Abstract
Characterization of third-generation-cephalosporin-resistant Klebsiella pneumoniae isolates originating mainly from one human hospital (n = 22) and one companion animal hospital (n = 25) in Bern (Switzerland) revealed the absence of epidemiological links between human and animal isolates. Human infections were not associated with the spread of any specific clone, while the majority of animal infections were due to K. pneumoniae sequence type 11 isolates producing plasmidic DHA AmpC. This clonal dissemination within the veterinary hospital emphasizes the need for effective infection control practices.
TEXT
The rapid spread of multidrug-resistant (MDR) Klebsiella pneumoniae has led to major concerns in hospitals (1). During the past few years, cases of infections caused by K. pneumoniae strains resistant to clinically important classes of antibiotics, including third-generation cephalosporins (3GCs), have also been reported in companion animals (2–6). 3GCs represent important antibiotics for the treatment of serious infections caused by K. pneumoniae before the use of last-resort carbapenems. Transmission of 3GC-resistant K. pneumoniae (3GC-R-Kp) between companion animals and humans represents a possible threat to both human and animal health (7). This prompted us to determine which resistance determinants and clonal lineages are associated with 3GC-R-Kp obtained primarily from one human hospital as well as from one companion animal clinic in Bern, Switzerland.
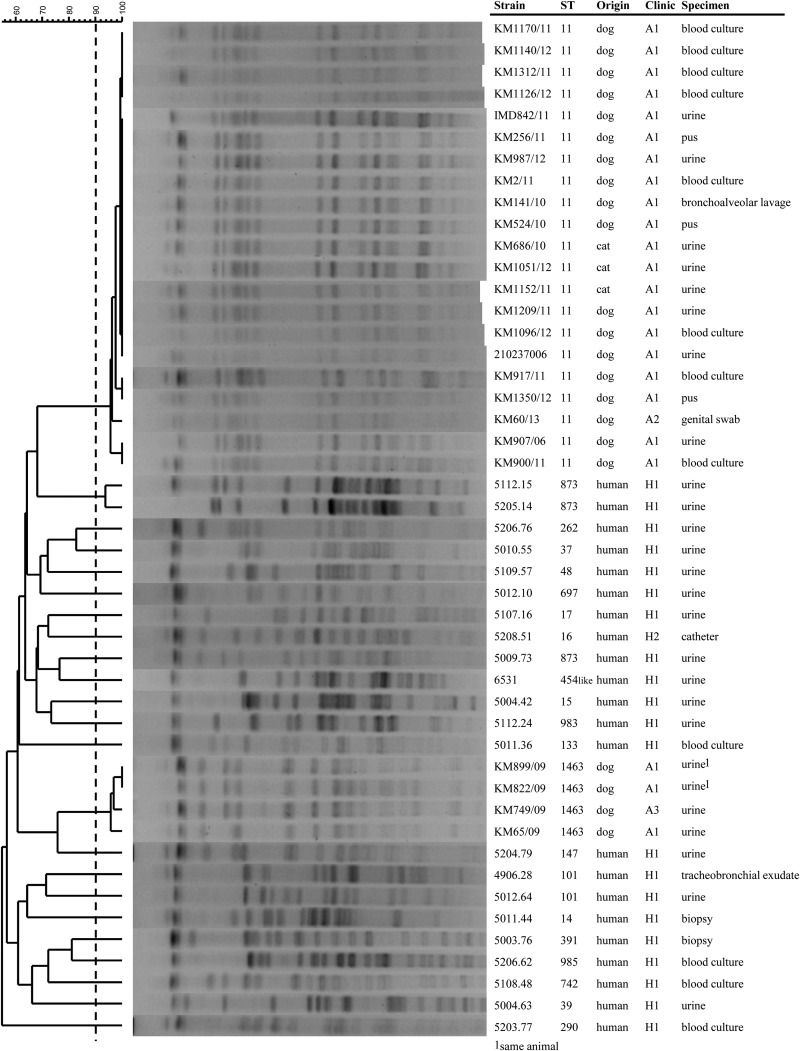
Isolates were selected based on decreased susceptibility to cefotaxime or ceftazidime (MICs of both, ≥1 μg/ml) (8). Species identification was confirmed by using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonik) (9). The human isolates consisted of all 3GC-R-Kp isolates from patients admitted to different wards of the same hospital (hospital 1 [H1]) between 2013 and 2014 (n = 21), except one (5208.51) which came from H2. K. pneumoniae isolates were predominantly recovered from urine and less frequently from blood and biopsy specimens (Table 1). Multilocus sequence typing (MLST) (http://www.pasteur.fr/recherche/genopole/PF8/mlst/) and XbaI pulsed-field gel electrophoresis (PFGE) profiles (contour-clamped homogeneous electric field [CHEF] DR-III apparatus [Bio-Rad]; run time, 18.5 h; gradient, 6 V/cm; initial switch time, 2.2 s; final switch time, 54.2 s; angle, 120°) (10) revealed that the human isolates differed genetically from each other; each patient was infected with a unique strain that exhibited a distinct PFGE profile, and all of the isolates belonged to different sequence types (ST) except two isolates that were of ST101 and three that were of ST873 (Fig. 1). The absence of clonal spread of K. pneumoniae, even within the same ward, is suggestive of infection control best practices at the hospital. Such diversity indicates that resistance may emerge independently of the wards in different K. pneumoniae lineages, e.g., through the acquisition of resistance to 3GCs.
TABLE 1.
Antimicrobial resistance profile and genetic characteristics of K. pneumoniae isolates from human infection sitesa
| Strain | Source | Hospitalization required? (ward) | Resistance phenotype | ST | Carbapenemase gene | ESBL/pAmpC gene(s) | Other bla gene(s)b | Other antibiotic resistance gene(s) and integronsc | Amino acid substitution(s) in: |
|
|---|---|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | |||||||||
| 5203.77 | Blood culture | Yes (emergency room) | 3GCs NAL TMP TET KAN SMX | 290 | blaCTX-M-15 | blaTEM-1, blaOXA-1 | aac(6′)-Ib-cr, sul1, dfrA12, mrx-mphA, tet(A), intI1 | |||
| 5204.79 | Urine | No (SOC; ambulatory) | 3GCs GEN CIP NAL TMP | 147 | blaCTX-M-15 | aac(3)-IIc, dfrA14 | Ser83Ile | Ser80Ile | ||
| 5107.16 | Urine | Yes (urology/gynecology) | 3GCs GEN TMP STR TET SMX | 17 | blaCTX-M-14 | blaTEM-1, blaLAP-2 | aac(3)-IIc, qnrS1, dfrA1, strB, sul2, tet(A) | |||
| 5009.73 | Urine | No (SOC; palliative medicine) | 3GCs CIP (I) TMP STR TET SMX KAN | 873 | blaCTX-M-15 | blaTEM-1, blaOXA-1 | qnrB, aac(6′)-Ib-cr, dfrA14, strA, strB, sul2, tet(A), intI1 | |||
| 5206.76 | Urine | Yes (urology) | AmpC TET SMX | 262 | ND | blaSHV-110 | ||||
| 5012.10 | Urine | Yes (medicine, abdominal surgery) | 3GCs STR SMX | 697 | blaCTX-M-3 | blaTEM-1 | strB, sul2 | |||
| 5208.51 | Catheter | Unknown (external laboratory) | AmpC FEP MERO GEN AMP CIP NAL KAN | 16 | blaNDM-1 | aac(6′)-Ib | Ser83Phe, Asp87Asn | Glu84Lys | ||
| 4906.28 | Tracheobronchial exudate | Yes (ICU) | 3GCs GEN CIP NAL TMP STR TET SMX | 101 | blaOXA-48 | blaCTX-M-15 | blaTEM-1, blaOXA-1 | aac(3)-IIc, aadA1-like, qnrB, aac(6′)-Ib-cr, dfrA14, strA, strB, sul2, tet(A), intI1 | Ser83Tyr, Asp87Ala | Ser80Ile |
| 5012.64 | Urine | Yes (emergency room) | 3GCs FEP GEN CIP NAL KAN | 101 | blaCTX-M-15 | blaTEM-1, blaOXA-9 | aadA1-like | Ser83Tyr, Asp87Gly | Ser80Ile | |
| 5011.44 | Biopsy specimen | Yes (orthopedics) | AmpC CHL TET SMX | 14 | ND | blaSHV-28 | tet(D), intI1, ΔompK36 | |||
| 5108.48 | Blood culture | Yes (oncology) | 3GCs FEP GEN TMP TET SMX | 742 | blaCTX-M-1, blaCTX-M-14 | blaLAP-2 | aac(3)-IIc, qnrS1, dfrA1, sul1, tet(A) | |||
| 5003.76 | Biopsy specimen | Yes (orthopedics) | 3GCs TMP STR TET SMX KAN | 391 | blaCTX-M-15 | aph(3′)-Ia, dfrA7, strA, strB, sul2, tet(D) | ||||
| 5109.57 | Urine | Yes (gynecology) | 3GCs CIP TMP TET SMX | 48 | blaCTX-M-15, blaCTX-M-14 | blaTEM-1, blaOXA-1 | qnrB, aac(6′)-Ib-cr, dfrA14, sul2, tet(A) | |||
| 5004.63 | Urine | Yes (cardiology) | 3GCs GEN CHL TMP STR TET SMX KAN | 39 | blaCTX-M-15 | blaTEM-1, blaOXA-1 | aac(3)-IIc, aph(3′)-Ia, aadA1-like, aadA4-like, aac(6′)-Ib-cr, catA1, dfrA1, dfrA17, strA, strB, sul1, sul2, mrx-mphA, tet(A), intI1 | |||
| 5206.62 | Blood culture | Yes (oncology) | 3GCs GEN CIP (I) TMP STR TET SMX | 985 | blaCTX-M-15 | blaTEM-1, blaOXA-1 | aac(3)-IIc, qnrB, aac(6′)-Ib-cr, dfrA14, strB, sul2, tet(A), intI1 | |||
| 5004.42 | Urine | Yes (ICU) | 3GCs CIP NAL | 15 | blaCTX-M-15 | Ser83Phe, Asp87Ala | Ser80Ile | |||
| 5011.36 | Blood culture | Yes (oncology) | AmpC TMP SMX CHL | 133 | blaDHA-1 | qnrB, catA1, sul1, intI1 | ||||
| 5112.24 | Urine | Yes (urology) | 3GCs GEN TMP STR TET SMX | 983 | blaCTX-M-15 | blaTEM-1, blaOXA-1 | aac(3)-IIc, qnrB, dfrA14, strB, sul2, tet(A) | |||
| 5010.55 | Urine | Yes (medicine) | 3GCs TMP STR TET SMX KAN | 37 | blaCTX-M-1 | aph(3′)-Ia, dfrA7, strA, strB, sul2, tet(D) | ||||
| 5112.15 | Urine | Yes (urology) | 3GCs GEN CHL STR SMX | 873 | blaCTX-M-14 | blaSHV-27, blaLAP-2 | aac(3)-IIc, qnrS1, strA, strB, sul2 | |||
| 5205.14 | Urine | Yes (medicine) | 3GCs CIP CHL NAL TMP STR SMX | 873 | blaCTX-M-14 | blaSHV-27, blaLAP-2 | qnrS1, dfrA12, dfrA13, strA, strB, sul2 | |||
| 6531 | Urine | Yes (urology/gynecology) | AmpC SMX | 454-like | blaDHA-1 | blaOXA-1 | qnrB, strA, strB, sul1 | |||
ST, sequence type; 454-like, ST was assigned based on the combination of six alleles, since the tonB allele could not be amplified by PCR; SOC, specialized outpatient clinic; ICU, intensive care unit; 3GCs, 3rd-generation cephalosporins; AmpC, 3rd-generation cephalosporins and β-lactamase inhibitors; FEP, cefepime (4th-generation cephalosporin); CIP, ciprofloxacin; CIP (I), ciprofloxacin intermediate; CHL, chloramphenicol; NAL, nalidixic acid; TMP, trimethoprim; SMX, sulfamethoxazole; KAN, kanamycin; GEN, gentamicin; STR, streptomycin; TET, tetracycline; ND, no gene detected.
Non-ESBL SHVs were not reported.
Genes and functions: aadA, streptomycin adenylyltransferase; aph(3′)-Ia, kanamycin O-phosphotransferase; aac(3)-IIc, gentamicin acetyltransferase; qnr, quinolone resistance protein; aac6′-Ib-cr, variant of aminoglycoside N(6′)-acetyltransferase–ciprofloxacin-modifying enzyme; aac(6′)-Ib, N-acetyltransferase; cat, chloramphenicol acetyltransferase; dfr, dihydrofolate reductase for trimethoprim resistance; sul, dihydropteroate synthetase for sulfonamide resistance; mrx-mphA, macrolide inactivation resistance protein-phosphotransferase; tet, tetracycline efflux; str, streptomycin phosphotransferase; intI1, integrase.
FIG 1.
Dendrogram of XbaI PFGE patterns of Klebsiella pneumoniae isolates from human and animal origins. Cluster analysis was made with BioNumerics software version 7.1 (Applied Maths, Belgium) by the unweighted-pair group method using average linkages (UPGMA) and with Dice comparison settings (optimization, 1.5%; position tolerance, 1.5%). The cutoff for determining clonality was ≥90% (21).
In contrast, only two different clonal lineages (ST11, n = 21, and ST1463, n = 4) were found among the animal isolates, which gathered into two PFGE clusters (Fig. 1). ST1463 represents a novel lineage, so far detected only in veterinary settings in Switzerland. All of the 3GC-R-Kp isolates from companion animals (22 dogs and 3 cats) admitted to the same clinic (A1) between 2006 and 2012 were included in the study, except for one of ST11 and one of ST1463, which came from two other clinics (A2 and A3) (Fig. 1). Only one dog was hospitalized because of pneumonia. The others were admitted for non-K. pneumoniae-associated diseases, including 7 outpatients (6 dogs and 1 cat) with one (n = 2) or multiple (n = 5) visits during the study period, 9 dogs hospitalized for leptospirosis treatment with dialysis, and 3 dogs and 2 cats hospitalized for injuries that required surgical interventions (Table 2). Infections developed after they had been treated with intensive care, including after placement and manipulation of indwelling venous and urinary catheters. Such invasive interventions are already known to be critical for the development of K. pneumoniae septicemia and urinary tract infections (UTIs) in humans (11–13). One dog hospitalized in another clinic was found to be a carrier of the same ST11 clone (KM60/13). This dog had a history of hospitalization in the first clinic (A1) 7 years before, where it may have acquired the K. pneumoniae strain and was still colonized; alternatively, this clone was also present in the second clinic (A2). Long-term persistence (for years) of Enterobacteriaceae is a well-known phenomenon which may contribute to the spread of nosocomial isolates into the community, as well as increase the risk of infection in cases requiring hospitalization and clinical intervention (14). In this regard, studies on risk factors for MDR bacterial infections are needed in order to apply specific population measures to prevent the dissemination of epidemic strains into animal clinics.
TABLE 2.
Antimicrobial resistance profiles and genetic characteristics of K. pneumoniae isolates from animal infection sitesa
| Strain | Source | Hospitalization required? (reason) | Resistance phenotype | ST | ESBL/pAmpC gene(s)c | Other antibiotic resistance genes and integronsd | Amino acid substitution in: |
|
|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | |||||||
| KM907/06 | Dog urine | Yes (esophagus perforation) | AmpC CIP CHL NAL SMX | 11 | blaDHA-1 | aadA2-like, qnrB, aac(6′)-Ib-cr, catA1, catB3-like, sul1, arr1, intI1 | Ser83Ile | Ser80Ile |
| KM60/13 | Dog genital swab | No (OPP) | AmpC CIP CHL NAL SMX KAN | 11 | blaDHA-1 | aph(3′)-Ia, qnrB, aac(6′)-Ib-cr, catB3-like, sul1, mrx-mphA, arr1, intI1 | Ser83Ile | Ser80Ile |
| KM1350/12 | Dog pus | Yes (cruciate ligament rupture) | Same as for KM60/13 | 11 | blaDHA-1 | Same as for KM60/13 | Ser83Ile | Ser80Ile |
| IMD842/11 | Dog urine | Yes (stomach ulcer) | AmpC CIP CHL NAL TMP SMX KAN | 11 | blaDHA-1 | aph(3′)-Ia, aadA2-like, qnrB, aac(6′)-Ib-cr, catA1, catB3-like, dfrA12, sul1, mrx-mphA, arr1, intI1 | Ser83Ile | Ser80Ile |
| KM256/11 | Dog pus | No (OPP; wound care after accident) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM2/11 | Dog blood culture | Yes (intoxication) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM141/10 | Dog BAL fluid | Yes (pneumonia) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1051/12 | Cat urine | Yes (urolithiasis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM900/11 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM917/11 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1152/11 | Cat urine | Yes (feline lower urinary tract disease) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1170/11 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1312/11 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1140/12 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1209/11 | Dog urine | OPP (immunosuppression) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM524/10 | Dog pus | Yes (hit by car) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1096/12 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM987/12 | Dog urine | No (OPP; protein-losing enteropathy) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM1126/12 | Dog blood culture | Yes (leptospirosis) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM686/10 | Cat urine | No (OPP; fibrosarcoma) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| 210237006 | Dog urine | Yes (hit by car) | Same as for IMD842/11 | 11 | blaDHA-1 | Same as for IMD842/11 | Ser83Ile | Ser80Ile |
| KM899/09b | Dog urine | Yes (leptospirosis) | 3GCs CIP CHL NAL TMP SMX KAN STR | 1463 | blaCTX-M-1 | aph(3′)-Ia, aadA2-like, dfrA12, strB, sul1, sul2, mrx-mphA | Ser83Ile | Ser80Ile |
| KM749/09 | Dog urine | No (OPP; UTI) | AmpC, CIP CHL NAL TMP SMX KAN STR | 1463 | blaCMY-2, blaCTX-M-1 | aph(3′)-Ia, aadA2-like, dfrA12, strB, su11 sul2 | Ser83Ile | Ser80Ile |
| KM65/09 | Dog urine | No (OPP; UTI) | AmpC CIP CHL NAL TMP SMX KAN STR TET | 1463 | blaCMY-2, blaCTX-M-1 | aph(3′)-Ia, aadA2-like, dfrA12, strB, sul1, sul2, tet(A) | Ser83Ile | Ser80Ile |
| KM822/09b | Dog urine | Yes (leptospirosis) | Same as for KM899/09 | 1463 | blaCMY-2, blaCTX-M-1 | Same as for KM899/09 | Ser83Ile | Ser80Ile |
OPP, outpatient pet; UTI, urinary tract infection; ST, sequence type; 3GCs, 3rd-generation cephalosporins; AmpC, 3rd-generation cephalosporins and beta-lactamase inhibitors; CIP, ciprofloxacin; CHL, chloramphenicol; NAL, nalidixic acid; TMP, trimethoprim; SMX, sulfamethoxazole; KAN, kanamycin; STR, streptomycin; TET, tetracycline.
KM899/09 and KM822/09 were isolated from the same animal.
All strains also had the blaOXA-1 gene.
Genes and functions: aadA, streptomycin adenylyltransferase; aph(3′)-Ia, kanamycin O-phosphotransferase; qnrB, quinolone resistance protein; aac6′-Ib-cr, variant of aminoglycoside N(6′)-acetyltransferase–ciprofloxacin-modifying enzyme; cat, chloramphenicol acetyltransferase; dfrA12, dihydrofolate reductase for trimethoprim resistance; sul, dihydropteroate synthetase for sulfonamide resistance; mrx-mphA, macrolide inactivation resistance protein-phosphotransferase; arr1, rifampin ADP ribosyltransferase; tet, tetracycline efflux; str, streptomycin phosphotransferase; intI1, integrase.
Antimicrobial susceptibility was determined using the microdilution ESB1F and EUMVS2 plates (TREK Diagnostic Systems) according to Clinical and Laboratory Standards Institute (CLSI) guidelines and interpretative criteria (8). Antibiotic resistance genes were detected using AMR08 ArrayStrip microarrays (15) together with the HybridizationPlus kit (Alere Technologies GmbH). Carbapenemases, extended-spectrum β-lactamases (ESBLs), plasmid-mediated AmpC (pAmpCs), and amino acid substitutions in the fluoroquinolone resistance-determining region (QRDR) of ParC and GyrA were further identified by sequence analysis of translated sequences obtained from PCR products (see Table S1 in the supplemental material). Eighteen of 21 isolates of ST11 had identical resistance profiles and contained the same resistance genes, including the pAmpC blaDHA-1 gene, emphasizing the spread of a specific clone in a veterinary setting. The three other ST11 isolates had the same genes but lacked the kanamycin resistance gene aph(3′)-Ia and/or the trimethoprim resistance gene dfrA12 (Table 2). Of note, isolate KM907/06, the least resistant strain, was isolated in 2006, whereas the other strains were isolated between 2010 and 2013. ST1463 isolates displayed resistance profiles similar to those of ST11 isolates, with additional resistance to streptomycin. However, resistance to 3GCs was associated with blaCMY-2 and/or blaCTX-M-1 (Table 2).
The resistance profiles of the 3GC-R-Kp isolates from humans were highly diverse. Resistance to 3GCs was associated predominantly with the presence of blaCTX-M genes. Only two isolates carried blaDHA-1. Two isolates were resistant to carbapenems and harbored blaNDM-1 and blaOXA-48 (Table 1). Unlike with the 3GC-R-Kp isolates from animals, all of which were resistant to quinolones, only 9 of 22 human isolates exhibited decreased susceptibility to this class of antibiotics (Table 1). These isolates also did not contain the rifampin resistance gene arr-1. However, tetracycline resistance associated with tet(A) and tet(D), as well as gentamicin resistance [aac(3)-IIc], was frequent in human isolates. Resistance to sulfonamides, trimethoprim, chloramphenicol, and streptomycin was also common in human isolates, associated either with the same resistance mechanisms as those detected in animal isolates or with others (Table 1).
Selected isolates were used to determine, by PCR (PBRT kit; Diatheva), the Inc group of plasmids carrying blaDHA-1, blaCTX-M-1, blaCTX-M-3, blaCTX-M-14, blaCTX-M-15, and blaOXA-48 after electroporation into Escherichia coli TOP10 (Invitrogen) and filter mating conjugation using either E. coli JF33 (Rifr) or E. coli J53 (sodium azide resistant; kindly provided by L. Poirel) as the recipient (16). Transformed cells were selected on Mueller-Hinton II agar containing either 70 μg/ml ampicillin, 2 μg/ml cefotaxime, or 0.5 μg/ml meropenem. The blaDHA-1 and blaCTX-M-1 genes from animal isolates were associated with nonconjugative R plasmids. R plasmids are commonly found in K. pneumoniae isolates (17) and play an important role as collectors of resistance genes in both human and companion animal settings (2, 3, 17, 18). The blaCMY-2 gene detected in ST1463 isolates was linked to I1 plasmids belonging to ST2 as determined by plasmid MLST (pMLST) (19), a combination already observed in E. coli from dogs in Denmark (20). CMY-2/IncI1 plasmids of ST2 were also found in E. coli from dogs hospitalized in clinic A1 (data not shown), suggesting a possible plasmid exchange between K. pneumoniae and E. coli within the same clinic. Otherwise, in the human isolates, the different bla genes were associated with diverse plasmids, further underlining the high genetic diversity of the isolates (Table 3). The blaNDM-1 gene could not be transferred by either electrotransformation or conjugation. Additional resistance genes, but not the blaCMY-2- and blaCTX-M-14-containing plasmids, were simultaneously transferred with the different bla genes (Table 3).
TABLE 3.
Characteristics of 3rd-generation-cephalosporin-resistant E. coli recipients after conjugation with K. pneumoniae strains or electrotransformation with K. pneumoniae plasmid DNAa
| K. pneumoniae donor | Origin | E. coli transformant | Transformation | Resistance phenotype | Carbapenemase gene | ESBL/pAmpC gene | Other bla gene(s) | Other antibiotic resistance genes and integronsb | Replicon | Inc group |
|---|---|---|---|---|---|---|---|---|---|---|
| 5012.10 | Human | NW5A | C | 3GCs STR SMX | blaCTX-M-3 | blaTEM-1 | sul2, strB | FII | IncFII | |
| 5109.57 | Human | NW8C | C | 3GCs | blaCTX-M-14 | FII | IncFII | |||
| KM749/09 | Dog | NW10A | C | AmpC STR | blaCMY-2 | I1 | IncI1-alpha | |||
| NW10F | CI, E | 3GCs CHL TMP STR SMX KAN | blaCTX-M-1 | aph(3′)-Ia, aadA2-like, dfrA12, sul1 | R | not assigned | ||||
| KM256/11 | Dog | NW11C | NC, E | AmpC STR SMX KAN | blaDHA-1 | blaOXA-1 | aph(3′)-Ia, qnrB, aac6′-Ib-cr, catB3-like, mrx-mphA, arr-1, intI1 | R | Not assigned | |
| KM899/09 | Dog | NW12A | NC, E | 3GCs TMP STR SMX KAN | blaCTX-M-1 | aph(3′)-Ia, aadA2-like, dfrA12, sul2 | R | Not assigned | ||
| 4906.28 | Human | NW15C | C | 3GCs GEN TMP STR TET SMX | blaCTX-M-15 | blaOXA-1 | aac(3)-IIc, aadA1-like, strB, qnrB, aac(6′)-Ib-cr, dfrA14, sul2, tet(A), intI1 | FIIk | IncF | |
| NW4906 | C | 3GCs | blaOXA-48 | L/M | IncL/M | |||||
| 5010.55 | Human | NW18A | C | 3GCs SMX | blaCTX-M-1 | sul2 | X1 | IncX1 | ||
| 5203.77 | Human | NW5203 | C | 3GCs TET TMP KAN SMX | blaCTX-M-15 | blaOXA-1, blaTEM-1 | aac(6′)-Ib-cr, dfrA12, sul1, mrx-mphA, tet(A), intI1 | FIIk | IncF |
C, conjugative; CI, conjugative only together with the blaCMY-2 plasmid (IncI1); NC, nonconjugative; E, plasmid transformation by electroporation; Inc, incompatibility group; ST, sequence type; 3GCs, 3rd-generation cephalosporins; AmpC, 3rd-generation cephalosporins and beta-lactamase inhibitors; CHL, chloramphenicol; TMP, trimethoprim; SMX, sulfamethoxazole; KAN, kanamycin; GEN, gentamicin; STR, streptomycin; TET, tetracycline.
Genes and functions: aadA, streptomycin adenylyltransferase; aac(3)-IIc, gentamicin acetyltransferase; aph(3′)-Ia, kanamycin O-phosphotransferase; qnrB, quinolone resistance protein; aac6′-Ib-cr, variant of aminoglycoside N(6′)-acetyltransferase–ciprofloxacin-modifying enzyme; cat, chloramphenicol acetyltransferase; dfr, dihydrofolate reductase for trimethoprim resistance; sul, dihydropteroate synthetase for sulfonamide resistance; mrx-mphA, macrolide inactivation resistance protein-phosphotransferase; arr-1, rifampin ADP ribosyltransferase; tet(A), tetracycline efflux; strB, streptomycin phosphotransferase; intI1, integrase.
Although exchange of MDR K. pneumoniae strains between humans and pets have been suggested by several studies (2, 6), we did not detect any clones or plasmids shared by isolates from humans and pets. The only common bla genes detected in both hosts consisted of blaCTX-M-1 and blaDHA-1. However, blaCTX-M-1 was found on different plasmids in isolates from animals and humans. Only blaDHA-1 seemed to be associated with the same plasmid type, R, in both human and animal isolates, but these plasmids were apparently not conjugative. Nevertheless, a K. pneumoniae isolate of ST11 producing DHA-1 appeared to have a particular affinity for veterinary settings, as it has also been found in hospitalized animals in Spain (3). Of note, none of the animals were infected with the zoonotic clones ST15-CTX-M-15 and ST101-CTX-M-15, recently reported in companion animals in France, Germany, and Italy (2, 4, 6). However, these clonal lineages were present among the human isolates of our study. The emergence of predominant K. pneumoniae clones in different animal clinics in different geographical regions indicates that several specific lineages are able to persist and disseminate in animal clinical environments.
In conclusion, this study revealed the absence of epidemiological links between the 3GC-R-Kp isolates from settings of human and veterinary medicine in Switzerland. However, the results clearly demonstrated nosocomial spread of MDR K. pneumoniae in veterinary settings, and they emphasize the importance of hospital infection control best practices being used as has been suggested for many years in human contexts.
(This study was conducted by Nadia Wohlwend in partial fulfillment of the requirements for an M.S. thesis from the Molecular Life Sciences program of the Science Faculty, University of Bern, Bern, Switzerland.)
Supplementary Material
ACKNOWLEDGMENTS
We thank Alexandra Collaud, Alexandra Rossano, and Debora Vogt for assistance or advice. We also thank the Center of Zoonosis, Bacterial Animal Diseases and Antibiotic Resistance (ZOBA), Laboratory Laupeneck, and the Institute for Infectious Diseases (IFIK) for providing isolates.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04408-14.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donati V, Feltrin F, Hendriksen RS, Svendsen CA, Cordaro G, García-Fernández A, Lorenzetti S, Lorenzetti R, Battisti A, Franco A. 2014. Extended-spectrum-β-lactamases, AmpC β-lactamases and plasmid mediated quinolone resistance in Klebsiella spp. from companion animals in Italy. PLoS One 9:e90564. doi: 10.1371/journal.pone.0090564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hidalgo L, Gutierrez B, Ovejero CM, Carrilero L, Matrat S, Saba CK, Santos-Lopez A, Thomas-Lopez D, Hoefer A, Suarez M, Santurde G, Martin-Espada C, Gonzalez-Zorn B. 2013. Klebsiella pneumoniae sequence type 11 from companion animals bearing ArmA methyltransferase, DHA-1 β-lactamase, and QnrB4. Antimicrob Agents Chemother 57:4532–4534. doi: 10.1128/AAC.00491-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haenni M, Ponsin C, Métayer V, Médaille C, Madec JY. 2012. Veterinary hospital-acquired infections in pets with a ciprofloxacin-resistant CTX-M-15-producing Klebsiella pneumoniae ST15 clone. J Antimicrob Chemother 67:770–771. doi: 10.1093/jac/dkr527. [DOI] [PubMed] [Google Scholar]
- 5.Stolle I, Prenger-Berninghoff E, Stamm I, Scheufen S, Hassdenteufel E, Guenther S, Bethe A, Pfeifer Y, Ewers C. 2013. Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J Antimicrob Chemother 68:2802–2808. doi: 10.1093/jac/dkt259. [DOI] [PubMed] [Google Scholar]
- 6.Ewers C, Stamm I, Pfeifer Y, Wieler LH, Kopp PA, Schonning K, Prenger-Berninghoff E, Scheufen S, Stolle I, Günther S, Bethe A. 2014. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J Antimicrob Chemother 69:2676–2680. doi: 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 7.Seiffert SN, Hilty M, Perreten V, Endimiani A. 2013. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Updat 16:22–45. doi: 10.1016/j.drup.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing. Twenty-fourth informational supplement M100-S24, vol 34, no. 1 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liesegang A, Tschäpe H. 2002. Modified pulsed-field gel electrophoresis method for DNA degradation-sensitive Salmonella enterica and Escherichia coli strains. Int J Med Microbiol 291:645–648. doi: 10.1078/1438-4221-00180. [DOI] [PubMed] [Google Scholar]
- 11.Meatherall BL, Gregson D, Ross T, Pitout JD, Laupland KB. 2009. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med 122:866–873. doi: 10.1016/j.amjmed.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 12.Ortega M, Marco F, Soriano A, Almela M, Martínez JA, Pitart C, Mensa J. 2013. Epidemiology and prognostic determinants of bacteraemic catheter-acquired urinary tract infection in a single institution from 1991 to 2010. J Infect 67:282–287. doi: 10.1016/j.jinf.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kola A, Holst M, Chaberny IF, Ziesing S, Suerbaum S, Gastmeier P. 2007. Surveillance of extended-spectrum β-lactamase-producing bacteria and routine use of contact isolation: experience from a three-year period. J Hosp Infect 66:46–51. doi: 10.1016/j.jhin.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Card R, Zhang J, Das P, Cook C, Woodford N, Anjum MF. 2013. Evaluation of an expanded microarray for detecting antibiotic resistance genes in a broad range of Gram-negative bacterial pathogens. Antimicrob Agents Chemother 57:458–465. doi: 10.1128/AAC.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey J, Ghersa P, Palacios PG, Belet M. 1986. Physical and genetic analysis of the ColD plasmid. J Bacteriol 166:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G, Décré D. 2014. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58:4207–4210. doi: 10.1128/AAC.02773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Yan JJ, Su IJ, Tsai SF, Lauderdale TL. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 50:3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother 61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 20.Damborg P, Gaustad IB, Olsen JE, Guardabassi L. 2011. Selection of CMY-2 producing Escherichia coli in the faecal flora of dogs treated with cephalexin. Vet Microbiol 151:404–408. doi: 10.1016/j.vetmic.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Woksepp H, Ryberg A, Billström H, Hällgren A, Nilsson LE, Marklund BI, Olsson-Liljequist B, Schön T. 2014. Evaluation of high-resolution melting curve analysis of ligation-mediated real-time PCR, a rapid method for epidemiological typing of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens. J Clin Microbiol 52:4339–4342. doi: 10.1128/JCM.02537-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.