Paraspeckle protein 1 (PSPC1), a component of nuclear paraspeckles, is identified as an importin α2 (IMPα2) binding partner in mouse spermatogenic cells. PSPC1-IMPα2 binding modulates PSPC1 delivery to paraspeckles, highlighting the potential for regulated importin synthesis to direct RNA metabolism and cellular differentiation.
Abstract
Importin (IMP) superfamily members mediate regulated nucleocytoplasmic transport, which is central to key cellular processes. Although individual IMPα proteins exhibit dynamic synthesis and subcellular localization during cellular differentiation, including during spermatogenesis, little is known of how this affects cell fate. To investigate how IMPαs control cellular development, we conducted a yeast two-hybrid screen for IMPα2 cargoes in embryonic day 12.5 mouse testis, a site of peak IMPα2 expression coincident with germ-line masculization. We identified paraspeckle protein 1 (PSPC1), the original defining component of nuclear paraspeckles, as an IMPα2-binding partner. PSPC1-IMPα2 binding in testis was confirmed in immunoprecipitations and pull downs, and an enzyme-linked immunosorbent assay–based assay demonstrated direct, high-affinity PSPC1 binding to either IMPα2/IMPβ1 or IMPα6/IMPβ1. Coexpression of full-length PSPC1 and IMPα2 in HeLa cells yielded increased PSPC1 localization in nuclear paraspeckles. High-throughput image analysis of >3500 cells indicated IMPα2 levels can directly determine PSPC1-positive nuclear speckle numbers and size; a transport-deficient IMPα2 isoform or small interfering RNA knockdown of IMPα2 each reduced endogenous PSPC1 accumulation in speckles. This first validation of an IMPα2 nuclear import cargo in fetal testis provides novel evidence that PSPC1 delivery to paraspeckles, and consequently paraspeckle function, may be controlled by modulated synthesis of specific IMPs.
INTRODUCTION
Cellular differentiation processes, including spermatogenesis, are mediated by an ordered series of gene expression and cell cycle events (reviewed in Eddy, 2002; Eddy and O'Brien, 1998) that require regulated trafficking of specific proteins between the nucleus and cytoplasm (Hogarth et al., 2005; Major et al., 2011). Passage between these two compartments occurs only through the nuclear envelope–localized nuclear pore complexes (NPCs), which permit active transport of cargo proteins >45 kDa by a group of transport proteins known as the importins (IMPs) and exportins (Macara, 2001; Major et al., 2011).
IMPα proteins are adaptor molecules in the classical nuclear import pathway, linking a cargo protein bearing a nuclear localization sequence (NLS) to an importin β1 (IMPβ1) molecule (Macara, 2001). IMPβ1 facilitates movement of this trimeric complex into the nucleus through its transient interactions with the nucleoporins lining the NPC. There are six known IMPα proteins in mice; these are categorized into three groups based on their structural similarities, with most known to have binding specificity for different cargoes (Miyamoto et al., 1997; Nigg, 1997; Tsuji et al., 1997; Jans et al., 2000; Goldfarb et al., 2004; Mason et al., 2009; Kelley et al., 2010). Here we employ the mouse nomenclature for naming IMPαs, with IMPα2 the product of the Kpna2 gene (Major et al., 2011). Using embryonic stem cells (ESCs) as a model differentiation system, IMPα subtype switching was previously shown to drive major differentiation outcomes through its impact on the nuclear import of specific transcription factor subsets (Yasuhara et al., 2007; Young et al., 2011). IMPα2 is essential for maintenance of ESCs in an undifferentiated state (Yasuhara et al., 2007) through its capacity to restrict the nuclear import of key transcription factors, such as OCT6, which drive differentiation into neural lineages (Yasuhara et al., 2013). Accumulating evidence that regulated synthesis of IMPα proteins is also of fundamental importance to spermatogenesis includes the identification of distinct cargoes bound to individual IMPαs in progressively maturing male germ cells of the mammalian testis (Miyamoto et al., 2013; Arjomand et al., 2014).
IMPα2 is required for germ cell differentiation in Drosophila melanogaster (Miyamoto et al., 2012). Mutation of IMPα2 in Drosophila causes a block in transport between nurse cells and the developing oocyte that results in female sterility (Gorjanacz et al., 2002), and IMPα2 is expressed in male mitotic and meiotic germ cells (Giarre et al., 2002). Despite recent establishment of genetically modified mouse lines targeting components of the nucleocytoplasmic transport system (Miyamoto et al., 2012), none is reported for IMPα2 at present. The IMPα2 expression profile during postnatal murine testis development is distinct from that of other IMPα mRNAs and proteins (Hogarth et al., 2006, 2007). Two peaks are apparent when IMPα2 mRNA levels are analyzed across a developmental murine testis time course, one at the onset of gonad differentiation, embryonic day (E) 12.5, and the second in the adult testis (Shima et al., 2004; Small et al., 2005; Major et al., 2011). These peaks coincide with important differentiation transitions in the male germ line, and identification of IMPα cargoes is predicted to reveal key control elements required for spermatogenesis.
The mammalian testis provides an excellent model in which to explore the relationship between selective cargo protein binding to IMPs and cellular differentiation outcomes. In mice, the bipotential primordial germ cells complete their migration into the urogenital ridge by E11.5, where their cell cycle and differentiation fate is determined by the surrounding somatic environment (Adams and McLaren, 2002; Bowles et al., 2006; Koubova et al., 2006). Embryonic testicular germ cells, termed “gonocytes,” proliferate until approximately E14.5, become quiescent, and then reenter the mitotic cell cycle shortly after birth. This last transition marks the beginning of spermatogenesis, a complex process involving three key stages: 1) mitotic divisions of spermatogonial stem cells and spermatogonia, 2) meiotic division of spermatocytes, and 3) morphological changes that transform haploid, round spermatids into the elongated, motile mature spermatozoa that deliver the male's genetic material to the oocyte (Kerr et al., 2006).
IMPα2 was used in our previous yeast two-hybrid (Y2H) analysis of the adult mouse testis, which identified chromatin remodeling factors in haploid spermatids as IMPα2 cargo (Ly-Huynh et al., 2011). Focus on the peak in IMPa2 mRNA level at E12.5 in the present study was chosen, as it corresponds to the time when the sexually indifferent gonad is first masculinized. This process is critical for establishment of the secondary sexual features by which an individual's gender is identified at birth, and its disturbance is considered to underlie testicular dysgenesis syndrome and an increased risk of testicular germ cell cancer developing in young adults (Looijenga et al., 2011). The expanding interest in IMPα2 as a biomarker for cancer reflects its strong expression, particularly within the nucleus, in many tumor types (Christiansen and Dyrskjot, 2013). Our screen has identified the first five candidate IMPα2 cargoes in the fetal testis. Among these, paraspeckle protein 1 (PSPC1) was selected for further analysis due to its previous characterization as an integral component of nuclear paraspeckles and its detection within germ cells of the adult testis in which IMPα2 is also present (Fox et al., 2002; Kuwahara et al., 2006).
Paraspeckles are currently understood to arise from the assembly of PSPC1 and at least two other core Drosophila behavior, human splicing (DBHS) proteins: PSF (splicing factor proline/glutamine rich [also named SFPQ and REP1]) and NONO (the non POU domain–containing, octamer-binding protein [also named nonA, NRB54, and P54NRB]) (Fox et al., 2002, 2005) around the specific long noncoding RNA transcript, nuclear-enriched abundant transcript 1 (NEAT1) (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). Paraspeckles are one of many distinct subnuclear bodies identified as containing discrete constituents (Fong et al., 2013). Our measurement of preferential binding by PSPC1 for IMPα2 and IMPα6 in the presence of IMPβ1 documents the importance of classical nucleocytoplasmic transport for paraspeckle formation. This study offers the new insight that PSPC1 delivery to paraspeckles can be controlled through regulated synthesis of nuclear transport machinery during development, with broader implications for cell biology and RNA metabolism based on the functional importance of this distinct subnuclear structure.
RESULTS
PSPC1 is a cargo of IMPα2
To identify IMPα2 cargoes in the fetal testis, we screened a Y2H library constructed using E12.5 mouse testes for binding partners of full-length IMPα2. We identified five candidate cargoes: PSPC1; rRNA-processing 1 homologue; Maged1 melanoma antigen, family D, 1; heterogeneous nuclear ribonucleoprotein C; and biorientation of chromosomes in cell division 1-like. All candidate IMPα2 cargoes were determined to contain putative NLSs using the cNLS Mapper program (Kosugi et al., 2009), and each was predicted to localize in the nucleus using PSORT II (Nakai and Horton, 1999; Supplemental Table S1). Publicly accessible microarray data (www.ncbi.nlm.nih.gov/geo; Barrett et al., 2013) indicate that the expression profile of each candidate IMPα2 cargo coincides with IMPα2 in the fetal and adult mouse testis (Supplemental Figure S1).
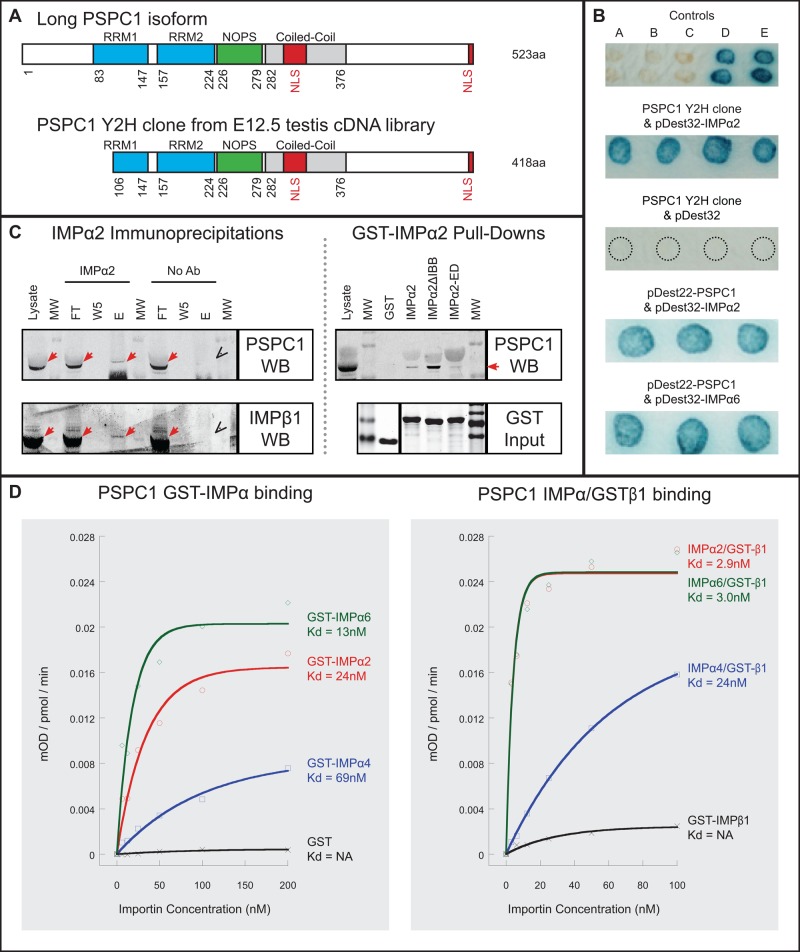
PSPC1 was selected for further investigation. It is a DBHS-containing protein family member; the full-length 523-amino-acid (aa) protein contains two RNA recognition motifs (RRMs), a NONA/paraspeckle (NOPS) domain, and two putative NLSs (Myojin et al., 2004; Passon et al., 2012; Figure 1A). The span of these domains was determined by alignment of the mouse PSPC1 sequence with the human sequence for which the PSPC1/NONO heterodimer structure has been reported (Passon et al., 2012). The two putative NLSs were previously identified in PSPC1 (Myojin et al., 2004) through homology with candidate NLSs identified in PSF (Dye and Patton, 2001). One NLS corresponds to two independent but overlapping potential bipartite NLSs in the aa 331–358 region, and the second is a monopartite NLS identified at the C-terminus (aa 517–525). A shorter PSCP1 isoform (391 aa) lacks the C-terminal putative NLS and is highly expressed in the kidney (Myojin et al., 2004). The longer isoform is expressed at low levels in many adult mouse tissues but is enriched in the adult testis and present in round spermatids, pachytene spermatocytes, and Sertoli cells (Myojin et al., 2004). The clone identified in this screen encodes a sequence lacking the first 105 aa (Figure 1A) but includes both putative NLSs, indicating it corresponds to the long PSPC1 isoform. The interaction between IMPα2 and PSPC1 was first tested in yeast, using Y2H library PSPC1 clone (Figure 1B) in addition to full-length PSPC1. Both interact with IMPα2 and with IMPα6 (Figure 1B).
FIGURE 1:
PSPC1 identified as a selective cargo of IMPα2. (A) Mouse PSPC1 schematics. Full-length PSPC1 (long isoform, 523 aa) contains two RRMs (blue), a NOPS (green), a coiled-coil domain (gray), and two putative NLSs (red). The first putative NLS is within the coiled-coil domain and the second is at the C-terminus (adapted from Myojin et al., 2004; Passon et al., 2012). The E12.5 testis cDNA library clone identified in this Y2H screen encodes aa 106–523 of the long PSPC1 isoform. (B) PSPC1 interacts with IMPα2 and IMPα6 in the Y2H system, as assessed by X-gal substrate blue signal intensity. Controls: yeast X-gal controls (A: no interaction; B: weak; C: intermediate; D: strong; E: very strong). PSPC1 Y2H clone + IMPα2: four blue spots indicate strong interactions between the PSPC1 Y2H clone and IMPα2 in cotransformants. PSPC1 Y2H Clone + pDEST32 plasmid (autoactivation control): four black dotted circles denote cotransformants in yeast showing no interaction. PSPC1 + IMPα2 and PSPC1 + IMPα6: X-gal signals illustrate strong interaction using full-length PSPC1. (C) Left, IMPα2 IPs using day 28 testis lysate. Samples are lysate, size markers (MW), flowthrough (FT), wash no. 5 (W5), and elution (E). PSPC1 detected by Western blotting (PSPC1 WB) is ∼59 kDa within the IMPα2 IP elution lane (red arrows) and is not detected in the IP-negative control (“V”-shaped arrowhead, lacking IP antibody; No Ab), demonstrating that PSPC1 is coimmunoprecipitated with IMPα2. IMPα2 IPs were confirmed by detection of IMPβ1, a known IMPα2 interactor (IMPβ1 WB). Red arrows indicate IMPβ1 (detected at ∼98 kDa); “V”-shaped arrowhead indicates the absence of IMPβ1 in negative control. Right, GST-tagged IMPα2 proteins “pull down” a PSPC1 band (detected at ∼57 kDa) from day 28 testis lysate (GST-IMPα2 pull downs). Western blotting detects PSPC1 (PSPC1 WB), sample lanes are day 28 testis lysate (lysate), size markers (MW), GST control (GST), GST-tagged full-length IMPα2 (IMPα2), GST-tagged truncated IMPα2 (IMPα2ΔIBB), and GST-tagged mutated IMPα2 (IMPα2-ED). Red arrow highlights the PSPC1 band present in GST‑IMPα2 samples that is absent from the GST control. The GST-tagged input proteins (3 μg) were separated by SDS–PAGE and stained with Coomasie (GST input), to show relative input equivalence (bands at ∼24, 77, 73, and 77 kDa, respectively). (D) Representative PSPC1 ELISA-based IMP-binding assays using GST-IMPα proteins and GST alone (left) and using tag-free IMPαs predimerized with GST‑IMPβ1 or GST‑ IMPβ1 alone (right). Kd estimates are indicated.
Two approaches demonstrated that IMPα2 and PSPC1 associate in testis cells. Endogenous IMPα2 and PSPC1 proteins were coimmunoprecipitated from a day 28 mouse testis lysate (Figure 1C, left-hand side). In addition, purified recombinant glutathione S-transferase (GST)-tagged IMPα2 proteins pulled down endogenous PSPC1 from a day 28 mouse testis lysate, while GST alone did not (Figure 1C, right-hand side). Two additional control proteins (IMPα2‑ED and IMPα2ΔIBB) were used for these experiments. The IMPα2‑ED mutant contains two point mutations within the NLS-binding groove, a lysine at amino acid 192 and an arginine at amino acid 396, severely decreasing the ability of IMPα2 to bind cargo protein NLSs, while IMPβ1-binding capacity is maintained (Giesecke and Stewart, 2010; Yasuda et al., 2012). The truncated IMPα2ΔIBB protein lacks the IMPβ-binding domain (Yasuda et al., 2012) and is considered to bind to cargo proteins more effectively than full-length IMPα2 protein, because it lacks the autoinhibitory activity present in the IBB domain (Giesecke and Stewart, 2010). The IMPα2ΔIBB protein pulled down more endogenous PSPC1 than the native protein did, while the IMPα2-ED protein pulled down less (Figure 1C), as predicted.
To further characterize the interaction between PSPC1 and IMPαs from each of the three structural clades (Kelley et al., 2010), we performed an enzyme-linked immunosorbent assay (ELISA)-based IMP-binding assay using affinity-purified HIS‑tagged recombinant PSPC1 protein with GST-tagged or GST-void IMPα (Supplemental Figure S2, A–C). Data from a representative experimental replicate are presented in Figure 1D; mean Kd and Bmax values from three independent experiments are shown in Table 1. Using GST‑IMPα proteins, we measured the binding affinities of IMPα2 and IMPα6 (mean Kd values of 15.0 nM and 5.9 nM, respectively) and found them to be greater than that of IMPα4 (mean Kd of 57.9 nM). The GST-alone control displayed negligible binding to PSPC1. Predimerization of GST-void IMPα2, IMPα4, or IMPα6 with GST‑IMPβ1 to form a canonical nuclear transport heterodimer increased the affinity of each IMPα for PSPC1. The observed mean Kd values for IMPα2 and IMPα6 were 1.9 nM and 2.1 nM, respectively, while the affinity of IMPα4 was lower, with a much larger mean Kd value of 21.3 nM. GST‑IMPβ1 on its own displayed low to negligible PSPC1 binding.
TABLE 1:
PSPC1 ELISA-based binding assay average estimated Kd and Bmax values.
| GST-IMPα2 | GST-IMPα4 | GST-IMPα6 | GST | IMPα2/GST-IMPβ1 | IMPα4/GST-IMPβ1 | IMPα6/GST-IMPβ1 | GST-IMPβ1 | |
|---|---|---|---|---|---|---|---|---|
| Kd ± SEM | 15.0 ± 4.3 | 57.9 ± 7.5 | 5.9 ± 3.3 | NA | 1.9 ± 0.5 | 21.3 ± 10.8 | 2.1 ± 0.5 | 35.5 ± 9.3 |
| Bmax ± SEM | 81.1 ± 7.3 | 52.4 ± 9.3 | 93.3 ± 5.8 | NA | 100.0 ± NA | 85.4 ± 3.2 | 95.0 ± 2.8 | 16.3 ± 3.1 |
Mean Kd and Bmax ± SEM from three independent ELISA-based IMP-binding assay experiments are shown. The Bmax values are displayed as comparative percentages relative to IMPα2/GST‑IMPβ1 (set to 100%).
Together these findings demonstrate that PSPC1 and IMPα2 interact and provide strong evidence that PSPC1 nuclear import is preferentially mediated through IMPα2/IMPβ1 or IMPα6/IMPβ1 heterodimer complexes using the classical nucleocytoplasmic transport pathway.
PSPC1 is expressed in distinct cell populations during testis development
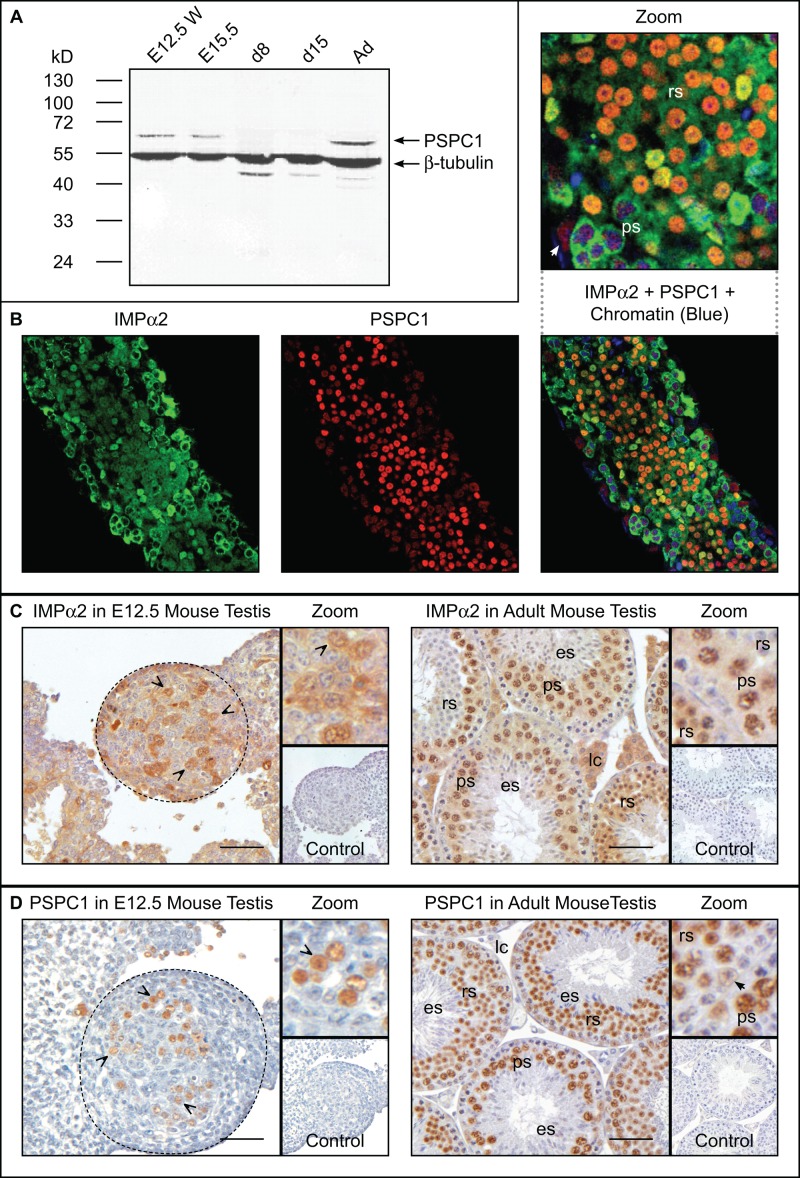
We next set out to document the pattern of PSPC1 expression throughout testis development, as this has only been shown previously in adult mouse testes (Myojin et al., 2004; Kuwahara et al., 2006). Western blotting was performed on mouse testis lysates from animals aged E13.5 through to adulthood using a mouse monoclonal antibody (mAb) that specifically binds the long, testis-enriched PSPC1 isoform (Figure 2A). The detected band of ∼60 kDa matches that previously reported for the longer isoform in adult mouse testis (Myojin et al., 2004). No band was detected in either 8 or 15 d postpartum (dpp) lysates, indicating up-regulation of PSPC1 occurs in postmitotic germ cells. The potential for the shorter PSPC1 isoform to be present in the testis remains to be established.
FIGURE 2:
PSPC1 coexpression with IMPα2 in germ cells. (A) Western blot detection of PSPC1 protein (60 kDa) and β-tubulin (protein loading control; 55 kDa) in E12.5 whole-embryo lysate (E12.5 WE) and in whole testis lysates at E15.5, 8 dpp (d8), 15 dpp (d15), and adult (Ad). (B) Confocal laser-scanning microscopy images of whole-mount immunofluorescence showing PSPC1 and IMPα2 in adult testis seminiferous tubules. Left-hand image, IMPα2 (green); middle, PSPC1 alone (red); right, merged image with TO-PRO staining (chromatin). ps: pachytene spermatocytes; rs: round spermatids; white arrow: Sertoli cell nucleus. (C) IMPα2 immunohistochemistry on E12.5 (1:50) and adult testes (1:100); control lacked primary antibody. (D) PSPC1 immunohistochemistry on E12.5 testis (1:50) and adult mouse testis (1:100). (C and D) ps: pachytene spermatocytes; rs: round spermatids; es: elongating spermatids; lc: Leydig cell; black arrow: Sertoli cell nucleus; “V”-shaped arrowhead: gonocyte (E12.5). Dashed line circles testis within E12.5 samples. Scale bar: 50 μm.
Whole-mount immunofluorescence identified IMPα2 and PSPC1 within overlapping germ cell subpopulations in the adult mouse testis (Figure 2B). Germ cell subtypes were identified by their size and shape, distinct chromatin morphology, and position within the seminiferous epithelium. PSPC1 was observed within the nucleus of pachytene spermatocytes, round spermatids, and their supporting Sertoli cells (Figure 2B). The IMPα2 signal was strongest within the pachytene spermatocyte cytoplasm and was readily detected in both cytoplasm and nucleus of round spermatids (Figure 2B).
Visualization of IMPα2 (Figure 2C) and PSPC1 (Figure 2D) cellular distribution used a newly available antibody for IMPα2 detection, yielding a more distinct and stronger signal than previously visualized by us in fixed-tissue sections (Ly-Huynh et al., 2011). In the E12.5 mouse testis, IMPα2 was detected in the cytoplasm and nucleus of gonocytes, the most primitive male germ cells. In adult mouse testis, the IMPα2 signal was most intense within the nucleus of pachytene spermatocytes and early round spermatids; a weaker, predominantly cytoplasmic signal was evident within other germ cell types and in Sertoli cells. PSPC1 was detected in the gonocyte nucleus and cytoplasm at E12.5 (Figure 2D) and in the nucleus of spermatocytes, round spermatids, and Sertoli cells in the adult testis (Figure 2D; as reported in Myojin et al., 2004; Kuwahara et al., 2006).
These results delineate a substantial overlap in the cellular and subcellular distribution of PSPC1 and IMPα2 in adult and embryonic mouse testes, supporting the hypothesis that IMPα2 serves as a transport factor for cargoes expressed in a developmentally regulated manner. We therefore assessed the potential for IMPα2 to influence PSPC1 nuclear import and paraspeckle formation.
IMPα2 targets PSPC1 to paraspeckles
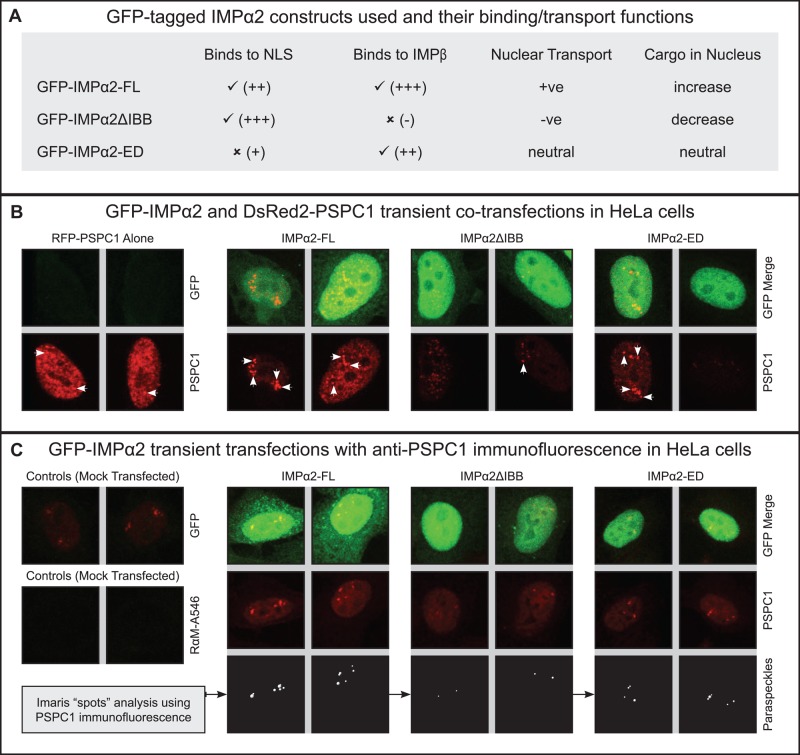
For modulation of both IMPα2 levels and functionality, constructs encoding green fluorescent protein (GFP)-tagged proteins corresponding to GST‑IMPα2 isoforms used in pull‑down experiments were transfected into COS‑7 and HeLa cells. Predicted nuclear transport outcomes for IMPα2 cargoes arising from overexpression of these proteins are outlined in Figure 3A. Full-length IMPα2 (GFP‑IMPα2‑FL) was expected to increase nuclear accumulation of IMPα2 transport cargoes, while GFP-IMPα2ΔIBB protein was predicted to compete with endogenous IMPα2 for cargo binding and reduce cargo nuclear transport. Finally, GFP‑IMPα2‑ED was expected to have minimal or no effect on endogenous cargo transport and was considered as a control for nonnuclear transport-related effects of GFP‑IMPα2 isoform overexpression.
FIGURE 3:
IMPα2 targets PSPC1 to paraspeckles in HeLa cells. Transient transfection with plasmids encoding GFP-tagged IMPα2-FL, IMPα2ΔIBB, or IMPα2-ED. Images are flattened Z‑series captured via confocal laser-scanning microscopy 2 d posttransfection. White arrows mark several paraspeckles (PSPC1 positive nuclear bodies). (A) GFP-tagged IMPα2 isoforms and their functional properties. (B) HeLa cells transfected to express DsRed2-PSPC1 alone or cotransfected to express DsRed2‑PSPC1 with GFP‑IMPα2 constructs. Bottom images depict DsRed2-PSPC1 localization alone, top panels depict both IMPα2 (green) and PSPC1 (red) localization. The DsRed2‑PSPC1 alone control is shown, with the red signal excluded from the top panel to clearly show the absence of a green signal. Each image corresponds to a single cell nucleus, with two separate images shown for each experimental group. (C) HeLa cells either mock transfected (controls) or transfected to express GFP‑IMPα2 constructs. Endogenous PSPC1 was visualized using immunofluorescence, with two cell images shown for each experimental group. The “GFP” control sample shows no detectable signal in mock-transfected cells, while the “RαM-A546” control lacking primary antibody shows the absence of secondary antibody binding. Examples of “spots” analysis using Imaris software to identify and quantify PSPC1 nuclear speckles are shown in the bottom panels.
The impact of each isoform on ectopic PSPC1 localization into paraspeckles was assessed following cotransfection with a DsRed2-tagged PSCP1 construct. Immunofluorescence to detect endogenous PSPC1 was performed for cells transfected with only GFP‑IMPα2 variants. Constructs in HeLa cells yielded qualitatively similar outcomes to those in COS‑7 cells (Figure 3B and Supplemental Figure S2D), with GFP-IMPα2-FL protein increasing speckle number and size. Fluorescent PSPC1-positive nuclear speckles were visible in 68% of COS‑7 cells transfected with PSPC1 alone and in 64% of cells cotransfected with IMPα2-FL and PSPC1, suggesting that paraspeckle formation was not augmented by ectopic PSPC1. However, IMPα2‑FL and PSPC1 cotransfection in COS‑7 cells resulted in qualitatively larger and more abundant PSPC1 nuclear speckles than did transfection with PSPC1 alone.
Several major experimental and analytical refinements enabled quantification of these outcomes in HeLa cells, the most common model for paraspeckle analyses (Fox and Lamond, 2010). Owing to variability in paraspeckle numbers detected within nontransfected HeLa cells, a minimum of 100 cells per sample was examined. Confocal three-dimensional z‑series images permitted identification and quantification of immunofluorescent PSPC1 punctate nuclear foci over the full cell volume. Imaris software image analysis was performed on samples into which only GFP‑IMPα constructs were introduced, with immunofluorescence used to detect endogenous PSPC1 (examples in Figure 3C).
Quantitative outcomes are summarized in Table 2; Supplemental Figures S3 and S4 contain detailed analyses. All data analyses assumed that outcomes for each cell arose from independent events. Because the data were skewed, the geometric mean (GM) rather than the arithmetic mean was examined. Nonparametric Mann-Whitney U-tests were initially used to test for significant differences between transfection groups on a per-cell basis, and all PSPC1 nuclear speckle–negative cells were included (Supplemental Figure S3). These pairwise comparisons identified significant differences in immunofluorescent PSPC1 nuclear speckles arising from transfection with IMPα2 constructs corresponding to differing nuclear transport capacities. Significantly lower numbers of immunofluorescent PSPC1 nuclear speckles, total volume of these nuclear foci, and total intensity of PSPC1 associated with nuclear speckles was measured within cells of the GFP‑IMPα2ΔIBB group in comparison with either GFP‑IMPα2‑FL or GFP‑IMPα2‑ED samples. Analysis using a parametric test, an analysis of variance with Games-Howell post hoc test, yielded similar outcomes (Supplemental Figure S3).
TABLE 2:
Outcomes of modulating IMPα2 expression and transport function on PSPC1-positive nuclear speckles.
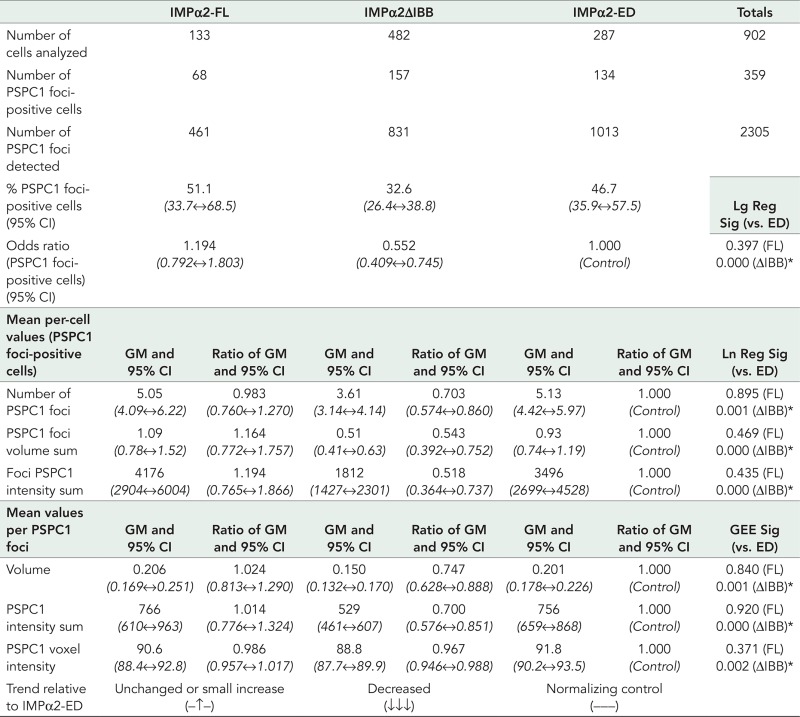 |
The analyzed cell numbers for each group, number of detected PSPC1-positive nuclear foci, and proportion of cells determined to contain PSPC1 nuclear foci are presented. Values are normalized against the IMPα2‑ED control (values set to 1), an “odds ratio” identifies the fold difference in number of PSPC1 nuclear foci-positive cells compared with the IMPα2‑ED sample. Data assessed on a per-cell or PSPC1 nuclear foci basis include GMs with 95% Wald confidence intervals (95% CI, range in italics) and the ratio of GMs. For determining significant differences between groups, a logistic regression (Lg Reg) model was used for PSPC1 foci-positive/negative cells, linear regression (Ln Reg) models were used for per-cell data (PSPC1 foci-positive cells) and GEEs were used for per-PSPC1 nuclear foci data. Significance (Sig) values compared with the IMPα2‑ED control are shown. Using Bonferroni correction, the significance threshold was reassigned from 0.05 to 0.016 (0.05 ÷ 3 transfection groups). Even with these stringent conditions, a significantly lower value is recorded for each assessed parameter within the IMPα2ΔIBB transfection group data set (*). Further details are provided in Supplemental Figures S3 and S4.
The properties of the identified nuclear PSPC1 speckles were analyzed in more detail, considering individual PSPC1 speckle–positive cells (Table 2 and Supplemental Figure S4). The most striking outcome was that GFP-IMPα2ΔIBB–transfected cells had significantly fewer endogenous PSPC1 nuclear foci per cell; these nuclear speckles were significantly smaller (volume), and the sum intensity of immunofluorescent PSPC1 signal present in nuclear speckles was lower, both within individual foci and as a cumulative value for each cell. The data summarized in Table 2 (extended in Supplemental Figures S3 and S4) highlight key impacts of IMPα2 transport capacity on PSPC1 localization to nuclear bodies. First, the proportion of cells containing immunofluorescent PSPC1 nuclear speckles was lower in the IMPα2ΔIBB (32.6%) in comparison with FL and ED isoform (51.1% and 46.7%, respectively) samples, as highlighted in the calculated odds ratio when the IMPα2-ED value was set at 1.000 and IMPα2‑FL and IMPα2ΔIBB groups were at 1.194 and 0.552, respectively. Second, when exclusively PSPC1 nuclear speckle–positive cells were examined on a per-cell basis to compare the ratio of GMs, with the IMPα2-ED value set to 1.000, the IMPα2ΔIBB sample value was consistently lower for the number of speckles per cell (FL: 0.983; ΔIBB: 0.703), sum speckle volume per cell (FL: 1.164; ΔIBB: 0.543), and sum of the immunofluorescence intensity associated with PSPC1 nuclear speckles (FL: 1.194; ΔIBB: 0.518). Finally, the same reduction in the ratio of GMs within the IMPα2ΔIBB group was identified for individual immunofluorescent PSPC1 nuclear speckle parameters of volume (FL: 1.024; ΔIBB: 0.747), sum PSPC1 intensity (FL: 1.014; ΔIBB: 0.700), and PSPC1 voxel intensity (FL: 0.986; ΔIBB: 0.967). These differences were all significant (Table 2).
For investigating the impact of modulating IMPα2 levels and functionality on another core paraspeckle component, PSF-positive nuclear speckles were analyzed based on preliminary data collected in HeLa cells using GFP-tagged IMPα2 constructs as described above for PSPC1 nuclear speckles. Summarized in Supplemental Table S2, the proportion of PSF nuclear speckle–positive cells followed a similar trend to PSPC1 nuclear speckles, with IMPα2-FL causing a significant increase and IMPα2ΔIBB a significant decrease. No significant changes to PSF nuclear speckle parameters was observed at the per-cell or per-speckle level.
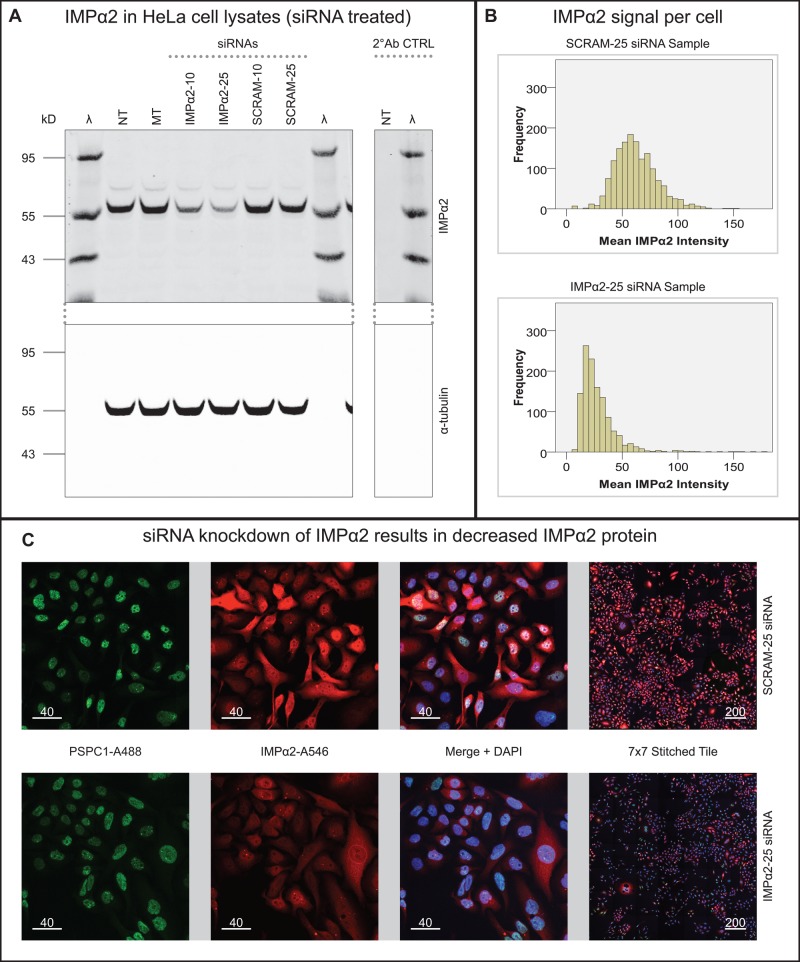
To complement these findings, we measured the impact on PSPC1-positive nuclear paraspeckles after reducing endogenous IMPα2 expression in HeLa cells using small interfering RNA (siRNA). Effective knockdown of IMPα2 with siRNA was confirmed on a population level and for individual cells using both Western blotting and immunofluorescence (Figure 4). The GM of the IMPα2 immunofluorescence signal demonstrated that the IMPα2 protein within cells in the siRNA-treated group dropped to 53.2% of the level in the cells treated with the scrambled (SCRAM) siRNA control oligo. PSPC1 nuclear speckle numbers, size, and PSPC1 signal intensity were then assessed by immunofluorescence for endogenous PSPC1 (summarized in Table 3). While the siRNA-induced decrease in IMPα2 protein levels did not cause a significant reduction in the percentage of cells containing PSPC1 nuclear speckles, it significantly reduced the mean (geometric) number of PSPC1 nuclear speckles per cell to 88.9% that of the SCRAM control value, the sum volume of speckles per cell to 81.8% of control, and the sum of PSPC1 immunofluorescence signal associated with nuclear speckles per cell to 79.2% of control values. The measurements of individual PSPC1 nuclear speckles also revealed statistically significant parameter reductions in the IMPα2 siRNA treatment group, although these were of a smaller magnitude, with per-PSPC1 nuclear foci measurements relative to SCRAM control value at 94.4% for speckle volume, 91.0% for speckle PSPC1 intensity sum, and 96.5% for PSPC1 mean voxel intensity per speckle.
FIGURE 4:
siRNA knockdown of IMPα2. (A) Dual fluorescent Western blot for IMPα2 protein and α-tubulin loading control detection. Lanes indicated are not transfected (NT), mock transfected (MT), transfected with IMPα2 siRNA at 10 or 25 nM final concentration (IMPα2‑10 or IMPα2‑25), or transfected with scrambled siRNA control at 10 or 25 nM final concentration (SCRAM‑10 or SCRAM‑25). Lanes with size markers (λ) have the molecular weight indicated on each blot in kilodaltons (kDa). (B) Histograms of the mean IMPα2 intensity (detected per cell) as determined by immunofluorescence in the IMPα2‑25 and SCRAM‑25 groups. (C) Overview of images collected from IMPα2‑25 and SCRAM‑25 samples with immunofluorescence for PSPC1 and IMPα2 with DAPI as a nuclear marker. Scale bars: 40 or 200 μM, as indicated.
TABLE 3:
Outcomes of siRNA knockdown of IMPα2 on PSPC1-positive nuclear speckles.
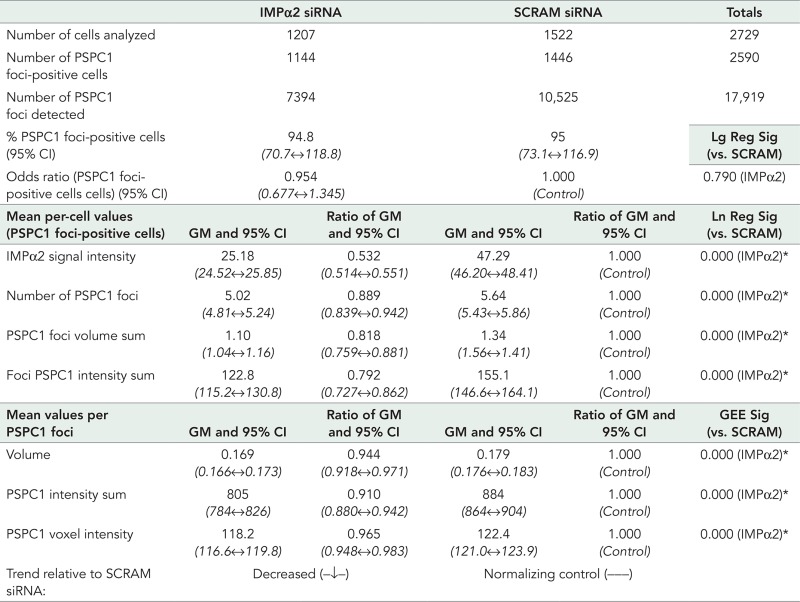 |
The analyzed cell numbers for each group, number of detected PSPC1-positive nuclear foci, and proportion of cells determined to contain PSPC1 nuclear foci are presented. Values are normalized against the SCRAM siRNA control (values set to 1), an “odds ratio” identifies the fold difference in number of PSPC1 nuclear foci-positive cells compared with the SCRAM siRNA sample. Data assessed on a per-cell or PSPC1 nuclear foci basis include GMs with 95% Wald confidence intervals (95% CI, range in italics), and the ratio of GMs. For determining significant differences between groups, a logistic regression (Lg Reg) model was used for PSPC1 foci-positive/negative cells, linear regression (Ln Reg) models were used for per-cell data (PSPC1 foci-positive cells), and GEEs were used for per PSPC1 nuclear foci data. Significance (Sig) values compared with the SCRAM siRNA control are shown. Using a threshold of 0.025, a significantly lower value is recorded for each assessed parameter within the IMPα2 siRNA group data set (*) for per-cell or per-PSPC1 foci data. The IMPα2 signal intensity per cell dropped significantly to 53.2% that of the SCRAM siRNA control sample. Further details are provided in Figure 4.
Overall these data indicate that the IMPα2 and PSPC1 interaction is relevant to the nuclear function of PSPC1 nucleocytoplasmic transport and subsequent localization to paraspeckles. They reveal for the first time that modulating the functional levels of IMPα2 can directly determine the number of cells that contain PSPC1 nuclear speckles and the number of speckles that these cells contain and influence the detectable size and intensity of PSPC1 staining within individual nuclear foci.
DISCUSSION
Changes in IMP protein levels and transport activity are now understood to be of central importance to cellular differentiation, although the mechanistic basis for this is poorly understood. As an approach to filling this knowledge gap, and because IMPα2 synthesis is dynamic during germ cell differentiation in the mammalian testis, we sought to identify IMPα2 cargoes in the E12.5 testis. Through a Y2H screen, we identified five potential IMPα2 cargoes, with each containing a cNLS for canonical nuclear transport via IMPα protein binding. Characterization of the potential interaction between PSPC1 and IMPα2 was of particular interest, because both are present in the same germ cells (Kuwahara et al., 2006; Major et al., 2011). Identification of PSPC1 as a component of the distinct subnuclear paraspeckle domain (Fox et al., 2002) provided a functional framework for our interest in this molecule. Paraspeckles are rich in A-to-I edited RNA transcripts; they have been implicated as a site for nuclear retention of RNAs and RNA metabolism and thereby influence cellular differentiation and stress responses (Fox and Lamond, 2010). PSPC1 contains two putative NLSs (Myojin et al., 2004), and notably, the C-terminal NLS lies outside the solved structure, while the putative bipartite NLS resides in the coiled-coil domain, meaning neither of the predicted NLSs are part of the reported core globular structure (Passon et al., 2012), and both are likely to be accessible to IMPα for binding. We demonstrated that PSPC1 and IMPα2 proteins colocalize in specific male germ cell types, and we obtained biochemical evidence that PSPC1 and IMPα2 interact in the testis.
The potential for IMPα6 to mediate PSPC1 nuclear import was indicated by both Y2H and ELISA-based binding assays (Figure 1). While IMPα2 and PSPC1 are coexpressed in several germ cell types spanning fetal through adult life, the expression of IMPα6 is maximal during the latest spermatogenic stage, spermiogenesis (Hogarth et al., 2006; Major et al., 2011). We propose that PSPC1 and IMPα6 interact during spermiogenesis, when IMPα2 levels are declining. The indication that PSPC1 nuclear import is preferentially mediated by IMPα2/IMPβ1 or IMPα6/IMPβ1 heterodimer complexes reinforces the functional importance of regulated synthesis of IMP proteins, as shown for IMPα and IMPβ proteins in fetal and postnatal germ-line cells (Hogarth et al., 2006, 2007; Loveland et al., 2006; Yamaguchi et al., 2006; Ly-Huynh et al., 2011; Major et al., 2011; Miyamoto et al., 2012, 2013; Tran et al., 2012; Whiley et al., 2012; Arjomand et al., 2014). We know that developmental switches can be affected by coordinated up- and down-regulation of specific cargoes and individual factors for nucleocytoplasmic transport. Recent studies have shown that IMPα2 must be down-regulated and IMPα1 up-regulated to enable ESCs to differentiate into neural linage progenitors (Yasuhara et al., 2007). This allows OCT6 and BRN2 to be released from IMPα2 sequestration (via a C-terminal acidic domain) in the cytoplasm for nuclear translocation by IMPα1/IMPβ1 or IMPα4/IMPβ1 (Yasuhara et al., 2013).
Our quantitative analysis approach has documented the highly significant effect of impaired IMPα2 cargo-binding capacity on PSPC1 subnuclear targeting to paraspeckles within individual cells. The software-assisted image analysis eliminated the inherent subjectivity associated with manually assessing paraspeckle number and size, enabling quantitative analysis of 2305 PSPC1-containing nuclear speckles in 902 cells within the GFP-tagged IMPα2 transfection experiment and 17,919 PSPC1 nuclear speckles in 2729 cells of the siRNA experiment. A reduced number of HeLa cells showed nuclear speckle localization of PSPC1 after transfection to express the dominant-negative IMPα2ΔIBB-encoding construct compared with the IMPα2‑FL or the IMPα2‑ED isoforms, with siRNA knockdown of IMPα2 also demonstrating similar reductions in detectable PSPC1 nuclear speckle numbers and associated parameters. These outcomes provide strong evidence that the binding relationship between IMPα2 and PSPC1 can modulate PSPC1 delivery to paraspeckles and thus determine paraspeckle-related RNA metabolic functions.
That IMPα2ΔIBB did not completely block paraspeckle localization of PSPC1 may reflect the presence of endogenous IMPα2, so although classical nuclear transport efficiency is reduced, it is not eliminated. Consistent with this concept, siRNA knockdown of IMPα2 also did not completely block PSPC1 localization to paraspeckles, although significant changes in the parameters of PSPC1 nuclear speckles were documented. While overexpression of IMPα2-FL did not lead to a significant increase in any of the assessed PSPC1 nuclear speckle attributes, small increases were measured for the proportion of speckle-positive cells and sum PSPC1 immunofluorescent speckle volume and intensity, parameters for which the odds ratio or the ratio of GMs was greater than 1.0 (Table 2). This suggests that the cellular level of PSPC1 is also an important factor limiting detectable PSPC1 nuclear body number and size. Modulating IMPα2 in HeLa cells changed the detectable proportion of cells containing PSF-positive nuclear speckles, but there were no significant changes observed at the per-cell or per-PSF nuclear speckle level. We therefore hypothesize that the underlying architectural scaffold/structure of the paraspeckle is not affected by perturbations of PSPC1 delivery; rather, we suspect the functional significance of the paraspeckle is changed by dictating which RNA transcripts and other paraspeckle components are recruited. While our results indicate that IMPα2 can facilitate PSPC1 nucleocytoplasmic transport, whether IMPα2 specifically targets PSPC1 to nuclear paraspeckles remains to be established. The potential for IMPα2 or other IMPs to deliver cargo directly to nuclear subcompartments is a focus of current work in our laboratory.
The adenosine deaminases acting on RNA (ADARs) perform A-to-I editing of selected RNA transcripts at specific residues, and the three present in mammalian cells have distinct interactions with IMPα proteins (Maas and Gommans, 2009a). ADAR3 and a broadly expressed ADAR2 splice variant found in testis (Maas and Gommans, 2009b) both bind to IMPα2 but not to IMPα1 or IMPα3. In contrast, the main splice form of ADAR2 will bind to both IMPα1 and IMPα3 (Maas and Gommans, 2009a). This suggests that regulated IMPα2 expression could modulate paraspeckle function in two ways, first directly, by mediating PSPC1 nuclear import and localization into paraspeckles, and second in an indirect manner, by controlling nuclear import of specific ADARs that impact on A-to-I editing of specific transcripts and their subsequent localization into paraspeckles. The capacity of IMPα2 to facilitate the nuclear transport of other key paraspeckle components or regulate the expression of key paraspeckle components either directly or indirectly has yet to be determined. The possibility that IMPα2 is regulating paraspeckles indirectly through effects upon the cell cycle or another mechanism has also not been excluded.
Our current working model, summarized in Figure 5, is that the regulated expression of IMPα2 within fetal and postnatal germ-line cells will drive PSPC1 nuclear import and localization into paraspeckles. This model highlights the need to develop a better functional understanding of the downstream effects of modulating the relative proportions of PSPC1 or other core DBHS proteins localized to the paraspeckle, observed here as changes in immunofluorescent PSPC1 nuclear speckle number and size. Given that NEAT1 and paraspeckles are absent from human ESCs and that both can be induced upon differentiation, it has been suggested that paraspeckles are indicative of a differentiated cell state (Chen and Carmichael, 2009). NEAT1 transcript levels increase during muscle differentiation, and paraspeckles (punctate NEAT1 foci) are enlarged and present in greater numbers upon differentiation of myoblasts into myotubes (Sunwoo et al., 2009). Interestingly, regulated expression of nuclear transport machinery is also implicated in muscle differentiation, with elevation of IMPα2 during differentiation linked to myoblast proliferation, myocyte migration, and myotube size (Hall et al., 2011). Thus the results of the present study, which document the capacity for IMPα2 to drive an increase in PSPC1 nuclear speckle size and numbers in HeLa cells, further suggest that developmental peaks in IMPα2 expression, such as in the germ cells of the E12.5 testis or during postmitotic spermatogenesis (Major et al., 2011), reflect its role in determining cellular differentiation. We have measured a decrease in IMPα2 levels as meiotic male germ cells transform into haploid round spermatids (Arjomand et al., 2014). This cellular transition is accompanied by major changes in RNA synthesis, storage, and turnover, and it should be informative to document what roles PSPC1 serves during these stages. The contribution of changing PSPC1/PSF levels will also be important to consider along with measuring the stoichiometry of these proteins as spermatogenesis proceeds. Given the current knowledge of paraspeckle function, we hypothesize that modulation of PSPC1 delivery to paraspeckles will manifest as changes that include nuclear retention of specific A-to-I edited RNA transcripts, altered RNA processing, and changes to general cellular stress responses (Prasanth et al., 2005; Chen and Carmichael, 2009; Fox and Lamond, 2010; Nakagawa et al., 2011; Nakagawa and Hirose, 2012; Naganuma et al., 2012; Hirose et al., 2014). This further highlights the need to understand how IMP levels and functional activities are determined.
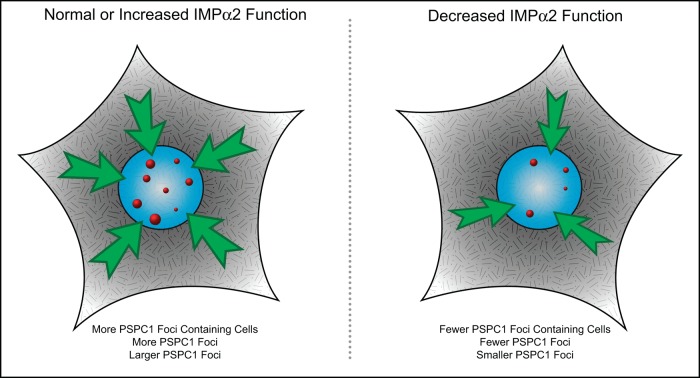
FIGURE 5:
Proposed model of canonical nucleocytoplasmic transport role in PSPC1 intracellular targeting to paraspeckles. Stylized here are the outcomes of transfection experiments performed on HeLa cells. It was observed that modulating functional IMPα2 levels and, subsequently, nucleocytoplasmic transport of IMPα2 cargoes (depicted as green arrows) influences the number and size of detected PSPC1 nuclear speckles (shown in red) and the proportion of cells containing these PSPC1-positive paraspeckles. We hypothesize downstream impacts on the paraspeckle functions, proposed in current literature to include the nuclear retention of RNAs, changes to RNA processing, and stress responses.
MATERIALS AND METHODS
Animals and tissue sample preparation
Postnatal testis were from Swiss and C57/BL6xCBA male mice, and fetal testes were collected from embryos staged by fore- and hindlimb morphology from time-mated pregnant Swiss female mice from both Monash University and the University of Queensland Animal Services. All investigations conformed to the NHMRC/CSIRO/AAC Code of Practice for the Care and Use of Animals for Experimental Purposes and were approved by the Monash University Standing Committee on Ethics in Animal Experimentation or the University of Queensland Animal Ethics Committee. Lysates of fetal and juvenile-age testis samples for Western blots were pooled from different mice; each adult testis lysate was from one animal. Tissues for coimmunoprecipitations (co-IPs), pull down, or whole-mount immunofluorescence were used immediately after collection, while tissues for all other protein analyses were snap frozen and stored at −80°C. Tissues for immunohistochemistry were fixed in Bouin's for 5 h, dehydrated through graded ethanols, paraffin embedded, and sectioned (3–5 μm).
E12.5 testis cDNA library construction and yeast two-hybrid screening
Total RNA was isolated from ∼500 E12.5 testes using the Microfast Track mRNA isolation kit (Invitrogen, Melbourne, Australia) according to the manufacturer's instructions. A cDNA library was generated using the CloneMiner cDNA Library Construction Kit (Invitrogen, Version B) by ligation into the pDONR222 vector. The plasmid library was transformed into ElectroMAXDH10β T1 cells and then recombined into the Y2H prey vector, pDEST22, using the Invitrogen Gateway system.
Putative IMPα2-binding partners were identified using the Invitrogen Y2H screening protocol. The pDEST22 library clones and full-length IMPα2-pDEST32 bait plasmid (Ly-Huynh et al., 2011) were cotransformed into MaV203-competent yeast. Cotransformed yeasts were screened for potential interactors using three different reporter genes, and plasmids were isolated from clones passing all three tests. The interaction was validated by a second yeast cotransformation into MaV203 cells with full-length IMPα2-pDEST32 or pDEST32 alone.
Constructs and vectors
The murine PSPC1 full-length cDNA construct (encoding aa 3–523) was amplified by PCR (mRNA Accession: NM_025682.3) with forward (5′ggggacaagtttgtacaaaaaagcaggcttcttaagaggaaacctgaagcaag3′) and reverse (5′ggggaccactttgtacaagaaagctgggtcttaatatctccgacgcttattag3′) primers. This was recombined into pDEST22 (for Y2H system), pDEST17 (for HIS-tagged bacterial expression), and dsRED2 (DsRed2-tagged mammalian cell expression) vectors using the Invitrogen Gateway System.
The Y2H constructs encoding IMPα2, IMPα4, and IMPα6 in the pDest32 vector (Invitrogen) were previously generated (Ly-Huynh et al., 2011). GFP-IMPα2 constructs for mammalian cell expression were generated previously (Ly-Huynh et al., 2011; Young et al., 2011) and encoded full-length IMPα2 (GFP‑IMPα2‑FL), ED mutant (GFP‑IMPα2‑ED), and truncated IMPα2 (GFP‑IMPα2ΔIBB). For bacterial cell expression and affinity-based purification, the same IMPα2 variants were cloned into pGEX‑6P-2 (GE Life Sciences, Rydalmere, Australia) for production of GST, GST‑IMPα2-FL, GST‑IMPα2-ED, and GST‑IMPα2ΔIBB (Yasuda et al., 2012; Miyamoto et al., 2013). The GST-IMPβ1 plasmid used the pGEX-2T vector (GE Life Sciences; Arjomand et al., 2014).
Purification of GST/HIS-tagged and tag-void recombinant proteins expressed in Escherichia coli
Bacterial proteins were induced in BL21-DE3 cells with 1.0 mM isopropyl β-d-1-thiogalactopyranoside; HIS-tagged: 28°C, 3.5 h; GST-tagged: 18°C, overnight (O/N). Bacteria were then harvested (6000 × g, 10 min, 4°C), washed with cold NaCl (0.9% wt/vol), and bacterial pellets stored at −80°C.
Bacterial pellets with GST-tagged proteins were resuspended on ice in GST lysis buffer (50 mM Tris-HCl, pH 8.3, 500 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol [DTT], and 0.2 mM phenylmethylsulfonyl fluoride [1 M stock in EtOH]) and lysed by freeze-thawing twice (alternating between liquid nitrogen and shaking at 20°C) followed by probe sonication (5 × 30 s, 1-min rests). Affinity-based purification was as previously described (Miyamoto et al., 1997, 2013) using glutathione Sepharose 4B slurry (GS4B; GE Life Sciences). GST-tagged proteins were eluted with GST elution buffer (100 mM Tris-HCl, pH 8.3, 100 mM NaCl, 1 mM EDTA [3Na], 2 mM DTT, 1 μg/ml 1:1000 protease inhibitor cocktail [PIC, Calbiochem cocktail set III, Merck-Millipore, Bayswater, Australia], 20 mM glutathione). GST-void IMPα proteins were generated by instead cleaving the pGEX-6P linker between the GST tag and the IMPα by O/N incubation with PreScission Protease (GE Life Sciences; 80 U/ml of GS4B slurry, 4°C). All proteins were reequilibrated in GST dialysis buffer (20 mM HEPES-NaOH, pH 7.3, 100 mM CH3COOK, 2 mM DTT, 1:1000 protease inhibitor cocktail [PIC]), concentrated (Amicon Ultra centrifugal filters; Merck-Millipore), and stored at −80°C. Protein concentrations were estimated by DC Protein Quant Assay (Bio-Rad, Gladesville, Australia) and SDS–PAGE against a bovine serum albumin (BSA) standard.
Bacterial pellets with HIS-tagged proteins were resuspended on ice in Ni‑NTA lysis buffer (300 mM NaCl, pH 8.0, 50 mM NaH2PO4, pH 8.0, 40 mM imidazole, 1:1000 PIC) and processed as above (freeze-thawing, sonication, centrifugation). Supernatant was rotated with prewashed Ni-NTA agarose (Qiagen, Doncaster, Australia) slurry (4°C, 2 h), washed (5×) with cold Ni-NTA wash buffer (300 mM NaCl, pH 8.0, 50 mM NaH2PO4, pH 8.0, 40 mM imidazole), then eluted in Ni-NTA elution buffer (300 mM NaCl, pH 8.0, 50 mM NaH2PO4, pH 8.0, 200 mM imidazole). HIS-tagged PSPC1 protein tended to precipitate over time, so a fresh preparation was used in every assay, and protein concentrations were estimated immediately (DC Protein Quant Assay).
Co-IPs
Adult mouse testis were homogenized on ice through needles (18 to 30 gauge) in IP lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X‑100, PIC 1:500) and then centrifuged. Lysate supernatant (200 μl [∼200 μg protein]) was incubated (4°C with rotation, 2 h) with 2.0 μg anti-IMPα2 (cat. no. sc6917, lot no. c1407; Santa Cruz Biotechnology) and added to a microspin column (GE Life Sciences) containing 60 μl prewashed protein A/G-PLUS-Agarose (Santa Cruz Biotechnology, distributed by ThermoFisher Scientific, Scoresby, Australia), and the volume was brought to 500 μl with IP lysis buffer for O/N incubation (4°C with rotation). Samples of flowthrough, washes (IP lysis buffer), and elution in 2× sample buffer (4% vol/vol SDS, 25% vol/vol glycerol, 12.5% vol/vol [0.5M Tris HCl, pH 6.8], 5% vol/vol β-mercaptoethanol, bromophenol blue) were collected for Western blot detection of PSPC1 and IMPβ1.
IMPα2 pull-down experiments
Testis lysates prepared as for co-IPs were precleared with GS4B slurry (60 μl/200 μl lysate, 4°C with rotation, ∼2 h). GS4B slurry aliquots (60 μl) were equilibrated with transport buffer (20 mM HEPES-NaOH, pH 7.3, 110 mM CH3COOK, 2 mM Mg(CH3COO)2, 5 mM CH3COONa, 0.5 mM EGTA, 2 mM DTT, PIC 1:500) in microspin columns (GE Life Sciences), and then GST-tagged protein (300 pmol) was added in 300 μl of transport buffer for binding to GS4B (4°C, 1 h with rotary agitation). Precleared testis lysate (200 μl) was added and incubated O/N (4°C, gentle rotation). Flowthrough, washes (transport buffer), and eluate (2× sample buffer) samples were collected for Western blot detection of PSPC1.
ELISA-based binding assay
An ELISA-based IMP-binding assay (Hubner et al., 1997) was performed using recombinant HIS-tagged PSPC1 (5 pmol/well) coated onto 96-well plates (cat. no. 260836; Nunc, distributed by ThermoFisher Scientific, Scoresby, Australia). Triplicate wells were incubated with different concentrations of GST, GST-tagged IMPs, or GST‑tag cleaved IMPs preincubated with GST‑IMPβ1. After washing, IMP binding was quantitated using an anti-GST antibody (cat. no. 27457701; GE Life Sciences) and a rabbit anti-goat secondary conjugated to alkaline phosphatase (cat. no. A-4187; Sigma-Aldrich). The absorbance change at 405 nm was recorded for 90 min after p-nitrophenyl phosphate substrate (Sigma-Aldrich, Castle Hill, Australia) addition. Kd and Bmax values were estimated by curve fitting using KaleidaGraph version 3.6 by Synergy.
Western blotting
Samples were homogenized in RIPA buffer with PIC at 4°C, as previously described (Loveland et al., 1999). Tissue lysates (30 μg per lane) were mixed with 2× sample buffer, run on 12% SDS–PAGE gels with protein-size standards (PageRuler Prestained Protein Ladder; Fermentas-Thermo Scientific, Scoresby, Australia), transferred to Amersham Hybond-C Extra nitrocellulose membrane (GE Life Sciences), and blocked in 5% skim milk/Tris-buffered saline (TBS: 50 mM Tris, 150 mM NaCl, pH 7.5) for 1 h. Washes were with 0.1% Tween 20/TBS, and incubations were at room temperature (RT) unless otherwise stated. The membrane was rotated with an anti-PSPC1 mouse mAb (Kuwahara et al., 2006; 1:200 in 5% skim milk/0.1% Tween-20/TBS, O/N, 4°C) and washed, and then rabbit anti-mouse IRDye800 secondary antibody (1:10,000; cat. no. 610431020; Rockland Immunochemicals, distributed by Jomar Bioscience, Kensington, Australia; in 5% skim milk/0.1% Tween-20/0.01% SDS/TBS) was applied (60 min) and detected using the Odyssey Imaging System (Li-Cor Biosciences, Lincoln, NE). Subsequent incubation with anti β-tubulin mouse mAb (1:10,000; Sigma-Aldrich) provided the loading control. At least two independent lysates were examined for each age and antibody, with consistent results. The IMPα2 pull-down and co-IP samples used the PSPC1 mAb diluted 1:1000 to detect PSPC1. Co-IP samples were subsequently blotted with IMPβ1 antibody (1:1000; cat. no. sc11367; Santa Cruz Biotechnology) and goat anti-rabbit IRDye800 secondary (1:10,000; cat. no. 611132122; Rockland Immunochemicals). The siRNA samples were simultaneously blotted with goat anti-IMPα2 (1:1000; cat. no. sc-6917; Santa Cruz Biotechnology) and mouse anti-α tubulin (cat. no. T5168; Sigma-Aldrich) antibodies with rabbit anti-goat Alexa Fluor 680 (1:10,000; cat. no. A21088; Molecular Probes–Invitrogen) and rabbit ant-mouse IRDye800 (1:10,000; cat. no. 610431020; Rockland Immunochemicals) secondary antibodies.
Whole-mount immunofluorescence of mouse seminiferous tubules
Tubules were collected in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) from decapsulated adult mouse testes (Szczepny et al., 2009). Within 1 h of testis collection, 1-cm tubule lengths were fixed simultaneously in 3.2% paraformaldehyde (PFA; Merck; 1.5 h, RT). Processing occurred in 24-well plate wells (cat. no. 353047; BD Falcon, North Ryde, Australia) containing >3 tubule fragments with gentle rocking. Tubules were incubated in PBS/0.5% BSA (Sigma-Aldrich; 30 min, RT) and then in 0.5% Triton X‑100 (Merck) in PBS (1 h, RT). Anti-PSPC1 mouse mAb (1:100) and goat polyclonal antibody to IMPα2 (1:100; cat. no. sc-6917; Santa Cruz Biotechnology) in 0.5%BSA/PBS were added (O/N, 4°C). After PBS washes (4×, 1–3 h each), secondary antibodies (rabbit anti-mouse Alexa Fluor 546 [cat. no. A11060; Molecular Probes–Invitrogen], donkey anti-goat Alexa Fluor 488 [cat. no. A11055; Molecular Probes–Invitrogen] and a nuclear marker [4′,6-diamidino-2-phenylindole, DAPI; Molecular Probes–Invitrogen]; TO-PRO [Molecular Probes–Invitrogen]; or Draq5 [BioStatus, distributed by Sapphire Bioscience, Waterloo, Australia]) were added (1:200 in 0.5% BSA/PBS, O/N, 4°C). Samples were washed in PBS (4×, 1–3 h, then O/N, 4°C) and then imaged either wet with an immersion lens or mounted in a “tape well” formed by placing two layers of double-sided tape with holes punched onto a slide. The well was filled to excess with mounting media (GVA Mount [Zymed–Invitrogen], SlowFade Gold, or ProLong Gold [Invitrogen]), and the coverslip was pressed to close the well. Excess mounting media was removed, samples were left flat to set (O/N, 4°C) and were then sealed with nail polish. Images were acquired with a Leica (Mannheim, Germany) SP5 laser-scanning confocal system with DMI6000 microscope using a 20×/1.0 NA water-immersion lens in the Monash Micro Imaging facility (MMI, Clayton, Australia).
Immunohistochemistry
Immunohistochemistry with anti-PSPC1 (Kuwahara et al., 2006) and anti‑IMPα2 (cat. no. ab84440; Abcam) antibodies was performed as previously described (Loveland et al., 1999) using 50 mM glycine for antigen retrieval (pH 3.5; >90°C for 8 min), and diluted primary antibodies (0.1% BSA/TBS) for O/N incubation at RT. Control sections lacked primary antibody. Secondary antibody incubations (1:500 in 0.1% BSA/TBS, 1 h), either biotinylated rabbit anti-mouse antibody (cat. no. E0354; DAKO, North Sydney, Australia) or biotinylated goat anti-rabbit antibody (cat. no. 656140; Invitrogen), were followed by Vectastain Elite ABC kit reagent (Vector Laboratories, distributed by Abacus Als, Waterford, Australia). Antibody binding was detected as a brown precipitate following development with 0.20 mg/ml 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich). Harris hematoxylin counterstain (Sigma-Aldrich) and DPX mountant (Sigma-Aldrich) were applied. Germ and somatic cell types were identified based on nuclear morphology and position within the developing gonad (Byskov, 1986; Russell, 1990). Slides were imaged using a Mirax Midi slide scanner (Carl Zeiss, Sydney, Australia) with a 20× objective.
Cell culture, transfection, and indirect immunofluorescence staining
COS‑7 and HeLa cells were maintained in DMEM with 5% (vol/vol) and 10% (vol/vol) fetal calf serum, respectively, penicillin-streptomycin, l-glutamine, and MEM nonessential amino acids in 5% CO2 at 37°C. Twenty-four hours before transfection, cells were seeded on round coverslips in medium lacking penicillin-streptomycin in 24-well plates for GFP/red fluorescent protein (RFP)-tagged transfections or 12-well plates for siRNA knockdown. PSPC1 and IMPα2 DNA constructs were transfected using Lipofectamine 2000 (Invitrogen) with 2.5 μg (single plasmid) or 1.25 μg (each cotransfection), following the manufacturer's method. The Dharmacon ON‑TARGETplus siRNA system (GE Life Sciences) was used as per the manufacturer's instructions for siRNA knockdown of IMPα2 (SMARTpool L‑004702-00) with a nontargeting (SCRAM siRNA) control pool (D-001810-10) as the siRNA negative control.
Cells were fixed 48 h later in 3.2% PFA/PBS for 10 min and then washed and stored in PBS at 4°C. For indirect immunofluorescence staining, all incubations and washes (4×, 5 min each) were at RT with rocking. Cells were blocked (0.5% BSA/PBS, 30 min) and permeabilized (0.1% Triton X‑100/PBS, 10 min), then anti-PSPC1/PSF (Kuwahara et al., 2006; 1:100 in 0.5% BSA/PBS) or anti-PSPC1/PSF and anti-IMPα2 (1:100; cat. no. ab84440; Abcam, Melbourne, Australia) were applied for O/N incubation. Coverslips were washed and then incubated with secondary antibodies (rabbit anti‑mouse Alexa Fluor 546 for GFP/RFP-tagged transfections and donkey anti-mouse Alexa Fluor 488 plus goat anti-rabbit Alexa Fluor 546 for siRNA knockdown samples [1:200 in 0.5% BSA/PBS, 1.5 h; cat. nos. A11060, A21202, A11010; Molecular Probes–Invitrogen]). Coverslips were washed, incubated with a nuclear marker (DRAQ5 [Biostatus] or DAPI [Invitrogen]), mounted on glass slides with GVA Mount (Zymed–Invitrogen) O/N at 4°C, and then sealed using nail polish.
COS‑7 and HeLa cell image acquisition
COS‑7 cells were imaged using a Leica (Mannheim, Germany) TCS/NT laser-scanning confocal system on a DMIR microscope at 60× magnification (MMI). HeLa cell imaging was performed with a Leica SP5 laser-scanning confocal system (DMI6000 microscope, motorized stage, 63× water/glycerol objective; MMI). Images were collected as Z-series and tiled in a 7 × 7 field-of-view grid (∼1.7 mm2).
Imaris-assisted PSPC1 nuclear speckle analysis of cells transfected with GFP/RFP-tagged constructs
Individual transfected cells were three-dimensionally cropped into new files from the larger image sets described above for analysis. Cells expressing GFP or GFP-tagged IMP constructs were first identified using the Imaris software package (Bitplane, version 7). The Imaris batch processor mapped a “surface” around all cells within every image with GFP signal above a threshold level set from mock-transfected cell images. During the subsequent cell cropping, manual assessment was also performed, so cells were selected for analysis only if the entire cell nucleus was clearly defined and distinguishable within the image (x, y, and z); cells with overlapping nuclei or multinucleated cells were excluded. Rare cells containing cytoplasmic PSPC1 speckles were also excluded. To assess PSPC1 speckle number, size, and PSPC1 intensity within each cell, we used the Imaris batch processor to perform a “spots” analysis on each cell (individual image). Results were combined using the CSV Merge command, and the output files were manipulated using Excel (Microsoft, version 2007) and analyzed using SPSS software (IBM, version 17.0).
Imaris-assisted image analysis to detect PSPC1 nuclear speckles in cells with siRNA knockdown or PSF nuclear speckles in cells transfected with GFP-tagged constructs
Individual cells, nuclei, and PSPC1/PSF nuclear foci were identified in the larger image sets described above using the Cells module of the Imaris software package (Bitplane, version 7). The results (output in CSV file formats) were combined and manipulated using Python scripts (Python Software Foundation, version 2.7) and then analyzed using SPSS software (IBM, versions 17 and 20).
Statistical analysis
For statistical testing, individual cells were assumed to be independent, but speckles within each cell were assumed to be correlated. When analyzing the individual cell or speckle data, three outcome types were generated: 1) binary responses based on whether or not a cell was positive for speckles, 2) counts data based on the number of speckles within each cell (including/excluding zeros), and 3) continuous data based on speckle volume sum and speckle PSPC1 intensity sum.
Comparisons between groups were made using generalized linear models; logistic regression for the binary data and linear regression for the count and continuous data. As the count and continuous data were both skewed, the data were transformed using the natural logarithm to allow valid statistical inference from the linear regression models. The p values are based on the transformed data; however, the results were then back-transformed to give estimates in the original scale for ease of interpretation. By taking the exponent of the mean of log-transformed data, the GM (and confidence intervals [CIs]) was obtained on the original linear scale. By taking the exponent of the linear regression coefficients obtained on the log-transformed scale, the ratio of the GMs (and their 95% CIs) was obtained on the original scale. The IMPα2-ED group and the SCRAM siRNA group were used as the reference groups for all models within the GFP/RFP-tagged transfections and the siRNA knockdowns, respectively. Odds ratios are given for logistic regression results. When assessing data on a per-speckle basis, continuous outcomes were examined, which again required log transformations. Generalized estimating equations (GEEs) were used to enable correlation between speckles originating from the same cell (Hanley et al., 2003). Mann-Whitney U-tests and plots with arithmetic means (and 95% CIs) identified statistically significant differences for the same groups.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the excellent support from MMI during confocal image acquisition and analysis. We thank Elizabeth A. Richards for assistance with immunohistochemisty, Catherine M. Browne for assistance with E12.5 gonad dissections, and Katharine F. Adcroft and Phoebe A. S. Kipen for assistance with Adobe software. This work was supported in part by grants from the Australian Research Council Centre of Excellence in Biotechnology and Development (CE0348239 to K.L.L., D.A.J., and P.K.), the Australian Research Council (DP0878102 to Y.M. and K.L.L.), the Australian National Health and Medical Research Council (ID545916 to K.L.L. and APP1002486 to D.A.J.), an Australian Postgraduate Award (to A.T.M.), and Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad and an Australian Research Council Fellowship to Y.M.
Abbreviations used:
- ADAR
adenosine deaminases acting on RNA
- BSA
bovine serum albumin
- CI
confidence interval
- co-IP
coimmunoprecipitation
- DAPI
4′,6-diamidino-2-phenylindole
- DBHS
Drosophila behavior, human splicing
- dpp
days postpartum
- DTT
dithiothreitol
- E
embryonic day
- ELISA
enzyme-linked immunosorbent assay
- ESC
embryonic stem cell
- GEE
generalized estimating equation
- GFP
green fluorescent protein
- GM
geometric mean
- GS4B
glutathione Sepharose 4B slurry
- GST
glutathione S-transferase
- IMP
importin
- mAb
monoclonal antibody
- MMI
Monash Micro Imaging
- NEAT1
nuclear-enriched abundant transcript 1
- NLS
nuclear localization signal
- NONO
non POU domain–containing, octamer-binding protein
- NOPS
NONA/paraspeckle
- NPC
nuclear pore complex
- O/N
overnight
- PFA
paraformaldehyde
- PIC
protease inhibitor cocktail
- PSF
splicing factor proline/glutamine rich
- PSPC1
paraspeckle protein 1
- RRM
RNA recognition motifs
- RT
room temperature
- SCRAM
scrambled
- siRNA
small interfering RNA
- TBS
Tris-buffered saline
- Y2H
yeast two-hybrid.
Footnotes
Author contributions: A.T.M., C.A.H., K.L.L., D.A.J., and Y.M. designed the experiments. A.T.M., C.A.H., and M.A.S. implemented the experiments. Y.M., P.K., and Y.K. created and supplied specialized reagents. C.L.S., A.T.M., and K.L.L. designed and performed statistical analysis. A.T.M., C.A.H., K.L.L., and D.A.J. had primary responsibility for manuscript writing, while all authors made significant contributions to the final document.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0678) on February 18, 2015.
REFERENCES
- Adams IR, McLaren A. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development. 2002;129:1155–1164. doi: 10.1242/dev.129.5.1155. [DOI] [PubMed] [Google Scholar]
- Arjomand A, Baker MA, Li C, Buckle AM, Jans DA, Loveland KL, Miyamoto Y. The alpha-importome of mammalian germ cell maturation provides novel insights for importin biology. FASEB J. 2014;28:3480–3493. doi: 10.1096/fj.13-244913. [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, Yashiro K, Chawengsaksophak K, Wilson MJ, Rossant J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- Byskov AG. Differentiation of mammalian embryonic gonad. Physiol Rev. 1986;66:71–117. doi: 10.1152/physrev.1986.66.1.71. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen A, Dyrskjot L. The functional role of the novel biomarker karyopherin alpha 2 (KPNA2) in cancer. Cancer Lett. 2013;331:18–23. doi: 10.1016/j.canlet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Patton JG. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Male germ cell gene expression. Recent Prog Horm Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- Eddy EM, O'Brien DA. Gene expression during mammalian meiosis. Curr Top Dev Biol. 1998;37:141–200. [PubMed] [Google Scholar]
- Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol. 2013;203:149–164. doi: 10.1083/jcb.201303145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giarre M, Torok I, Schmitt R, Gorjanacz M, Kiss I, Mechler BM. Patterns of importin-alpha expression during Drosophila spermatogenesis. J Struct Biol. 2002;140:279–290. doi: 10.1016/s1047-8477(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Giesecke A, Stewart M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J Biol Chem. 2010;285:17628–17635. doi: 10.1074/jbc.M110.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Gorjanacz M, Adam G, Torok I, Mechler BM, Szlanka T, Kiss I. Importin-alpha 2 is critically required for the assembly of ring canals during Drosophila oogenesis. Dev Biol. 2002;251:271–282. doi: 10.1006/dbio.2002.0827. [DOI] [PubMed] [Google Scholar]
- Hall MN, Griffin CA, Simionescu A, Corbett AH, Pavlath GK. Distinct roles for classical nuclear import receptors in the growth of multinucleated muscle cells. Dev Biol. 2011;357:248–258. doi: 10.1016/j.ydbio.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell . 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth C, Itman C, Jans DA, Loveland KL. Regulated nucleocytoplasmic transport in spermatogenesis: a driver of cellular differentiation? BioEssays. 2005;27:1011–1025. doi: 10.1002/bies.20289. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Calanni S, Jans DA, Loveland KL. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Dev Dyn. 2006;235:253–262. doi: 10.1002/dvdy.20569. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Jans DA, Loveland KL. Subcellular distribution of importins correlates with germ cell maturation. Dev Dyn. 2007;236:2311–2320. doi: 10.1002/dvdy.21238. [DOI] [PubMed] [Google Scholar]
- Hubner S, Xiao CY, Jans DA. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? BioEssays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Kelley JB, Talley AM, Spencer A, Gioeli D, Paschal BM. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 2010;11:63. doi: 10.1186/1471-2121-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JB, Loveland KL, O'Bryan MK, de Kretser DM. Cytology of the testis and intrinsic control mechanisms. In: Neill JD, editor. In: Knobil and Neill's Physiology of Reproduction. Houston, TX: Gulf Professional Publishing; 2006. pp. 827–947. [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci USA. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara S, Ikei A, Taguchi Y, Tabuchi Y, Fujimoto N, Obinata M, Uesugi S, Kurihara Y. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol Reprod. 2006;75:352–359. doi: 10.1095/biolreprod.106.051136. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Stoop H, Biermann K, Oosterhuis JW. Dissecting the molecular pathways of (testicular) germ cell tumour pathogenesis; from initiation to treatment-resistance. Int J Androl. 2011;34:e234–e251. doi: 10.1111/j.1365-2605.2011.01157.x. [DOI] [PubMed] [Google Scholar]
- Loveland KL, Herszfeld D, Chu B, Rames E, Christy E, Briggs LJ, Shakri R, de Kretser DM, Jans DA. Novel low molecular weight microtubule-associated protein-2 isoforms contain a functional nuclear localization sequence. J Biol Chem. 1999;274:19261–19268. doi: 10.1074/jbc.274.27.19261. [DOI] [PubMed] [Google Scholar]
- Loveland KL, Hogarth C, Szczepny A, Prabhu SM, Jans DA. Expression of nuclear transport importins beta 1 and beta 3 is regulated during rodent spermatogenesis. Biol Reprod. 2006;74:67–74. doi: 10.1095/biolreprod.105.042341. [DOI] [PubMed] [Google Scholar]
- Ly-Huynh JD, Lieu KG, Major AT, Whiley PA, Holt JE, Loveland KL, Jans DA. Importin alpha2-interacting proteins with nuclear roles during mammalian spermatogenesis. Biol Reprod. 2011;85:1191–1202. doi: 10.1095/biolreprod.111.091686. [DOI] [PubMed] [Google Scholar]
- Maas S, Gommans WM. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. Nucleic Acids Res. 2009a;37:5822–5829. doi: 10.1093/nar/gkp599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Gommans WM. Novel exon of mammalian ADAR2 extends open reading frame. PLoS One. 2009b;4:e4225. doi: 10.1371/journal.pone.0004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major AT, Whiley PA, Loveland KL. Expression of nucleocytoplasmic transport machinery: clues to regulation of spermatogenic development. Biochim Biophys Acta. 2011;1813:1668–1688. doi: 10.1016/j.bbamcr.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Mason DA, Stage DE, Goldfarb DS. Evolution of the metazoan-specific importin alpha gene family. J Mol Evol. 2009;68:351–365. doi: 10.1007/s00239-009-9215-8. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Baker MA, Whiley PA, Arjomand A, Ludeman J, Wong C, Jans DA, Loveland KL. Towards delineation of a developmental alpha-importome in the mammalian male germline. Biochim Biophys Acta. 2013;1833:731–742. doi: 10.1016/j.bbamcr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Boag PR, Hime GR, Loveland KL. Regulated nucleocytoplasmic transport during gametogenesis. Biochim Biophys Acta. 2012;1819:616–630. doi: 10.1016/j.bbagrm.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- Myojin R, Kuwahara S, Yasaki T, Matsunaga T, Sakurai T, Kimura M, Uesugi S, Kurihara Y. Expression and functional significance of mouse paraspeckle protein 1 on spermatogenesis. Biol Reprod. 2004;71:926–932. doi: 10.1095/biolreprod.104.028159. [DOI] [PubMed] [Google Scholar]
- Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Hirose T. Paraspeckle nuclear bodies–useful uselessness. Cell Mol Life Sci. 2012;69:3027–3036. doi: 10.1007/s00018-012-0973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, Fox AH, Bond CS. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci USA. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Russell LD. Histological and Histopathological Evaluation of the Testis. St. Louis, MO: Cache River Press; 1990. [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MEN ε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepny A, Hogarth CA, Young J, Loveland KL. Identification of Hedgehog signaling outcomes in mouse testis development using a hanging drop-culture system. Biol Reprod. 2009;80:258–263. doi: 10.1095/biolreprod.108.067926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MH, Aul RB, Xu W, van der Hoorn FA, Oko R. Involvement of classical bipartite/karyopherin nuclear import pathway components in acrosomal trafficking and assembly during bovine and murid spermiogenesis. Biol Reprod. 2012;86:84. doi: 10.1095/biolreprod.111.096842. [DOI] [PubMed] [Google Scholar]
- Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- Whiley PA, Miyamoto Y, McLachlan RI, Jans DA, Loveland KL. Changing subcellular localization of nuclear transport factors during human spermatogenesis. Int J Androl. 2012;35:158–169. doi: 10.1111/j.1365-2605.2011.01202.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi YL, Tanaka SS, Yasuda K, Matsui Y, Tam PP. Stage-specific importin13 activity influences meiosis of germ cells in the mouse. Dev Biol. 2006;297:350–360. doi: 10.1016/j.ydbio.2006.04.465. [DOI] [PubMed] [Google Scholar]
- Yasuda Y, Miyamoto Y, Yamashiro T, Asally M, Masui A, Wong C, Loveland KL, Yoneda Y. Nuclear retention of importin alpha coordinates cell fate through changes in gene expression. EMBO J. 2012;31:83–94. doi: 10.1038/emboj.2011.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y, et al. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Yamagishi R, Arai Y, Mehmood R, Kimoto C, Fujita T, Touma K, Kaneko A, Kamikawa Y, Moriyama T, et al. Importin alpha subtypes determine differential transcription factor localization in embryonic stem cells maintenance. Dev Cell. 2013;26:123–135. doi: 10.1016/j.devcel.2013.06.022. [DOI] [PubMed] [Google Scholar]
- Young JC, Major AT, Miyamoto Y, Loveland KL, Jans DA. Distinct effects of importin α2 and α4 on Oct3/4 localization and expression in mouse embryonic stem cells. FASEB J. 2011;25:3958–3965. doi: 10.1096/fj.10-176941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.