MiR-1188 at the imprinted Dlk1-Dio3 domain is a novel player in hepatocellular carcinoma. MiR-1188 expression was found to be decreased in hepatoma cells, and, when overexpressed, miR-1188 inhibited Bcl2 and Sp1 expression, suppressed cell proliferation and migration, promoted apoptosis in vitro, and inhibited tumor growth in vivo.
Abstract
The aberrant expression of microRNAs (miRNAs) has frequently been reported in cancer studies; miRNAs play roles in development, progression, metastasis, and prognosis. Recent studies indicate that the miRNAs within the Dlk1-Dio3 genomic region are involved in the development of liver cancer, but the role of miR-1188 in hepatocellular carcinoma (HCC) and the pathway by which it exerts its function remain largely unknown. Here we demonstrate that miR-1188 is significantly down-regulated in mouse hepatoma cells compared with normal liver tissues. Enhanced miR-1188 suppresses cell proliferation, migration, and invasion in vitro and inhibits the tumor growth of HCC cells in vivo. Moreover, overexpressed miR-1188 promotes apoptosis, enhances caspase-3 activity, and also up-regulates the expression of Bax and p53. MiR-1188 directly targets and negatively regulates Bcl-2 and Sp1. Silencing of Bcl-2 and Sp1 exactly copies the proapoptotic and anti-invasive effects of miR-1188, respectively. The expression of apoptosis- and invasion-related genes, such as Vegfa, Fgfr1, and Rprd1b, decreases after enhancement of miR-1188, as determined by gene expression profiling analysis. Taken together, our results highlight an important role for miR-1188 as a tumor suppressor in hepatoma cells and imply its potential role in cancer therapy.
INTRODUCTION
MicroRNAs (miRNAs) are conserved short RNAs that suppress protein expression through base pairing with the 3′-untranslated region (UTR) of target mRNAs (Jewer et al., 2012). Increasing evidence suggests that miRNAs play important roles in multiple pathological processes and have significant functions as both tumor suppressors and oncogenes (Ren et al., 2014; Shen et al., 2014; Su et al., 2014). As a new class of genomic information, miRNA dysregulation can provide insight into new carcinogenic pathways and opportunities for biomarker and therapeutic target discovery (Denoyelle et al., 2014; Roderburg and Luedde, 2014).
Hepatocellular carcinoma (HCC) is one of the most common cancers and contributes markedly to increased cancer-related death (Cha and Dematteo, 2005; Aravalli et al., 2008). Altered miRNA expression is observed in HCCs collected from cohorts in different studies. Furthermore, several dysregulated miRNAs have been shown to regulate cell growth, apoptosis, migration, or invasion (Xiong et al., 2010; Lu et al., 2011; Xie et al., 2013; Liu et al., 2014; Wang et al., 2014). These findings suggest that dysregulation of miRNA may be associated with hepatocarcinogenesis. More extensive investigations are required to elucidate the role of miRNAs in the development of HCC in order to identify which ones might be used as novel prognosis predictors or as therapeutic targets for HCC (Zhang et al., 2012; Yang et al., 2013b).
Sp1 is a member of the Sp/Kruppel-like factor (KLF) family (Yasuda et al., 2002). A previous study reported that Sp1 was critically involved in cell growth and tumorigenesis (Lou et al., 2005). Aberrant expression of Sp1 is regulated by miRNAs; this has been observed in various cancers, including hepatocellular carcinomas, breast cancer, and gastric cancer (Xu et al., 2011a, 2012; Banerjee et al., 2012). Bcl-2, as an oncogene, has been implicated in apoptosis and tumorigenesis (Janumyan et al., 2003; Kelly and Strasser, 2011). It has been shown that miR-15 and miR-16 can down-regulate Bcl-2 in many types of cancer, including gastric cancer and chronic lymphocytic leukemia (Cimmino et al., 2005; Xia et al., 2008). Moreover, the introduction of miR-29 dramatically repressed the ability of HCC cells to form tumors and promoted HCC cells apoptosis by targeting Bcl-2 (Xiong et al., 2010).
The Mus musculus intron-encoded miR-1188 is located in the Dlk1-Dio3 imprinted miRNA cluster (Glazov et al., 2008). Previous research showed that overexpression of this miRNA cluster is positively correlated with HCC stem cell markers and associated with a conventional biomarker for liver cancer (Luk et al., 2011). In the present study, low miR-1188 expression was found in a hepatoma cell line when compared with normal liver tissue. Cell proliferation, Transwell cell migration and invasion assays, and in vitro colony formation assays demonstrated that overexpression of miR-1188 suppressed hepatoma cell proliferation and invasion. Moreover, overexpressed miR-1188 promoted apoptosis and enhanced caspase-3 activity; it also up-regulated Bax and p53 protein expression. In addition, miR-1188 suppressed the ability of HCC cells to form tumors in vivo. Furthermore, we used a luciferase reporter assay and Western blot to confirm that miR-1188 might function as a tumor suppressor in hepatoma by targeting Sp1 and Bcl-2. Gene expression profiling analysis of enhanced miR-1188 cells revealed changes in apoptosis- and invasion-related genes such as Vegfa, Fgfr1, and Rprd1b. Our findings will help to elucidate the functions of miR-1188 and its roles in HCC.
RESULTS
The expression of miR-1188 was down-regulated and inversely correlated with Bcl-2/Sp1 in Hepa1-6 cells
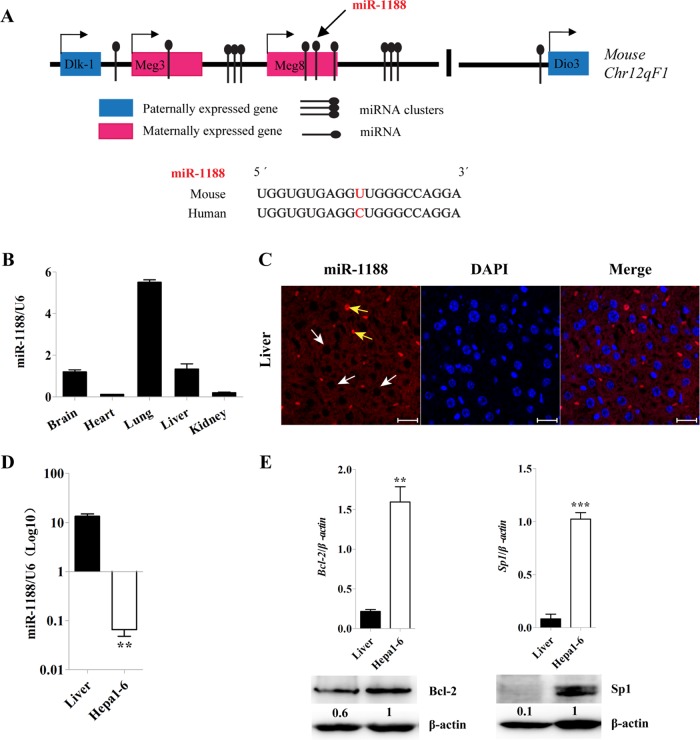
MiR-1188 is located in the Meg8 transcripts from a maternally expressed gene in the Dlk1-Dio3 imprinted cluster on mouse chromosome 12qF1. According to miRBase data and sequence alignment, there is only one base difference between human and mouse miR-1188 (Figure 1A).
FIGURE 1:
The location and expression of miR-1188 in mouse. (A) M. musculus miR-1188 is located on the maternally expressed gene in a Dlk1-Dio3 imprinted cluster. (B, D) qRT-PCR analysis showed the relative levels of miR-1188 in 6-wk-old mice and Hepa1-6 cells. Relative miRNA levels were determined after normalization with U6. (C) Expression of miR-1188 in liver tissue using in situ hybridization (viewed by confocal microscopy). White arrows indicate mature miR-1188 localization in the cytoplasm, and yellow arrows represent blood cells. DAPI staining (blue) was used to visualize the nuclei; scale bars, 10 μm. (E) qRT-PCR and immunoblot analyses showed the relative levels of Bcl-2 and Sp1 in Hepa1-6 cells and liver tissue. Each value was determined in triplicate; **p < 0.01, ***p < 0.001.
Expression of miR-1188 was determined in hepatoma cells and the major organs of mice. Quantitative real-time PCR (qRT-PCR) showed that miR-1188 was widely expressed in the major organs (Figure 1B), and markedly down-regulated in Hepa1-6 cells, when compared with normal liver tissue (Figure 1D). We analyzed miR-1188 in the liver by in situ hybridization; mature miR-1188 localized in the cytoplasm (Figure 1C). We hypothesized that enhanced Bcl-2 and Sp1 expression in hepatoma cells could be a result of reduced miR-1188 expression, so we examined Bcl-2 and Sp1 mRNA and protein levels in Hepa1-6 and normal liver tissue. Figure 1E shows that, compared with normal liver tissue, the expression of Bcl-2 and Sp1 was significantly higher in Hepa1-6 cells.
MiR-1188 suppressed cell proliferation, migration, and invasion in vitro
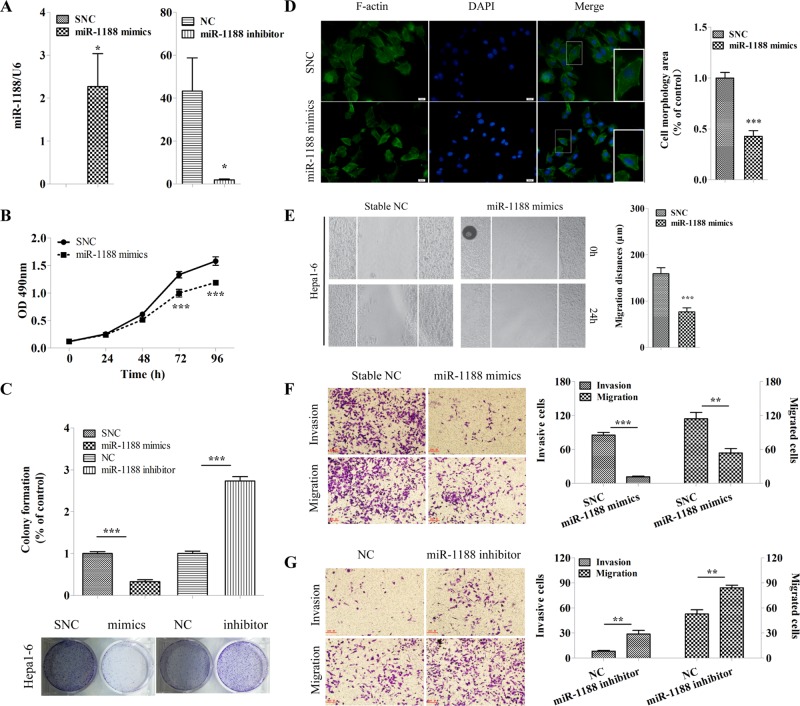
To investigate whether miR-1188 plays a role in the development and progression of liver cancer, we transfected cells with miR-1188 mimics, stable negative controls (SNCs), inhibitor, or negative control (NC). Figure 2A shows that miR-1188 expression in cells transfected with mimics was up-regulated hundredsfold, whereas in cells transfected with inhibitor, the expression decreased by almost 95%. Use of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays demonstrated that cell viability was reduced by 25% in cells transfected with mimics compared with SNC at 96 h (Figure 2B), suggesting a proliferation-suppressive function of miR-1188. Moreover, colony formation assays showed that enhanced miR-1188 levels suppressed colony formation, whereas miR-1188 inhibitor increased colony formation compared with the NC group (Figure 2C). These data indicate that miR-1188 could inhibit the growth of hepatoma cells.
FIGURE 2:
MiR-1188 suppressed cell proliferation, migration, and invasion in Hepa1-6 cells in vitro. (A) qRT-PCR analysis showed the relative levels of miR-1188 after Hepa1-6 cells were transfected with mimics or inhibitor. Relative miRNA levels were determined after normalization with U6. (B) Representative growth curves corresponding to Hepa1-6 cells transfected with SNC or miR-1188 mimics. (C) Colonies grown from Hepa1-6 cells transfected with mimics, SNC, inhibitor, or NC were counted. Percentage of control means the ratio of the experimental group to the control group, with each control defined at a value of 1 for statistical analysis. (D) Immunofluorescence staining of Alexa Fluor 488–conjugated phalloidin, which is a high-affinity probe for F-actin (green), in Hepa1-6 cells transfected with SNC or mimics for 24 h (viewed by fluorescence microscope). Enlarged images of the boxed area are shown in the bottom right corner of the merge image; scale bars, 20 μm. Cell areas (morphology) were calculated and analyzed by image analysis. Ten randomly selected regions were analyzed. (E) Confluent monolayers of cells transfected with SNC or mimics were wounded and incubated for an additional 24 h. The migration distances were calculated. (F, G) Migration and invasion of Hepa1-6 cells transfected with SNC, mimics, NC, or inhibitor. Error bars represent the SE obtained from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001.
Changes in cell morphology are important parameters for cancer invasion and migration. We measured and analyzed the morphology (area) of cells transfected with mimics or SNC in each well of a 24-well plate. The cell size distribution showed significant differences between mimics and SNC-treated cells (Figure 2D). MiR-1188, remarkably, inhibited the cell area by twofold overall compared with SNC, showing that miR-1188 triggers morphological changes in hepatoma cells.
To evaluate whether miR-1188 is biologically involved in the modulation of tumor cell migration and invasion in Hepa1-6 cells, we performed wound-healing and Transwell matrix penetration assays. Figure 2E shows that Hepa1-6 cells transfected with the miR-1188 mimics displayed lower migratory speed than cells transfected with SNC. Consistent with this result, Transwell migration assays showed that the number of cells that migrated after transfection with miR-1188 inhibitor was ∼1.6-fold higher than for NC. Furthermore, the invasion capability of Hepa1-6 cells increased 3.5-fold as a result of miR-1188 inhibition. Opposite results were clearly observed in the mimics group (Figure 2, F and G), indicating an inhibitory effect of miR-1188 on migration and invasion of hepatoma cells. These results suggest that miR-1188 represses traits key to the development of metastasis in hepatoma cells in vitro.
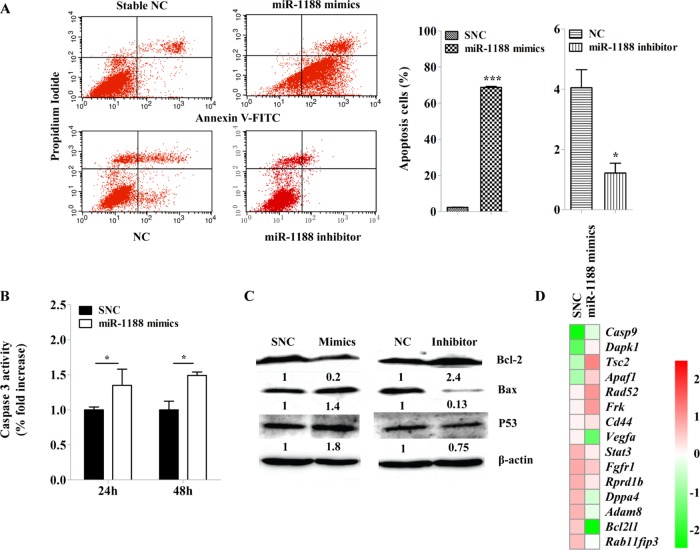
Up-regulation of miR-1188 induced apoptosis and activated caspase-3
Figure 3B shows that transient transfection of miR-1188 mimics resulted in significantly increased caspase-3 activation, which is a key indication of cells undergoing apoptosis. We next observed that up-regulation of miR-1188 enhanced apoptosis by nearly 70-fold compared with SNC in Hepa1-6 cells (Figure 3A). Taken together, these data indicate that up-regulation of miR-1188 induced hepatoma cell apoptosis. To explore the molecules involved in apoptosis, we measured the protein expressions of Bax, Bcl-2, and p53 in enhanced miR-1188 cells. Figure 3C shows that overexpression of miR-1188 significantly suppressed the expression of Bcl-2 in Hepa1-6 cells. However, the expression of Bax and p53 was up-regulated. Taken together, these results indicate that miR-1188 might induce apoptosis partially by targeting Bcl-2.
FIGURE 3:
MiR-1188 induces apoptosis in Hepa1-6 cells in vitro. (A) Annexin V–FITC/PI staining of cells treated with scramble or miR-1188 mimics or inhibitor for 20 h. (B) MiR-1188 increased the activation of caspase-3. (C) Immunoblot analysis of Bcl-2, Bax, and P53 in Hepa1-6 cells transfected with miR-1188 mimics or inhibitor. (D) Heat map of gene expression profile data for 15 genes from Hepa1-6 cells transfected with SNC or miR-1188 mimics. The values were centered and scaled columnwise using the pheatmap R package and sorted by the ratio of mimics and SNC. Color from red to green represents genes that were down-regulated. Error bars represent the SE obtained from three independent experiments; *p < 0.05, ***p < 0.001.
However, the expression of other genes regulated by miR-1188 might also play important roles in tumorigenesis. Gene expression profiling analysis of miR-1188–overexpressing Hepa1-6 cells revealed changes in levels of apoptosis- and tumorigenesis-related genes. Functional grouping analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) showed that 15 of the genes differentially regulated by enhanced miR-1188 were related to apoptosis and migration. The expression of these genes was changed in enhanced miR-1188 cells, indicating that they are in potential relationship with miR-1188 (Figure 3D). Many related genes, such as Vegfa, Fgfr1, and Rprd1b, were down-regulated after enhanced miR-1188 expression. Genes up-regulated by enhanced miR-1188 included tumor suppressors CD44, Dapk1, and Apaf1 (Haraguchi et al., 2000; Lee et al., 2005; Williams et al., 2013).
MR-1188 directly targets Bcl-2 and Sp1
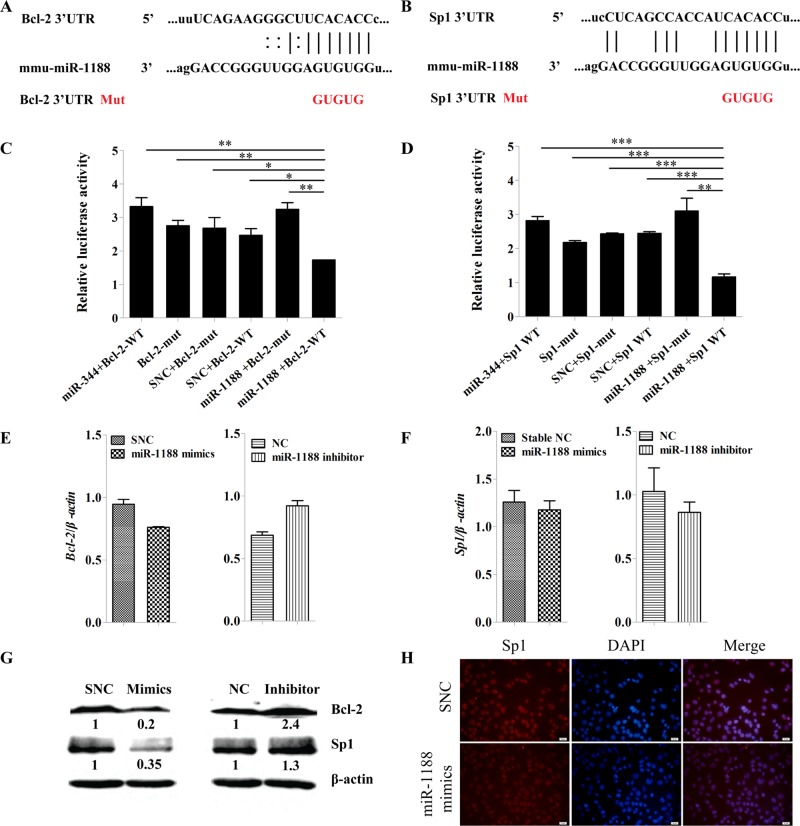
To begin to understand the molecular mechanisms underlying the proapoptotic and anti–migratory/invasive effects of miR-1188, we postulated that Bcl-2 and Sp1 might represent targets of miR-1188, containing in their 3′-UTRs potential binding sites for the miRNA (Figure 4, A and B).
FIGURE 4:
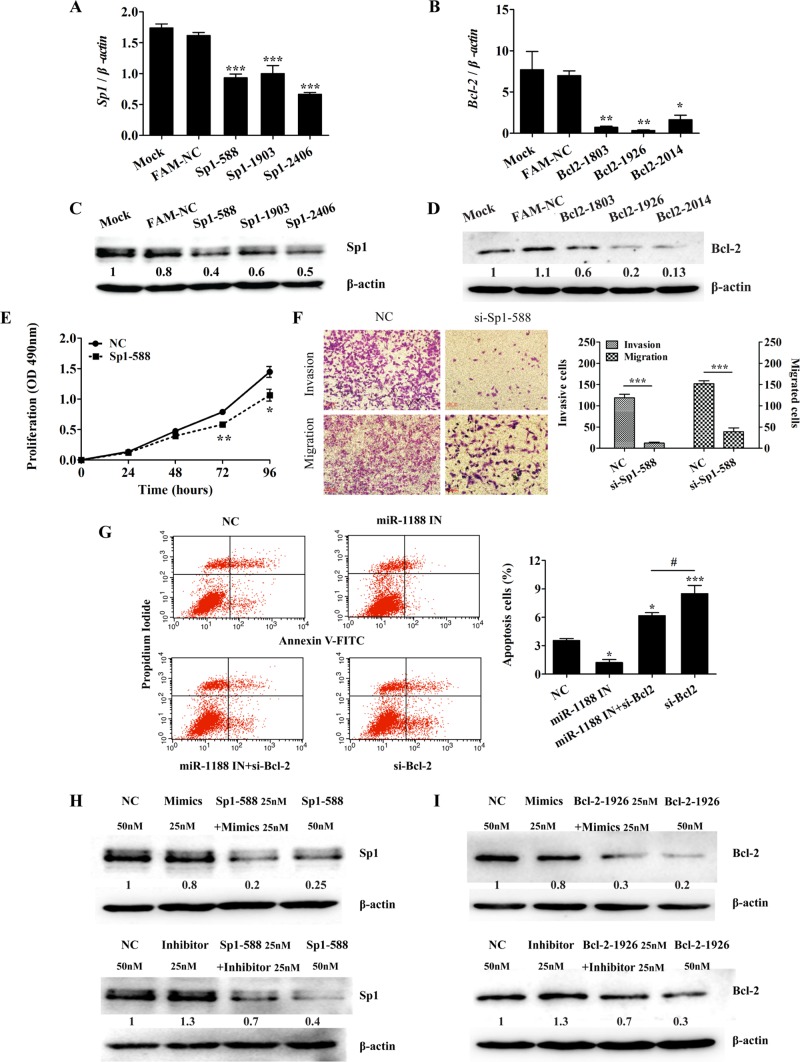
Bcl-2 and Sp1 are direct targets of miR-1188. (A, B) Computer prediction of the binding of miR-1188 to the 3′-UTRs of mouse Bcl-2 and Sp1. The mutated base sequences in the luciferase reporter assays are shown in red. (C, D) Relative luciferase activity of the indicated Bcl-2 or Sp1 reporter constructs. MiR-344 was used as a positive control and does not target Bcl-2/Sp1. (E, F) qRT-PCR analysis shows the relative levels of Bcl-2 and Sp1 after transfection of miR-1188 mimics or miR-1188 inhibitor into Hepa1-6 cells. (G) Immunoblot analysis of Bcl-2 and Sp1 proteins in Hepa1-6 cells transfected with miR-1188 mimics or inhibitor. (H) Immunofluorescence staining showed that Sp1 expression was regulated by miR-1188. Overexpression of miR-1188 suppressed Sp1 expression in Hepa1-6 cells. DAPI staining (blue) was used to visualize nuclei; scale bars, 20 μm. Error bars represent the SE obtained from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001.
To address whether these two genes were directly regulated by miR-1188, we performed luciferase reporter assays. The wild-type Bcl-2 and Sp1 3′-UTRs, as well as mutant forms, were cloned into a pMIR-REPORT luciferase vector. Hepa1-6 cells were transiently transfected with Bcl-2 or Sp1 3′-UTRs and miR-1188 mimics or SNC. Renilla luciferase vector was used for normalization. The results showed that miR-1188 inhibited the luciferase activity of the wild-type Bcl-2 and Sp1 3′-UTRs but not the mutant Bcl-2 and Sp1 3′-UTRs, suggesting that miR-1188 could directly target Bcl-2 and Sp1 (Figure 4, C and D).
To assess further whether miR-1188 repressed Bcl-2 or Sp1 expression in the mouse hepatoma intracellular environment, we analyzed Bcl-2 and Sp1 expression in Hepa1-6 cells after transfection with miR-1188 mimics or inhibitor. MiR-1188 changed the expression of Bcl-2 and Sp1 at the posttranscriptional level (Figure 4, E and F). Western blot assays showed that Bcl-2 and Sp1 protein levels were dramatically decreased in the miR-1188 mimics–treated group. Conversely, levels of Bcl-2 and Sp1 were increased in miR-1188 inhibitor groups compared with NC treatment (Figure 4G). To further confirm that Sp1 was negatively regulated by miR-1188, we analyzed Sp1 expression by immunofluorescence. Sp1 was predominantly located in the nucleus 48 h after the transfection of miR-1188 mimics, and the level of Sp1 was significantly reduced compared with SNC (Figure 4H). Collectively these results suggest that Bcl-2 and Sp1 are direct targets of miR-1188, and miR-1188 affects Bcl-2 and Sp1 expression by directly binding to their 3′-UTR target regions.
MiR-1188 mediated apoptosis and invasion by modulating Bcl-2 and Sp1 levels
To investigate further the relationship between miR-1188 and its targets, we knocked down Bcl-2 and Sp1 by RNA interference. Expression of Bcl-2 and Sp1 was down-regulated, as shown by qRT-PCR and Western blot (Figure 5, A– D). To confirm further that Sp1 was regulated by small interfering RNA (siRNA), we analyzed Sp1 expression using immunofluorescence (unpublished data). Subsequently, we used Sp1–5′-UTR 588 and Bclx2-5′-UTR 1926 siRNAs for further experiments. As expected, knockdown of Sp1 markedly inhibited cell proliferation as determined by MTT assays; the suppression rate was 25 and 34% at 72 and 96 h, respectively (Figure 5E). Similarly, Sp1 knockdown reduced cell migration and invasion in Hepa1-6 cells (Figure 5F). These results suggested that knockdown of Sp1 changed the biological behavior of Hepa1-6 cells in much the same pattern as miR-1188 up-regulation. In addition, Figure 5G shows that suppression of Bcl-2 by siRNA promoted apoptosis and miR-1188 inhibitor led to reduced apoptosis, whereas miR-1188 inhibitor rescued apoptosis caused by suppression of Bcl-2 in cells cotransfected with inhibitor and siRNA compared with cells cotransfected with siRNA alone.
FIGURE 5:
MiR-1188 mediates invasion and apoptosis by modulating Bcl-2 and Sp1 levels. (A, B) qRT-PCR analysis showed the relative levels of Bcl-2 and Sp1 in Hepa1-6 cells transfected with NC or siRNA, respectively. (C, D) Immunoblot analysis of Bcl-2 and Sp1 in Hepa1-6 cells transfected with siRNA. (E) Representative growth curves corresponding to Hepa1-6 cells transfected with NC or si-Sp1-588. (F) Migration and invasion of Hepa1-6 cells transfected with NC or si-Sp1-588. (G) Annexin V–FITC/PI staining of cells cotransfected with NC or miR-1188 inhibitor and/or si-Bcl-2 for 20 h. (H, I) Immunoblot analysis showed that the levels of Bcl-2 and Sp1 were changed in Hepa1-6 cells transfected with miR-1188 mimics, miR-1188 inhibitor, si-Bcl-2, or si-Sp1. More significantly, miR-1188 inhibitor rescued the level of Bcl-2 or Sp1 in cells cotransfected with inhibitor and siRNAs compared with cells transfected with siRNA alone. Error bars represent the SE obtained from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001.
Western blots showed that the levels of Bcl-2 and Sp1 were reduced in hepatoma cells transfected with either miR-1188 mimics or Bcl-2/Sp1 siRNAs (Figures 5, H and I). In contrast, down-regulation of miR-1188 elevated the expression of both Bcl-2 and Sp1. More significantly, miR-1188 inhibitor rescued the protein expression of Bcl-2 or Sp1 in cells cotransfected with inhibitor and siRNA compared with cells cotransfected with siRNA alone (Figure 5, H and I). These results indicate that Bcl-2 or Sp1 might be involved in mediating the tumor-suppressive function of miR-1188.
MiR-1188 inhibits tumor growth in vivo
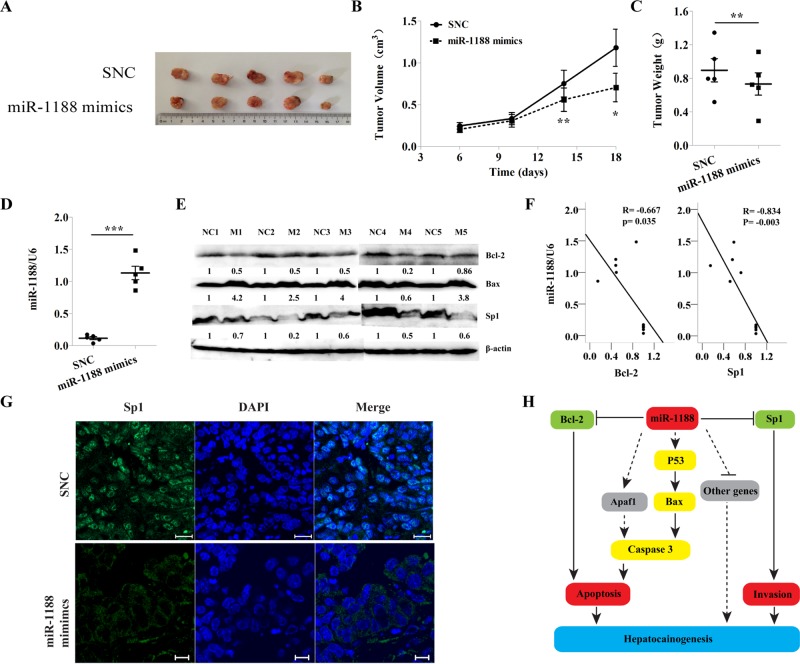
The data showing that miR-1188 negatively modulated hepatoma cell proliferation in vitro prompted us to ask whether the miRNA acted as an antitumor factor in vivo. To address this issue, we measured dynamic growth of subcutaneously xenografted hepatoma tumors in BALB/c-nu mice over 18 d. Our results showed that mice bearing control and miR-1188–overexpressing Hepa1-6 cells on the left and right flanks, respectively, began to exhibit differences in tumor growth between the two sides in the first week, and the difference increased until the experimental endpoint (Figure 6, A and B). This trend was confirmed by the weights of the dissected tumors (Figure 6C), strongly suggesting a marked deceleration of tumor cell proliferation by miR-1188.
FIGURE 6:
MiR-1188 inhibited hepatoma tumor growth in vivo. (A) Hepa1-6 cells were injected subcutaneously into the left and right flanks of nude mice and the transplanted tumors harvested (n = 5). (B, C) The weight and volume of the harvested transplanted tumors. (D) The expression of miR-1188 in the tumors as found by qRT-PCR. (E) The expression of Bax, Bcl-2, and Sp1 in the tumors as revealed by immunoblot. (F) Inverse correlation between miR-1188 and Sp1 or Bcl-2 protein level in the tumors. Statistical analysis was performed using Pearson's R, and R and P values are as shown. (G) Immunohistochemistry showed the Sp1 expression (viewed by confocal microscopy). DAPI staining (blue) was used to visualize nuclei; Scale bars, 20 μm. (H) Schematic diagram showing the signaling network in which miR-1188 is involved. Error bars represent the SE obtained from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001.
qRT-PCR revealed that the expression of miR-1188 was increased in hepatoma xenograft tumor tissues treated with miR-1188 mimics compared with those treated with SNC (Figure 6D). Western blot showed that expression of Bcl-2 and Sp1 was decreased in tumor tissues containing mimics compared with SNC (Figure 6E). Correlation analysis indicated that there was a significant inverse correlation between miR-1188 and Bcl-2/Sp1 protein expression, with R = –0.834 and –0.667, respectively (Figure 6F). Furthermore, tumor tissues were embedded in paraffin and then stained for histological examination. To observe the localization of Sp1 protein in tumor tissues, we performed fluorescence immunohistochemical staining. Green fluorescence indicated Sp1 protein expression (Figure 6G), which was located in the nucleus. Together these data support our in vitro findings that miR-1188 was inversely correlated with Bcl-2/Sp1 and that Bcl-2 and Sp1 were potential target genes for miR-1188.
DISCUSSION
MiR-1188 is located in transcripts of Meg8, which has been identified as a maternally expressed gene in the mouse Dlk1-Dio3 imprinted cluster. The imprinted miRNA cluster is involved in the pathogenesis of different diseases (Benetatos et al., 2013) and modulates important pathways such as mitogen-activated protein kinase (MAPK), Wnt, JAK-STAT, P53, and many more (Iliopoulos et al., 2009; Swarbrick et al., 2010; Gattolliat et al., 2011). For example, miR-127 inhibited cell proliferation and migration by targeting MAPK4 in gastric cancer cells (Guo et al., 2013). Down-regulated miR-136 targets AEG-1 and Bcl-2 in human glioma, which promotes the apoptosis of glioma (Yang et al., 2012). Other evidence suggests that miRNAs at the imprinted Dlk1-Dio3 cluster are also involved in the development of liver cancer. MiR-433 is a potent inhibitor of HCC cell migration through its action on CREB1 (Yang et al., 2013a). MiR-127 can result in the formation of hepatocellular carcinoma by decreasing apoptosis rates (Tryndyak et al., 2009). MiR-370, which is also located among the Meg8 transcripts, can inhibit the growth and metastasis of HCC cells in vivo (Xu et al., 2013). In our study, miR-1188 could regulate cell proliferation, apoptosis, migration, and invasion and suppress tumor growth by directly targeting Bcl-2 and Sp1 in hepatoma cells, suggesting that miR-1188 may also participate in the development of liver cancer.
The present study reports, for the first time, the functional effect of miR-1188 as a novel tumor suppressor in hepatoma cells. MiR-1188 was expressed at a low level in hepatoma cells compared with normal liver tissue, which suggested that down-regulation of the expression level of miR-1188 was correlated with cancer progression. Overexpression of miR-1188 in hepatoma cell lines reduced cell viability and invasion ability, promoted apoptosis, and suppressed tumorigenicity, suggesting a tumor suppressor activity. Therefore, miR-1188 might serve an important function in apoptosis and invasion pathways. Furthermore, we characterized Bcl-2 and Sp1 as functional targets of miR-1188 and proved their involvement (Figure 6H), thereby indicating a fundamental role of miR-1188 in tumorigenesis and suggesting the potential application of miR-1188 in prognosis prediction and cancer therapy.
Most cases of HCC arise in cirrhotic livers with persistent inflammation. Further understanding of the mechanistic link between inflammation and HCC would help to identify potential therapeutic targets for HCC. Sp1 is a key transcriptional regulator that exerts a variety of biological functions. Elevated expression of Sp1 has been observed in HCC (Lietard et al., 1997; Yang et al., 2001; Fang et al., 2007). Of interest, Sp1 could regulate miR-365, which is involved in the NF-κB pathway (Xu et al., 2011b), whereas NF-κB has been shown to play a major role in lipopolysaccharide-induced expression of interleukin-6 and other inflammatory cytokines (Luedde and Schwabe, 2011). In our study, knockdown of Sp1 expression decreased cell proliferation, migration, and invasion, the same as observed with increased levels of the miR-1188. More significantly, the relationship between miR-1188 and Sp1 was identified by Western blot of cells cotransfected with mimics or inhibitor and siRNAs. In the inhibitor groups, even if there was an Sp1 interference fragment, miR-1188 knockdown still rescued the expression of Sp1. Moreover, we analyzed the negative correlation of the expression levels of miR-1188 and Sp1 in vivo. Collectively our data suggest that in HCC, miR-1188 mediates invasion by the modulation of Sp1 levels. Thus miR-1188 might be linked with inflammatory pathways by targeting Sp1 as well as other pathways in liver cells.
Next we found that enhanced miR-1188 could dramatically induce apoptosis of hepatoma cells. Previous studies showed that proteins in the Bcl-2 family are central regulators of programmed cell death (Lessene et al., 2008), and p53 acts as a tumor suppressor in many tumor types. First, we detected the expression of Bcl2, Bax, and p53. Of interest, enhanced miR-1188 decreased Bcl-2 protein levels and increased Bax and p53 protein levels. By using a bioinformatic analysis, we confirmed that Bcl-2 was a target of miR-1188. Furthermore, suppression of Bcl-2 by siRNA promoted apoptosis, and miR-1188 inhibitor led to reduced apoptosis, whereas miR-1188 inhibitor rescued apoptosis caused by suppression of Bcl-2 in cells cotransfected with inhibitor and siRNA. Moreover, the relationship between miR-1188 and Bcl-2 was also identified by Western blot in cells cotransfected with mimics or inhibitor and siRNAs. This confirmed that miR-1188 indirectly targeted Bcl-2 and was involved in hepatoma cell apoptosis. The mechanism by which miR-1188 regulates p53 in hepatocellular carcinoma needs to be addressed in further studies. We speculate that p53 is involved in another apoptosis pathway that causes miR-1188 to induce hepatoma cell apoptosis (Figure 6H).
Although we found that Bcl-2 and Sp1 are involved in miR-1188–mediated hepatocellular carcinoma progression, Bcl-2 or Sp1 knockdown did not completely abolish the effects of miR-1188 knockdown, suggesting that other genes regulated by miR-1188 may contribute to inhibition of apoptosis and invasion by miR-1188. Therefore we further determined the expression profile of miR-1188–regulated genes. Many tumorigenesis-related genes involved in apoptosis and invasion were induced at enhanced levels in cells with elevated miR-1188, including Vegfa, Fgfr1, and Rprd1b. They are all proto-oncogenes involved in cell proliferation, migration, and apoptosis (Wang et al., 2013; Zhou et al., 2013). Rprd1b was shown to enhance the transcription of CCND1 and promote the transcriptional activity of the β-catenin–TCF4 complex in response to Wnt signaling (Zhang et al., 2014). In the present study, enhanced miR-1188 decreased the expression of CCND1 (unpublished data), which suggested that Rprd1b might be a putative miR-1188 target. Certainly, genes up-regulated by enhanced miR-1188, such as CD44, Dapk1, and Apaf1, have a tumor suppressor function (Haraguchi et al., 2000; Lee et al., 2005; Williams et al., 2013). In addition, Bcl-2 suppressed Apaf1-independent cell death (Haraguchi et al., 2000). The correlation between miR-1188 and these genes during the course of hepatocellular carcinoma needs to be addressed in further studies.
In summary, this work investigated the potential role of miR-1188 in tumorigenesis and its underlying mechanisms. The data suggest that down-regulation of miR-1188 might play an important role in the development of liver cancer and that miR-1188 might be used as a prognostic marker and therapeutic target for hepatocellular carcinoma.
MATERIALS AND METHODS
Cell line and cell culture
Mouse HCC cell line Hepa1-6 was provided by the Institute of Biochemistry and Cell Biology of the Chinese Academy of Science (Shanghai, China) and originated from the American Type Culture Collection (Manassas, VA). Cells were cultured in DMEM with 10% fetal bovine serum and 2 mM l-glutamine (Corning, Corning, NY). The cell line was incubated at 37°C under a 5% CO2 atmosphere.
RNA isolation and qRT-PCR
Total RNAs were isolated from mouse liver tissues and Hepa1-6 cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA) and a mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA). The expression of miR-1188 was analyzed by TaqMan miRNA assays according to the manufacturer's protocols (Applied Biosystems) using RNU6B as an endogenous control. The mRNA expression of Bcl-2/Sp1 was analyzed by SYBR qPCR assays (Applied Biosystems) using β-actin as an endogenous control. The comparative threshold cycle (CT) method was used to quantify the target genes relative to the endogenous control. Primers used for PCR are listed in Supplemental Table S1.
MiRNA and siRNA transfections
All miRNAs and siRNAs, including negative controls (scrambled RNAs), were chemically synthesized by GenePharma (Shanghai, China) and transfected into Hepa1-6 cells at a final concentration of 25, 50, or 100 nM with Lipofectamine 2000 (Invitrogen). Sequences are listed in Supplemental Table S2.
MTT and colony formation assays
Hepa1-6 cells at 24 h posttransfection were seeded at 2 × 103 cells/well in 96-well plates and the procedure was repeated after 24, 48, 72, and 96 h. Ten microliters of MTT was added to 100 μl of culture medium/well. After 4 h of incubation at 37°C, the medium was removed and 150 μl of dimethyl sulfoxide was added. The absorbance was measured at a wavelength of 490 nm in a plate reader (Bio-Rad, Hercules, CA). Soft agar colony formation was performed as described previously and measured after 10 d (Stephenson et al., 1971). Each assay was performed in triplicate.
Wound-healing, cell migration, and invasion assays
Confluent monolayer cells were scratched with a 200-μl pipette tip and then washed with phosphate-buffered saline (PBS) to clear cell debris. Fresh serum-free medium was added, and the cells were allowed to close the wound for 24 h.
Cells (3 × 104) with the indicated transfections were plated on the top side of Transwell chambers (Corning) coated with or without Matrigel (BD Biosciences, San Diego, CA). After incubation for 48 h, cells were removed from the upper chamber with cotton swabs while the lower surface cells were fixed with 4% paraformaldehyde (PFA) at room temperature for 30 min and stained with 0.1% crystal violet for 20 min. Five independent fields were counted under the microscope (BX51; Olympus, Tokyo, Japan). Each assay was performed in triplicate.
Caspase-3 activity assay
The activity of caspase-3 was determined using a caspase-3 activity kit (Beyotime, Beijing, China) according to the manufacturer's instructions. Cell lysates at 24 or 48 h posttransfection were assessed in 96-well microtiter plates by incubating 10 μl of protein of cell lysate/sample in 80 μl of reaction buffer containing 10 μl of caspase-3 substrate (Ac-DEVD-pNA). Lysates were incubated at 37°C for 4 h, and then the samples were measured at a wavelength of 405 nm in a plate reader (Bio-Rad). Each assay was performed in triplicate.
Flow cytometric assay
Hepa1-6 cells at 20 h posttransfection were washed twice with cold PBS and then resuspended. Cells were suspended in 1× binding buffer (BD Biosciences) at a concentration of 1 × 106 cells/ml. Then 100 μl of the solution (1 × 105 cells) was transferred to a 5-ml culture tube; 5 μl of fluorescein isothiocyanate (FITC)–Annexin V and 5 μl of propidium iodide (PI) were added, and the mixture was incubated for 15 min at room temperature in the dark. Finally, 400 μl of 1× binding buffer was added to the tube, and the samples were analyzed by flow cytometry (FACSCantoII; BD Biosciences). The data were analyzed using FlowJo software. Each assay was performed in triplicate.
Plasmid construction and luciferase reporter assays
Wild-type and mutant 3′-UTRs were amplified from mouse cDNA using the primers listed in Supplemental Table S1. Mutations in miRNA target sites were generated using the KOD-Plus-Mutagenesis Kit (Toyobo, Japan). Target fragments were cloned into the pMIR-REPORT luciferase vector (Ambion, Austin, TX), to produce pMIR-Bcl-2/Sp1-3′UTR-WT and pMIR-Bcl-2/Sp1-3′UTR-mut. Hepa1-6 cells were seeded into 24-well plates and cotransfected with 0.5 μg of the appropriate pMIR-REPORT luciferase vector, 0.05 μg of pRL-TK Renilla luciferase vector, and 100 nM miR-1188 mimics or stable negative control, using Lipofectamine 2000. After 48 h of transfection, luciferase activity was measured using the dual luciferase reporter assay system (Promega, Madison, WI). Each assay was performed in triplicate.
Western blot analysis
Cells in six-well plates at 48 h posttransfection were lysed with RIPA buffer. A total of 20–40 μg of whole protein was run on 7.5% SDS–PAGE and transferred onto polyvinylidene fluoride membrane. The membrane was blocked and then incubated with primary antibodies overnight at 4°C. In this study, we used antibodies including Bcl-2, Bax, P53, β-actin (Cell Signaling Technology, Beverly, MA), and Sp1 (Abcam, Cambridge, United Kingdom). Quantitative western blot was performed using an Odyssey infrared imaging system (LI-COR, Lincoln, NE).
Immunofluorescence
Hepa1-6 cells were cultured on 24-well chamber slides and fixed with 4% PFA for 30 min. The slides were washed in PBS three times and incubated at 4°C overnight with antibody against Sp1 or F-actin diluted 1:2500 in 3% bovine serum albumin (BSA)/PBS. After washing with PBS and incubation at 37°C for 2 h with anti-FITC antibodies diluted 1:1000 in 1% BSA/PBS, the slides were incubated with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and examined using a fluorescence microscope (Olympus).
Immunohistochemistry and in situ hybridization
Tissues were fixed in 4% PFA and embedded in paraffin blocks. Sections (3 μm) were used for examination. Sp1 expression was detected following the protocol described in Immunofluorescence. In situ hybridization was performed using methods described previously (Huang et al., 2013). MiR-1188 LAN probes (Exiqon; http://Exiqon.com) were labeled with a DIG-3′-end labeling kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. An antisense LNA probe for miR-1188, with sequence TGGTGTGAGGTTGGGCCAGGA, was purchased from Exiqon. Sense probes were used as negative controls for all experiments. The signal was detected by incubation with anti-FITC antibodies (1:1000; Invitrogen). Nuclei were stained with DAPI. The images were recorded with a confocal microscope (Zeiss, Jena, Germany).
In vivo assay
Six-week-old female BALB/C athymic mice were used for examining tumorigenicity. Cells (5 × 106) at 48 h posttransfection were resuspended in 100 μl of PBS and implanted into the left and right flanks by subcutaneous injection. The volume of the tumor developed was measured with a caliper every 4 d for 18 d, and the tumor volume was calculated according to volume = (length × width2)/2. All animal procedures were performed according to protocols approved by the Rules for Animal Experiments published by the Chinese Government (Beijing, China).
Microarray analysis
Total RNA was extracted from Hepa1-6 cells transfected with miR-1188 mimics and stable negative control using TRIzol. Microarray analysis was performed by Genergy Bio (JingNeng, Shanghai, China). The DAVID Bioinformatics Resources database was used to perform gene function enrichment analysis.
Statistical analysis
Data are presented as the mean ± SEM of at least three independent experiments. Two-tailed Student's t test and one-way analysis of variance were used to analyze significant differences. We performed Spearman's correlation analysis between miRNAs and their corresponding targets. p < 0.05, p < 0.01, and p < 0.001 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Natural Science Foundation of China (Grants 31371478, 31171383, 31100934, and 31100928), the State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (No. 2013DX07), the Natural Science Foundation of Heilongjiang Province (Grant QC2014C016), and the Innovation and Technology Special Fund for Researchers of Harbin City (Grant 2013RFXXJ010).
Abbreviations used:
- HCC
hepatocellular carcinoma
- miRNA
microRNA
- PFA
paraformaldehyde
- qRT-PCR
quantitative real-time PCR
- UTR
untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1576) on February 18, 2015.
REFERENCES
- Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- Banerjee N, Talcott S, Safe S, Mertens-Talcott SU. Cytotoxicity of pomegranate polyphenolics in breast cancer cells in vitro and vivo: potential role of miRNA-27a and miRNA-155 in cell survival and inflammation. Breast Cancer Res Treat. 2012;136:21–34. doi: 10.1007/s10549-012-2224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I, Briasoulis E. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814. doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha C, Dematteo RP. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol. 2005;19:25–37. doi: 10.1016/j.bpg.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle C, Lambert B, Meryet-Figuiere M, Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH, Gauduchon P, et al. miR-491–5p-induced apoptosis in ovarian carcinoma depends on the direct inhibition of both BCL-XL and EGFR leading to BIM activation. Cell Death Dis. 2014;5:e1445. doi: 10.1038/cddis.2014.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Fu Y, Liang Y, Li Z, Zhang W, Jin J, Yang Y, Zha X. Increased expression of integrin beta1 subunit enhances p21WAF1/Cip1 transcription through the Sp1 sites and p300-mediated histone acetylation in human hepatocellular carcinoma cells. J Cell Biochem. 2007;101:654–664. doi: 10.1002/jcb.21223. [DOI] [PubMed] [Google Scholar]
- Gattolliat CH, Thomas L, Ciafre SA, Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P, Valteau-Couanet D, et al. Expression of miR-487b and miR-410 encoded by 14q32.31 locus is a prognostic marker in neuroblastoma. Br J Cancer. 2011;105:1352–1361. doi: 10.1038/bjc.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, McWilliam S, Barris WC, Dalrymple BP. Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol Biol Evol. 2008;25:939–948. doi: 10.1093/molbev/msn045. [DOI] [PubMed] [Google Scholar]
- Guo LH, Li H, Wang F, Yu J, He JS. The tumor suppressor roles of miR-433 and miR-127 in gastric cancer. Int J Mol Sci. 2013;14:14171–14184. doi: 10.3390/ijms140714171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi M, Torii S, Matsuzawa S, Xie Z, Kitada S, Krajewski S, Yoshida H, Mak TW, Reed JC. Apoptotic protease activating factor 1 (Apaf-1)-independent cell death suppression by Bcl-2. J Exp Med. 2000;191:1709–1720. doi: 10.1084/jem.191.10.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Han Z, Cui W, Zhang F, He H, Zeng T, Sugimoto K, Wu Q. Dynamic expression pattern of Pde4d and its relationship with CpG methylation in the promoter during mouse embryo development. Biochem Biophys Res Commun. 2013;441:982–987. doi: 10.1016/j.bbrc.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Bimpaki EI, Nesterova M, Stratakis CA. MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Res. 2009;69:3278–3282. doi: 10.1158/0008-5472.CAN-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janumyan YM, Sansam CG, Chattopadhyay A, Cheng N, Soucie EL, Penn LZ, Andrews D, Knudson CM, Yang E. Bcl-xL/Bcl-2 coordinately regulates apoptosis, cell cycle arrest and cell cycle entry. EMBO J. 2003;22:5459–5470. doi: 10.1093/emboj/cdg533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewer M, Findlay SD, Postovit LM. Post-transcriptional regulation in cancer progression: microenvironmental control of alternative splicing and translation. J Cell Commun Signal. 2012;6:233–248. doi: 10.1007/s12079-012-0179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Strasser A. The role of Bcl-2 and its pro-survival relatives in tumourigenesis and cancer therapy. Cell Death Differ. 2011;18:1414–1424. doi: 10.1038/cdd.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Rho SB, Chun T. Programmed cell death 6. (PDCD6) protein interacts with death-associated protein kinase 1 (DAPk1): additive effect on apoptosis via caspase-3 dependent pathway. Biotechnol Lett. 2005;27:1011–1015. doi: 10.1007/s10529-005-7869-x. [DOI] [PubMed] [Google Scholar]
- Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov. 2008;7:989–1000. doi: 10.1038/nrd2658. [DOI] [PubMed] [Google Scholar]
- Lietard J, Musso O, Theret N, L'Helgoualc'h A, Campion JP, Yamada Y, Clement B. Sp1-mediated transactivation of LamC1 promoter and coordinated expression of laminin-gamma1 and Sp1 in human hepatocellular carcinomas. Am J Pathol. 1997;151:1663–1672. [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Lin XJ, Yang XJ, Zhou L, He S, Zhuang SM, Yang J. A novel AP-1/miR-101 regulatory feedback loop and its implication in the migration and invasion of hepatoma cells. Nucleic Acids Res. 2014;42:12041–12051. doi: 10.1093/nar/gku872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, O'Reilly S, Liang H, Maher VM, Sleight SD, McCormick JJ. Down-regulation of overexpressed sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 2005;65:1007–1017. [PubMed] [Google Scholar]
- Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, Blazar BR, Zeng Y, Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T, Schwabe RF. NF-kappaB in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk JM, Burchard J, Zhang C, Liu AM, Wong KF, Shek FH, Lee NP, Fan ST, Poon RT, Ivanovska I, et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem. 2011;286:30706–30713. doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren LH, Chen WX, Li S, He XY, Zhang ZM, Li M, Cao RS, Hao B, Zhang HJ, Qiu HQ, Shi RH. MicroRNA-183 promotes proliferation and invasion in oesophageal squamous cell carcinoma by targeting programmed cell death 4. Br J Cancer. 2014;111:2003–2013. doi: 10.1038/bjc.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol. 2014;61:1434–1437. doi: 10.1016/j.jhep.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Shen G, Rong X, Zhao J, Yang X, Li H, Jiang H, Zhou Q, Ji T, Huang S, Zhang J, Jia H. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;35:2748–2755. doi: 10.1093/carcin/bgu208. [DOI] [PubMed] [Google Scholar]
- Stephenson JR, Axelrad AA, McLeod DL, Shreeve MM. Induction of colonies of hemoglobin-synthesizing cells by erythropoietin in vitro. Proc Natl Acad Sci USA. 1971;68:1542–1546. doi: 10.1073/pnas.68.7.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, Zhai PF, Gong JN, Shen C, Song L, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene, doi: 10.1038/onc.2014.274. 2014 doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- Swarbrick A, Woods SL, Shaw A, Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ, Judson RL, et al. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16:1134–1140. doi: 10.1038/nm.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48:479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- Wang J, Li J, Wang X, Zheng C, Ma W. Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem Biophys Res Commun. 2013;439:47–53. doi: 10.1016/j.bbrc.2013.08.032. [DOI] [PubMed] [Google Scholar]
- Wang SC, Lin XL, Li J, Zhang TT, Wang HY, Shi JW, Yang S, Zhao WT, Xie RY, Wei F, et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS One. 2014;9:e101330. doi: 10.1371/journal.pone.0101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Motiani K, Giridhar PV, Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Exp Biol Med (Maywood) 2013;238:324–338. doi: 10.1177/1535370213480714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- Xie C, Jiang XH, Zhang JT, Sun TT, Dong JD, Sanders AJ, Diao RY, Wang Y, Fok KL, Tsang LL, et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene. 2013;32:2282–2291. doi: 10.1038/onc.2012.251. 2291.e1–7. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011a;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WP, Yi M, Li QQ, Zhou WP, Cong WM, Yang Y, Ning BF, Yin C, Huang ZW, Wang J, et al. Perturbation of MicroRNA-370/Lin-28 homolog A/nuclear factor kappa B regulatory circuit contributes to the development of hepatocellular carcinoma. Hepatology. 2013;58:1977–1991. doi: 10.1002/hep.26541. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, Jiang L, Sun Z, Miao Z, Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Xiao SB, Xu P, Xie Q, Cao L, Wang D, Luo R, Zhong Y, Chen HC, Fang LR. miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-kappaB. J Biol Chem. 2011b;286:21401–21412. doi: 10.1074/jbc.M110.198630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Huang ZZ, Wang J, Lu SC. The role of c-Myb and Sp1 in the up-regulation of methionine adenosyltransferase 2A gene expression in human hepatocellular carcinoma. FASEB J. 2001;15:1507–1516. doi: 10.1096/fj.01-0040com. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013a;288:28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang Y, Wang L. A feedback inhibition between miRNA-127 and TGFbeta/c-Jun cascade in HCC cell migration via MMP13. PLoS One. 2013b;8:e65256. doi: 10.1371/journal.pone.0065256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wu J, Guan H, Cai J, Fang L, Li J, Li M. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett. 2012;586:3608–3612. doi: 10.1016/j.febslet.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Hirayoshi K, Hirata H, Kubota H, Hosokawa N, Nagata K. The Kruppel-like factor Zf9 and proteins in the Sp1 family regulate the expression of HSP47, a collagen-specific molecular chaperone. J Biol Chem. 2002;277:44613–44622. doi: 10.1074/jbc.M208558200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu C, Duan X, Ren F, Li S, Jin Z, Wang Y, Feng Y, Liu Z, Chang Z. CREPT/RPRD1B, a recently identified novel protein highly expressed in tumors, enhances the beta-catenin.TCF4 transcriptional activity in response to Wnt signaling. J Biol Chem. 2014;289:22589–22599. doi: 10.1074/jbc.M114.560979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu L, Wang Y, Zhang M, Li L, Zhu D, Li X, Gu H, Zhang CY, Zen K. Protective role of estrogen-induced miRNA-29 expression in carbon tetrachloride-induced mouse liver injury. J Biol Chem. 2012;287:14851–14862. doi: 10.1074/jbc.M111.314922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, Wang Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.