Abstract
Thirty-nine Klebsiella pneumoniae carbapenemase (KPC)-producing Pseudomonas aeruginosa isolates, all exhibiting high-level resistance to carbapenems and other β-lactam antibiotics, were isolated in Hangzhou, China. Molecular epidemiology analysis indicated the presence of two dominant clones, namely, clones A and B, both of which belong to sequence type 463 (ST463). A genetic environment analysis demonstrated that both clones harbor an ISKpn8 transposase, blaKPC-2, and an ISKpn6-like transposase. These findings depict the features of clonal expansion and transmission of KPC-2-producing P. aeruginosa strains in Hangzhou, China.
TEXT
Pseudomonas aeruginosa is one of the most common and clinically important pathogens worldwide, causing both high morbidity and mortality among infected patients (1). According to an antimicrobial resistance surveillance of bacterial pathogens in China in 2012 (CHINET), the isolation rate of P. aeruginosa ranked second among the nonfermentative bacteria (2). Carbapenems are considered to be the most efficient antibiotics for the treatment of serious infections caused by multidrug-resistant Gram-negative bacilli. However, with the widespread use of such agents, the resistance rates of P. aeruginosa to carbapenems have increased rapidly. The findings of CHINET also showed that the rates of resistance of P. aeruginosa to imipenem and meropenem in 2013 reached 29.0% and 27.0%, respectively (2). The common resistance mechanisms of P. aeruginosa to carbapenems are the loss of the outer membrane protein OprD and the overexpression of efflux pumps and/or the intrinsic chromosomally encoded AmpC β-lactamase (3). The production of carbapenemases, which is recognized as another mechanism for carbapenem resistance, varies between countries. A survey conducted in the United States on 452 carbapenem-resistant P. aeruginosa isolates revealed that 90.0% of the isolates displayed the loss of OprD, 55.0% exhibited the overexpression of efflux pumps, and 25.0% produced AmpC β-lactamase; yet only four isolates were observed to produce carbapenemases (3). Conversely, longitudinal surveillance of carbapenem-resistant P. aeruginosa isolates in Belarus, Kazakhstan, and Russia demonstrated that the incidence of carbapenemase production increased yearly, from 4.5% in 2002 to 2004 to 28.7% in 2008 to 2010 (4).
In our study, 398 carbapenem-resistant P. aeruginosa isolates were collected from 10 hospitals in Zhejiang Province, China, in 2013. The geographical distribution of P. aeruginosa isolates was as follows: 55 isolates from the Second Affiliated Hospital of Zhejiang University (Hangzhou), 30 isolates from the First Affiliated Hospital of Zhejiang University (Hangzhou), 21 isolates from the Sir Run Run Shaw Hospital of Zhejiang University (Hangzhou), 20 isolates from the Red Cross Hospital (Hangzhou), 108 isolates from Zhejiang Provincial People's Hospital (Hangzhou), 20 isolates from the Taizhou Hospital of Zhejiang Province (Taizhou), 45 isolates from the Cixi People's Hospital (Ningbo), 48 isolates from the Third People's Hospital of Wenzhou City (Wenzhou), and 42 isolates from the Second People's Hospital of Jiaxing City (Jiaxing). Species identification was performed using the Vitek 2 compact system (bioMérieux, Marcy l'Etoile, France).
All 389 imipenem-resistant isolates detected by the Kirby-Bauer disk diffusion method, as recommended by Clinical and Laboratory Standards Institute (CLSI) guidelines (5), were screened for the most common carbapenemase genes, including blaKPC (6), blaNDM-1 (7), blaVIM, and blaIMP (8). For the blaKPC-positive isolates, analyses of the other β-lactamase genes, including blaOXA-50, blaCTX-M, blaTEM, blaSHV, blaPER, and blaVEB, were performed. DNA sequence analysis indicated that 38 P. aeruginosa isolates harbored the blaKPC-2 gene only, one harbored both the blaKPC-2 and blaPER-1 genes, and the blaVIM-2 gene and blaIMP-4 gene were each detected in one strain. None of the other β-lactamases genes were detected. All of the 39 KPC-producing P. aeruginosa isolates were collected from Hangzhou (10 isolates from the 2nd Affiliated Hospital of Zhejiang University, 6 isolates from Sir Run Run Shaw Hospital of Zhejiang University, and 23 isolates from Zhejiang Provincial People's Hospital). The features of the geographical spread of the KPC-2 carbapenemase-producing P. aeruginosa isolates were similar to those of Enterobacteriaceae in Zhejiang Province, as reported previously (9, 10), which showed that such resistant isolates were first found in a big city and subsequently spread rapidly to the surrounding cities. The resistance mechanisms of the remaining 350 non-KPC-producing carbapenem-resistant P. aeruginosa isolates remain unknown but are most likely due to a loss of the OprD porin and/or the overexpression of efflux pump genes.
An analysis of the genetic environment of the blaKPC-2 gene in P. aeruginosa isolates was analyzed using primer-walking sequencing, as previously described (11). Twenty-six isolates were completely identical to the plasmid PE1, which was extracted from a KPC-2 carbapenemase-producing Escherichia coli isolate from our previous study (11). The same five major genes, encoding the Tn3 transposase, Tn3 resolvase, ISKpn8 transposase, KPC-2, and an ISKpn6-like transposase, were present in both the P. aeruginosa and E. coli isolates (Table 1). The identical blaKPC-2 nucleotide segments recovered in both P. aeruginosa and E. coli isolates indicates the probable transmission of blaKPC-2 from Enterobacteriaceae to P. aeruginosa.
TABLE 1.
Summary of antimicrobial susceptibility, β-lactamase production characteristics, and results of molecular typing of selected P. aeruginosa isolates
| Isolate(s)a | Molecular typing results |
β-lacta mase(s) present | Clone | MIC (mg/liter) forb: |
Genetic environment of blaKPC-2 gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PFGE | No. of isolates | ST | IPM | MEM | CAZ | FEP | LEV | CI | AK | GM | ||||
| SRM9–SRM13, SRM15–SRM22 | A1 | 13 | 463 | KPC-2 | A1 | 64 to 512 | 128 to >512 | 128 to 512 | >512 | 1 to 16 | 0.125 to 6 | 3 to 8 | 2 to 6 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| ZE2, ZE3 | A2 | 2 | 463 | KPC-2 | A2 | 128 to 512 | 32 to >512 | 128 | 256 to >512 | 1 to 8 | 0.38 to 4 | 0.75 to 4 | 0.19 to 4 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| SRM4–SRM6 | A3 | 3 | 463 | KPC-2 | A3 | 128 to 256 | 512 | 128 to 256 | >512 | 8 to 16 | 4 | 4 | 2 to 3 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| ZE4 | A4 | 1 | 463 | KPC-2 | A4 | 256 | >512 | 64 | >512 | 8 | 2 | 2 | 3 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| SRM23 | A5 | 1 | 463 | KPC-2 | A5 | 512 | >512 | 512 | >512 | 16 | 3 | 6 | 6 | ISKpn8-blaKPC-2-ISKpn6 |
| SYF2, SYF3, SYF5 | B1 | 3 | 463 | KPC-2 | B1 | 128 to 256 | 512 to >512 | 64 to 128 | 512 | 64 | >256 | 3 to 4 | 1.5 to 3 | ISKpn8-blaKPC-2-ISKpn6 |
| ZE7 | B2 | 1 | 463 | KPC-2 | B2 | 128 | 512 | 64 | 256 | 16 | 4 | 4 | 4 | ISKpn8-blaKPC-2-ISKpn6 |
| SYF6 | B3 | 1 | 463 | KPC-2 | B3 | 256 | 512 | 64 | 512 | 16 | 6 | 6 | 3 | ISKpn8-blaKPC-2-ISKpn6 |
| SYF1, SRM2 | C | 2 | 1076 | KPC-2 | C | 128 to 256 | 512 | 32 to 64 | 256 to 512 | 8 | 3 | 4 to 6 | 3 | ISKpn8-blaKPC-2-ISKpn6 |
| ZE1 | D | 1 | 209 | KPC-2 | D | 256 | >512 | 256 | >512 | 0.5 | 0.25 | 8 | 4 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| SRM1 | E | 1 | 1755 | KPC-2 | E | 32 | 64 | 32 | 256 | 32 | >256 | 2 | 1.0 | ISKpn8-blaKPC-2-ISKpn6 |
| ZE5 | F | 1 | 836 | KPC-2 | F | 128 | >512 | 128 | >512 | 0.5 | 0.125 | 4 | 1.5 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| ZE10 | G | 1 | 463 | KPC-2 | G | 256 | >512 | 256 | >512 | 8 | 3 | 4 | 3 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| ZE6 | H | 1 | 463 | KPC-2 | H | 64 | 32 | 64 | 128 | 8 | 2 | 2 | >256 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| ZE8 | I | 1 | 463 | KPC-2 | I | 128 | >512 | 512 | >512 | 8 | 4 | 4 | 3 | ISKpn8-blaKPC-2-ISKpn6 |
| SYF4 | J | 1 | 463 | KPC-2, PER-1 | J | 64 | 64 | >512 | >512 | 8 | 3 | 0.50 | >256 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| ZE9 | K | 1 | 463 | KPC-2 | K | 16 | 4 | 256 | 256 | 1 | 0.19 | 3 | 2 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| SRM14 | L | 1 | 244 | KPC-2 | L | 128 | >512 | 512 | >512 | 16 | 2 | 4 | 3 | ISKpn8-blaKPC-2-ISKpn6 |
| SRM7 | M | 1 | 463 | KPC-2 | M | 128 | 512 | 128 | >512 | 16 | 4 | 6 | 4 | ISKpn8-blaKPC-2-ISKpn6 |
| SRM8 | N | 1 | 357 | KPC-2 | N | 16 | 32 | 16 | 64 | 8 | 1.0 | 6 | 4 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
| SRM3 | O | 1 | 850 | KPC-2 | O | 8 | 16 | 16 | 64 | 4 | 0.75 | 12 | 4 | Tn3-ISKpn8-blaKPC-2-ISKpn6 |
ZE1 to ZE10, 10 strains of KPC-2-producing P. aeruginosa isolated from 2nd Affiliated Hospital of Zhejiang University; SYF1 to SYF6, 6 strains of KPC-2-producing P. aeruginosa isolated from Sir Run Run Shaw Hospital of Zhejiang University; SRM1 to SRM23, 23 strains of KPC-2-producing P. aeruginosa isolated from Zhejiang Provincial People's Hospital.
IPM, imipenem; MEM, meropenem; CAZ, ceftazidime; FEP, cefepime; LEV, levofloxacin; CI, ciprofloxacin; AK, amikacin; GM, gentamicin. The MICs of colistin and polymyxin B for all P. aeruginosa isolates were 1 to 2 mg/liter. The MICs of aztreonam were >256 mg/liter, except for strain SRM3, with an MIC of 128 mg/liter. The MICs of cefoperazone-sulbactam and piperacillin-tazobactam were >256 mg/liter, except for SRM3 and SRM8 (MICs, 32 to 128 mg/liter).
For the 39 blaKPC-positive isolates, the MICs were determined using Etest (bioMérieux, Marcy l'Etoile, France) for amikacin, ciprofloxacin, and gentamicin. The MICs for all other antibiotics were determined by the agar dilution method, as recommended by the CLSI (12). None of the isolates were susceptible to carbapenems, with MICs ranging from 8 mg/liter to 512 mg/liter for imipenem and 4 mg/liter to >512 mg/liter for meropenem (Table 1). All isolates were nonsusceptible to cephems and β-lactam–β-lactamase inhibitor combinations. All P. aeruginosa isolates were susceptible to polymyxin B, colistin, and amikacin, and 92.3% of the isolates were susceptible to gentamicin.
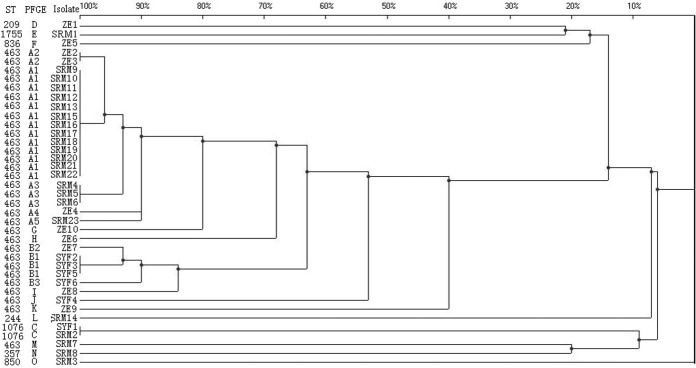
To investigate the molecular epidemiology of KPC-producing P. aeruginosa isolates, pulsed-field gel electrophoresis (PFGE) was performed as previously described but with a slight modification (13). Genomic DNA was digested using the restriction enzyme SpeI, and isolates with a Dice similarity index of ≥85% were defined as belonging to the same PFGE group (14). Multilocus sequence typing (MLST) was performed as recommended by the P. aeruginosa PubMLST website (http://pubmlst.org/paeruginosa/). The 39 carbapenem-resistant P. aeruginosa isolates were found to belong to different clones, designated clones A to O. The most prevalent clones were A (51.3% [20/39]) and B (12.8% [5/39]). The isolates of clones A and B were then divided into five subclonal groups (A1 to A5) and 3 subclonal groups (B1 to B3), respectively, on the basis of genetic similarity (Fig. 1). Isolates from clones A and B were found in several different hospitals, suggesting interhospital clonal spread. Thirty-one of the 39 carbapenem-resistant P. aeruginosa isolates were found to belong to sequence type 463 (ST463). The remaining eight isolates belonged to various single-sequence types, with ST1755 (11-5-5-11-4-4-7) among them being the most notable. To our best knowledge, this is the first report of the discovery of ST1755 (11-5-5-11-4-4-7). A large-scale emergence of clonally related KPC-2-producing P. aeruginosa ST463 isolates has never been reported elsewhere. Considering the experience given by previous reports (9, 10), the KPC-producing P. aeruginosa ST463 isolates we identified surely have a high chance of spreading from Hangzhou to the surrounding cities.
FIG 1.
PFGE profile of SpeI-digested DNA from 39 KPC-producing P. aeruginosa isolates. An unweighted-pair group method using average linkages (UPGMA) dendrogram based on Dice similarity coefficients was generated using the UVIBand software (Bio-Rad). Eighty-five percent similarity was used as the cutoff point.
In summary, the present study provides the first report of the clonal spread of KPC-2-producing ST463 P. aeruginosa isolates. In view of the rapid emergence and transmission of the KPC-producing P. aeruginosa isolates in Zhejiang, China, carbapenem-resistant P. aeruginosa isolates should be carefully monitored, and increased care must be taken to prevent the spread of KPC-producing P. aeruginosa isolates in China.
ACKNOWLEDGMENTS
We thank Qing Yang, Jie Lin, Ya-ping Pan, Huo-xiang Lv, Su-fei Yu, Kai Zhao, Yang-fang Chen, and Xiao-yan Wu for the kind collection of the carbapenem-resistant P. aeruginosa isolates from 1st Affiliated Hospital of Zhejiang University, Sir Run Run Shaw Hospital of Zhejiang University, Red Cross Hospital, Zhejiang Provincial People's Hospital, Taizhou Hospital of Zhejiang Province, Cixi People's Hospital, 3rd People's Hospital of Wenzhou City, and 2nd People's Hospital of Jiaxing City, respectively.
We declare no conflicts of interest.
REFERENCES
- 1.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Zhu DM, Hu FP, Jiang XF, Hu ZD, Li Q, Sun ZY, Chen ZJ, Xu YC, Zhang XJ, Wang CQ, Wang AM, Ni YX, Sun JY, Zhu YZ, Yu YS, Lin J, Xu YH, Shen JL, Su DH, Zhuo C, Wei LH, Wu L, Zhang ZX, Ji P, Zhang H, Kong J, Hu YJ, Ai XM, Shan B, Du Y. 2013. CHINET 2012 surveillance of bacterial resistance in China. Chin J Infect Chemother 13:321–330. [Google Scholar]
- 3.Davies TA, Queenan AM, Morrow BJ, Shang W, Amsler K, He W, Lynch AS, Pillar C, Flamm RK. 2011. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007–09. J Antimicrob Chemother 66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 4.Edelstein MV, Skleenova EN, Shevchenko OV, D'Souza JW, Tapalski DV, Azizov IS, Sukhorukova MV, Pavlukov RA, Kozlov RS, Toleman MA, Walsh TR. 2013. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis 13:867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Hu YY, Yang XF, Gu DX, Zhou HW, Hu QF, Zhao K, Yu SF, Chen GX. 2014. Emergence of NDM-producing non-baumannii Acinetobacter spp. isolated from China. Eur J Clin Microbiol Infect Dis 33:853–860. doi: 10.1007/s10096-013-2024-4. [DOI] [PubMed] [Google Scholar]
- 8.Yan JJ, Hsueh PR, Ko WC, Luh KT, Tsai SH, Wu HM, Wu JJ. 2001. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother 45:2224–2228. doi: 10.1128/AAC.45.8.2224-2228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai JC, Zhou HW, Zhang R, Chen GX. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother 52:2014–2018. doi: 10.1128/AAC.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. 2009. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai JC, Zhang R, Hu YY, Zhou HW, Chen GX. 2014. Emergence of Escherichia coli sequence type 131 isolates producing KPC-2 carbapenemase in China. Antimicrob Agents Chemother 58:1146–1152. doi: 10.1128/AAC.00912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing: 15th informational supplement. CLSI document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 13.Hu YY, Cai JC, Zhang R, Zhou HW, Sun Q, Chen GX. 2012. Emergence of Proteus mirabilis harboring blaKPC-2 and qnrD in a Chinese hospital. Antimicrob Agents Chemother 56:2278–2282. doi: 10.1128/AAC.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carriço JA, Pinto FR, Simas C, Nunes S, Sousa NG, Frazão N, de Lencastre H, Almeida JS. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J Clin Microbiol 43:5483–5490. doi: 10.1128/JCM.43.11.5483-5490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]