Aneuploidy causes DNA replication defects and premature adaptation to DNA damage, with profound consequences for genome stability. Such abnormalities provide the substrate for translocations and deletions that are a hallmark of cancer.
Abstract
Genetic instability is a hallmark of aneuploidy in budding and fission yeast. All aneuploid yeast strains analyzed to date harbor elevated levels of Rad52-GFP foci, a sign of DNA damage. Here we investigate how continuously elevated levels of DNA damage affect aneuploid cells. We show that Rad52-GFP foci form during S phase, consistent with the observation that DNA replication initiation and elongation are impaired in some aneuploid yeast strains. We furthermore find that although DNA damage is low in aneuploid cells, it nevertheless has dramatic consequences. Many aneuploid yeast strains adapt to DNA damage and undergo mitosis despite the presence of unrepaired DNA leading to cell death. Wild-type cells exposed to low levels of DNA damage exhibit a similar phenotype, indicating that adaptation to low levels of unrepaired DNA is a general property of the cell's response to DNA damage. Our results indicate that by causing low levels of DNA damage, whole-chromosome aneuploidies lead to DNA breaks that persist into mitosis. Such breaks provide the substrate for translocations and deletions that are a hallmark of cancer.
INTRODUCTION
Changes in chromosome number, a condition known as aneuploidy, have a profound effect on the fitness of an organism. In humans, for example, all autosomal monosomies and most autosomal trisomies are lethal. The few trisomies that are viable lead to early childhood lethality (trisomies 13 and 18) or developmental abnormalities and mental retardation (trisomy 21; reviewed in Pfau and Amon, 2012). Aneuploidy is also a hallmark of cancer. It is estimated that between 75 and 90% of solid human tumors are aneuploid (Holland and Cleveland, 2009; Schvartzman et al., 2010).
Given the profound effect of the aneuploid condition on human health, it is essential to understand how chromosome copy number alterations affect cellular and organismal physiology. Comprehensive whole-genome gene expression and proteome analyses revealed that changes in gene copy number lead to a corresponding change in gene expression of ∼80% of genes (Torres et al., 2007, 2010; Pavelka et al., 2010; Stingele et al., 2012; Dephoure et al., 2014). Studies in budding yeast showed that it is these changes in gene expression that are responsible for the phenotypes seen in aneuploid cells. Introduction of chromosome-size amounts of human or mouse DNA that exhibit little transcriptional and translational activity in yeast have, unlike duplicated yeast chromosomes, little or no effect on fitness (Torres et al., 2007).
The demonstration that changes in relative gene expression are the primary source of the adverse effects of aneuploidy on cells and organisms prompts the question of whether the phenotypes that are observed in aneuploid organisms are due to changes in the gene dosage of a small number of specific genes or are caused by the sum of changes in gene expression of many genes that on their own have little or no effect on fitness. The answer appears to be that both effects contribute to the aneuploid condition. For example, duplication of the APP gene is believed to be responsible for the early onset of Alzheimer's-like pathologies observed in individuals with Down syndrome (Rovelet-Lecrux et al. 2006). However, aneuploid yeast and mammalian cells also share phenotypes, collectively called the aneuploidy-associated stresses, which appear to be caused by concomitant changes in dosage of many genes (Torres et al. 2007; Williams et al. 2008; Tang et al. 2011). Aneuploidy impairs proliferation of budding yeast, fission yeast, and mammalian cells, with a G1 delay being especially prominent (Baker et al., 2004; Niwa et al., 2006; Torres et al., 2007; Williams et al., 2008; Li et al., 2009; Thompson and Compton, 2010; Stingele et al., 2012; Thorburn et al., 2013). Whole-chromosomal aneuploidies also lead to a transcriptional response. A gene expression signature similar to the environmental stress response (Gasch et al., 2000) in budding yeast has been observed in aneuploid budding and fission yeast strains, Arabidopsis, and mouse and human cells (Torres et al., 2007; Sheltzer et al., 2012; Sheltzer, 2013). Finally, aneuploid cells exhibit phenotypes characteristic of the disruption of protein homeostasis. Aneuploid yeast and mammalian cells harbor higher levels of protein aggregates and exhibit sensitivity to compounds that interfere with protein folding and turnover (Torres et al., 2007, 2010; Tang et al., 2011; Oromendia et al., 2012; Stingele et al., 2012; Donnelly et al., 2014).
Here we investigate the molecular basis of one consequence of aneuploidy—genome instability. We previously generated 13 budding yeast strains harboring an additional copy of one of the yeast chromosomes (henceforth disomes; Torres et al., 2007). These strains exhibit increased genomic instability compared to euploid control strains (Sheltzer et al., 2011). Genomic instability was also observed in budding yeast strains harboring multiple aneuploidies and in fission yeast, indicating that genomic instability is a widespread consequence of the aneuploid condition (Sheltzer et al., 2011; Zhu et al., 2012). Analysis of individual disomes revealed that different chromosomal aneuploidies elicit different forms of genomic instability. Some disomes exhibited increased chromosome loss rates; others, increased mutation rates or mitotic recombination. However, of interest, all aneuploid yeast strains analyzed to date harbor elevated levels of Rad52 foci, a sign of DNA damage and ongoing homologous recombination (Lisby et al., 2001). Why Rad52-GFP foci accumulated in aneuploid cells was not understood.
We find that Rad52-GFP foci form during S phase and persist for prolonged periods of time in aneuploid yeast strains, indicating that replication defects cause increased double-strand-break formation and/or DNA-repair defects. Indeed, our studies show that DNA replication initiation and elongation are impaired in several disomic yeast strains. We further show that the degree of DNA damage that the disomic yeast strains experience is not high, but nevertheless has dramatic consequences. Many disomic yeast strains analyzed entered mitosis inappropriately in the presence of DNA damage. This mitosis in the presence of unrepaired DNA was preceded by prolonged cell cycle arrest, demonstrating that the disomes initially respond to the DNA damage but then adapt and enter mitosis in the presence of DNA lesions. Wild-type cells exposed to low levels of DNA damage exhibit a similar phenotype, indicating that adaptation to low levels of unrepaired DNA is a general property of the way in which cells respond to DNA damage. The consequences are dramatic. Haploid yeast cells permanently cease to divide within the next one or two divisions. However, in diploid cells, death may not always be the outcome of such an aberrant mitosis. It could lead to deletions and translocations. Thus whole-chromosome aneuploidy could facilitate the generation of structural chromosomal abnormalities, another hallmark of cancer.
RESULTS
Rad52-GFP foci form during S phase and persist in disomic yeast strains
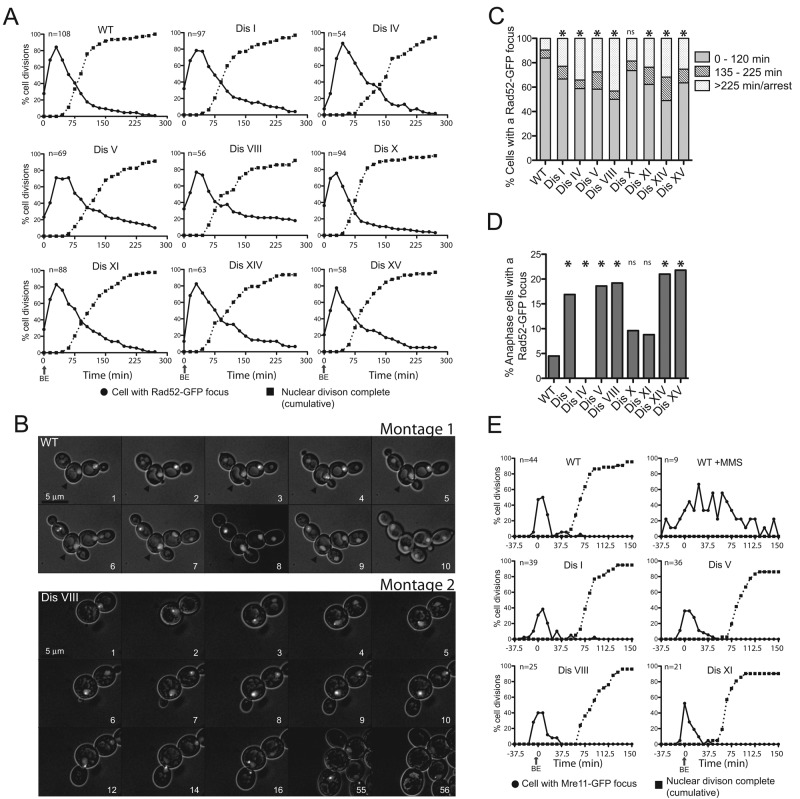
We previously analyzed the localization of the recombination protein Rad52–green fluorescent protein (GFP) in seven disomic yeast strains (disomes IV, V, VIII, X, XI, XIV, and XV) and yeast strains harboring multiple chromosome gains or losses (Sheltzer et al., 2011). All aneuploid strains analyzed harbored a higher percentage of cells containing Rad52-GFP foci than euploid control cells. To investigate the molecular basis of this phenotype, we first asked when during the cell cycle Rad52-GFP foci form in the various disomic yeast strains. To this end, we followed Rad52-GFP focus formation by time-lapse microscopy in strains disomic for chromosome I, IV, V, VIII, X, XI, XIV, or XV using the CellASIC microfluidics system.
In the experimental setup we used, nearly every cell of wild-type and disomic strains experienced DNA damage, as judged by the appearance of at least one Rad52 focus in the nucleus (Figure 1A). This is presumably due to the continuous exposure to short-wavelength light. We found that Rad52-GFP foci appeared within a few frames of when a bud was first detected and resolved before nuclear division (Figure 1, A and B, montage 1, arrowhead). However, Rad52-GFP foci persisted for longer periods of time in disomes than in wild type (Figure 1, A and B, montage 2). Whereas most wild-type cells harbored Rad52-GFP foci for 0–120 min, a significant fraction of disomic cells contained Rad52-GFP foci for >135 min (135–225 min), and many arrested with persistent Rad52-GFP foci (>225 min; Figure 1C). Our previous studies demonstrated that the Rad52-GFP foci seen in the aneuploid yeast strains indeed represent sites of double-strand-break (DSB) repair because disomes that harbor a RAD52 deletion and thus cannot repair DSBs spawn daughter cells that are inviable at an increased frequency (Sheltzer et al., 2011). Of interest, the disomic yeast strains exhibited an additional abnormality. In all but one disome analyzed, Rad52-GFP foci were also observed in cells undergoing anaphase (Figure 1D), a phenotype indicative of DNA damage checkpoint failure or adaptation. We conclude that disomic yeast strains harbor higher levels of damaged DNA and undergo mitosis in the presence of Rad52-GFP foci.
FIGURE 1:
DNA damage occurs during S phase and can persist into anaphase in aneuploid yeast strains. (A) Wild-type (A24352), disome I (A35868), disome IV (A26532), disome V (A26533), disome VIII (A25342), disome X (A25343), disome XI (A25421), disome XIV (A25344), and disome XV (A25345) cells containing a RAD52-GFP fusion were analyzed using time-lapse microscopy to analyze cellular morphology and the presence of Rad52-GFP foci in cells. The graphs show percentage of cells that contain one or more Rad52-GFP foci (closed circles) or cumulative cell divisions (closed squares) over time. Cell divisions were synchronized so that the time of bud emergence (BE) occurred at the zero time point. (B) Montage 1, example of a wild-type cell (black arrowhead) acquiring a Rad52-GFP focus during S phase and resolving it before undergoing anaphase. A Rad52-GFP focus was considered to be present in frames 3–9. The focus is weakly present in frame 3 and seen in the bud in frame 9. Montage 2, example of a disome VIII cell (arrowhead) acquiring a Rad52-GFP focus during S phase and undergoing anaphase in the presence of a Rad52-GFP focus. The cell subsequently dies. A Rad52-GFP focus was considered to be present in all frames except frame 56. (C) Percentage of cells analyzed in A harboring a Rad52-GFP focus for the indicated time brackets. WT, n = 136; disome I, n = 144; disome IV, n = 85; disome V, n = 120; disome VIII, n = 102; disome X, n = 140; disome XI, n = 114; disome XIV, n = 104; disome XV, n = 107. The asterisk above the column indicates statistical significance (chi-squared test; p < 0.005). ns, no statistically significant difference between WT and disome X. (D) Percentage of cells analyzed in A that proceed aberrantly into anaphase despite the presence of a Rad52-GFP focus. WT, n = 192; disome I, n = 126; disome IV, n = 85; disome V, n = 93; disome VIII, n = 80; disome X, n = 123; disome XI, n = 102; disome XIV, n = 85; disome XV, n = 81. The asterisk above the column indicates statistical significance (chi-squared test; p < 0.05). ns, no statistically significant difference between WT and disomes X and XI. (E) Wild-type (A35954), disome I (A35955), disome V (A35957), disome VIII (A35958), and disome XI (A35959) cells containing a MRE11-GFP fusion were analyzed using time-lapse microscopy to analyze cellular morphology and the presence of Mre11-GFP foci in cells. Percentage of cells that contain one or more Mre11-GFP foci (closed circles) or cumulative cell divisions (closed squares) over time. The duration of each time point was 7.5 min. Cell divisions were synchronized so that the time of BE occurred at zero time point. Wild-type cells treated with 0.1% MMS for 30 min before the start of imaging were analyzed as a positive control.
DNA damage occurs during DNA replication in the disomes
We first investigated why disomic yeast strains harbor higher levels of Rad52-GFP foci. To this end, we assessed when during the cell cycle DNA damage occurs. Rad52-GFP foci appeared concomitantly with bud formation, indicating that DNA was damaged during DNA replication. However, Rad52 requires cyclin-dependent kinase (CDK) activity to form repair foci (Aylon et al., 2004; Ira et al., 2004; Huertas et al., 2008). It was therefore possible that the Rad52-GFP foci accumulated at sites of damage that occurred during the preceding mitosis or G1. To address this possibility, we examined Mre11 localization. Mre11 is part of the MRX complex, which functions at an early step in the repair of DSBs and does not require CDK activity to associate with sites of DNA damage (reviewed in Stracker and Petrini, 2011)
When wild-type cells are treated with the DNA-damaging agent methyl methanesulfonate (MMS), Mre11-GFP foci are seen throughout the cell cycle (Figure 1E), indicating that Mre11-GFP can bind to DSBs in all cell cycle stages. In disomes I, V, VIII, and XI, Mre11-GFP foci appeared at the time of budding (Figure 1E). In a few cells, the focus was visible shortly before the appearance of the bud. We believe that this is unlikely to be a sign of DNA damage occurring during G1 but, instead, of DNA damage occurring during very early stages of S phase in cells where the bud forms outside of the plane of focus. Mre11-GFP foci appeared earlier than Rad52-GFP foci with respect to budding, which is consistent with the known residence time of these two proteins at DSBs (Stracker and Petrini, 2011). Because Mre11-GFP foci are rarely seen outside of S phase in the disomes, we conclude that DNA damage occurs during DNA replication. The analysis of histone H2A phosphorylation supports this conclusion. Phosphorylation of histone H2A, a very early response to DNA damage, was not detected in G1-arrested wild-type and disomic yeast strains (Supplemental Figure S1, 0 time point). However, all cells were capable of phosphorylating histone H2A when challenged with MMS during the G1 arrest (Supplemental Figure S1, 30- and 60-min time points).
Disomes V and VIII exhibit DNA replication defects
Our previous studies indicated that levels of Rad52-GFP foci or any other manifestation of genomic instability correlated with neither degree of aneuploidy nor with any other phenotype shared between the disomic strains (Sheltzer et al., 2011). This observation prompted us to pursue the hypothesis that different disomic strains exhibit different defects in DNA replication and repair. Some disomic yeast strains could experience more DNA damage during S phase, whereas others could have difficulties repairing the damaged DNA.
We first considered the possibility that DNA replication was impaired in some of the disomes, causing increased formation of DSBs. To test this idea, we analyzed DNA replication in disomes V and VIII because both strains are exquisitely sensitive to the DNA replication inhibitor hydroxyurea (HU; Sheltzer et al., 2011). We synchronized cells in G1 with α-factor pheromone and monitored DNA replication after release from the G1 block. We included a strain lacking the gene encoding the S phase cyclin Clb5 in this analysis because clb5Δ cells show both replication initiation and elongation defects (Epstein and Cross, 1992; Schwob and Nasmyth, 1993).
Most disomes exhibit delays in cell cycle entry due to defects in the G1–S phase transition (Torres et al., 2007; Thorburn et al., 2013). To distinguish these cell cycle entry defects from DNA replication initiation defects, we designated the time point when DNA replication was first noticeable as the 0 time point and closely followed DNA replication for the next 50 min (Figure 2A). This analysis showed that cells disomic for chromosome V were slow to both initiate and complete DNA replication. Whereas the majority of wild-type cells had initiated DNA replication within 5 min of the first sign of DNA replication, as judged by increased DNA content in the entire population (the entire distribution shifted to the right), only a small fraction of disome V cells had initiated DNA replication, as judged by an extending of the DNA content distribution to the right (Figure 2A). This delay was especially obvious when comparing the 10-min time points. Disome V cells were also slow to complete DNA replication. Whereas the majority of wild-type cells had completed DNA replication within 25 min of initiation, replication was not completed until 35 min after initiation in disome V cells (Figure 2). This replication-elongation defect is best seen when the replication profiles of the later time points are superimposed (Figure 2B). We conclude that disome V cells exhibit replication-initiation and -elongation defects. These defects are subtle, as judged by the fact that they are less pronounced than those of cells lacking CLB5 (Figure 2, A and B).
FIGURE 2:
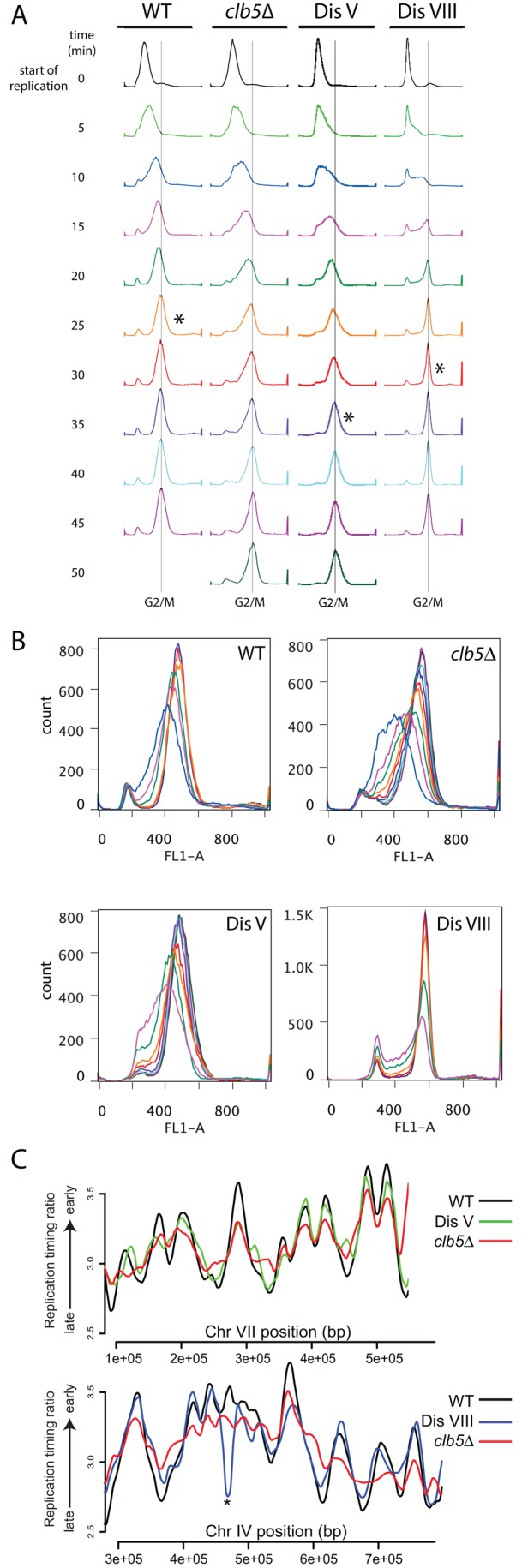
Disome V cells exhibit defects in DNA replication initiation and elongation. Wild-type (A11311), cells deleted for CLB5 (A35992), disome V (A28265), and disome VIII (A27036) cells were arrested in G1 with α-factor (5 μg/ml) for ∼165 min. Cells were then washed and transferred into medium lacking pheromone. Samples were taken every 5 min to determine DNA content by fluorescence-activated cell sorting (A, B) and DNA copy number (C). The DNA profiles shown in A and B were normalized so that time zero represents the first histogram with noticeable replication. The asterisk in A denotes the time point when replication was considered complete. DNA samples from the start of replication to the time point with the asterisk were pooled for each strain for the analysis shown in C and Supplemental Figure S2. The graph in B shows time points color coded so as to correspond to the histogram profiles from A superimposed to illustrate that completion of DNA replication is slow in disome V cells but not in disome VIII cells. The replication profiles shown in C are the average of three biological replicates and plotted relative to a G1 sample for each strain. The complete replication profiles are shown in Supplemental Figure S2.
To examine further the kinetics of DNA replication initiation and elongation in disome V cells, we pooled DNA samples from the start of replication until its completion and determined DNA copy number by deep sequencing. DNA copy number was assessed relative to a G1 sample (Figure 2C and Supplemental Figure S2). In this analysis, regions of the genome that replicate early, such as origins of DNA replication, will appear as peaks, whereas regions that replicate late will appear as valleys (Yabuki et al., 2002). The DNA replication-initiation and -elongation defects of clb5Δ cells were readily observable in this analysis, with peak height reduced and slopes less steep (Figure 2C and Supplemental Figure S2). Consistent with our DNA content analysis, the replication defect in disome V cells was less pronounced than that of cells lacking CLB5. However, decreased peak height and slope steepness were nevertheless evident in a significant fraction of the genome (Figure 2C and Supplemental Figure S2). We conducted this analysis from three biological replicates, making us confident that the subtle replication defect that we observe is indeed a biological property of disome V cells rather than due to technical variability. We conclude that cells harboring an additional copy of chromosome V exhibit DNA replication-initiation and -elongation defects. We also observed earlier replication in a small number of regions of the genome in disome V cells (Figure 1C). Whether these few regions represent cryptic origins that are fired when replication proceeds slowly remains to be determined.
Like disome V, disome VIII exhibited a subtle DNA replication-initiation defect (Figure 2A). However, once initiation had occurred, replication elongation appeared to proceed efficiently (Figure 2, A and B). The analysis of replication profiles substantiated this finding. Initiation of replication at some origins appeared delayed, as judged by a decrease in peak height, but alterations in slope, which are indicative of changes in replication elongation speed, were not evident (Figure 2C and Supplemental Figure S2).
Subtle changes in replication could be due to changes in the balance between firing of origins in the ribosomal DNA (rDNA) and early origins. For example, cells lacking the rDNA silencing factor Sir2 exhibit a delay in early origin firing (Yoshida et al., 2014). rDNA replication was not significantly altered in disome V and VIII cells (unpublished data), indicating that alterations in rDNA replication were not responsible for the delay in early origin firing in the two disomes.
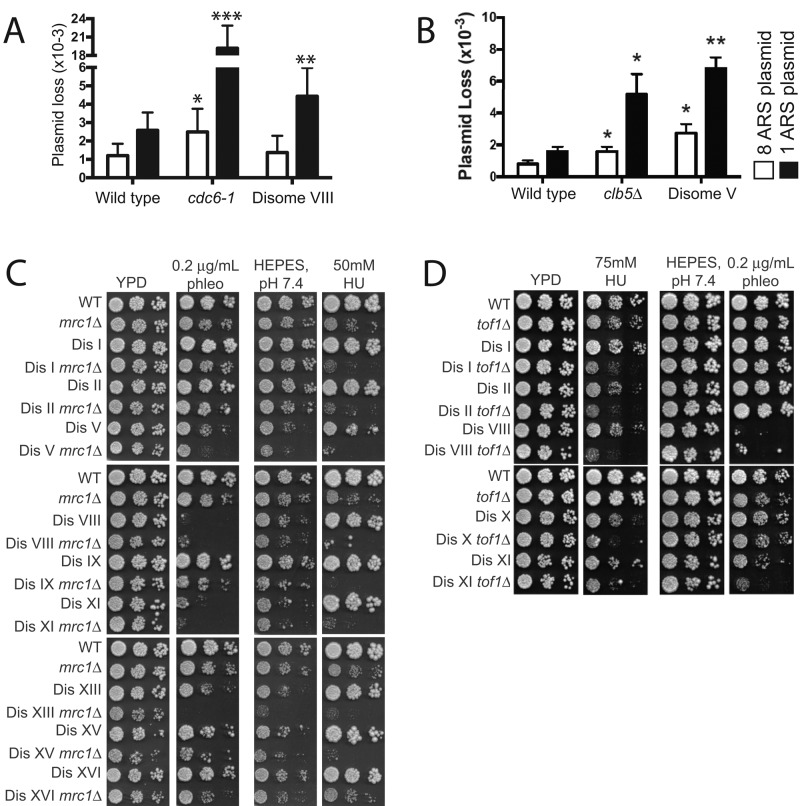
Replication-initiation defects can be detected by comparing the frequency of loss of a plasmid carrying one origin of replication (ARS) with that of a plasmid carrying eight ARS sequences (Hogan and Koshland, 1992; Cheng et al., 2010). Indeed, cells harboring a temperature-sensitive allele in the gene encoding the replication initiation factor Cdc6 lose plasmids with a single ARS at a much higher frequency than plasmids with eight ARSs (Figure 3A; Hogan and Koshland, 1992). Disome V and VIII cells also exhibited an increased frequency in the loss of a single ARS plasmid, which was suppressed when additional ARS sequences were present on the plasmid (Figure 3, A and B). The degree of plasmid loss seen was similar to that of cells lacking CLB5 (Figure 3B). We conclude that cells carrying an extra copy of chromosome V or VIII exhibit defects in DNA replication initiation. We note that disomes XI, XV, and XVI also exhibit similar replication initiation defects in this assay (Supplemental Figure S3).
FIGURE 3:
Disome VIII cells exhibit DNA replication-initiation defects. (A) Plasmid loss rate was determined in wild-type (A35934, A35933), cdc6-1 (A35944, A35943), and disome VIII (A35937, A35938) cells harboring a plasmid bearing either one or eight ARS sequences as described in Materials and Methods. Graphs indicate the mean and SD of at least 12 independent cultures. Statistical tests were performed between the wild-type strain harboring the eight-ARS plasmid and the mutant or disomic strains harboring the eight-ARS plasmid, or between the wild-type strain harboring the one-ARS plasmid and the mutant or disomic strains harboring the one-ARS plasmid. *p < 0.05, **p < 0.005, ***p < 0.0005 (Wilcoxon rank-sum test). (B) Plasmid loss rate was determined in wild-type (A35934, A35933), clb5::URA3 (A36737, A36739), and disome V (A36896, A36897) cells harboring a plasmid bearing either one or eight ARS sequences as described in Materials and Methods. Graphs indicate the mean and SD of three independent cultures. Statistical tests were performed between the wild-type strain harboring the eight-ARS plasmid and the mutant or disomic strains harboring the eight-ARS plasmid, or between the wild-type strain harboring the one-ARS plasmid and the mutant or disomic strains harboring the one-ARS plasmid. *p < 0.05; **p < 0.005 (Wilcoxon rank-sum test). (C) Tenfold dilutions of euploid and disomic cells wild type or mutant for MRC1 on YPD plates, YPD plates + 50 mM hydroxyurea, YPD + HEPES, pH 7.4, or YPD + HEPES, pH 7.4, + 0.2 μg/ml phleomycin. (D) Tenfold dilutions of euploid and disomic cells wild type or mutant for TOF1 on YPD plates, YPD plates + 75 mM hydroxyurea, YPD + HEPES, pH 7.4, or YPD + HEPES, pH 7.4, + 0.2 μg/ml phleomycin.
Genetic interactions between disomes V and VIII and deletions in genes required for efficient DNA replication support the idea that the two disomes are defective in DNA replication. Tof1, Mrc1, and Csm3 function in a complex that associates with DNA polymerase during replication elongation and are believed to convey processivity to DNA polymerase and mediate DNA damage checkpoint surveillance (Tourrière et al., 2005; Hodgson et al., 2007; Bando et al., 2009). Deleting MRC1 significantly enhanced the sensitivity of disome V and VIII to the DNA replication inhibitor HU and the sensitivity of disome V cells to the DNA-damaging agent phleomycin or MMS (Figure 3B; Sheltzer et al., 2011; note that disome VIII cells are so sensitive to phleomycin that it was not possible to detect enhancement at the phleomycin concentrations used in this analysis). The HU sensitivity of disome VIII cells was also enhanced by deleting TOF1 (Figure 3C).
We observed similar genetic interactions between deletions of TOF1 or MRC1 and other disomes. Almost all disomes analyzed exhibited increased HU sensitivity when combined with a deletion in MRC1 or TOF1, and many showed increased sensitivity to phleomycin (Figure 3, B and C). We conclude that disomes V and VIII are defective in DNA replication initiation. As judged by the genetic interactions with mrc1Δ or tof1Δ and plasmid loss assays, this defect is not restricted to these two disomes but is a widespread phenomenon among the disomic yeast strains. Disome V cells also exhibit replication-elongation defects. Whether this is a common occurrence among the disomes remains to be determined.
Repair of an HO-induced double-strand break is normal in disomes I, V, VIII, X, and XI
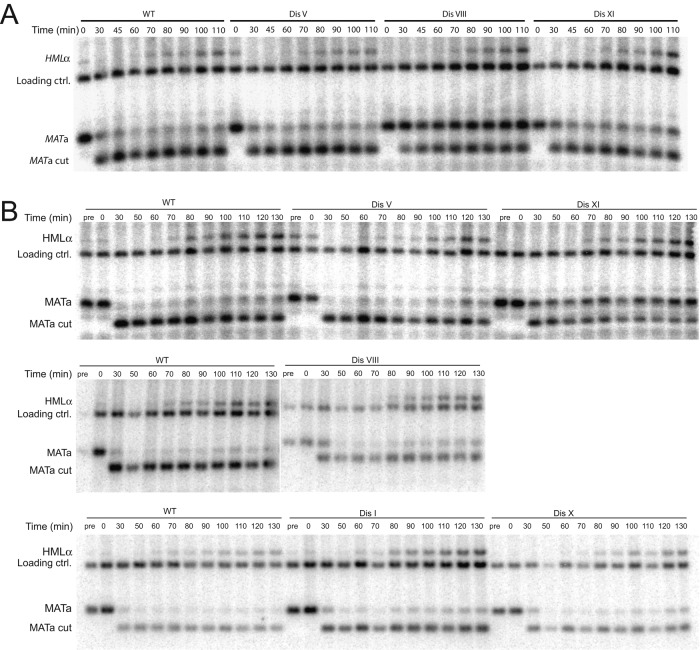
The extended presence of Rad52-GFP foci in some of the disomic strains could be due to defects in repair of damaged DNA. To test this possibility, we introduced a galactose-inducible HO endonuclease construct into euploid and some disomic yeast strains. HO's only target site in the yeast genome is within the MAT locus. Cleavage in the MAT locus facilitates mating-type conversion. Using appropriate restriction enzymes, one can follow HO cleavage and repair of the HO-induced DSB from the silent mating-type locus encoding the opposite mating type over time by Southern blot analysis (reviewed in Sugawara and Haber, 2006).
We added galactose to exponentially growing cells for 40 min, which led to efficient cleavage in the mating-type locus (MATa cut; Figure 4A). On repression of HO expression by glucose addition, the double-strand break was efficiently repaired from the silent HMLα locus, as judged by the appearance of the recombinant product termed HMLα (Figure 4A). The kinetics of repair in disomes V, VIII, and XI were indistinguishable from that of the wild-type strain (Figure 4A). Note that, for unknown reasons, the disome V strain harboring the GAL-HO construct was unstable. Only 80% of cells harbored the additional copy of chromosome V despite continuous selection for the extra chromosome (Supplemental Figure S4).
FIGURE 4:
Repair of an HO-induced double-strand break occurs normally in disomes I, V, VIII, X, and XI. (A) Wild-type (A35884), disome V (HB392), disome VIII (A35886), and disome XI (A35885) cells containing a GAL-HO fusion were grown to exponential phase in YP medium containing 2% lactate. Galactose (2%) was then added (t = 0) and MATa cleavage (MATa cut) and repair from the HMLα locus (HMLα) were analyzed at the indicated times. At 40 min after HO induction, 2% glucose was added to repress HO expression. Note that 20% of disome V cells had lost the additional chromosome despite continuous selection for both copies of chromosome V (Supplemental Figure S4). We therefore did not keep this strain. (B) Wild-type (A35884), disome I (A35892), disome V (HB392), disome VIII (A35886), disome X (A35891), and disome XI (A35885) cells containing a GAL-HO fusion were grown as in A, but cells were treated with phleomycin (10 μg/mL) in HEPES-buffered medium for 1 h before addition of galactose and throughout the duration of the experiment, with the exception of the top set of results, for which phleomycin was only present in the 1 h before the addition of galactose.
Given that the increase in Rad52-GFP focus formation was subtle in many of the disomes, we hypothesized that the disomes are capable of efficiently repairing minor DNA damage, such as a single HO-induced DSB, but may have difficulties repairing a HO break in the presence of high levels of DNA damage. To test this hypothesis, we examined the repair kinetics of the HO-induced break in the MATa locus in the presence of 10 μg/ml phleomycin at 1 h before repair (Figure 4B, top) or 1 h before and during repair (Figure 4B, middle and bottom). We analyzed disomes I, V, VIII, X, and XI using this experimental strategy and found all disomes to repair the HO break efficiently (Figure 4B). We conclude that repair of an endonuclease-induced DSB is not affected in disomes I, V, VIII, X, and XI. However, it is important to note that this finding does not exclude the possibility that the disomes have difficulty repairing other types of DNA damage. In fact, the finding that the disomes are sensitive to DNA-damaging agents strongly argues that chemically induced DNA damage that could involve combinations of complex DNA breaks, cross-links, and adducts are not effectively repaired in the disomes.
Disomic yeast strains undergo mitosis in the presence of DNA damage
The live-cell analysis described in Figure 1 showed that in addition to harboring Rad52-GFP foci that persist longer in cells, many disomes entered anaphase in their presence. This observation raised the interesting possibility that these disomic yeast strains either fail to recognize damaged DNA or adapt to the damage and enter mitosis without having repaired the damage.
In response to DNA damage, cells activate the DNA damage checkpoint, which in turn halts cell cycle progression before entry into mitosis in mammals and the onset of anaphase in budding yeast (reviewed in Zhou and Elledge, 2000). Our previous studies showed that the DNA damage response was functional in the disomic yeast strains as judged by the ability of the disomic strains to activate the DNA damage checkpoint kinase Rad53 in response to treatment of cells with the DNA-damaging agent phleomycin (Sheltzer et al., 2011). However, the amount of damage caused by this treatment is high. We therefore considered the possibility that the low degree of DNA damage generated in the disomic yeast strains was insufficient to generate a robust DNA damage response and cell cycle arrest. To test this hypothesis, we determined whether cells arrested before mitosis before initiating anaphase with a Rad52-GFP focus. This analysis revealed that the disomes arrested for long periods of time, during which a Rad52-GFP focus was continuously present, before finally entering anaphase in the presence of a Rad52-GFP focus (Figure 5A; disome I, 276 min; disome II, 306 min; disome V, 261 min; disome VIII, 346 min; disome X, 345 min; disome XI, 270 min; disome XIV, 344 min; disome XV, 265 min). Thus the disomes initially respond to the DNA damage but eventually enter mitosis despite the presence of a Rad52-GFP focus.
FIGURE 5:
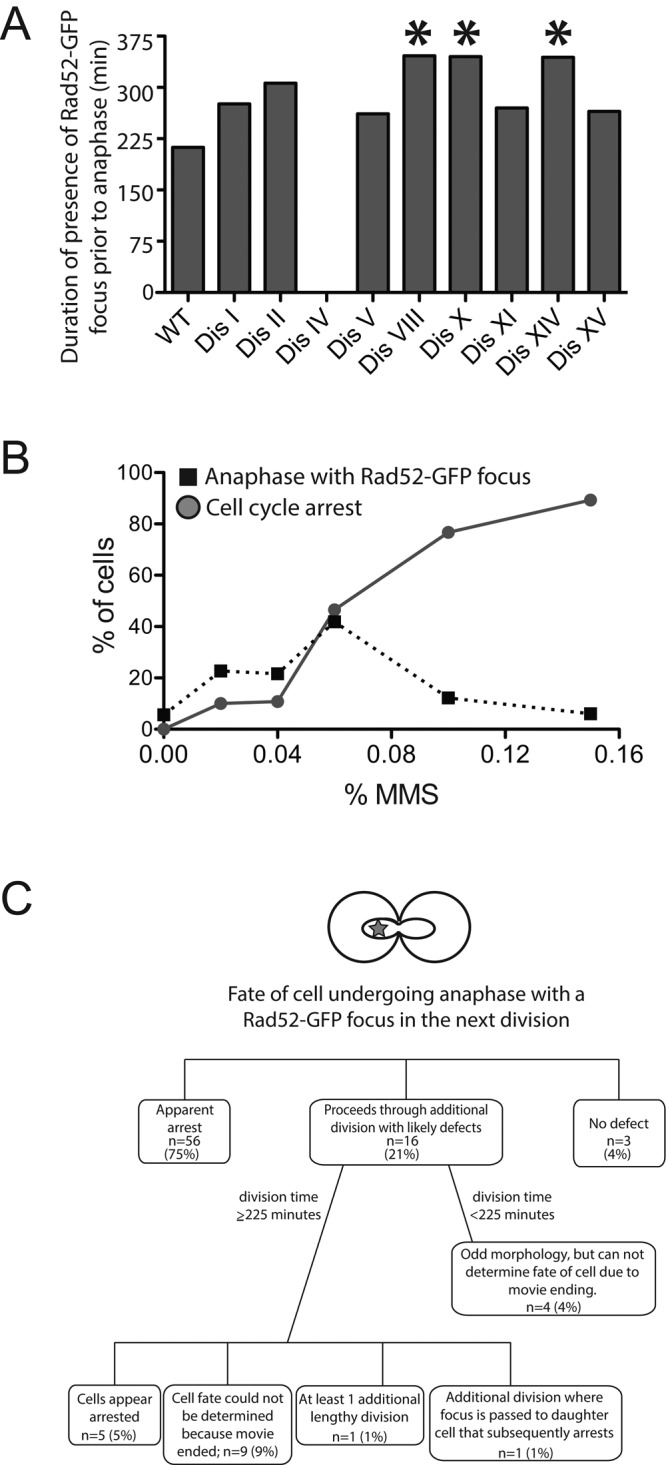
Disomic yeast strains undergo anaphase in the presence of DNA damage. (A) Time- lapse microscopy movies described in Figure 1 were analyzed to determine the amount of time the WT and disomic yeast strains harbored a Rad52-GFP focus before entering anaphase despite the presence of a Rad52-GFP fusion. WT, n = 18; disome 1, n = 23; disome II, n = 12; disome IV, not analyzed; disome V, n = 14; disome VIII, n = 11; disome X, n = 10; disome XI, n = 9; disome XIV, n = 20; disome XV, n = 18. Statistical differences between WT and disomes were determined by a one-way ANOVA of all disomes except disome IV followed by a posthoc Tukey honest significant difference test of significance. This analysis showed that the difference between WT and disomes VIII (p = 0.0174), X (p = 0.0267), and XIV (p = 0.0026) was statistically significant (indicated with an asterisk). The other differences were not. (B) Wild-type cells (A24352) were treated with the indicated amounts of MMS and analyzed by time-lapse microscopy. The percentage of cells undergoing anaphase in the presence of a Rad52-GFP focus (black squares) or that arrested in metaphase (closed circles) was determined. For 0% MMS, n = 81; 0.02% MMS, n = 88; 0.04% MMS, n = 37; 0.06% MMS, n = 43; 0.08% MMS, n = 90; 0.1% MMS, n = 90; 0.15% MMS, n = 84. (C) Schematic describing the fate of wild-type cells treated with MMS (data from all concentrations pooled) analyzed in B that underwent anaphase in the presence of a Rad52-GFP focus.
Is entry into anaphase in the presence of Rad52-GFP foci a characteristic of the disomic yeast strains or a more general property of cells responding to low levels of DNA damage? To address this question, we treated wild-type cells with increasing doses of MMS. This analysis revealed that at doses between 0.04 and 0.08% MMS, as many as 40% of wild-type cells underwent anaphase despite the presence of a Rad52-GFP focus (Figure 5B). At higher MMS concentrations, cells permanently arrested in metaphase. As in the disomes, anaphase entry in the presence of a Rad52 focus was preceded by prolonged cell cycle arrest (211 min), during which time a Rad52-GFP focus was continuously present (Figure 5A). It thus appears that wild-type cells, too, detect low levels of DNA damage and arrest before anaphase for significant periods of time but eventually segregate their chromosomes despite not having resolved Rad52-GFP foci.
Before concluding that low levels of DNA damage may not be an absolute barrier to anaphase entry, it was important to establish that the Rad52 foci observed in cells undergoing anaphase indeed represented DNA damage and were not simply Rad52-GFP remnants on successfully repaired DNA. To address this possibility, we followed the fate of wild-type cells challenged with low doses of MMS that had undergone mitosis despite harboring a Rad52-GFP focus. This analysis revealed that only 4% of cells continued to divide without any apparent defect (Figure 5C). All other cells showed significant proliferation defects after the anaphase during which a Rad52-GFP focus was present. Seventy-five percent of such cells arrested in the subsequent cell cycle (Figure 5C). An additional 21% proceeded through one additional division and then did not divide within the time frame of the movie (Figure 5C). This analysis demonstrates that the presence of a Rad52-GFP focus during anaphase indeed reflects mitosis occurring in the presence of unrepaired DNA, with dramatic detrimental consequences for the cell. We conclude that when low levels of DNA damage are not successfully repaired within 4–6 h, cells adapt and proceed through mitosis in the presence of damaged DNA. Adaptation occurred faster in our experimental setup than in previous studies, in which a single DSB was generated during G1 when a repair template is absent. Under these conditions, cells arrested for ≥8 h before entering mitosis in the presence of this double-strand break (Pellicioli et al., 2001). We do not know why adaptation occurs more quickly in our experimental setup but speculate that it is due to differences in the nature of the DNA damage. A single DSB created during G1 could generate long tracks of resected single-stranded DNA that emit a strong and persistent DNA damage checkpoint signal. The DNA damage elicited by MMS in wild-type cells or by fork collapse in the disomes may generate a weaker or shorter-lived checkpoint signal.
DISCUSSION
Genetic instability is a hallmark of aneuploidy in budding and fission yeast. Every aneuploid yeast strain that we and others have analyzed exhibits some form of genomic instability. Chromosome loss rate, mutation rate, and microsatellite instability were observed in many different yeast strains harboring single additional chromosomes. A very prominent phenotype among aneuploid budding and fission yeast strains is an increase in the percentage of cells harboring Rad52-GFP foci. Here we investigate the molecular basis of this phenotype. We find that Rad52-GFP foci form during S phase, indicating that replication defects cause increased DSB formation. The levels of DNA damage that the disomic strains experience are not high, but they have a significant effect on cellular fitness. Many disomic yeast strains enter mitosis in the presence of DNA damage after a prolonged cell cycle arrest. Wild-type cells exposed to low levels of DNA damage exhibit a similar phenotype, indicating that adaptation to low levels of DNA damage is a general property of the manner in which cells respond to DNA damage. If genomic instability is also a property of aneuploid mammalian cells, this aspect of the DNA damage response could very well contribute to the structural abnormalities that are so frequently observed in cancer.
In our live-cell analysis, wild-type and aneuploid yeast strains accumulated Rad52-GFP foci during S phase, but they persisted for longer periods of time in the aneuploid strains. We did not detect DNA damage repair defects in the disomes that we analyzed, but DNA replication-initiation and -elongation defects were detected in many disomic yeast strains. From these results, we conclude that an increased number of DSBs contributes to the prolonged presence of Rad52-GFP foci in the disomic strains. Determining how many more DSBs form in disomic strains than in wild type was not possible, however, because Rad52-GFP foci cluster within cells (Lisby et al., 2001).
A detailed analysis of disomes V and VIII revealed replication-initiation problems in both strains and replication-elongation defects in disome V cells. It thus appears that various aspects of DNA replication are sensitive to aneuploidy. Why DNA replication is affected by so many different aneuploidies is not clear. Chromatin structure, as well as DNA replication initiation and elongation, are mediated by many large, multisubunit complexes. Stoichiometric imbalances in one or several of the complexes required for error-free DNA replication could arise in many different aneuploidies. This in turn could cause the formation of partially assembled complexes or interfere with the function of these complexes in some other manner. Thus far we have not been able to identify the genes that cause the DNA replication defects in the different disomes. We screened through a centromere-based plasmid library that harbors 75% of the genes encoded on chromosome VIII but were not able to identify a single gene that, when introduced in single copy into wild-type cells, confers hydroxyurea or phleomycin sensitivity (H.B., unpublished data). It thus appears that changes in copy number of multiple genes on chromosome 8 are responsible for the sensitivity to genotoxic agents and presumably the DNA replication defects that are observed in disome VIII cells.
Although replication defects appear to be widespread among the disomes, we have not obtained any evidence to indicate that DSB repair is impaired in the disomes. Repair of an HO-induced DSB occurred with wild-type kinetics in all of the disomic strains we analyzed irrespective of whether it was the only DNA damage that was induced or was generated in the context of additional genotoxin-induced DNA damage. However, this result does not necessarily mean that these disomic yeast strains can repair all forms of DNA damage effectively. The fact that most aneuploid yeast strains are sensitive to DNA-damaging agents such as phleomycin or MMS in fact strongly argues that some forms of chemically induced DNA damage are not repaired efficiently in the disomes. Further studies are needed to determine the identity of this DNA damage.
The perhaps most-striking phenotype we observed is that a fraction of disome I, II, V, VIII, XIV, and XV cells entered mitosis despite the presence of DNA damage. The percentage of cells entering anaphase in the presence of a Rad52-GFP focus was also elevated in disomes X and XI but did not reach significance. Disome IV cells never entered anaphase despite harboring a Rad52-GFP focus. Disome IV cells proliferate extremely poorly (Torres et al., 2007), which could account for the fact that entry into mitosis in the presence of DNA damage does not occur. Wild-type (WT) cells treated with low doses of the DNA-damaging agent MMS also progressed through mitosis with Rad52-GFP foci.
Our live-cell imaging analysis showed that cells initially arrested in metaphase but eventually adapted and then entered anaphase without having repaired their damaged DNA. Adaptation to DNA damage has been proposed to serve as a last-ditch effort at survival after all repair options have been exhausted (Vidanes et al., 2010). However, in haploid cells, this effort is largely futile. The vast majority of cells that underwent anaphase with a Rad52-GFP focus ceased to divide within one or two cell divisions. It is worth noting, however, that adaptation in the presence of low levels of DNA damage may provide a survival benefit in diploid cells. The presence of two chromosomal copies likely protects cells from dying even when significant amounts of genetic information are lost as the result of chromosome segregation in the face of unrepaired DSBs. Thus it will be very interesting to uncover the molecular mechanisms that either quench DNA damage signaling or override it to mediate adaptation. The Polo kinase Cdc5 has been shown to be required for adaptation to DNA damage in yeast and mammals (Toczyski et al., 1997; Pellicioli et al., 2001; Yoo et al., 2004; reviewed in Serrano and D'Amours, 2014). Cdc5 down-regulates the DNA damage checkpoint pathway by inhibiting the DNA damage checkpoint kinase Rad53 (Vidanes et al., 2010; reviewed in Serrano and D'Amours, 2014). Determining whether the kinase responds differently to low and high levels of DNA damage will be important questions in the future.
Our findings raise the remarkable possibility that in multicellular organisms, low levels of DNA damage may in fact be more detrimental than high levels of damage. Mitosis in the presence of DNA damage can lead to translocations, deletions, and other types of mutagenic events. Because such genomic alterations have all been implicated in tumorigenesis, low levels of DNA damage could have substantial protumorigenic effects. Given that mitosis in the presence of DNA damage is a key characteristic of aneuploidy, it is furthermore tempting to speculate that this aspect of aneuploidy could contribute to tumor evolution, thus explaining why the aneuploid condition, despite its antiproliferative effects, is a hallmark of cancer.
MATERIALS AND METHODS
Strains and plasmids
Strains used in this study are described in Supplemental Table S1 and are derivatives of W303. Strains were constructed using PCR-based methods described by Longtine et al. (1998). The generation of disomic strains has been described previously (Torres et al. 2007). Karyotypes of all disomic strains were confirmed by comparative genome hybridization (Torres et al. 2007). Growth conditions are described in the figure legends.
Live-cell microscopy
Cells harboring a GFP-tagged copy of RAD52 at the endogenous locus were grown to log phase in SC-HIS+G418 (synthetic complete medium lacking histidine, plus geneticin G418) medium and transferred to a microfluidic chamber (CellASIC, Hayward, CA). Cells were imaged every 15 min using a Zeiss (Oberkochen, Germany) Axio Observer-Z1 with a 100× objective equipped with a Hamamatsu (Bridgewater, NJ) ORCA-ER digital camera. Eleven Z-stacks (0.6 μm apart), with 100-ms exposure, full gain, and 2 × 2 binning, were acquired and maximally projected. A single transmitted-light 20-ms exposure image was acquired for each Z-stack. MetaMorph software was used for image acquisition and processing.
For MMS treatment and imaging, cells were grown overnight in SC-HIS+G418 medium and then transferred to yeast extract/peptone/dextrose (YPD) and arrested with α-factor for ∼105 min. MMS was added at the indicated concentration for ∼25 min while cells were still arrested in G1. Cells were then washed and transferred to fresh selective medium lacking pheromone and imaged on a microfluidic chamber as described.
Cells harboring a GFP-tagged copy of MRE11 at the endogenous locus were imaged similarly to cells carrying a tagged copy of RAD52, but instead of a microfluidic chamber, cells were placed on an agarose pad slide and imaged every 7.5 min. In addition, we used Definite Focus (Zeiss) to ensure that images remained in focus.
For Figure 1, A and C, only cells that had a focus and divided in ≤25 time frames were included in our analysis. The analysis in Figure 1D included only the first division after release from the G1 arrest to avoid artifacts caused by cells experiencing DNA damage due to their being imaged for various lengths of time.
DNA copy number analysis
DNA was extracted according to Blitzblau et al. (2012).
Samples were sequenced on an Illumina (San Diego, CA) HiSeq to produce paired end reads 40 nucleotides in length. Reads were aligned to the W303 genome and processed with module add bwa version 0.7.5a, samtools version 0.1.19, bedtools version 2.17.0, and ucsc-tools version 20120530. Alignment data were expressed as counts of first in pair reads from properly aligned pairs (sam flags 99 and 83) with mapping quality ≥10 per 50–base pair window per 5 million accepted reads.
For each strain, the number of counts for each replicate was summed and then divided by the G1 count values corresponding to that strain (e.g., (WTa + WTb + WTc)/WTG1). For the clb5Δ strain, the WT G1 count values were used for normalization. A LOESS smoothing function was performed on each chromosome, where span was equal to 0.025 × max(largest chromosome length/length of chromosome x), where chromosome x is the chromosome of interest. This normalized the amount of smoothing to the size of each particular chromosome. A loess.predict command was then used to give values for any data that were missing. The loess.predict values versus chromosome position were plotted.
Mating-type-switching time courses
Time courses were carried out as described in Hicks et al. (2011). Briefly, cells were grown in YP-lactate, and 2% galactose was added (0 time point) to induce HO expression. At 40 min thereafter, HO expression was repressed by the addition of 2% glucose. For time courses involving the addition of phleomycin, cells were resuspended in YP-lactate buffered with 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4. Phleomycin was added to a final concentration of 10 μg/ml for 60 min before the addition of galactose and either washed out before repair or left in the medium for the remainder of the experiment, as indicated.
Genomic DNA was isolated at the indicated time points and digested with StyI. DNA was separated on a 1% agarose gel made in 1× TBE (Tris/borate/EDTA) and run in 1× TBE buffer at 150 V. Neutral transfer to the membrane (Hybond-N+; GE Healthcare Amersham, Pittsburgh, PA) was carried out according to the manufacturer's protocol using a basic capillary transfer apparatus using 20× saline-sodium citrate as transfer buffer. After overnight transfer, the membrane was cross-linked using the preset ultraviolet exposure on a GE Healthcare UVC 500 cross-linker (70,000 μJ/cm2).
The 32P-labeled probe was the PCR product of primers AW264 and MAT10, described in Hicks et al. (2011), using the Megaprime DNA Labeling System and ProbeQuant G-50 Micro Columns (Amersham). The blot was exposed to a storage phosphor screen and imaged using a Typhoon Trio (Amersham). The HMLα fragment served as loading control for the experiment because the concentration remains unchanged throughout the time course.
γ-H2A immunoblot analysis
Strains were grown overnight in SC-HIS+G418 medium and transferred to YPD at an OD600nmn of ∼0.18. Cells were arrested in G1 with α-factor (5 μg/ml) for the duration of the experiment. At 160 min into arrest, MMS (0.3%) was added. Samples were collected before MMS addition and at the indicated time points.
Protein samples were trichloroacetic acid extracted as described in Attner and Amon (2012) and separated by PAGE, 15% acrylamide gel, and transferred onto a nitrocellulose membrane. The membrane was blocked in 5% bovine serum albumin in Tris-buffered saline/Tween 20 for 30 min. γ-H2A was detected by incubation with the primary antibody (rabbit polyclonal 15083; Abcam, Cambridge, MA) at 1:1000 in blocking buffer at room temperature for 1 h, followed by washing and incubation with the secondary antibody anti-rabbit horseradish peroxidase at 1:10,000 in blocking buffer at room temperature for 40 min. The anti-Pgk1 antibodies (Molecular Probes, Eugene, OR) were used at a 1:10,000 dilution.
Flow cytometry
Cells were resuspended in 70% ethanol. After fixation, cells were washed once and resuspended in 50 mM sodium citrate, pH 7.0, containing 0.25 mg/ml RNase A. Samples were incubated at 37°C overnight. Samples were then pelleted and resuspended in 50 mM sodium citrate containing 1 μM Sytox Green (Life Technologies, Grand Island, NY). Cells were kept in the dark for at least 1 h and sonicated before analysis on the flow cytometer model BD (San Jose, CA) FACSCalibur running CellQuest Pro software.
Plasmid loss experiments
The loss rates of plasmids harboring one or eight ARS sequences, as well as a selectable LEU2 marker, were determined as described in Zhang et al. (2002). In brief, cells were first grown overnight in –HIS-LEU+G418 medium. Subsequently, 200 cells were plated on –HIS+G418 plates, and 200,000 cells were inoculated into –HIS+G418 medium and allowed to grow for an additional 24 h. After this time, 200 cells were plated on –HIS+G418 plates. After 3–5 d of growth, the –HIS+G418 plates were replica plated to –HIS-LEU+G418 plates to determine the fraction of cells that had lost the plasmid. In the experiment shown in Figure 3A and Supplemental Figure S3, 12 biological replicates were analyzed; in the experiment shown in Figure 3B, 3 biological replicates were analyzed.
Supplementary Material
Acknowledgments
We thank Steve Bell for the cdc6-1 mutant strain, Jim Haber for the GAL-HO construct, and Chun Liang for the p1ARS and p8ARSs plasmids. We thank Charlie Whittaker of the Barbara K. Ostrom (1978) Bioinformatics and Computing Facility, Swanson Biotechnology Center (Cambridge, MA), and Theresa and Clara Weis for technical support. We thank members of the Amon lab for suggestions. This work was supported by National Institutes of Health Grant GM056800 to A.A., a Ruth L. Kirschstein NRSA Fellowship to H.B., and a National Science Foundation Graduate Fellowship to J.S. This work was supported in part by Koch Institute Support (Core) Grant P30-CA14051 from the National Cancer Institute. A.A. is an investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- ARS
autonomously replicating sequence
- BE
bud emergence
- CDK
cyclin-dependent kinase
- DSB
double strand break
- GFP
green fluorescent protein
- HU
hydroxyurea
- MMS
methyl methane sulfonate
- rDNA
ribosomal DNA
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1442) on February 18, 2015.
REFERENCES
- Attner MA, Amon A. Control of the mitotic exit network during meiosis. Mol Biol Cell. 2012;16:3122-3132. doi: 10.1091/mbc.E12-03-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, Shirahige K. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 2009;284:34355–34365. doi: 10.1074/jbc.M109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau HG, Chan CS, Hochwagen A, Bell SP. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS Genet. 2012;8:e1002643. doi: 10.1371/journal.pgen.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Xu Z, Wang J, Zhai Y, Lu Y, Liang C. ATP-dependent pre-replicative complex assembly is facilitated by Adk1p in budding yeast. J Biol Chem. 2010;285:29974–29980. doi: 10.1074/jbc.M110.161455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Passerini V, Durrbaum M, Stingele S, Storchova Z. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 2014;33:2374–2387. doi: 10.15252/embj.201488648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci USA. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Calzada A, Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell. 2007;18:3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Tange Y, Kurabayashi A. Growth arrest and chromosome instability in aneuploid yeast. Yeast. 2006;23:937–950. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. APP locus duplication causes autosomal dominant early-onset alzheimer disease with cerebral amyloid antipathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Schvartzman J-M, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Serrano D, D'Amours D. When genome integrity and cell cycle decisions collide: roles of polo kinases in cellular adaptation to DNA damage. Syst Synth Biol. 2014;8:195–203. doi: 10.1007/s11693-014-9151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 2013;73:6401–6412. doi: 10.1158/0008-5472.CAN-13-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox JUR, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:1–12. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell. 2013;24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Tourrière H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Vidanes GM, Sweeney FD, Galicia S, Cheung S, Doyle JP, Durocher D, Toczyski DP. CDC5 inhibits the hyperphosphorylation of the checkpoint kinase Rad53, leading to checkpoint adaptation. PLoS Biol. 2010;8:e1000286. doi: 10.1371/journal.pbio.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004;117:575–588. doi: 10.1016/s0092-8674(04)00417-9. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Bacal J, Desmarais D, Padioleau I, Tsaponina O, Chabes A, Pantesco V, Dubois E, Parrinello H, Skrzypczak M, et al. The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol Cell. 2014;54:691–697. doi: 10.1016/j.molcel.2014.04.032. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yu Z, Fu X, Liang C. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell. 2002;109:849–860. doi: 10.1016/s0092-8674(02)00805-x. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet. 2012;8:e1002719. doi: 10.1371/journal.pgen.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.