Abstract
The role of mecA mutations in conferring resistance to ceftobiprole and ceftaroline, cephalosporins with anti-methicillin-resistant Staphylococcus aureus (MRSA) activity, was determined with MRSA strains COL and SF8300. The SF8300 ceftaroline-passaged mutant carried a single mecA mutation, E447K (E-to-K change at position 447), and expressed low-level resistance. This mutation in COL conferred high-level resistance to ceftobiprole but only low-level resistance to ceftaroline. The COL ceftaroline-passaged mutant, which expressed high-level resistance to ceftobiprole and ceftaroline, had mutations in pbp2, pbp4, and gdpP but not mecA.
TEXT
Treatment for methicillin-resistant Staphylococcus aureus (MRSA) has been complicated by increasing rates of antibiotic resistance. MRSA strains express penicillin binding protein 2a (PBP2a), encoded by mecA, which provides resistance to β-lactams by protecting the reactive serine in a narrow, inaccessible cleft, allowing cell wall synthesis to continue in the presence of antibiotic (1, 2). Ceftobiprole and ceftaroline are anti-MRSA cephalosporins that inhibit PBP2a at therapeutically useful concentrations. The R2 group of ceftobiprole extends into the narrow cleft of PBP2a to access the active site, whereas ceftaroline binding causes an allosteric change in PBP2a, revealing the active site for binding by a second molecule (3, 4). Ceftobiprole has been evaluated in clinical trials, and ceftaroline is FDA approved for treatment of skin and skin structure infections, including those caused by MRSA (5–10).
Previous studies from our group have shown that passage of the COL strain in ceftobiprole selects for resistant mutants with mutations in PBP2a (11). This study examines whether ceftaroline passage also selects for similar mutations. Passage experiments were conducted with two different MRSA backgrounds: COL, an archaic homogeneously resistant isolate from 1961, and SF8300, a heterogeneously resistant contemporary USA300 MRSA strain. COLnex and SF8300ex, mecA-negative derivative strains of COLn and SF8300, were obtained by (12) excising chromosomal staphylococcal cassette chromosome mec (SCCmec) (13). Wild-type mecA was reintroduced on plasmid pYK20 (derived from plasmid pAW8 and containing wild-type mecA cloned from COL [14]) into COLnex and SF8300ex for passage studies. If mecA was mediating resistance, then either curing a resistant mutant of the plasmid or introducing it into a susceptible background should result in loss or gain of phenotype, respectively, allowing determination of the contribution of mecA to resistance. Strains and plasmids used in this study are listed in Tables 1 and 2. COLnex(pYK20) and SF8300ex(pYK20) were serially passaged in tryptic soy broth containing increasing concentrations of ceftaroline (Forest Laboratories) as previously described (11).
TABLE 1.
List of parental and mutant strains used in mecA-positive passage studiesa
| Strain | Description | Phenotype |
|---|---|---|
| COLn | Parental strain | Mcr |
| COLnex | SCCmec excision strain derived from COLn | Mcs |
| COLnex(pAW8) | COLnex with an empty plasmid | Mcs |
| COLnex (pYK20) | COLnex with mecA on a plasmid | Mcr |
| COLnexpB(pYK20COLB*) | COLnex(pYK20) mutant passaged in ceftobiprole | BPRr Mcr |
| COLnexpT(pYK20COLT*) | COLnex(pYK20) mutant passaged in ceftaroline | CPTr Mcr |
| COLnexpB | COLnexpB(pYK20COLB*) cured of mecA plasmid | Mcs |
| COLnexpT | COLnexpT(pYK20COLT*) cured of mecA plasmid | Mcs |
| SF8300 | USA300 MRSA clinical isolate | Mcr Emr |
| SF8300ex | SCCmec excision strain derived from SF8300 ES | Mcs Ems |
| SF8300ex(pAW8) | SF8300ex with an empty plasmid | Mcs Ems |
| SF8300ex(pYK20) | SF8300ex with mecA on a plasmid | Mcr |
| SF8300expT(pYK208300T*) | SF8300ex(pYK20) mutant passaged in ceftaroline | CPTr Mcr |
| SF8300expT | SF8300expT(pYK208300T*) cured of mecA plasmid | Mcs |
An asterisk indicates a plasmid generated during ceftobiprole (B) or ceftaroline (T) selection in a COL or SF8300 background.
TABLE 2.
Plasmids used in mecA-positive passage studies
| Plasmida | Origin |
|---|---|
| pAW8 | Empty plasmid (see reference 14) |
| pYK20 | pAW8 containing mecA |
| pYK20COLB* | pYK20 in COLnex passaged in ceftobiprole |
| pYK20COLT* | pYK20 in COLnex passaged in ceftaroline |
| pYK208300T* | pYK20 in SF8300ex passaged in ceftaroline |
An asterisk indicates a passaged plasmid isolated during ceftobiprole (B) or ceftaroline (T) selection in a COL or SF8300 background.
After 28 days, COLnex(pYK20) and SF8300ex(pYK20) passaged in ceftaroline yielded two mutants, COLnexpT(pYK20COLT*) and SF8300expT(pYK208300T*) (an asterisk indicates a plasmid generated during ceftobiprole [B] or ceftaroline [T] selection in a COL or SF8300 background) (Table 1). MICs to ceftaroline, ceftobiprole, and other β-lactams (Sigma-Aldrich) were determined by the broth dilution method according to CLSI standards (Table 3) (15). COLnexpT(pYK20COLT*) expressed high-level resistance to both ceftaroline and ceftobiprole, with MICs of 64 μg/ml and 32 μg/ml, respectively. SF8300expT(pYK208300T*) expressed low-level resistance by passage day 4, with a MIC of 4 μg/ml, which remained unchanged despite 24 more passages (Table 3 and Fig. 1). The previously described ceftobiprole-passaged mutant of COLnex(pYK20) (11), COLnexpB(pYK20COLB*), showed high-level resistance to both ceftobiprole and ceftaroline, with a MIC of 64 μg/ml to each.
TABLE 3.
MICs of drugs for mecA-positive parent and passaged mutant strains
| Strain | MIC (μg/ml) ofa: |
||||||
|---|---|---|---|---|---|---|---|
| NAF | AMP | CFZ | CXT | CTX | CPT | BPR | |
| COLn | 256 | 16 | >256 | >256 | >256 | 1 | 2 |
| COLnex | 1 | 0.5 | 1 | 4 | 4 | <0.25 | 1 |
| COLnex(pAW8) | 0.5 | <0.25 | 0.5 | 4 | 4 | <0.25 | 1 |
| COLnex(pYK20) | 128 | 8 | 256 | 256 | >265 | 1 | 0.5 |
| COLnexpT(pYK20COLT*) | 128 | 64 | >256 | 8 | >256 | 64 | 32 |
| COLnexpB(pYK20COLB*) | >256 | 128 | >256 | >256 | >256 | 64 | 64 |
| SF8300 | 32 | 128 | 32 | 64 | 128 | 1 | 1 |
| SF8300ex | 0.5 | <0.25 | 1 | 4 | 4 | <0.25 | 0.5 |
| SF8300ex(pAW8) | <0.25 | <0.25 | 0.5 | 4 | 4 | <0.25 | 0.5 |
| SF8300ex(pYK20) | 32 | 8 | 128 | 128 | >256 | 0.5 | 1 |
| SF8300pT(pYK208300T*) | 128 | 16 | 128 | 128 | 256 | 4 | 4 |
NAF, nafcillin; AMP, ampicillin; CFZ, cefazolin; CXT, cefoxitin; CTX, ceftriaxone; CPT, ceftaroline; BRP, ceftobiprole.
FIG 1.
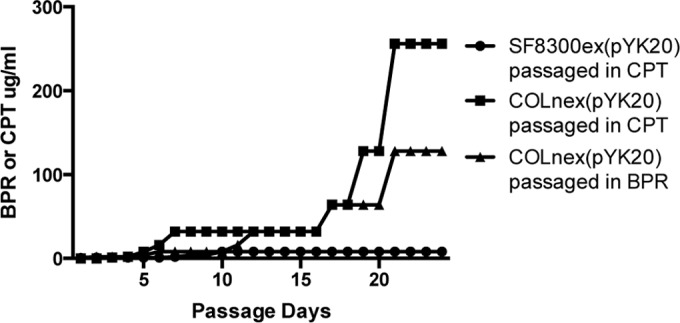
Emergence of ceftobiprole- and ceftaroline-passaged mecA-positive mutants. Resistance to ceftobiprole (BPR) and ceftaroline (CPT) was generated by passaging strains in subinhibitory concentrations of each antibiotic in broth. The highest concentration of drug in which strains grew each day is shown on the y axis.
Plasmid mecA was sequenced for mutations in passaged strains. Since PBPs are the target of β-lactams, pbp1, -2, -3, and -4 were also sequenced. Lastly, gdpP and acrB were sequenced, as mutations in these genes had been identified in a mecA-negative ceftobiprole-resistant mutant of COL (16). Plasmid pYK208300T* (pYK20 from SF8300ex passaged in ceftaroline), had a single point mutation (G to A at coding sequence [CDS] position 1339) in mecA, resulting in a nonsynonymous amino acid change, E447K. No mutations were present in other PBP genes, acrB, or gdpP (Table 4).
TABLE 4.
Mutations in mecA-positive mutant strains passaged with ceftobiprole and ceftaroline
| Strain | Mutation(s) ina: |
||||||
|---|---|---|---|---|---|---|---|
| Plasmid-borne mecA | pbp1 | pbp2 | pbp3 | pbp4 | gdpP | acrB | |
| COLnexpB(pYK20COLB*) | E150K, Y446L, E447K, F467Y, R589K, S649A | None | None | None | None | None | None |
| COLnexpT(pYK20COLT*) | None | None | D156N | None | T201A, F241L | H443Y | None |
| SF8300expT(pYK208300T*) | E447K | None | None | None | None | None | None |
None, gene sequence was wild type.
Plasmid pYK20COLT* (pYK20 from COLnex passaged in ceftaroline) had no mutations in mecA. A point mutation (C to T at CDS position 1327) introducing the H443Y mutation into GdpP, a point mutation (G to A at CDS position 466) introducing the D156N mutation into PBP2, and two point mutations (A to G at CDS position 601 and C to G at CDS position 723) introducing T201A and F241L mutations into PBP4 were found (Table 4). Of note, ceftaroline passage of COL containing mecA in its natural chromosomal location also resulted in a mutant with high-level resistance but lacking mutations in mecA (data not shown). Sequence analysis of that passaged mutant revealed a point mutation in pbp2 (G to A at CDS position 1891), resulting in the amino acid change G631S, and one in pbp4 (T to A at CDS position 414), introducing a N138K mutation. These data indicate that, even in the presence of mecA, ceftaroline can select for mutations in other genes, resulting in resistance.
Curing COLnexpB(pYK20COLB*) and SF8300expT(pYK208300T*) of their plasmids, each of which had mecA mutations, decreased the MICs for all β-lactams tested (Tables 5 and 6). Curing COLnexpT(pYK20COLT*) of its plasmid, which lacked mecA mutations, had no effect on MICs (Table 5). Transforming pYK20COLB*, which contains 6 substitution mutations in mecA, into the susceptible COLnex parent, yielding the transformant COLnex(pYK20COLB*), resulted in high-level ceftaroline and ceftobiprole resistance. Transforming pYK208300T*, which contains the single mutation E447K in mecA, into COLnex, yielding the transformant COLnex(pYK208300T*), resulted in high-level resistance to ceftobiprole (MIC of 64 μg/ml) but only low-level resistance to ceftaroline (MIC of 4 μg/ml) (Table 5). Transforming pYK20COLB* into SF8300ex, yielding the transformant SF8300ex(pYK20COLB*), resulted in high-level resistance to ceftaroline and ceftobiprole (MIC of 32 μg/ml to each) (Table 6). Multiple attempts to transform SF8300ex with pYK208300T* were unsuccessful.
TABLE 5.
MICs of drugs for mecA-positive passaged mutant strains cured of plasmid and parental strains transduced with plasmids from passaged strains in the COL background
| Strain | MIC (μg/ml) ofa: |
||||||
|---|---|---|---|---|---|---|---|
| NAF | AMP | CFZ | CXT | CTX | CPT | BPR | |
| COLn | 256 | 16 | >256 | >256 | >256 | 1 | 2 |
| COLnex | 1 | 0.5 | 1 | 4 | 4 | <0.25 | 1 |
| COLnex(pAW8) | 0.5 | <0.25 | 0.5 | 4 | 4 | <0.25 | 1 |
| COLnex(pYK20) | 128 | 8 | 256 | 256 | >265 | 1 | 0.5 |
| COLnex(pYK208300T*) | >256 | 32 | >256 | 64 | 32 | 4 | 64 |
| COLnex(pYK20COLB*) | >256 | 32 | >256 | 128 | >256 | 32 | 64 |
| COLnexpB(pYK20COLB*) | >256 | 128 | >256 | >256 | >256 | 64 | 64 |
| COLnexpB, plasmid cured | 0.5 | <0.25 | 0.5 | 4 | 4 | <0.25 | 1 |
| COLnexpT(pYK20COLT*) | 128 | 64 | >256 | 8 | >256 | 64 | 32 |
| COLnexpT, plasmid cured | 256 | 128 | 256 | 16 | >256 | 64 | 32 |
NAF, nafcillin, AMP, ampicillin; CFZ, cefazolin; CXT, cefoxitin; CTX, ceftriaxone; CPT, ceftaroline; BRP, ceftobiprole.
TABLE 6.
MICs of drugs for mecA-positive passaged mutant strains cured of plasmid and parental strains transduced with plasmids from passaged strains in the USA300 strain SF8300 background
| Strain | MIC (μg/ml) ofa: |
||||||
|---|---|---|---|---|---|---|---|
| NAF | AMP | CFZ | CXT | CTX | CPT | BPR | |
| SF8300 | 32 | 128 | 32 | 64 | 128 | 1 | 1 |
| SF8300ex | 0.5 | <0.25 | 1 | 4 | 4 | <0.25 | 0.5 |
| SF8300ex(pAW8) | <0.25 | <0.25 | 0.5 | 4 | 4 | <0.25 | 0.5 |
| SF8300ex(pYK20) | 32 | 8 | 32 | 32 | 256 | 0.5 | 1 |
| SF8300ex(pYK20COLB*) | 32 | 16 | 64 | 64 | 128 | 32 | 32 |
| SF8300pT(pYK208300T*) | 128 | 16 | 128 | 128 | 256 | 4 | 4 |
| SF8300pT, plasmid cured | 0.5 | 1 | 1 | 4 | 8 | 2 | 1 |
NAF, nafcillin; AMP, ampicillin; CFZ, cefazolin; CXT, cefoxitin; CTX, ceftriaxone; CPT, ceftaroline; BRP, ceftobiprole.
A single amino acid change at E447 in mecA appears to play a key role in resistance. It is present in ceftobiprole-passaged COL (among other mutations) (11), it is the only mecA mutation in the ceftaroline-passaged SF8300, and it has been reported in clinical isolates (17, 18). Structurally, E447 resides in the penicillin-binding domain of PBP2a and interacts with the R2 group of ceftobiprole and other β-lactams (3, 19). E447K conferred high-level ceftobiprole resistance and low-level ceftaroline resistance in the COLnex background, likely due to structural differences between the two compounds. Multiple mutations in mecA yield high-level resistance to both antibiotics (20). The genetic background also plays a role. Heterogeneous SF8300 passaged in ceftaroline developed low-level resistance to ceftobiprole and ceftaroline with E447K (MIC 4 μg/ml). This mutation has been associated with low-level resistance to ceftaroline in clinical isolates (17).
In conclusion, passage in either ceftaroline or ceftobiprole selects for an E447K mutation in PBP2a, a mutation found in ceftaroline-resistant clinical isolates, underscoring its importance in mediating the resistance phenotype (17, 18). Although whole-genome sequencing was not performed, which is a limitation of this study, genes other than mecA play a role because the level of resistance differed between COL and SF8300 backgrounds upon introduction of the E447K mutation and the highly resistant passaged COL mutant had no mecA mutations. Although there are likely others, mutations in genes encoding PBP4 and GdpP seem to be particularly important, as these have been repeatedly identified in ceftobiprole- and ceftaroline-passaged mutants. The role of these genes and others is under active investigation.
ACKNOWLEDGMENT
This work was funded by NIH grant 5R01 AI100291 to H.F.C. (principal investigator).
REFERENCES
- 1.Chambers HF. 1988. Methicillin-resistant staphylococci. Clin Microbiol Rev 1:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuda C, Suvorov M, Vakulenko SB, Mobashery S. 2004. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J Biol Chem 279:40802–40806. doi: 10.1074/jbc.M403589200. [DOI] [PubMed] [Google Scholar]
- 3.Lovering AL, Gretes MC, Safadi SS, Danel F, de Castro L, Page MG, Strynadka NC. 2012. Structural insights into the anti-methicillin-resistant Staphylococcus aureus (MRSA) activity of ceftobiprole. J Biol Chem 287:32096–32102. doi: 10.1074/jbc.M112.355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otero LH, Rojas-Altuve A, Llarrull LI, Carrasco-Lopez C, Kumarasiri M, Lastochkin E, Fishovitz J, Dawley M, Hesek D, Lee M, Johnson JW, Fisher JF, Chang M, Mobashery S, Hermoso JA. 2013. How allosteric control of Staphylococcus aureus penicillin binding protein 2a enables methicillin resistance and physiological function. Proc Natl Acad Sci U S A 110:16808–16813. doi: 10.1073/pnas.1300118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrison MW, Kawamura NM, Wen MM. 2012. Ceftaroline fosamil: a new cephalosporin active against resistant Gram-positive organisms including MRSA. Expert Rev Anti Infect Ther 10:1087–1103. doi: 10.1586/eri.12.112. [DOI] [PubMed] [Google Scholar]
- 6.Anderson SD, Gums JG. 2008. Ceftobiprole: an extended-spectrum anti-methicillin-resistant Staphylococcus aureus cephalosporin. Ann Pharmacother 42:806–816. doi: 10.1345/aph.1L016. [DOI] [PubMed] [Google Scholar]
- 7.Barbour A, Derendorf H. 2010. Resistance and the management of complicated skin and skin structure infections: the role of ceftobiprole. Ther Clin Risk Manag 6:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers HF. 2006. Ceftobiprole: in-vivo profile of a bactericidal cephalosporin. Clin Microbiol Infect 12(Suppl s2):17–22. doi: 10.1111/j.1469-0691.2006.01404.x. [DOI] [PubMed] [Google Scholar]
- 9.Bazan JA, Martin SI. 2010. Ceftaroline fosamil: a novel broad-spectrum cephalosporin. Drugs Today (Barc) 46:743–755. doi: 10.1358/dot.2010.46.10.1519172. [DOI] [PubMed] [Google Scholar]
- 10.Bush K. 2012. Improving known classes of antibiotics: an optimistic approach for the future. Curr Opin Pharmacol 12:527–534. doi: 10.1016/j.coph.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee R, Gretes M, Basuino L, Strynadka N, Chambers HF. 2008. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 52:2089–2096. doi: 10.1128/AAC.01403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama Y, Zhang HZ, Chambers HF. 2003. Effect of disruption of Staphylococcus aureus PBP4 gene on resistance to beta-lactam antibiotics. Microb Drug Resist 9:329–336. doi: 10.1089/107662903322762752. [DOI] [PubMed] [Google Scholar]
- 14.Katayama Y, Robinson DA, Enright MC, Chambers HF. 2005. Genetic background affects stability of mecA in Staphylococcus aureus. J Clin Microbiol 43:2380–2383. doi: 10.1128/JCM.43.5.2380-2383.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. CLSI document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level beta-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother 54:4900–4902. doi: 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alm RA, McLaughlin RE, Kos VN, Sader HS, Iaconis JP, Lahiri SD. 2014. Analysis of Staphylococcus aureus clinical isolates with reduced susceptibility to ceftaroline: an epidemiological and structural perspective. J Antimicrob Chemother 69:2065–2075. doi: 10.1093/jac/dku114. [DOI] [PubMed] [Google Scholar]
- 18.Long SW, Olsen RJ, Mehta SC, Palzkill TG, Cernoch PL, Perez KK, Musick WL, Rosato AE, Musser JM. 2014. PBP2a mutations causing high-level ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 58:6668–6674. doi: 10.1128/AAC.03622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim D, Strynadka NC. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol 9:870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 20.Mendes RE, Tsakris A, Sader HS, Jones RN, Biek D, McGhee P, Appelbaum PC, Kosowska-Shick K. 2012. Characterization of methicillin-resistant Staphylococcus aureus displaying increased MICs of ceftaroline. J Antimicrob Chemother 67:1321–1324. doi: 10.1093/jac/dks069. [DOI] [PubMed] [Google Scholar]


