Motile cilia have nine doublet microtubules, with hundreds of associated proteins that repeat in modules. Each module contains three radial spokes, which differ in their architecture, protein composition, and function. The conserved proteins FAP61 and FAP251 are crucial for the assembly and stable docking of RS3 and cilia motility.
Abstract
Dynein motors and regulatory complexes repeat every 96 nm along the length of motile cilia. Each repeat contains three radial spokes, RS1, RS2, and RS3, which transduct signals between the central microtubules and dynein arms. Each radial spoke has a distinct structure, but little is known about the mechanisms of assembly and function of the individual radial spokes. In Chlamydomonas, calmodulin and spoke-associated complex (CSC) is composed of FAP61, FAP91, and FAP251 and has been linked to the base of RS2 and RS3. We show that in Tetrahymena, loss of either FAP61 or FAP251 reduces cell swimming and affects the ciliary waveform and that RS3 is either missing or incomplete, whereas RS1 and RS2 are unaffected. Specifically, FAP251-null cilia lack an arch-like density at the RS3 base, whereas FAP61-null cilia lack an adjacent portion of the RS3 stem region. This suggests that the CSC proteins are crucial for stable and functional assembly of RS3 and that RS3 and the CSC are important for ciliary motility.
INTRODUCTION
Cilia are evolutionarily conserved, microtubule-based cell protrusions that are ubiquitously present in eukaryotes, with the exception of most fungi and higher plants. There are two types of cilia: solitary, immotile primary cilia (9 + 0 microtubule organization), which perform sensory and signaling functions, and motile cilia (9 + 2), which are often present in large numbers and propel cells or extracellular fluid. In humans, ciliary motility drives the clearance of mucus and bacteria out of the airways, circulation of the cerebrospinal fluid in brain ventricles and spinal cord, transport of oocytes in the Fallopian tube, and sperm motility. Defects in the assembly or function of motile cilia cause multisymptom and heterogeneous human disorders, including primary ciliary dyskinesia (PCD; Horani et al., 2013).
Motile cilia contain structures that are not present in primary cilia: the central pair complex (CPC), with two singlet microtubules and associated projections (Carbajal-Gonzalez et al., 2013); and peripheral doublet microtubule-associated structures, including the rows of inner and outer dynein arms (IDAs and ODAs, respectively), radial spokes (RSs), the nexin-dynein regulatory complex (N-DRC), the calmodulin (CaM)- and radial spoke-associated complex (CSC), and the modifier of inner arms complex (Dymek and Smith, 2007; Heuser et al., 2009, 2012; Pigino et al., 2011; Bui et al., 2012; Yamamoto et al., 2013). These complexes are distributed on the surface of the doublet microtubules, forming 96-nm repeat units. The activity of the dynein motors is believed to be regulated by the mechanochemical signals transmitted from the CPC through the radial spokes to the IDAs, ODAs, and associated regulatory complexes, such as the N-DRC (Smith and Yang, 2004; Heuser et al., 2009; Lin et al., 2011; Oda et al., 2014).
Radial spokes are T-shaped complexes that are anchored perpendicularly to the doublets with their bases and extend their stems and heads toward the CPC (Goodenough and Heuser, 1985; Nicastro et al., 2006). Each 96-nm repeat unit contains a triplet of nonidentical radial spokes (RS1, RS2, and RS3), except that in some species, including Chlamydomonas reinhardtii, RS3 is reduced, with only a base and a knob-like stem remnant (Pigino et al., 2011; Lin et al., 2012). In Chlamydomonas, RS1 and RS2 contain at least 23 polypeptides (Piperno et al., 1977, 1981; Yang et al., 2001, 2006). The size, architecture, and protein composition of the stems and heads of RS1 and RS2 are similar, but there are structural and compositional differences in their bases; for example, the Chlamydomonas CSC subunits (Dymek et al., 2011; Heuser et al., 2012) and flagellar associated protein 206 (FAP206; DUF3508 domain containing) in Tetrahymena (Vasudevan et al., 2015) contribute to the microtubule docking of RS2 but not of RS1.
Based on classical transmission electron microscopy (TEM), all three radial spokes per axonemal repeat were believed to be identical and reduced to only two spokes in Chlamydomonas flagella. However, cryo-electron tomography (cryo-ET) studies showed both that the structure of RS3 is markedly different from that of RS1 and RS2 and that Chlamydomonas flagella contain a truncated RS3 that is very similar in shape to the corresponding basal portion of full-length RS3 (Pigino et al., 2011; Lin et al., 2012). In Chlamydomonas, the activity of the dynein arms is at least in part regulated by direct physical contact between the projections of the CPC and the heads of RS1 and RS2 (Oda et al., 2014). However, the existence of the naturally headless RS3 in Chlamydomonas opens the question of whether radial spokes in general or RS3 specifically can function without direct mechanical interactions with the CPC. Thus further investigations of both the full-length and headless RS3 could bring clues to the functions of individual radial spokes and spokes in general.
Of interest, the short RS3 and the CSC are present in the axonemes of the pf14 Chlamydomonas mutant that is called “spokeless” because RS1 and RS2 are missing (Piperno et al., 1981; Dymek and Smith, 2007; Pigino et al., 2011; Barber et al., 2012). In Chlamydomonas, the CSC is located near the microtubule surface, within the broad region that interconnects the bases of RS2, RS3, and the N-DRC (Dymek and Smith, 2007; Dymek et al., 2011; Heuser et al., 2012). The CSC consists of CaM and three proteins that are conserved in organisms with motile cilia: FAP61, FAP91, and FAP251 (also known as CaM-IP3, CaM-IP2, and CaM-IP4), which are orthologues of the human C20orf26, AAT-1, and WDR66 proteins, respectively. FAP91 is an A-kinase anchoring protein (AKAP)–binding protein and interacts with RSP3 (Dymek and Smith, 2007), a conserved ciliary AKAP (Gaillard et al., 2001), a base/stem protein of RS1, and RS2 (Piperno et al., 1981). In Chlamydomonas, knockdown of either FAP61 or FAP91 by artificial microRNAs results in reduced cell swimming speed, abnormal flagellar waveform, and ultrastructural defects in RS2 and RS3, with frequent loss of both spokes and partial destabilization of the N-DRC (Heuser et al., 2012).
Here we address the function of two CSC subunits—FAP61 and the mostly unstudied FAP251—in Tetrahymena, a ciliated model organism that, unlike Chlamydomonas, assembles three full-length radial spokes. We take advantage of DNA homologous recombination–based gene targeting to generate strains lacking either FAP61 or FAP251. We show that the knockout of each CSC subunit in Tetrahymena leads to abnormal ciliary motility and destabilizes RS3 but not RS2 or RS1. In both null mutants, RS3 is either missing or incompletely assembled, and the distinct structural defects in RS3 reveal the likely locations of FAP251 and FAP61. Overall our study identifies two CSC components as RS3-specific building blocks and show that RS3, despite its natural truncation in some species, is important for ciliary motility.
RESULTS
The CSC proteins localize to cilia in Tetrahymena cells
A phylogenetic survey revealed that the CSC proteins are present in species with motile cilia and absent in organisms that lack cilia or assemble only nonmotile, sensory cilia, such as Caenorhabditis elegans (Supplemental Figures S1 and S2; Dymek and Smith, 2007). One exception is Drosophila melanogaster, which forms motile sperm flagella but lacks a FAP251 orthologue. However, there are FAP251 orthologues in other insects: Tribolium castaneum, Apis mellifera, and Anopheles gambiae (Supplemental Figure S2). The genome of Tetrahymena thermophila encodes single orthologues of each of the Chlamydomonas CSC proteins: FAP61p (TTHERM_00641200), FAP91p (TTHERM_00578560), and FAP251p (TTHERM_01262850). The amino acid sequence of the predicted Tetrahymena FAP61p is 24% identical and 41% similar to that of the Chlamydomonas FAP61 and 26% identical and 47% similar to that of the human orthologue, C20orf26, whereas the sequence of FAP91p is 31% identical and 51% similar to that of the human orthologue. The sequence of the WD40 containing protein FAP251p is 29% identical and 48% similar to the sequence of the Chlamydomonas and human orthologues. In ciliates, including Tetrahymena, the FAP61 orthologues have a lineage-specific stretch of ∼400 amino acids that is predicted to form numerous helices (Supplemental Figure S1).
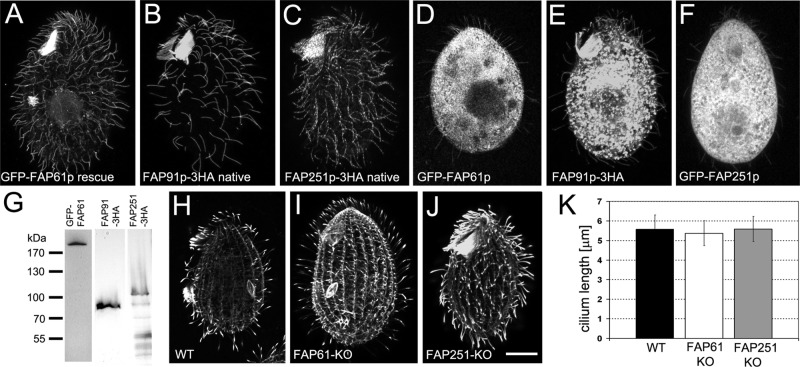
When tagged in their native loci, FAP91–3× hemagglutinin (3HA) or FAP251-3HA were detected in cilia (Figure 1, B and C). We could not detect a FAP61-3HA tagged at its native locus; however, the GFP–FAP61 (GFP, green fluorescent protein), which rescued the motility of FAP61-KO cells (see later discussion) localized to the locomotory and oral cilia (Figure 1, A and G). Similarly, overexpressed transgenic GFP-FAP61, GFP-FAP251, and FAP91-3HA localized to the locomotory and oral cilia in addition to their strong presence in the cell body (Figure 1, D–F).
FIGURE 1:
The CSC components FAP61, FAP91, and FA251 localize to Tetrahymena cilia but do not affect the length of cilia. (A–G) Localization of CSC proteins. FAP61 (A, D), FAP91 (B, E) and FAP251 (C, F) were expressed in FAP61-KO–rescued cells tagged with GFP (A) or tagged with 3HA in the native loci in wild-type background (B, C) or overexpressed as GFP-tagged (D, F) or 3HA-tagged (E) proteins. Note that all proteins localize to cilia; as expected, in the case of overexpression, the proteins also accumulated in the cell body. (G) Western blot of cilia (isolated from GFP-FAP61–rescued cells) and total cell extract (isolated from FAP91-3HA– and FAP251-3HA–expressing cells) probed with anti-GFP or anti-HA antibodies, respectively. Predicted molecular weight: GFP-FAP61, 223 kDa; FAP91-3HA, 79 kDa; and FAP251-3HA, 119 kDa. (H–J) Ciliary phenotype in knockout strains. FAP61-KO (I) and FAP251-KO (J) cells assemble cilia of normal length compared with wild-type cells (H) based on immunofluorescence with anti–α-tubulin antibodies. Bar, 10 μm. (K) Graph showing the average length of cilia. At least 50 cilia were measured for each genotype. Bars represent standard errors.
FAP251 contains WD40 repeats near its N-terminus and in the central region and a predicted EF hand–like domain near the C-terminus (D821–F982). The WD40 repeats facilitate protein–protein interactions (Li and Roberts, 2001). To assess which part(s) of FAP61 and FAP251 are required for their targeting to cilia, we overexpressed GFP-tagged, truncated variants (Supplemental Figure S3). The FAP61 truncations lacking the N-terminal (F330–D1699) or C-terminal (M1–D1400) fragments (Supplemental Figure S3, A and B), as well as FAP251 truncations missing various WD40 repeat–containing regions (W179–D1009, A342–D1009, M1–D370:E787–D1009), failed to be targeted to cilia (Supplemental Figure S3, D–F). A FAP251 truncation lacking the C-terminal region with the predicted EF hand–like domain (GFP-FAP251-M1–G792) was successfully targeted to cilia (Supplemental Figure S3G). Thus the potential calcium regulation of FAP251 is not required for its ciliary localization. Cells overexpressing truncated FAP61 or FAP251 in the wild-type background swam, proliferated, and formed food vacuoles at similar rates to those of wild-type cells.
FAP61 and FAP251 are important for ciliary motility
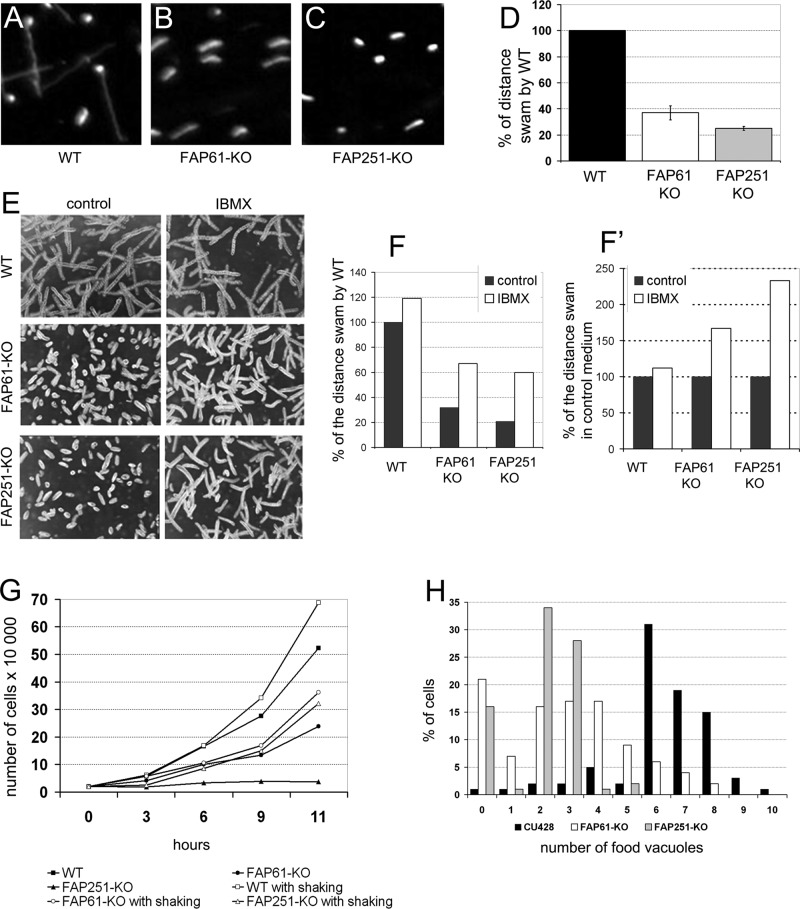
To learn about the function of FAP61 and FAP251, we generated FAP61 and FAP251 gene knockout (KO) strains (Supplemental Figure S4). Both FAP61-KO and FAP251-KO cells showed deficiencies in cell multiplication and ciliary functions, but the deletion of FAP251 consistently resulted in a more severe phenotype. High-speed video recording of live cells showed that the FAP61-KO and FAP251-KO mutants moved with greatly reduced velocities (Figure 2, A–D, and Supplemental Movies S1–S3). Immunofluorescence with anti–α-tubulin antibodies showed that the FAP61-KO and FAP251-KO cells assemble cilia that are normal in the number and length (Figure 1, H–K). The rate of cilium assembly after deciliation was also unaffected. Thus the reduced swimming velocity is likely a result of abnormal ciliary motility. Indeed, video recordings using a high-speed camera revealed that deletion of either FAP251 or FAP61 severely disturbed the waveform of beating cilia; the knockout cilia appeared stiffer (and therefore likely had a reduced bend amplitude), and the cells lacked proper metachrony across the ciliary rows (Supplemental Movies S4 and S5).
FIGURE 2:
Deletions of FAP61 or FAP251 strongly affect ciliary motility. (A–C) Video captures of paths of swimming wild-type (A), FAP61-KO (B), and FAP251-KO (C) cells recorded for 2.5 s. (D) Average swimming distances per 2.5 s normalized to the wild-type value (n = 100 for each strain); knockout cells swam 37 ± 5.5% (FAP61-KO) and 25 ± 1.5% (FAP251-KO) of the wild-type distance. (E) Paths of swimming cells untreated or treated with 0.5 mM IBMX. (F, F′) Average distance of swimming paths normalized to either untreated wild-type cells (F) or untreated cells with the same genetic background (F′); note that IBMX treatment partially rescues the swimming defect in the mutants. (G) Growth curves of wild-type and knockout strains grown at 30°C in SPP medium with or without shaking. (H) Evaluation of the efficiency of phagocytosis based on the number of India ink–labeled food vacuoles per cell formed during 10 min (averages of three experiments). Wild type, 6.2 food vacuoles, n = 322 cells; FAP61-KO, 2.8, n = 477; and FAP251-KO, 2.1, n = 452.
The reduced rate of the velocity of the knockouts compared with wild type was also observed under conditions that stimulate rapid ciliary beating. 3-Isobutyl-1-methylxanthine (IBMX), an inhibitor of cyclical nucleotide phosphodiesterases, increases the levels of cAMP and cGMP (Torphy, 1998), resulting in an increase of ciliary beat frequency in Tetrahymena (Hennessey et al., 1985; Geary et al., 1995; Wyatt et al., 1998). Treatment with 0.5 mM IBMX increased swimming velocity in both wild-type and knockout strains (Figure 2, F and F′). Of interest, IBMX sped up wild-type cells by 20%, whereas FAP61-KO and FAP251-KO cells sped up 60 and 140%, respectively, compared with untreated cells (Figure 2F′). However, even with IBMX, the swimming velocity of mutants did not return to wild-type level (Figure 2, E and F).
Proper beating of the locomotory cilia plays an important role in the final stage of Tetrahymena cell division (rotokinesis), during which daughter cells split by pulling apart (Brown et al., 1999). Strains lacking either FAP61 or FAP251 multiplied more slowly than wild-type cells when cultured in the standard medium SPP (Figure 2G) or in the enriched medium MEPP (data not shown), on which cilia-less mutants can multiply. Shaking of the cell culture can substitute for rotokinesis by providing the mechanical force needed for scission of daughter cells (Brown et al., 2003). As expected for a deficiency in ciliary motility, the rates of multiplication of the FAP61-KO and FAP251-KO cells increased with shaking (Figure 2G), and the calculated doubling times of the knockout strains approached that of the wild type (wild type, 2.2 h; FAP61-KO, 2.6 h; and FAP251-KO, 2.7 h).
At their anterior end, Tetrahymena cells have an oral apparatus that contains four groups of cilia, called the membranelles. The synchronous beating of the membranelles directs food particles into the oral cavity, and this motility is required for phagocytosis (Nilsson, 1979). When new food vacuoles were labeled with India ink added to the medium, both knockout strains showed greatly reduced rate of phagocytosis, indicating malfunction of the oral cilia (Figure 2H).
Introduction of the transgenes encoding either for FAP61 or FAP251 rescued the cell multiplication and motility phenotypes of the respective gene deletions (Supplemental Movies S6 and S7), and the rescuing tagged proteins were detected in the cilia. We conclude that the observed mutant phenotypes of the FAP61-KO and FAP251-KO cells are caused by the absences of the targeted proteins, showing that FAP61 and FAP251 are important for ciliary motility.
FAP61 and FAP251 are required for stable assembly of RS3
Studies in Chlamydomonas established that the CSC interacts with three structures: RS2, RS3, and the N-DRC (Dymek et al., 2011; Heuser et al., 2012). Longitudinal views of wild-type Tetrahymena cilia imaged by both classical TEM and cryo-ET show that all doublets have radial spoke triplets that repeat with 96-nm periodicity (Supplemental Figure S5), as described previously (Goodenough and Heuser, 1985; Pigino et al., 2011; Lin et al., 2012). On similar views of FAP61-KO cilia, we frequently observed 96-nm repeats with only a radial spoke pair; the spacing of 32 nm between the two remaining spokes suggests that they are RS1 and RS2, whereas RS3 is missing (Supplemental Figure S5, orange dots). A similar defect was observed only sporadically in the cilia of FAP251-KO (Supplemental Figure S5).
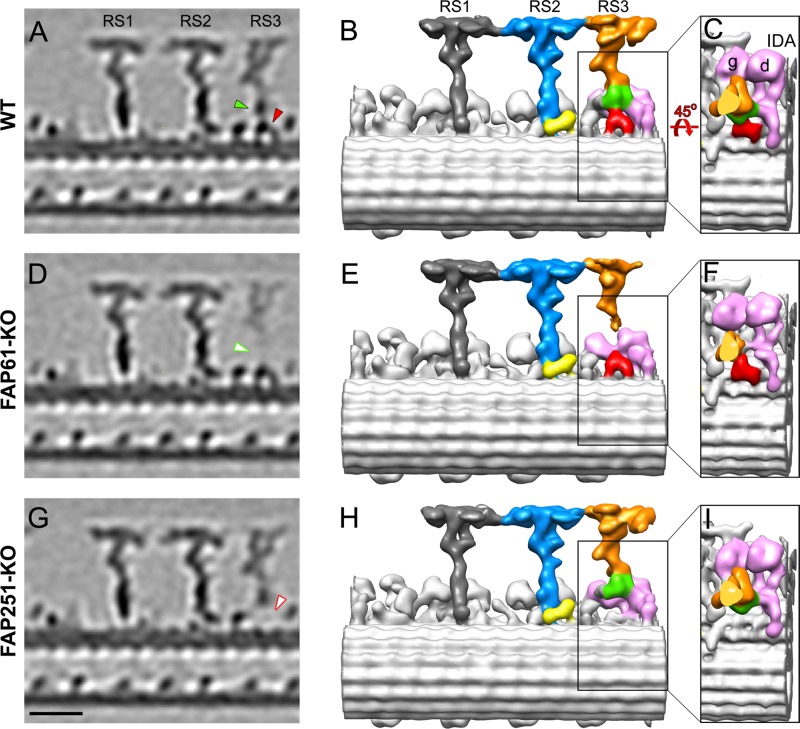
We used cryo-ET and subtomogram averaging to increase the resolution of the three-dimensional (3D) reconstructions of the 96-nm repeats of wild-type and mutant cilia (Figure 3). In the averaged 96-nm repeats of both knockouts, RS1 and RS2 appeared unaffected, including the back prong of RS2 (Figure 3, E and H, in yellow), which was reduced in the Chlamydomonas strains with depletion of either FAP61 or FAP91 (Heuser et al., 2012). In contrast, the electron density of RS3 was clearly reduced in both the FAP61-KO and FAP251-KO axonemes (Figure 3, D–I, and Supplemental Movies S8–S10). This indicates that a subset of the averaged 96-nm repeats lacks RS3, which agrees with our classical TEM and raw cryotomogram observations (Supplemental Figure S5). In addition to the overall reduced density of RS3, two distinct regions were completely missing from the remaining RS3 structure in the mutant cells; FAP61-KO lacked a part of the stem region (Figure 3, D and E), and FAP251-KO lacked an arch-like density at the spoke base where RS3 docks to the microtubule surface (Figure 3, G and H). The FAP61-dependent stem region (Figure 3, green) and the FAP251-dependent, arch-like RS base (Figure 3, red) are directly adjacent, which is consistent with the biochemical evidence that the three CSC proteins form a complex (Dymek and Smith, 2007).
FIGURE 3:
Comparison of the 3D structures of the 96-nm repeats from Tetrahymena WT, FAP61-KO, and FAP251-KO axonemes. Tomographic slices (A, D, G) and isosurface renderings (B, C, E, F, H, I) show the averaged 96-nm repeats in longitudinal views in WT (A–C) and the two CSC-knockout mutants, FAP61-KO (D–F) and FAP251-KO (G–I). In the WT strain, the central stem region of RS3 (green) connects to the arch-like basal region (red), the main distal part of RS3 (orange), and the tails of the two dynein IDAs g and d (pink; B, C). Both CSC-knockout mutants show structural defects in the basal half of RS3: in FAP61-KO (D, E), the central stem region (green arrowhead in A), and in FAP251-KO (G, H), the base (red arrowhead in A) are barely visible. Note that the main distal part of RS3 (orange) is present in all strains, but in FAP61-KO, its electron microscopic density is significantly weaker (D). The other two radial spokes, RS1 (dark gray) and RS2 (blue/yellow), appear structurally normal in the mutants compared with WT; note that also the back prong of RS2 (yellow), which was reduced in the artificial microRNAi mutants of FAP61 and FAP91 in Chlamydomonas (Heuser et al., 2012), seems unaffected in the Tetrahymena FAP61-KO and FAP251-KO mutants studied here. Bar, 20 nm.
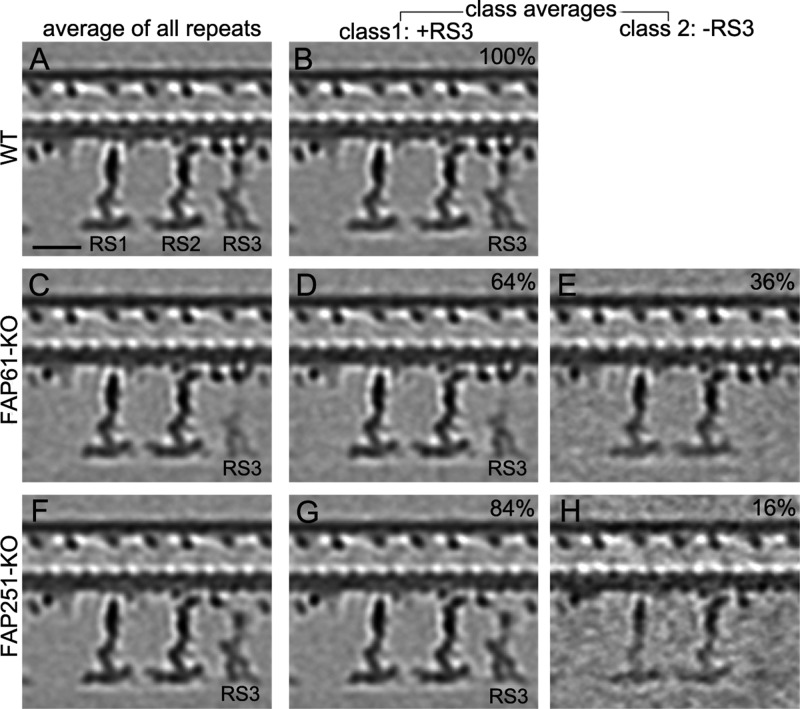
In addition to the lack of specific structures in the knockout axonemes, most of the remaining RS3 density was reduced in the total averages of the knockout repeats, indicating heterogeneity among the repeats (Figure 4, left column: average of all repeats). We used automatic image classification (Heumann et al., 2011) to sort the individual subtomograms into homogeneous subclasses that had RS3 either present (+RS3) or absent (–RS3; Figure 4). Similar to our observations made using classical TEM and raw cryotomograms, the fraction of 96-nm repeats lacking the entire RS3 was larger in the FAP61-KO (36%) than in the FAP251-KO (16%) data set (–RS3; Figure 4, E and H). The class averages with RS3 present (+RS3; Figure 4, D and G) confirmed that even when RS3 assembled in the knockout 96-nm repeats, it was incomplete, missing either a part of the stem in FAP61-KO (Figure 4D) or the arch-like base in FAP251-KO (Figure 4G). A classification analysis specifically focused on the FAP251-dependent, arch-like RS3 base revealed that in the FAP251-KO cilia, in about half of the 96-nm axonemal repeats, small remnants of the arch were visible, but in different subregions (Supplemental Figure S6, classes 1–4, red arrows and isosurface color). This suggests that apart from FAP251, additional protein(s) assemble the arch-like region of the RS3 base, but without FAP251, the remaining arch-forming components are unstable or flexible.
FIGURE 4:
Classification analysis of RS3 in cilia from FAP61-KO and FAP251-KO. Longitudinal slices of the averaged 96-nm repeats show the presence (class 1: +RS3) or absence (class 2: –RS3) of the main distal part of RS3 in WT (A, B) and the CSC-knockout mutants FAP61-KO (C–E) and FAP251-KO (F–H). Whereas RS3 is present in all WT 96-nm repeats (100%, B), it is missing from 36% and 16% of the 96-nm repeats from FAP61-KO (E) and FAP251-KO (H), respectively. The loss of RS3 on some of the 96-nm repeats resulted in reduced RS3 density in the mutant averages, including all repeats (C, F), especially in FAP61-KO (C). Even in the class average in which RS3 is present (D, G), the mutant RS3 lacks parts of the stem region in FAP61-KO (D) or the arch-like RS base in FAP251-KO (G), respectively. Note that although the distal part of RS3 was present in 64% of FAP61-KO repeats (D), the class 1 (+RS3) average still shows a slightly reduced distal RS3 density; this could be because a large part of the RS3 stem is missing in the FAP61-KO, so that the stem connection between the distal and basal parts of RS3 is greatly reduced and likely less stable, leading to a somewhat flexible position of the distal part of RS3 in individual repeats (positional heterogeneity) and blurring of the structure in averages as compared with WT. Bar, 20 nm.
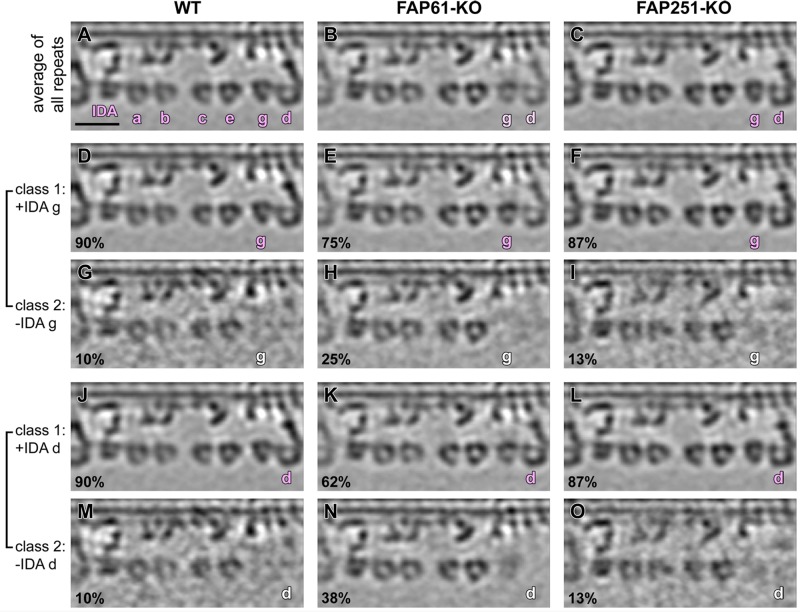
Most of the 96-nm repeats contain seven IDA arms: the two-headed I1 dynein (dynein f) and six single-headed dyneins (dyneins a–e and g). In wild-type Tetrahymena, the tails of the IDAs that localize to the positions IA5 (dynein g) and IA6 (dynein d) connect to the stem density that is missing in the axonemes of the FAP61-KO mutant (Figure 3, compare C and F; FAP61-dependent structure in green). Subtomogram averages of all 96-nm repeats showed that compared with wild type (Figure 5A), the electron densities of dyneins g and d were weaker in FAP61-KO (Figure 5B) but not in FAP251-KO (Figure 5C). A classification analysis focused on dyneins g and d revealed that even in wild-type axonemes, both dyneins g and d are present only in a subset (90%) of the repeats (Figure 5, D, G, J, and M). This could be due to natural variation among the 96-nm repeats or a result of partial extraction during the axoneme isolation procedure. The classification result for FAP251-KO was similar to that for wild type, with only 13% missing dyneins g and d (Figure 5, F, I, L, and O), whereas in FAP61-KO, the loss of dyneins g and d was elevated to 25–38% of the 96-nm repeats (Figure 5, E, H, K, and N).
FIGURE 5:
Classification analysis of IDAs g and d in axonemes from FAP61-KO and FAP251-KO. Longitudinal slices of averaged 96-nm repeats show the presence (g/d in pink) and absence (g/d in white) of IDAs g and d in WT (A, D, G, J, M), FAP61-KO (B, E, H, K, N), and FAP251-KO (C, F, I, L, O). The densities of IDAs g and d, in the average of all 96-nm repeats from FAP61-KO (B), are slightly weaker than those of WT (A), whereas FAP251-KO (C) shows a density similar to that of WT. Classification analysis of WT 96-nm repeats revealed that they lack IDA g (G) and IDA d (M) in a small subset (10%) of repeats. A similar fraction (13%) of 96-nm repeats in FAP251-KO lacks IDA g (I) and IDA d (O), whereas in FAP61-KO mutants, two to three times more repeats lacked IDA g (25%; H) and IDA d (38%; N) compared with WT. Bar, 20 nm.
FAP251 interacts with other CSC subunits and radial spoke proteins
To identify proteins that interact with FAP251, we performed immunoprecipitations with anti-GFP antibodies and radial spoke–enriched axonemal extracts (Dymek and Smith, 2007) from either FAP251-KO cells rescued with GFP-FAP251 or GFP-expressing wild-type cells as a negative control (Supplemental Figure S7). In addition to GFP-FAP251 and the partner CSC subunits FAP61 and FAP91, the mass spectrometry analysis of the GFP-FAP251 immunoprecipitate revealed two dynein heavy chains, Dyh15p and Dyh22p (Table 1). Dyh15p and Dyh22p are orthologous to the Chlamydomonas DHC7, which forms the dynein g heavy chain (position IA5; Wickstead and Gull, 2007; Wilkes et al., 2008). This finding agrees with the cryotomography results, showing that dynein g is reduced in the subtomogram average of the 96-nm repeat in FAP61-KO (Figure 5B). In addition, among the GFP-FAP251 coimmunoprecipitated proteins were four adenylate kinases (TTHERM_00148750, TTHERM_00558060, TTHERM_00227800, TTHERM_00317200), that is, phosphotransferases that catalyze the reversible conversion of adenine nucleotides (Panayiotou et al., 2014).
TABLE 1:
Mass spectrometry analysis of proteins coimmunoprecipitated with GFP-FAP251.
| Protein name | TGD genome ID | Number of identified peptides |
|---|---|---|
| FAP251 | TTHERM_01262850 | 70 |
| FAP91 | TTHERM_00578560 | 36 |
| FAP61 | TTHERM_00641200 | 134 |
| RSP3 | TTHERM_00566810 | 29 |
| RSP3 | TTHERM_00418270 | 59 |
| RSP8 | TTHERM_00313520 | 21 |
| RSP16 | TTHERM_00471260 | 17 |
| RSP16 | TTHERM_00238810 | 13 |
| RSP4/6 | TTHERM_00427590 | 16 |
| RSP4/6 | TTHERM_00502580 | 11 |
| RSP9 | TTHERM_00430020 | 23 |
| Dpy-30 motif–containing protein RSP2-like | TTHERM_00394410 | 63 |
| Dpy-30 domain–containing protein | TTHERM_000569069 | 20 |
| DHC15 (homologue of Chlamydomonas Dhc7) | TTHERM_00433800 | 41 |
| DHC22 (homologue of Chlamydomonas Dhc7) | TTHERM_00565600 | 29 |
| LRR-containing protein 34 | TTHERM_00695730 | 10 |
| LRR-containing protein 23 | XP_001014710.1 | 11 |
| Adenylate kinase 9 | TTHERM_00148750 | 170 |
| Adenylate kinase 1 | TTHERM_00317200 | 112 |
| Adenylate kinase 7 | TTHERM_00558060 | 29 |
| Adenylate kinase 8 | TTHERM_00227800 | 42 |
| Casein kinase | TTHERM_00938880 | 19 |
| ATU1 (α-tubulin) | TTHERM_00558620 | 24 |
| BTU1 (β-tubulin) | TTHERM_00348510 | 52 |
| FAP206 (DUF3508) | TTHERM_00820660 | 25 |
| FAP50 = CCDC135 (DRC7 of the N-DRC) | TTHERM_00760390 | 15 |
Table contains proteins that were identified by ≥10 peptides.
In Chlamydomonas, the stems and bases of RS1 and RS2 contain proteins RSP3, 5, 7, 8, 11–15, and 17–-22 (Piperno et al., 1977, 1981; Yang et al., 2006; Pigino et al., 2011), and RSP3 interacts with the CSC via FAP91 (Dymek and Smith, 2007). Among the proteins that coimmunoprecipitated with GFP-FAP251 were the orthologues of RSP8 (TTHERM_00313520) and RSP3. The Tetrahymena genome encodes three proteins with homology to RSP3 (TTHERM_00566810, TTHERM_01044600, TTHERM_00418270). Only two of these (TTHERM_00566810, TTHERM_00418270) were identified as interactors of GFP-FAP251. Chlamydomonas studies also indicated that RSP4 and the similar RSP6 are located in the RS heads, which connects to the stem through a bifurcated neck composed of the RSP2, RSP16, and RSP23 proteins (Piperno et al., 1977, 1981; Yang et al., 2006; Pigino et al., 2011; Barber et al., 2012). The Tetrahymena genome encodes three proteins with homology to RSP4/6 (TTHERM_00444180, TTHERM_00502580, TTHERM_00427590). Again, two of these RSP4/6 proteins (TTHERM_00502580, TTHERM_00427590), as well as two proteins with homology to RSP16 (TTHERM_00471260) and a protein with limited homology to the weakly evolutionarily conserved RSP2 (TTHERM_00394410), copurified with GFP-FAP251. At this point, our data are insufficient to determine whether the immunoprecipitated radial spoke proteins interact with the CSC directly or indirectly (see the Discussion). Recently we found that the Tetrahymena homologue of the Chlamydomonas FAP206, a protein containing a DUF3508 domain, functions as a microtubule-docking adapter at the base of RS2 (Vasudevan et al., 2015). Of importance, FAP206 also coprecipitated with GFP-FAP251, indicating that there is a bridge between RS2 and RS3, as previously proposed based on the structural analysis of CSC knockdowns in Chlamydomonas (Heuser et al., 2012).
DISCUSSION
The CSC is a recently discovered component of motile axonemes that plays an important but poorly understood role in ciliary motility. Here we took advantage of the Tetrahymena model to generate CSC subunit–null mutants and address the function of CSC in cilia with three full-length radial spokes per axonemal repeat. We found that the CSC subunits FAP61 and FAP251 are important for ciliary motility, and the knockout of either protein results in specific structural defects in RS3. The effect of RS3 defects on ciliary motility is surprising because RS3 is naturally truncated in some species with motile cilia, including the widely studied flagellated model Chlamydomonas. Similarly intriguing is that in Tetrahymena, the deletion of FAP251 results in a more severe phenotype than the deletion of FAP61, even though the structure of RS3 is more affected in FAP61-KO than FAP251-KO. This suggests that the primary reasons for abnormal ciliary motility in the CSC mutants, especially in FAP251 knockouts, are defects at the RS3 base (e.g., interruption of signal transduction), whereas instability of the RS3 stem and head and thus reduced mechanical interactions between the RS3 head and CPC projections have relatively minor effects on motility.
FAP61 and FAP251 build the basal substructures of RS3
In Tetrahymena, the 96-nm repeat contains three T-shaped radial spokes, RS1–3, which are similar in length. In contrast, the third spoke in Chlamydomonas, called short RS3 or RS3S, is limited to a knob that lacks a spoke head and most of the stem (Pigino et al., 2011; Barber et al., 2012; Lin et al., 2012). Structural analyses of the 96-nm repeats from Chlamydomonas strains with knockdown of either FAP61 or FAP91 suggest that the CSC localizes near the bases of RS2 and RS3S and connects to the N-DRC (Dymek et al., 2011; Heuser et al., 2012). In the Chlamydomonas knockdown strains, both RS2 and RS3 are frequently lost, whereas RS1 is unaffected, which provided the first evidence that the individual radial spokes have nonidentical requirements for their assembly (Dymek et al., 2011; Heuser et al., 2012).
Here we show that deletion of either FAP61 or FAP251 in Tetrahymena destabilizes only RS3. More precisely, FAP251 is needed to assemble an arch-like structure at the RS3 base, and FAP61 is needed to assemble a globular structure in the stem of RS3 adjacent to the arch-like structure. Moreover, in Tetrahymena, assembly of the FAP251-dependent, arch-like structure does not require FAP61, and, conversely, assembly of the FAP61-dependent stem part is independent of FAP251. In contrast, knockdown of FAP61 in Chlamydomonas led to the loss of RS3S, including the arch-like structure. Another difference to Chlamydomonas is that in the Tetrahymena CSC knockouts, no structural defects were detected in RS2, the N-DRC, or the B-tubule inner junction near the N-DRC base (for detailed comparison see Supplemental Table S1; Dymek et al., 2011; Heuser et al., 2012).
Taking together the data obtained in Tetrahymena and Chlamydomonas, we can conclude that the CSC components play an evolutionarily conserved role in the assembly of RS3 substructures. Because the basal part of RS3 is very similar in shape to RS3S (Pigino et al., 2011; Lin et al., 2012), and the CSC knockdowns in Chlamydomonas destabilize RS3S, it is most likely that FAP61 and FAP251 build the same structures in the bases of RS3/RS3S in all motile axonemes. However, the depletions of CSC subunits seemed to have stronger destabilizing effects on the structures connected to the CSC in Chlamydomonas than in Tetrahymena. Two factors that could reduce the destabilizing effects on RS2 and RS3 in Tetrahymena are the connection between the spoke heads of RS2 and RS3 and a stronger RS2 base in Tetrahymena. In Chlamydomonas, RS3S is too short to connect to the RS2 head, and the RS2 base has only two prongs (front and back), whereas the RS2 base in Tetrahymena has an additional, third microtubule-docking site (side prong), which could make RS2 attachment more resistant to defects in neighboring structures.
CSC forms a bridge between RS2 and RS3
A bridge that connects RS2 and RS3S and includes the CSC was suggested previously based on comparisons of the CSC knockdown and wild-type structures in Chlamydomonas (Heuser et al., 2012). Given that we can now place two subunits of the CSC—FAP61 and FAP251—within the base of RS3, it is very likely that the remaining subunit of CSC—FAP91—contacts the base of RS2 on the side facing RS3 (the back prong of RS2). This model is supported by the following additional Tetrahymena data for FAP206, a component of the RS2 base (Vasudevan et al., 2015): 1) loss of FAP206 destabilizes the front and back prongs of RS2 (but not the side prong) and simultaneously reduces the levels of FAP91 in axonemes; 2) in 11% of the FAP206-KO axonemal repeats, the RS3 was missing; and 3) FAP206 coimmunoprecipitates with FAP251, providing direct biochemical evidence for the existence of an RS2–RS3 bridge. Although there is no such structure visible by cryo–electron microscopy, such a bridge could be located close to the microtubule surface, or its conformation could be too dynamic to be revealed at a sufficient resolution by cryo-ET.
FAP61 stabilizes IDAs that assemble in the vicinity of RS3
In both Chlamydomonas and Tetrahymena, subtomogram averaging shows that the depletion of FAP61 leads to the reduction of a subset of IDAs. In both species, the depletion results in a weak reduction of IA6/dynein d. However in Chlamydomonas, there was also a strong reduction of IA4/dynein e and a weak reduction of IA2/dynein a (Heuser et al., 2012), whereas in Tetrahymena, IA5/dynein g was reduced. The interaction between the Tetrahymena CSC and dynein g was confirmed by coimmunoprecipitation (Table 1). Consistently, in Tetrahymena, the loss of FAP61 selectively affected the IDAs present in the vicinity of RS3 (dyneins d and g), whereas the major IDA defect in Chlamydomonas—IA4 reduction—seems to be related to the N-DRC defect (Heuser et al., 2012). In particular, in Tetrahymena, dynein g is likely sensitive to the loss of CSC because its tail is attached to the base of RS3. Furthermore, in Tetrahymena, the stem of RS3, which includes FAP61, may interact more closely with the IDAs in its vicinity, that is, IA5/dynein g and IA6/dynein d.
ODAs are believed to control primarily the ciliary beat frequency, whereas IDAs are believed to control primarily the ciliary waveform (Brokaw and Kamiya, 1987; Kamiya, 2002). Both CSC knockouts swim with reduced velocity and display abnormal ciliary waveform; however, the defects are more severe in FAP251-KO. In contrast to Tetrahymena FAP61-KO cells, which show a partial loss of dyneins d and g, FAP251-KO cells contain IDAs at wild-type level. Therefore it is likely that the motility defects in CSC-deficient cells result from malfunction of IDAs in the vicinity of RS3 (dyneins d and g)—for example, through misregulation—rather than from the loss of dynein arms.
A recent study showed that radial spokes transduce mechanical signals generated by direct mechanical interactions between the central pair projections and the radial spoke heads of RS1 and RS2, down to dynein arms (Oda et al., 2014). In Tetrahymena, the full-length RS3 also interacts with the CPC, and proper signal transduction by RS3 to IDAs located nearby could be affected by the loss of FAP61 or FAP251. On the other hand, the CSC is also important for cilia motility in axonemes with a headless RS3S (Dymek and Smith, 2007). It is therefore possible that in Chlamydomonas, IDAs associated with RS3S receive signals that originate from interactions between CPC and RS2 and are transferred by the CSC bridge to RS3. Thus there might be not only a physical but also a functional connection between RS2 and RS3 involving the CSC. In the future, the function of the RS3 radial spoke head/stem in organisms with full-length RS3 could be studied by generating headless RS3 mutants. Such experiments may be feasible if proteins specific to the stem or head of RS3 are identified (see later discussion).
Heterogeneity of radial spoke protein composition
We show that in Tetrahymena, similar to Chlamydomonas, CSC coimmunoprecipitates with radial spoke proteins (Table 1). Although all three radial spokes in the Tetrahymena 96-nm repeat are full-length structures, the morphology of the head and stem of RS3 is markedly different than that of RS1 and RS2, suggesting that there are differences in protein composition (Pigino et al., 2011; Lin et al., 2012). RSP3 is a major component of the stems of RS1 and RS2, whereas RSP4 and RSP6 are located in the heads of RS1 and RS2. The Tetrahymena macronuclear genome encodes three paralogues of both RSP3 and RSP4/6 proteins. Intriguingly, we found only two paralogues for both RSP3 and RSP4/6 in a complex with FAP251. The missing paralogues could be specific to RS1, which does not seem to interact with the CSC. Future proteomic analyses could reveal paralogues or even novel ciliary proteins that are specific to the head, neck, and stem of RS3. Even if the differential usage of certain proteins to build individual radial spokes is an adaptive feature of ciliates, in the future, the knockouts of differentially used radial spoke proteins could create a valuable tool to dissect the functions of individual radial spokes or treat RS-related ciliopathies.
The presence of FAP50/DRC7 (Yang et al., 2011; Bower et al., 2013) in the GFP-FAP251 immunoprecipitate suggests that also in Tetrahymena cilia the CSC interacts with the N-DRC, as shown for Chlamydomonas (Heuser et al., 2012).
Of interest, among the proteins that coprecipitated with GFP-FAP251, we found four adenylate kinases that catalyze the reversible enzymatic reaction 2 ADP ↔ ATP + AMP, producing ATP and maintaining the balance between ADP and ATP. Our observations agree with reports on the tight association of adenylate kinases with the axoneme in Tetrahymena (Nakamura et al., 1999) and other organisms (Noguchi et al., 2001). A mutation in the adenylate kinase 7 causes PCD in a mouse model (Fernandez-Gonzalez et al., 2009; Mata et al., 2012). In Chlamydomonas, one of the central pair proteins, CPC1, has an adenylate kinase domain, and its loss causes a ciliary deficiency that is rescued by excess of ATP (Zhang and Mitchell, 2004). Moreover, in Chlamydomonas, mutation in either ODA5 or ODA10 protein, both of which are required for assembly of ODAs, is associated with reduction in the level of an unidentified adenylate kinase–related protein, suggesting that Oda5/Oda10 anchor an adenylate kinase in the proximity of dynein arms (Wirschell et al., 2004). Thus the localized production of ATP could play a role in selective activation (binding to catalytic AAA1 domain of dynein) or regulation (binding to noncatalytic AAA2-4 domains; Inoue and Shingyoji, 2007; DeWitt et al., 2015) of the axonemal dynein ATPases. Previous studies also suggested that the activity of some IDA subtypes is affected by the concentration of ADP (Yagi, 2000). Thus the ciliary adenylate kinases could be important for establishing a location-specific composition of the adenine nucleotide pool (Dzeja and Terzic, 2003). It remains to be determined whether these adenylate kinases are associated specifically with RS3. If this is the case, one way in which RS3 could function is by controlling the production of ATP in its vicinity, which in turn could affect the preferential activity of neighboring IDAs.
MATERIALS AND METHODS
Protein sequence analysis
The Chlamydomonas reinhardtii amino acid sequences of FAP61 and FAP251 were used as bait to search for homologues in the Tetrahymena Genome Database (TGD; Eisen et al., 2006), Flybase, and the National Center for Biotechnology Information database. The sequences were aligned in the ClustalX2 program (Jeanmougin et al., 1998) and edited in SeaView (Galtier et al., 1996). Some predicted protein sequences were manually corrected (see the legends of Supplemental Figures S1 and S2). Domain analyses of FAP61 and FAP251 were performed using SMART (http://smart.embl-heidelberg.de/), InterProScan (www.ebi.ac.uk), HHpred (toolkit.tuebingen.mpg.de/hhpred), COILS (www.ch.embnet.org), and WDSP (wu.scbb.pkusz.edu.cn/wdsp; Wang et al., 2013, 2015).
Tetrahymena strains and culture
Wild-type CU428.2 and B2086.2 strains were obtained from the Tetrahymena Stock Center (Cornell University, Ithaca, NY). The CU522 strain, which carries a mutation (K350M) in the BTU1 coding region resulting in paclitaxel sensitivity, was used for introduction of the transgenes, enabling protein overexpression (Gaertig et al., 1994). With the exception of FAP251-KO cells, all strains were grown in the standard medium SPP with shaking at 30°C (Gorovsky et al., 1975) unless otherwise indicated. The FAP251-KO cells were grown in the MEPP medium, in which cells are not dependent on cilium-based phagocytosis (Orias and Rasmussen, 1976). All media were supplemented with an antibiotic-antimycotic mix at 1:500 (Sigma-Aldrich, St. Louis, MO). The knockout cells were grown in 25–50 ml of medium volume in 300-ml Erlenmeyer flasks with shaking (80 rpm) at room temperature.
Phenotypic studies
To measure proliferation rate, cells at initial density of (2–2.5) × 104 cells/ml were grown at 30°C with or without shaking in SPP or MEPP and counted every 3 h. To measure the rate of phagocytosis, cells were fed 0.1% India ink for 10 min at 30°C and fixed with 2% paraformaldehyde. The number of vacuoles per cell was scored in ≥100 cells. To compare cell motilities, swimming cells were recorded using a Zeiss spinning disk microscope with 10× objective. To measure the length of swimming paths, cells were diluted to a density of 7500 cells/ml and recorded in dark field for 2.5 s using a Sony Alpha1 camera and 10× objective.
To assay the effect of IBMX, cells were grown in SPP and washed in wash solution (10 mM Tris-HCl, 50 μM CaCl2, and 10 mM 3-(N-morpholino)propanesulfonic acid [MOPS], pH 7.2). Cells were preincubated in wash solution for 1 h and placed in 80-μl drops. The motility of the IBMX-treated cells was recorded using a Moticam 480 imaging system and a 40× objective. The length of the recorded swim paths was measured using ImageJ, version1.42 (Schneider et al., 2012).
To visualize the ciliary beating of the faster-swimming wild-type strain, the cells were video recorded at 500 frames/s by a Photronics 1280 PCI FastCam on a Nikon Eclipse E600 microscope at 600× total magnification. Beating of cilia in the mutant cells were recorded with a Leica DMIR2 inverted microscope (660× magnification) equipped with a QImaging Retiga EXi charge-coupled device camera at 60 frames/s. To compare the ciliary beating of mutants and wild type, the digital videos were converted to the viewing rate of 30 frames/s.
Protein tagging and domain analysis
To overexpress (untagged or GFP- or HA-tagged) proteins in the nonessential BTU1 locus, the coding regions of FAP61 (TTHERM_00641200), FAP91 (TTHERM_00578560), and FAP251 (TTHERM_01262850) were amplified from the CU428 strain genomic DNA by PCR using PfuUltraII Fusion (Agilent Technologies, Santa Clara, CA) or Phusion Hot Start II (Thermo Scientific, Waltham, MA) polymerases with the addition of MluI and BamHI sites using the primers listed in Supplemental Table S2 and cloned into the pMTT1-GFP plasmid (Wloga et al., 2006). For expression of the HA-tagged proteins, pMTT1-GFP was modified by replacing the GFP coding region positioned between the HindIII and MluI sites with a fragment encoding an HA epitope tag to generate pMTT1-HA. For expression of the C-terminally 3HA-tagged proteins, the sequence encoding 3HA was inserted using the BamHI site.
For domain truncation analyses, fragments of the FAP61 or FAP251 open reading frames (Supplemental Figure S3) were amplified from the genomic DNA with the addition of MluI and BamHI restriction sites (primers in Supplemental Table S2) and cloned into pMTT1-GFP or pMTT1-HA. A 15-μg amount of a transgene plasmid was digested with ApaI and SacII to separate the targeting fragment from the plasmid, precipitated onto DNAdel Gold Carrier Particles (Seashell Technology, La Jolla, CA) according to the manufacturer's instructions, and biolistically transformed into the CU522 strain. Transformants were selected for 3–4 d on SPP with 20 μM paclitaxel (BioShop Canada) at 30°C.
To express C-terminally 3HA-tagged proteins in their native loci, 1.8-kb fragments of the 3′ untranslated region of FAP61, FAP91, or FAP251 were amplified from the genomic DNA with the addition of PstI and SacI sites (primers in Supplemental Table S2) and cloned on one side of the rpl29 marker in pMTT1-RPL29 (Tsao and Gorovsky, 2008). Next 1.5-kb fragments of the corresponding C-terminal coding regions without a stop codon but with a 3HA epitope tag and a 0.5 kb of BTU1 transcription terminator were cloned (in this order) between the ApaI and ClaI sites of the pMTT1-RPL29 derivatives made in the previous step. About 15–20 μg of the final plasmids was digested with ApaI and SacI and used for biolistic transformation, and transformants were selected for 3–4 d at 30°C on SPP or MEPP with 2.5 μg/ml CdCl2 and 10 μg/ml cycloheximide and grown under cycloheximide selection to promote phenotypic assortment.
Germline knockouts of FAP61 and FAP251 and transgene-based rescues
Fragments of either the FAP61 or FAP251 gene were amplified with primers listed in Supplemental Table S2 and subcloned on the sides of the neo4 plasmid (Mochizuki, 2008) to obtain transgenes with neo4 positioned in a reverse transcriptional orientation to the targeted gene coding region. The targeting fragments containing homologous sequences flanking neo4 were separated from the plasmid by digestion with ApaI and SacII, and 20 μg of each digested plasmid was used for biolistic transformation of conjugating CU428 and B2086 cells. The FAP61 and FAP251 gene knockout transformants were selected, and heterokaryons and homokaryons (FAP61-KO or FAP251-KO strain) were made as described (Cassidy-Hanley et al., 1997; Dave et al., 2009). The deletion of part of the FAP61 or FAP251 coding region was confirmed by PCR (primers in Supplemental Table S2).
For rescue, the knockout cells were biolistically transformed with 15 μg of pMTT1-GFP-FAP251 or pMTT-GFP-FAP61 digested with ApaI and SacII. After 3 d of selection on SPP with 100 μg/ml paromomycin and 2.5 μg/ml CdCl2, vigorously swimming rescued transformants were present in nearly all wells; such cells were not observed in a mock biolistic transformation (without plasmid DNA).
Immunofluorescence and Western blot
For all fluorescence imaging, cells were handled as drops on coverslips. For the transgenic protein visualization, cells at initial density of (1–2) × 105 cells/ml were grown in SPP, and expression of the transgene was induced with 2.5 μg/ml CdCl2 for 2–24 h at 30°C. To localize the HA-tagged proteins, 10 μl of cells was fixed by combining with a 10-μl mixture of 0.5% Triton X-100 and 1% paraformaldehyde in PHEM (60 mM 1,4-piperazinediethanesulfonic acid [PIPES], 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 10 mM ethylene glycol tetraacetic acid [EGTA], 2 mM MgCl2, pH 6.9; Schliwa and van Blerkom, 1981). The GFP signal was observed in live cells or after fixation with 0.5% Triton X-100 and 1% paraformaldehyde. For tubulin detection, 10 μl of cells was fixed with 10–15 μl of 0.5% NP-40 and 2% paraformaldehyde in PHEM. After drying, the coverslips with cells were blocked for 10 min with 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS; 130 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, 10 mM EGTA, 2 mM MgCl2, pH 7.2) and incubated overnight at 4°C with the following primary antibodies diluted in 3% BSA in PBS: mouse anti-HA monoclonal antibody (16B12; Covance, Berkeley, CA) at 1:300 dilution, rabbit anti-HA polyclonal antibody (Covance) at 1:300 dilution, or mouse anti–α-tubulin 12G10 antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) diluted 1:200 (concentrate) or 1:20 (culture supernatant). After washing for 3 × 5 min in PBS, coverslips were incubated with secondary antibodies, either anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG, conjugated with Alexa 488 or Alexa 555 (Invitrogen, Eugene, OR) diluted 1:300 in 3% BSA/PBS for 1.5 h at room temperature. After washing, coverslips were mounted in Fluoromount-G (Southern Biotech, Birmingham, AL) and imaged using one of the following confocal microscopes: Zeiss LSM780, Leica TCS SP5, or Leica TCS SP8.
For Western blot analysis, cilia were isolated as previously described (Wloga et al., 2008). To extract the total cell protein, 107 cells were collected by centrifugation, washed with 10 mM Tris-HCl (pH 7.5), and resuspended in cold 10 mM Tris-HCl (pH 7.5) with protease inhibitors to a 150-μl final volume; after addition of 5× SDS, sample buffer cell suspensions were heated at 95°C for 5 min.
Immunoprecipitation and mass spectrometry
Cilia from the Tetrahymena GFP-FAP251 rescue cells and GFP-expressing cells (negative control) were harvested as described (Wloga et al., 2008). To isolate the axonemes, cilia were suspended in 350 μl of 10 mM Tris-HCl buffer, pH 7.5, with protease inhibitors (Complete; Roche, Indianapolis, IN), combined with an equal volume of 2% NP-40 and 1 M NaCl in 80 mM Tris-HCl buffer, pH 7.5, and incubated for 15 min on ice. The axonemes were pelleted at 20,000 × g and treated with 0.5 M KI, 30 mM NaCl, 5 mM MgSO4, 0.5 M EDTA, 1 mM dithiothreitol, and 10 mM HEPES, pH 7.5, for 30 min on ice (Dymek and Smith, 2007). The extracted axonemes were collected by centrifugation at 16,000 × g for 15 min at 4°C and concentrated to 1.8 mg/ml by ultrafiltration in Vivaspin columns (Sartorius, Goettingen, Germany).
A 50-μl amount of the KI axonemal extract was diluted in 10 mM Tris-HCl, pH 7.5, to a final volume of 0.5 ml and incubated with 50 μl of GFP-TRAP or control beads (Chromotek, Planegg-Martinsried, Germany) with rotation for 1 h at 4°C according to the manufacturer's instructions. Beads were washed six times for 5 min with 1 ml of 40 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.2% NP-40 at 4°C. Proteins were eluted from beads with 0.2 M glycine, pH 2.2, precipitated with cold acetone, suspended in the SDS sample buffer, separated on 8% SDS–PAGE gel, and silver stained according to Shevchenko et al. (1996).
Classical transmission electron microscopy
Tetrahymena cells were grown to a mid log phase. A 5-ml amount of cells was collected, washed with 10 mM Tris-HCl, pH 7.5, and fixed with 2% glutaraldehyde (Sigma-Aldrich; diluted from 8% with 0.1 M cacodylate buffer, pH 7.2) on ice with gentle rocking. After 1 h, 10 μl of freshly prepared 1% tannic acid was added, and samples were incubated for an additional 1 h. After five washes for 10 min each wash in cold 0.1 M cacodylate buffer, pH 7.2, samples were postfixed on ice for 1 h with 1% osmium tetroxide (Sigma-Aldrich) in ultrapure water. After five washes for 10 min each wash with water at room temperature, samples were embedded in 2% low–melting point agarose, and chunks were cut into 2- to 3-mm fragments and dehydrated in a percentage series of ethanol followed by infiltration in Durcupan ACM Fluka (Sigma-Aldrich) according to the manufacturer's instructions.
To study isolated cilia, 150 ml of wild-type or knockout culture was grown to a density of 3 × 105 cells/ml and deciliated with a pH shock, and cilia were recovered (Wloga et al., 2008). Cilia were fixed as described and sectioned (40- to 50-nm-thick sections) as described (Angus et al., 2001). All samples were analyzed using a JEM 1200 EX transmission electron microscope (JOEL, Japan).
Axoneme isolation and cryosample preparation
Axonemes were isolated from Tetrahymena strains CU428 (wild type [WT]), FAP61-KO, and FAP251-KO as described (Wloga et al., 2008). After deciliation, cell bodies were removed by three centrifugation steps—once at 1500 × g for 5 min and twice at 1860 × g for 5 min at 4°C. Then cilia in the supernatant were pelleted by centrifugation at 10,000 × g for 15 min at 4°C. The ciliary membrane was removed by detergent treatment using HMEEK buffer (30 mM HEPES, 25 mM KCl, 5 mM MgSO4, 0.1 mM EDTA, and 1.0 mM EGTA, pH 7.4) with 1% IGEPAL CA-630 (Sigma-Aldrich) for 30 min at 4°C. The axonemes were collected by centrifugation at 10,000 × g for 10 min and resuspended in HMEEK buffer. A 3-μl amount of axonemes and 1 μl of a 5×-concentrated 10-nm colloidal gold solution (Sigma-Aldrich) were applied to a glow-discharged Quantifoil holey carbon grid (Quantifoil Micro Tools, Jena, Germany). Excess liquid was blotted for ∼2 s with filter paper, and then the grid was immediately plunged into liquid ethane using a home-made plunge freezer to achieve sample vitrification. The frozen samples were stored in liquid nitrogen until further examination.
Cryo–electron tomography
The vitrified cryosample was transferred into a Tecnai F30 transmission electron microscope (FEI, Eindhoven, Netherlands) equipped with a field emission gun and a postcolumn energy filter (Gatan, Pleasanton, CA). The axonemes were imaged at 300 keV under low-dose conditions and in the zero-loss mode of the energy filter (20-eV slit width); images were recorded with a 2k × 2k charge-coupled device camera (Gatan, Pleasanton, CA) at −8 μm defocus and magnification of 13,500, resulting in a pixel size of 1 nm. Tilt series of axonemes were collected by rotating the sample from about −65 to +65° with 1.5–2.5° angular increments using SerialEM software (Mastronarde, 2005). The cumulative electron dose per tilt series was restricted to ∼100 e/Å2.
Image processing
The two-dimensional images of tilt series were aligned using the 10-nm gold fiducials and reconstructed into 3D tomograms by weighted backprojection using IMOD software (Kremer et al., 1996). Only tomograms of intact, noncompressed or mildly compressed axonemes (12, 11, and 11 tomograms for WT, FAP61-KO, and FAP251-KO strains, respectively) were further processed. To increase resolution, subtomograms containing individual 96-nm repeats were extracted from the raw cryotomograms, aligned, and averaged using PEET software (Nicastro et al., 2006). To analyze heterogeneity concerning the presence/absence of RS3, inner dyneins g and d, and the arch-like structure at the RS3 base, subtomograms were further analyzed using the classification approach provided in PEET (Heumann et al., 2011) and as previously described (Heuser et al., 2012). The University of California, San Francisco, Chimera package (Pettersen et al., 2004) was used for coloring and isosurface rendering visualization of the averaged 3D axonemal structures.
Supplementary Material
Acknowledgments
We are very grateful to the staff of the Laboratory of Imaging Tissue Structure and Function, Nencki Institute of Experimental Biology, Warsaw, Poland; Henryk Bilski and Szymon Suski (Laboratory of Electron Microscopy, Nencki Institute); and Chen Xu (Electron Microscopy Facility, Brandeis University, Waltham, MA) for skilful technical help, training, and facility management. The mass spectrometry analyses were done in the Mass Spectrometry Laboratory, Institute of Biochemistry and Biophysics, PAS, Warsaw, Poland. The monoclonal anti–α-tubulin 12G10 antibody developed by J. Frankel and E. M. Nelsen was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology, University of Iowa, Iowa City, IA. This research was supported by Ministry of Science and Higher Education Grant N301706640 and EMBO Installation Grant 2331 to D.W., National Institutes of Health Grants GM089912 to J.G. and GM083122 to D.N., and a March of Dimes Foundation grant to D.N.
Abbreviations used:
- CaM
calmodulin
- CPC
central pair complex
- cryo-ET
cryo–electron tomography
- CSC
calmodulin- and spoke-associated complex
- FAP
flagellar associated protein
- IDA
inner dynein arm
- N-DRC
nexin-dynein regulatory complex
- ODA
outer dynein arm
- RS
radial spoke
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1545) on February 18, 2015.
*These authors contributed equally.
REFERENCES
- Angus SP, Edelmann RE, Pennock DG. Targeted gene knockout of inner arm 1 in Tetrahymena thermophila. Eur J Cell Biol. 2001;80:486–497. doi: 10.1078/0171-9335-00178. [DOI] [PubMed] [Google Scholar]
- Barber CF, Heuser T, Carbajal-Gonzalez BI, Botchkarev VV, Jr, Nicastro D. Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol Biol Cell. 2012;23:111–120. doi: 10.1091/mbc.E11-08-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell. 2013;24:1134–1152. doi: 10.1091/mbc.E12-11-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Brown JM, Fine NA, Pandiyan G, Thazhath R, Gaertig J. Hypoxia regulates assembly of cilia in suppressors of Tetrahymena lacking an intraflagellar transport subunit gene. Mol Biol Cell. 2003;14:3192–3207. doi: 10.1091/mbc.E03-03-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Hardin C, Gaertig J. Rotokinesis, a novel phenomenon of cell locomotion-assisted cytokinesis in the ciliate Tetrahymena thermophila. Cell Biol Int. 1999;23:841–848. doi: 10.1006/cbir.1999.0480. [DOI] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajal-Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken) 2013;70:101–120. doi: 10.1002/cm.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D, Wloga D, Gaertig J. Manipulating ciliary protein-encoding genes in Tetrahymena thermophila. Methods Cell Biol. 2009;93:1–20. doi: 10.1016/S0091-679X(08)93001-6. [DOI] [PubMed] [Google Scholar]
- DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, Yildiz A. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat Struct Mol Biol. 2015;22:73–80. doi: 10.1038/nsmb.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Heuser T, Nicastro D, Smith EF. The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell. 2011;22:2520–2531. doi: 10.1091/mbc.E11-03-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja PP, Terzic A. Phosphotransfer networks and cellular energetics. J Exp Biol. 2003;206:2039–2047. doi: 10.1242/jeb.00426. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A, Kourembanas S, Wyatt TA, Mitsialis SA. Mutation of murine adenylate kinase 7 underlies a primary ciliary dyskinesia phenotype. Am J Respir Cell Mol Biol. 2009;40:305–313. doi: 10.1165/rcmb.2008-0102OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, Gu L, Gorovsky MA. Electroporation-mediated replacement of a positively and negatively selectable beta-tubulin gene in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1994;91:4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard AR, Diener DR, Rosenbaum JL, Sale WS. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP) J Cell Biol. 2001;153:443–448. doi: 10.1083/jcb.153.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Geary CA, Davis CW, Paradiso AM, Boucher RC. Role of CNP in human airways: cGMP-mediated stimulation of ciliary beat frequency. Am J Physiol. 1995;268:L1021–L1028. doi: 10.1152/ajplung.1995.268.6.L1021. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985;100:2008–2018. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA, Yao MC, Keevert JB, Pleger GL. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Hennessey T, Machemer H, Nelson DL. Injected cyclic AMP increases ciliary beat frequency in conjunction with membrane hyperpolarization. Eur J Cell Biol. 1985;36:153–156. [PubMed] [Google Scholar]
- Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol. 2011;175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Dymek EE, Lin J, Smith EF, Nicastro D. The CSC connects three major axonemal complexes involved in dynein regulation. Mol Biol Cell. 2012;23:3143–3155. doi: 10.1091/mbc.E12-05-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Brody SL, Ferkol TW. Picking up speed: advances in the genetics of primary ciliary dyskinesia. Pediatr Res. 2013;75:158–164. doi: 10.1038/pr.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Shingyoji C. The roles of noncatalytic ATP binding and ADP binding in the regulation of the dynein motile activity in flagella. Cell Motil Cytoskeleton. 2007;64:690–704. doi: 10.1002/cm.20216. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lin J, Heuser T, Carbajal-Gonzalez BI, Song K, Nicastro D. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 2012;69:88–100. doi: 10.1002/cm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, Nicastro D. Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J Biol Chem. 2011;286:29175–2991. doi: 10.1074/jbc.M111.241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mata M, Lluch-Estelles J, Armengot M, Sarrion I, Carda C, Cortijo J. New adenylate kinase 7 (AK7) mutation in primary ciliary dyskinesia. Am J Rhinol Allergy. 2012;26:260–264. doi: 10.2500/ajra.2012.26.3784. [DOI] [PubMed] [Google Scholar]
- Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene. 2008;425:79–83. doi: 10.1016/j.gene.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Iitsuka K, Fujii T. Adenylate kinase is tightly bound to axonemes of Tetrahymena cilia. Comp Biochem Physiol B Biochem Mol Biol. 1999;124:195–199. doi: 10.1016/s0305-0491(99)00114-5. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Nilsson JR. Phagotrophy in Tetrahymena. In: Biochemistry and Physiology of Protozoa. In: Levandowsky M, Hutner SH, editors. New York: Academic Press; 1979. pp. 2, 329–379. [Google Scholar]
- Noguchi M, Sawada T, Akazawa T. ATP-regenerating system in the cilia of Paramecium caudatum. J Exp Biol. 2001;204:1063–1071. doi: 10.1242/jeb.204.6.1063. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Biol. 2014;204:807–819. doi: 10.1083/jcb.201312014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Rasmussen L. Dual capacity for nutrient uptake in Tetrahymena. IV. Growth without food vacuoles and its implications. Exp Cell Res. 1976;102:127–137. doi: 10.1016/0014-4827(76)90307-4. [DOI] [PubMed] [Google Scholar]
- Panayiotou C, Solaroli N, Karlsson A. The many isoforms of human adenylate kinases. Int J Biochem Cell Biol. 2014;49:75–83. doi: 10.1016/j.biocel.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol. 2011;195:673–687. doi: 10.1083/jcb.201106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Luck DJ. Two-dimensional analysis of flagellar proteins from wild-type and paralyzed mutants of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1977;74:1600–1604. doi: 10.1073/pnas.74.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, van Blerkom J. Structural interaction of cytoskeletal components. J Cell Biol. 1981;90:222–235. doi: 10.1083/jcb.90.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torphy TJ. Phosphodiesterase isozymes: molecular targets for novel antiasthma agents. Am J Respir Crit Care Med. 1998;157:351–370. doi: 10.1164/ajrccm.157.2.9708012. [DOI] [PubMed] [Google Scholar]
- Tsao CC, Gorovsky MA. Different effects of Tetrahymena IFT172 domains on anterograde and retrograde intraflagellar transport. Mol Biol Cell. 2008;19:1450–1461. doi: 10.1091/mbc.E07-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan KK, Song KK, Alford LM, Sale WS, Dymek EE, Smith EF, Hennessey T, Joachimiak E, Urbanska P, Wloga D, et al. FAP206 is a microtubule-docking adapter for ciliary radial spoke 2 and dynein c. Mol Biol Cell. 2015;26:696–710. doi: 10.1091/mbc.E14-11-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu XJ, Zou XD, Wu XH, Ye ZQ, Wu YD. WDSPdb: a database for WD40-repeat proteins. Nucleic Acid Res. 2015;43(Database issue):D339–D344. doi: 10.1093/nar/gku1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang F, Zhou Z, Wu XH, Wu YD. A method for WD40 repeat detection and secondary structure prediction. PLoS One. 2013;8:e65705. doi: 10.1371/journal.pone.0065705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. Twenty-five dyneins in Tetrahymena: A re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton. 2008;65:342–351. doi: 10.1002/cm.20264. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Pazour G, Yoda A, Hirono M, Kamiya R, Witman GB. Oda5p, a novel axonemal protein required for assembly of the outer dynein arm and an associated adenylate kinase. Mol Biol Cell. 2004;15:2729–2741. doi: 10.1091/mbc.E03-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol Biol Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Rogowski K, Sharma N, Van Dijk J, Janke C, Edde B, Bre MH, Levilliers N, Redeker V, Duan J, et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008;7:1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt TA, Spurzem JR, May K, Sisson JH. Regulation of ciliary beat frequency by both PKA and PKG in bovine airway epithelial cells. Am J Physiol. 1998;275:L827–L835. doi: 10.1152/ajplung.1998.275.4.L827. [DOI] [PubMed] [Google Scholar]
- Yagi T. ADP-dependent microtubule translocation by flagellar inner-arm dyneins. Cell Struct Funct. 2000;25:263–267. doi: 10.1247/csf.25.263. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa HA, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, Sale WS. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol. 2013;201:263–278. doi: 10.1083/jcb.201211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cochran DA, Gargano MD, King I, Samhat NK, Burger BP, Sabourin KR, Hou Y, Awata J, Parry DA, et al. Regulation of flagellar motility by the conserved flagellar protein CG34110/Ccdc135/FAP50. Mol Biol Cell. 2011;22:976–987. doi: 10.1091/mbc.E10-04-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153:1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mitchell DR. Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J Cell Sci. 2004;117:4179–4188. doi: 10.1242/jcs.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.