Abstract
Background
Impaired bioenergetics is a prominent feature of the failing heart, but the underlying metabolic perturbations are poorly understood.
Methods and Results
We compared metabolomic, gene transcript, and protein data from six paired failing human left ventricular (LV) tissue samples obtained during left ventricular assist device (LVAD) insertion (heart failure (HF) samples) and at heart transplant (post-LVAD samples). Non-failing left ventricular (NFLV) wall samples procured from explanted hearts of patients with right HF served as novel comparison samples. Metabolomic analyses uncovered a distinct pattern in HF tissue: 2.6 fold increased pyruvate concentrations coupled with reduced Krebs cycle intermediates and short-chain acylcarnitines, suggesting a global reduction in substrate oxidation. These findings were associated with decreased transcript levels for enzymes that catalyze fatty acid oxidation and pyruvate metabolism and for key transcriptional regulators of mitochondrial metabolism and biogenesis, peroxisome proliferator-activated receptor gamma co-activator1α (PGC1A, 1.3 fold) and estrogen-related receptor α (ERRA, 1.2 fold) and γ (ERRG, 2.2 fold). Thus, parallel decreases in key transcription factors and their target metabolic enzyme genes can explain the decreases in associated metabolic intermediates. Mechanical support with LVAD improved all of these metabolic and transcriptional defects.
Conclusions
These observations underscore an important pathophysiologic role for severely defective metabolism in HF, while the reversibility of these defects by LVAD suggests metabolic resilience of the human heart.
Keywords: heart failure, metabolism, left ventricular assist device, cardiac remodeling, mitochondria
Introduction
Although neurohormonal blockade and mineralocorticoid antagonism have improved survival in heart failure (HF),1, 2 the five-year mortality rate of this disorder (59%) still equals that of the most common forms of cancer (58%).3 Thus, better treatments for advanced HF are urgently needed. Because multiple metabolic defects are present in the myocardium during HF, approaches that target myocardial metabolism to improve cardiac energy provision are an attractive therapeutic strategy. However, interpretation of extant human studies is complicated by difficulties associated with procuring sufficient functioning left ventricular (LV) myocardium in a timely manner and with obtaining comparable non-failing LV (NFLV) tissue. In the present investigation, we performed metabolomic, transcriptomic and protein analyses on rapidly and identically procured NFLV and paired LV from HF patients before and after left ventricular assist device (LVAD) support. Our results demonstrate that the metabolic alterations and accompanying changes in gene expression that occur in HF are reversed with mechanical unloading of the LV.
Methods
Patients and Tissue Collection
LV tissue was collected from 12 HF patients during LVAD insertion and from the same 12 patients at the time of LVAD removal (post-LVAD). Six of the paired samples were subjected to metabolomics, gene and protein analyses (Group 1, Table 1, Supplemental Table 1) and 6 different paired samples were subjected to transcriptomic analyses (Group 2, Supplemental Table 2). The 2 groups of patients were chosen as the first 6 available for Group 1and the next 6 available for Group 2. Pre and post-LVAD echocardiograms were interpreted in a blinded fashion by a single experienced echocardiographer (Fig. 1). LV size and systolic function were defined in accord with current recommendations.4, 5 NFLV wall samples were acquired from 6 patients during heart-double lung transplantation for severe right HF (Table 1). All participants signed written informed consent approved by the Houston Methodist Hospital Institutional Review Board.
Table 1.
Table of demographics (GROUP 1)
| NFLV | HF | p-value* | PostLVAD† | p-value‡ | |
|---|---|---|---|---|---|
| Number (n) | n=6 | n=6 | n=6 | ||
| Age - yr | 40 ± 14 | 58 ± 9 | 0.03 | 59 ± 9 | 0.8 |
| Sex - no. (%) | |||||
| Male | 1 (17) | 6 (100) | 6 (100) | ||
| Female | 5 (83) | 0 (0) | 0 (0) | ||
| Race - no. (%) | |||||
| White | 1 (17) | 4 (67) | 4 (67) | ||
| Black | 3 (50) | 0 (0) | 0 (0) | ||
| Other | 2 (33) | 2 (33) | 2 (33) | ||
| Etiology - no. (%) | |||||
| PHTN | 6 (100) | N/A | N/A | ||
| Ischemic | N/A | 5 (83) | 5 (83) | ||
| Non-Ischemic | N/A | 1 (17) | 1 (17) | ||
| Echocardiography | |||||
| EF (%) | 58 ± 11 | 20.6 ± 5 | <0.0001 | 30.3 ± 14.9 | 0.1 |
| LVEDd (cms) | 3.6 ± 0.7 | 6.4 ± 0.5 | <0.0001 | 5.2 ± 0.9 | 0.01 |
| LV size (ml) | N/A | 273 ± 67 | 187 ± 71 | 0.05 | |
| Median Days on LVAD | N/A | N/A | 245.5 | ||
| DM (%) | 1 (17) | 3 (50) | 3 (50) | ||
| BMI (kg/m2) | 25.6 ± 4 | 30.1 ± 5 | 0.1 | 30.4 ± 5 | 0.9 |
| PO2 (mmHg) | 151 ± 180 | 203 ± 158 | 0.08 | 367 ± 80 | 0.04 |
| LDH (IU/L) | 820 ± 88 | 578 ± 197 | 0.009 | 726 ± 181 | 0.2 |
NFLV vs. HF,
paired samples HF-PostLVAD,
HF vs. postLVAD
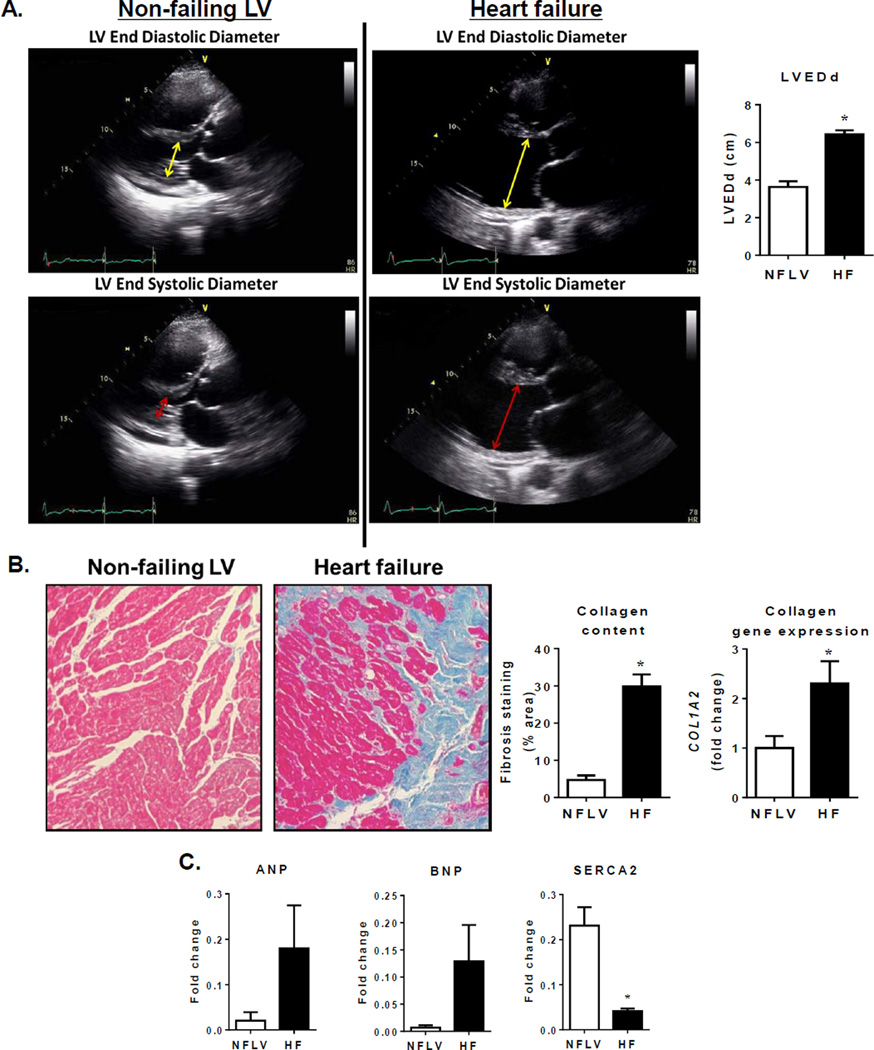
Figure 1.
NFLV tissue serves as a unique comparator for failing LV. Panel A: Representative echocardiography images of NFLV and failing LV for size and function comparison. Non-failing LV: Upper figure illustrates normal LV size (yellow double headed arrow defines the left ventricular end diastolic diameter (LVEDd) 3cm). Lower figure illustrates normal LV systolic function (double headed red arrow defines the left ventricular end systolic diameter (LVESd) 1.8 cm, equates to a fractional shortening of 40% and a LV ejection fraction (EF, ~ 68%). Failing LV: Upper figure illustrates a severely dilated LV (yellow double headed arrow notes a LVEDd ~ 7 cm). Lower figure illustrates severely depressed LV systolic function (double headed red arrow LVESd ~ 6.3 cm equates to a fractional shortening of 10% and a LVEF ~17%). Quantitation of LV end diastolic dimension (LVEDd) indicating normal size of NFLVs and LV hypertrophy in HF (Mean±SEM; *P<0.001 versus NFLV by Mann Whitney test, NFLV: N=6, HF: N=6). Panel B: images for Masson’s trichrome staining, quantitation for collagen content and gene expression of collagen 1α2 (COL1A2) indicative of absence of fibrosis in NFLV but increased fibrosis in LV of HF patients (Mean±SEM; *P<0.001 versus NFLV by Mann Whitney test, NFLV: N=6, HF: N=6). Panel C: changes in expression of characteristic heart failure genes ANP, BNP and SERCA (Mean±SEM; *P<0.01 versus NFLV by Mann Whitney test; NFLV: N=6, HF: N=6).
Fresh tissue was obtained directly from the surgeon. Scar free, reddish LV apex samples (~2 grams) were immediately dissected and tissue was freeze-clamped in liquid nitrogen for RNA, protein and metabolomic analyses within 1–2 minutes following the hand-off. Additional sections were paraffin embedded for histology.
Fibrosis measurement
Masson’s trichrome stained LV sections were analyzed by optical microscopy imaging with collagen content (blue staining) expressed as a percentage of the analyzed LV area.
Metabolomic Analyses
Mass spectrometry-based metabolic profiling was used to measure acylcarnitines, amino acids and organic acids (Fig. 2) as described previously and as detailed in Supplemental Methods.6–8 Metabolites were correlated to echocardiographic parameters for Group 1 patients (Fig. 3).
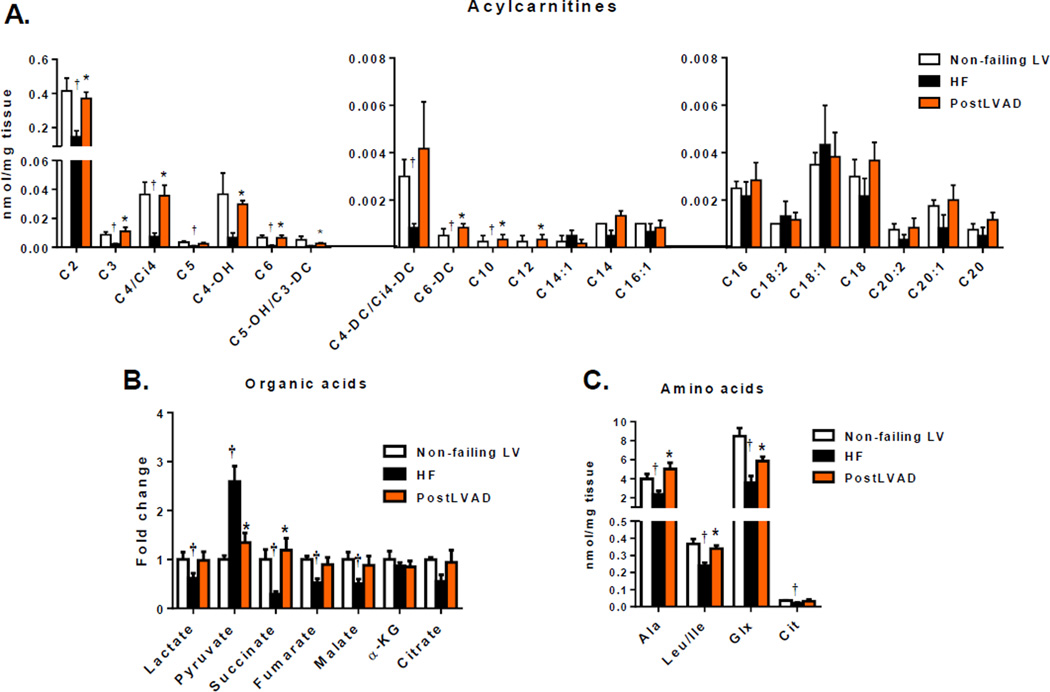
Figure 2.
Metabolomic changes in HF are rescued by LVAD. Levels of acylcarnitine species (A), organic acids (B), amino acids (C) for LV samples from NFLV (N=4), HF (N=6) and post-LVAD (N=6) patients were measured. (Mean±SEM; *P<0.05 versus HF, †P<0.05 versus NFLV; by multiple testing analyses using Benjamini and Hochberg method, FDR 12.5%).
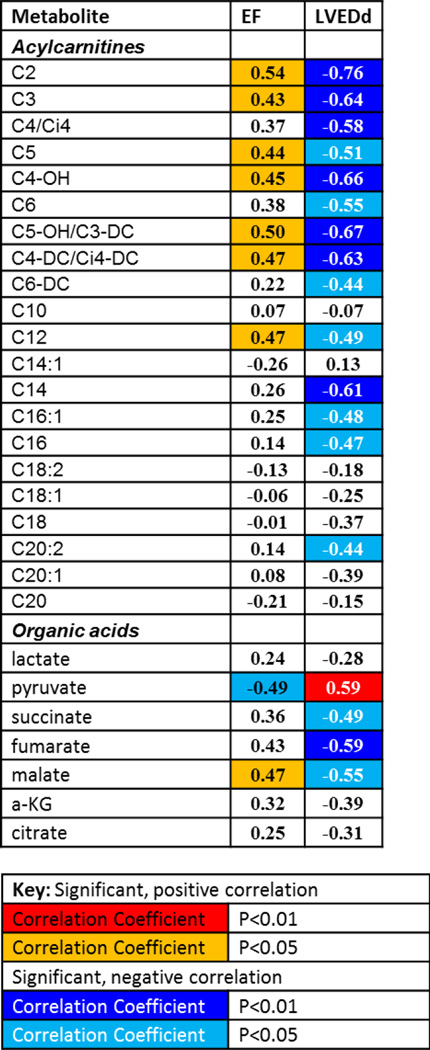
Figure 3.
Correlation analyses between echocardiography parameters (EF and LVEDd) and LV tissue metabolites.
Gene and protein expression studies
Quantitative real-time polymerized chain reaction (qRT-PCR) with Taqman probes and primers (Applied Biosystems) was conducted as previously described.9, 10 Candidate genes were selected based on their important metabolic function in each of the categories in Fig. 4 (A–D, F–G) such as the regulation of a rate-limiting step or master regulation of the activity of a pathway. All gene expression values were normalized against the house-keeping gene, PPIA (cyclophilin A), which revealed nearly identical expression among the 3 groups. PPIA was chosen after comparative analysis with glyceraldehyde 3-phosphate dehydrogenase (GAPDH), Actin B (ACTB), tubulin 1A (TUB1A), ribosomal gene L32 (RPL32), beta-2 microglogulin (B2M) and 18S ribosomal RNA. For protein analyses, LV tissue lysates were analyzed using standard Western blot techniques detailed in Supplementary Methods. Telomere length was measured using qRT-PCR to determine telomere repeat sequence copy number in relation to the single copy gene 36b4, as previously described.11
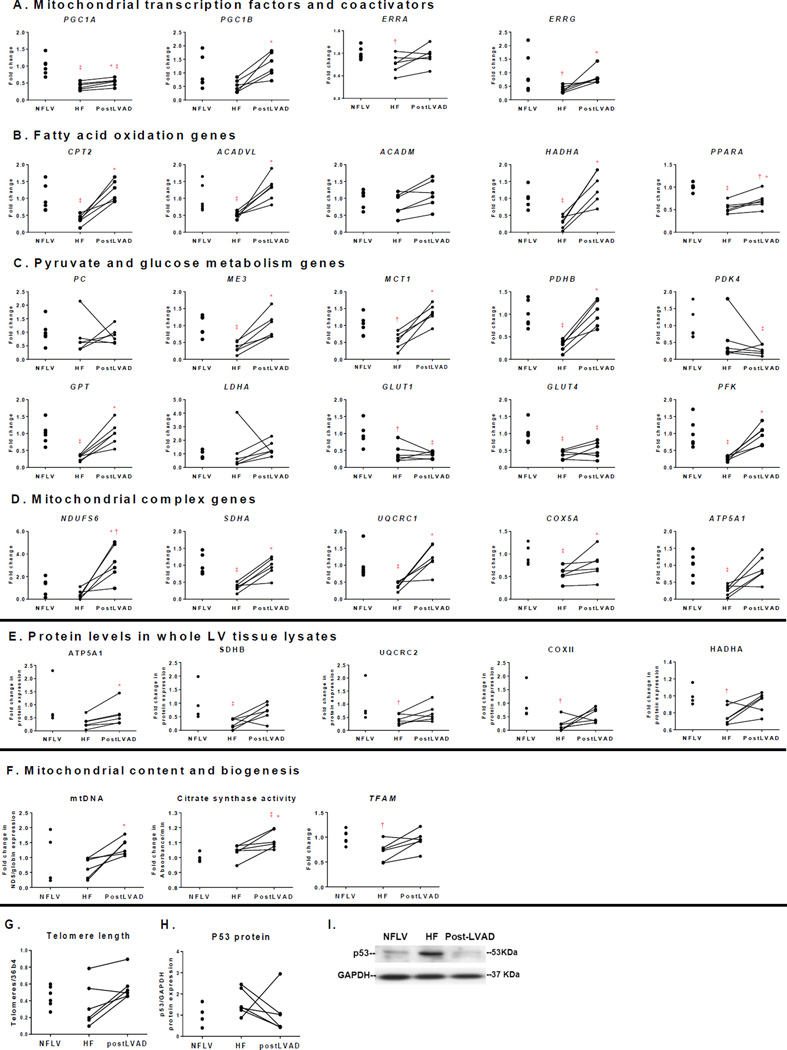
Figure 4.
Expression levels of metabolic genes and proteins reduced in HF are increased by LVAD. Candidate genes include (A): mitochondrial transcription factors and coactivators; (B): fatty acid oxidation; (C): pyruvate and glucose metabolism; and (D): mitochondrial complex proteins. All genes are normalized to PPIA. (E): mitochondrial protein expression in whole heart lysates, normalized to GAPDH. (F): mitochondrial DNA, CS activity and TFAM expression, indicators of mitochondrial biogenesis. (G): telomere length, (H): quantitation of P53 protein expression and (I): representative blot for P53 protein. Lines connect data points for the same subject before and after LVAD implantation. (Mean±SEM; *P<0.05 versus HF by paired Wilcoxon signed rank test; †P<0.05, ‡P<0.01 versus NFLV by Mann-Whitney test. NFLV: N=6 for gene expression and N=4 for protein expression, HF: N=6, post-LVAD: N=6). Gene abbreviations: ACADVL: Acyl-CoA dehydrogenase, very long chain; ACADM: Acyl-CoA dehydrogenase, medium chain; ATP5A1: ATP synthase alpha1; CPT2: Carnitine palmitoyltransferase II; COX2/5A: Cytochrome c oxidase II/Va; ERRA and G: Estrogen-related receptor α/γ; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; GLUT1: Glucose transporter 1; GPT: Glutamic Pyruvate Transaminase; HADHA: 3-hydroxyacyl-coenzyme A dehydrogenase; LDHA: Lactate Dehydrogenase; ME3: Malic Enzyme; MCT1: Monocarboxylate Transporter; NDUFS6: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6; PC: Pyruvate Carboxylase; PDHB: Pyruvate Dehydrogenase B; PDK4: Pyruvate dehydrogenase kinase 4; PFK: Phosphofructokinase; PGC1A and B: Peroxisome proliferator-activated receptor-γ coactivator 1 α/β; PPARA: Peroxisome proliferator-activated receptor α; PPIA: Peptidylprolyl isomerase A (cyclophilin A); SDHA/B: Succinate dehydrogenase A/B; TFAM: Transcription factor A, mitochondrial; UQCRC1/2: Cytochrome b-c1 complex 1/2.
Microarray analyses were performed by the Genomic and RNA Profiling Core Facility of Baylor College of Medicine. RNA samples were analyzed using HG-U133 arrays scanned at 6-um resolution with an Agilent G2500A Technologies Gene Array scanner. The entire microarray dataset was analyzed using Gene Set Enrichment Analysis (GSEA) v2.0.10 software (www.broadinstitute.org/gsea/index.jsp)12. The GSEA was performed with all 186 human Kyoto Encylopedia of Genes and Genomes (KEGG) pathways to identify gene function pathways significantly enriched with up-regulated or down-regulated genes post-LVAD support. The 12 pathways that changed most significantly according to the GSEA calculations are shown in Table 2.
Table 2.
Top 12 KEGG pathways most changed post-LVAD as detected by GSEA
| Pathway Name | Size | ES | NES | Nominal p-value |
False Discovery Rate q-value |
Family wise-error rate p-value |
|
|---|---|---|---|---|---|---|---|
| 1 | KEGG_Ribosome | 79 | 0.83 | 3.91 | 0.00 | 0.00 | 0.00 |
| 2 | KEGG_Oxidative_Phosphorylation | 110 | 0.66 | 3.33 | 0.00 | 0.00 | 0.00 |
| 3 | KEGG_Parkinsons_Disease | 106 | 0.64 | 3.22 | 0.00 | 0.00 | 0.00 |
| 4 | KEGG_Spliceosome | 107 | 0.63 | 3.20 | 0.00 | 0.00 | 0.00 |
| 5 | KEGG_Huntingtons_Disease | 164 | 0.53 | 2.87 | 0.00 | 0.00 | 0.00 |
| 6 | KEGG_Proteasome | 42 | 0.67 | 2.76 | 0.00 | 0.00 | 0.00 |
| 7 | KEGG_Protein_Export | 20 | 0.70 | 2.40 | 0.00 | 0.00 | 0.00 |
| 8 | KEGG_Alzheimers_Disease | 149 | 0.45 | 2.40 | 0.00 | 0.00 | 0.00 |
| 9 | KEGG_Fatty_Acid_Metabolism | 41 | 0.56 | 2.29 | 0.00 | 0.00 | 0.00 |
| 10 | KEGG_Prion_Diseases | 35 | 0.57 | 2.27 | 0.00 | 0.00 | 0.00 |
| 11 | KEGG_Valine_Leucine_And_Isoleucine_Degradation | 43 | 0.54 | 2.24 | 0.00 | 0.00 | 0.00 |
| 12 | KEGG_Citrate_Cycle Cycle | 29 | 0.58 | 2.18 | 0.00 | 0.00 | 0.00 |
Key:
Pathways with primarily mitochondrial genes are indicated in bold.
Size: Number of genes in the gene set after filtering out those genes not in the expression dataset
ES: Enrichment score for the gene set; i.e., the degree to which this gene set is overrepresented at the top or bottom of the ranked list of genes in the expression dataset
NES: Normalized enrichment score; i.e., the enrichment score for the gene set after it has been normalized across analyzed gene sets.
Mitochondrial DNA content and citrate synthase (CS) assay
DNA was isolated from LV samples (Qiagen DNA extraction kit) and analyzed by qRT-PCR to assess the ratio of the mitochondrial-encoded NADH dehydrogenase-5 (ND5) to the nuclear-encoded βglobin genes. CS activity, a measure of mitochondrial content, was analyzed in LV protein lysates using a microplate assay from MitoSciences (Abcam).
Statistical Analysis
GraphPad Prism 5.0 was used for all statistical analyses. Mann-Whitney non-parametric tests were employed to identify significant differences in mRNA expression, protein expression, and metabolomic assay differences between NFLV vs. HF (all unpaired samples). For paired HF vs. post-LVAD samples, all differences were identified using Wilcoxon signed rank tests. Two-tailed tests were performed with a significance level of α=0.05. Sample sizes for each measurement are indicated in Table 1 and figure legends. False discovery rates were determined for all acylcarnitine species, organic acids and amino acids using the Benjamini and Hochberg method.13 For the 19 acylcarnitine species significant in Figure 2 and S1, approximately 2 acylcarnitine species will be false positive at the 12.5% false discovery rate level when comparing NFLV vs. HF or HF vs. post-LVAD. Spearman rank correlation coefficients were computed between acylcarnitines, organic acids, and the patient-specific echocardiography parameters LV end-diastolic diameter at diastole (LVEDd) and ejection fraction (EF).
Results
Patient Characteristics
Clinical characteristics for NFLV, Group 1 HF and post-LVAD subjects are summarized in Table 1. These HF patients, who had both ischemic and non-ischemic etiology, had significantly depressed EF and remodeling as shown by increased LVEDd. All six post-LVAD subjects received continuous-flow pumps: three received an axial flow HeartMate II™ (HM II; Thoratec Corp, Pleasanton, CA), and three received a centrifugal VentrAssist™ (VentraCore, Ltd., Australia) pump implanted as a bridge to cardiac transplantation. The median duration of LVAD support was 245.5 days (range 215–325). The HeartMate II™ operating speeds ranged from 9,000 to 10,000 rpm and VentrAssist™ from 2200 to 2400 rpm, which provided partial to full circulatory support capability. Perioperative arterial pO2 and post-operative continuous pulse oximetry did not indicate significant hypoxemia in any of the groups.
NFLV samples were procured from 6 patients with severe right ventricular failure (mean age 40 years, 83% female), due to either primary or secondary pulmonary hypertension as the indication for heart/lung transplant (Table 1). These LV samples had no evidence of failure based on multiple criteria: normal EF and LVEDd assessed by echocardiography (Fig. 1A), lack of fibrosis (Fig. 1B), and atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP) mRNA levels generally below, and sarco/endoplasmic reticulum Ca2+-ATPase (SERCA2) mRNA levels above, HF levels (Fig. 1C). The 6 paired HF and post-LVAD and 4 of 6 NFLV samples were used for protein and metabolomics analyses, and the 6 paired samples and 6 NFLV samples were used for gene expression analyses.
Medications taken include anti-hypertension drugs (angiotensin converting enzyme inhibitors, beta blockers, diuretics, aldosterone antagonists), anti-atrial fibrillation (Digoxin), anti-cholesterol (statins), anti-diabetic (insulin), anti-coagulants (warfarin) and others. Detailed medications for each group are listed in Supplemental Table 1.
Clinical characteristics for Group 2 patients are summarized in Supplemental Table 2. In this group, 3 patients received the Novacor LVAD (pulsatile-flow pumps) and 3 patients received the DeBakey LVAD (continuous-flow pumps), with a median duration of LVAD support of 54.5 days. These 6 paired HF and post-LVAD were used for transcriptomic analyses.
Metabolomic analyses reveal improved substrate utilization with LVAD
Tandem mass spectrometry (MS/MS) was used to analyze acylcarnitine species ranging in size from 2 to 22 carbon atoms (Figs. 2A, S1A–B). The abundance of short- and medium-chain acylcarnitines from C2–C10 inclusively was reduced in failing versus non-failing LV samples. HF samples had reduced levels of acetylcarnitine (C2), a surrogate for acetyl-CoA, and a product of glucose, amino acid and fatty acid oxidation; propionylcarnitine (C3) and isovalerylcarnitine (C5), which derive from amino acid catabolism; and succinylcarnitine (C4-DC) and butyrylcarnitine (C4), products of amino acid and fatty acid oxidation. There was little difference in concentrations of longer chain acylcarnitines (C14 and above) as a function of HF status. LVAD universally restored C2–C10 acylcarnitines to NFLV values, suggesting a global restoration of substrate oxidation.
Several organic acids were assayed by Gas Chromatography-Mass Spectrometry as a measure of the influx of metabolic fuels into the Krebs cycle (Fig. 2B). Pyruvate levels were increased in HF samples (p<0.05), whereas nearly all measured Krebs cycle intermediates, including citrate, succinate, fumarate and malate were reduced, consistent with reduced entry of pyruvate and anaplerotic substrates into the mitochondrial matrix. However, alpha-ketoglutarate levels were not decreased in HF samples, possibly due anaplerotic metabolism of glutamate.14, 15 LVAD normalized levels of pyruvate and all Krebs cycle intermediates, suggesting restoration of normal metabolism of pyruvate and other mitochondrial fuels.
Amino acids constitute a major source of anaplerotic substrates provided to the Krebs cycle.16 Concentrations of alanine, leucine/isoleucine, glutamine/glutamic acid and citrulline were reduced in failing heart muscle, but increased after LVAD support (Fig. 2C, S1C). These data provide further evidence for restoration of metabolic homeostasis in response to mechanical unloading of the failing heart.
Diabetes is a common comorbidity in HF. Ability of glucose metabolism and effect of insulin treatment may affect the cardiac metabolic profile. However, we did not find any significant differences in cardiac organic-acid profiles or acylcarnitine species in HF patients with or without diabetes (Fig. S2).
Multivariate correlation analyses indicated that EFs were positively correlated with short chain acylcarnitines including C2, C3, C5, C4-OH, C5-OH/C3-DC, C4-CD/Ci4-DC and medium chain C12 (Fig. 3). On the other hand, LVEDd values negatively correlated with C2, C3, C4/i4, C5, C4-OH, C6, C5-OH/C3-DC, C4-CD/Ci4-DC, C6-DC, C12, C14, C16-1, C16 and C20-2. While pyruvate was negatively correlated with EF, it was positively correlated with LVEDd. In contrast, succinate, fumarate and malate were negatively correlated with LVEDd and malate was positively correlated with EF. These data suggest that decrease in EF or increase in LVEDd is associated with lower levels of short-chain break down products of acylcarnitines, indicative of impaired substrate oxidation in HF. The data also suggest that reduction in EF or increase in LVEDd is associated with accumulation of pyruvate but reduction in Krebs cycle intermediates, providing additional evidence that pyruvate metabolism is suppressed in HF.
Metabolic gene changes in HF and post-LVAD samples parallel metabolite changes
In order to delineate potential mechanisms underlying the metabolomic changes observed in HF and post-LVAD, we measured transcripts encoding metabolic transcription factors and their target mitochondrial enzymes. Transcript levels of peroxisome proliferator-activated receptor (PPAR)-γ coactivator 1 (PGC1A), a master regulator of mitochondrial biogenesis and metabolism, and estrogen-related receptors (ERRA and ERRG), which partner with PGC1A in transcriptional regulation, were reduced in failing versus non-failing LV (Fig. 4A), although related PGC1B expression did not change significantly. Consistent with the decrease in transcription factor expression, mRNA levels of key enzymes of fatty acid oxidation (Carnitine palmitoyltransferase II (CPT2), Acyl-CoA dehydrogenase, very long chain (ACADVL), 3-hydroxyacyl-coenzyme A dehydrogenase (HADHA), peroxisome proliferator-activated receptor α (PPARA) were sharply decreased in HF versus NFLV samples (Figs. 4B and S3).
Expression of pyruvate carboxylase (PC), pyruvate dehydrogenase kinase 4 (PDK4), and lactate dehydrogenase (LDHA) was similar in HF and NFLV samples. However, expression of the monocarboxylase pyruvate transporter (MCT1) and other enzymes involved in pyruvate metabolism, including pyruvate dehydrogenase (PDHB), malic enzyme (ME3) and both isoforms of pyruvate/alanine aminotransferases (GPT) decreased in HF versus NFLV tissue (Fig. 4C). Concurrent with pyruvate accumulation in the failing heart, these gene expression changes imply impaired mitochondrial pyruvate transport and metabolism. The transcripts encoding glucose transporters (GLUT1 and GLUT4) and the glycolytic enzyme, phosphofructokinase (PFK), were also reduced in failing heart, suggesting impaired glucose uptake and metabolism.
Transcript and protein levels of representative components of all five complexes of the mitochondrial electron transport chain were decreased in HF samples, while mRNA expression of the uncoupling protein (UCP3), was not different among the groups (Figs. 4D–E, S3). Mitochondrial transcription factor A (TFAM), which regulates mitochondrial DNA copy number was also reduced in HF tissue, although mitochondrial-to-genomic DNA ratios and CS activity were not significantly lower than in NFLV tissue (Fig. 4F).
Mirroring the tissue metabolomic results, LVAD-mediated ventricular unloading reversed nearly all the transcript changes associated with HF (Figs. 4A and S3). Expression of key transcription factors and co-regulators, including PGC1A, PGC1B, and ERRG were up-regulated after mechanical unloading (Figs. 4A). Expression of genes involved in mitochondrial biogenesis, mitochondrial respiration, fatty acid and pyruvate metabolism were also globally increased, and several of these changes were confirmed at the protein level (Fig. 4E and S4). Mitochondrial DNA and CS activity also increased with ventricular unloading (Fig. 4F). Although GLUT1 and GLUT4 expression remained low, PFK increased, suggesting at least partial restoration of glycolysis and glucose metabolism. Taken together, these data indicate that LVAD restores or improves multiple pathways of substrate utilization in the failing human heart.
P53 is a senescence gene that down-regulates metabolic transcription factors, particularly PGC1A and B.17, 18 Telomere dysfunction is associated with increased P53 levels.18 There were no differences between P53 protein levels and telomere length between NFLV and HF ventricles, however, P53 levels decreased post-LVAD in 5 of 6 subjects and LVAD support trended to increase cardiac telomere length (Fig. 4G–I).
Mitochondrial gene pathways are enriched by LVAD
Microarray analyses were performed on six paired HF and post-LVAD samples, obtained from Group 2 HF patients (Supplemental Table 2) in order to evaluate unbiased changes in cardiac gene expression after LVAD support. GSEA found that 7 of the 12 pathways most enriched with up-regulated genes post-LVAD insertion were associated with mitochondrial function (Table 2). These included the KEGG oxidative phosphorylation pathway (ranked #2 by GSEA) and neurodegenerative disease pathways associated with mitochondrial dysfunction and oxidative stress (Parkinson’s, Alzheimer’s and Huntington’s disease sets; ranked #3, 5 and 8). These pathways shared 74 genes in common with the oxidative phosphorylation pathway. Several related metabolic pathways (fatty acid metabolism, valine-leucine-isoleucine degradation, and the Krebs/tricarboxylic acid cycle; ranked #9, 11, 12) were also increased in post-LVAD samples.
These results correspond well with Group 1 data, which demonstrated increased expression of genes involved in mitochondrial-mediated fatty acid oxidation, pyruvate metabolism, and electron transport chain activity after LVAD insertion (Fig. 4). The enrichment of metabolic and mitochondrial pathways in the Group 2 samples suggests that these are among the most significant functional gene expression changes that occur following LVAD support and are thus likely to explain the metabolomic improvements observed following LVAD placement.
Discussion
In a broad investigation combining targeted metabolomics, transcriptomics and protein analyses, we observed metabolite and expression profiles indicative of global impairment in mitochondrial substrate utilization in failing human LV tissue. We found substantially decreased medium and short chain acylcarnitines, including C2 acylcarnitine, a surrogate for acetyl-CoA; increased pyruvate levels with low levels of downstream Krebs cycle intermediates; and decreased C3 and C5 acylcarnitines, products of amino acid catabolism. Correlation analyses showed that short chain acylcarnitines, which are breakdown products of fatty acids, glucose and amino acids, correlated negatively with echocardiography parameter EF and positively with LVEDd. These alterations were accompanied by reduced expression of key transcription factors, PGC1A, PGC1B, and ERRG, as well as their target genes, which encode multiple mitochondrial metabolic enzymes. These expression patterns were validated by transcriptomic analyses of paired pre- and post-LVAD LV tissue from a second group of subjects. These metabolite and gene changes provide a plausible explanation for the multiple defects in mitochondrial substrate utilization seen in HF tissue. Mechanical unloading converted the metabolomic and transcriptomic profiles of HF to resemble NFLV. The failing heart is considered “an engine out of fuel” with defects in energetics that could arise at several steps including substrate utilization, oxidative phosphorylation or ATP delivery.19 Our data show that failing human LVs have increased levels of some substrates compared to NFLVs, but decreased levels of their metabolites, suggesting that a major defect in substrate utilization leads to impaired LV function.
Although we did not measure metabolic fluxes, a major strength of our study is the paired pre-/post-LVAD samples from the same patients, which provides unique insight into the mechanisms of metabolic changes associated with HF. In patients with HF, LVAD support enhances myocyte performance, calcium handling, microvascular flow, and myocardial architecture,20–23 and prolongs life. In our subjects LVAD improved cardiac function and quality of life, and the pre and post-LVAD paired analysis confirmed that LV volume and LVEDd decreased, suggesting a more favorable ventricular environment. Beneficial hemodynamic effects of continuous flow LVAD support have been documented by us and others,4, 5 and we have also observed that pulsatile LVAD support improves clinical markers of HF.24 We found that ventricular unloading substantially induced expression of transcription factors that regulate multiple downstream mitochondrial enzymes to enhance levels of intermediary metabolites in key energy-yielding metabolic pathways (Fig. 5). Similar metabolic outcomes were suggested by proteomic profiling of pre- and post-LVAD myocardium by de Weger et al.,25 in which the authors found reduced expression of cytoskeleton proteins suggestive of ‘reverse remodeling’ but increased expression of proteins involved in mitochondrial metabolism. Our findings support the de Weger study, but provide more extensive understanding of metabolic alterations in HF by comparing changes in transcriptional regulation and tissue metabolites in paired pre-/post-LVAD myocardium.
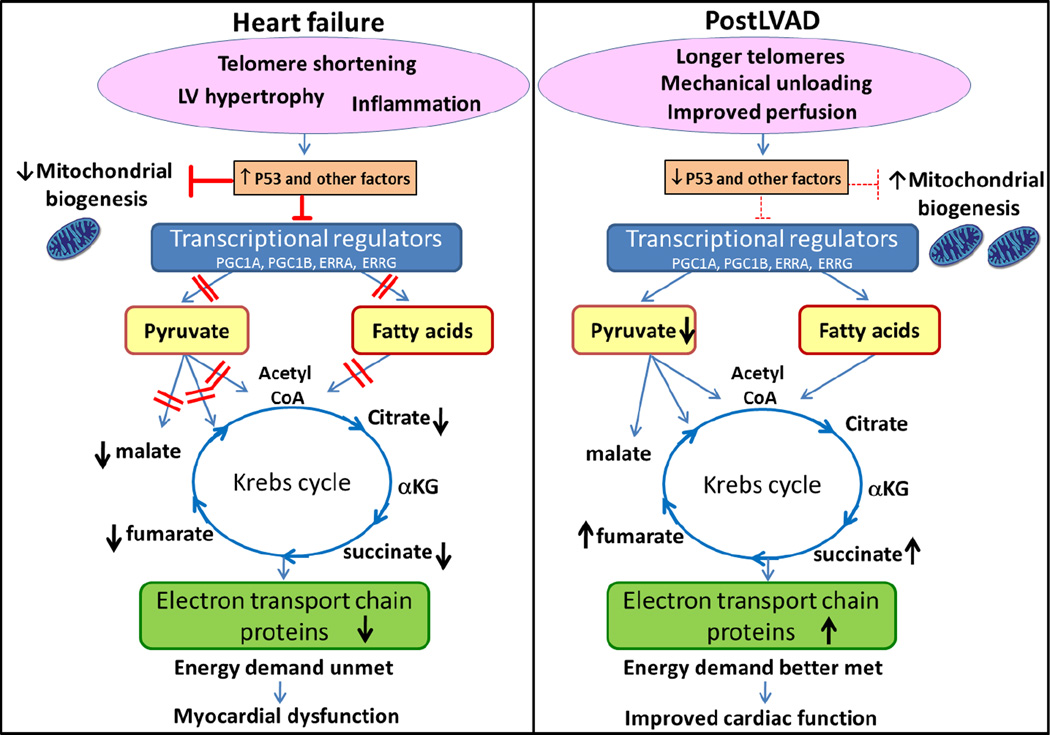
Figure 5.
Mechanical unloading reverses abnormal metabolic profile in failing human heart. In HF, telomere shortening and other deleterious presentations induce one or more regulatory proteins such as P53 which influence the myocardial metabolism. For example, P53 suppresses expression of metabolic transcription factors and inhibits mitochondrial biogenesis in HF. Loss of transcriptional regulation results in impaired β-oxidation and blockage of the various catabolic pathways of pyruvate causing accumulation of pyruvate and reduced Krebs cycle activity. Generalized defects in substrate utilization results in an inability to meet energy demands and ultimately in myocardial dysfunction. LVAD increased length of telomeres which reversed the action of P53, enhancing expression of key transcriptional regulators, and increasing both mitochondrial biogenesis and metabolic enzymes to enhance utilization of pyruvate and fatty acid. These changes post-LVAD enable the heart to better meet energy demands.
One potential criticism of our results is whether the NFLV we studied are an appropriate comparator for failing LV. These NFLV had no evidence of failure, based on multiple measures, and were processed in a manner identical to the HF and post-LVAD samples. However, NFLV were obtained primarily from women, who were generally younger than HF subjects and received different medications than the HF subjects due to their having RV rather than LV dysfunction. On the other hand, our NFLV were not exposed to the death-related catecholamine surge or placed in cardioplegic solution, the fate of commonly used LV obtained from hearts collected, but ultimately not used, for transplantation. Thus, despite the limitations outlined above, we believe our NFLV may provide useful insight as a comparator for metabolic and gene expression differences between failing and NFLV.
Defective metabolism of pyruvate is a key feature of HF unveiled in this investigation. Our gene expression and metabolomics results suggest that all routes of pyruvate metabolism are attenuated in failing LV including decarboxylation of pyruvate to acetyl-CoA by PDH, carboxylation to form oxaloacetate by PC and ME, transamination to alanine by GPT, and reduction to lactate by LDHA. Indeed, positive correlation of LVEDd and pyruvate, and negative correlation of Krebs cycle intermediates with LVEDd support our findings that HF is associated with impaired pyruvate metabolism, which is reversed with improvement of LVEDd with LVAD support. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila and humans has recently been discovered by Rutter and his group.26 Yeast and Drosophila mutants lacking this activity had decreased levels of Krebs cycle intermediates, similar to our results. These changes are consistent with our finding of attenuated expression of genes involved in pyruvate metabolism in HF. In addition, we find mitochondrial pyruvate carriers, MPC1 and MCP2, increased with LVAD by 2.653 and 1.984 fold respectively in our microarray analyses. The importance of impaired pyruvate metabolism is underscored by the classical concept that “fats burn in the fire of carbohydrates”27, since, in addition to conversion to acetyl-CoA via PDH, pyruvate is also carboxylated to malate and oxaloacetate via ME and PC.28 Oxaloacetate is a substrate for the CS reaction that allows acetyl-CoA derived from fatty acid oxidation to enter Krebs cycle. Thus, normal pyruvate metabolism is critical for both glucose and fatty acid oxidation in the heart.29 Our study demonstrates that human HF involves metabolic reprogramming that limits fatty acid oxidation per se, but also impairs entry of pyruvate and potentially other anaplerotic substrates into the Krebs cycle. This effect limits energy provision not only from glucose, but also from fatty acids. The over-arching phenotype of the failing heart is therefore one of energy starvation, due to impaired oxidation of all of the major macronutrients (Fig. 5).
Mitochondrial biogenesis and multiple genes involved in mitochondrial metabolism were decreased in HF compared to NFLV and improved with LVAD. Our unbiased GSEA analyses also showed enrichment of mitochondrial metabolic pathways with LVAD in a separate cohort of patients. To determine the factors regulating these mitochondrial changes, we measured expression of PGC1A, PGC1B, ERRAand ERRG, key transcriptional regulators of mitochondrial substrate utilization and biogenesis. Decreased PGC1A and ERRA have been seen in human HF, but the metabolic consequences were not reported.30, 31 One recent study of human heart suggested that ERRA, but not PGC1A, was down regulated in failing human heart,32 and that impaired mitochondrial biogenesis precedes HF in right ventricular hypertrophy due to congenital heart disease.33 PGC1A, ERRA, and ERRG were reduced in HF compared to NFLV in our study, and mechanical unloading increased expression of three of the four key transcription factors (PGC1A, PGC1B, ERRG) measured in our paired samples. These factors are redundant in their activity, but require interaction for full activity. Thus, HF may be associated with a generalized deficit in transcriptional regulation of metabolism.34
Indeed, impaired cardiac transcriptional regulation can lead to altered cardiac function and enhanced failure in response to stress.35 PGC1A is pivotal for efficient mitochondrial fatty acid oxidation, and cardiac-specific PGC1A deletion results in accelerated HF following aortic banding.36 Interestingly, genes down regulated in hearts of ERRα knockout mice following cardiac stress are similar to those decreased in our human HF tissue, including mitochondrial complex genes, enzymes involved in fatty acid oxidation, and ERRA targets such as CPT2, GLUT1 and GLUT4.37 Low CPT2, accompanied by decreases in fatty acid oxidation enzymes, can contribute to reduced catabolism of long and medium chain acyl CoAs, which equilibrate with the acylcarnitines measured in the current study.38 Mice with cardiac ERRG and PGC1B deletion also develop cardiac pathology in response to stress,39, 40 and simultaneous loss of multiple transcription factors has additive effects to impair the cardiac response to stress.41 Similar to these models of transcription factor knockout in the mouse, our results suggest that the failing human heart loses expression of key transcription factors that orchestrate normal fuel catabolism and mitochondrial biogenesis, thereby seriously impairing cardiac metabolism and myocardial function (Fig. 5).
The transcription factor P53 can suppress expression of PGC1A and PGC1B, and is increased in human and mouse failing myocardium.42, 43 Telomere dysfunction, such as loss of telomerase, leads to telomere shortening, which is reported in human failing as well as in aging hearts.44 Telomere shortening can lead to increased cardiac P53 activity, decreased PGC1A/B and ERRA expression, decreased levels of mtDNA, and attenuated expression of genes in oxidative phosphorylation, mitochondrial dysfunction, gluconeogenesis, and oxidative stress.18 Knockdown of P53 in the short telomere mouse and in other mouse models of increased cardiac P53 reversed these patterns and improved cardiac function.18, 43, 45 Thus, it is possible that HF-induced increases in cardiac P53 may promote the global transcription factor and downstream metabolomic changes reported herein, thereby exacerbating cardiac dysfunction. While we did not find significant P53 differences between HF and NFLV samples in our patients, P53 levels decreased in 5/6 patients and telomere lengths trended to increase post-LVAD. This reversal may be important to the LVAD-induced re-vitalization of a cardiac metabolic cascade in which increased transcription factor activity stimulates mitochondrial biogenesis to increase fatty acid oxidation through increased Krebs cycle activity (Fig. 5). However, P53 may not be the only protein regulating these metabolic transcription factors, and further studies are required to completely understand the nature of the upstream regulators.
We conclude that deciphering the mechanisms by which ventricular unloading increases expression of key metabolic transcriptional regulators may help to guide novel therapeutic strategies for HF. Our observations in paired pre- and post LVAD ventricles indicates that the defect in cardiac fuel utilization in HF is reversible. Therefore, releasing the transcriptional brake on substrate utilization and biogenesis may emerge as adjuvant treatment in HF. Small molecules or gene therapies that enhance transcriptional regulators and decrease P53 activity may be a novel strategy to re-energize the heart to prevent progression of this deadly disease.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Dale Abel (University of Iowa) for helpful scientific discussions about this work, and Yuelan Ren and Tuo Deng for technical assistance.
Funding Sources: Our study was supported by the NIH (R21 CA133153 to DJH, P01DK58398 to CBN, and R01HL061483 to HT); generous gifts from MacDonald Foundation and Zucker Family to WAH; Methodist DeBakey Heart & Vascular Center Texans Grant, generous gifts from John Kotts Family, Rodney Bradley Family and Stedman-West foundation to DJH, American Heart Association fellowship to AAG, and John S. Dunn research foundation to STCW.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Jhund PS, Macintyre K, Simpson CR, Lewsey JD, Stewart S, Redpath A, et al. Long-term trends in first hospitalization for heart failure and subsequent survival between 1986 and 2003: a population study of 5.1 million people. Circulation. 2009;119:515–523. doi: 10.1161/CIRCULATIONAHA.108.812172. [DOI] [PubMed] [Google Scholar]
- 2.Zannad F, McMurray JJ, Krum H, van Veldhuisen DJ, Swedberg K, Shi H, et al. Eplerenone in patients with systolic heart failure and mild symptoms. N Engl J Med. 2011;364:11–21. doi: 10.1056/NEJMoa1009492. [DOI] [PubMed] [Google Scholar]
- 3.Stewart S, Ekman I, Ekman T, Oden A, Rosengren A. Population impact of heart failure and the most common forms of cancer: a study of 1 162 309 hospital cases in Sweden (1988 to 2004) Circ Cardiovasc Qual Outcomes. 2010;3:573–580. doi: 10.1161/CIRCOUTCOMES.110.957571. [DOI] [PubMed] [Google Scholar]
- 4.Estep JD, Chang SM, Bhimaraj A, Torre-Amione G, Zoghbi WA, Nagueh SF. Imaging for ventricular function and myocardial recovery on nonpulsatile ventricular assist devices. Circulation. 2012;125:2265–2277. doi: 10.1161/CIRCULATIONAHA.111.040238. [DOI] [PubMed] [Google Scholar]
- 5.Ambardekar AV, Walker JS, Walker LA, Cleveland JC, Jr, Lowes BD, Buttrick PM. Incomplete recovery of myocyte contractile function despite improvement of myocardial architecture with left ventricular assist device support. Circ Heart Fail. 2011;4:425–432. doi: 10.1161/CIRCHEARTFAILURE.111.961326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turer AT, Stevens RD, Bain JR, Muehlbauer MJ, van der Westhuizen J, Mathew JP, et al. Metabolomic profiling reveals distinct patterns of myocardial substrate use in humans with coronary artery disease or left ventricular dysfunction during surgical ischemia/reperfusion. Circulation. 2009;119:1736–1746. doi: 10.1161/CIRCULATIONAHA.108.816116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Caglayan E, Stauber B, Collins AR, Lyon CJ, Yin F, Liu J, et al. Differential roles of cardiomyocyte and macrophage peroxisome proliferator-activated receptor gamma in cardiac fibrosis. Diabetes. 2008;57:2470–2479. doi: 10.2337/db07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001;104:2923–2931. doi: 10.1161/hc4901.100526. [DOI] [PubMed] [Google Scholar]
- 11.Brouilette SW, Moore JS, McMahon AD, Thompson JR, Ford I, Shepherd J, et al. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet. 2007;369:107–114. doi: 10.1016/S0140-6736(07)60071-3. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. 1995;57:289–300. Series B. [Google Scholar]
- 14.McDaniel HG, Jenkins R, Yeh M, Razzaque A. Glutamic dehydrogenase from rat heart mitochondria. II. Kinetic characteristics. J Mol Cell Cardiol. 1984;16:303–309. doi: 10.1016/s0022-2828(84)80601-x. [DOI] [PubMed] [Google Scholar]
- 15.McDaniel HG, Jenkins R, McDaniel R. Conditions for glutamate dehydrogenase activity in heart mitochondria. Biochem Med Metab Biol. 1993;50:75–84. doi: 10.1006/bmmb.1993.1048. [DOI] [PubMed] [Google Scholar]
- 16.Russell RR, 3rd, Taegtmeyer H. Changes in citric acid cycle flux and anaplerosis antedate the functional decline in isolated rat hearts utilizing acetoacetate. J Clin Invest. 1991;87:384–390. doi: 10.1172/JCI115008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villeneuve C, Guilbeau-Frugier C, Sicard P, Lairez O, Ordener C, Duparc T, et al. p53-PGC-1alpha pathway mediates oxidative mitochondrial damage and cardiomyocyte necrosis induced by monoamine oxidase-A upregulation: role in chronic left ventricular dysfunction in mice. Antioxid Redox Signal. 2013;18:5–18. doi: 10.1089/ars.2011.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 20.Birks EJ, George RS, Hedger M, Bahrami T, Wilton P, Bowles CT, et al. Reversal of severe heart failure with a continuous-flow left ventricular assist device and pharmacological therapy: a prospective study. Circulation. 2011;123:381–390. doi: 10.1161/CIRCULATIONAHA.109.933960. [DOI] [PubMed] [Google Scholar]
- 21.Drakos SG, Kfoury AG, Selzman CH, Verma DR, Nanas JN, Li DY, et al. Left ventricular assist device unloading effects on myocardial structure and function: current status of the field and call for action. Curr Opin Cardiol. 2011;26:245–255. doi: 10.1097/HCO.0b013e328345af13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambardekar AV, Buttrick PM. Reverse remodeling with left ventricular assist devices: a review of clinical, cellular, and molecular effects. Circ Heart Fail. 4:224–233. doi: 10.1161/CIRCHEARTFAILURE.110.959684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slaughter MS, Rogers JG, Milano CA, Russell SD, Conte JV, Feldman D, et al. Advanced heart failure treated with continuous-flow left ventricular assist device. N Engl J Med. 2009;361:2241–2251. doi: 10.1056/NEJMoa0909938. [DOI] [PubMed] [Google Scholar]
- 24.Razeghi P, Young ME, Ying J, Depre C, Uray IP, Kolesar J, et al. Downregulation of metabolic gene expression in failing human heart before and after mechanical unloading. Cardiology. 2002;97:203–209. doi: 10.1159/000063122. [DOI] [PubMed] [Google Scholar]
- 25.de Weger RA, Schipper ME, Siera-de Koning E, van der Weide P, van Oosterhout MF, Quadir R, et al. Proteomic profiling of the human failing heart after left ventricular assist device support. J Heart Lung Transplant. 2011;30:497–506. doi: 10.1016/j.healun.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenfeld G. Ueber die Behandlung der Zuckerkrankheit. Berliner Klinische Wockenschrift. 1909;46:957–961. [Google Scholar]
- 28.Russell RR, 3rd, Taegtmeyer H. Pyruvate carboxylation prevents the decline in contractile function of rat hearts oxidizing acetoacetate. Am J Physiol. 1991;261:H1756–H1762. doi: 10.1152/ajpheart.1991.261.6.H1756. [DOI] [PubMed] [Google Scholar]
- 29.Taegtmeyer H, Hems R, Krebs HA. Utilization of energy-providing substrates in the isolated working rat heart. Biochem J. 1980;186:701–711. doi: 10.1042/bj1860701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garnier A, Zoll J, Fortin D, N'Guessan B, Lefebvre F, Geny B, et al. Control by circulating factors of mitochondrial function and transcription cascade in heart failure: a role for endothelin-1 and angiotensin II. Circ Heart Fail. 2009;2:342–350. doi: 10.1161/CIRCHEARTFAILURE.108.812099. [DOI] [PubMed] [Google Scholar]
- 31.Sihag S, Cresci S, Li AY, Sucharov CC, Lehman JJ. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res. 2010;106:1541–1548. doi: 10.1161/CIRCRESAHA.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karamanlidis G, Bautista-Hernandez V, Fynn-Thompson F, Del Nido P, Tian R. Impaired mitochondrial biogenesis precedes heart failure in right ventricular hypertrophy in congenital heart disease. Circ Heart Fail. 2011;4:707–713. doi: 10.1161/CIRCHEARTFAILURE.111.961474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubert G, Vega RB, Kelly DP. Perturbations in the gene regulatory pathways controlling mitochondrial energy production in the failing heart. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schilling J, Kelly DP. The PGC-1 cascade as a therapeutic target for heart failure. J Mol Cell Cardiol. 2011;51:578–583. doi: 10.1016/j.yjmcc.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, et al. The transcriptional coactivator PGC-1alpha is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol. 2008;295:H185–H196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, et al. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Violante S, Ijlst L, van Lenthe H, de Almeida IT, Wanders RJ, Ventura FV. Carnitine palmitoyltransferase 2: New insights on the substrate specificity and implications for acylcarnitine profiling. Biochim Biophys Acta. 2010;1802:728–732. doi: 10.1016/j.bbadis.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Riehle C, Wende AR, Zaha VG, Pires KM, Wayment B, Olsen C, et al. PGC-1beta deficiency accelerates the transition to heart failure in pressure overload hypertrophy. Circ Res. 2011;109:783–793. doi: 10.1161/CIRCRESAHA.111.243964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubert G, Vega RB, Kelly DP. Perturbations in the gene regulatory pathways controlling mitochondrial energy production in the failing heart. Biochim Biophys Acta. 2013;1833:840–847. doi: 10.1016/j.bbamcr.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leri A, Franco S, Zacheo A, Barlucchi L, Chimenti S, Limana F, et al. Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. Embo J. 2003;22:131–139. doi: 10.1093/emboj/cdg013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toko H, Takahashi H, Kayama Y, Oka T, Minamino T, Okada S, et al. Ca2+/calmodulin-dependent kinase IIdelta causes heart failure by accumulation of p53 in dilated cardiomyopathy. Circulation. 2010;122:891–899. doi: 10.1161/CIRCULATIONAHA.109.935296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oeseburg H, de Boer RA, van Gilst WH, van der Harst P. Telomere biology in healthy aging and disease. Pflugers Arch. 2010;459:259–268. doi: 10.1007/s00424-009-0728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.