Abstract
Although both tetracycline and tigecycline inhibit protein synthesis by sterically hindering the binding of tRNA to the ribosomal A site, tigecycline shows increased efficacy in both in vitro and in vivo activity assays and escapes the most common resistance mechanisms associated with the tetracycline class of antibiotics. These differences in activities are attributed to the tert-butyl-glycylamido side chain found in tigecycline. Our structural analysis by X-ray crystallography shows that tigecycline binds the bacterial 30S ribosomal subunit with its tail in an extended conformation and makes extensive interactions with the 16S rRNA nucleotide C1054. These interactions restrict the mobility of C1054 and contribute to the antimicrobial activity of tigecycline, including its resistance to the ribosomal protection proteins.
INTRODUCTION
The ribosome, a central component of the protein synthesis machinery, is one of the major targets of clinically relevant antibiotics (1–3). In the last decade, crystal structures of a broad variety of antibiotics bound to either the large (50S) or the small (30S) subunit of the bacterial ribosome have been reported, unraveling their mechanism of action and demonstrating that they interact at a few distinct but functionally important sites (1, 2). For example, on the 50S subunit, antibiotics target primarily the peptidyl transferase center, the GTPase-associated center, or the ribosomal exit tunnel and hamper protein synthesis by interfering with the incorporation of new amino acids into the growing peptide chain (1). On the 30S subunit, antibiotics have thus far been observed at or near mRNA and tRNA binding sites and generally interfere with correct tRNA binding to the A site or with translocation of the tRNA/mRNA from the A site to the P site (1, 2). Tetracycline (TET) is an example of a 30S subunit binding antibiotic, with both structural and biochemical studies indicating that it binds the ribosome primarily in a pocket formed by the 16S rRNA helices 31 (h31) and 34, although secondary binding sites have also been observed (4–6). The significance of these secondary sites is unclear as binding to the primary site correlates best with the antimicrobial activity of the drug and resistance mutations (7).
Upon their introduction into medicine in 1948, tetracyclines were quickly accepted because they offered a broad spectrum of activity (8). However, given the widespread use of “legacy” tetracyclines for more than 60 years, resistance in clinically important bacterial pathogens is common (8, 9). Accordingly, modern tetracycline derivatives, like tigecycline (TIG), omadacycline, and eravacycline (TP-434, Erv), have been developed and display activity against bacterial strains resistant to the legacy tetracyclines (3). TIG was the first representative of these derivatives to be approved for use by the FDA (10). Omadacycline is currently under clinical development for the treatment of acute bacterial skin infections (11), community-acquired bacterial pneumonia, and complicated urinary tract infections, and eravacycline is under clinical development for complicated intraabdominal and complicated urinary tract infections (12).
These modern tetracycline derivatives are generally distinguished by a C-7 or C-9 substituent of the D ring; for example, TIG is structurally derived from minocycline by attaching a tert-butyl-glycylamido side chain at position C-9 of the tetracycline four-ring backbone (Fig. 1). Interestingly, biochemical and structural studies have shown that these tetracycline derivatives target the primary tetracycline binding site, but in the case of TIG, the C-9 substituent confers it with an ∼10- to 100-fold higher affinity for the ribosome, an ∼10- to 30-fold increase in inhibitory activity in in vitro translation assays, and, most importantly, the ability to evade many of the most common tetracycline resistance determinants (3, 6, 9, 13, 14). The binding of tetracycline and derivatives like TIG to the primary site interferes with the accommodation of incoming aminoacyl-tRNA into the ribosomal A site via a steric clash with the anticodon loop (4–6). A systematic analysis of various tetracycline derivatives (6) has suggested that the increased potency of TIG stems from additional interactions made between the tert-butyl-glycylamido side chain and the ribosome, in particular, a stacking interaction between the glycylamido moiety and C1054 (h34). The same study (6) also showed a correlation between the bulkiness of the side chain, but not the increased affinity it potentially confers, and enhanced activity against bacterial strains resistant to the legacy tetracyclines. To complement recent investigations of TIG bound to the 70S ribosome (6) and facilitate the increasing development of modern tetracycline derivatives characterized by a C-9 substituent, we have investigated TIG's mode of interaction with the Thermus thermophilus 30S ribosomal subunit using X-ray crystallography. We observed an alternative conformation for the functionally important tert-butyl-glycylamido side chain and, consequently, novel interactions with the 16S rRNA that may be critical for the rational design of newer tetracycline derivatives.
FIG 1.
Chemical structures of tetracycline, minocycline, and tigecycline, drawn schematically with their common backbone ring structures (A, B, C, D) colored distinctly. Carbon atom assignments for the 4-ring backbone are indicated on tigecycline.
MATERIALS AND METHODS
Structure determination.
The 30S ribosomal subunit was purified from Thermus thermophilus HB8 and crystallized (15, 16). Crystals were soaked for 12 to 24 h with 100 μM tigecycline and flash-frozen in liquid nitrogen while cryoprotected with 2-methyl-2,4-pentanediol. Diffraction data were collected at beam lines X06SA of the Swiss Light Source (Villigen, Switzerland), ID29 of the European Synchrotron Radiation Facility (Grenoble, France), and BL13 of ALBA (Barcelona, Spain). Data were processed with the XDS (17) and CCP4 (18) program packages. The native structure of the 30S subunit (PDB code 2ZM6) was refined against the structure factor amplitudes of the 30S-TIG complex using the PHENIX program package (19). TIG was modeled for residual electron density using COOT (20), and the resulting 30S-TIG model was further refined to convergence. For the calculation of the free R factor, 5% of the data were omitted throughout refinement. Figures containing structures were illustrated with MarvinSketch (ChemAxon) and PyMOL (http://pymol.sourceforge.net). rRNA residues were numbered according to the Escherichia coli scheme, and helices are indicated using the standard nomenclature (21) throughout the article.
Protein structure accession number.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession no. 4YHH.
RESULTS
Overall structure.
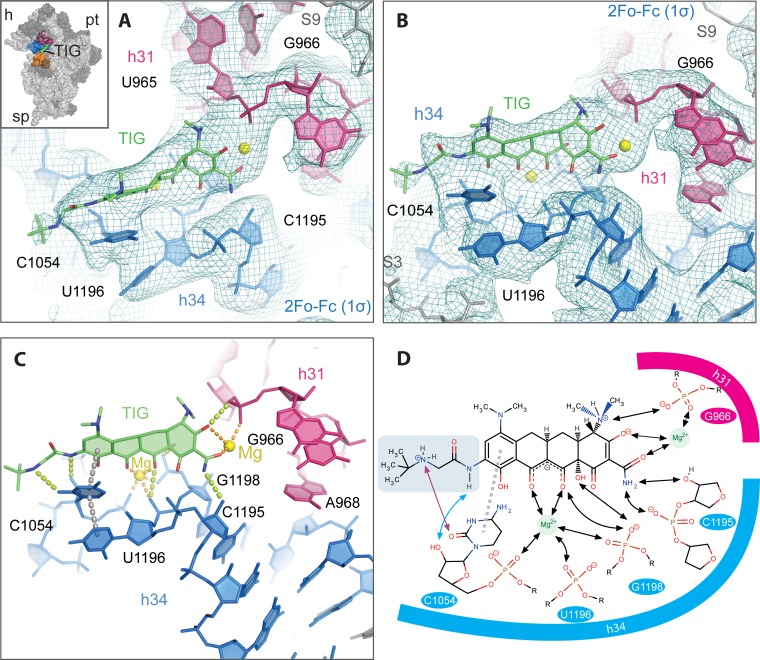
To provide an atomic-level description of the ribosomal subunit-TIG interaction, we determined the structure of the antibiotic bound to the T. thermophilus 30S ribosomal subunit at 3.4 Å resolution. The native structure of the 30S ribosomal subunit crystallized in the absence of TIG (PDB code 2ZM6) was refined against the structure factor amplitudes of the 30S-TIG complex (Table 1; see also Materials and Methods). The unbiased Fo-Fc difference Fourier map (see Fig. S1A in the supplemental material) obtained after the initial round of refinement using the ligand-free 30S subunit structure (2ZM6) indicated that the 585-Da TIG molecule was bound in a position that overlaps with the primary tetracycline binding site, a pocket formed by h34 (1196 to 1198, 1052 to 1055) and the loop of h31 (965, 966). In this initial difference map, electron density was clearly observed for the 4-member ring moiety of TIG as well as the glycylamido moiety of its derivatized side chain (see Fig. S1A in the supplemental material). Although the 30S-TIG complex was formed using antibiotic concentrations 4 orders of magnitude greater than its dissociation constant, TIG was not found bound to any of the secondary binding sites previously observed for the parent compound tetracycline (4, 5). This is consistent with the recent X-ray crystallographic studies using 70S ribosomes and biochemical studies, suggesting a single saturable binding site for TIG on the ribosome and the inability of TIG to protect A892, an rRNA residue in one of the secondary binding sites, from modification by dimethyl sulfate (6, 14). Addition of the TIG molecule and subsequent refinement at a resolution of 3.4 Å led to a final model with crystallographic Rwork/Rfree values of 22.8%/28.7% (Table 1). As seen in Fig. 2A and B, density is visible for the entire TIG model, including the terminal methyl group present in the tert-butyl-glycylamido side chain in the fully refined 2Fo-Fc map (Fig. S1B in the supplemental material shows the same map at a higher threshold). The density is consistent with TIG being in the zwitterionic extended conformation characteristic of the tetracyclines when bound to the ribosome or the TET repressor tet(R) (22).
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s)a |
|---|---|
| Data collection statistics | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 409.590, 409.590, 171.730 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50–3.4 (3.58–3.4) |
| Rsymb (%) | 16.2 |
| I/σI | 4.45 (0.3) |
| Completeness (%) | 96.7 (84.3) |
| Multiplicity | 4.5 (3.8) |
| Refinement | |
| Resolution (Å) | 48.7–3.4 |
| No. of reflections | 171,310 |
| Rwork/Rfreec (%) | 22.8/28.7 |
| Total no. of atoms | 51,732 |
| Ribosome | 51,585 |
| Ligand | 42 |
| Ions | 105 |
| RMSd deviations | |
| Bond length (Å) | 0.010 |
| Bond angle (°) | 1.308 |
Values in parentheses refer to the highest-resolution shell.
Rsym = Σ|I − 〈I〉|/ΣI, where I is a measured and 〈I〉 is the average intensity of the reflection.
Rwork/Rfree = Σ|Fo − Fc|/ΣFo, where Fo is the observed and Fc is the calculated structure factor amplitude of the reflection. A randomly selected 5% of the reflections was set aside from the beginning and the rest were used in the refinement.
RMS, root mean square.
FIG 2.
The environment of the tigecycline-binding site on the 30S ribosomal subunit. TIG binds the 30S ribosomal subunit in a pocket formed by h34 (blue) and h31 (red). The final model with 2Fo-Fc electron density map (1.0σ) surrounding the TIG-binding site is shown from two perspectives in panels A and B. A similar view is shown in Fig. S1B in the supplemental material with the final 2Fo-Fc map rendered at a slightly higher contour level (1.4σ). The relatively weak density for the terminal tert-butyl moiety probably results from the fact that it is connected to the rest of the molecule by two freely rotatable bonds and does not make direct interactions with the binding pocket. TIG is shown in an orientation similar to that of Fig. 1 where the A ring is on the right and the D ring with the tert-butyl-glycylamido side chain is on the left. The two Mg2+ ions are drawn as yellow spheres. The inset in panel A shows the position of the TIG binding pocket on the 30S subunit (viewed from the subunit interface side) with subunit landmarks (pt, platform; h, head; sp, spur). 16S rRNA helices 18, 31, and 34 are colored orange, pink, and blue, respectively. (C) The TIG binding pocket is shown from the same perspective as that in panel B with its putative interactions indicated with dotted lines, including hydrogen bonds (yellow), coordination of the Mg2+ ions (orange), and stacking interactions (gray). These interactions are summarized schematically in panel D. Here the tert-butyl-glycylamido side chain is delineated in light green, and its interactions with C1054 are indicated with cyan and violet arrows. Note that the hydrogen bond between the secondary amine of the side chain and O2 of C1054 is chemically possible but ambiguous in the electron density map. The hydroxyl moiety at position 3 is drawn in a deprotonated form as predicted by nuclear magnetic resonance (NMR) and molecular dynamics (MD) studies (22, 25).
TIG binding site.
Compared to the results of previous studies on TET (4, 5), the 4 backbone rings of TIG interact similarly with rRNA (Fig. 2C and D; see also Fig. S2 in the supplemental material), including (i) the coordination of a Mg2+ ion between the polar groups of rings B and C of TIG, (ii) putative hydrogen bonds to residues G1198 and C1195, and (iii) a possible stacking interaction between C1054 and ring D of TIG. Furthermore, in agreement with the results of a recent study showing TIG bound to 70S ribosomes (6), we observe that a second Mg2+ ion is coordinated by ring A of TIG and the phosphate backbone of G966 in h31 (Fig. 2). Relative to this previous structure (6), there is a slight shift of the 4-ring backbone and second Mg2+ ion (∼1 Å), such that the predicted coordination pattern is different in the two structures. Here the Mg2+ is potentially coordinated by both the hydroxyl oxygen at position 3 and the acetamide moiety at position 2 of TIG, while the previous study indicated only a potential interaction with the hydroxyl oxygen. Given the resolution of the data, the significance of this difference is unclear.
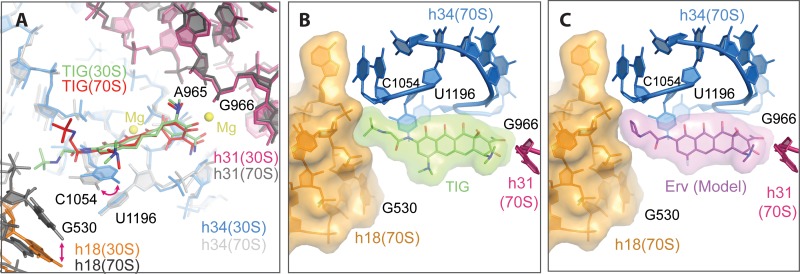
The electron density maps (Fig. 2A and B) support the placement of the tert-butyl-glycylamido side chain in an extended conformation reaching toward h18 (G530). This differs from the orientation seen in the 70S-TIG structure (Fig. 3A, green versus red) where this side chain has a bent conformation and the amido moiety forms stacking interactions with the base of C1054 (6). In the extended conformation, observed in the 30S-TIG structure (Fig. 3A, green), C1054 is shifted by ∼2.3 Å relative to the 70S-TIG structure and C1054 is more favorably positioned to stack with the D ring rather than the side chain (Fig. 2C and D). Furthermore, when the tert-butyl-glycylamido side chain is in the extended conformation, it is characterized by (i) the peptide bond being maintained in a planar conformation consistent with its partial double bond character, (ii) a potential hydrogen bond between the 2′-hydroxyl of C1054 and the peptide bond amide nitrogen (Fig. 2D, blue arrow), and (iii) a potential hydrogen bond between the secondary amine, which is most likely protonated (theoretical pKa = 10.7), and the base of C1054 (Fig. 2D, violet arrow). It is interesting to note that in the 70S-TIG structure h18 moves closer to the A site, a conformational switch (closed form) seen when, for example, the head and shoulder rotate inward toward the 30S subunit center during decoding (23). As seen in Fig. 3B, h18 in the “closed” conformation does not clash with the side chain of TIG in the extended conformation. Therefore, the conformational switch of h18 is likely not the main source of the differences observed in the tert-butyl-glycylamido side chain when the 70S and 30S structures are compared (Fig. 3A).
FIG 3.
Comparison of TIG's binding mode in the 30S and 70S structures. (A) The 30S-TIG structure (this study) and the 70S-TIG structure (6) have been superimposed using 16S rRNA residues surrounding the TIG binding pocket as the guide. TIG as seen in the 30S structure is colored green, and TIG as seen in the 70S structure is red. h18, h34, and h31 of the 30S-TIG complex are in orange, blue, and red, respectively, and those from the 70S-TIG complex are in shades of gray. Although for the most part the two structures are superimposable, there are notable differences in the placement of h18 (shifts closer to the A site by ∼5 Å in the 70S structure), the position of the base in C1054 (it shifts by 2.5 Å, favoring a parallel shifted π stacking interaction with the heteroaromatic D ring on the 30S structure) and the conformation of the tert-butyl-glycylamido side chain (extended versus bent in the 30S and 70S structures, respectively). (B) To illustrate that the extended side chain conformation of TIG is permissible when the binding pocket is configured as seen in the 70S structure (specifically the shifting of h18), TIG has been aligned to the 70S structure (6) and rendered as a van der Waals surface. This shows that the side chain of TIG in the extended conformation does not significantly clash with h18. (C) Likewise, if the newer tetracycline derivative eravacycline (Erv) (9) is modeled with its side chain extended similar to TIG, it would also not clash with h18 as seen in the 70S structure.
DISCUSSION
Our investigation into the mode of interaction between TIG and the 30S subunit complements the previous studies with 70S ribosomes (6) and supports a model in which TIG binding to the primary tetracycline binding site involves two Mg2+ ions (Fig. 2). Importantly, however, we did observe some differences in the conformation of the functionally important tert-butyl-glycylamido side chain of TIG (Fig. 3A). These differences indicate that TIG has two alternative modes of interaction with the ribosome. The first mode seen in the 70S structure (6) is characterized by a bent conformation of the side chain and stacking interactions between its amide and C1054. The second mode, seen on the 30S subunit, is characterized by an extended conformation where the peptide bond in the tert-butyl-glycylamido side chain of TIG maintains its planar nature, and extensive polar interactions are potentially formed with both the base and ribose of C1054 (Fig. 2C and D). We speculate that the different binding modes observed in the 30S and 70S structures result from the fact that the primary tetracycline binding site in particular residues in h18 and C1054 (h34) is conformationally variable; namely its architecture can change when, for example, the head and shoulder rotate inward toward the 30S subunit center during decoding (23). As seen in Fig. 3A, h18, in particular, is in two distinct positions in the 30S and 70S structures. TIG must be able to accommodate these changes in order to remain tightly bound to the ribosome. The alignment of the 30S and 70S structures indicates that the extended conformation of TIG seen in the 30S structure is compatible with the closed positioning of h18 as seen in the 70S structure, and, therefore, this change alone is not sufficient to explain the differences in the conformation of the side chain (Fig. 3B). In addition, alignments with models of newer tetracycline derivatives, like eravacycline, suggest that they can also be accommodated on the 70S structure with their side chains modeled in an extended conformation (Fig. 3C).
The increased affinity and efficacy of TIG (compared to those of minocycline and tetracycline) in both in vivo and in vitro activity assays as well as TIG's ability to evade the most common tetracycline resistance mechanism are generally attributed to the bulky tert-butyl-glycylamido side chain. When TIG binds the ribosome with its side chain in the extended conformation, as seen in the 30S-TIG structure, it clashes noticeably more with the accommodating A-tRNA than tetracycline (see Fig. S2A in the supplemental material) and displays extensive interactions (polar and Van der Waals contacts) with its binding pocket; the buried surface area increases from 1,261 Å2 in the TET structure to 1,367 Å2 in the TIG structure. These characteristics alone might explain TIG's higher affinity for the ribosome and increased ability to interfere with A-site binding. Moreover, TIG interacts extensively with C1054 (Fig. 2C and D) and therefore restricts the mobility of this nucleotide. This might have functional consequences as this nucleotide is known to be highly dynamic and important for A-site decoding (6, 23). Therefore, the mode of action of TIG may be 2-fold: it extensively clashes with the anticodon loop of the accommodating A-tRNA, and it fixes C1054 in a conformation incompatible with the decoding reaction (see Fig. S3 in the supplemental material). Additionally, the flexibility of C1054 is important for the action of the ribosomal protection proteins (RPPs) that confer resistance to tetracyclines (24). In a recent cryo-electron microscopy (EM) structure study, it was reported that the RPP, Tet(M), interacts with the tetracycline binding pocket around C1054 and dislodges TET by perturbing the interaction between C1054 and the drug (3, 24). Therefore, as TIG interacts more extensively with C1054 via its D ring and tert-butyl-glycylamido side chain, it may evade the action of the RPPs by (i) sterically hindering access to C1054 (3, 24) and/or (ii) further locking C1054 in a conformation that is incompatible with the action of the RPP (see Fig. S2B in the supplemental material). Although this is not observed at the current resolution, it is interesting to speculate that the conformation of C1054 might indirectly affect the coordination/positioning of the Mg2+ ion interacting with the phosphate groups of C1054, C1196, and C1198 and the hydrophilic face of tetracycline and its derivatives (Fig. 2D). Accordingly, the drugs would fix C1054 in a conformation with a Mg2+ ion coordination favorable for drug binding, while the RPPs would work to distort this coordination by manipulating C1054; this would allow the RPPs to indirectly interfere with a significant number of interactions seen in the tetracycline binding pocket (Fig. 2D) to dislodge the drug.
Supplementary Material
ACKNOWLEDGMENTS
Support was provided by Bizkaia: Talent and the European Union's Seventh Framework Programme (Marie Curie actions [COFUND] to S.R.C., A.S., and T.K.), the Cluster of Excellence Macromolecular Complexes at Goethe University Frankfurt (DFG project EXC 115 to P.F. and S.C.), the Deutsche Forschungsgemeinschaft (FU579 1-3 to P.F.), and a Marie Curie career integration grant (PCIG14-GA-2013-632072 to P.F.).
These studies could not have been performed without the expert assistance of the staff at the X06SA, ID29, and BL13 beam lines (Swiss Light Source [SLS], European Synchrotron Radiation Facility [ESRF], and ALBA, respectively).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04895-14.
REFERENCES
- 1.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DN. 2009. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol 44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen F, Starosta AL, Arenz S, Sohmen D, Donhofer A, Wilson DN. 2014. Tetracycline antibiotics and resistance mechanisms. Biol Chem 395:559–575. doi: 10.1515/hsz-2013-0292. [DOI] [PubMed] [Google Scholar]
- 4.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodersen DE, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143–1154. doi: 10.1016/S0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 6.Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, Blanchard SC, Wilson DN, Yusupova G. 2013. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci U S A 110:3812–3816. doi: 10.1073/pnas.1216691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connell S, Tracz D, Nierhaus K, Taylor D. 2003. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother 47:3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossman TH, Starosta AL, Fyfe C, O'Brien W, Rothstein DM, Mikolajka A, Wilson DN, Sutcliffe JA. 2012. Target- and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibiotic. Antimicrob Agents Chemother 56:2559–2564. doi: 10.1128/AAC.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Testa RT, Petersen PJ, Jacobus NV, Sum PE, Lee VJ, Tally FP. 1993. In vitro and in vivo antibacterial activities of the glycylcyclines, a new class of semisynthetic tetracyclines. Antimicrob Agents Chemother 37:2270–2277. doi: 10.1128/AAC.37.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noel GJ, Draper MP, Hait H, Tanaka SK, Arbeit RD. 2012. A randomized, evaluator-blind, phase 2 study comparing the safety and efficacy of omadacycline to those of linezolid for treatment of complicated skin and skin structure infections. Antimicrob Agents Chemother 56:5650–5654. doi: 10.1128/AAC.00948-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomkin JS, Ramesh MK, Cesnauskas G, Novikovs N, Stefanova P, Sutcliffe JA, Walpole SM, Horn PT. 2014. Phase 2, randomized, double-blind study of the efficacy and safety of two dose regimens of eravacycline versus ertapenem for adult community-acquired complicated intra-abdominal infections. Antimicrob Agents Chemother 58:1847–1854. doi: 10.1128/AAC.01614-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson MW, Ruzin A, Feyfant E, Rush TS III, O'Connell J, Bradford PA. 2006. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob Agents Chemother 50:2156–2166. doi: 10.1128/AAC.01499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer G, Berens C, Projan SJ, Hillen W. 2004. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J Antimicrob Chemother 53:592–599. doi: 10.1093/jac/dkh125. [DOI] [PubMed] [Google Scholar]
- 15.Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal RK. 2005. Interaction of era with the 30S ribosomal subunit: implications for 30S subunit assembly. Mol Cell 18:319–329. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102:615–623. doi: 10.1016/S0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 22.Aleksandrov A, Simonson T. 2009. Molecular mechanics models for tetracycline analogs. J Comput Chem 30:243–255. doi: 10.1002/jcc.21040. [DOI] [PubMed] [Google Scholar]
- 23.Ogle JM, Carter AP, Ramakrishnan V. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci 28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 24.Dönhöfer A, Franckenberg S, Wickles S, Berninghausen O, Beckmann R, Wilson DN. 2012. Structural basis for TetM-mediated tetracycline resistance. Proc Natl Acad Sci U S A 109:16900–16905. doi: 10.1073/pnas.1208037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Othersen OG, Waibel R, Lanig H, Gmeiner P, Clark T. 2006. SCRF-DFT and NMR comparison of tetracycline and 5a,6-anhydrotetracycline in solution. J Phys Chem B 110:24766–24774. doi: 10.1021/jp064457s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.