Abstract
Genetic diversity in drug metabolism and disposition is mainly considered as the outcome of the inter-individual genetic variation in polymorphism of drug-xenobiotic metabolizing enzyme (XME). Among the XMEs, glutathione-S-transferases (GST) gene loci are an important candidate for the investigation of diversity in allele frequency, as the deletion mutations in GST M1 and T1 genotypes are associated with various cancers and genetic disorders of all major Population Affiliations (PAs). Therefore, the present population based phylogenetic study was focused to uncover the frequency distribution pattern in GST M1 and T1 null genotypes among 45 Geographically Assorted Human Populations (GAHPs). The frequency distribution pattern for GST M1 and T1 null alleles have been detected in this study using the data derived from literatures representing 44 populations affiliated to Africa, Asia, Europe, South America and the genome of PA from Gujarat, a region in western India. Allele frequency counting for Gujarat PA and scattered plot analysis for geographical distribution among the PAs were performed in SPSS-21. The GST M1 and GST T1 null allele frequencies patterns of the PAs were computed in Seqboot, Gendist program of Phylip software package (3.69 versions) and Unweighted Pair Group method with Arithmetic Mean in Mega-6 software. Allele frequencies from South African Xhosa tribe, East African Zimbabwe, East African Ethiopia, North African Egypt, Caucasian, South Asian Afghanistan and South Indian Andhra Pradesh have been identified as the probable seven patterns among the 45 GAHPs investigated in this study for GST M1-T1 null genotypes. The patternized null allele frequencies demonstrated in this study for the first time addresses the missing link in GST M1-T1 null allele frequencies among GAHPs.
Introduction
Metabolic activities play an important role in shaping the livelihood of an organism. Drug-xenobiotic compound metabolizing enzyme (XME) systems are the most investigated pathways that are involved in equilibrating the health status of an individual. Among the numerous drug related genes investigated, Glutathione-S-transferases (GST) of Phase II XMEs were found to play an important role in cellular protection and in cellular resistance to drugs by glutathione conjugation reactions. GST classes convert the active endogenous and/or exogenous carcinogenic compounds to their detoxified form. Among the GST classes, GST M1 and GST T1 were found associated to a loss of function with a structural deletion (null mutation); moreover, they were also found to modify the detoxification ability of the individual exposed to tobacco or carcinogenic pollutants in the environment [1]. Genotoxins such as aromatic hydrocarbon epoxides and products of oxidative stress such as DNA hydroperoxides, polycyclic aromatic hydrocarbon diol epoxides are catalyzed and detoxified by GST M1 while, the constituents of cigarette smoke such as alkyl halides, bezo (a) pyrene diol epoxide, acrolein are catalyzed and detoxified by GST T1 [1, 2]. Several factors such as environmental pollution, dietary habits and activity-dependent genetic differences have been reported as modulators of GST expression and susceptibility to xenobiotic compound detoxification [3]. Numerous studies in the recent past have hypothesized the difference in metabolic rate of M1 and T1 classes of GST as the risk factor associated to cancers of bladder, pancreas, upper aero digestive tract, lung, esophageal, head-neck, melanoma and also in Balken endemic nephropathy patients [4–9]. Further, the inter-individual difference in drug disposition and efficacy has been investigated by various authors [10] and the observed frequency distributions of GST M1-T1 genotypes among different populations are reported as ethnic or PAs dependent [10, 11]. Drugs are the major hope of remedy for the people around the globe with various metabolic and genetic disorders but the scenario in the past was found distressed as the effectiveness of the drugs were reported by the influence of the unidentified polymorphic patterns observed in drug metabolism genes among different ethnics or PAs [11–13]. Though researchers from different PAs are interested in analyzing the frequencies of GST M1 and T1 null genotypes and their possible risk association with various disorders, they are not able to report conclusive association in all major PAs [14, 15]. Recent advances in molecular techniques have opened a new era of pharmacogenomics and several researchers are inclined towards investigating the relationship in genetic diversity and allelic frequency of GST classes to insight genetic predisposition or susceptibility among various ethnics or PAs. In this context, probing the genetic variation in GST classes is inevitable for genomic epidemiological studies and to develop new common drugs in future to majority of PAs [9, 16, 17]. The allele frequency pattern in GST M1-T1 null genotypes of different PAs are yet to be explored to unlock several phenomenons related to a risk association with genetic diseases and drug dispositions [10]. A study including statistically valid number of subjects from various major PAs could address the issue of understanding the phenomenon for frequency distribution pattern in geographically assorted human populations (GAHPs); however, it will be tedious and might require huge population size [16]. Therefore, the present study was focused to uncover the genetic distance based ancestral origin or genetic affinity among GAHPs to address the paradigm for GST M1-T1 null allele frequency diversity. We are currently exploring how best to do this for the large number of populations in the present analysis to understand this phenomenon of frequency distribution pattern in GAHPs. GST M1 and GST T1 loci investigated in this study have been derived from literatures representing 44 different populations affiliated from Africa, Asia, Europe, South America and the genome of Gujarat PA, a region in western India. GST M1-T1 null allele frequency of 45 GAHPs were computed for phylogenesis with pair wise genetic distance based unweighted pair group method with arithmetic mean (UPGMA) and the findings of seven patterns for GST M1-T1 null allele frequency in this study have been demonstrated for the first time with highest genetic affinity. The patterns of null allele frequencies reported in this study add insights to determine a conclusive risk association of GST M1-T1 loci with several cancer or genetic disorders.
Materials and Methods
Subjects
The present investigation includes GST M1 and GST T1 null allele frequency of 45 GAHPs from 39 studies. Null allele frequency of Gujarat population was investigated in this study from 504 healthy unrelated volunteers of Gujarati origin with a mean age of 60 years. After signing the informed consent to participate in the study, blood samples of 2 mL were collected from each subject. Data of the remaining 44 populations were collected from different populations investigated by various authors (Table 1). Several studies of same ethnicity were also gathered in the study to fulfill the statistical significance of the study and to minimize the varying frequency of polymorphism among the ethnic groups while, the data gathered from Naveen et al. [18] had allele frequencies of combined Tamilnadu and Pondicherry PAs. The study was approved by the institutional ethics committee of Shrimathi Vasantben Ratilal Desai Cancer Research Centre, Rajkot Cancer Society - India.
Table 1. GST M1-T1 null allele frequency of geographically assorted human populations.
| GST M1 Null | GST T1 Null | ||||
|---|---|---|---|---|---|
| Geographic Region (short form) | Frequency | Sample | Frequency | Sample | Reference |
| I. Eastern Asia (eAs) | 0.521 | 8931 | 0.476 | 7875 | [10] |
| 1. Japan (eAs_Jap) | 0.501 | 2215 | 0.496 | 1518 | [10] |
| 2. Korea (eAs_Kor) | 0.527 | 3704 | 0.509 | 3641 | [10] |
| 3. China (eAs_Chi) | 0.535 | 2467 | 0.443 | 2355 | [10] |
| 4. Mongolia (eAs_Mon) | 0.464 | 207 | 0.256 | 207 | [21] |
| II. South Eastern Asia (seAs) | 0.562 | 1666 | 0.351 | 890 | [10] |
| 5. Vietnam (seAs_Vie) | 0.420 | 100 | 0.300 | 100 | [22] |
| 6. Philippines (seAs_Phi) | 0.517 | 60 | 0.333 | 60 | [23] |
| 7. Indonesia (seAs_Ids) | 0.556 | 162 | 0.414 | 162 | [24] |
| 8. Singapore - Malay (seAs_S_M) | 0.653 | 167 | 0.383 | 167 | [25] |
| III. Southern Asia (sAs) | |||||
| a. India (sAs_Ind) | 0.296 | 4720 | 0.182 | 4570 | [14, 15, 18, 26–35], Present Study |
| 9. Uttar Pradesh (Ind_Up) | 0.327 | 1107 | 0.174 | 1107 | [14, 29–32] |
| 10. West Bengal (Ind_Wb) | 0.270 | 67 | 0.130 | 67 | [34] |
| 11. Gujarat (Ind_Guj) | 0.200 | 504 | 0.355 | 504 | Present Study |
| 12. Maharashtra (Ind_Mah) | 0.299 | 2060 | 0.138 | 2060 | 26–28, 31] |
| 13. Andhra Pradesh (Ind_Ap) | 0.359 | 370 | 0.254 | 370 | [18, 33] |
| 14. Karnataka (Ind_Kar) | 0.258 | 260 | 0.191 | 110 | [15, 18] |
| 15. Tamilnadu (Ind_Tn) | 0.235 | 200 | 0.188 | 200 | [18] |
| 16. Kerala (Ind_Ker) | 0.324 | 182 | 0.128 | 182 | [18, 35] |
| b. Afghanistan (sAs_Afg) | 0.460 | 656 | 0.186 | 656 | [36] |
| c. Iran (sAs_Iran) | 0.406 | NA | 0.382 | NA | [37] |
| d. Pakistan (sAs_Pak) | 0.450 | 111 | 0.230 | 111 | [38] |
| IV. Northern Europe (nEu) | 0.533 | 3686 | 0.165 | 2291 | [10] |
| 17. Sweden (nEu_Swd) | 0.546 | 747 | 0.147 | 626 | [5, 39] |
| 18. Finland (nEu_Fin) | 0.469 | 482 | 0.130 | 385 | [5] |
| 19. Denmark (nEu_Dn) | 0.536 | 537 | 0.129 | 358 | [5] |
| 20. UK (nEu_Uk) | 0.578 | 1122 | 0.205 | 922 | [5] |
| V. Western Europe (wEu) | 0.515 | 6486 | 0.183 | 5562 | [10] |
| 21. Netherlands (wEu_Ned) | 0.504 | 419 | 0.229 | 419 | [5] |
| 22. Germany (wEu_Ger) | 0.516 | 3054 | 0.173 | 3054 | [4] |
| 23. France (wEu_Fra) | 0.534 | 1184 | 0.168 | 512 | [5] |
| VI. Southern Europe (sEu) | 0.509 | 3770 | 0.195 | 2660 | [10] |
| 24. Italy (sEu_Ita) | 0.494 | 810 | 0.163 | 553 | [5] |
| 25. Spain (sEu_Spa) | 0.504 | 1132 | 0.221 | 1121 | [7] |
| 26. Slovenia (sEu_Sln) | 0.520 | 102 | 0.255 | 102 | [5] |
| 27. Greece (sEu_Gr) | 0.520 | 171 | 0.099 | 171 | [6] |
| VII. Eastern Europe (eEu) | 0.511 | 1184 | 0.188 | 1169 | [10] |
| 28. Czech Republic (eEu_Cze) | 0.567 | 67 | 0.224 | 67 | [40] |
| 29. Bulgaria (eEu_Bul) | 0.518 | 112 | 0.161 | 112 | [9] |
| 30. Poland (eEu_Pol) | 0.511 | 321 | 0.193 | 321 | [41] |
| 31. Slovakia (eEu_Slk) | 0.512 | 332 | 0.180 | 322 | [39] |
| 32. Russia (eEu_Rus) | 0.497 | 352 | 0.193 | 352 | [42] |
| VIII. Africa (Af) | |||||
| 33. North African Egypt (nAf_Egp) | 0.555 | 200 | 0.295 | 200 | [21] |
| 34. West African Nigeria (wAf_Nig) | 0.300 | 300 | 0.370 | 300 | [43] |
| 35. South African Xhosa (sAf_Xho) | 0.211 | 128 | 0.406 | 128 | [44] |
| 36. South African Namibia (sAf_Nam) | 0.112 | 134 | 0.358 | 134 | [45] |
| 37. Middle African Cameroon (mAf_Cam) | 0.278 | 126 | 0.468 | 126 | [46] |
| 38. East African Ethiopia (eAf_Eth) | 0.435 | 153 | 0.373 | 153 | [46] |
| 39. East African Somalia (eAf_Som) | 0.400 | 100 | 0.440 | 100 | [47] |
| 40. East African Zimbabwe (eAf_Zim) | 0.240 | 150 | 0.260 | 150 | [48] |
| IX. Caucasian (wAs_Cau) | 0.529 | 2714 | 0.197 | 1223 | [10] |
| X. South American Brazil (sAm_Brz) | 0.397 | 794 | 0.267 | 794 | [49, 50] |
| Total number of Samples | 36009 | 29092 | |||
DNA isolation and Genotyping
Lahiri and Nurnberger method was used to isolate genomic DNA from whole blood [19]; the Huang et al., method of multiplex polymerase chain reaction was performed to identify GST M1 and T1 polymorphism with albumin gene as internal control [20]. Amplified products of PCR were visualized in 2% agarose gel and the band patterns were analyzed for polymorphism.
Geographically assorted human populations (GAHPs)
On the basis of interest in allele frequency patterns and availability of GST M1-T1 null allele frequency data shared by all populations, we choose 45 representative geographically assorted human populations around the world from 38 investigations reported by various authors from Asia [10, 14, 15, 18, 21–38], Europe [4–7, 9, 10, 39–42], Africa [21, 37, 43–50] and South America [49, 50] as summarized in Table 1. Of these 45 GAHPs, 4 were chosen from Eastern Asia (Japan, Korea, China and Mongolia [10, 21]), 4 from South Eastern Asia (Vietnam, Philippines, Indonesia and Singapore-Malay [10, 22–25]), 8 from Southern Asia India (Tamilnadu, Kerala, Karnataka, Andhra Pradesh, Maharashtra, West Bengal, Uttar Pradesh and Gujarat [14, 15, 18, 26–35, present study]), 3 from Southern Asia (Afghanistan, Iran, Pakistan [36–38]), 4 from Northern Europe (Sweden, Finland, Denmark and UK [5, 10, 39]), 4 from Southern Europe (Italy, Spain, Slovenia and Greece [5–7, 10]), 5 from Eastern Europe (Czech Republic, Bulgaria, Poland, Slovakia and Russia [9, 10, 39–42]), 3 from Western Europe (Netherlands, Germany and France [4, 5, 10]), 8 from Africa (Egypt, Nigeria, Xhosa tribe, Namibia, Cameroon, Ethiopia, Somalia and Zimbabwe [21, 37, 43–50]), 1 from South American Brazil [49, 50] and Caucasian (Americans and Canadians [10]). The “Caucasian” population used in this study was arbitrarily termed as “West Asian Caucasians” (wAs_Cau) to precise the geographical region and allele frequency patterns. All the PAs were grouped into continental regions as per the guidelines of Statistics Division of the United Nations (http://unstats.un.org/unsd/methods/m49/m49regin.htm [51]). Initially, 20 different continental region populations, as summarized in Table 2, were used in phylogenetic analysis for GST M1-T1 null allele frequency to minimize the effect of inbreeding [52]. Of these 20 continental region populations, the null allele frequency of thirteen populations such as Afghanistan, Iran, Pakistan (3 from South Asia), Brazil (1 from South America), Egypt (1 from North Africa), Nigeria (1 from West Africa), Xhosa tribe, Namibia (2 from South Africa), Cameroon (1 from Middle Africa), Ethiopia, Somalia and Zimbabwe (3 from East Africa), Caucasian (West Asia, as described earlier) and the average null allele frequency of seven continental regions such as East Asia (Japan, Korea, China, Mongolia), South East Asia (Vietnam, Philippines, Indonesia, Singapore-Malay), South Asia India (Tamilnadu, Kerala, Karnataka, Andhra Pradesh, Maharashtra, West Bengal, Uttar Pradesh, Gujarat), North Europe (Sweden, Finland, Denmark, UK), West Europe (Netherlands, Germany, France), South Europe (Italy, Spain, Slovenia, Greece) and East Europe (Czech Republic, Bulgaria, Poland, Slovakia, Russia) were used in the phylogenetic analyses, as summarized in Table 1 for GST M1-T1 null allele frequency. Since, some of the populations have considerable gene admixture from other PAs [52] another phylogenetic analysis was performed using all the 45 GAHPs for GST M1-T1 null allele frequency (Table 3).
Table 2. GST M1-T1 null allele frequency based genetic distance between 20 different continental region populations.
| S. No. | Populations | eAf_Zim | sAf_Nam | sAf_Xho | mAf_Cam | wAf_Nig | sAs_Ind | sAm_Brz | eAf_Som | sAs_Iran | eAf_Eth | sAs_Pak | sAs_Afg | sEu | eEu | wEu | eAs | wAs_Cau | nEu | nAf_Egt | seAs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | eAf_Zim | ||||||||||||||||||||
| 2 | sAf_Nam | 0.019641 | |||||||||||||||||||
| 3 | sAf_Xho | 0.018002 | 0.007702 | ||||||||||||||||||
| 4 | mAf_Cam | 0.036741 | 0.028132 | 0.006642 | |||||||||||||||||
| 5 | wAf_Nig | 0.011698 | 0.025018 | 0.007576 | 0.009135 | ||||||||||||||||
| 6 | sAs_Ind | 0.007203 | 0.050393 | 0.046757 | 0.068305 | 0.027352 | |||||||||||||||
| 7 | sAm_Brz | 0.019686 | 0.071517 | 0.047429 | 0.050077 | 0.017980 | 0.012453 | ||||||||||||||
| 8 | eAf_Som | 0.047449 | 0.069856 | 0.031371 | 0.014241 | 0.013198 | 0.062987 | 0.027019 | |||||||||||||
| 9 | sAs_Iran | 0.033673 | 0.067958 | 0.033278 | 0.022080 | 0.010096 | 0.040405 | 0.011558 | 0.003247 | ||||||||||||
| 10 | eAf_Eth | 0.041147 | 0.083845 | 0.045069 | 0.031565 | 0.016495 | 0.043599 | 0.010910 | 0.005520 | 0.000882 | |||||||||||
| 11 | sAs_Pak | 0.037341 | 0.107807 | 0.078333 | 0.079443 | 0.037756 | 0.020062 | 0.003628 | 0.042121 | 0.021916 | 0.017784 | ||||||||||
| 12 | sAs_Afg | 0.044754 | 0.124665 | 0.097123 | 0.102102 | 0.052050 | 0.021304 | 0.008620 | 0.060075 | 0.035118 | 0.029735 | 0.001511 | |||||||||
| 13 | sEu | 0.064578 | 0.155621 | 0.119309 | 0.117913 | 0.066300 | 0.036737 | 0.015131 | 0.064593 | 0.039712 | 0.031804 | 0.003921 | 0.002064 | ||||||||
| 14 | eEu | 0.066210 | 0.158879 | 0.122818 | 0.122007 | 0.069013 | 0.037348 | 0.016296 | 0.067829 | 0.042182 | 0.034056 | 0.004511 | 0.002175 | 0.000038 | |||||||
| 15 | wEu | 0.068642 | 0.163151 | 0.126834 | 0.126154 | 0.072004 | 0.038764 | 0.017691 | 0.070734 | 0.044494 | 0.036072 | 0.005254 | 0.002530 | 0.000130 | 0.000031 | ||||||
| 16 | eAs | 0.112148 | 0.157404 | 0.093422 | 0.056769 | 0.057067 | 0.119908 | 0.055200 | 0.015755 | 0.021559 | 0.017606 | 0.060443 | 0.079037 | 0.071492 | 0.074740 | 0.077050 | |||||
| 17 | wAs_Cau | 0.114150 | 0.162684 | 0.097673 | 0.060689 | 0.059434 | 0.120082 | 0.054782 | 0.017359 | 0.022368 | 0.017846 | 0.058754 | 0.076677 | 0.068473 | 0.071609 | 0.073799 | 0.000099 | ||||
| 18 | nEu | 0.079650 | 0.181776 | 0.143916 | 0.143304 | 0.084726 | 0.045716 | 0.023990 | 0.082505 | 0.054067 | 0.044384 | 0.008885 | 0.004708 | 0.001084 | 0.000751 | 0.000480 | 0.085835 | 0.082097 | |||
| 19 | nAf_Egt | 0.087395 | 0.174577 | 0.121158 | 0.102434 | 0.066203 | 0.066198 | 0.023311 | 0.043045 | 0.028058 | 0.019145 | 0.013337 | 0.017263 | 0.009810 | 0.010731 | 0.011240 | 0.032074 | 0.029501 | 0.013382 | ||
| 20 | seAs | 0.098593 | 0.178065 | 0.117995 | 0.091480 | 0.065434 | 0.083794 | 0.031293 | 0.033343 | 0.024411 | 0.015989 | 0.023986 | 0.031810 | 0.022755 | 0.024282 | 0.025180 | 0.016697 | 0.014669 | 0.028655 | 0.002821 |
The values represented in the table were computed between the population affiliations by Nei’s (1972) standard genetic distance (DST) method and were used in phylogenetic tree of 20 different continental regions population affiliations for GST M1-T1 null allele frequency (Fig. 1). Abbreviations used were same as those in Table 1.
Table 3. GST M1-T1 null allele frequency based genetic distance between 45 geographically assorted human populations (GAHPs).
| S. No. | GAHPs | eAf_Zim | sAf_Nam | Ind_Guj | sAf_Xho | Ind_Tn | Ind_Kar | Ind_Wb | mAf_Cam | Ind_Mah | wAf_Nig | Ind_Ker | Ind_Up | Ind_Ap | sAm_Brz |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | eAf_Zim | ||||||||||||||
| 2 | sAf_Nam | 0.019641 | |||||||||||||
| 3 | Ind_Guj | 0.008566 | 0.004982 | ||||||||||||
| 4 | sAf_Xho | 0.018002 | 0.007702 | 0.002144 | |||||||||||
| 5 | Ind_Tn | 0.003500 | 0.033450 | 0.022055 | 0.037140 | ||||||||||
| 6 | Ind_Kar | 0.003729 | 0.037771 | 0.023632 | 0.038408 | 0.000355 | |||||||||
| 7 | Ind_Wb | 0.012420 | 0.058122 | 0.041785 | 0.061245 | 0.003252 | 0.002526 | ||||||||
| 8 | mAf_Cam | 0.036741 | 0.028132 | 0.014999 | 0.006642 | 0.063507 | 0.062461 | 0.090338 | |||||||
| 9 | Ind_Mah | 0.013628 | 0.064129 | 0.044263 | 0.063067 | 0.004934 | 0.003307 | 0.000574 | 0.088816 | ||||||
| 10 | wAf_Nig | 0.011698 | 0.025018 | 0.007718 | 0.007576 | 0.026876 | 0.025085 | 0.042346 | 0.009135 | 0.040441 | |||||
| 11 | Ind_Ker | 0.018509 | 0.075817 | 0.052630 | 0.072183 | 0.008654 | 0.006243 | 0.002038 | 0.096709 | 0.000540 | 0.045207 | ||||
| 12 | Ind_Up | 0.011897 | 0.062884 | 0.039784 | 0.055683 | 0.006375 | 0.003826 | 0.003100 | 0.075196 | 0.001229 | 0.031146 | 0.001306 | |||
| 13 | Ind_Ap | 0.011155 | 0.056511 | 0.029903 | 0.039082 | 0.013492 | 0.009762 | 0.014788 | 0.047057 | 0.011066 | 0.014791 | 0.011238 | 0.005068 | ||
| 14 | sAm_Brz | 0.019686 | 0.071517 | 0.039623 | 0.047429 | 0.023229 | 0.018123 | 0.023228 | 0.050077 | 0.017729 | 0.017980 | 0.016579 | 0.009579 | 0.001318 | |
| 15 | eAf_Som | 0.047449 | 0.069856 | 0.039132 | 0.031371 | 0.071354 | 0.065680 | 0.087771 | 0.014241 | 0.080505 | 0.013198 | 0.082462 | 0.062745 | 0.031855 | 0.027019 |
| 16 | sAs_Iran | 0.033673 | 0.067958 | 0.035776 | 0.033278 | 0.050368 | 0.044624 | 0.060441 | 0.022080 | 0.053470 | 0.010096 | 0.054000 | 0.038840 | 0.015631 | 0.011558 |
| 17 | seAs_Vie | 0.027183 | 0.078906 | 0.043887 | 0.048612 | 0.034281 | 0.028238 | 0.035694 | 0.045118 | 0.028849 | 0.017602 | 0.027294 | 0.018065 | 0.004738 | 0.001318 |
| 18 | eAf_Eth | 0.041147 | 0.083845 | 0.047123 | 0.045069 | 0.056782 | 0.049923 | 0.064060 | 0.031565 | 0.055751 | 0.016495 | 0.054832 | 0.040296 | 0.016788 | 0.010910 |
| 19 | sAs_Pak | 0.037341 | 0.107807 | 0.067666 | 0.078333 | 0.036617 | 0.029669 | 0.029982 | 0.079443 | 0.022419 | 0.037756 | 0.018481 | 0.013929 | 0.007683 | 0.003628 |
| 20 | sAs_Afg | 0.044754 | 0.124665 | 0.082681 | 0.097123 | 0.039341 | 0.032172 | 0.028470 | 0.102102 | 0.020874 | 0.052050 | 0.015882 | 0.014104 | 0.012496 | 0.008620 |
| 21 | eAs_Mon | 0.041766 | 0.111479 | 0.069331 | 0.077606 | 0.043779 | 0.036213 | 0.038385 | 0.074231 | 0.029918 | 0.036336 | 0.025780 | 0.019540 | 0.009564 | 0.004097 |
| 22 | nEu_Fin | 0.056413 | 0.147166 | 0.103438 | 0.122646 | 0.045284 | 0.037978 | 0.029516 | 0.132931 | 0.022041 | 0.072547 | 0.015927 | 0.017497 | 0.021698 | 0.018273 |
| 23 | sEu_Ita | 0.061485 | 0.153594 | 0.106118 | 0.122436 | 0.053407 | 0.045069 | 0.038414 | 0.126364 | 0.029524 | 0.070064 | 0.022816 | 0.022503 | 0.022062 | 0.016456 |
| 24 | eEu_Rus | 0.059262 | 0.147482 | 0.099679 | 0.113354 | 0.054266 | 0.045694 | 0.041407 | 0.113525 | 0.032070 | 0.062336 | 0.025631 | 0.023486 | 0.019501 | 0.013120 |
| 25 | eAs_Jap | 0.110350 | 0.146520 | 0.099722 | 0.084594 | 0.142570 | 0.132315 | 0.158417 | 0.048140 | 0.144938 | 0.053266 | 0.143253 | 0.118214 | 0.072885 | 0.059379 |
| 26 | wEu_Ned | 0.059823 | 0.144141 | 0.095375 | 0.105798 | 0.058632 | 0.049635 | 0.048197 | 0.101024 | 0.038141 | 0.055834 | 0.031893 | 0.027464 | 0.018923 | 0.011362 |
| 27 | sEu_Spa | 0.060246 | 0.145849 | 0.097044 | 0.108186 | 0.058185 | 0.049224 | 0.047092 | 0.104342 | 0.037126 | 0.057711 | 0.030768 | 0.026857 | 0.019275 | 0.011877 |
| 28 | eEu_Pol | 0.065713 | 0.157598 | 0.107503 | 0.121115 | 0.060599 | 0.051493 | 0.046757 | 0.119726 | 0.036775 | 0.067624 | 0.029742 | 0.027621 | 0.023028 | 0.015739 |
| 29 | eEu_Slk | 0.067544 | 0.161727 | 0.111408 | 0.126171 | 0.061054 | 0.051968 | 0.046119 | 0.126173 | 0.036234 | 0.071687 | 0.028989 | 0.027678 | 0.024546 | 0.017452 |
| 30 | wEu_Ger | 0.070266 | 0.166596 | 0.115586 | 0.130954 | 0.063009 | 0.053801 | 0.047246 | 0.131318 | 0.037254 | 0.075319 | 0.029750 | 0.028878 | 0.026377 | 0.019164 |
| 31 | seAs_Phi | 0.070253 | 0.140613 | 0.090160 | 0.090823 | 0.080995 | 0.070962 | 0.078774 | 0.072213 | 0.066801 | 0.045336 | 0.061492 | 0.049833 | 0.027442 | 0.016743 |
| 32 | eEu_Bul | 0.072759 | 0.171473 | 0.120080 | 0.136502 | 0.064240 | 0.055011 | 0.047413 | 0.138030 | 0.037460 | 0.079764 | 0.029721 | 0.029583 | 0.028364 | 0.021260 |
| 33 | sEu_Sln | 0.066941 | 0.151257 | 0.099936 | 0.107876 | 0.068535 | 0.058783 | 0.059143 | 0.098202 | 0.048016 | 0.056728 | 0.041428 | 0.035289 | 0.022831 | 0.013657 |
| 34 | sEu_Gre | 0.084181 | 0.192766 | 0.140787 | 0.162602 | 0.069434 | 0.060400 | 0.047888 | 0.171015 | 0.038494 | 0.101709 | 0.029929 | 0.033419 | 0.038936 | 0.032905 |
| 35 | eAs_Kor | 0.131072 | 0.171461 | 0.120063 | 0.103009 | 0.165454 | 0.153899 | 0.180971 | 0.061388 | 0.165744 | 0.067925 | 0.162999 | 0.136593 | 0.087477 | 0.071632 |
| 36 | wAs_Cau | 0.114150 | 0.162684 | 0.110741 | 0.097673 | 0.142911 | 0.131528 | 0.153814 | 0.060689 | 0.139138 | 0.059434 | 0.135556 | 0.112713 | 0.069730 | 0.054782 |
| 37 | wEu_Fra | 0.079786 | 0.181720 | 0.127681 | 0.143431 | 0.071836 | 0.061962 | 0.054405 | 0.142357 | 0.043640 | 0.084247 | 0.035309 | 0.034781 | 0.032085 | 0.023807 |
| 38 | eAs_Chi | 0.107068 | 0.162350 | 0.109199 | 0.098764 | 0.132097 | 0.120617 | 0.139605 | 0.064352 | 0.125031 | 0.057523 | 0.120667 | 0.100165 | 0.060995 | 0.046306 |
| 39 | nEu_Dnk | 0.086463 | 0.194885 | 0.140372 | 0.159569 | 0.074495 | 0.064731 | 0.053988 | 0.162804 | 0.043567 | 0.097621 | 0.034687 | 0.036500 | 0.038115 | 0.030493 |
| 40 | nEu_Swd | 0.088823 | 0.197255 | 0.141108 | 0.158695 | 0.078575 | 0.068343 | 0.058687 | 0.158719 | 0.047608 | 0.096026 | 0.038515 | 0.039288 | 0.038475 | 0.029849 |
| 41 | nAf_Egt | 0.087395 | 0.174577 | 0.117293 | 0.121158 | 0.093461 | 0.082045 | 0.085005 | 0.102434 | 0.071600 | 0.066203 | 0.063981 | 0.055069 | 0.035784 | 0.023311 |
| 42 | seAs_Ids | 0.110068 | 0.176501 | 0.119385 | 0.111506 | 0.131239 | 0.118996 | 0.134139 | 0.077907 | 0.118852 | 0.064351 | 0.112960 | 0.094840 | 0.058800 | 0.043301 |
| 43 | eEu_Cze | 0.093352 | 0.195596 | 0.135981 | 0.146458 | 0.091159 | 0.079672 | 0.075757 | 0.134876 | 0.062724 | 0.084693 | 0.053635 | 0.049608 | 0.038780 | 0.027114 |
| 44 | nEu_Uk | 0.100862 | 0.209262 | 0.147632 | 0.159813 | 0.096473 | 0.084651 | 0.078719 | 0.149197 | 0.065400 | 0.094779 | 0.055589 | 0.052818 | 0.043805 | 0.031824 |
| 45 | seAs_S_M | 0.171196 | 0.272027 | 0.195663 | 0.189285 | 0.188418 | 0.171818 | 0.180613 | 0.146069 | 0.160261 | 0.120939 | 0.148990 | 0.133165 | 0.096023 | 0.074061 |
The values represented in the table were computed between the population affiliations by Nei’s (1972) standard genetic distance (DST) method and were used in phylogenetic tree of 45 geographically assorted human populations for GST M1-T1 null allele frequency (Fig. 2). Abbreviations used were same as those in Table 1.
Statistical analysis
Distributions of GST M1 and GST T1 null alleles in Gujarati population were calculated by frequency counting method in SPSS-21 (4-27AEA) for windows. The standard genetic distance (DST) between different PAs for GST M1-T1 null allele frequencies were calculated by Nei’s (1972) method in Phylip 3.69 version [52, 53]. Least DST values between the PAs were used to compute clades with more than 50% of 1000 bootstrap replicates by Felsenstein (1989) method and then the phylogenetic trees were constructed in Mega-6 software by UPGMA method [54–56]. Finally, the clusters of PAs split found among the geographically assorted human populations in phylogenetic tree were used in the scattered plot to analyze their geographical distribution. The longitude (X-axis) and latitude (Ys-axis) of different continental regions were used to construct the scattered plot in SPSS-21 as summarized in Table 4. The online web source world atlas was used to compute the latitude and longitude of the respective geographic locations (http://www.worldatlas.com/aatlas/latitude_and_longitude_finder.htm [57]).
Table 4. Geographical location of population affiliations from 20 different continental regions used in scattered plot analyses.
| Location a | Latitude | Longitude |
|---|---|---|
| sAf_Xho | - 30.33 | 22.56 |
| eAf_Zim | - 19.0 | 29.9 |
| nAf_Egp | 26.49 | 30.48 |
| eAf_Eth | 9.8 | 40.29 |
| wAf_Nig | 9.4 | 8.4 |
| mAf_Cam | 7.22 | 12.21 |
| eAf_Som | 5.9 | 46.11 |
| sAf_Nam | - 22.57 | 18.29 |
| sAs_Iran | 32.25 | 53.41 |
| sAs_Pak | 30.22 | 69.20 |
| sAs_Af | 33.56 | 67.42 |
| sAs_Ind | 20.35 | 78.57 |
| eAs | 22.16 | 114.14 |
| seAs | 11.35 | 121.37 |
| wAs_Cau | 43 | 43.45 |
| nEu | 62.16 | 12.20 |
| wEu | 46.12 | 1.15 |
| sEu | 41.16 | - 1.12 |
| eEu | 59.80 | 36.29 |
| sAm _Brz | -14.14 | - 51.55 |
a Latitudes in the northern hemisphere were listed with positive values, as were longitudes in the eastern hemisphere; Latitudes in the southern hemisphere were listed with negative values, as in longitudes of western hemisphere. Abbreviations used were same as those in Table 1.
Results
Phylogenetic tree for GST M1-T1 null allele frequency in GAHPs
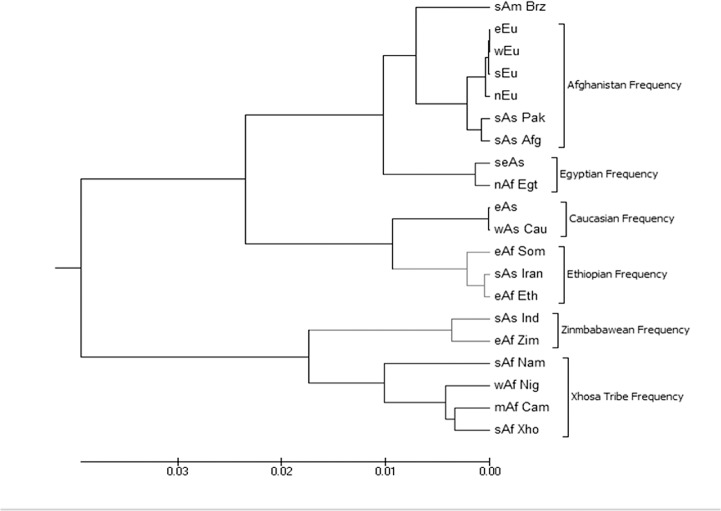
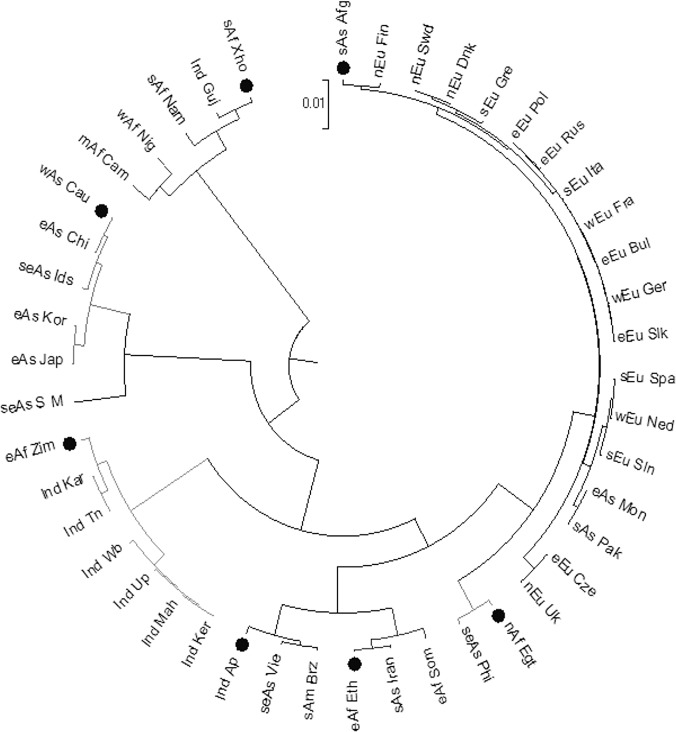
The frequency of GST M1 and T1 null genotypes in Gujarat populations of India was observed as 0.200 and 0.355 respectively in this present study. The pair wise genetic distance matrix computed for GST M1-T1 null allele frequency between the respective PAs using Nei’s (1972) standard genetic distance method (DST) in gendist program of Phylip-3.69 software for the 20 different continental regions PAs and the 45 GAHPs described elsewhere were summarized in Tables 2 and 3 respectively. The phylogenetic analyses of 20 different continental regions (Fig. 1) and 45 GAHPs (Fig. 2) for GST M1-T1 null allele frequency were performed using the pair wise genetic distance matrix by UPGMA method in MEGA-6 software [52, 56]. Consense program that clustered more than 50% of 1000 bootstrap replicates was used to assess the reliability of the constructed phylogenetic trees [53, 54]. The Nei’s DST value varies from 0.0001 to 0.007 (Table 2) and 0.0006 to 0.008 (Table 3) for GST M1-T1 null allele frequency among the 20 different continental regions PAs and the 45 GAHPs respectively. The phylogenetic trees in Figs. 1 and 2 showed consistent clusters or patterns for GST M1-T1 null allele frequency from populations such as South African Xhosa tribe, South African Zimbabwe, East African Ethiopia, North African Egypt, Caucasian and South Asian Afghanistan. The least DST value of PAs from South Asian Iran (0.000882) and East African Somalia (0.00552) with East African Ethiopian allele frequency grouped them as one of the clusters in the phylogenetic trees for GST M1-T1 null allele frequency. Few of the African PAs such as West African Nigeria, Middle African Cameroon and South African Namibia were clustered with South African Xhosa tribe allele frequency with least DST value that ranged from 0.006642 to 0.007702. In addition, the South Asian Indian Gujarat PAs (0.00214) was observed with highest affinity to South African Xhosa cluster than any other clusters (Fig. 2). The average GST M1-T1 null allele frequency in South Asian Indian PA was observed as a cluster to East African Zimbabwe allele frequency (Fig. 1) with least DST value (0.007203) out of 20 continental regions. Further, the least DST value to East African Zimbabwe allele frequency was observed with South Asian Indian PAs (Table 3) from Tamilnadu (0.0035) and Karnataka (0.003729). Furthermore, PA from Karnataka was observed with least DST value to Maharashtra allele frequency (0.003307), which was clustered with allele frequency from West Bengal (0.000574), Uttar Pradesh (0.003826) and Kerala PAs of South Asian India (0.006243) as shown in Fig. 2. Nevertheless, the South Asian Indian Andhra Pradesh allele frequency was observed with least DST value only to Uttar Pradesh PA (0.005068) than any other Indian PAs for GST M1-T1 null allele frequency and stood as a separate cluster in the phylogenetic tree with highest genetic affinity to South America (Brazil, 0.001318) and South East Asia allele frequency (Vietnam, 0.004738) from geographically distant continental regions.
Fig 1. Phylogenetic tree of 20 different continental regions population affiliations for GST M1-T1 null allele frequency.
The tree was produced by the UPGMA method from DST values in Table 2 and cluster with more than 50% of 1000 bootstrap replicates were included in the consensus tree obtained by Felsenstein (1989) phylogeny interference package. Major group of GST M1-T1 null allele frequencies were from population of Xhosa tribe, Zimbabwe, Ethiopia, Egypt, Afghanistan and Caucasian. Abbreviations used were same as those in Table 1.
Fig 2. Phylogenetic tree of 45 geographically assorted human populations for GST M1-T1 null allele frequency.
This tree was based on DST values in Table 3. Other aspects were same as those in Fig. 1. Major group of GST M1-T1 null allele frequencies were from population of Xhosa tribe, Zimbabwe, Ethiopia, Egypt, Afghanistan, Caucasian and Andhra Pradesh. Abbreviations used were same as those in Table 1.
PAs from China (0.000757), Japan (0.001457) and Korea (0.001522) of East Asia and Pakistan (0.001511) of South Asia were observed as another cluster with least DST value to Caucasians [Americans and Canadians (10)] for GST M1-T1 null allele frequency out of 45 combinations (Fig. 2). However, Mongolia of East Asia was observed with least DST value to Pakistan allele frequency (0.000686) than the Caucasians (0.046671). The least DST value between European continental regions and South Asian Afghanistan allele frequency that ranged from 0.002064 to 0.004708 was clustered together for GST M1-T1 null allele frequency in the phylogenetic tree of 20 different continental regions (Fig. 1). Nevertheless, the phylogenetic tree of 45 GAHPs (Fig. 2) clustered only 13 European PAs (Sweden, Finland, Denmark, Netherlands, Germany, France, Italy, Spain, Greece, Bulgaria, Poland, Slovakia and Russia) out of 16 investigated in this study with South Asian Afghanistan allele frequency (least DST value that ranged from 0.001176 to 0.00723) while, the other 3 PAs [Slovenia (0.002462), Czech Republic (0.004116) and UK (0.006718)] were clustered with North African Egypt allele frequency. Singapore-Malay and Indonesia PAs from South East Asia were observed with least DST value to East Asia (China, 0.001246) and Caucasian (0.003788) respectively for GST M1-T1 null allele frequency. Nevertheless, the other counter parts from same continental region were observed as the most diverse PAs with PA admixture from North Africa (Egypt, 0.002571) and East Africa (Ethiopia, 0.007943) for Philippines; South Asia India (Andhra Pradesh, 0.004738) and East Africa (Ethiopia, 0.004922) for Vietnam among the 45 GAHPs investigated in this study for GST M1-T1 null allele frequency as shown in Table 3 and Fig. 2 respectively.
GST M1-T1 null allele frequency patterns among the GAHPs
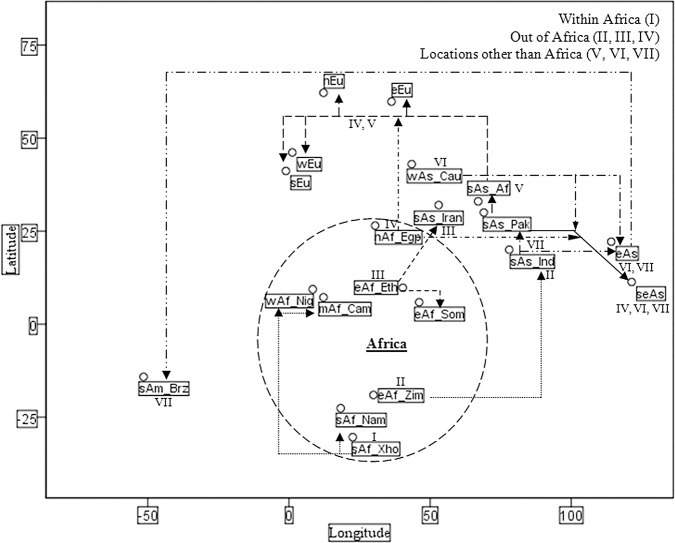
The effect of isolation by geographical distance in population differentiation [51] was validated in a scattered plot with respect to the phylogenetic clusters of 45 GAHPs for GST M1-T1 null allele frequency that corresponds to the latitudes and longitudes of 20 different continental regions representing PAs from Africa, Asia, Europe and America (Table 4). The scattered plot illustrated in Fig. 3 suggest three major geographical split for the seven GST M1-T1 null allele frequency clusters or patterns observed in the phylogenetic tree of 45 GAHPs (Fig. 2). South African Xhosa allele frequency pattern (I) observed mostly in continental regions of Africa suggest an “Africa” split in the scattered plot with least population differentiation to Nigeria (West Africa), Cameroon (Middle Africa) and Namibia (South Africa). However, the GST M1-T1 null allele frequency patterns of PAs from other African continental region such as East African Zimbabwe (II), East African Ethiopia (III) and North African Egypt (IV) were observed with least population differentiation to PAs from non African continental regions such as South Asia (India and Iran), South East Asia (Philippines and Vietnam), Southern Europe (Slovenia), Eastern Europe (Czech Republic) and Northern Europe (UK) irrespective of the geographical isolation suggest an “out of Africa” split. Finally, the remaining three non-African GST M1-T1 null allele frequency patterns observed from Caucasian (V), South Asian Afghanistan (VI) and South Asian Indian Andhra Pradesh (VII) were geographically distributed in different continental regions such as Asia, Europe and America with the exception of Africa suggest an “other than Africa” split among the 45 GAHPs in the scattered plot.
Fig 3. Geographic Location of GST M1-T1 null allele frequency patterns in Scattered Plot.
This scenario is largely a speculation based on Nei’s DST based phylogenetic trees in Figs. 1 and 2, which suggest seven patterns for GST M1-T1 null allele frequency among the 45 geographically assorted human populations. The scattered plot shows the genetic affinity and geographical distribution of the seven probable patterns with three major split among the continental regions such as an “African” split with South African Xhosa pattern (I), an “out of Africa” split with East African Zimbabwe (II), East African Ethiopia (III), North African Egypt (IV) patterns and an “other than Africa” split with South Asian Afghanistan (V), West Asian Caucasian (VI), South Asian Indian Andhra Pradesh patterns (VII). Abbreviations used were same as those in Table 1.
Discussion
Understanding the genetic variation and diversity among the geographically assorted human populations (GAHPs) is an interesting topic in population genetics with a wide range of neutral genetic markers and adaptive markers being employed to uncover the patterns of genetic diversity [58]. To address the extent of diversity in allele frequency distribution among populations from different ethnicity, region, country or continent is difficult, nevertheless understanding this phenomenon is inevitable [8, 11, 16, 17] and recent advances in the molecular techniques excel the perspective of inter-individual genetic variations in GAHPs. Allele frequencies of large number of neutral markers or of even few candidate markers that duplicate or decay to favor new environments and lead to rapid adaptations are often used for investigating the patterns [58]. The paradigm of allele frequency among the populations holds the key to unlock the existing problem of inter-individual genetic variation in xenobiotic metabolizing enzymes (XMEs) and in particular the decay or null allele frequency of Glutathione-S-transferase’s classes such as Mu 1 (GST M1) and Theta 1 (GST T1), which are considered as the major risk factor for various diseases including several types of cancers [8–10]. Therefore, the present investigation analyzed the pattern for GST M1-T1 null allele frequency among GAHPs using a phylogenetic approach. A set of 20 different continental region PAs (Table 2) and 45 GAHPs (Table 3) were recruited for GST M1-T1 null allele frequency data from 38 previously reported works and genomic data of Indian Gujarat PA in this study (Table 1) and the respective phylogenetic trees (Figs. 1 and 2) have been constructed by UPGMA method based on Nei’s (1972) standard genetic distance (DST) with clusters more than 50% of 1000 bootstrap replicates obtained by Felsenstein (1989) program [52, 54, 56]. In addition to the ancestral origin or genetic affinity based clusters for GST M1-T1 null allele frequency demonstrated in the phylogenetic trees, a positive correlation between the genetic distance and geographical distance were analyzed for the effect of isolation in population differentiation by distance in a scattered plot (Fig. 3). Indeed, the observations from phylogenetic trees and scattered plot of different PAs constructively reveals the findings of seven probable patterns for GST M1-T1 null allele frequency among the GAHPs in concordance to the reports of archeological signatures, ancient gene flows and sex-specific components [59–62].
The genetic affinity and geographical distribution of 20 different continental regions that included 45 GAHPs investigated in this study (Figs. 1–3) revealed the findings of an allele frequency pattern for GST M1-T1 null genotypes among Namibia (South Africa), Nigeria (West Africa), Cameroon (Middle Africa), Gujarat (South Asian Indian) and Xhosa tribe (South Africa) for the first time. We report here, the findings of Xhosa allele frequency (I) with major genetic affinity towards populations from Africa (Namibia, Nigeria, Cameroon) as an “Africa” split pattern for GST M1-T1 null allele frequency in agreement to the reports of linkage disequilibrium computed for loss of variants in GST classes by Polimanti et al. (2013). The observations in phylogenetic trees (Figs. 1 and 2) and scattered plot analysis (Fig. 3) demonstrated the findings of another three patterns such as East Africa Zimbabwean allele frequency - II in population from India (South Asia), East Africa Ethiopian allele frequency - III in populations from Iran (South Asia) and Somalia (East Africa) and North Africa Egyptian allele frequency - IV in populations from Slovenia (Southern Europe), Czech Republic (Eastern Europe) and UK (Northern Europe) for GST M1-T1 null genotypes. The findings of Ethiopian - III and Egyptian - IV allele frequency pattern for GST M1-T1 null genotypes in this study are in concordance to the earlier reports of genome wide diversity study in the Levant by Haber et al., (2013), who found two major groups with one close to Africans and Middle Easterners and the other closer to modern day Europeans [61]. Further, the findings of these Zimbabwean, Ethiopian and Egyptian patterns from African populations with high genetic affinity towards non-African populations for GST M1-T1 null allele frequency have been reported as an “Out of Africa” split in this study in corroborate to the findings of Templeton (2002), who reported the out-of-Africa theory of migration and the ancestral root of allele frequency admixture [63].
The allele frequency of population from South Asian Afghanistan with high genetic affinity to majority of European PAs investigated in this study (Tables 2 and 3 and Figs. 2 and 3) has been reported as pattern - V for GST M1-T1 null genotypes in accordance to the reports of various authors [60, 62, 64]. Further, population in Pakistan (South Asia) has been reported with Afghanistan (South Asia) pattern for GST M1-T1 null allele frequency (Table 3) in this study, though it was found with genetic affinity to PAs from Mongolia (East Asia), Europe (South, East and West) and Andhra Pradesh (South India) in corroborate to the earlier reports of Templeton (2002), who stated the findings of considerable overlap among East Asians, Europeans and South Indian populations [64]. Moreover, the pattern of allele frequency from Caucasians (Americans and Canadians) found among East Asians (Fig. 1) in this study has been identified as pattern - VI for GST M1-T1 null allele frequency and reported for the first time. Finally, the allele frequency from South Indian Andhra Pradesh PA was found with least genetic distance (Table 3) to populations from Pakistan (South Asia), Vietnam (South East Asia) and Brazil (South America) irrespective of the phenomenon of population differentiation by geographical isolation [52] and has been reported as pattern - VII for GST M1-T1 null genotypes (Figs. 2 and 3) among the 45 GAHPs in this study. These observations of South East Asian and South American PAs with the null allele frequency pattern from South Indian Andhra Pradesh PA are in agreement to the reports of agro-pastoral system in South India that acted as agricultural center and source of dispersion to lineages from different preexisting populations [60]. Furthermore, the reported East Africa patterns from Zimbabwe (II), Ethiopia (III) among India, Iran (South Asia) and South Asian pattern from South Indian Andhra Pradesh (VII) among Vietnam (South East Asia) populations in this study for GST M1-T1 null allele frequency are in concordance to the reports of migration pattern of Homo sapiens from East Africa with the demographic expansions by Field and Lahr (2006), who investigated the geographic information systems during oxygen isotope stage 4 [62]. GST M1-T1 null allele frequency from South East Asian PAs has been reported as the complex admixture of Zimbabwe (II), Ethiopia (III) and Andhra Pradesh (VII) patterns in this study. Finally, the scattered plot analysis (Fig. 3), clearly demonstrates the findings of allele frequency patterns from South Asian Afghanistan - V, Caucasian - VI and South Indian Andhra Pradesh - VII as an “Other than Africa” split among 45 GAHPs for GST M1-T1 null genotypes with respect to their geographical distribution. This observation of other than Africa split in this study has been reported here in agreement to the concepts of later migration of the populations in regions other than Africa [60, 64]. In conclusion, the data of seven patterns for GST M1-T1 null allele frequency from Xhosa tribe (I), Zimbabwe (II), Ethiopia (III), Egypt (IV), Afghanistan (V), Caucasian (VI) and South Indian Andhra Pradesh (VII) reported in this study compare constructively with the earlier studies that suggested the PAs of relatively recent origin show comparatively small genetic differences and high genetic affinity among them [11, 46, 52, 58]. Findings of these seven patterns (I-VII) for GST M1-T1 null allele frequency reported here, would shed some light to address the missing link in most of the genomic epidemiological studies that lacks conclusive risk association [9, 16, 17]. The “Africa” (I), “Out of Africa” (II, III and IV) and “Other than Africa” (V, VI and VII) split among the 45 GAHPs reported in this study have to be explored further to rationalize the GST M1-T1 null allele’s frequency patterns in world populations.
Acknowledgments
The authors wish to thank the participants of this study for their cooperation; Dr. V. K. Gupta, Director Shree Gulabkuvar Talakchand Sheth Cancer hospital, Rajkot for providing technical guidance; Shree Manibhai Virani and Shremathi Navalben Virani Science College, Rajkot, Gujarat, India and Department of Animal Science, Bharathidasan University, Tiruchirappalli, Tamilnadu, India for providing the support in sample collection and facilities respectively. The Authors are grateful to Dr. Akbar Ali Khan Pathan, Assistant Professor, Genome Research Chair, Department of Biochemistry, College of Science, King Saud University, Riyadh-11451, Kingdom of Saudi Arabia for facilitating data analysis and for Dr. K Balakrishnan, Associate Professor, Department of Immunology, Madurai Kamaraj University, Madurai, Tamilnadu - 625 021, India for his critical suggestions.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1. Strange RC, Spiter MA,Ramachandran S,Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21–26. [DOI] [PubMed] [Google Scholar]
- 2. Kim WJ, Kim H,Kim CH,Lee MS,Oh BR,Lee HM, et al. GSTT1-null genotype is a protective factor against bladder cancer. Urology 2002;60:913–918. [DOI] [PubMed] [Google Scholar]
- 3. Thier R, Bruning T, Roos PH, Rihs H, Golka K, Ko Y, et al. Markers of genetic susceptibility in human environmental hygiene and toxicology: the role of selected CYP, NAT and GST genes. Int J Hyg Environ Health 2003;206:149–171. [DOI] [PubMed] [Google Scholar]
- 4. Abbas A, Delvinquiere K, Lechevrel M, Lebailly P, Gauduchon P, Launoy G, et al. GSTM1, GSTT1, GSTP1 and Cyp1A1 genetic polymorphisms and susceptibility to esophageal cancer in a French population: different pattern of squamous cell carcinoma and adenocarcinoma. World J Gastroenterol 2004;10:3389–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bu H, Rosdahl I, Holmdahl-Kallen K, Sun XF, Zhang. Significance of glutathione S-transferase M1, T1 and P1 polymorphisms in Swedish melanoma patients. Oncol Rep. 2007;17:859–864. [DOI] [PubMed] [Google Scholar]
- 6. Dialyna IA, Miyakis S, Georgatou N, Spandidos DA. Genetic polymorphisms of CYP1A1, GSTM1 and GSTT1 genes and lung cancer risk. Oncol Rep. 2003;10:1829–1835. [PubMed] [Google Scholar]
- 7. Garcia-Closas M, Malats N, Silverman D, Dosemeci M,Kogevinas M,Hein DW, et al. NAT2 slow Acetylation, GSTM1 null genotype and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 2005;366:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Senthilkumar KP, Thirumurugan R. Risk Modulation of GSTM1 - GSTT1 interactions to Head and Neck Cancer in tobacco users. Mol Bio Rep. 2014;41:5635–5644. 10.1007/s11033-014-3433-x [DOI] [PubMed] [Google Scholar]
- 9. Toncheva DL, Von AN, Atanasova SY, Dimitrow TG, Armstrong VW, Oellerich M. Identification of NQ01 and GSTs genotype frequencies in Bulgarian patients with Balken endemic nephropathy. J Nephrol. 2004;17:384–389. [PubMed] [Google Scholar]
- 10. Kurose K, Sugiyama E, Saito Y. Population Differences in Major Functional Polymorphisms of Pharmacokinetics / pharmacodynamics-related Genes in Eastern Asians and Europeans: Implications in the Clinical Trials for Novel Drug Development. Drug Metab Pharmacokinetics 2012;27:9–54. [DOI] [PubMed] [Google Scholar]
- 11. Polimanti R, Carboni C, Baesso I, Piacentini S, Iorio A, Stefano DGF, et al. Genetic variability of glutathione S-transferase enzymes in human populations: Functional inter-ethnic differences in detoxification systems. Gene 2013;512:102–107. 10.1016/j.gene.2012.09.113 [DOI] [PubMed] [Google Scholar]
- 12. Evans WE, Johnson JA. Pharmacogenomics: The Inherited Basis for Interindividual Differences in Drug Response. Annu Rev Genomics Hum Genet. 2001;2:9–39. [DOI] [PubMed] [Google Scholar]
- 13. O'Donnell PH, Dolan ME. Cancer Pharmacoethnicity: Ethnic Differences in Susceptibility to the Effects of Chemotherapy. Clin Cancer Res. 2009;15:4806–4814. 10.1158/1078-0432.CCR-09-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghosh T, Gupta S, Bajpai P, Agarwal D, Agarwal M, Gupta OP, et al. Association of CYP1A1, GSTM1, and GSTT1 gene polymorphism with risk of oral submucous fibrosis in a section of North Indian population. Mol Biol Rep. 2012;39:9383–9389. 10.1007/s11033-012-1802-x [DOI] [PubMed] [Google Scholar]
- 15. Shukla D, Kale AD, Hallikerimath S, Vivekanandhan S, Venkatakanthaiah Y. Genetic polymorphism of drug metabolizing enzymes (GSTM1 and CYP1A1) as risk factors for oral premalignant lesions and oral cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2012;156:253–259. 10.5507/bp.2012.013 [DOI] [PubMed] [Google Scholar]
- 16. Saijo N. The Role of Pharmacoethnicity in the Development of Cytotoxic and Molecular Targeted Drugs in Oncology. Yonsei Med J. 2013;54:1–14. 10.3349/ymj.2013.54.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salah GB, Kallabi F, Maatoug S, Mkaouar-Rebai E, Fourati A, Fakhfakh F, et al. Polymorphisms of glutathione S-transferases M1, T1, P1 and A1 genes in the Tunisian population: An intra and interethnic comparative approach. Gene 2012;498:317–322. 10.1016/j.gene.2012.01.054 [DOI] [PubMed] [Google Scholar]
- 18. Naveen AT, Adithan C, Padmaja N, Shashindran CH, Abraham BK, Sathyanarayanamoorthy K, et al. Glutathione S-transferase M1 and T1 null genotype distribution in South Indians. Eur J Clin Pharmacol. 2004;60:403–406. [DOI] [PubMed] [Google Scholar]
- 19. Lahiri DK, Nurnberger JI. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang K, Sandler RS, Millikan RC, Schroeder JC, North KE, Hu J. GSTM1 and GSTT1 polymorphisms, cigarette smoking, and risk of colon cancer: a population-based case-control study in North Carolina (United States). Cancer causes & control 2006;17:385–394. [DOI] [PubMed] [Google Scholar]
- 21. Hamdy SI, Hiratsuka M, Narahara K, Endo N, El-Enany M, Moursi N, et al. Genotype and allele frequencies of TPMT, NAT2, GST, SULT1A1 and MDR-1 in the Egyptian population. Br J Clin Pharmacol. 2003;55:560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Agusa T, Iwata H, Fujihara J, Kunito T, Takeshita H, Minh TB, et al. Genetic polymorphisms in glutathione S-transferase (GST) superfamily and arsenic metabolism in residents of the Red River Delta, Vietnam. Toxicol Appl Pharmacol. 2010;242:353–362. [DOI] [PubMed] [Google Scholar]
- 23. Rimando MG, Chu MN, Yuson E, de Castro-Berna G, Okamoto T. Prevalence of GSTT1, GSTM1 and NQ01(609C>T) in Filipino children with ALL (acute lymphoblastic leukemia). Biosci Rep. 2008;28:117–124. 10.1042/BSR20070010 [DOI] [PubMed] [Google Scholar]
- 24. Amtha R, Ching CS, Zang R, Razak IA, Basuki B, Roeslan BO, et al. GSTM1, GSTT1 and CYP1A1 polymorphisms and risk of oral cancer: a case-contro study in Jakarta, Indonesia. Asian Pac J Cancer Prev. 2009;10:21–26. [PubMed] [Google Scholar]
- 25. Lee EJ, Wong JY, Yoeh PN, Gong NH. Glutathione S transferase-theta (GSTT1) genetic polymorphism among Chinese, Malaya and Indians in Singapore. Pharmacogenetics 1995;5:332–334. [DOI] [PubMed] [Google Scholar]
- 26. Anantharaman D, Chaubal PM, Kannan S, Bhisey RA, Mahimkar MB. Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: tobacco exposure as a risk modulator. Carcinogenesis 2007;28:1455–1462. [DOI] [PubMed] [Google Scholar]
- 27. Buch S, Kotekar A, Kawle D, Bhisey R (2001). Polymorphisms at CYP and GST gene loci. Prevalence in the Indian population. Eur J Clin Pharmacol 57 (6–7), 553–555. [DOI] [PubMed] [Google Scholar]
- 28. Buch SC, Notani PN, Bhisey RA. Polymorphism at GSTM1, GSTM3 and GSTT1 gene loci and susceptibility to oral cancer in an Indian population. Carcinogenesis 2002;23:803–807. [DOI] [PubMed] [Google Scholar]
- 29. Jain M, Kumar S, Rastogi N, Lal P, Ghoshal UC, Tiwari A, et al. GSTT1, GSTM1 and GSTP1 genetic polymorphisms and interaction with tobacco, alcohol and occupational exposure in esophageal cancer patients from North India. Cancer Lett. 2006;242:60–67. [DOI] [PubMed] [Google Scholar]
- 30. Konwar R, Manchanda PK, Chaudhary P, Nayak VL, Singh V, Bid HK. Glutathione S-transferase (GST) gene variants and risk of benign prostatic hyperplasia: a report in a North Indian population. Asian Pac J Cancer Prev. 2010;11:1067–1072. [PubMed] [Google Scholar]
- 31. Mishra DK, Kumar A, Srivastava DS, Mittal RD. Allelic variation of GSTT1, GSTM1 and GSTP1 genes in North Indian population. Asian Pac J Cancer Prev. 2004;5:362–365. [PubMed] [Google Scholar]
- 32. Singh M, Shah PP, Singh AP, Ruwali M, Mathur N, Pant MC, et al. Association of genetic polymorphisms in glutathione S-transferases and susceptibility to head and neck cancer. Mutat Res. 2008;638:184–194. [DOI] [PubMed] [Google Scholar]
- 33. Sabitha K, Reddy MVV, Jamil K. GST genotypes in head and neck cancer patients and its clinical implications. Afr J Biotechnol. 2008;7:3853–3859. [Google Scholar]
- 34. Sikdar N, Datta S, Dey B, Paul RR, Panda CK, Roy B. Homozygous Null Genotype at Glutathione S-transferase M1 Locus as a Risk Factor for Oral Squamous Cell Carcinoma in Indian Tobacco Users. Int J Hum Genet. 2005;5:37–44. [Google Scholar]
- 35. Sreelekha TT, Ramadas K, Pandey M, Thomas G, Nalinakumari KR, Pillai MR. Genetic polymorphism of CYP1A1, GSTM1 and GSTT1 genes in Indian oral cancer. Oral Oncol. 2001;37:593–598. [DOI] [PubMed] [Google Scholar]
- 36. Saify K, Saadat I, Saadat M. Genetic polymorphisms of glutathione S-transferase T1 (GSTT1) and M1 (GSTM1) in selected populations of Afghanistan. Mol Biol Rep. 2012;39:7855–7859. 10.1007/s11033-012-1628-6 [DOI] [PubMed] [Google Scholar]
- 37. Saadat M. GSTM1 null genotype associated with age standardized cancer mortality rate in 45 countries from five continents: an ecologic study. Int J Cancer Res. 2007;3:74–91. [Google Scholar]
- 38. Shaikh RS, Amir M, Masood AI, Sohail A, Athar HU, Siraj S, et al. Frequency distribution of GSTM1 and GSTT1 null allele in Pakistani population and risk of disease incidence. Environ Toxicol Pharmacol. 2010;30:76–9 10.1016/j.etap.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 39. Garte S, Gaspari L, Alexandrie A, Ambrosone C, Autrup H, Autrup JL, et al. Metabolic Gene Polymorphism Frequencies in Control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–1248. [PubMed] [Google Scholar]
- 40. Binkova B, Smerhovsky Z, Strejc P, Boubelik O Stavkova Z, Chvatalova I, et al. DNA-adducts and atherosclerosis: a study of accidental and sudden death males in the Czech Republic. Mutat Res. 2002;501:115–128. [DOI] [PubMed] [Google Scholar]
- 41. Gajecka M, Rydzaniacz M, Jaskula-Sztul R, Kujawski M, Szyfter W, Szyfter K. CYP1A1, CYP2D6, CYP2E1, NAT2, GSTM1 and GSTT1 polymorphisms or their combinations are associated with increased risk of the laryngeal squamous cell carcinoma. Mutat Res. 2005;574:112–123. [DOI] [PubMed] [Google Scholar]
- 42. Gra O, Mityaeva O, Berdichevets I, Kozhekbaeva Z, Fesenko D, Goldenkova-Pavlova I, et al. Microarray-Based Detection of CYP1A1, CYP2C9, CYP2C19, CYP2D6, GSTT1, GSTM1, MTHFR, MTRR, NQ01, NAT2, HLA-DQA1, and ABO allele frequencies in native Russians. Genet Test Mol Biomarkers 2010;14:329–342. 10.1089/gtmb.2009.0158 [DOI] [PubMed] [Google Scholar]
- 43. Ebeshi BU, Bolaji OO, Masimirembwa CM. Glutothione-S-transferase (M1 and T1) polymorphisms in Nigerian populations. J Med Genet Genomics 2011;3:56–60. [Google Scholar]
- 44. Adams CH, Werely CJ, Victor TC, Hoal EG, Rossouw G, Heldin VPD. Allele frequencies for glutathione S-transferase and N-acetyltransferase 2 differ in African population groups and may be associated with oesophageal cancer or tuberculosis incidence. Clin Chem Lab Med. 2003;41:600–605. [DOI] [PubMed] [Google Scholar]
- 45. Fujihara J, Yasuda T, Iida R, Takatsuka H, Fujii Y, Takeshita H. Cytochrome P450 1A1, glutathione S-transferases M1 and T1 polymorphisms in Ovambos and Mongolians. Leg Med (Tokyo) 2009;Suppl 1:S408–S410. 10.1016/j.legalmed.2009.01.073 [DOI] [PubMed] [Google Scholar]
- 46. Piacentini S, Polimanti R, Porreca F, Martı´nez-Labarga C, Stefano GFD, Fuciarelli M. GSTT1 and GSTM1 gene polymorphisms in European and African populations. Mol Biol Rep. 2011;38:1225–1230. 10.1007/s11033-010-0221-0 [DOI] [PubMed] [Google Scholar]
- 47. Buchard A, Sanchez JJ, Dalhoff K, Morling N. Multiplex PCR detection of GSTM1, GSTT1, and GSTP1 gene variants: simultaneously detecting GSTM1 and GSTT1 gene copy number and the allelic status of the GSTP1 Ile105Val genetic variant. J Mol Diagn. 2007;9:612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dandara C, Sayi J, Masimirembwa CM, Magimba A, Kaaya S, Sommers DK, et al. Genetic polymorphism of cytochrome P450 1A1 (Cyp1A1) and glutathione transferases (M1, T1 and P1) among Africans. Clin Chem Lab Med. 2002;40:953–957. [DOI] [PubMed] [Google Scholar]
- 49. Rossini A, Rapozo CM, Amorim MF, Macedo JM, Medina R, Neto JF, et al. Frequencies of GSTM1, GSTT1, and GSTP1 polymorphisms in a Brazilian population. Genet Mol Res. 2002;1:233–240. [PubMed] [Google Scholar]
- 50. Magno LAV, Talbot J, Talbot T, Santos BAM, Souza RP, Marin LJ, et al. Glutathione-S-Transferase variants in a Brazilian Population. Pharmacology 2009;83:231–236. 10.1159/000205823 [DOI] [PubMed] [Google Scholar]
- 51.Composition of macro geographical (continental) regions, geographical sub-regions, and selected economic and other groupings. Available: http://unstats.un.org/unsd/methods/m49/m49regin.htm
- 52. Nei M. Genetic distance between populations. Am Nat. 1972;106:283–292. [Google Scholar]
- 53. Felsenstein J. Phylip - Phylogeny Inference Package (Version 3.6). Distributed by the author, Department of Genome Sciences, University of Washington, Seattle; 2004. [Google Scholar]
- 54. Felsenstein J. Phylip - Phylogeny Inference Package (Version 3.2). Cladistics 1989;5: 164–166. [Google Scholar]
- 55. Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sneath PHA and Sokal RR. Numerical Taxonomy. Freeman, San Francisco; 1973.
- 57.Latitude and Longitude Finder. Available: http://www.worldatlas.com/aatlas/latitude_and_longitude_finder.htm
- 58. Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nat Rev Genet. 2008;9:421–432. 10.1038/nrg2339 [DOI] [PubMed] [Google Scholar]
- 59. Jorde L, Wooding S. Genetic variation, classification and’race'. Nat Genet. 2004;36:28–33. [DOI] [PubMed] [Google Scholar]
- 60. ArunKumar G, Soria-Hernanz DF, Kavitha VJ, Arun VS, Syama A, Ashokan KS, et al. Population Differentiation of Southern Indian Male Lineages Correlates with Agricultural Expansions Predating the Caste System. PLoS ONE 2012;7: e50269 10.1371/journal.pone.0050269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Haber M, Gauguier D, Youhanna S, Patterson N, Moorjani P, Botigue LR, et al. Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture. PLoS Genet. 2013;9: e1003316 10.1371/journal.pgen.1003316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Field J, Lahr MM. Assessment of the Southern Dispersal: GIS-Based Analyses of Potential Routes at Oxygen Isotopic Stage 4. J World Prehist. 2006;19:1–45. [Google Scholar]
- 63. Templeton A. Out of Africa again and again. Nature 2002;416:45–51. [DOI] [PubMed] [Google Scholar]
- 64. Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Naidu JM, et al. Genetic Evidence on the Origins of Indian Caste Populations. Genome Research 2001;11:994–1104. 10.1101/gr.173301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.