida T, Tokunaga T, Hatanaka H, Kijima H, Yamazaki H, Ueyama Y, Tamaoki N, Nakamura M. (1998) Brain-specific angiogenesis inhibitor 1 expression is inversely correlated with vascularity and distant metastasis of colorectal cancer. Int J Oncol 13:967–970. [DOI] [PubMed] [Google Scholar]
- Fukuzawa T, Hirose S. (2006) Multiple processing of Ig-Hepta/GPR116, a G protein-coupled receptor with immunoglobulin (Ig)-like repeats, and generation of EGF2-like fragment. J Biochem 140:445–452. [DOI] [PubMed] [Google Scholar]
- Fukuzawa T, Ishida J, Kato A, Ichinose T, Ariestanti DM, Takahashi T, Ito K, Abe J, Suzuki T, Wakana S, et al. (2013) Lung surfactant levels are regulated by Ig-Hepta/GPR116 by monitoring surfactant protein D. PLoS ONE 8:e69451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle J, Sittig D, Hanisch I, Wobus M, Wandel E, Loeffler M, Aust G. (2006) Individual cell-based models of tumor-environment interactions: Multiple effects of CD97 on tumor invasion. Am J Pathol 169:1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN. (1999) Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev 13:2549–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. (2014) miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun 446:179–186. [DOI] [PubMed] [Google Scholar]
- Geng FS, Abbas L, Baxendale S, Holdsworth CJ, Swanson AG, Slanchev K, Hammerschmidt M, Topczewski J, Whitfield TT. (2013) Semicircular canal morphogenesis in the zebrafish inner ear requires the function of gpr126 (lauscher), an adhesion class G protein-coupled receptor gene. Development 140:4362–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber PA, Hevezi P, Buhren BA, Martinez C, Schrumpf H, Gasis M, Grether-Beck S, Krutmann J, Homey B, Zlotnik A. (2013) Systematic identification and characterization of novel human skin-associated genes encoding membrane and secreted proteins. PLoS ONE 8:e63949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn TD, Talbot WS. (2013) Analysis of Gpr126 function defines distinct mechanisms controlling the initiation and maturation of myelin. Development 140:3167–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloriam DE, Fredriksson R, Schiöth HB. (2007) The G protein-coupled receptor subset of the rat genome. BMC Genomics 8:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Hamann J, Lin HH, Stacey M. (2011) F4/80 and the related adhesion-GPCRs. Eur J Immunol 41:2472–2476. [DOI] [PubMed] [Google Scholar]
- Gouveia LO, Sobral J, Vicente AM, Ferro JM, Oliveira SA. (2011) Replication of the CELSR1 association with ischemic stroke in a Portuguese case-control cohort. Atherosclerosis 217:260–262. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte J, Swaminath G, Danao J, Tian H, Li Y, Wu X. (2012) Signaling property study of adhesion G-protein-coupled receptors. FEBS Lett 586:1214–1219. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Sheward WJ, Harmar AJ, de Galan L, Hoovers JMN, Little PFR. (1997) Celsr1, a neural-specific gene encoding an unusual seven-pass transmembrane receptor, maps to mouse chromosome 15 and human chromosome 22qter. Genomics 45:97–104. [DOI] [PubMed] [Google Scholar]
- Haitina T, Olsson F, Stephansson O, Alsiö J, Roman E, Ebendal T, Schiöth HB, Fredriksson R. (2008) Expression profile of the entire family of Adhesion G protein-coupled receptors in mouse and rat. BMC Neurosci 9:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitina T, Fredriksson R, Foord SM, Schiöth HB, Gloriam DE. (2009) The G protein-coupled receptor subset of the dog genome is more similar to that in humans than rodents. BMC Genomics 10:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda S, Suzuki T. (2013) Golden goal controls dendrite elongation and branching of multidendritic arborization neurons in Drosophila. Genes Cells 18:960–973. [DOI] [PubMed] [Google Scholar]
- Hamann J, Eichler W, Hamann D, Kerstens HM, Poddighe PJ, Hoovers JM, Hartmann E, Strauss M, van Lier RA. (1995) Expression cloning and chromosomal mapping of the leukocyte activation antigen CD97, a new seven-span transmembrane molecule of the secretion receptor superfamily with an unusual extracellular domain. J Immunol 155:1942–1950. [PubMed] [Google Scholar]
- Hamann J, Hartmann E, van Lier RAW. (1996a) Structure of the human CD97 gene: exon shuffling has generated a new type of seven-span transmembrane molecule related to the secretin receptor superfamily. Genomics 32:144–147. [DOI] [PubMed] [Google Scholar]
- Hamann J, Vogel B, van Schijndel GMW, van Lier RAW. (1996b) The seven-span transmembrane receptor CD97 has a cellular ligand (CD55, DAF). J Exp Med 184:1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann J, Stortelers C, Kiss-Toth E, Vogel B, Eichler W, van Lier RA. (1998) Characterization of the CD55 (DAF)-binding site on the seven-span transmembrane receptor CD97. Eur J Immunol 28:1701–1707. [DOI] [PubMed] [Google Scholar]
- Hamann J, Kwakkenbos MJ, de Jong EC, Heus H, Olsen AS, van Lier RAW. (2003) Inactivation of the EGF-TM7 receptor EMR4 after the Pan-Homo divergence. Eur J Immunol 33:1365–1371. [DOI] [PubMed] [Google Scholar]
- Hamann J, Koning N, Pouwels W, Ulfman LH, van Eijk M, Stacey M, Lin HH, Gordon S, Kwakkenbos MJ. (2007) EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. Eur J Immunol 37:2797–2802. [DOI] [PubMed] [Google Scholar]
- Hamann J, Veninga H, de Groot DM, Visser L, Hofstra CL, Tak PP, Laman JD, Boots AM, van Eenennaam H. (2010) CD97 in leukocyte trafficking. Adv Exp Med Biol 706:128–137. [DOI] [PubMed] [Google Scholar]
- Hamoud N, Tran V, Croteau LP, Kania A and Coté JF (2014) G-Protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc Natl Acad Sci USA 111:3745–3750. [DOI] [PMC free article] [PubMed]
- Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. (2010) Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 42:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle J, Kurozumi K, Dmitrieva N, Sayers MP, Ahmad S, Waterman P, Weissleder R, Chiocca EA, Kaur B. (2010) Enhanced antitumor efficacy of vasculostatin (Vstat120) expressing oncolytic HSV-1. Mol Ther 18:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Shinohara K, Wang J, Ikeuchi S, Yoshiba S, Meno C, Nonaka S, Takada S, Hatta K, Wynshaw-Boris A, et al. (2010) Planar polarization of node cells determines the rotational axis of node cilia. Nat Cell Biol 12:170–176. [DOI] [PubMed] [Google Scholar]
- Hatanaka H, Oshika Y, Abe Y, Yoshida Y, Hashimoto T, Handa A, Kijima H, Yamazaki H, Inoue H, Ueyama Y, et al. (2000) Vascularization is decreased in pulmonary adenocarcinoma expressing brain-specific angiogenesis inhibitor 1 (BAI1). Int J Mol Med 5:181–183. [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, Call JA, Klibanov AL, Yan Z, Mandell JW, Ravichandran KS. (2013) Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497:263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Kean J, Bortolato A, Cheng RK, Doré AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. (2013) Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499:438–443. [DOI] [PubMed] [Google Scholar]
- Hu QX, Dong JH, Du HB, Zhang DL, Ren HZ, Ma ML, Cai Y, Zhao TC, Yin XL, Yu X, et al. (2014) Constitutive Gαi coupling activity of very large G protein-coupled receptor 1 (VLGR1) and its regulation by PDZD7 protein. J Biol Chem 289:24215–24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Chiang NY, Hu CH, Hsiao CC, Cheng KF, Tsai WP, Yona S, Stacey M, Gordon S, Chang GW, et al. (2012) Activation of myeloid cell-specific adhesion class G protein-coupled receptor EMR2 via ligation-induced translocation and interaction of receptor subunits in lipid raft microdomains. Mol Cell Biol 32:1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarcaya Najarro E, Ackley BD. (2013) C. elegans fmi-1/flamingo and Wnt pathway components interact genetically to control the anteroposterior neurite growth of the VD GABAergic neurons. Dev Biol 377:224–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P, van Roy F. (2009) Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol 41:349–369. [DOI] [PubMed] [Google Scholar]
- Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. (2008) Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem 283:14469–14478. [DOI] [PubMed] [Google Scholar]
- Izadyar F, Wong J, Maki C, Pacchiarotti J, Ramos T, Howerton K, Yuen C, Greilach S, Zhao HH, Chow M, et al. (2011) Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod 26:1296–1306. [DOI] [PubMed] [Google Scholar]
- Izutsu T, Konda R, Sugimura J, Iwasaki K, Fujioka T. (2011) Brain-specific angiogenesis inhibitor 1 is a putative factor for inhibition of neovascular formation in renal cell carcinoma. J Urol 185:2353–2358. [DOI] [PubMed] [Google Scholar]
- Jaspars LH, Vos W, Aust G, Van Lier RA, Hamann J. (2001) Tissue distribution of the human CD97 EGF-TM7 receptor. Tissue Antigens 57:325–331. [DOI] [PubMed] [Google Scholar]
- Jeong BC, Kim MY, Lee JH, Kee HJ, Kho DH, Han KE, Qian YR, Kim JK, Kim KK. (2006) Brain-specific angiogenesis inhibitor 2 regulates VEGF through GABP that acts as a transcriptional repressor. FEBS Lett 580:669–676. [DOI] [PubMed] [Google Scholar]
- Jeong SJ, Luo R, Li S, Strokes N, Piao X. (2012) Characterization of G protein-coupled receptor 56 protein expression in the mouse developing neocortex. J Comp Neurol 520:2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SJ, Luo R, Singer K, Giera S, Kreidberg J, Kiyozumi D, Shimono C, Sekiguchi K, Piao X. (2013) GPR56 functions together with α3β1 integrin in regulating cerebral cortical development. PLoS ONE 8:e68781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321:1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamesh N, Aradhyam GK, Manoj N. (2008) The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis. BMC Evol Biol 8:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. (2010) Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466:869–873. [DOI] [PubMed] [Google Scholar]
- Kang G, Hwang WC, Do IG, Wang K, Kang SY, Lee J, Park SH, Park JO, Kang WK, Jang J, et al. (2013) Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PLoS ONE 8:e82770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Xiao X, Harata M, Bai Y, Nakazaki Y, Soda Y, Kurita R, Tanaka T, Komine F, Izawa K, et al. (2006) Antiangiogenic activity of BAI1 in vivo: implications for gene therapy of human glioblastomas. Cancer Gene Ther 13:385–392. [DOI] [PubMed] [Google Scholar]
- Karpus ON, Veninga H, Hoek RM, Flierman D, van Buul JD, Vandenakker CC, vanBavel E, Medof ME, van Lier RA, Reedquist KA, et al. (2013) Shear stress-dependent downregulation of the adhesion-G protein-coupled receptor CD97 on circulating leukocytes upon contact with its ligand CD55. J Immunol 190:3740–3748. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, et al. Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium (2009) Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 41:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur B, Brat DJ, Calkins CC, Van Meir EG. (2003) Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am J Pathol 162:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur B, Brat DJ, Devi NS, Van Meir EG. (2005) Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene 24:3632–3642. [DOI] [PubMed] [Google Scholar]
- Kaur B, Cork SM, Sandberg EM, Devi NS, Zhang Z, Klenotic PA, Febbraio M, Shim H, Mao H, Tucker-Burden C, et al. (2009) Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res 69:1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausar T, Sharma R, Hasan MR, Tripathi SC, Saraya A, Chattopadhyay TK, Gupta SD, Ralhan R. (2011) Clinical significance of GPR56, transglutaminase 2, and NF-κB in esophageal squamous cell carcinoma. Cancer Invest 29:42–48. [DOI] [PubMed] [Google Scholar]
- Ke N, Sundaram R, Liu G, Chionis J, Fan W, Rogers C, Awad T, Grifman M, Yu D, Wong-Staal F, et al. (2007) Orphan G protein-coupled receptor GPR56 plays a role in cell transformation and tumorigenesis involving the cell adhesion pathway. Mol Cancer Ther 6:1840–1850. [DOI] [PubMed] [Google Scholar]
- Kee HJ, Koh JT, Kim MY, Ahn KY, Kim JK, Bae CS, Park SS, Kim KK. (2002) Expression of brain-specific angiogenesis inhibitor 2 (BAI2) in normal and ischemic brain: involvement of BAI2 in the ischemia-induced brain angiogenesis. J Cereb Blood Flow Metab 22:1054–1067. [DOI] [PubMed] [Google Scholar]
- Kee HJ, Ahn KY, Choi KC, Won Song J, Heo T, Jung S, Kim JK, Bae CS, Kim KK. (2004) Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett 569:307–316. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Park YM, Baik KH, Choi HY, Yang GS, Koh I, Hwang JA, Lee J, Lee YS, Rhee H, et al. (2012) Exome sequencing and subsequent association studies identify five amino acid-altering variants influencing human height. Hum Genet 131:471–478. [DOI] [PubMed] [Google Scholar]
- Kim YK, Moon S, Hwang MY, Kim DJ, Oh JH, Kim YJ, Han BG, Lee JY, Kim BJ. (2013) Gene-based copy number variation study reveals a microdeletion at 12q24 that influences height in the Korean population. Genomics 101:134–138. [DOI] [PubMed] [Google Scholar]
- Kirchhoff C, Osterhoff C, Samalecos A. (2008) HE6/GPR64 adhesion receptor co-localizes with apical and subapical F-actin scaffold in male excurrent duct epithelia. Reproduction 136:235–245. [DOI] [PubMed] [Google Scholar]
- Klenotic PA, Huang P, Palomo J, Kaur B, Van Meir EG, Vogelbaum MA, Febbraio M, Gladson CL, Silverstein RL. (2010) Histidine-rich glycoprotein modulates the anti-angiogenic effects of vasculostatin. Am J Pathol 176:2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh JT, Lee ZH, Ahn KY, Kim JK, Bae CS, Kim HH, Kee HJ, Kim KK. (2001) Characterization of mouse brain-specific angiogenesis inhibitor 1 (BAI1) and phytanoyl-CoA alpha-hydroxylase-associated protein 1, a novel BAI1-binding protein. Brain Res Mol Brain Res 87:223–237. [DOI] [PubMed] [Google Scholar]
- Koh JT, Kook H, Kee HJ, Seo YW, Jeong BC, Lee JH, Kim MY, Yoon KC, Jung S, Kim KK. (2004) Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Exp Cell Res 294:172–184. [DOI] [PubMed] [Google Scholar]
- Koirala S, Jin Z, Piao X, Corfas G. (2009) GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci 29:7439–7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou I, Takahashi Y, Johnson TA, Takahashi A, Guo L, Dai J, Qiu X, Sharma S, Takimoto A, Ogura Y, et al. (2013) Genetic variants in GPR126 are associated with adolescent idiopathic scoliosis. Nat Genet 45:676–679. [DOI] [PubMed] [Google Scholar]
- Krasnoperov V, Deyev IE, Serova OV, Xu C, Lu Y, Buryanovsky L, Gabibov AG, Neubert TA, Petrenko AG. (2009) Dissociation of the subunits of the calcium-independent receptor of alpha-latrotoxin as a result of two-step proteolysis. Biochemistry 48:3230–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, Little AR, Plotnikov AN, Wu D, Holz RW, et al. (1997) alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron 18:925–937. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Almén MS, Fredriksson R, Schiöth HB. (2012) The origin of GPCRs: identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS ONE 7:e29817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Almén MS, Fredriksson R, Schiöth HB. (2013) Remarkable similarities between the hemichordate (Saccoglossus kowalevskii) and vertebrate GPCR repertoire. Gene 526:122–133. [DOI] [PubMed] [Google Scholar]
- Kudo S, Konda R, Obara W, Kudo D, Tani K, Nakamura Y, Fujioka T. (2007) Inhibition of tumor growth through suppression of angiogenesis by brain-specific angiogenesis inhibitor 1 gene transfer in murine renal cell carcinoma. Oncol Rep 18:785–791. [PubMed] [Google Scholar]
- Kuhnert F, Mancuso MR, Shamloo A, Wang HT, Choksi V, Florek M, Su H, Fruttiger M, Young WL, Heilshorn SC, et al. (2010) Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science 330:985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Chang GW, Lin HH, Pouwels W, de Jong EC, van Lier RA, Gordon S, Hamann J. (2002) The human EGF-TM7 family member EMR2 is a heterodimeric receptor expressed on myeloid cells. J Leukoc Biol 71:854–862. [PubMed] [Google Scholar]
- Kwakkenbos MJ, Kop EN, Stacey M, Matmati M, Gordon S, Lin HH, Hamann J. (2004) The EGF-TM7 family: a postgenomic view. Immunogenetics 55:655–666. [DOI] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Matmati M, Madsen O, Pouwels W, Wang Y, Bontrop RE, Heidt PJ, Hoek RM, Hamann J. (2006) An unusual mode of concerted evolution of the EGF-TM7 receptor chimera EMR2. FASEB J 20:2582–2584. [DOI] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Pouwels W, Matmati M, Stacey M, Lin HH, Gordon S, van Lier RA, Hamann J. (2005) Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J Leukoc Biol 77:112–119. [DOI] [PubMed] [Google Scholar]
- Lagerström MC, Schiöth HB. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov 7:339–357. [DOI] [PubMed] [Google Scholar]
- Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, Schmitt A, Vernier P, Lesch KP, Bally-Cuif L. (2012) The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry 17:946–954. [DOI] [PubMed] [Google Scholar]
- Langenhan T, Prömel S, Mestek L, Esmaeili B, Waller-Evans H, Hennig C, Kohara Y, Avery L, Vakonakis I, Schnabel R, et al. (2009) Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Dev Cell 17:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan T, Aust G, Hamann J. (2013) Sticky signaling—adhesion class G protein-coupled receptors take the stage. Sci Signal 6:re3. [DOI] [PubMed] [Google Scholar]
- Lanoue V, Usardi A, Sigoillot SM, Talleur M, Iyer K, Mariani J, Isope P, Vodjdani G, Heintz N, Selimi F. (2013) The adhesion-GPCR BAI3, a gene linked to psychiatric disorders, regulates dendrite morphogenesis in neurons. Mol Psychiatry 18:943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA. (1973) The development of spatial patterns in the integument of insects, in Developmental Systems: Insects (Counce SJ, Waddington CH, eds) pp 157–209, Academic Press, London, New York. [Google Scholar]
- Lawrence PA, Shelton PM. (1975) The determination of polarity in the developing insect retina. J Embryol Exp Morphol 33:471–486. [PubMed] [Google Scholar]
- Leemans JC, te Velde AA, Florquin S, Bennink RJ, de Bruin K, van Lier RA, van der Poll T, Hamann J. (2004) The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J Immunol 172:1125–1131. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Cotecchia S, Samama P, Costa T. (1993) Constitutive activity of receptors coupled to guanine nucleotide regulatory proteins. Trends Pharmacol Sci 14:303–307. [DOI] [PubMed] [Google Scholar]
- Legrand F, Tomasevic N, Simakova O, Lee CC, Wang Z, Raffeld M, Makiya MA, Palath V, Leung J, Baer M, et al. (2014) The eosinophil surface receptor epidermal growth factor-like module containing mucin-like hormone receptor 1 (EMR1): a novel therapeutic target for eosinophilic disorders. J Allergy Clin Immunol 133:1439–1447, e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF, Ushkaryov YA. (1997) Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem 272:21504–21508. [DOI] [PubMed] [Google Scholar]
- Li S, Jin Z, Koirala S, Bu L, Xu L, Hynes RO, Walsh CA, Corfas G, Piao X. (2008) GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci 28:5817–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao FB. (2003) Actin filament-stabilizing protein tropomyosin regulates the size of dendritic fields. J Neurosci 23:6171–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Roszko I, Sepich DS, Ni M, Hamm HE, Marlow FL, Solnica-Krezel L. (2013) Gpr125 modulates Dishevelled distribution and planar cell polarity signaling. Development 140:3028–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebscher I, Schöneberg T, Prömel S. (2013) Progress in demystification of adhesion G protein-coupled receptors. Biol Chem 394:937–950. [DOI] [PubMed] [Google Scholar]
- Lin HH, Stacey M, Hamann J, Gordon S, McKnight AJ. (2000) Human EMR2, a novel EGF-TM7 molecule on chromosome 19p13.1, is closely related to CD97. Genomics 67:188–200. [DOI] [PubMed] [Google Scholar]
- Lin HH, Stacey M, Saxby C, Knott V, Chaudhry Y, Evans D, Gordon S, McKnight AJ, Handford P, Lea S. (2001) Molecular analysis of the epidermal growth factor-like short consensus repeat domain-mediated protein-protein interactions: dissection of the CD97-CD55 complex. J Biol Chem 276:24160–24169. [DOI] [PubMed] [Google Scholar]
- Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S. (2004) Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem 279:31823–31832. [DOI] [PubMed] [Google Scholar]
- Lin HH, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J, Kerley M, Mucenski ML, Gordon S, Stein-Streilein J. (2005) The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med 201:1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Stacey M, Yona S, Chang GW. (2010) GPS proteolytic cleavage of adhesion-GPCRs. Adv Exp Med Biol 706:49–58. [DOI] [PubMed] [Google Scholar]
- Lisé MF, El-Husseini A. (2006) The neuroligin and neurexin families: from structure to function at the synapse. Cell Mol Life Sci 63:1833–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little KD, Hemler ME, Stipp CS. (2004) Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol Biol Cell 15:2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Trojanowicz B, Ye L, Li C, Zhang L, Li X, Li G, Zheng Y, Chen L. (2012) The invasion and metastasis promotion role of CD97 small isoform in gastric carcinoma. PLoS ONE 7:e39989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. (2006) Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet B Neuropsychiatr Genet 141B:918–925. [DOI] [PubMed] [Google Scholar]
- LopezJimenez ND, Sainz E, Cavenagh MM, Cruz-Ithier MA, Blackwood CA, Battey JF, Sullivan SL. (2005) Two novel genes, Gpr113, which encodes a family 2 G-protein-coupled receptor, and Trcg1, are selectively expressed in taste receptor cells. Genomics 85:472–482. [DOI] [PubMed] [Google Scholar]
- Lu YY, Sweredoski MJ, Huss D, Lansford R, Hess S, Tirrell DA. (2014) Prometastatic GPCR CD97 is a direct target of tumor suppressor microRNA-126. ACS Chem Biol 9:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. (2011) G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci USA 108:12925–12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerker T, van Wijk E, Overlack N, Kersten FF, McGee J, Goldmann T, Sehn E, Roepman R, Walsh EJ, Kremer H, et al. (2008) A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum Mol Genet 17:71–86. [DOI] [PubMed] [Google Scholar]
- Magalhaes AC, Dunn H, Ferguson SS. (2012) Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br J Pharmacol 165:1717–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F, Pfeufer A, Aulchenko YS, Franklin CS, Isaacs A, Pichler I, Wild SH, Oostra BA, Wright AF, Campbell H, et al. EUROSPAN Consortium (2009) A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the EUROSPAN project. Circ Cardiovasc Genet 2:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero M, Simões FC, Han HD, Snell C, Peterkin T, Bridges E, Mangala LS, Wu SY, Pradeep S, Li D, et al. (2013) A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell 24:229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matmati M, Pouwels W, van Bruggen R, Jansen M, Hoek RM, Verhoeven AJ, Hamann J. (2007) The human EGF-TM7 receptor EMR3 is a marker for mature granulocytes. J Leukoc Biol 81:440–448. [DOI] [PubMed] [Google Scholar]
- Matsubara D, Horiuchi SY, Shimono K, Usui T, Uemura T. (2011) The seven-pass transmembrane cadherin Flamingo controls dendritic self-avoidance via its binding to a LIM domain protein, Espinas, in Drosophila sensory neurons. Genes Dev 25:1982–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaheri F, Breus O, Durdu S, Haas P, Wittbrodt J, Gilmour D, Peri F. (2014) Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat Commun 5:4046. [DOI] [PubMed] [Google Scholar]
- McGee J, Goodyear RJ, McMillan DR, Stauffer EA, Holt JR, Locke KG, Birch DG, Legan PK, White PC, Walsh EJ, et al. (2006) The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci 26:6543–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, White PC. (2010) Studies on the very large G protein-coupled receptor: from initial discovery to determining its role in sensorineural deafness in higher animals. Adv Exp Med Biol 706:76–86. [DOI] [PubMed] [Google Scholar]
- Michalski N, Michel V, Bahloul A, Lefèvre G, Barral J, Yagi H, Chardenoux S, Weil D, Martin P, Hardelin JP, et al. (2007) Molecular characterization of the ankle-link complex in cochlear hair cells and its role in the hair bundle functioning. J Neurosci 27:6478–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkowska P, Hofmann A, Sedek L, Slamova L, Mejstrikova E, Szczepanski T, Schmitz M, Cario G, Stanulla M, Schrappe M, et al. (2013) Leukemia surfaceome analysis reveals new disease-associated features. Blood 121:e149–e159. [DOI] [PubMed] [Google Scholar]
- Moers A, Nürnberg A, Goebbels S, Wettschureck N, Offermanns S. (2008) Galpha12/Galpha13 deficiency causes localized overmigration of neurons in the developing cerebral and cerebellar cortices. Mol Cell Biol 28:1480–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogha A, Benesh AE, Patra C, Engel FB, Schöneberg T, Liebscher I, Monk KR. (2013) Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J Neurosci 33:17976–17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, Moens CB, Talbot WS. (2009) A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science 325:1402–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Oshima K, Jörs S, Heller S, Talbot WS. (2011) Gpr126 is essential for peripheral nerve development and myelination in mammals. Development 138:2673–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarro EH, Wong L, Zhen M, Carpio EP, Goncharov A, Garriga G, Lundquist EA, Jin Y, Ackley BD. (2012) Caenorhabditis elegans flamingo cadherin fmi-1 regulates GABAergic neuronal development. J Neurosci 32:4196–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Fu YH, Clark AM, Nakahara S, Hamano K, Iwasaki N, Matsui A, Arinami T, Ptácek LJ. (2002) A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann Neurol 52:654–657. [DOI] [PubMed] [Google Scholar]
- Nie T, Hui X, Gao X, Li K, Lin W, Xiang X, Ding M, Kuang Y, Xu A, Fei J, et al. (2012) Adipose tissue deletion of Gpr116 impairs insulin sensitivity through modulation of adipose function. FEBS Lett 586:3618–3625. [DOI] [PubMed] [Google Scholar]
- Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K, Yoshida S, Ono M, Kuwano M, Nakamura Y, et al. (1997) A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene 15:2145–2150. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Honda H, Takeichi M. (2012) Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149:1084–1097. [DOI] [PubMed] [Google Scholar]
- Nordström KJ, Fredriksson R, Schiöth HB. (2008) The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJ, Lagerström MC, Wallér LM, Fredriksson R, Schiöth HB. (2009) The Secretin GPCRs descended from the family of Adhesion GPCRs. Mol Biol Evol 26:71–84. [DOI] [PubMed] [Google Scholar]
- Nordström KJ, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB. (2011) Independent HHsearch, Needleman—Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol 28:2471–2480. [DOI] [PubMed] [Google Scholar]
- Oda K, Shiratsuchi T, Nishimori H, Inazawa J, Yoshikawa H, Taketani Y, Nakamura Y, Tokino T. (1999) Identification of BAIAP2 (BAI-associated protein 2), a novel human homologue of hamster IRSp53, whose SH3 domain interacts with the cytoplasmic domain of BAI1. Cytogenet Cell Genet 84:75–82. [DOI] [PubMed] [Google Scholar]
- Okajima D, Kudo G, Yokota H. (2010) Brain-specific angiogenesis inhibitor 2 (BAI2) may be activated by proteolytic processing. J Recept Signal Transduct Res 30:143–153. [DOI] [PubMed] [Google Scholar]
- Okajima D, Kudo G, Yokota H. (2011) Antidepressant-like behavior in brain-specific angiogenesis inhibitor 2-deficient mice. J Physiol Sci 61:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, 3rd, Ghosh A. (2012) FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron 73:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola KJ, Hall RA. (2012) Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol Pharmacol 82:777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. (2011) The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem 286:28914–28921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. (2014) Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci Signal 7:ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Ravichandran KS. (2010) Emerging roles of brain-specific angiogenesis inhibitor 1. Adv Exp Med Biol 706:167–178. [DOI] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450:430–434. [DOI] [PubMed] [Google Scholar]
- Patra C, van Amerongen MJ, Ghosh S, Ricciardi F, Sajjad A, Novoyatleva T, Mogha A, Monk KR, Mühlfeld C, Engel FB. (2013) Organ-specific function of adhesion G protein-coupled receptor GPR126 is domain-dependent. Proc Natl Acad Sci USA 110:16898–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YM, van de Garde MD, Cheng KF, Baars PA, Remmerswaal EB, van Lier RA, Mackay CR, Lin HH, Hamann J. (2011) Specific expression of GPR56 by human cytotoxic lymphocytes. J Leukoc Biol 90:735–740. [DOI] [PubMed] [Google Scholar]
- Piao X, Basel-Vanagaite L, Straussberg R, Grant PE, Pugh EW, Doheny K, Doan B, Hong SE, Shugart YY, Walsh CA. (2002) An autosomal recessive form of bilateral frontoparietal polymicrogyria maps to chromosome 16q12.2-21. Am J Hum Genet 70:1028–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, et al. (2004) G protein-coupled receptor-dependent development of human frontal cortex. Science 303:2033–2036. [DOI] [PubMed] [Google Scholar]
- Piao X, Chang BS, Bodell A, Woods K, Benzeev B, Topcu M, Guerrini R, Goldberg-Stern H, Sztriha L, Dobyns WB, et al. (2005) Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann Neurol 58:680–687. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Hofmann K, Bork P. (1999) A latrophilin/CL-1-like GPS domain in polycystin-1. Curr Biol 9:R585–R588. [DOI] [PubMed] [Google Scholar]
- Prömel S, Frickenhaus M, Hughes S, Mestek L, Staunton D, Woollard A, Vakonakis I, Schöneberg T, Schnabel R, Russ AP, et al. (2012a) The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Reports 2:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prömel S, Waller-Evans H, Dixon J, Zahn D, Colledge WH, Doran J, Carlton MB, Grosse J, Schöneberg T, Russ AP, et al. (2012b) Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Dev Dyn 241:1591–1602. [DOI] [PubMed] [Google Scholar]
- Prömel S, Langenhan T, Araç D. (2013) Matching structure with function: the GAIN domain of adhesion-GPCR and PKD1-like proteins. Trends Pharmacol Sci 34:470–478. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. (2007) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94. [DOI] [PubMed] [Google Scholar]
- Qi L, Ma J, Qi Q, Hartiala J, Allayee H, Campos H. (2011) Genetic risk score and risk of myocardial infarction in Hispanics. Circulation 123:374–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian F, Boletta A, Bhunia AK, Xu H, Liu L, Ahrabi AK, Watnick TJ, Zhou F, Germino GG. (2002) Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc Natl Acad Sci USA 99:16981–16986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Glasco DM, Zhou L, Sawant A, Ravni A, Fritzsch B, Damrau C, Murdoch JN, Evans S, Pfaff SL, et al. (2010) Atypical cadherins Celsr1-3 differentially regulate migration of facial branchiomotor neurons in mice. J Neurosci 30:9392–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Huang Y, Feng J, Alvarez-Bolado G, Grove EA, Yang Y, Tissir F, Zhou L, Goffinet AM. (2014) Genetic evidence that Celsr3 and Celsr2, together with Fzd3, regulate forebrain wiring in a Vangl-independent manner. Proc Natl Acad Sci USA 111:E2996–E3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M, Masuram S, Schiöth HB. (2014) The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol 54:9–26. [DOI] [PubMed] [Google Scholar]
- Ravni A, Qu Y, Goffinet AM, Tissir F. (2009) Planar cell polarity cadherin Celsr1 regulates skin hair patterning in the mouse. J Invest Dermatol 129:2507–2509. [DOI] [PubMed] [Google Scholar]
- Regard JB, Sato IT, Coughlin SR. (2008) Anatomical profiling of G protein-coupled receptor expression. Cell 135:561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J, van Wijk E, Märker T, Zimmermann U, Jürgens K, te Brinke H, Overlack N, Roepman R, Knipper M, Kremer H, et al. (2005) Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum Mol Genet 14:3933–3943. [DOI] [PubMed] [Google Scholar]
- Ribasés M, Ramos-Quiroga JA, Sánchez-Mora C, Bosch R, Richarte V, Palomar G, Gastaminza X, Bielsa A, Arcos-Burgos M, Muenke M, et al. (2011) Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav 10:149–157. [DOI] [PubMed] [Google Scholar]
- Robinson A, Escuin S, Doudney K, Vekemans M, Stevenson RE, Greene ND, Copp AJ, Stanier P. (2012) Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat 33:440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaee M, Clark AJ, Oh MC, Ivan ME, Bloch O, Kaur G, Sun MZ, Kim JM, Oh T, Berger MS, et al. (2013) Overexpression of CD97 confers an invasive phenotype in glioblastoma cells and is associated with decreased survival of glioblastoma patients. PLoS ONE 8:e62765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Kaneda K, Suekane A, Ichihara E, Nakahata S, Yamakawa N, Nagai K, Mizuno N, Kogawa K, Miura I, et al. (2013) Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia 27:1637–1649. [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, et al. (2007) Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature 449:346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert JR, Mlodzik M. (2007) Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet 8:126–138. [DOI] [PubMed] [Google Scholar]
- Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. (2009) Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol 7:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer B, Onishi K, Lo C, Colakoglu G, Zou Y. (2011) Vangl2 promotes Wnt/planar cell polarity-like signaling by antagonizing Dvl1-mediated feedback inhibition in growth cone guidance. Dev Cell 20:177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidhar S, Lorente G, Nagavarapu U, Nelson A, Kuo J, Cummins J, Nikolich K, Urfer R, Foehr ED. (2005) GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene 24:1673–1682. [DOI] [PubMed] [Google Scholar]
- Shima Y, Copeland NG, Gilbert DJ, Jenkins NA, Chisaka O, Takeichi M, Uemura T. (2002) Differential expression of the seven-pass transmembrane cadherin genes Celsr1-3 and distribution of the Celsr2 protein during mouse development. Dev Dyn 223:321–332. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kawaguchi SY, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, Hirano T, Uemura T. (2007) Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci 10:963–969. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Usui T, Yanagawa S, Takeichi M, Uemura T. (2001) Asymmetric colocalization of Flamingo, a seven-pass transmembrane cadherin, and Dishevelled in planar cell polarization. Curr Biol 11:859–863. [DOI] [PubMed] [Google Scholar]
- Shin D, Lin ST, Fu YH, Ptácek LJ. (2013) Very large G protein-coupled receptor 1 regulates myelin-associated glycoprotein via Gαs/Gαq-mediated protein kinases A/C. Proc Natl Acad Sci USA 110:19101–19106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratsuchi T, Nishimori H, Ichise H, Nakamura Y, Tokino T. (1997) Cloning and characterization of BAI2 and BAI3, novel genes homologous to brain-specific angiogenesis inhibitor 1 (BAI1). Cytogenet Cell Genet 79:103–108. [DOI] [PubMed] [Google Scholar]
- Shukla AK, Xiao K, Lefkowitz RJ. (2011) Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci 36:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Lelianova V, Hopkins C, Volynski KE, Ushkaryov Y. (2009) Functional cross-interaction of the fragments produced by the cleavage of distinct adhesion G-protein-coupled receptors. J Biol Chem 284:6495–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O, Rahman MA, Zangrandi A, Fidalgo S, Tonevitsky AG, et al. (2011) Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci USA 108:12113–12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer K, Luo R, Jeong SJ, Piao X. (2013) GPR56 and the developing cerebral cortex: cells, matrix, and neuronal migration. Mol Neurobiol 47:186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skradski SL, Clark AM, Jiang H, White HS, Fu YH, Ptácek LJ. (2001) A novel gene causing a mendelian audiogenic mouse epilepsy. Neuron 31:537–544. [DOI] [PubMed] [Google Scholar]
- Sleckman BP, Khan WN, Xu W, Bassing CH, Malynn BA, Copeland NG, Bardon CG, Breit TM, Davidson L, Oltz EM, et al. (2000) Cloning and functional characterization of the early-lymphocyte-specific Pb99 gene. Mol Cell Biol 20:4405–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski JD, Nobles SL, Heffron DS, Park D, Ravichandran KS, Mandell JW. (2011) Brain-specific angiogenesis inhibitor-1 expression in astrocytes and neurons: implications for its dual function as an apoptotic engulfment receptor. Brain Behav Immun 25:915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N, Stolk L, Nica A, Inouye M, Hofman A, et al. (2009) Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet 5:e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern C, Cook JM, Neetoo-Isseljee Z, Taylor DL, Kettleborough CA, Merritt A, Bassoni DL, Raab WJ, Quinn E, Wehrman TS, et al. (2013) Screening β-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors. J Biomol Screen 18:599–609. [DOI] [PubMed] [Google Scholar]
- Specht D, Wu SB, Turner P, Dearden P, Koentgen F, Wolfrum U, Maw M, Brandstätter JH, tom Dieck S. (2009) Effects of presynaptic mutations on a postsynaptic Cacna1s calcium channel colocalized with mGluR6 at mouse photoreceptor ribbon synapses. Invest Ophthalmol Vis Sci 50:505–515. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey M, Lin HH, Gordon S, McKnight AJ. (2000) LNB-TM7, a group of seven-transmembrane proteins related to family-B G-protein-coupled receptors. Trends Biochem Sci 25:284–289. [DOI] [PubMed] [Google Scholar]
- Stacey M, Chang GW, Sanos SL, Chittenden LR, Stubbs L, Gordon S, Lin HH. (2002) EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J Biol Chem 277:29283–29293. [DOI] [PubMed] [Google Scholar]
- Stacey M, Chang GW, Davies JQ, Kwakkenbos MJ, Sanderson RD, Hamann J, Gordon S, Lin HH. (2003) The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood 102:2916–2924. [DOI] [PubMed] [Google Scholar]
- Steimel A, Wong L, Najarro EH, Ackley BD, Garriga G, Hutter H. (2010) The Flamingo ortholog FMI-1 controls pioneer-dependent navigation of follower axons in C. elegans. Development 137:3663–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Wobus M, Boltze C, Schütz A, Wahlbuhl M, Hamann J, Aust G. (2002) Expression and regulation of CD97 in colorectal carcinoma cell lines and tumor tissues. Am J Pathol 161:1657–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson JR, Paavola KJ, Schaefer SA, Kaur B, Van Meir EG, Hall RA. (2013) Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J Biol Chem 288:22248–22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson JR, Purcell RH, Hall RA. (2014) The BAI subfamily of adhesion GPCRs: synaptic regulation and beyond. Trends Pharmacol Sci 35:208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann R, Schröck K, Böselt I, Stäubert C, Russ A, Schöneberg T. (2011) Evolution of GPCR: change and continuity. Mol Cell Endocrinol 331:170–178. [DOI] [PubMed] [Google Scholar]
- Struhl G, Casal J, Lawrence PA. (2012) Dissecting the molecular bridges that mediate the function of Frizzled in planar cell polarity. Development 139:3665–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D, Johnson R, Cooper K, Bray S. (2002) Asymmetric localization of frizzled and the determination of notch-dependent cell fate in the Drosophila eye. Curr Biol 12:813–824. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ohno S. (2006) The PAR-aPKC system: lessons in polarity. J Cell Sci 119:979–987. [DOI] [PubMed] [Google Scholar]
- Tang X, Jin R, Qu G, Wang X, Li Z, Yuan Z, Zhao C, Siwko S, Shi T, Wang P, et al. (2013) GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res 73:6206–6218. [DOI] [PubMed] [Google Scholar]
- Tissir F, Bar I, Jossin Y, De Backer O, Goffinet AM. (2005) Protocadherin Celsr3 is crucial in axonal tract development. Nat Neurosci 8:451–457. [DOI] [PubMed] [Google Scholar]
- Tissir F, Qu Y, Montcouquiol M, Zhou L, Komatsu K, Shi D, Fujimori T, Labeau J, Tyteca D, Courtoy P, et al. (2010) Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat Neurosci 13:700–707. [DOI] [PubMed] [Google Scholar]
- Tobaben S, Südhof TC, Stahl B. (2002) Genetic analysis of α-latrotoxin receptors reveals functional interdependence of CIRL/latrophilin 1 and neurexin 1 α. J Biol Chem 277:6359–6365. [DOI] [PubMed] [Google Scholar]
- Tönjes A, Koriath M, Schleinitz D, Dietrich K, Böttcher Y, Rayner NW, Almgren P, Enigk B, Richter O, Rohm S, et al. (2009) Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Hum Mol Genet 18:4662–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Kenzelmann D, Trzebiatowska A, Chiquet-Ehrismann R. (2007) Teneurins: transmembrane proteins with fundamental roles in development. Int J Biochem Cell Biol 39:292–297. [DOI] [PubMed] [Google Scholar]
- Urano T, Shiraki M, Yagi H, Ito M, Sasaki N, Sato M, Ouchi Y, Inoue S. (2012) GPR98/Gpr98 gene is involved in the regulation of human and mouse bone mineral density. J Clin Endocrinol Metab 97:E565–E574. [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. (1999) Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98:585–595. [DOI] [PubMed] [Google Scholar]
- Vallon M, Essler M. (2006) Proteolytically processed soluble tumor endothelial marker (TEM) 5 mediates endothelial cell survival during angiogenesis by linking integrin alpha(v)beta3 to glycosaminoglycans. J Biol Chem 281:34179–34188. [DOI] [PubMed] [Google Scholar]
- Valtcheva N, Primorac A, Jurisic G, Hollmén M, Detmar M. (2013) The orphan adhesion G protein-coupled receptor GPR97 regulates migration of lymphatic endothelial cells via the small GTPases RhoA and Cdc42. J Biol Chem 288:35736–35748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pel M, Hagoort H, Hamann J, Fibbe WE. (2008a) CD97 is differentially expressed on murine hematopoietic stem-and progenitor-cells. Haematologica 93:1137–1144. [DOI] [PubMed] [Google Scholar]
- van Pel M, Hagoort H, Kwakkenbos MJ, Hamann J, Fibbe WE. (2008b) Differential role of CD97 in interleukin-8-induced and granulocyte-colony stimulating factor-induced hematopoietic stem and progenitor cell mobilization. Haematologica 93:601–604. [DOI] [PubMed] [Google Scholar]
- van Wijk E, van der Zwaag B, Peters T, Zimmermann U, Te Brinke H, Kersten FF, Märker T, Aller E, Hoefsloot LH, Cremers CW, et al. (2006) The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum Mol Genet 15:751–765. [DOI] [PubMed] [Google Scholar]
- Veninga H, Becker S, Hoek RM, Wobus M, Wandel E, van der Kaa J, van der Valk M, de Vos AF, Haase H, Owens B, et al. (2008) Analysis of CD97 expression and manipulation: antibody treatment but not gene targeting curtails granulocyte migration. J Immunol 181:6574–6583. [DOI] [PubMed] [Google Scholar]
- Veninga H, de Groot DM, McCloskey N, Owens BM, Dessing MC, Verbeek JS, Nourshargh S, van Eenennaam H, Boots AM, Hamann J. (2011) CD97 antibody depletes granulocytes in mice under conditions of acute inflammation via a Fc receptor-dependent mechanism. J Leukoc Biol 89:413–421. [DOI] [PubMed] [Google Scholar]
- Volynski KE, Silva JP, Lelianova VG, Atiqur Rahman M, Hopkins C, Ushkaryov YA. (2004) Latrophilin fragments behave as independent proteins that associate and signal on binding of LTX(N4C). EMBO J 23:4423–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller-Evans H, Prömel S, Langenhan T, Dixon J, Zahn D, Colledge WH, Doran J, Carlton MB, Davies B, Aparicio SA, et al. (2010) The orphan adhesion-GPCR GPR126 is required for embryonic development in the mouse. PLoS ONE 5:e14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, Wellman PJ, Setlow B. (2012) Initial characterization of mice null for lphn3, a gene implicated in ADHD and addiction. Brain Res 1463:85–1492. [DOI] [PubMed] [Google Scholar]
- Wandel E, Saalbach A, Sittig D, Gebhardt C, Aust G. (2012) Thy-1 (CD90) is an interacting partner for CD97 on activated endothelial cells. J Immunol 188:1442–1450. [DOI] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. (2004) Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA 101:15603–15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JJ, Zhang LL, Zhang HX, Shen CL, Lu SY, Kuang Y, Wan YH, Wang WG, Yan HM, Dang SY, et al. (2013a) Gpr97 is essential for the follicular versus marginal zone B-lymphocyte fate decision. Cell Death Dis 4:e853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ward Y, Tian L, Lake R, Guedez L, Stetler-Stevenson WG, Kelly K. (2005) CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 105:2836–2844. [DOI] [PubMed] [Google Scholar]
- Wang W, Da R, Wang M, Wang T, Qi L, Jiang H, Chen W, Li Q. (2013b) Expression of brain-specific angiogenesis inhibitor 1 is inversely correlated with pathological grade, angiogenesis and peritumoral brain edema in human astrocytomas. Oncol Lett 5:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Thekdi N, Smallwood PM, Macke JP, Nathans J. (2002) Frizzled-3 is required for the development of major fiber tracts in the rostral CNS. J Neurosci 22:8563–8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cho SG, Wu X, Siwko S, Liu M. (2014) G-protein coupled receptor 124 (GPR124) in endothelial cells regulates vascular endothelial growth factor (VEGF)-induced tumor angiogenesis. Curr Mol Med 14:543–554. [DOI] [PubMed] [Google Scholar]
- Wansleeben C, Meijlink F. (2011) The planar cell polarity pathway in vertebrate development. Dev Dyn 240:616–626. [DOI] [PubMed] [Google Scholar]
- Ward Y, Lake R, Yin JJ, Heger CD, Raffeld M, Goldsmith PK, Merino M, Kelly K. (2011) LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res 71:7301–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward Y, Lake R, Martin PL, Killian K, Salerno P, Wang T, Meltzer P, Merino M, Cheng SY, Santoro M, et al. (2012) CD97 amplifies LPA receptor signaling and promotes thyroid cancer progression in a mouse model. Oncogene 32:2726–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington SJ, Strutt H, Strutt D. (2013) The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Development 140:1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warschkau H, Kiderlen AF. (1999) A monoclonal antibody directed against the murine macrophage surface molecule F4/80 modulates natural immune response to Listeria monocytogenes. J Immunol 163:3409–3416. [PubMed] [Google Scholar]
- Weinsheimer S, Brettman AD, Pawlikowska L, Wu DC, Mancuso MR, Kuhnert F, Lawton MT, Sidney S, Zaroff JG, McCulloch CE, et al. (2012) G Protein-Coupled Receptor 124 (GPR124) Gene Polymorphisms and Risk of Brain Arteriovenous Malformation. Transl Stroke Res 3:418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T, et al. (2013) Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23:159–170. [DOI] [PubMed] [Google Scholar]
- Weston MD, Luijendijk MW, Humphrey KD, Möller C, Kimberling WJ. (2004) Mutations in the VLGR1 gene implicate G-protein signaling in the pathogenesis of Usher syndrome type II. Am J Hum Genet 74:357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus M, Vogel B, Schmücking E, Hamann J, Aust G. (2004) N-glycosylation of CD97 within the EGF domains is crucial for epitope accessibility in normal and malignant cells as well as CD55 ligand binding. Int J Cancer 112:815–822. [DOI] [PubMed] [Google Scholar]
- Wolfrum U (2012) Usher Syndrome: Pathogenesis, Diagnosis and Therapy (Ahuja S, ed) p 458, NOVA Science Publishers, Hauppauge NY. [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318:1108–1113. [DOI] [PubMed] [Google Scholar]
- Wu J, Lei L, Wang S, Gu D, Zhang J. (2012) Immunohistochemical expression and prognostic value of CD97 and its ligand CD55 in primary gallbladder carcinoma. J Biomed Biotechnol 2012:587672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MP, Doyle JR, Barry B, Beauvais A, Rozkalne A, Piao X, Lawlor MW, Kopin AS, Walsh CA, Gussoni E. (2013) G-protein coupled receptor 56 promotes myoblast fusion through serum response factor- and nuclear factor of activated T-cell-mediated signalling but is not essential for muscle development in vivo. FEBS J 280:6097–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Begum S, Hearn JD, Hynes RO. (2006) GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci USA 103:9023–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Takamura Y, Yoneda T, Konno D, Akagi Y, Yoshida K, Sato M. (2005) Vlgr1 knockout mice show audiogenic seizure susceptibility. J Neurochem 92:191–202. [DOI] [PubMed] [Google Scholar]
- Yagi H, Tokano H, Maeda M, Takabayashi T, Nagano T, Kiyama H, Fujieda S, Kitamura K, Sato M. (2007) Vlgr1 is required for proper stereocilia maturation of cochlear hair cells. Genes Cells 12:235–250. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Fuku N, Tanaka M, Aoyagi Y, Sawabe M, Metoki N, Yoshida H, Satoh K, Kato K, Watanabe S, et al. (2009) Identification of CELSR1 as a susceptibility gene for ischemic stroke in Japanese individuals by a genome-wide association study. Atherosclerosis 207:144–149. [DOI] [PubMed] [Google Scholar]
- Yang L, Chen G, Mohanty S, Scott G, Fazal F, Rahman A, Begum S, Hynes RO, Xu L. (2011) GPR56 Regulates VEGF production and angiogenesis during melanoma progression. Cancer Res 71:5558–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Friedland S, Corson N, Xu L. (2014) GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res 74:1022–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MY, Hilton MB, Seaman S, Haines DC, Nagashima K, Burks CM, Tessarollo L, Ivanova PT, Brown HA, Umstead TM, et al. (2013) Essential regulation of lung surfactant homeostasis by the orphan G protein-coupled receptor GPR116. Cell Reports 3:1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LL, Papakrivopoulou J, Long DA, Goggolidou P, Connolly JO, Woolf AS, Dean CH. (2010a) The planar cell polarity gene Vangl2 is required for mammalian kidney-branching morphogenesis and glomerular maturation. Hum Mol Genet 19:4663–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LL, Schnatwinkel C, Murdoch JN, Bogani D, Formstone CJ, Townsend S, Greenfield A, Niswander LA, Dean CH. (2010b) The PCP genes Celsr1 and Vangl2 are required for normal lung branching morphogenesis. Hum Mol Genet 19:2251–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Lin HH, Dri P, Davies JQ, Hayhoe RP, Lewis SM, Heinsbroek SE, Brown KA, Perretti M, Hamann J, et al. (2008a) Ligation of the adhesion-GPCR EMR2 regulates human neutrophil function. FASEB J 22:741–751. [DOI] [PubMed] [Google Scholar]
- Yona S, Lin HH, Siu WO, Gordon S, Stacey M. (2008b) Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem Sci 33:491–500. [DOI] [PubMed] [Google Scholar]
- Zallocchi M, Delimont D, Meehan DT, Cosgrove D. (2012) Regulated vesicular trafficking of specific PCDH15 and VLGR1 variants in auditory hair cells. J Neurosci 32:13841–13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendman AJW, Cornelissen IMHA, Weidle UH, Ruiter DJ, van Muijen GNP. (1999) TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett 446:292–298. [DOI] [PubMed] [Google Scholar]
- Zohrabian VM, Nandu H, Gulati N, Khitrov G, Zhao C, Mohan A, Demattia J, Braun A, Das K, Murali R, et al. (2007) Gene expression profiling of metastatic brain cancer. Oncol Rep 18:321–328. [PubMed] [Google Scholar]
- Zyryanova T, Schneider R, Adams V, Sittig D, Kerner C, Gebhardt C, Ruffert H, Glasmacher S, Hepp P, Punkt K, et al. (2014) Skeletal muscle expression of the adhesion-GPCR CD97: CD97 deletion induces an abnormal structure of the sarcoplasmatic reticulum but does not impair skeletal muscle function. PLoS ONE 9:e100513. [DOI] [PMC free article] [PubMed] [Google Scholar]