Abstract
A 9-year surveillance for multidrug-resistant (MDR) Pseudomonas aeruginosa in the Hiroshima region showed that the number of isolates harboring the metallo-β-lactamase gene blaIMP-1 abruptly increased after 2004, recorded the highest peak in 2006, and showed a tendency to decline afterwards, indicating a history of an epidemic. PCR mapping of the variable regions of the integrons showed that this epidemic was caused by the clonal persistence and propagation of an MDR P. aeruginosa strain harboring the blaIMP-1 gene and an aminoglycoside 6′-N-acetyltransferase gene, aac(6′)-Iae in a class I integron (In113), whose integrase gene intl1 was disrupted by an IS26 insertion. Sequence analysis of the representative strain PA058447 resistance element containing the In113-derived gene cassette array showed that the element forms an IS26 transposon embedded in the chromosome. It has a Tn21 backbone and is composed of two segments sandwiched by three IS26s. In Japan, clonal nationwide expansion of an MDR P. aeruginosa NCGM2.S1 harboring chromosomally encoded In113 with intact intl1 is reported. Multilocus sequence typing and genomic comparison strongly suggest that PA058447 and NCGM2.S1 belong to the same clonal lineage. Moreover, the structures of the resistance element in the two strains are very similar, but the sites of insertion into the chromosome are different. Based on tagging information of the IS26 present in both resistance elements, we suggest that the MDR P. aeruginosa clone causing the epidemic in Hiroshima for the past 9 years originated from a common ancestor genome of PA058447 and NCGM2.S1 through an IS26 insertion into intl1 of In113 and through IS26-mediated genomic rearrangements.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic human pathogen and a leading cause of nosocomial infections. It also causes mastitis in dairy cows as mild chronic persistent inflammation (1). Carbapenems have been used to treat P. aeruginosa infections; however, P. aeruginosa isolates resistant to carbapenems are reported in many countries and an acquired resistance to carbapenems by metallo-β-lactamase (MBL) genes causes a serious therapeutic problem in clinical settings. Carbapenem-resistant P. aeruginosa strains producing MBLs were first discovered in Japan (2, 3) and have since been reported in many countries all over the world (4).
Specifically, IMP, VIM, SPM, and NDM are the most important types of MBL with worldwide distribution, and their genes are frequently carried on class 1 integrons (4, 5). Class 1 integrons are often found on plasmids or as a part of transposons or resistance genetic elements that are able to integrate into chromosomal elements, resulting in an increase in the number of resistant Gram-negative bacilli (6). MBL-producing P. aeruginosa strains often behave as multidrug-resistant (MDR) strains. The recent increase of nosocomial infections caused by MDR P. aeruginosa has raised a serious concern.
In the Hiroshima prefecture, we have conducted a multihospital surveillance for drug-resistant P. aeruginosa in nine hospitals and reported an increase of MDR P. aeruginosa carrying blaIMP-1 from 2004 to 2006 (7). According to PCR mapping of the variable regions of the integrons, we classified the isolates into six blaIMP-1 integron cassette types and designated the isolate types A through F. We reported an increase of type F MDR P. aeruginosa harboring the blaIMP-1 and a unique aminoglycoside N-acetyltransferase gene, aac(6′)-Iae, in the class I integron named In113, whose integrase gene intl1 was disrupted by IS26 insertion. In113 was first discovered in isolate NCGM2.S1 (previously named IMCJ2.S1) (8) from a nosocomial P. aeruginosa outbreak occurring in the neurosurgery ward of a hospital in Sendai, Miyagi prefecture, 600 km from Hiroshima, in 2002 (9). Further epidemiology studies by the same research group demonstrated that MDR P. aeruginosa carrying aac(6′)-Iae with a pulse type identical to that of NCGM2.S1 was prevalent in Miyagi (10) and Tokyo (11), suggesting that MDR P. aeruginosa NCGM2.S1 was widely disseminated in Japan. Our initial surveillance from 2004 to 2006 also identified MDR P. aeruginosa harboring In113 with an intact intl1 gene, classified as type E (7).
Here, we describe further longitudinal molecular epidemiology studies of drug-resistant P. aeruginosa in the Hiroshima region and demonstrate an epidemic by a single clonal type F MDR P. aeruginosa that has continued for 9 years. Sequence studies of the type F representative strain PA058447 resistance element carrying blaIMP-1 showed that the element forms an IS26 transposon with a Tn21 backbone. Our data suggest that type F MDR P. aeruginosa, prevalent in the Hiroshima region, and strain NCGM2.S1, carrying In113 and prevalent in the Tohoku region, share the same clonal origin.
MATERIALS AND METHODS
Collection of P. aeruginosa isolates and antimicrobial susceptibility testing.
Nonrepetitive P. aeruginosa isolates were obtained from patients from eight major hospitals in Hiroshima during July 2004 to December 2012. Identification of P. aeruginosa was performed by the submitting hospital laboratories. They were cultured on nalidixic acid-cetrimide (NAC) agar (Eiken, Tokyo, Japan) and verified using PCR amplification with primers specific for P. aeruginosa 16S rRNA. Imipenem- or ceftazidime-resistant P. aeruginosa isolates were selected when the MIC of imipenem was ≥8 mg/liter or that of ceftazidime was ≥16 mg/liter. MICs were determined using the broth microdilution method from the Clinical and Laboratory Standards Institute (CLSI) (12). The antibiotics tested were ciprofloxacin (Meiji Seika Pharma, Ltd., Tokyo), imipenem (Banyu Pharmaceutical Co., Ltd., Tokyo), and amikacin (Banyu Pharmaceutical Co., Ltd., Tokyo).
Criteria for MDR P. aeruginosa were in accordance with the Law Concerning the Prevention of Infections and Medical Care for Patients with Infections from the Japanese Ministry of Health, Labor, and Welfare, whereby the criteria are resistance to imipenem (MIC ≥ 16 mg/liter), amikacin (MIC ≥ 32 mg/liter), and ciprofloxacin (MIC ≥ 4 mg/liter). The criterion for amikacin resistance (MIC ≥ 64 mg/liter) was different from that given in the CLSI guidelines (MIC ≥ 32 mg/liter) (more stringent).
MBL screening test with an inhibitor using a double-disc synergy test with two Kirby-Bauer discs.
All of the imipenem- or ceftazidime-resistant P. aeruginosa isolates were screened for the presence of MBL using sodium mercaptoacetic acid (SMA) as previously described (13).
PCR detection and characterization of the variable regions of the blaIMP-1-containing integrons.
The primers used were described previously (7). PCR amplification was performed using TaKaRa Ex Taq DNA polymerase (TaKaRa, Tokyo, Japan) with 25 cycles as follows: denaturing at 98°C for 10 s, annealing at 50°C for 30 s, and polymerization at 72°C for 1 min. The structure of the variable regions of the blaIMP-1-containing integrons was determined using a PCR mapping approach with primers designed from the conserved integron sequences flanking the cassette array as previously described (7).
Fosmid library and screening.
A fosmid library of P. aeruginosa PA058447 was constructed using a CopyControl fosmid library production kit (Epicentre) according to the manufacturer's instructions. The genomic DNA was sheared using a 26-gauge syringe. After blunting and phosphorylation of the DNA fragments, the fragments were separated by pulsed-field gel electrophoresis with 1% certified low-melt agarose (Bio-Rad Laboratories, Inc., Tokyo, Japan). Fragments of ∼40 kb were recovered from the gel using GELase (Epicentre). After the DNA solution was concentrated with a Microcon YM-100 filter (Millipore), the fragments were ligated to the pCC1FOS vector, packaged into phage particles, and transfected into Escherichia coli EPI300-T1 cells. We collected 2,880 chloramphenicol-resistant clones. Fosmid clones containing blaIMP-1 were further selected on LB agar plates containing ampicillin (30 mg/liter).
Nucleotide sequencing, assembly, and genome comparison.
The nucleotide sequence of the cloned fragment into the fosmid vector was determined using the random shotgun sequencing method described previously (14). Collected sequences were assembled using the Sequencher DNA sequencing software (v3.0; Gene Codes). Gaps were closed by direct sequencing of the PCR products amplified with oligonucleotide primers designed to anneal to each end of the neighboring contigs. Initially, potential protein-encoding regions (open reading frames [ORFs]) that were ≥150 bp long were identified using MetaGeneAnnotator (15) and the In Silico Molecular Cloning software package, Genomics Edition (InSilico Biology Inc., Yokohama, Japan). Each ORF was reviewed manually for the presence of a ribosomal binding sequence. Functional annotation was assigned based on homology searches against the GenBank nonredundant protein sequence database using the program BLASTP (16). Protein and nucleotide sequences were compared with those in the sequence databases using the BLAST and FASTA programs at the DDBJ (DNA Data Bank of Japan; http://www.ddbj.nig.ac.jp/). The draft genome sequence of PA058447 was determined using Illumina MiSeq (Nextera paired-end library; 3,525,294-bp sequences; 66.49-fold coverage) sequencing platform. The standard protocol with the Nextera XT DNA sample preparation kit was used. Whole-genome comparison of PA058447 and NCGM2.S1 was performed with BRIG (17) using unassembled Illumina reads of PA058447 and the genome sequence of NCGM2.S1 mapping with reference to the genome of PAO1.
Nucleotide sequence accession number.
The nucleotide sequence described here has been deposited in GenBank under accession number AB983593.
RESULTS AND DISCUSSION
Screening for drug-resistant strains.
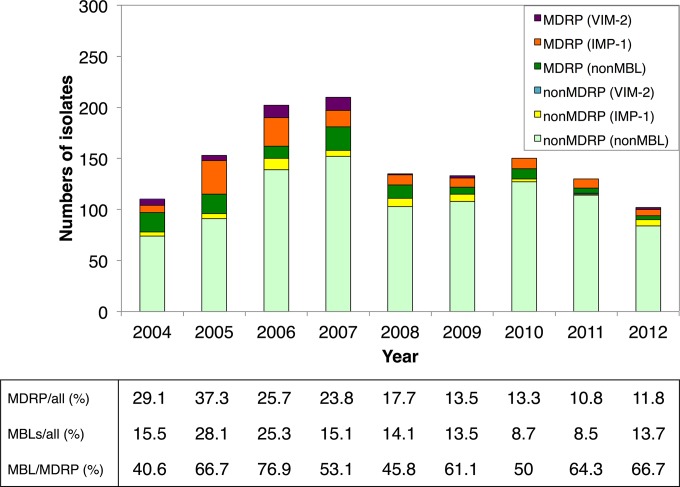
A total of 1,434 P. aeruginosa isolates resistant to imipenem or ceftazidime found during July 2004 to December 2012 were from nine hospitals (identified by the letters “a” to “i”) in Hiroshima. The strains were isolated most commonly from either sputum or urine samples (619 [43.2%] or 345 [24.1%], respectively). The annual numbers of imipenem- or ceftazidime-resistant P. aeruginosa strains continuously increased during the first 4 years of the study, from 110 in 2004 to 206 in 2007, and then fluctuated from 2008 to 2012 (Fig. 1). The prevalence of MDR P. aeruginosa in imipenem- or ceftazidime-resistant isolates continued at a higher ratio of over 20% in the first 4 years, with a maximum of 37.3% in 2005. After 2008, the rate started to gradually decline and reached 12% in 2012, but further follow-up may be necessary to infer the prevalence of MDR P. aeruginosa in this region. Similarly the prevalence of MBL producers among the imipenem- or ceftazidime-resistant isolates reached a maximum of 28.1% in 2005 and gradually decreased and was 13.2% in 2012. The prevalence of MBL producers among MDR P. aeruginosa strains dramatically increased during the first 3 years, from 40.6 to 76.9%, but this high rate did not recede thereafter and remained at 50 to 60%, even though the prevalence of MBL in imipenem- or ceftazidime-resistant isolates significantly decreased. Our previous studies demonstrated that all of the suggested MBL-producing strains were positive for either blaIMP-1 or blaVIM-2 (7). Among them, blaIMP-1-positive strains have been isolated from various hospitals. Follow-up studies indicated that this tendency continued to 2012. We detected six integron polymorphs in blaIMP-1 integron (types A to F) during 2004 to 2006, and another type (denoted as type G) was added in 2012 (18) (Fig. 2). Cumulative data of annual isolation of blaIMP-1-positive strains indicated that they reached a maximum in 2006 and gradually decreased to the level of ca. 15 strains per year, which is still higher than that in 2004 (Fig. 2A). After 2005, type F has been overrepresented in blaIMP-1-positive strains. Type F was first isolated in four of eight hospitals in 2005, but the number of hospitals with a history of isolation of this type reached seven within the 9-year period (Fig. 2B).
FIG 1.
Annual number of imipenem- or ceftazidime-resistant P. aeruginosa isolates. The annual number of MDR P. aeruginosa isolates (MDRP) or metallo-β-lactamase-producing isolates (MBL) with imipenem- or ceftazidime-resistant P. aeruginosa and the annual number of MBL in MDRP are shown.
FIG 2.
(A) Annual survey of the number and the integron type of P. aeruginosa isolates carrying blaIMP-1 integron. (B) Annual survey of isolation incidence of P. aeruginosa carrying the blaIMP-1 integron in each hospital (a to i) participating in this study. Letters A to H indicate the integron types shown in Fig. S1 in the supplemental material.
Resistance element type F.
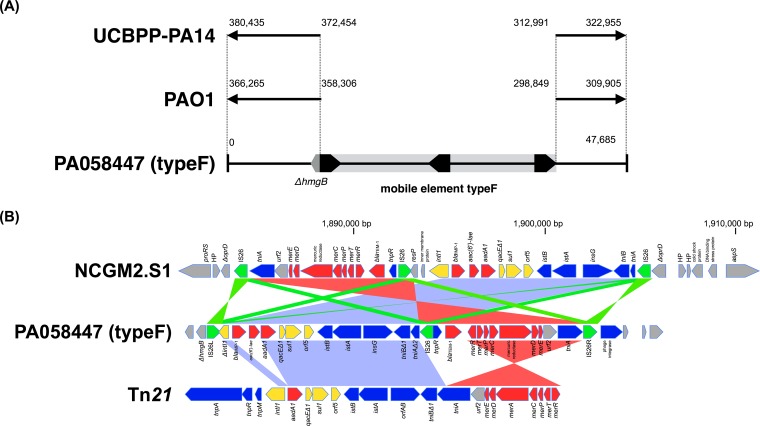
The type F isolates shared identical blaIMP-1 integron gene cassettes and further downstream sequences with the type E, except that the 5′-intl1 gene is truncated by an IS26 insertion in type F (see Fig. S1 in the supplemental material). The insertion of IS26 into intl1 resulted in the formation of a composite transposon in which two IS26s are bracketing the variable region of In113 and flanking genes derived from In2 (see Fig. S1 in the supplemental material) (19). To further delineate the overall structure of the type F resistance element containing blaIMP-1, a fosmid library for strain PA058447 was constructed. From 2,880 fosmid clones, 26 clones including the blaIMP-1 gene were selected on LB agar plates containing ampicillin. Complete sequencing of one of the cloned 47.7-kb fragments showed that the overall structure of the resistance element is composed of two segments sandwiched by three IS26 elements embedded in the chromosome disrupting the gene fumarylacetoacetate hydrolase (hmgB) (Fig. 3A and B). Flanking sequences of the resistance element clearly demonstrated that the element's 3′ downstream region had an orientation opposite to that of PAO1 and UCBPP-PA14 sequences, suggesting that there was a process of genomic inversion during the acquisition of the type F resistance element (Fig. 3A). The left segment (3′) of the resistance element corresponds to the genetic content of the type F integron (7) (Fig. 3B). The right segment (5′) has 11 ORFs, most of which constitute the mer operon of Tn21 (19) (Table 1). A homology search of nucleotide sequences of the overall resistance element indicated that its backbone is part of Tn21, especially In2 and the mer gene operon segmented by IS26s (Fig. 3B). Previously, the mobile element of P. aeruginosa carrying blaIMP-1 (strain 101/1477) isolated in Japan has been characterized (20). It is carried on a plasmid and located on an integron, In31. In2 and In31 share identical sets of 25-bp inverted repeats (IRi and IRt) sandwiching integrons, but the nucleotide sequence flanking IRt of In31 is different from that of In2 and that found in the type F right segment, suggesting that the type F element and In31 are evolutionary distinct.
FIG 3.
(A) Schematic representation of genomic rearrangement due to mobilization of the type F resistance element. Black pentagons indicate IS26s and their orientations. hmgB, fumarylacetoacetate hydrolase gene. Alignments of the complete nucleotide sequence of the cloned fragment from PA058447 and genome sequences of PAO1 and UCBPP-PA14 are shown. The gray bar indicates the type F resistance element. (B) Structural comparison of PA058447 type F resistance element (IS26 transposon), NCGM2.S1 resistance element (IS26 transposon), and Tn21. Color shading indicates homologous regions. Coding sequences are shown as pentagons (green, IS26; blue, transposon; yellow, integron; red, antibiotic resistance gene; gray, other function or hypothetical protein).
TABLE 1.
Features of ORFs around type F integron (GenBank accession no. AB983593)
| ORF | Location (bp) | Strand | Gene | Size (bp) | Length (aa)a | Translation signal sequenceb | Source | Homologue as determined by BLAST and/or FASTA |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Description(s) | Identity (%) | Overlapc (aa) | Accession no. | ||||||||
| 1 | 595–1121 | − | ΔhmgB | 527 | P. aeruginosa (PAO1) | Fumarylacetoacetate hydrolase (5′-side 139 bp truncated) | 99 | 174/221 | AE004091 | ||
| 2 | 1185–1889 | + | Transposase gene | 705 | 234 | GGAGCTGCACATG | P. aeruginosa | Insertion sequence 26 transposase | 100 | 234/234 | AB104852 |
| 3 | 1942–2316 | − | ΔintI1 | 375 | P. aeruginosa | Integrase1 (3′-side 639 bp truncated) | 100 | 125/337 | AF313472 | ||
| 4 | 2469–3209 | + | blaIMP-1 | 741 | 246 | AAGGAAAAGTATG | P. aeruginosa | Metallo-β-lactamase blaIMP-1 | 100 | 246/246 | AB104852 |
| 5 | 3363–3914 | + | aac6′-Iae | 552 | 183 | AAGAGGTTTTATG | P. aeruginosa | Aminoglycoside acetyltransferase 6′-Iae (aac6′-Iae) | 100 | 183/183 | AB104852 |
| 6 | 3989–4780 | + | aadA1 | 792 | 263 | TAAACATCATG | P. aeruginosa | Aminoglycoside adenyltransferase A1 (aadA1) | 100 | 263/263 | AB104852 |
| 7 | 4944–5291 | + | qacEΔ1 | 348 | 115 | GGAGATATATCATG | P. aeruginosa | Quaternary ammonium compound reistance (qacEΔ1) | 100 | 115/115 | AB104852 |
| 8 | 5285–6124 | + | sul1 | 840 | 279 | GGAGGCCGACGCCATG | P. aeruginosa | Sulfonamide resistance protein (sul1) | 100 | 279/279 | AB104852 |
| 9 | 6252–6752 | + | orf5 | 501 | 166 | AGGGGAGCGAATG | P. aeruginosa | Acetyltransferase (GNAT) family protein | 100 | 166/166 | AB104852 |
| 10 | 6928–7710 | − | istB | 783 | 260 | AAGGAGAGCCCATGATG | P. aeruginosa | IstB-like ATP binding family protein | 100 | 260/260 | AB104852 |
| 11 | 7700–9223 | − | istA | 1,524 | 507 | GGAGAAATCAAGGAGTG | P. aeruginosa | Possible transposase of IS1326 | 100 | 507/507 | AB104852 |
| 12 | 9346–10890 | + | insG | 1,545 | 514 | AGGAGTAGTTCATG | P. aeruginosa | Transposase for IS1353 | 100 | 514/514 | CP008739 |
| 13 | 10941–11801 | − | tniB | 861 | 286 | GAGGAGTGGTAGCCGTG | P. aeruginosa | Transposition protein TniB | 100 | 286/286 | AB104852 |
| 14 | 11804–12231 | − | ΔtniA | 428 | K. pneumoniae | Transposition protein TniA-C-terminal part | KF976462 | ||||
| 15 | 12292–12996 | − | Transposase gene | 705 | 234 | GGAGCTGCACATG | P. aeruginosa | Insertion sequence 26 transposase | 100 | 234/234 | AB104852 |
| 16 | 13060–13452 | + | tnpRΔ2 | 393 | Salmonella enterica | Resolvase tnpRΔ2 | EU219534 | ||||
| 17 | 13635–14495 | + | blaTEM-1 | 861 | 286 | AAGGAAGAGTATG | P. aeruginosa | Extended-spectrum β-lactamase blaTEM-1 | 100 | 286/286 | X54607 |
| 18 | 14777–15211 | − | merR | 435 | 144 | GGAGTCAAGCGATATG | Salmonella enterica | Activator/repressor of mer operon (merR) | 100 | 144/144 | AM991977 |
| 19 | 15283–15633 | + | merT | 351 | 116 | AAGGGCCAACGTATG | Salmonella enterica | Mercuric ion transport protein (merT) | 100 | 116/116 | AM991977 |
| 20 | 15647–15922 | + | merP | 276 | 91 | TGGATTTCCCTATG | Salmonella enterica | Periplasmic mercuric ion binding protein (merP) | 100 | 91/91 | AM991977 |
| 21 | 15958–16380 | + | merC | 423 | 140 | GAGAGCCGCTTCATG | Salmonella enterica | Transmembrane protein (merC) | 100 | 140/140 | AM991977 |
| 22 | 16432–18126 | + | merA | 1,695 | 564 | AAGGAACGATGGTATG | Salmonella enterica | Mercuric ion reductase (merA) | 100 | 564/564 | AM991977 |
| 23 | 18144–18506 | + | merD | 363 | 120 | AAGGAGGTGTGCGATG | Salmonella enterica | Putative secondary regulatory protein (merD) | 100 | 120/120 | AM991977 |
| 24 | 18503–18739 | + | merE | 237 | 78 | AGGAGGCATTGCCGTG | Salmonella enterica | Mercuric resistance protein (merE) | 100 | 78/78 | AM991977 |
| 25 | 18736–19443 | + | urf2 | 708 | 235 | Salmonella enterica | EAL domain protein Urf2 (urf2) | 100 | 235/235 | AM991977 | |
| 26 | 19482–20786 | + | tniAΔ1 | 1,305 | 434 | GAGGTGAGCATG | Salmonella enterica | Transposition protein TniAΔ1 | 100 | 434/434 | EU219534 |
| 27 | 20833–21537 | + | Transposase gene | 705 | 234 | GGAGCTGCACATG | P. aeruginosa | Insertion sequence 26 | 100 | 234/234 | AB104852 |
| 28 | 21733–22893 | + | Phage integrase gene | 1,161 | 386 | GAGGCATTTGCCATG | P. aeruginosa | Phage integrase | 97 | 386/387 | EU595745 |
aa, amino acids.
Underlining indicates ribosome binding site; boldface indicates start codon.
Overlap indicates the number of overlapping amino acids/total number of amino acids.
Class I integron plays an important role in the dissemination of antimicrobial-resistance genes and horizontal gene transfer. The recent emergence and prevalence of drug-resistant Acinetobacter baumannii carrying class I integron with a gene array identical to that of P. aeruginosa suggests the efficient transfer of class I integrons among P. aeruginosa and A. baumannii. So far, In113 has not been identified in A. baumannii, but the prevalence of MDR P. aeruginosa carrying In113 in Japan may pose a threat, indicating a potential for the emergence of A. baumannii carrying In113 in future.
Comparison of PA058447 type F resistance element and NCGM2.S1 In113 resistance element.
We compared the overall structure of the type F resistance element to the NCGM2.S1 In113 resistance element using the genome sequence of NCGM2.S1 (8). Both elements showed high similarity in gene organization: the In113 resistance element is also composed of two segments sandwiched by three IS26 elements and embedded in the chromosome (Fig. 3B). The left segment of the In113 resistance element has inversion symmetry to the right segment of the type F resistance element. Conversely, the right segment of the NCGM2.S1 resistance element contains the left segment of type F resistance element and a few additional 5′ flanking genes. The In113 resistance element of NCGM2.S1 has three copies of IS26. Like those of the type F resistance element, two genes at both ends are in direct relative orientation, flanking two interstitial segments forming a composite transposon of the IS6 family (21). This transposon is inserted into oprD, resulting in complete disruption of the gene. Conversely, PCR analysis revealed that oprD in strain PA058447 carrying the type F resistance element was intact. Therefore, the type F resistance element is not a direct sibling of the In113 resistance element by simple insertion of additional IS26 into the intl1 gene of In113.
Hypothesis for the molecular evolution of type F.
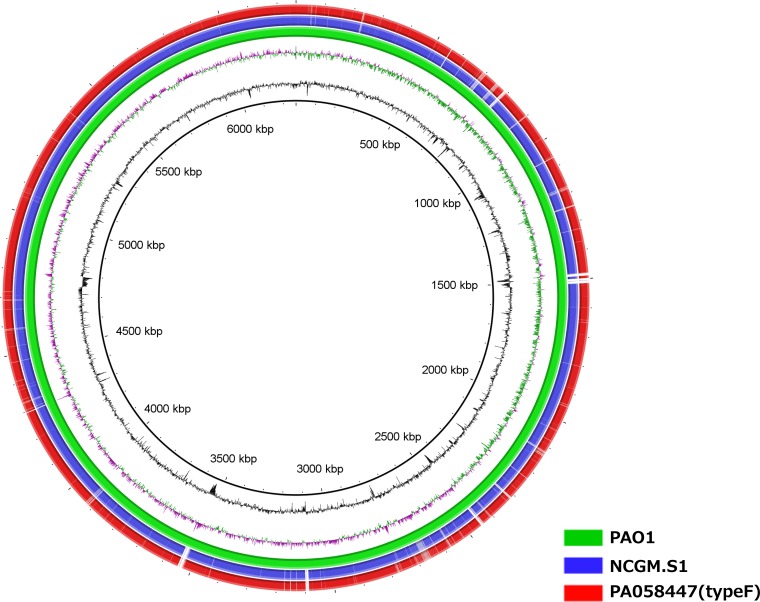
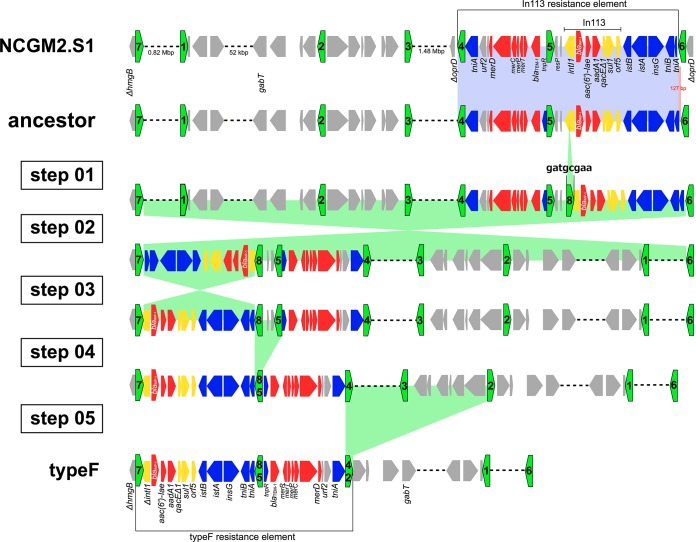
Multilocus sequence typing (MLST) indicated that MDR P. aeruginosa type E (carrying In113) and type F, isolated in Hiroshima, and those strains carrying the In113 resistance element, isolated in the Tohoku area, belonged to the same ST, ST235. To further compare the chromosomal backgrounds of type F and MDR P. aeruginosa carrying the In113 resistance element isolated in Tohoku, we obtained a draft sequence of strain type F (PA058447) using a new-generation sequencer. Figure 4 shows BLASTn comparisons between the draft genome sequence of strain PA058447 and the complete genome sequence of NCGM2.S1 compared to the PAO1 genome depicted by BRIG (17). The data suggest that both genome sequences are very similar. In northern Hiroshima, prior to the abrupt increase of type F, a large outbreak of MDR P. aeruginosa carrying In113 (type E) occurred in 2003. We therefore postulate that type F originated from a similar MDR P. aeruginosa carrying In113 (i.e., type E) by IS26 insertion into intl1 gene and subsequent dynamic chromosomal rearrangements. In the NCGM2.S1 genome, seven IS26s (green pentagons) are present (Fig. 5). Three IS26s are within the In113-carrying resistance element inserted into oprD, and the other four are in other sites on the genome. Eight-base-pair target duplication flanking sequences in each IS26 known as direct inverted repeats show a signature of history of mobility and integration mediated by IS26 (22). In the NCGM2.S1 genome, the hmgB gene is disrupted by insertion of IS26-7 (green pentagon numbered 7 in Fig. 5), whose upstream sequence is identical to that of IS26L in the type F resistance element in PA058447 (Fig. 3B). Conversely, the downstream sequence of IS26-2 in NCGM2.S1 (Fig. 5) is identical to that of IS26R in the type F resistance element (Fig. 3B). Based on the information obtained by checking the identity of direct inverted-repeat 8-bp sequences flanking both sides of IS26s present in the type F resistance element and in NCGM2.S1 (data not shown), we assumed that the ancestor strain possessed the same framework of the genome as NCGM2.S1, but the In113 resistance element is present somewhere other than in the oprD (Fig. 5). We propose that several steps in genomic rearrangements through IS26 insertion into intl1 gene (step 01), IS26-mediated inversions (steps 02 and 03), and deletions (steps 04 and 05) in the ancestor genome may have generated PA058447 carrying the type F resistance element (Fig. 5). IS26 has been implicated in disseminating resistance genes in many ways (19). Two IS26s in direct repeats forming a transposon cause replicon fusion to transfer the transposon from the donor replicon to the target replicon (21, 23). Further, it appears to facilitate the mobilization of resistance genes on the chromosome. Mobility of the integrons by insertion of IS26 into or proximally to the 5′- or 3′ conserved sequence (CS) is reported (24–30). For In53, insertion of IS26 into intl1 gene resulted in a knockout of the integrase function (30) as suggested in the type F resistance element. Further, multiple IS26s could contribute to formation of multiresistant loci by RecA-dependent homologous recombination. In this study, insertion of IS26 into intl1 gene probably facilitated the genomic rearrangement to generate the type F resistance element.
FIG 4.
Comparison of genome sequences of PA058447 and NCGM2.S1. Unassembled sequences of P. aeruginosa PA058447 or P. aeruginosa NCGM2.S1 genome sequences were mapped and compared to the complete genome sequence of P. aeruginosa PAO1. After assembly, contigs were ordered and compared to the complete P. aeruginosa PAO1 genome. The innermost rings represent GC skew (purple/green) and GC content (black).
FIG 5.
Hypothetical model of the genesis of P. aeruginosa PA058447 carrying the type F resistance element from an ancestor P. aeruginosa strain carrying the In113 resistance element. Green pentagons indicate IS26s and their orientations. Color shading indicates homologous regions. Coding sequences are shown as pentagons (green, IS26; blue, transposon; yellow, integron; red, antibiotic resistance gene; gray, other function or hypothetical protein). The In113 resistance element in an ancestor P. aeruginosa isolate is an IS26-transposon. The IS26 transposon can be translocated into oprD to create NCGM2.S1. Genesis of P. aeruginosa PA058447 carrying the type F resistance element from the ancestor P. aeruginosa strain carrying the In113 resistance element can be achieved through IS26-8 insertion into intl1 (step 01), genomic inversions (steps 02 and 03), and genomic deletions (steps 04 and 05) (the numbering of the steps does not imply a particular order).
Type F and NCGM2.S1 belong to the same clonal lineage, ST235. ST235 is an international clone detected in Europe (31–41), Russia (42), Asia (7, 43–49), and the Middle East (50). ST235, ST111, and ST175 are regarded as epidemic high-risk clones, and ST235 lacks pyocyanin and pyoverdine production (51). The association of ST235 with a variety of carbapenem resistance acquired by horizontal transfer of the genes such as blaPER (31, 36, 40, 41, 50), blaOXA (36, 43, 50), blaGES (31, 39), blaSPM (36), blaVIM (31–34, 36–38, 42, 44, 52), blaIMP (36, 43–49), blaFIM (53), and blaNDM (54) suggests its importance in public health and in P. aeruginosa's flexibility in adaptation. Recently, IMP-6-producing P. aeruginosa ST235 has become endemic in Korea (48). Bae et al. (43) reported the integron cassette structures of ST235, and one of them had a gene array identical to those of type F, except that the metallo-β-lactamase gene was blaIMP-6 instead of blaIMP-1. Since IMP-6 differs from IMP-1 by only 1 amino acid at position 196 due to one nucleotide substitution, it may be reasonable to assume that they are derived from a common original integron structure. Further study may be necessary to delineate the evolutional relationship of these ST235s with very similar integron cassette structures. Of note, ST235 frequently shows multidrug resistance and the virulence genotype ΔexoS exoU+ (where exoS is absent) (35), which is the case in type F and NCGM2.S1. The reason why MDR P. aeruginosa type F showed a higher dissemination potential for a long time in Hiroshima remains to be explained. Recently, Skurnik et al. (55) demonstrated that carbapenem-resistant oprD mutants show enhanced in vivo fitness through enhanced mucosal colonization potential, pathogenesis, and resistance to innate immunity. This is a supportive piece of evidence to explain the nationwide dissemination of NCGM2.S1, since it lacks functional OprD. On the other hand, MDR P. aeruginosa carrying the type F resistant element has an intact OprD. It should be noted that OprD is also acting as a specific channel for basic amino acids and small peptides (56, 57) and is possibly important for survival in the environment. Further molecular study is necessary to see how the bacteria increase their prevalence through the deployment of the cost-benefit selection of gene expression in infection process.
In conclusion, our longitudinal surveillance shows that a MDR P. aeruginosa strain carrying blaIMP-1 on an IS26 transposon with a Tn21 backbone became epidemic and showed continued persistence in Hiroshima for more than 9 years. We suggest that this clone originated from a P. aeruginosa isolate carrying In113 through an IS26-mediated insertional inactivation of the intl1 gene and genomic reorganization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Nelson for editorial assistance.
This paper is dedicated to Takashi Yokoyama, who significantly contributed to the promotion of research in nosocomial infectious diseases in Hiroshima.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04207-14.
REFERENCES
- 1.Hogan J, Larry Smith K. 2003. Coliform mastitis. Vet Res 34:507–519. doi: 10.1051/vetres:2003022. [DOI] [PubMed] [Google Scholar]
- 2.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother 38:71–78. doi: 10.1128/AAC.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 35:147–151. doi: 10.1128/AAC.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh TR. 2010. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents 36(Suppl 3):S8–S14. doi: 10.1016/S0924-8579(10)70004-2. [DOI] [PubMed] [Google Scholar]
- 5.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 6.Cambray G, Guerout A-M, Mazel D. 2010. Integrons. Annu Rev Genet 44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 7.Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, Kashiyama S, Hayashida S, Harino T, Tsuji T, Itaha H, Gotoh N, Matsubara A, Usui T, Sugai M. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother 64:46–51. doi: 10.1093/jac/dkp142. [DOI] [PubMed] [Google Scholar]
- 8.Miyoshi-Akiyama T, Kuwahara T, Tada T, Kitao T, Kirikae T. 2011. Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J Bacteriol 193:7010–7010. doi: 10.1128/JB.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekiguchi J-I, Asagi T, Miyoshi-Akiyama T, Fujino T, Kobayashi I, Morita K, Kikuchi Y, Kuratsuji T, Kirikae T. 2005. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob Agents Chemother 49:3734–3742. doi: 10.1128/AAC.49.9.3734-3742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekiguchi J-I, Asagi T, Miyoshi-Akiyama T, Kasai A, Mizuguchi Y, Araake M, Fujino T, Kikuchi H, Sasaki S, Watari H, Kojima T, Miki H, Kanemitsu K, Kunishima H, Kikuchi Y, Kaku M, Yoshikura H, Kuratsuji T, Kirikae T. 2007. Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J Clin Microbiol 45:979–989. doi: 10.1128/JCM.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiguchi J-I, Teruya K, Horii K, Kuroda E, Konosaki H, Mizuguchi Y, Araake M, Kawana A, Kuratsuji T, Kirikae T, Yoshikura H, Miyazaki H. 2007. Molecular epidemiology of outbreaks and containment of drug-resistant Pseudomonas aeruginosa in a Tokyo hospital. J Infect Chemother 13:418–422. doi: 10.1007/s10156-007-0560-5. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement (M100-S23). CLSI, Wayne, PA. [Google Scholar]
- 13.Toleman MA. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J Antimicrob Chemother 50:673–679. doi: 10.1093/jac/dkf210. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi T, Hayashi T, Takami H, Nakasone K, Ohnishi M, Nakayama K, Yamada S, Komatsuzawa H, Sugai M. 2000. Phage conversion of exfoliative toxin A production in Staphylococcus aureus. Mol Microbiol 38:694–705. doi: 10.1046/j.1365-2958.2000.02169.x. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi H, Taniguchi T, Itoh T. 2008. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res 15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi K, Hayashi I, Kouda S, Kato F, Fujiwara T, Kayama S, Hirakawa H, Itaha H, Ohge H, Gotoh N, Usui T, Matsubara A, Sugai M. 2013. Identification and characterization of a novel aac(6′)-Iag associated with the blaIMP-1–integron in a multidrug-resistant Pseudomonas aeruginosa. PLoS One 8:e70557. doi: 10.1371/journal.pone.0070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laraki N, Galleni M, Thamm I, Riccio ML, Amicosante G, Frère JM, Rossolini GM. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother 43:890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollet B, Iida S, Shepherd J, Arber W. 1983. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res 11:6319–6330. doi: 10.1093/nar/11.18.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida S, Mollet B, Meyer J, Arber W. 1984. Functional characterization of the prokaryotic mobile genetic element IS26. Mol Gen Genet 198:84–89. doi: 10.1007/BF00328705. [DOI] [PubMed] [Google Scholar]
- 24.Curiao T, Cantón R, Garcillán-Barcia MP, de la Cruz F, Baquero F, Coque TM. 2011. Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-β-lactamase-producing Escherichia coli clones from humans. Antimicrob Agents Chemother 55:2451–2457. doi: 10.1128/AAC.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uemura S, Yokota S-I, Mizuno H, Sakawaki E, Sawamoto K, Maekawa K, Tanno K, Mori K, Asai Y, Fujii N. 2010. Acquisition of a transposon encoding extended-spectrum β-lactamase SHV-12 by Pseudomonas aeruginosa isolates during the clinical course of a burn patient. Antimicrob Agents Chemother 54:3956–3959. doi: 10.1128/AAC.00110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullik A, Pfeifer Y, Prager R, von Baum H, Witte W. 2010. A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J Med Microbiol 59:580–587. doi: 10.1099/jmm.0.016188-0. [DOI] [PubMed] [Google Scholar]
- 27.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. 2010. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J 24:1160–1166. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]
- 28.Doublet B, Praud K, Weill F-X, Cloeckaert A. 2009. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J Antimicrob Chemother 63:282–289. doi: 10.1093/jac/dkn500. [DOI] [PubMed] [Google Scholar]
- 29.Miriagou V, Carattoli A, Tzelepi E, Villa L, Tzouvelekis LS. 2005. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob Agents Chemother 49:3541–3543. doi: 10.1128/AAC.49.8.3541-3543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naas T, Mikami Y, Imai T, Poirel L, Nordmann P. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol 183:235–249. doi: 10.1128/JB.183.1.235-249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guzvinec M, Izdebski R, Butic I, Jelic M, Abram M, Koscak I, Baraniak A, Hryniewicz W, Gniadkowski M, Tambic Andrasevic A. 2014. Sequence types 235, 111, and 132 predominate among multidrug-resistant Pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob Agents Chemother 58:6277–6283. doi: 10.1128/AAC.03116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liakopoulos A, Mavroidi A, Katsifas EA, Theodosiou A, Karagouni AD, Miriagou V, Petinaki E. 2013. Carbapenemase-producing Pseudomonas aeruginosa from central Greece: molecular epidemiology and genetic analysis of class I integrons. BMC Infect Dis 13:505. doi: 10.1186/1471-2334-13-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koutsogiannou M, Drougka E, Liakopoulos A, Jelastopulu E, Petinaki E, Anastassiou ED, Spiliopoulou I, Christofidou M. 2013. Spread of multidrug-resistant Pseudomonas aeruginosa clones in a university hospital. J Clin Microbiol 51:665–668. doi: 10.1128/JCM.03071-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sardelic S, Bedenic B, Colinon-Dupuich C, Orhanovic S, Bosnjak Z, Plecko V, Cournoyer B, Rossolini GM. 2012. Infrequent finding of metallo-β-lactamase VIM-2 in carbapenem-resistant Pseudomonas aeruginosa strains from Croatia. Antimicrob Agents Chemother 56:2746–2749. doi: 10.1128/AAC.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, Do T, Lanotte P, Mastouri M, Elghmati MS, Rojo F, Mejdi S, Giske CG. 2011. Population structure of Pseudomonas aeruginosa from five mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 6:e25617. doi: 10.1371/journal.pone.0025617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cholley P, Thouverez M, Hocquet D, van der Mee-Marquet N, Talon D, Bertrand X. 2011. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol 49:2578–2583. doi: 10.1128/JCM.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glupczynski Y, Bogaerts P, Deplano A, Berhin C, Huang TD, Van Eldere J, Rodriguez-Villalobos H. 2010. Detection and characterization of class A extended-spectrum-β-lactamase-producing Pseudomonas aeruginosa isolates in Belgian hospitals. J Antimicrob Chemother 65:866–871. doi: 10.1093/jac/dkq048. [DOI] [PubMed] [Google Scholar]
- 38.Samuelsen O, Toleman MA, Sundsfjord A, Rydberg J, Leegaard TM, Walder M, Lia A, Ranheim TE, Rajendra Y, Hermansen NO, Walsh TR, Giske CG. 2010. Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother 54:346–352. doi: 10.1128/AAC.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libisch B, Poirel L, Lepsanovic Z, Mirovic V, Balogh B, Pászti J, Hunyadi Z, Dobák A, Füzi M, Nordmann P. 2008. Identification of PER-1 extended-spectrum β-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol Med Microbiol 54:330–338. doi: 10.1111/j.1574-695X.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- 41.Empel J, Filczak K, Mrówka A, Hryniewicz W, Livermore DM, Gniadkowski M. 2007. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum β-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J Clin Microbiol 45:2829–2834. doi: 10.1128/JCM.00997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edelstein MV, Skleenova EN, Shevchenko OV, D'souza JW, Tapalski DV, Azizoc ISV, Sukhorukova MV, Pavlukov RA, Kozlov RS, Toleman MA, Walsh TR. 2013. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis 13:867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 43.Bae IK, Suh B, Jeong SH, Wang KK, Kim YR, Yong D, Lee K. 2014. Molecular epidemiology of Pseudomonas aeruginosa clinical isolates from Korea producing β-lactamases with extended-spectrum activity. Diagn Microbiol Infect Dis 79:373–377. doi: 10.1016/j.diagmicrobio.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Kim MJ, Bae IK, Jeong SH, Kim SH, Song JH, Choi JY, Yoon SS, Thamilkitkul V, Hsueh PR, Yasin RM, Lalitha MK, Lee K. 2013. Dissemination of metallo-β-lactamase-producing Pseudomonas aeruginosa of sequence type 235 in Asian countries. J Antimicrob Chemother 68:2820–2824. doi: 10.1093/jac/dkt269. [DOI] [PubMed] [Google Scholar]
- 45.Lee JY, Peck KR, Ko KS. 2013. Selective advantages of two major clones of carbapenem-resistant Pseudomonas aeruginosa isolates (CC235 and CC641) from Korea: antimicrobial resistance, virulence and biofilm-forming activity. J Med Microbiol 62:1015–1024. doi: 10.1099/jmm.0.055426-0. [DOI] [PubMed] [Google Scholar]
- 46.Cho HH, Kwon KC, Sung JY, Koo SH. 2013. Prevalence and genetic analysis of multidrug-resistant Pseudomonas aeruginosa ST235 isolated from a hospital in Korea, 2008-2012. Ann Clin Lab Sci 43:414–419. [PubMed] [Google Scholar]
- 47.Kitao T, Tada T, Tanaka M, Narahara K, Shimojima M, Shimada K, Miyoshi-Akiyama T, Kirikae T. 2012. Emergence of a novel multidrug-resistant Pseudomonas aeruginosa strain producing IMP-type metallo-β-lactamases and AAC(6′)-Iae in Japan. Int J Antimicrob Agents 39:518–521. doi: 10.1016/j.ijantimicag.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 48.Yoo JS, Yang JW, Kim HM, Byeon J, Kim HS, Yoo JI, Chung GT, Lee YS. 2012. Dissemination of genetically related IMP-6-producing multidrug-resistant Pseudomonas aeruginosa ST235 in South Korea. Int J Antimicrob Agents 39:300–304. doi: 10.1016/j.ijantimicag.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. 2011. Dissemination of IMP-6 metallo-β-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother 66:2791–2796. doi: 10.1093/jac/dkr381. [DOI] [PubMed] [Google Scholar]
- 50.Fazeli H, Sadighian H, Esfahani BN, Pourmand MR. 2014. Molecular epidemiology and mechanisms of antimicrobial resistance in Pseudomonas aeruginosa isolates causing burn wound infection in Iran. J Chemother 26:222–228. doi: 10.1179/1973947813Y.0000000132. [DOI] [PubMed] [Google Scholar]
- 51.Mulet X, Cabot G, Ocampo-Sosa AA, Dominguez MA, Zamorano L, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2013. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother 57:5527–5535. doi: 10.1128/AAC.01481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giske CG, Libisch B, Colinon C, Scoulica E, Pagani L, Füzi M, Kronvall G, Rossolini GM. 2006. Establishing clonal relationships between VIM-1-like metallo-β-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J Clin Microbiol 44:4309–4315. doi: 10.1128/JCM.00817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier J-D, Rossolini GM. 2013. FIM-1, a new acquired metallo-β-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob Agents Chemother 57:410–416. doi: 10.1128/AAC.01953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carattoli A, Fortini D, Galetti R, Garcia-Fernandez A, Nardi G, Orazi D, Capone A, Majolino I, Proia A, Mariani B, Parisi G, Morrone A, Petrosillo N. 2013. Isolation of NDM-1-producing Pseudomonas aeruginosa sequence type ST235 from a stem cell transplant patient in Italy, May 2013. Euro Surveill 18(46):pii=20633 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20633. [DOI] [PubMed] [Google Scholar]
- 55.Skurnik D, Roux D, Cattoir V, Danilchanka O, Lu X, Yoder-Himes DR, Han K, Guillard T, Jiang D, Gaultier C, Guerin F, Aschard H, Leclercq R, Mekalanos JJ, Lory S, Pier GB. 2013. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc Natl Acad Sci U S A 110:20747–20752. doi: 10.1073/pnas.1221552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang H, Hancock RE. 1996. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J Bacteriol 178:3085–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trias J, Nikaido H. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem 265:15680–15684. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.