Abstract
Chronic periodontitis is one of the most prevalent human diseases and is caused by dysbiosis of the subgingival microbiota. Treatment involves primarily mechanical disruption of subgingival biofilms and, in certain cases, adjunctive use of systemic antibiotic therapy. In vitro biofilm models have been developed to study antimicrobial agents targeting subgingival species. However, these models accommodate a limited number of taxa, lack reproducibility, and have low throughput. We aimed to develop an in vitro multispecies biofilm model that mimics subgingival plaque, to test antimicrobial agents. Biofilms were cultivated using the Calgary Biofilm Device and were exposed to amoxicillin (AMX), metronidazole (MTZ), azithromycin (AZM), and AMX-MTZ at four different concentrations for 12, 24, or 36 h. Chlorhexidine (CHX) (0.12%) was used as the positive control. The compositions of the biofilms were analyzed by checkerboard DNA-DNA hybridization, and the percent reduction in biofilm metabolic activity was determined using 2,3,5-triphenyltetrazolium chloride and spectrophotometry. Thirty-five of the 40 species used in the inoculum were consistently recovered from the resulting in vitro biofilms. After 36 h of exposure at the 1:27 dilution, AMX-MTZ reduced metabolic activity 11% less than CHX (q = 0.0207) but 54% more than AMX (q = 0.0031), 72% more than MTZ (q = 0.0031), and 67% more than AZM (q = 0.0008). Preliminary evidence of a synergistic interaction between AMX and MTZ was also observed. In summary, we developed reproducible biofilms with 35 subgingival bacterial species, and our results suggested that the combination of AMX and MTZ had greater antimicrobial effects on these in vitro multispecies biofilms than expected on the basis of the independent effects of the drugs.
INTRODUCTION
Periodontitis is a persistent health problem that affects the U.S. population in epidemic proportions. The most recent data from the National Health and Nutrition Examination Survey (NHANES) (2009 to 2010) suggest that the prevalence of chronic periodontitis among U.S. adults is over 47%, representing 64.7 million adults (1). Chronic periodontitis is the main cause of tooth loss in adults. Treatment costs U.S. taxpayers $4.4 billion each year (2), but existing therapies do not eradicate the disease. Frequent posttreatment follow-up visits are needed and are costly, which may be one reason why the prevalence is higher in populations of low socioeconomic status (3).
A single periodontal pocket harbors a complex polymicrobial community of up to hundreds of taxa, and periodontal diseases are triggered by dysbiosis of subgingival organisms. Organization of the subgingival microbiota in biofilms makes it challenging to control periodontal infections, since biofilms help protect resident organisms from both antimicrobial agents and immune mechanisms (4). Still, the use of systemic and local antibiotics as an adjunct to mechanical treatment provides clinical benefit beyond that achieved by scaling and root planing alone (5–15).
However, the threat of bacterial resistance to currently used antibiotics raises the need to develop new agents. The first step in such an endeavor is to demonstrate efficacy against subgingival microorganisms in vitro, ideally in models involving organized biofilms that respond like those in vivo. Currently used in vitro oral multispecies biofilm models all suffer from one or more limitations, generally comprising only up to 5 or 10 species (16–21). In addition, existing models lack reproducibility and have low throughput (17, 19).
The primary aim of this study was to develop an in vitro multispecies biofilm model that can be used to test antimicrobial agents for their inhibition of bacterial metabolic activity. Specifically, we aimed to develop an in vitro model that mimics the composition and structure of subgingival plaque while also exhibiting variability among individual biofilms and allowing for high throughput. In part to validate the system, we compared the antimicrobial activities of different concentrations of amoxicillin (AMX), metronidazole (MTZ), and azithromycin (AZM), using chlorhexidine (CHX) as the gold standard and positive control. Using the system also allowed us to test the much-debated hypothesis (22–24) that MTZ and AMX act synergistically, rather than independently, in their antimicrobial activity against periodontal pathogens.
MATERIALS AND METHODS
Bacterial strains.
The following strains were used: Actinomyces gerencseriae ATCC 23840, Actinomyces israelii ATCC 12102, Actinomyces naeslundii ATCC 12104, Actinomyces oris ATCC 43146, Actinomyces odontolyticus ATCC 17929, Veillonella parvula ATCC 10790, Streptococcus gordonii ATCC 10558, Streptococcus intermedius ATCC 27335, Streptococcus mitis ATCC 49456, Streptococcus oralis ATCC 35037, Streptococcus sanguinis ATCC 10556, Streptococcus anginosus ATCC 33397, Streptococcus mutans ATCC 25175, Aggregatibacter actinomycetemcomitans ATCC 29523, Capnocytophaga gingivalis ATCC 33624 (27), Capnocytophaga ochracea ATCC 33596 (25), Capnocytophaga sputigena ATCC 33612 (4), Eikenella corrodens ATCC 23834, Campylobacter concisus ATCC 33237 (484), Campylobacter gracilis ATCC 33236 (1084), Campylobacter rectus ATCC 33238 (371), Campylobacter showae ATCC 51146, Eubacterium nodatum ATCC 33099, Eubacterium saburreum ATCC 33271, Fusobacterium nucleatum subsp. nucleatum ATCC 25586, Fusobacterium nucleatum subsp. polymorphum ATCC 10953, Fusobacterium nucleatum subsp. vincentii ATCC 49256, Fusobacterium periodonticum ATCC 33693, Parvimonas micra ATCC 33270, Prevotella intermedia ATCC 25611, Prevotella nigrescens ATCC 33563, Prevotella melaninogenica ATCC 25845, Streptococcus constellatus ATCC 27823 (M32b), Tannerella forsythia ATCC 43037 (338), Porphyromonas gingivalis ATCC 33277, Gemella morbillorum ATCC 27824, Leptotrichia buccalis ATCC 14201, Neisseria mucosa ATCC 19696, Propionibacterium acnes ATCC 11827, and Selenomonas noxia ATCC 43541.
Media and culture conditions.
Most species, including Actinomyces subsp., Streptococcus subsp., and Fusobacterium subsp., were cultured on tryptic soy agar with 5% sheep blood, under anaerobic conditions (85% nitrogen, 10% carbon dioxide, and 5% hydrogen), while Eubacterium subsp. and N. mucosa were cultivated on fastidious anaerobe agar with 5% sheep blood, P. melaninogenica and P. gingivalis were cultured on tryptic soy agar with yeast extract enriched with 1% hemin, 5% menadione, and 5% sheep blood, and T. forsythia was grown on tryptic soy agar with yeast extract enriched with 1% hemin, 5% menadione, 5% sheep blood, and 1% N-acetylmuramic acid. After 48 h of growth, all species were transferred to glass tubes with brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD) supplemented with 1% hemin.
Biofilm formation.
After 24 h of growth in BHI broth with 1% hemin, the optical density (OD) at 600 nm was adjusted to 0.1, corresponding to approximately 108 cells/ml of each species. Individual cell suspensions of each species were diluted to 107 cells/ml, with adjustment for their respective cell sizes. Aliquots of 100 μl containing 106 cells of each species were mixed to yield a final biofilm inoculum. Thirty-three milliliters of BHI broth with 1% hemin and 5% sheep blood was added to yield a final volume of 45 ml of inoculum. The multispecies biofilm model was developed using the Calgary Biofilm Device (CBD) (25). Three 96-well plates (Nunc; Thermo Scientific, Roskilde, Denmark) were seeded with 150 μl of inoculum per well, containing 104 cells of each of the 40 species, and covers with 96 polystyrene pegs were applied (Nunc TSP system; Thermo Scientific, Roskilde, Denmark). The covered plates were then incubated at 37°C under anaerobic conditions, using the BD GasPak EZ system (Becton Dickinson, Sparks, MD). After 72 h of incubation, the covers were transferred daily to new 96-well plates with fresh broth (BHI broth with 1% hemin and 5% sheep blood). This procedure was repeated for the next 4 days to yield 7-day biofilms on each of the 96 polystyrene pegs in each plate. Separate plates were used for different times of exposure to antimicrobials (see below).
Exposure to antibiotics.
The 7-day biofilms were washed twice in 200 μl of rinse solution (1% phosphate-buffered saline [PBS]), and pegs from the first column of each plate were removed to establish the baseline values for biofilm metabolic activity and biofilm composition, as determined with the checkerboard DNA-DNA hybridization technique (see below). The remaining 88 pegs from each plate were exposed to different concentrations of AMX (catalog no. 190145; MP Biomedicals), MTZ (catalog no. 155710; MP Biomedicals), AZM (catalog no. A2076; TCI), and the combination of AMX and MTZ. Starting at 108 μg/ml of AMX, 270 μg/ml of MTZ, and 216 μg/ml of AZM, the antibiotics were serially 3-fold diluted to yield 4 concentrations of each antibiotic and of AMX-MTZ (1:1, 1:3, 1:9, and 1:27). CHX (Peridex [0.12% chlorhexidine gluconate]; National Drug Code [NDC] 488788-0620-1) was used as a positive control. Pegs coated with biofilms were washed twice with 200 μl of rinse solution (1% PBS), transferred to 96-well plates containing the different concentrations of antibiotics, and incubated for 12, 24, or 36 h (one plate for each time point). Each condition was tested in triplicate.
Biofilm metabolic activity.
The percent reduction in biofilm metabolic activity was determined using 2,3,5-triphenyltetrazolium chloride (TTC) (catalog no. 17779; Fluka Analytical) and spectrophotometry. TTC is used to differentiate between metabolically active and inactive cells. The white substrate is enzymatically reduced to red 1,3,5-triphenylformazan (TFP) by living bacterial cells, due to the activity of various dehydrogenases. The change in the substrate color is read by spectrophotometry to determine the rate of reduction, which is used as an indirect measure of bacterial metabolic activity. To measure the metabolic activity of the biofilms, the remaining pegs were washed twice with rinse solution and transferred to plates with 200 μl per well of fresh BHI broth containing 1% hemin with 10% of a 1% TTC solution. Plates were then incubated under anaerobic conditions for 24 h at 37°C. TTC conversion was read at 485 nm using a fluorescence spectrophotometer (POLARstar Optima; BMG Labtech).
Checkerboard DNA-DNA hybridization.
After the TTC assay, 9 pegs (3 pegs from each 96-well plate) covered with 7-day biofilms were washed twice with rinse solution, extracted from the cover, and transferred to separate Eppendorf tubes containing 100 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.6]), and then 100 μl of 0.5 M NaOH was added. The tubes containing the pegs and the final solution were boiled for 10 min, the pegs were removed, and the solution was subsequently neutralized with the addition of 0.8 ml of 5 M ammonium acetate. Samples were then individually analyzed for their contents of 40 bacterial species, using the checkerboard DNA-DNA hybridization technique (26, 27). In brief, after the samples were lysed, the DNA was placed in lanes on a nylon membrane using a Minislot device (Immunetics, Cambridge, MA). After fixation of the DNA to the membrane, the membrane was placed in a Miniblotter 45 (Immunetics), with the lanes of DNA at a 90° angle with respect to the lanes of the device. Digoxigenin-labeled whole-genome DNA probes for 40 subgingival species were hybridized in individual lanes of the Miniblotter 45. After hybridization, the membranes were washed at high stringency, and the DNA probes were detected using a digoxigenin-specific antibody conjugated with alkaline phosphatase. Signals were detected using AttoPhos substrate (Amersham Life Sciences, Arlington Heights, IL), and results were read using a Typhoon Trio Plus variable mode imager (Molecular Dynamics, Sunnyvale, CA). Two lanes in each run contained standards with 105 or 106 cells of each species. Signals evaluated using the Typhoon Trio Plus variable mode imager were converted to absolute counts by comparison with the standards on the same membrane. Failure to detect a signal was recorded as zero. Pegs were analyzed before the antibiotic tests, to establish baseline values.
In vivo biofilm reference values.
To obtain reference values for the composition of in vivo biofilms, we examined the database from a previous study (28). In that study, checkerboard DNA-DNA hybridization was used to measure microbial counts of 38 of 40 species cultured in the in vitro biofilms (C. concisus and S. mutans were not included in that study). Up to 28 subgingival plaque samples from 178 subjects were analyzed. Samples obtained from deep periodontal pockets (pocket depth of ≥5 mm) were selected from the database, and the counts (×105) for each species were averaged within subjects and then across subjects. The microbial profile obtained served as a reference for the microbial composition of mature biofilms present in deep periodontal pockets.
Calculation of minimum similarity coefficient values.
We examined the consistency of the microbial profiles across the 9 in vitro biofilm samples (3 from each 96-well plate) and between them and the in vivo reference values by using the minimum similarity coefficient (29, 30). For this analysis, the percentage of the total DNA probe count was calculated for each species in each in vitro biofilm sample and for the mean reference values for in vivo biofilms. The minimum similarity coefficient was determined using the equation
where S is the minimum similarity coefficient, n is the number of species, x and y are individual samples, and min(xiyi) is the smallest proportion of a species common to both samples. For example, if a species was present at 1% in one sample and at 0.1% in a second sample, then the minimum similarity for that species is 0.1%. Once the minimum similarity value for each species in a pair of samples is calculated, the values are summed to give a summary measurement for the entire microbial community.
Scanning electron microscopy.
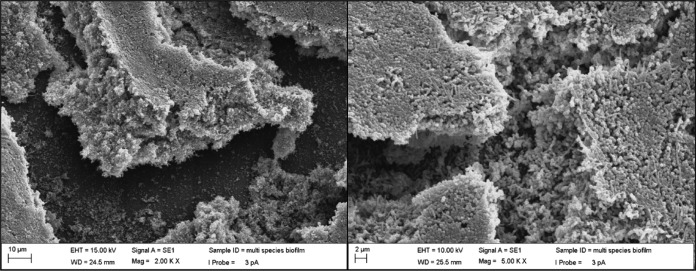
For scanning electron microscopy (SEM), biofilms were washed with PBS, fixed with 3% (vol/vol) paraformaldehyde and 4% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer at 4°C, washed twice with PBS, stained with 2% osmium tetroxide, incubated for 10 min in 50%, 70%, 80%, 95%, and 100% ethanol, air dried, and sputter coated. Images of gold-coated specimens (Denton V sputter coater; Denton, Moorestown, NJ) were obtained with a Zeiss Evo LS 10 scanning electron microscope (Carl Zeiss, Peabody, MA) in secondary electron mode, using 30 mA for 30 s. Imaging was performed under high vacuum at 10.00 kV, with a probe current of 3 pA and a working distance of 25.5 mm.
Data analysis.
We performed all data analyses other than calculation of the minimum similarity index with Stata version 12 (StataCorp, College Station, TX). For each time point, we averaged the three negative-control OD values. We divided each assay result by this number, which yielded the proportion of activity remaining in the presence of the antibiotic. We then subtracted this proportion from 1 and multiplied the result by 100 to derive the percent inhibition. We present means and standard deviations (SDs) for each antibiotic (or combination) at each dilution and time point. For each dilution and time point, we also performed t tests comparing each pair of treatments (both at the same dilution and time point). However, we considered the primary analyses to be those for the 1:27 dilutions at 36 h, because these concentrations are likely to be closest to those achieved in periodontal pockets when antibiotics are administered systemically. We applied a stepdown false-discovery correction for these 10 multiple comparisons by calculating q values, which can be interpreted like P values (31). The analyses for the remaining dilutions and time points are considered exploratory, but we also calculated corrected q values over all 144 comparisons, for benchmarking purposes (data not shown).
To estimate potential synergy between AMX and MTZ, the observations in each triplicate determination were treated as separate independent observations in a linear regression model in which percent inhibition was the dependent variable, separate indicators were used to specify the presence or absence of AMX and MTZ, and the combination of antibiotics was entered as a multiplicative interaction term. One way to interpret the interaction term is as the difference in AMX effects in the presence versus absence of MTZ, or vice versa. The interaction P value is from a test of the null hypothesis that AMX effects (AMX versus placebo) are the same regardless of whether MTZ is present or absent, and vice versa. Combined exposure to two effective agents should always increase the effects, even in the absence of synergy; the question posed here is whether the combined effects are greater than would be expected on the basis of two independent effects acting concurrently. For these analyses, we also estimated 95% confidence intervals (CIs) and performed two-sided tests with alpha at 0.05. We did not adjust for multiple comparisons, again considering the primary analyses to be those for the 1:27 dilutions at 36 h and analyses for all other time points and concentrations to be secondary.
RESULTS
Minimum similarity coefficient values.
There was a high degree of similarity in the compositions of in vitro biofilms across pegs and plates (Fig. 1). For instance, the minimum similarity coefficients among pegs from plate I were 79, 80, and 97, depending on which pairs of pegs were being compared. Plates II and III had minimum similarity coefficient values ranging from 77 to 93. When pegs from different plates were compared, minimum similarity coefficient values were also high, ranging from 70 to 88. Lower minimum similarity coefficient values were obtained when the compositions of in vitro biofilms were compared with the reference values for in vivo biofilms; however, values were consistent across the 9 pegs examined, ranging from 37 to 46.
FIG 1.
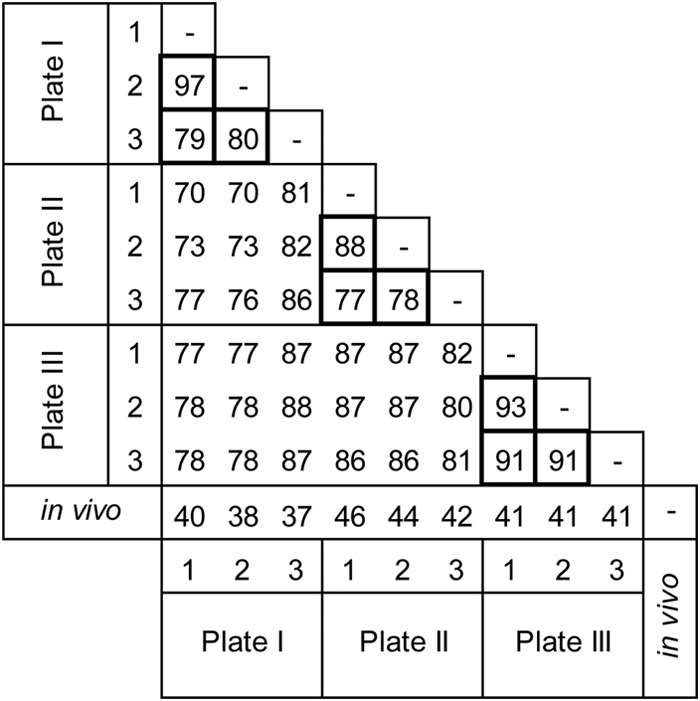
Matrix of minimum similarity coefficient values for the microbial profiles for 9 in vitro biofilms from three 96-well plates (plate I, plate II, and plate III) and for these in vitro biofilms versus mean values for in vivo biofilms from 178 subjects. Cells with thicker borders include minimum similarity coefficient values for comparisons among biofilms from the same plate.
Compositions of 7-day in vitro biofilms and in vivo biofilms.
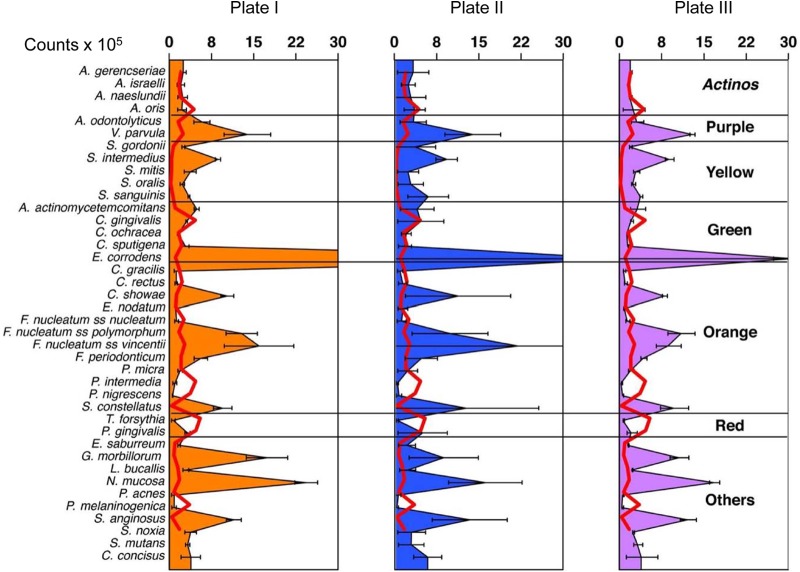
Five of the 40 species used in the inoculum, i.e., P. intermedia, P. nigrescens, T. forsythia, P. acnes, and P. melaninogenica, could not be consistently recovered from the biofilms that formed on the pegs (Fig. 2). The remaining 35 species, including putative periodontal pathogens such as A. actinomycetemcomitans, E. nodatum, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, F. periodonticum, P. micra, and P. gingivalis, were consistently recovered. The reference values for the in vivo biofilms demonstrated that several species clearly outgrew others in levels not observed in vivo (Fig. 2). For instance, V. parvula, S. mitis, S. sanguinis, C. showae, F. nucleatum subsp. polymorphum, and F. nucleatum subsp. vincentii achieved counts that were >5 times greater than those observed in periodontitis subjects. In addition, S. oralis, G. morbillorum, and N. mucosa achieved levels at least 10-fold greater than those observed in humans, while S. intermedius, E. corrodens, and S. anginosus reached counts that were at least 20 times higher than those in in vivo subgingival samples. The density and complexity of the biofilms were also demonstrated by the SEM images; different cell shapes were observed (Fig. 3).
FIG 2.
Mean levels (counts × 105) of the 40 subgingival species in 7-day biofilms grown in the CBD. Values represent the means for 3 pegs processed with checkerboard DNA-DNA hybridization for each 96-well plate, and the error bars indicate the standard errors of the mean. The species were ordered according to the microbial complexes described by Socransky et al. (45). The red lines illustrate the mean microbial profile of mature in vivo biofilms present in deep periodontal pockets.
FIG 3.
Scanning electron micrographs of biofilms formed on the pegs of the CBD.
Reductions in metabolic activity with antibiotics, as measured by TTC conversion.
In the experiments to measure antimicrobial activity using TTC conversion, the positive control, 0.12% CHX, decreased metabolic activity by approximately 95% at all three exposure times (i.e., 12, 24, and 36 h) (Table 1). All of the antibiotics induced dose-dependent decreases in microbial activity at all three time points; the only exception was AMX-MTZ at the 1:27 dilution at 24 h. Chlorhexidine reduced metabolic activity more strongly than did most antimicrobials tested at the higher dilutions (i.e., 1:9 and 1:27) (Table 1). In comparisons of the antibiotics or the combination versus others at 36 h, at dilutions of 1:27, AMX-MTZ reduced metabolic activity 11% less than CHX (q = 0.0207) but 54% more than AMX (q = 0.0031), 72% more than MTZ (q = 0.0031), and 67% more than AZM (q = 0.0008) (Table 2).
TABLE 1.
Reduction of metabolic activity after exposure to antibiotics at different concentrations and incubation times
| Time and dilution | % reduction of metabolic activity (mean ± SD) after exposure toa: |
||||
|---|---|---|---|---|---|
| CHX | MTZ | AMX | AZM | AMX-MTZ | |
| 12 h | |||||
| 1:1 | 94.2 ± 1.2 | 40.8 ± 2.6 | 90.4 ± 1.9 | 84.9 ± 7.7 | 93.4 ± 0.5 |
| 1:3 | 22.0 ± 17.4 | 37.3 ± 20.2 | 48.4 ± 32.4 | 71.5 ± 33.4 | |
| 1:9 | 18.5 ± 7.3 | 13.0 ± 32.5 | 21.6 ± 37.4 | 77.4 ± 13.5 | |
| 1:27 | 14.5 ± 9.4 | 0.4 ± 32.0 | −0.5 ± 5.7 | 67.3 ± 25.1 | |
| 24 h | |||||
| 1:1 | 94.8 ± 0.6 | 53.7 ± 3.3 | 64.0 ± 31.8 | 58.6 ± 28.8 | 60.8 ± 46.1 |
| 1:3 | 37.7 ± 8.7 | 37.8 ± 27.2 | 33.8 ± 15.5 | 69.3 ± 23.6 | |
| 1:9 | 23.8 ± 3.4 | 8.5 ± 9.2 | 21.5 ± 7.9 | 56.9 ± 45.6 | |
| 1:27 | 31.0 ± 8.1 | 25.3 ± 10.3 | 13.3 ± 6.4 | 16.0 ± 12.9 | |
| 36 h | |||||
| 1:1 | 95.3 ± 0.2 | 40.7 ± 18.6 | 93.7 ± 0.2 | 90.1 ± 5.4 | 94.3 ± 0.2 |
| 1:3 | 20.6 ± 42.2 | 88.8 ± 5.5 | 50.4 ± 60.7 | 92.5 ± 0.4 | |
| 1:9 | 39.7 ± 9.7 | 43.7 ± 11.8 | 13.0 ± 6.4 | 82.5 ± 2.5 | |
| 1:27 | 12.0 ± 16.4 | 29.8 ± 11.4 | 17.3 ± 7.0 | 84.1 ± 4.6 | |
CHX, chlorhexidine; MTZ, metronidazole; AMX, amoxicillin; AZM, azithromycin; SD, standard deviation.
TABLE 2.
Results of pairwise t tests comparing one drug (or combination) to another
| Antibiotic(s) compared and parametera | CHX | AMX | MTZ | AZM |
|---|---|---|---|---|
| AMX | ||||
| % mean difference (95% CI) | 65 (47–84) | |||
| q | 0.0020 | |||
| MTZ | ||||
| % mean difference (95% CI) | 83 (57–109) | 18 (−14 to 50) | ||
| q | 0.0023 | 0.2188 | ||
| AZM | ||||
| % mean difference (95% CI) | 78 (67–89) | 13 (−9 to 34) | −5 (−34 to 23) | |
| q | 0.0004 | 0.2188 | 0.6319 | |
| AMX-MTZ | ||||
| % mean difference (95% CI) | 11 (4–18) | −54 (−74 to −35) | −72 (−99 to −45) | −67 (−80 to −53) |
| q | 0.0207 | 0.0031 | 0.0031 | 0.0008 |
All results were assessed at 36 h, at dilutions of 1:27. CHX, chlorhexidine; AMX, amoxicillin; MTZ, metronidazole; AZM, azithromycin; CI, confidence interval.
Interaction between metronidazole and amoxicillin.
At 36 h, at the dilution of 1:27, AMX in the absence of MTZ induced inhibition of 30% (95% confidence interval [CI], 12% to 47%). In the presence of MTZ, this effect increased to 72% (95% CI, 55% to 90%; interaction P = 0.01). MTZ exhibited little to no inhibitory activity in the absence of amoxicillin (95% CI, 6% to 30%) but achieved 54% inhibition in the presence of amoxicillin (95% CI, 37% to 72%; interaction P = 0.01) (Table 3).
TABLE 3.
Interactions between AMX and MTZ at different concentrations at 36 h of incubation
| Dilution |
||||
|---|---|---|---|---|
| Drug treatment and parametera | 1:1 | 1:3 | 1:9 | 1:27 |
| AMX in absence of MTZ | ||||
| Difference in % inhibition (95% CI) | 0.94 (0.78–1.10) | 0.89 (0.54–1.23) | 0.44 (0.30–0.58) | 0.30 (0.12–0.47) |
| P | <0.0005 | <0.0005 | <0.0005 | 0.01 |
| AMX in presence of MTZ | ||||
| Difference in % inhibition (95% CI) | 0.54 (0.37–0.70) | 0.72 (0.37–1.07) | 0.43 (0.29–0.57) | 0.72 (0.55–0.90) |
| P | <0.0005 | <0.0005 | <0.0005 | <0.0005 |
| Interaction Pb | 0.01 | 0.52 | 0.93 | 0.01 |
| MTZ in absence of AMX | ||||
| Difference in % inhibition (95% CI) | 0.41 (0.25–0.57) | 0.21 (−0.14 to 0.55) | 0.40 (0.26–0.54) | 0.12 (−0.06 to 0.30) |
| P | <0.0005 | 0.28 | <0.0005 | 0.22 |
| MTZ in presence of AMX | ||||
| Difference in % inhibition (95% CI) | 0.01 (−0.16 to 0.17) | 0.04 (−0.31 to 0.38) | 0.39 (0.25–0.53) | 0.54 (0.37–0.72) |
| P | 0.94 | 0.84 | <0.0005 | <0.0005 |
| Interaction Pb | 0.01 | 0.52 | 0.93 | 0.01 |
A linear regression model was used, and the dependent variable was percent inhibition. AMX, amoxicillin; MTZ, metronidazole; CI, confidence interval.
Interaction P value comparing absence or presence of the second drug.
Both the dose-response relationships for individual antibiotics and the synergistic interaction were observed at 36 h (Tables 2 and 3). At their highest concentrations, we observed an antagonistic interaction between the two antibiotics at 36 h (Table 3); that is, each antibiotic exhibited less inhibitory activity in the presence of the other antibiotic than when used alone.
DISCUSSION
We described an in vitro multispecies biofilm model that mimics the composition and structure of subgingival plaque. The model presents several advantages over previously existing models (16–21), in that we were able to consistently establish 35 species, compared with the previous maximum of 5 to 10 species (18, 20, 21). Further, the Calgary Biofilm Device afforded a high-throughput system to grow several repeats of biofilms with good consistency across pegs and plates, as evidenced by the high minimum similarity coefficient values. We used growth conditions (specifically regarding the medium and atmosphere) designed to reproduce the environmental conditions found in the subgingival habitat of deep periodontal pockets. Parameters such as the presence of saliva or some of its constituents, pH, and surface coatings (e.g., l-lysine) were tested during optimization of the model. We also provided evidence that the in vitro biofilms share characteristics of in vivo biofilms. For example, SEM images demonstrated dense complex biofilms on the pegs of the CBD. Well-established periodontal pathogens such as A. actinomycetemcomitans, E. nodatum, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, F. periodonticum, P. micra, and P. gingivalis were consistently recovered from the formed biofilms, which, in addition to validating the model conditions, allows for testing of antimicrobials targeting these species.
Some features of the multispecies biofilm limit its potential uses, however. For example, we did not recover several known periodontal pathogens, specifically P. intermedia, P. nigrescens, T. forsythia, P. acnes, and P. melaninogenica, presumably because they failed to grow in the biofilms. Furthermore, in comparing counts of species recovered to levels reported for 178 subjects (28), we inferred that several species acted as microbial weeds and clearly outgrew other species at levels not observed in vivo. These differences resulted in relatively low minimum similarity coefficient values for the microbial profiles of in vitro and in vivo biofilms. In addition to raising general concerns about the applicability of the model to humans, these differences might have influenced the results of the specific experiment we conducted. For instance, the failure to recover several Prevotella species might have increased the tolerance of the biofilms to metronidazole, which specifically targets strict anaerobic Gram-negative bacteria. Furthermore, Treponema species were also excluded from the in vitro model, due to difficulties in the growth of these strict anaerobes.
At any given time, a single periodontal pocket harbors an average of 100 species, of 300 possible colonizers. Many of these species necessarily occur in very small proportions (32, 33). We selected 40 species that are among the most abundant and that collectively account for approximately 60% of the subgingival biofilm mass (34), and the number of species recovered, i.e., 35, is much greater than that achieved in other currently used biofilm models.
Conversion of TTC has been used as a test to measure the levels of metabolic activity of bacterial cells grown planktonically or as biofilms and can be used as a surrogate for cell viability (35–39). Chlorhexidine decreased the metabolic activity of the biofilms by approximately 95% ± 0.8% (mean ± SD) at all exposure times (12, 24, and 36 h), demonstrating its appropriateness as a positive control. At the primary time point (36 h) and the most physiologically relevant concentration (1:27 dilution), the combination of AMX and MTZ had greater antimicrobial effects than would be expected if the two drugs were working independently. Even at concentrations for which there was little statistical evidence of synergy, the additive effects still afforded enhanced antimicrobial activity. For most dilutions and time points tested, the combination of AMX and MTZ gave results similar to those obtained with CHX.
We identified the 1:27 dilutions as being of primary interest because they corresponded to levels obtained in gingival crevicular fluid (GCF) with the systemic use of these agents, i.e., 4, 10, and 8 μg/ml for AMX, MTZ, and AZM, respectively (40–42). However, we also tested higher concentrations to examine whether they had increased efficacy. It has been argued that the superiority of the combination of AMX and MTZ over MTZ alone observed in clinical trials (6, 8, 12–14) could be the consequence of an overall higher dosage of antibiotics, rather than a synergistic antimicrobial mechanism. Even at levels 27-fold higher than those typically obtained clinically, however, MTZ did not decrease the metabolic activity of the biofilms to levels achieved with the combination of AMX and MTZ. Similarly, AMX alone required concentrations 9 to 27 times higher than the combination to achieve greater reductions in the metabolic activity of the biofilms. Thus, these data are inconsistent with additive effects of AMX and MTZ and are more consistent with synergistic effects of these antimicrobials.
A possible explanation for the synergistic effects of metronidazole and amoxicillin may be increased uptake of metronidazole in the presence of amoxicillin, as has been described for A. actinomycetemcomitans (23). Furthermore, metronidazole is effective against species from the anaerobic genus Prevotella, which are capable of producing beta-lactamases (43). Therefore, another potential synergistic mechanism would be enhancement of amoxicillin activity in the presence of metronidazole due to decreases in the levels of beta-lactamases. Our findings provide additional support for the use of such drugs in the treatment of periodontal diseases.
In vitro biofilm models have been proposed as a means to examine the higher tolerance to antimicrobials that this mode of growth confers to bacteria (4). It has been argued that, due to greater tolerance to antimicrobials, MICs calculated using bacterial cells grown planktonically would bear little relevance to in vivo situations (25). The higher tolerance of biofilms to antimicrobials has also led periodontists to recommend that the use of these agents be accompanied or preceded by mechanical disruption of the subgingival biomass. This notion was challenged by a clinical study that demonstrated that the use of systemic AMX-MTZ therapy without subgingival debridement could lead to clinical improvements and changes in the subgingival microbiota similar to those obtained with mechanical therapy (i.e., scaling and root planing) (44). Here we provide additional support for the notion that AMX-MTZ, at concentrations observed in GCF after systemic administration, can significantly reduce the metabolic activity of subgingival bacterial species even when they are organized in complex biofilms. In conclusion, reproducible biofilms with 35 subgingival bacterial species were developed, and the combination of AMX and MTZ had greater antimicrobial effects on these in vitro multispecies biofilms than expected on the basis of the independent effects of the drugs.
ACKNOWLEDGMENT
This work was partially funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Ministério da Educação (Brazil) (grant 4986/10-5).
REFERENCES
- 1.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ, CDC Periodontal Disease Surveillance Workgroup. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res 91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 2.Brown LJ, Johns BA, Wall TP. 2002. The economics of periodontal diseases. Periodontology 2000 29:223–234. doi: 10.1034/j.1600-0757.2002.290111.x. [DOI] [PubMed] [Google Scholar]
- 3.Borrell LN, Talih M. 2012. Examining periodontal disease disparities among U.S. adults 20 years of age and older: NHANES III (1988–1994) and NHANES 1999–2004. Public Health Rep 127:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD. 2002. Dental biofilms: difficult therapeutic targets. Periodontology 2000 28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 5.Winkel EG, van Winkelhoff AJ, Timmerman MF, van der Velden U, van der Weijden GA. 2001. Amoxicillin plus metronidazole in the treatment of adult periodontitis patients: a double-blind placebo controlled study. J Clin Periodontol 28:296–305. doi: 10.1034/j.1600-051x.2001.028004296.x. [DOI] [PubMed] [Google Scholar]
- 6.Rooney J, Wade WG, Sprague SV, Newcombe RG, Addy M. 2002. Adjunctive effects to non-surgical periodontal therapy of systemic metronidazole and amoxicillin alone and combined: a placebo controlled study. J Clin Periodontol 29:342–350. doi: 10.1034/j.1600-051X.2002.290410.x. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho LH, D'Avila GB, Leao A, Haffajee AD, Socransky SS, Feres M. 2004. Scaling and root planing, systemic metronidazole and professional plaque removal in the treatment of chronic periodontitis in a Brazilian population. I. Clinical results. J Clin Periodontol 31:1070–1076. doi: 10.1111/j.1600-051X.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 8.Matarazzo F, Figueiredo LC, Cruz SE, Faveri M, Feres M. 2008. Clinical and microbiological benefits of systemic metronidazole and amoxicillin in the treatment of smokers with chronic periodontitis: a randomized placebo-controlled study. J Clin Periodontol 35:885–896. doi: 10.1111/j.1600-051X.2008.01304.x. [DOI] [PubMed] [Google Scholar]
- 9.Haffajee AD, Torresyap G, Socransky SS. 2007. Clinical changes following four different periodontal therapies for the treatment of chronic periodontitis: 1-year results. J Clin Periodontol 34:243–253. doi: 10.1111/j.1600-051X.2006.01040.x. [DOI] [PubMed] [Google Scholar]
- 10.Herrera D, Contreras A, Gamonal J, Oteo A, Jaramillo A, Silva N, Sanz M, Botero JE, León R. 2008. Subgingival microbial profiles in chronic periodontitis patients from Chile, Colombia and Spain. J Clin Periodontol 35:106–113. doi: 10.1111/j.1600-051X.2008.01263.x. [DOI] [PubMed] [Google Scholar]
- 11.Cionca N, Giannopoulou C, Ugolotti G, Mombelli A. 2009. Amoxicillin and metronidazole as an adjunct to full-mouth scaling and root planing of chronic periodontitis. J Periodontol 80:364–371. doi: 10.1902/jop.2009.080540. [DOI] [PubMed] [Google Scholar]
- 12.Silva MP, Feres M, Sirotto TA, Soares GM, Mendes JA, Faveri M, Figueiredo LC. 2011. Clinical and microbiological benefits of metronidazole alone or with amoxicillin as adjuncts in the treatment of chronic periodontitis: a randomized placebo-controlled clinical trial. J Clin Periodontol 38:828–837. doi: 10.1111/j.1600-051X.2011.01763.x. [DOI] [PubMed] [Google Scholar]
- 13.Feres M, Soares GMS, Mendes JAV, Silva MP, Faveri M, Teles R, Socransky SS, Figueiredo LC. 2012. Metronidazole alone or with amoxicillin as adjuncts to nonsurgical treatment of chronic periodontitis: a 1-year double-blinded, placebo-controlled, randomized clinical trial. J Clin Periodontol 39:1149–1158. doi: 10.1111/jcpe.12004. [DOI] [PubMed] [Google Scholar]
- 14.Soares GM, Mendes JA, Silva MP, Faveri M, Teles R, Socransky SS, Wang X, Figueiredo LC, Feres M. 2014. Metronidazole alone or with amoxicillin as adjuncts to non-surgical treatment of chronic periodontitis: a secondary analysis of microbiological results from a randomized clinical trial. J Clin Periodontol 41:366–376. doi: 10.1111/jcpe.12217. [DOI] [PubMed] [Google Scholar]
- 15.Matesanz-Pérez P, García-Gargallo M, Figuero E, Bascones-Martínez A, Sanz M, Herrera D. 2013. A systematic review on the effects of local antimicrobials as adjuncts to subgingival debridement, compared with subgingival debridement alone, in the treatment of chronic periodontitis. J Clin Periodontol 40:227–241. doi: 10.1111/jcpe.12026. [DOI] [PubMed] [Google Scholar]
- 16.Sissons CH. 1997. Artificial dental plaque biofilm model systems. Adv Dent Res 11:110–126. doi: 10.1177/08959374970110010201. [DOI] [PubMed] [Google Scholar]
- 17.Guggenheim B, Giertsen E, Schüpbach P, Shapiro S. 2001. Validation of an in vitro biofilm model of supragingival plaque. J Dent Res 80:363–370. doi: 10.1177/00220345010800011201. [DOI] [PubMed] [Google Scholar]
- 18.Guggenheim B, Gmür R, Galicia JC, Stathopoulou PG, Benakanakere MR, Meier A, Thurnheer T, Kinane DF. 2009. In vitro modeling of host-parasite interactions: the ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol 9:280. doi: 10.1186/1471-2180-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez MC, Llama-Palacios A, Blanc V, León R, Herrera D, Sanz M. 2011. Structure, viability and bacterial kinetics of an in vitro biofilm model using six bacteria from the subgingival microbiota. J Periodont Res 46:252–260. doi: 10.1111/j.1600-0765.2010.01341.x. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez MC, Llama-Palacios A, Marín MJ, Figuero E, León R, Blanc V, Herrera D, Sanz M. 2013. Validation of ATP bioluminescence as a tool to assess antimicrobial effects of mouthrinses in an in vitro subgingival-biofilm model. Med Oral Patol Oral Cir Bucal 18:e86–e92. doi: 10.4317/medoral.18376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanc V, Isabal S, Sánchez MC, Llama-Palacios A, Herrera D, Sanz M, León R. 2014. Characterization and application of a flow system for in vitro multispecies oral biofilm formation. J Periodont Res 49:323–332. doi: 10.1111/jre.12110. [DOI] [PubMed] [Google Scholar]
- 22.Pavicić MJ, van Winkelhoff AJ, de Graaff J. 1991. Synergistic effects between amoxicillin, metronidazole, and the hydroxymetabolite of metronidazole against Actinobacillus actinomycetemcomitans. Antimicrob Agents Chemother 35:961–966. doi: 10.1128/AAC.35.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavicić MJ, van Winkelhoff AJ, Pavicić-Temming YA, de Graaff J. 1994. Amoxycillin causes an enhanced uptake of metronidazole in Actinobacillus actinomycetemcomitans: a mechanism of synergy. J Antimicrob Chemother 34:1047–1050. doi: 10.1093/jac/34.6.1047. [DOI] [PubMed] [Google Scholar]
- 24.Kulik Kunz EM, Lenkeit K, Waltimo T, Weiger R, Walter C. 2014. Combinatorial effects of amoxicillin and metronidazole on selected periodontal bacteria and whole plaque samples. Arch Oral Biol 59:608–615. doi: 10.1016/j.archoralbio.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Socransky SS, Smith C, Martin L, Paster BJ, Dwehirst FE, Levin AE. 1994. Checkerboard DNA-DNA hybridization. Biotechniques 17:788–792. [PubMed] [Google Scholar]
- 27.Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, Goodson JM. 2004. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol 19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 28.Socransky SS, Haffajee AD, Teles R, Wennstrom JL, Lindhe J, Bogren A, Hasturk H, van Dyke T, Wang X, Goodson JM. 2013. Effect of periodontal therapy on the subgingival microbiota over a 2-year monitoring period. I. Overall effect and kinetics of change. J Clin Periodontol 40:771–780. doi: 10.1111/jcpe.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teles FR, Haffajee AD, Socransky SS. 2008. The reproducibility of curet sampling of subgingival biofilms. J Periodontol 79:705–713. doi: 10.1902/jop.2008.070424. [DOI] [PubMed] [Google Scholar]
- 30.Socransky SS, Tanner AC, Goodson JM, Haffajee AD, Walker CB, Ebersole JL, Sornberger GC. 1982. An approach to the definition of periodontal disease syndromes by cluster analysis. J Clin Periodontol 9:460–471. doi: 10.1111/j.1600-051X.1982.tb02107.x. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 32.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontology 2000 38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 35.Schaule G, Flemming HC, Ridgway HF. 1993. Use of 5-cyano-2,3-ditolyl tetrazolium chloride for quantifying planktonic and sessile respiring bacteria in drinking water. Appl Environ Microbiol 59:3850–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, Kim MJ, Kang HY, Seol SY, Cho DT, Kim J. 2010. A simple colorimetric method for testing antimicrobial susceptibility of biofilmed bacteria. J Microbiol 48:709–711. doi: 10.1007/s12275-010-0299-z. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczuk D, Ginalska G, Piersiak T, Miazga-Karska M. 2012. Prevention of biofilm formation on urinary catheters: comparison of the sparfloxacin-treated long-term antimicrobial catheters with silver-coated ones. J Biomed Mater Res B Appl Biomater 100:1874–1882. doi: 10.1002/jbm.b.32755. [DOI] [PubMed] [Google Scholar]
- 38.González OA, Escamilla C, Danaher RJ, Dai J, Ebersole JL, Mumper RJ, Miller CS. 2013. Antibacterial effects of blackberry extract target periodontopathogens. J Periodont Res 48:80–86. doi: 10.1111/j.1600-0765.2012.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown HL, van Vliet AH, Betts RP, Reuter MJ. 2013. Tetrazolium reduction allows assessment of biofilm formation by Campylobacter jejuni in a food matrix model. J Appl Microbiol 115:1212–1221. doi: 10.1111/jam.12316. [DOI] [PubMed] [Google Scholar]
- 40.Tenenbaum H, Jehl F, Gallion C, Dahan M. 1997. Amoxicillin and clavulanic acid concentrations in gingival crevicular fluid. J Clin Periodontol 24:804–807. doi: 10.1111/j.1600-051X.1997.tb01193.x. [DOI] [PubMed] [Google Scholar]
- 41.Pähkla ER, Koppel T, Saag M, Pähkla R. 2005. Metronidazole concentrations in plasma, saliva and periodontal pockets in patients with periodontitis. J Clin Periodontol 32:163–166. doi: 10.1111/j.1600-051X.2005.00653.x. [DOI] [PubMed] [Google Scholar]
- 42.Lai PC, Ho W, Jain N, Walters JD. 2011. Azithromycin concentrations in blood and gingival crevicular fluid after systemic administration. J Periodontol 82:1582–1586. doi: 10.1902/jop.2011.110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handal T, Olsen I, Walker CB, Caugant DA. 2004. β-Lactamase production and antimicrobial susceptibility of subgingival bacteria from refractory periodontitis. Oral Microbiol Immunol 19:303–308. doi: 10.1111/j.1399-302x.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 44.Lopez NJ, Socransky SS, Da Silva I, Japlit MR, Haffajee AD. 2006. Effects of metronidazole plus amoxicillin as the only therapy on the microbiological and clinical parameters of untreated chronic periodontitis. J Clin Periodontol 33:648–660. doi: 10.1111/j.1600-051X.2006.00957.x. [DOI] [PubMed] [Google Scholar]
- 45.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. [DOI] [PubMed] [Google Scholar]