Abstract
Background
In plants, 14-3-3 proteins are encoded by a large multigene family and are involved in signaling pathways to regulate plant development and protection from stress. Although twelve Populus 14-3-3s were identified based on the Populus trichocarpa genome V1.1 in a previous study, no systematic analysis including genome organization, gene structure, duplication relationship, evolutionary analysis and expression compendium has been conducted in Populus based on the latest P. trichocarpa genome V3.0.
Principal Findings
Here, a comprehensive analysis of Populus 14-3-3 family is presented. Two new 14-3-3 genes were identified based on the latest P. trichocarpa genome. In P. trichocarpa, fourteen 14-3-3 genes were grouped into ε and non-ε group. Exon-intron organizations of Populus 14-3-3s are highly conserved within the same group. Genomic organization analysis indicated that purifying selection plays a pivotal role in the retention and maintenance of Populus 14-3-3 family. Protein conformational analysis indicated that Populus 14-3-3 consists of a bundle of nine α-helices (α1-α9); the first four are essential for formation of the dimer, while α3, α5, α7, and α9 form a conserved peptide-binding groove. In addition, α1, α3, α5, α7, and α9 were evolving at a lower rate, while α2, α4, and α6 were evolving at a relatively faster rate. Microarray analyses showed that most Populus 14-3-3s are differentially expressed across tissues and upon exposure to various stresses.
Conclusions
The gene structures and their coding protein structures of Populus 14-3-3s are highly conserved among group members, suggesting that members of the same group might also have conserved functions. Microarray and qRT-PCR analyses showed that most Populus 14-3-3s were differentially expressed in various tissues and were induced by various stresses. Our investigation provided a better understanding of the complexity of the 14-3-3 gene family in poplars.
Introduction
Plant growing in nature constantly sense their environment and adapt to changes by using a range of biochemical and molecular mechanism [1]. And all these biological processes are controlled by signal transduction and metabolism regulation that have been known to occur via phosphorylation-mediated transition of protein states [2]. In many cases, to complete their regulatory actions, these phosphorylated proteins must physically associate with the specialized adapter proteins, which are known as 14-3-3 phosphoserine/phosphothreonine (pSer/pThr) binding protein [3–5].
14-3-3 genes encode a ubiquitous family of highly conserved eukaryotic proteins from fungi to humans and plants with several molecular and cellular functions [6]. It was discovered in 1976 during a study of the soluble acidic proteins of the mammalian brain and was named on the basis of fraction number during DEAE-cellulose chromatography and location after starch gel electrophoresis [7]. They regulate activities of a wide array of target proteins via protein-protein interactions which involves binding with pSer/pThr residues in the target proteins [8]. Through the functional modulation of a wide range of binding partners, 14-3-3 proteins are involved in many biologically important process, including cell cycle regulation, metabolism control, apoptosis, protein trafficking, stress response, and control of gene transcription [6,9–13].
The 14-3-3 proteins can form both homo- and heterodimers [14]. Structural studies confirmed a dimeric nature of 14-3-3 proteins and revealed that each monomer consists of nine antiparallel α-helices [15,16]. A large 40 Å wide deep channel located in the center of a cup-shaped 14-3-3 protein dimer contains two amphipathic grooves [12]. All isoforms recognize two high-affinity phosphorylation-dependent 14-3-3 binding motifs: RSXpSXP (mode 1) and RX(F/Y)XpSXP (mode 2), where X is any amino acid and pS represents a phosphoserine [17–19]. Each monomer in the dimer is capable of interacting with a separate target protein [13]. The dimeric property of 14-3-3s allows them to serve as scaffolds by bringing two different regions of the same protein into proximity within a single complex or two different proteins together [8].
In plants, the 14-3-3 isoform was identified as part of a protein/G-box complex and therefore named GF14 (G-box Factor 14-3-3 homologs) or GRF (G-box Regulatory Factor, or General Regulatory Factor) [20]. In the last decades, a large amount of evidence has begun to accumulate for the role of 14-3-3 proteins in plant development and stress response [6]. The first Arabidopsis 14-3-3 isoform GRF2 (GF14omega) was discovered as a constituent of a protein/G-box complex and implicated to be involved in regulation of gene transcription [21], subsequent studies have also shown that plant 14-3-3s have a broad range of functions [6,22,23]. To date, an increasing body of work is beginning to clarify the roles of 14-3-3s in stress response pathways in plants. There are two main ways by which environmental inputs may affect 14-3-3 activity or be affected by 14-3-3s. Firstly, environmental stimuli may activate signaling pathways that causes the phosphorylation of client proteins to which 14-3-3s then recognize and bind. Secondly, these stimuli may affect 14-3-3s themselves by means of transcriptional regulation of specific 14-3-3s or by affecting the levels of signaling molecules such as divalent cations or AMP to which 14-3-3s can bind. The post-translational modification of specific 14-3-3 isoforms may also be affected [23]. A direct evidence that 14-3-3s play a major functional role in environmental stress responses comes from the overexpression of Arabidopsis 14-3-3λ in cotton enhanced drought tolerance in transgenic plants [24]. 14-3-3s may also exert their effects by interacting with components of hormone signaling pathways [25].
Recently the whole genome sequencing of plants has assisted for a survey of plant 14-3-3 proteins and possible implications for their role in plant growth and developmental processes [8]. Because of the economic importance in pulp and biofuel production, the studies on the genus Populus have been hotspots for many years [26]. The completion of Populus trichocarpa genome sequence in 2006 makes it as a model tree for other tree species [27]. Despite that much is learnt about 14-3-3s in non-woody plants, the diversity of this group of proteins in woody plants is not yet known. To determine the structure and function of 14-3-3 in the Populus genus, we performed detailed systematic analyses of genome organization, gene structure, protein structure, and expression compendium. In this study, we report the comprehensive genomic identification and phylogenetic analysis of all fourteen members of the 14-3-3 gene family in Populus, as well as their expression profiles in different tissues and their responses under various abiotic stresses. Our preliminary results may provide insights to further investigate the functions of these genes.
Materials and Methods
Characteristics of Populus 14-3-3 genes and phylogenetic analysis
To identify potential members of the Populus 14-3-3 gene family, we performed multiple database searches. Published Arabidopsis and rice 14-3-3 protein sequences were retrieved and used as queries in tBLASTn searches against the P. trichocarpa genome database (http://www.phytozome.net/poplar.php, release 3.0). WoLF PSORT (http://wolfpsort.org) was used to predict protein subcellular localization [28]. The pI and molecular weight were estimated using the Compute pI/Mw tool from ExPASy (http://web.expasy.org/compute_pi).
Sequences of Medicago trucatula, Sorghum bicolor, Brachypodium distachyon, Vitis vinifera, Glycine max, and Physcomitrella patens were obtained from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) by tBLASTn searches using Arabidopsis and rice 14-3-3 protein sequences as queries. Multiple Sequences Alignment (MSA) of 14-3-3 protein sequences from P. trichocarpa and other eight plant species were performed using the Clustal X program (V2.1) [29]. A maximum likelihood (ML) phylogenetic tree was constructed using PhyML (V3.0) with 1,000 bootstrap replicates [30].
Gene structure, chromosomal location and synteny analysis
The exon and intron structures were illustrated using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn) [31] by aligning the cDNA sequences with the corresponding genomic DNA sequences from Phytozome (http://www.phytozome.net/). The chromosomal locations of the 14-3-3 genes were determined using the P. trichocarpa genome browser (http://www.phytozome.net/poplar). Tandem duplicated genes were defined as adjacent homologous 14-3-3 genes on the P. trichocarpa chromosomes, with no more than one intervening gene. For synteny analysis, synteny blocks within the P. trichocarpa genome and between P. trichocarpa and Arabidopsis thaliana genomes were downloaded from the Plant Genome Duplication Database (PGDD, http://chibba.agtec.uga.edu/duplication/) [32] and those containing Populus 14-3-3 genes were identified. The chromosomal locations of 14-3-3 genes were drawn using Circos software [33].
Calculation of Ka/Ks values
Amino acid sequences from segmentally duplicated pairs were aligned first by Clustal X program (V2.1) [29] and the aligned sequences were subsequently transferred into original cDNA sequences using the PAL2NAL program (http://www.bork.embl.de/pal2nal/) [34], which uses the CODEML program of PAML [35] to estimate synonymous (Ks) and nonsynonymous (Ka) substitution rates. Divergence time (T) was calculated using a synonymous mutation rate of λ substitutions per synonymous site per year as T = Ks/2λ (λ = 9.161029 for Populus) [36].
Protein modeling, molecular conservation and structural analysis
To better understand the molecular mechanism of Populus 14-3-3 protein, the deduced Populus 14-3-3 protein sequences were modelled using the top 10 PDB closed templates structures by I-Tasser [37]. The predicted organic binding site was based on the identification of analogs with similar binding sites taking into account their BS-scores, TM-scores (a scale for measuring the structural similarity between two structures), IDEN (percentage sequence identity in the structurally aligned region), the coverage of the alignment by TM-align, the COV of the model, and the structural alignment (which is equal to the number of structurally aligned residues divided by their length). A BS-score value of >0.5 signifies a binding site prediction with high confidence. The ligand(s) in the analog structure were then transferred onto the model and the fitness of the ligand-model complex (BS-score) was calculated by comparing the local structure and sequence similarity in the binding site region.
EST profiling and publicly available microarray data analyses
The expression profiles for each gene was obtained by evaluating its EST representation among 18 cDNA libraries derived from different tissues and/or developmental stages available at PopGenIE (http://www.popgenie.org/) [38].
The microarray data for various tissues and developmental stages available at NCBI Gene Expression Omnibus (GEO) database (http://www.ncibi.nlm.nih.gow/geo/) under the series accession numbers GSE13990 (from P. balsamifera), GSE30507 (from P. maximowiczii × nigra), and GSE13043 (from P. trichocarpa) were used for the tissue-specific expression analysis. The series GSE13990 includes Affymetrix microarray data from nine different tissue samples representing three biological replicates [39], series GSE30507 includes Affymetrix microarray data from six tissue samples representing two biological replicates [40], whereas series GSE13043 contains NimbleGen microarray data from five stem internodes (IN) from the apical bud to the base of the shoot (IN2 to IN5, IN9) in two biological replicates [41].
For abiotic and hormonal treatments, Affymetrix microarray data available in the NCBI GEO database under the series accession numbers GSE13109 (hypoxia), GSE26199 (heat) and GSE16786 were analyzed [42,43]. GSE16786 is composed of the following five subsets: GSE14893 (nitrogen limitation, genotype 1979), GSE14515 (nitrogen limitation, genotype 3200), GSE16783 (1 week after leaf wounding), GSE16785 (90 h after leaf wounding), and GSE16773 (methyl jasmonate-elicited suspension cell cultures). Probe sets corresponding to Populus 14-3-3 genes were identified using the online Probe Match tool POParray (http://aspendb.uga.edu/poparray). For genes with more than one probe sets, the median of expression values was considered. The expression data were gene-wise normalized. The data was normalized by the Gene Chip Robust Multiarray Analysis (GCRMA) algorithm followed by log transformation and average calculation. After normalization and log transformation of data for all the Populus genes present on the chip, the log signal intensity values for Populus probe set IDs corresponding to 14-3-3 genes were extracted for further analyses.
Plant material, RNA isolation and real-time qRT-PCR
Plant materials were collected from clonally propagated 1-year-old hybrid poplar (P. alba × P.glandulosa) clone (84K) grown in a growth chamber under long-day conditions (16 h light/8 h dark) at 23–25°C. Six vegetative tissues (SAM-shoot apical meristem, YL-young leaf, ML-mature leaf, PS-primary stem, SS-secondary stem, and R-root) were collected from 84K. Samples were frozen immediately in liquid nitrogen, and stored at -80°C for further analysis. Three biological replicates were performed.
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) with on-column treatment with RNase-free DNase I (Qiagen) to remove any contamination of genomic DNA. First-strand cDNA synthesis was carried out with approximately 1 μg RNA using the SuperScript III reverse transcription kit (Invitrogen) according to the manufacturer’s procedure. Primers with melting temperatures of 58–62°C and amplicon lengths of 150–250 bp were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm). qRT-PCR was conducted on LightCycler 480 Detection System (Roche, Penzberg, Germany) using SYBR Premix Taq Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Relative expression was calculated by the 2-ΔΔCt method [44]. The PtActin and PtTubulin were used as internal controls.
Results and Discussion
Identification of 14-3-3 gene family in Populus and other plant species
To identify 14-3-3 genes in Populus, we performed a tBLASTn search against P. trichocarpa genome release 3.0 (http://www.phytozome.net/poplar/) using 14-3-3 protein sequences in Arabidopsis and rice as queries and the resulting sequences were used as secondary queries [2,45]. After manual reannotation and confirmation of the motif, total of fourteen putative 14-3-3 genes were identified. In previous study, a total of twelve 14-3-3 genes were identified in Populus by a genome-wide bioinformatics survey based on the P. trichocarpa genome V1.1 [46]. In this study, we further revealed two additional 14-3-3 genes in Populus and extended the total member to fourteen. We designated Populus 14-3-3 genes as PtGRF following the nomenclature proposed in the previous study [45]. The 14-3-3 genes identified in P. trichocarpa encode proteins ranging from 238 to 306 amino acids (aa) in length, with predicted isoelectric points (pIs) ranging from 4.7 to 5.9. The detailed information of 14-3-3 genes in Populus were listed in Table 1.
Table 1. Populus 14-3-3 gene family.
| Group | Gene Name | Gene Locus | CDS (bp) | ORF (aa) | pI | MW (kDa) | PSORT predictions* |
|---|---|---|---|---|---|---|---|
| non-ε group | PtGRF1/2/4a | Potri.002G099800 | 789 | 262 | 4.7 | 29.38 | nucl_plas: 5.5, plas: 5.0, nucl: 4.0, chlo: 2.0, cyto: 2.0 |
| PtGRF1/2/4b | Potri.005G162400 | 786 | 261 | 4.7 | 29.30 | plas: 5.0, nucl_plas: 5.0, chlo: 4.0, nucl: 3.0, cyto: 2.0 | |
| PtGRF3/5/7a | Potri.004G101700 | 813 | 270 | 5.2 | 30.47 | cyto: 7.0, nucl: 3.0, chlo: 2.0, plas: 2.0 | |
| PtGRF3/5/7b | Potri.017G113300 | 792 | 263 | 4.8 | 29.57 | nucl_plas: 5.0, nucl: 4.0, cyto: 4.0, plas: 4.0, chlo: 2.0 | |
| PtGRF3/5/7c | Potri.T147900 | 759 | 252 | 4.9 | 28.47 | nucl_plas: 5.5, nucl: 5.0, cyto: 4.0, plas: 4.0 | |
| PtGRF6/8a | Potri.002G103800 | 768 | 255 | 4.7 | 28.60 | plas: 5.0, nucl_plas: 5.0, chlo: 4.0, nucl: 3.0, mito: 1.0 | |
| PtGRF6/8b | Potri.005G157700 | 783 | 260 | 4.9 | 29.10 | chlo: 5.0, plas: 4.0, nucl_plas: 4.0, nucl: 2.0, cyto: 2.0 | |
| ε group | PtGRF9a | Potri.001G392200 | 834 | 277 | 5 | 31.58 | chlo: 6.0, nucl_plas: 4.0, nucl: 3.0, plas: 3.0, cysk: 2.0 |
| PtGRF9b | Potri.011G110900 | 786 | 261 | 4.9 | 29.51 | nucl_plas: 5.5, nucl: 5.0, plas: 4.0, chlo: 3.0, cyto: 1.0 | |
| PtGRF11a | Potri.002G097500 | 762 | 253 | 4.9 | 28.97 | plas: 5.0, nucl_plas: 5.0, nucl: 3.0, cyto: 3.0, mito: 1.0, extr: 1.0 | |
| PtGRF11b | Potri.005G164500 | 921 | 306 | 5.1 | 34.76 | cyto: 4.0, chlo: 3.0, plas: 2.0, nucl: 1.0, mito: 1.0, extr: 1.0, cysk: 1.0 | |
| PtGRF12a | Potri.008G095000 | 789 | 262 | 4.8 | 29.63 | nucl: 7.0, nucl_plas: 6.5, plas: 4.0, cyto: 2.0 | |
| PtGRF12b | Potri.010G159300 | 789 | 262 | 4.8 | 29.63 | nucl_plas: 5.0, nucl: 4.0, plas: 4.0, cyto: 3.0, chlo: 1.0, mito: 1.0 | |
| PtGRF13 | Potri.001G125600 | 717 | 238 | 5.9 | 27.59 | nucl: 7.0, cyto: 5.0, mito: 1.0 |
Gene loci are obtained from the Phytozome website (http://www.phytozome.net).
*PSORT predictions: plas (plasma membrane), cyto (cytosol), chlo (chloroplast), cysk (cytoskeleton), nucl (nuclear), mito (mitochondrion), extr (extracellular). The numbers indicate the number of nearest neighbors to the query which localize to each site.
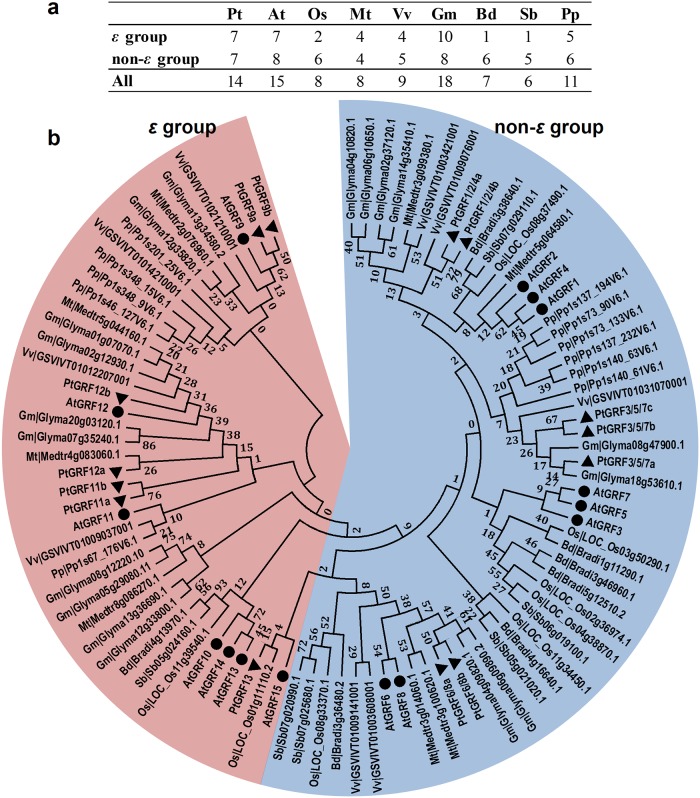
In order to gain insight into the evolutionary relationships among plant 14-3-3 proteins, we identified 14-3-3 genes from eight other plant species with whole genome sequences available, including moss (Physcomitrella patens), the monocotyledonous angiosperms Oryza sativa, Sorghum bicolor and Brachypodium distachyon, and the dicotyledonous angiosperms Arabidopsis thaliana, Medicago truncatula, Glycine max, and Vitis vinifera (Fig 1). All angiosperm genomes as well as the genome of the moss contained genes encoding 14-3-3 proteins. A complete list of all 14-3-3 genes identified in this study was provided in S1 Table. As comparative genomic study revealed a ratio of 1.4~1.6 putative poplar homologs for each Arabidopsis gene [27], it was hypothesized that 14-3-3 genes in Populus would be a large multi-gene family as fifteen 14-3-3s were identified in Arabidopsis [45]. However, according to this study, the number of Populus 14-3-3 genes was even smaller than that of Arabidopsis (Fig 1b and Table 1). It can be hypothesized that fourteen 14-3-3 genes would be sufficient for Populus to mediate signal transduction and a subset of 14-3-3 genes might have lost during the evolutionary process due to the functional redundancy. We named the Populus 14-3-3s based on their phylogenetic relationships with the Arabidopsis 14-3-3s. For example, PtGRF3/5/7a, PtGRF3/5/7b, and PtGRF3/5/7c were clustered in the same clade with AtGRF3, AtGRF5, and AtGRF7 (Fig 2a).
Fig 1. 14-3-3 family members (a) and their phylogenetic relationships (b) from nine plant species.
a. 14-3-3 family members of A. thaliana (At), P. trichocarpa (Pt), O. sativa (Os), M. truncatula (Mt), S. bicolor (Sb), B. distachyon (Bd), V. vinifera (Vv), G. max (Gm), and P. patens (Pp). b. Multiple alignment of 14-3-3 proteins from nine plant species was performed using Clustal X2.1. Phylogenetic tree was constructed using full-length protein sequences by the maximum likelihood (ML) method with 1,000 bootstrap replicates. Bootstrap support values are shown on each node. The two major groups are marked with different background colors. Detail information of 14-3-3s from nine plant species were listed in S1 Table.
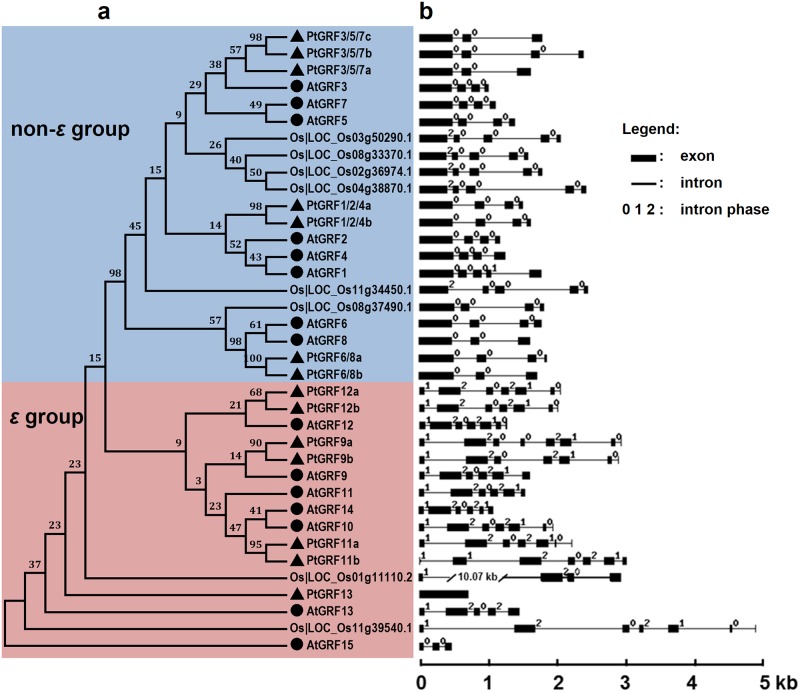
Fig 2. Phylogenetic analysis (a) and exon-intron structures (b) of P. trichocarpa, Arabidopsis, and rice 14-3-3s.
a. A multiple alignment of full-length 14-3-3 protein sequences from three species was executed using Clustal X2.1 and a phylogenetic tree was constructed by the maximum likelihood (ML) method with 1,000 bootstrap replicates. Bootstrap support values are shown on each node. The two major groups are marked with different background colors. b. Exon/intron structures of the 14-3-3 genes. Black boxes represent exons and lines represent introns. The numbers indicate the splicing phases of the 14-3-3s: 0, phase 0; 1, phase 1; and 2, phase 2.
Phylogenetic and gene structure analysis of Populus 14-3-3 gene family
To examine the evolutionary relationships of 14-3-3 proteins from different organisms, we constructed a phylogenetic tree by maximum likelihood (ML) method using the full-length 14-3-3 protein sequence alignments of nine plant species (P. trichocarpa, A. thaliana, O. sativa, M. truncatula, V. vinifera, G. max, B. distachyon, S. bicolor, and P. patens, Fig 1). The 14-3-3 proteins of all nine plant species were classified into two major groups (ε group and non-ε group, Fig 1a), the same as described previously [47]. Noticeably, the ratios of ε group members to total 14-3-3 genes in monocotyledon plants (25% in O. sativa, 14.3% in B. distachyon, and 16.7% in S. bicolor) were relatively less than that in moss (45.5% in P. patens) and in dicotyledon plants (50% in P. trichocarpa, 46.7% in A. thaliana, 50% in M. truncatula, 44.4% in V. vinifera, and 55.6% in G. max) (Fig 1a). This implying that some ε group 14-3-3 genes in monocotyledon plants might be lost during the evolution. The retained one or two ε group 14-3-3 genes might play the conserved function in monocotyledon plants.
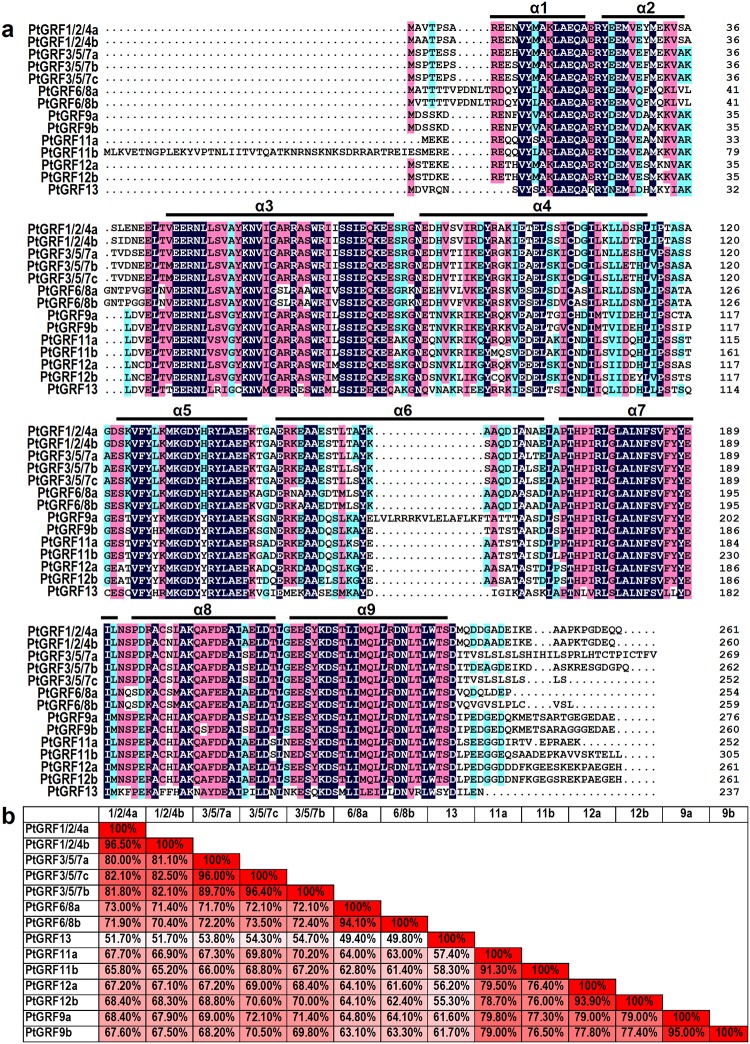
To gain further insights into the structural diversity of Populus 14-3-3 genes, we then analyzed the exon-intron organization in the coding sequence of the P. trichocarpa, Arabidopsis, and rice 14-3-3 genes (Fig 2). Exon-intron structural divergence within families plays a pivotal role in the evolution of multiple gene families. Generally, the positions of some spliceosomal introns are conserved in orthologous genes. In many cases, conservation of exon-intron organization in paralogous genes is high and sufficient to reveal the evolutionary relationship between introns [48]. As shown in Fig 2b, most members in each group have the similar number of exons and also exhibited nearly identical exon lengths. The intron phases are remarkably well conserved among family members, while the intron arrangements and intron phases are distinct between subfamilies (Fig 2b). An alignment of deduced Populus 14-3-3 proteins is shown in Fig 3a. The amino acid sequences are highly conserved except in the C-terminal and N-terminal regions. The sequence conservation among Populus 14-3-3 proteins was also supported by the percentage identity at amino acid level (49.4 ~ 96.5%, Fig 3b).
Fig 3. Sequence alignment and identity of Populus 14-3-3 proteins.
a. Amino acid sequence alignment of the fourteen Populus 14-3-3 proteins. Nine α-helices were marked as α1-α9. b. Amino acid identity among Populus 14-3-3 proteins was analyzed in pairwise fashion.
Chromosomal location and duplication analysis of Populus 14-3-3 genes
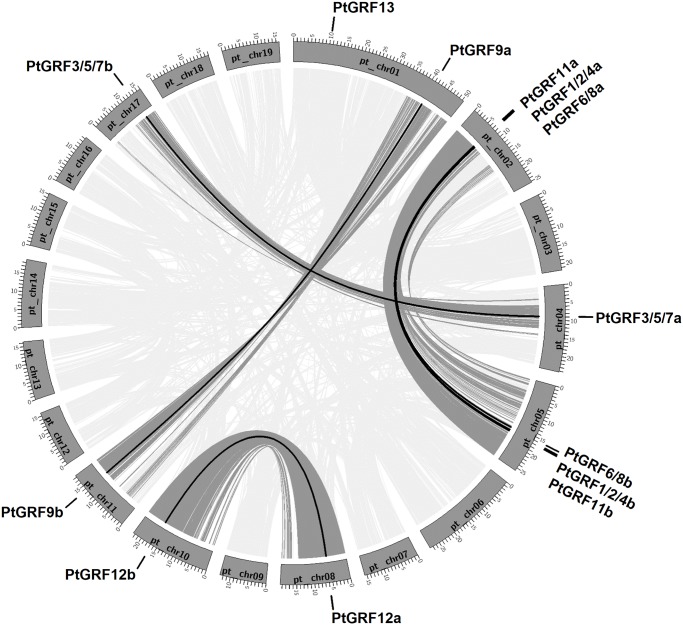
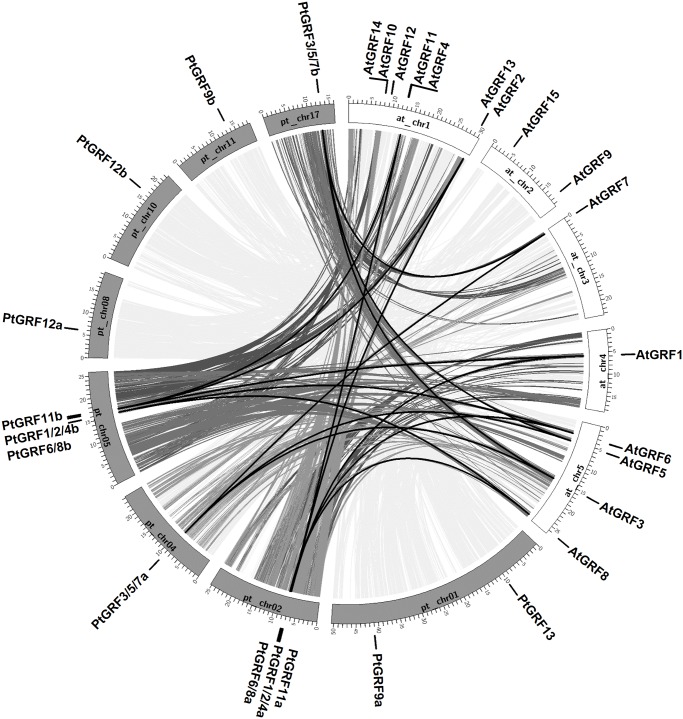
In silico mapping of the gene locus showed that thirteen of fourteen Populus 14-3-3 genes were mapped to eight of nineteen chromosomes (chr) unevenly, with only one gene (PtGRF3/5/7c) remained on as yet unmapped scaffolds (Fig 4). Previous analysis indicated that Populus genome has undergone at least three rounds of genome wide duplications followed by multiple segmental duplication, tandem duplication, and transposition events [49]. To determine the possible evolutionary relationship between Populus 14-3-3 genes and potential segmental duplications, Populus 14-3-3 genes were mapped to the nine duplicated blocks established in the previous study [27]. The distributions of 14-3-3 genes relative to the corresponding duplicated blocks were illustrated in Fig 4. Within the identified duplicated blocks associated with the recent salicoid duplication event, twelve of fourteen (85.7%) Populus 14-3-3 genes were preferentially retained duplicates that located in both duplicated regions. In contrasts, PtGRF13 was located on one block and lacked duplicates on their corresponding blocks. The results indicated that dynamic rearrangement may have occurred following the segmental duplication which results in the loss of some genes.
Fig 4. Distribution and synteny of 14-3-3s on Populus chromosomes.
Nineteen Populus chromosomes (chr01-chr19) are depicted as grey bars. Chromosome numbers are indicated in centers of each bar. Populus 14-3-3s are indicated by vertical black lines outer the circles. Duplicate pairs formed by whole or segmental genome duplication are connected by bold black lines.
Based on the genomic organization of Populus 14-3-3 genes, we found that all of the six Populus 14-3-3 paralogous gene pairs were generated by segmental duplications (Fig 4 and Table 2). PtGRF1/2/4a - PtGRF1/2/4b, PtGRF6/8a - PtGRF6/8b, and PtGRF11a - PtGRF11b were three of the paralogous gene pairs identified in this study. Interestingly, PtGRF1/2/4a, PtGRF6/8a, and PtGRF11a together with PtGRF1/2/4b, PtGRF6/8b, and PtGRF11b, were found within the same duplication blocks with identities higher than 60% (Fig 4 and Fig 3b). This arrangement, along with the closely related location in the phylogenetic tree, suggested that the three paralogous gene pairs might originate from a common ancestor which firstly underwent tandem duplication prior to the segmental duplication. Moreover, the results indicated that Populus 14-3-3 genes had been preferentially retained at a relatively high rate of 85.7%, which was much higher than the average rate (33%) following the salicoid genome wide duplication in the Populus lineage [27]. This also corroborated previous reports showing that genes involved in transcription regulations and signal transductions were preferentially retained following duplications [50–53].
Table 2. Divergence between paralogous 14-3-3 gene pairs in Populus.
| No. | Gene 1 | Gene 2 | Duplication | Ka | Ks | Ka/Ks | Date (million years ago) |
|---|---|---|---|---|---|---|---|
| 1 | PtGRF1/2/4a | PtGRF1/2/4b | W | 0.015 | 0.281 | 0.053 | 15.46 |
| 2 | PtGRF3/5/7a | PtGRF3/5/7b | W | 0.081 | 0.373 | 0.217 | 20.51 |
| 3 | PtGRF6/8a | PtGRF6/8b | W | 0.034 | 0.202 | 0.167 | 11.08 |
| 4 | PtGRF9a | PtGRF9b | W | 0.022 | 0.280 | 0.079 | 15.40 |
| 5 | PtGRF11a | PtGRF11b | W | 0.042 | 0.201 | 0.209 | 11.07 |
| 6 | PtGRF12a | PtGRF12b | W | 0.031 | 0.162 | 0.191 | 8.92 |
To verify whether Darwinian positive selection was involved in Populus 14-3-3 genes divergence after duplication, the substitution rate ratio of nonsynonymous (Ka) versus synonymous (Ks) were calculated for Populus 14-3-3 gene pairs. Generally, Ka/Ks = 1 means neutral selection, Ka/Ks>1 means accelerated evolution with positive selection, and Ka/Ks<1 means purifying selection [54]. As the results showed that the Ka/Ks ratios of total six Populus 14-3-3 gene pairs were less than 0.3 (Table 2), we could conclude the Populus 14-3-3 gene family had undergone great purifying selection pressure with limited functional divergence after segmental duplications. Based on the divergence rate of 9.1×10-9 synonymous mutations per synonymous site year proposed for Populus [55], duplications of the paralogous gene pairs were estimated to occur between 8.92 and 20.51 million years ago (Table 2).
Evolutionary relationship of 14-3-3 family between Populus and Arabidopsis
Comparing the sequences of all genes between genomes from different taxa provide the possibly to reconstruct the evolutionary history of each gene in its entirety [56]. To further investigate the origin and evolutionary process of Populus 14-3-3 genes, we analyzed the comparative synteny map between P. trichocarpa and Arabidopsis genomes (Fig 5). Since Arabidopsis is one of the most important model plant species and the functions of some Arabidopsis 14-3-3 genes have been well characterized [24,57–59]. Thus, through comparative genomics analysis we might infer the functions of Populus 14-3-3 genes based on their Arabidopsis homologous.
Fig 5. Synteny analysis of 14-3-3 genes between Populus and Arabidopsis.
Populus and Arabidopsis chromosomes are depicted as grey and blank boxes, respectively. 14-3-3s are indicated by vertical black lines outer the circles. Syntenic relationship are connected by lines between Populus and Arabidopsis chromosomes.
There are one Populus 14-3-3 gene pair was syntenic with corresponding one Arabidopsis 14-3-3 gene (PtGRF11a-PtGRF11b corresponding to AtGRF11), and this pair was duplicated within the recent large-scale genome duplication event (13 Ma) in Populus (Table 2). More challenging for syntenic interpretation are cases where duplicated Populus genes corresponded to two (PtGRF6/8a-PtGRF6/8b corresponding to AtGRF6 and AtGRF8) or three (PtGRF1/2/4a-PtGRF1/2/4b corresponding to AtGRF1, AtGRF2, and AtGRF4; PtGRF3/5/7a-PtGRF3/5/7b corresponding to AtGRF3, AtGRF5, and AtGRF7) Arabidopsis genes (Fig 5). While there are still two pairs (PtGRF9a-PtGRF9b and PtGRF12a-PtGRF12b) could not be mapped into any synteny blocks. It might because the genomes have undergone multiple rounds of significant chromosomal rearrangement, fusions, and following selective gene loss after speciation of Populus and Arabidopsis [27].
Structural characterization of Populus 14-3-3 proteins
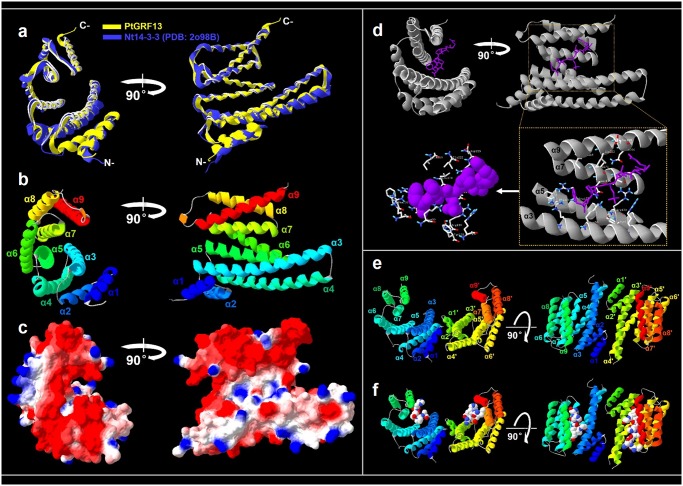
Because of the important role of 14-3-3 in plant development and environmental stress response, some reported crystal structures of 14-3-3 proteins from different plant species have been deposited in the Protein Data Bank (PDB) database (http://www.rcsb.org/pdb/home/home.do) up to date [60,61]. In order to understand the functional mechanism of Populus 14-3-3 proteins, we analyzed in detail the conformational features of Populus 14-3-3s using computational biology. We obtained the best predicted model of totally fourteen Populus 14-3-3 proteins based on the ten best structural templates and the crystal structures of 14-3-3 proteins from different organisms deposited in the Protein Database (Fig 6 and S1 Fig). The quality of the modeled protein was estimated by the C-score values generated by I-TASSER software, which reflects the coverage parameters in the structural simulations and the sequence alignment with the template [37]. C-score is a confidence scoring function to assessing the quality of a prediction and estimate the accuracy of the I-TASSER software predictions, which is based on the quality of the threading alignments and the convergence of I-TASSER’s structural assembly refinement simulations. Typically, a good predicted model is obtained when the estimated level of confidence (C-score) is between -5 and 2. The level of confidence for all our predicted Populus 14-3-3 models were in the range of -1.57 to 1.62 (Table 3), indicating that the protein structures were constructed with high accuracy. Other parameters like TM-score and root mean square deviation (RMSD) were used to check the topology and structural similarity of the models [62].
Fig 6. Structural analysis of Populus 14-3-3 protein.
a. Structural comparison of PtGRF13 (yellow) and Nt14-3-3(PDB: 2o98B) (blue). b. Monomeric structure of Populus 14-3-3 protein. The helices are labelled α1-α9. c. Electrostatic surface representation of the Populus 14-3-3 protein. The protein surface is colored according to its electrostatic potential: blue for positive and red for negative charge. d. Predicted binding model of Populus 14-3-3 monomer and other molecule. e. Structure of Populus 14-3-3 dimer. The helices are labelled α1-α9. f. Predicted binding model of Populus 14-3-3 dimer and other molecules. Each structure is rotated 90° to show different view side of the protein.
Table 3. Structural-dependent modeling parameters for the Populus 14-3-3 protein family.
| Gene Name | Gene ID | C-score | TM-score | RMSD (Å) | Template (higher Z-score) | PSI-BLAST % Identity with the template | Norm. Z-score |
|---|---|---|---|---|---|---|---|
| PtGRF1/2/4a | Potri.002G099800 | 0.31 | 0.75±0.10 | 5.2±3.4 | 2o98B | 91 | 2.73 |
| PtGRF1/2/4b | Potri.005G162400 | 0.26 | 0.75±0.10 | 5.3±3.4 | 2o98B | 91 | 2.73 |
| PtGRF3/5/7a | Potri.004G101700 | 0.01 | 0.71±0.11 | 5.9±3.7 | 2o98B | 88 | 2.69 |
| PtGRF3/5/7b | Potri.017G113300 | 0.04 | 0.72±0.11 | 5.8±3.6 | 2o98B | 86 | 2.74 |
| PtGRF3/5/7c | Potri.T147900 | 0.46 | 0.77±0.10 | 4.9±3.2 | 2o98B | 86 | 2.81 |
| PtGRF6/8a | Potri.002G103800 | 1.21 | 0.88±0.07 | 3.4±2.4 | 2o98B | 75 | 2.73 |
| PtGRF6/8b | Potri.005G157700 | 0.48 | 0.78±0.10 | 4.9±3.2 | 2o98B | 75 | 2.69 |
| PtGRF9a | Potri.001G392200 | 0.18 | 0.74±0.11 | 5.6±3.6 | 2o98B | 72 | 2.47 |
| PtGRF9b | Potri.011G110900 | 0.32 | 0.76±0.10 | 5.2±3.3 | 2o98B | 72 | 2.70 |
| PtGRF11a | Potri.002G097500 | 0.77 | 0.82±0.09 | 4.2±2.8 | 2o98B | 71 | 2.76 |
| PtGRF11b | Potri.005G164500 | -1.57 | 0.52±0.15 | 9.8±4.6 | 2o98B | 71 | 2.36 |
| PtGRF12a | Potri.008G095000 | 0.37 | 0.76±0.10 | 5.1±3.3 | 2o98B | 73 | 2.72 |
| PtGRF12b | Potri.010G159300 | 0.5 | 0.78±0.10 | 4.9±3.2 | 2o98B | 74 | 2.70 |
| PtGRF13 | Potri.001G125600 | 1.62 | 0.94±0.05 | 2.5±1.9 | 2o98B | 52 | 2.80 |
The determined 14-3-3 crystal structures were all homo- or heterodimers. Each monomer consists of a bundle of nine α-helices (α1-α9) organized into groups of two, two, two, and three helices (Fig 3 and Fig 6). The first four are essential for formation of the dimer, which has a sizeable aperture at the subunit interface (Fig 6e). Helices α3, α5, α7, and α9 form a conserved peptide-binding groove, which has a positively charged patch on one side and a hydrophobic patch on the other (Fig 6d and 6f). We also constructed the separate phylogenetic trees based on each of α-helices. We found that the domain of helices α1, α3, α5, α7, and α9 were evolving at a low rate, while α2, α4, and α6 were evolving at a relatively fast rate (S2 Fig). These results implying that the conserved α1, α3, α5, α7, and α9 domains might play conserved functions during the evolution. The predicted Populus 14-3-3 structures were similar to the 14-3-3 proteins in other species [13]. Although 14-3-3 proteins commonly form dimers, the monomers are sometimes functional, depending on the target protein, 14-3-3 proteins may use the dimerization process to control their cellular activities [13]. Generally, the 14-3-3 proteins bind to their binding partners mostly in a phosphorylation-dependent manner by recognizing specific pSer/pThr containing motifs [17]. In the binding event, two processes likely occur: binding of the phosphorylated peptide to the conserved groove (α3, α5, α7, and α9; primary interaction) and interaction of the globular domain of the protein with the remaining sections of the 14-3-3 (secondary interaction) [13]. The phosphorylation and other posttranslational modifications of 14-3-3 isoforms can efficiently modulate their interaction and dimerization [63]. In addition, 14-3-3 proteins not only bind a large and diverse set of phosphoproteins but also seem to coordinate their targets in different ways [64].
Populus 14-3-3 genes were differentially expressed across different tissues and involved in abiotic stress responses
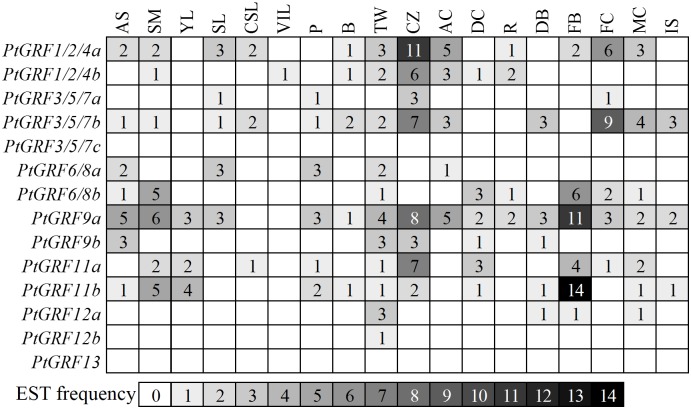
Publicly available Expression Sequence Tags (ESTs) by Digital Northern and whole genome microarray provide the useful tool to survey gene expression profiles in Populus [65,66]. We firstly conducted a preliminary analysis of Populus 14-3-3s expression across different tissues and under various growth conditions by counting the frequencies of ESTs in different poplar cDNA libraries obtained from PopGenIE (http://www.popgenie.org/) (Fig 7). Most of the Populus 14-3-3s had a broad expression pattern across different tissues. Noticeable, PtGRF1/2/4a with high abundance in cambial zone while PtGRF9a and PtGRF11b with high abundance in floral buds. A study in lily has suggested 14-3-3s may play a role in the germination and elongation of pollen [67]. It was suggested that the mechanisms by which 14-3-3s act in pollen grain development may include regulation of ion channels, osmoregulation, transport of proteins from the ER and Golgi to the plasma membrane, and also in mitochondrial energy generation for tube growth [68].
Fig 7. In silico EST analysis of Populus 14-3-3 genes.
EST frequency for each gene was calculated by evaluating its EST representation among 18 cDNA libraries available at PopGenIE (http://www.popgenie.org/) [38]. Color bar at bottom represents the frequencies of EST counts. AS: apical shoot, SM: shoot meristem, YL: young leaves, SL: senescing leaves, CSL: cold stressed leaves, VIL: Virus/fungus-infected leaves, P: petioles, B: bark, TW: tension wood, CZ: cambial zone, AC: active cambium, DC: dormant cambium, R: roots, DB: dormant buds, FB: flower buds, FC: female catkins, MC: male catkins, IS: imbibed seeds
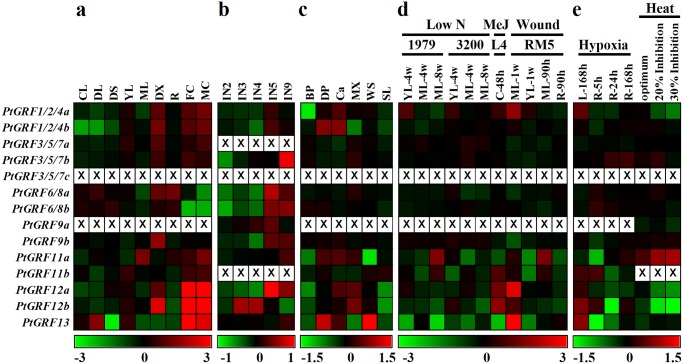
To gain more insights into the expression of Populus 14-3-3s in different tissues, a comprehensive analysis was conducted based on two Affymetrix (GSE13990, GSE30507) and a Nimblegen (GSE13043) microarray data [39–41]. Although these microarray datasets were performed in different platforms, they largely represented the Populus 14-3-3s presenting in this study (Fig 8a–8c). Five Populus 14-3-3s (PtGRF1/2/4a, 1/2/4b, 6/8a, 9b, and 12b) showed high transcript abundance in the differentiating xylem. Three Populus 14-3-3s (PtGRF12a, 12b, and 13) were preferentially expressed in male and female catkins (Fig 8a). During stem development, PtGRF3/5/7b had high accumulation in the basal stem undergoing secondary growth (internode 9), while PtGRF12b showed high expression level in the upper stem (internode 3 and 4) (Fig 8b). In addition, PtGRF1/2/4b and 13 were highly expressed in developing phloem (Fig 8c). The tissue specific expression implied that members of Populus 14-3-3s involved in special developmental processes. It has been proposed that 14-3-3s may allow the growth and development of cells to be co-ordinated with the metabolic status of the plants [69]. The expression of all six cotton 14-3-3 isoforms were increased during the elongation phases of fiber development. These included the regulation of hormone signaling pathways, regulation of Myb transcription factors and alterations in activity of the H+-ATPase activity leading to pH changes and subsequently structural changes of the cell wall [70].
Fig 8. Expression profiles of Populus 14-3-3 genes across different tissues and various abiotic stresses in different Populus species.
a. Expression profiles of Populus 14-3-3 genes across various tissues in P. balsamifera. The Affymetrix microarray data were obtained from NCBI Gene Expression Omnibus (GEO) database under the series accession number GSE13990. CL, continuous light-grown seedling; DL, etiolated dark-grown seedling transferred to light for 3 h; DS, dark-grown seedlings; YL, young leaf; ML, mature leaf; DX, differentiating xylem; R, root; FC, female catkins; MC, male catkins. b. Expression profiles of Populus 14-3-3 genes at different stem development stages in P. trichocarpa. The NimbleGen microarray data were obtained from GSE17230. IN2-IN5 and IN9, stem internodes 2 to 5 and internodes 9. c. Expression profiles of Populus 14-3-3 genes in different tissues of actively growing young poplar (P. maximowiczii × nigra) stems (one year old). The Affymetrix microarray data were obtained from GSE30507. BP, bark and mature phloem; DP, developing phloem; Ca, cambial zone; MX, mature xylem; WS, whole stem; SL, shoot apical meristem and leaf primordial. d. Expression profiles of Populus 14-3-3 genes across various stresses and genotypes analyzed. Microarray data was obtained from GSE16786. Genotypes analyzed included: Populus fremontii × angustifolia clones 1979, 3200, and RM5, Populus tremuloides clones 271 and L4, and Populus deltoids clones Soligo and Carpaccio. Tissues analyzed included: YL, young leaves; ML, mature leaves; R, root tips; C, suspension cell cultures. Stress treatments included: low N, nitrogen limitation; MeJ, Methyl Jasmonate elicitation; Wound, sampled either one week or 90 hours after wounding. e. Expression profiles of Populus 14-3-3 genes under hypoxia and heat stresses. Microarray data were obtained from GSE13109 (hypoxia) and GSE26199 (heat). Stress treatments included: Hypoxia, the leaves and root system of grey poplar (Populus × canescens) were flooded for up to 168 h; Heat, fully expanded leaf samples of P. trichocarpa were harvested at 4 physiological states as determined from prior gas exchange measurements (growth temperature: 22°C—baseline, 31.75°C—photosynthetic optimum, 38.4°C—20% inhibition of optimum and 40.5°C—30% inhibition of optimum). Color scale represents log2 expression values, green represents low level and red indicates high level of transcript abundances. Probe sets of Populus 14-3-3s were listed in S2 Table.
We then compared the expression patterns between Arabidopsis 14-3-3s and Populus 14-3-3s in various tissues. The PtGRF6/8a and PtGRF6/8b were syntenic with corresponding AtGRF6 and AtGRF8 (Fig 5), and they all had low abundance in floral organs in Populus and Arabidopsis separately (Fig 8a and S3 Fig). Although PtGRF12a and PtGRF12b could not mapped into Arabidopsis synteny blocks, they were highly expressed in floral organs as their orthologous AtGRF12 (Fig 8a and S3 Fig). The similar expression patterns of these orthologous between Populus and Arabidopsis implying that they might keep some conserved functions during the evolution.
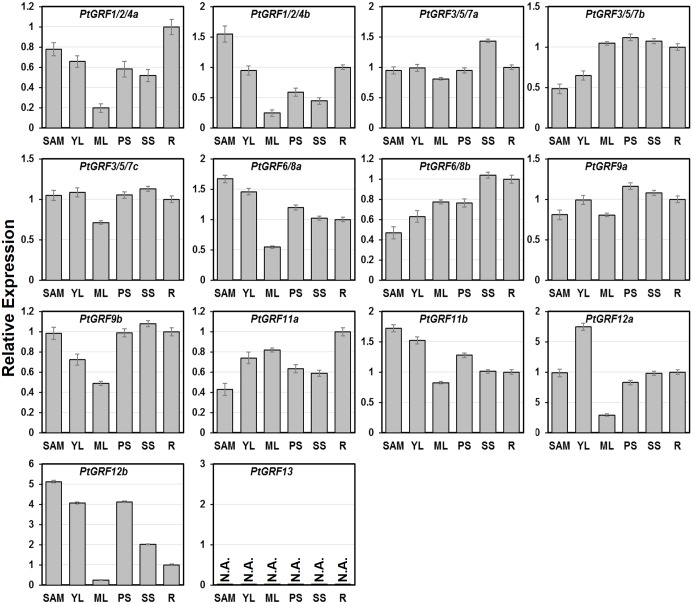
To verify the expression patterns of Populus 14-3-3s obtained by the microarray analysis, qRT-PCR analysis was performed on six different vegetative tissues for fourteen PtGRFs (Fig 9). Sequence specific primers were used to distinguish the amplicons of the paralogous pairs. Noticeably, PtGRF13 was relatively high expressed in male and female catkins by microarray analysis (Fig 8a), but it was not be detected by in silico EST analysis including male and female catkins (Fig 7). Our result also indicated that PtGRF13 cannot be detected in the six selected vegetative tissues by qRT-PCR (Fig 9). The inconsistency is thought to be due to the very low abundance of PtGRF13 and the old version of Populus genome were used in EST analysis. Taken together, the qRT-PCR results were in good agreement with the microarray data sets in this study, although the species (P. alba × P. glandulosa) used for qRT-PCR were different from the ones (P. balsamifera, P. trichocarpa, and P. maximowiczii × nigra) producing microarray data (see Materials and Methods). For example, PtGRF1/2/4a and PtGRF6/8a had low abundance in ML (Fig 9 and Fig 8a), while PtGRF12b and PtGRF9b were highly expressed in PS and SS respectively (Fig 9 and Fig 8b). The similar expression patterns between the qRT-PCR and microarray data suggested the conserved functions within the four species.
Fig 9. Expression profiles of Populus 14-3-3 genes in different tissues using qRT-PCR.
The relative mRNA abundance of Populus 14-3-3 genes were quantified in six vegetative tissues (SAM-shoot apical meristem, YL-young leaves, ML-mature leaves, PS-primary stem, SS-secondary stem, R-root). The average expression of each gene was calculated relatively to the first biological replicate of roots ± standard error (SE) (n≥3). Relative expression represents log2 expression values. All primer sequences are listed in S3 Table.
To further investigate the response of Populus 14-3-3s to abiotic stresses, we examined their expression patterns under low nitrogen limitation, methyl jasmonate (MeJ) treatment, mechanical wounding, hypoxia, and heat (Fig 8d and 8e). PtGRF11a was up-regulated both under nitrogen deprivation stress in eight-week-old mature leaves of genotype 1979 and at 90h after mechanical wounding in mature leaves of genotype RM5. In response to MeJ treatment, three genes (PtGRF11b, 12a, and 12b) were shown to be up-regulated (Fig 8d). In response to hypoxia, PtGRF1/2/4a, 12b, and 13 were significantly up-regulated at 168h after hypoxia in leaves. Under heat stress, only PtGRF11a was up-regulated when the photosynthesis was inhibited by 20% and 30% (Fig 8e). 14-3-3s are induced by various stresses have been identified in many plant species, indicating that they may play critical roles in plant adaptation to these stresses [1,23]. One mechanism by 14-3-3s could act in the regulation of such environmental stress responses is through the regulation of ion channels. In sugar beet cells exposed to cold or osmotic stress, 14-3-3 proteins increase at the plasma membrane and are associated with increased activity the H+-ATPase [1]. 14-3-3s may also exert their effects by interacting with components of hormone signaling pathways. A major pathway activated by stresses such as drought, temperature, and salt stress is the abscisic acid (ABA) signaling pathway [71]. 14-3-3s appear to be involved in the ABA signal pathway by interaction with the AREB/ABF/ABI5-like transcription factors that bind to ABA-response elements [25]. While the exact biochemical roles of Populus 14-3-3s in developmental processes and stress responses need to be further study.
Conclusion
In this present study, we carried out a detailed survey of the 14-3-3 gene family in Populus and characterized them on the bases of phylogenetic relationship, gene structures, gene duplication, and expression profiles across different tissues and abiotic stresses. A total of fourteen 14-3-3 genes were identified in the Populus genome, all of which are clustered into two distinct groups (ε group and non-ε group). Exon/intron structure analyses indicated that the gene structures are relatively conserved in each subgroup. The Populus genome contains six paralogous 14-3-3 gene pairs, and all of them are located in conserved positions in duplicated blocks, suggesting that the segmental duplication play the key role during evolution. Moreover, comparative expression pattern analysis of Populus 14-3-3s revealed that 14-3-3s might play various key roles in different development processes and various abiotic stresses in plants. Although the precise functions of Populus 14-3-3s remain largely unknown, our phylogenetic and expression analyses establishes a solid foundation for future comprehensive function analyses of Populus 14-3-3s.
Supporting Information
Structural comparison of superimposition of fourteen Populus 14-3-3 proteins (yellow) and Nicotiana tabacum 14-3-3 protein (blue, PDB:2o98B). 2D structural elements comparison, show a small deviations (RMSD) between protein conformations (Table 3).
(TIF)
The nine α-helices motifs were illustrated using WEBLOGO (http://weblogo.berkeley.edu/logo.cgi). The phylogenetic trees were constructed by the neighbor-joining method with 1,000 bootstrap replicates based on the separate nine α-helices domains of Populus 14-3-3 proteins. Numbers at each branch indicate bootstrap values. Scale bar corresponds to the estimated number of amino acid substitutions per site.
(TIF)
AtGRF6 and AtGRF8 had low abundance in floral organs. AtGRF12 was highly expressed in flowers and flora organs. The microarray data was obtained from AtGenExpress Visualization Tool (AVT, http://jsp.weigelworld.org/expviz/).
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Suppporting Information files.
Funding Statement
This work was supported by National Nonprofit Institute Research Grant of CAF [TGB2013009] and [CAFYBB2014ZX001-4] to JJH and the China Postdoctoral Science Foundation [2014M550104] to JZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Roberts MR, Salinas J, Collinge DB. 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol Biol. 2002;50(6):1031–9. [DOI] [PubMed] [Google Scholar]
- 2. Chen F, Li Q, Sun L, He Z. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 2006;13(2):53–63. [DOI] [PubMed] [Google Scholar]
- 3. Sehnke PC, DeLille JM, Ferl RJ. Consummating signal transduction the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell. 2002;14(suppl 1):S339–S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fulgosi H, Soll J, de Faria Maraschin S, Korthout HA, Wang M, Testerink C. 14-3-3 proteins and plant development. Plant Mol Biol. 2002;50(6):1019–29. [DOI] [PubMed] [Google Scholar]
- 5. Chung H-J, Sehnke PC, Ferl RJ. The 14-3-3 proteins: cellular regulators of plant metabolism. Trends Plant Sci. 1999;4(9):367–71. [DOI] [PubMed] [Google Scholar]
- 6. Darling DL, Yingling J, Wynshaw-Boris A. Role of 14-3-3 Proteins in Eukaryotic Signaling and Development. Curr Top Dev Biol. 2005;68:281–315. [DOI] [PubMed] [Google Scholar]
- 7. Moore B. Specific acidic proteins of the nervous system. Physiological and Biochemical Aspects of Nervous Integratin. 1967:343–59. [Google Scholar]
- 8. Li X, Dhaubhadel S. Soybean 14-3-3 gene family: identification and molecular characterization. Planta. 2011;233(3):569–82. 10.1007/s00425-010-1315-6 [DOI] [PubMed] [Google Scholar]
- 9. Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40(1):617–47. [DOI] [PubMed] [Google Scholar]
- 10. Hermeking H, Benzinger A. 14-3-3 proteins in cell cycle regulation. Semin Cancer Biol. 2006;16(3):183–92. [DOI] [PubMed] [Google Scholar]
- 11. Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16(3):162–72. [DOI] [PubMed] [Google Scholar]
- 12. Obšlova V, Šilhan J, BouŔa E, Teisinger J, Obšil T. 14-3-3 proteins: a family of versatile molecular regulators. Physiol Res. 2008:S11–S21. [DOI] [PubMed] [Google Scholar]
- 13. Yang X, Lee WH, Sobott F, Papagrigoriou E, Robinson CV, Grossmann JG, et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. P Natl Acad Sci USA. 2006;103(46):17237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3-3sigma functional specificity. J Biol Chem. 2005;280(19):18891–8. [DOI] [PubMed] [Google Scholar]
- 15. Ferl RJ. 14-3-3 proteins and signal transduction. Annu Rev Plant Biol. 1996;47(1):49–73. [DOI] [PubMed] [Google Scholar]
- 16. Jones DH, Martin H, Madrazo J, Robinson KA, Nielsen P, Roseboom PH, et al. Expression and structural analysis of 14-3-3 proteins. J Mol Biol. 1995;245(4):375–84. [DOI] [PubMed] [Google Scholar]
- 17. Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84(6):889–97. [DOI] [PubMed] [Google Scholar]
- 18. Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, et al. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4(2):153–66. [DOI] [PubMed] [Google Scholar]
- 19. Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, et al. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91(7):961–71. [DOI] [PubMed] [Google Scholar]
- 20. DeLille JM, Sehnke PC, Ferl RJ. The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001;126(1):35–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu G, DeLisle AJ, de Vetten NC, Ferl RJ. Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. P Natl Acad Sci USA. 1992;89(23):11490–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finnie C, Borch J, Collinge DB. 14-3-3 proteins: eukaryotic regulatory proteins with many functions. Plant Mol Biol. 1999;40(4):545–54. [DOI] [PubMed] [Google Scholar]
- 23. Denison FC, Paul AL, Zupanska AK, Ferl RJ. 14-3-3 proteins in plant physiology. Semin Cell Dev Biol. 2011;22(7):720–7. 10.1016/j.semcdb.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 24. Yan J, He C, Wang J, Mao Z, Holaday SA, Allen RD, et al. Overexpression of the Arabidopsis 14-3-3 protein GF14λ in cotton leads to a “stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 2004;45(8):1007–14. [DOI] [PubMed] [Google Scholar]
- 25. Schoonheim PJ, Sinnige MP, Casaretto JA, Veiga H, Bunney TD, Quatrano RS, et al. 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 2007;49(2):289–301. [DOI] [PubMed] [Google Scholar]
- 26. Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ. Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol. 2012;195(3):585–95. 10.1111/j.1469-8137.2012.04196.x [DOI] [PubMed] [Google Scholar]
- 27. Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313(5793):1596–604. [DOI] [PubMed] [Google Scholar]
- 28. Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larkin M, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. [DOI] [PubMed] [Google Scholar]
- 30. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. [DOI] [PubMed] [Google Scholar]
- 31. Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29(8):1023–6. [PubMed] [Google Scholar]
- 32. Lee T-H, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(D1):D1152–D8. 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(suppl 2):W609–W12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. [DOI] [PubMed] [Google Scholar]
- 36. Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 37. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S. The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol. 2009;182(4):1013–25. 10.1111/j.1469-8137.2009.02807.x [DOI] [PubMed] [Google Scholar]
- 39. Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149(2):981–93. 10.1104/pp.108.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ko JH, Kim HT, Hwang I, Han KH. Tissue-type-specific transcriptome analysis identifies developing xylem-specific promoters in poplar. Plant Biotechnol J. 2012;10(5):587–96. 10.1111/j.1467-7652.2012.00690.x [DOI] [PubMed] [Google Scholar]
- 41. Dharmawardhana P, Brunner AM, Strauss SH. Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa . BMC Genomics. 2010;11(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, et al. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 2009;149(1):461–73. 10.1104/pp.108.125989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weston DJ, Karve AA, Gunter LE, Jawdy SS, Yang X, Allen SM, et al. Comparative physiology and transcriptional networks underlying the heat shock response in Populus trichocarpa, Arabidopsis thaliana and Glycine max . Plant Cell Environ. 2011;34(9):1488–506. 10.1111/j.1365-3040.2011.02347.x [DOI] [PubMed] [Google Scholar]
- 44. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2− ΔΔCT Method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 45. Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001;127(1):142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y, Wang C, Lu H, Yang H. Molecular evolution and expression pattern of the Populus 14-3-3 gene family. J Beijing Forest Univ. 2010;32(3):1–7 (in Chinese). [Google Scholar]
- 47. Ferl RJ, Manak MS, Reyes MF. The 14-3-3s. Genome Biol. 2002;3(7):reviews3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hardison RC. A brief history of hemoglobins: plant, animal, protist, and bacteria. P Natl Acad Sci USA. 1996;93(12):5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brunner AM, Busov VB, Strauss SH. Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends Plant Sci. 2004;9(1):49–56. [DOI] [PubMed] [Google Scholar]
- 50. Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, et al. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep. 2013;40(3):2645–62. 10.1007/s11033-012-2351-z [DOI] [PubMed] [Google Scholar]
- 51. Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16(7):1667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, et al. Modeling gene and genome duplications in eukaryotes. P Natl Acad Sci USA. 2005;102(15):5454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang X, Tuskan GA, Cheng MZ. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 2006;142(3):820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–5. [DOI] [PubMed] [Google Scholar]
- 56. Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38. [DOI] [PubMed] [Google Scholar]
- 57. Gampala SS, Kim T-W, He J-X, Tang W, Deng Z, Bai M-Y, et al. An Essential Role for 14-3-3 Proteins in Brassinosteroid Signal Transduction in Arabidopsis . Dev Cell. 2007;13(2):177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 2000;19(12):2869–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, et al. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 2009;60(3):539–50. 10.1111/j.1365-313X.2009.03978.x [DOI] [PubMed] [Google Scholar]
- 60. Ottmann C, Marco S, Jaspert N, Marcon C, Schauer N, Weyand M, et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell. 2007;25(3):427–40. [DOI] [PubMed] [Google Scholar]
- 61. Würtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003;22(5):987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57(4):702–10. [DOI] [PubMed] [Google Scholar]
- 63. Obsil T. 14-3-3 proteins—a family of universal scaffolds and regulators. Semin Cell Dev Biol. 2011;22(7):661–2. 10.1016/j.semcdb.2011.09.011 [DOI] [PubMed] [Google Scholar]
- 64. Oecking C, Jaspert N. Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr Opin Plant Biol. 2009;12(6):760–5. 10.1016/j.pbi.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 65. Zhang J, Li J, Liu B, Zhang L, Chen J, Lu M. Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics. 2013;14(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa . BMC Plant Biol. 2010;10:145 10.1186/1471-2229-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pertl H, Gehwolf R, Obermeyer G. The distribution of membrane-bound 14-3-3 proteins in organelle-enriched fractions of germinating lily pollen. Plant Biol. 2005;7(2):140–7. [DOI] [PubMed] [Google Scholar]
- 68. Pertl H, Pöckl M, Blaschke C, Obermeyer G. Osmoregulation in Lilium pollen grains occurs via modulation of the plasma membrane H+-ATPase activity by 14-3-3 proteins. Plant Physiol. 2010;154(4):1921–8. 10.1104/pp.110.165696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bai M-Y, Zhang L-Y, Gampala SS, Zhu S-W, Song W-Y, Chong K, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. P Natl Acad Sci USA. 2007;104(34):13839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang Z-T, Zhou Y, Li Y, Shao S-Q, Li B-Y, Shi H-Y, et al. Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J Exp Bot. 2010;61(12):3331–44. 10.1093/jxb/erq155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chandler PM, Robertson M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Biol. 1994;45(1):113–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural comparison of superimposition of fourteen Populus 14-3-3 proteins (yellow) and Nicotiana tabacum 14-3-3 protein (blue, PDB:2o98B). 2D structural elements comparison, show a small deviations (RMSD) between protein conformations (Table 3).
(TIF)
The nine α-helices motifs were illustrated using WEBLOGO (http://weblogo.berkeley.edu/logo.cgi). The phylogenetic trees were constructed by the neighbor-joining method with 1,000 bootstrap replicates based on the separate nine α-helices domains of Populus 14-3-3 proteins. Numbers at each branch indicate bootstrap values. Scale bar corresponds to the estimated number of amino acid substitutions per site.
(TIF)
AtGRF6 and AtGRF8 had low abundance in floral organs. AtGRF12 was highly expressed in flowers and flora organs. The microarray data was obtained from AtGenExpress Visualization Tool (AVT, http://jsp.weigelworld.org/expviz/).
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Suppporting Information files.