Abstract
The Kerguelen archipelago, isolated in the Southern Ocean, shelters a blue mussel Mytilus metapopulation far from any influence of continental populations or any known hybrid zone. The finely carved coast leads to a highly heterogeneous habitat. We investigated the impact of the environment on the genetic structure in those Kerguelen blue mussels by relating allele frequencies to habitat descriptors. A total sample comprising up to 2248 individuals from 35 locations was characterized using two nuclear markers, mac-1 and Glu-5′, and a mitochondrial marker (COI). The frequency data from 9 allozyme loci in 9 of these locations were also reanalyzed. Two other nuclear markers (EFbis and EFprem's) were monomorphic. Compared to Northern Hemisphere populations, polymorphism in Kerguelen blue mussels was lower for all markers except for the exon Glu-5′. At Glu-5′, genetic differences were observed between samples from distinct regions (FCT = 0.077), as well as within two regions, including between samples separated by <500 m. No significant differentiation was observed in the AMOVA analyses at the two other markers (mac-1 and COI). Like mac-1, all allozyme loci genotyped in a previous publication, displayed lower differentiation (Jost's D) and FST values than Glu-5′. Power simulations and confidence intervals support that Glu-5′ displays significantly higher differentiation than the other loci (except a single allozyme for which confidence intervals overlap). AMOVA analyses revealed significant effects of the giant kelp Macrocystis and wave exposure on this marker. We discuss the influence of hydrological conditions on the genetic differentiation among regions. In marine organisms with high fecundity and high dispersal potential, gene flow tends to erase differentiation, but this study showed significant differentiation at very small distance. This may be explained by the particular hydrology and the carved coastline of the Kerguelen archipelago, together with spatially variable selection at Glu-5′.
Keywords: AMOVA, environment, F-statistics, local adaptive differentiation, mac-1, neutral structure
Introduction
In marine benthic organisms, a long planktonic larval stage generally allows gene flow between remote populations and consequently neutral genetic differentiation increases only slightly with geographical distance (Launey et al. 2002). Physical isolation (e.g., large distances, oceanic fronts, gyres) enhances genetic differences among populations. Differentiation may also arise locally through adaptation to localized environmental conditions (Maynard Smith 1966; Barton and Hewitt 1985). However, detecting adaptation through natural selection is difficult mainly because gene flow counters its effects at each generation (Kawecki and Ebert 2004; Sanford and Kelly 2011). Also, large variance in reproductive success (see Hedgecock's sweepstake reproduction hypothesis: Hedgecock 1994; Hedgecock and Pudovkin 2011) can generate transient chaotic patterns of genetic structure, known as ‘chaotic genetic patchiness’ (Johnson and Black 1984; Broquet et al. 2013) that sometimes resembles local adaptation. Differentiation between neighboring populations may be initiated by localized spatial heterogeneity in the environment such as hydrological characteristics (currents, exposition to wave action, salinity, temperature) or a complex topography (coastal shape, depth). Under conditions of fine-grained environmental heterogeneity, genetic differentiation at a selected locus may be higher between populations that differ environmentally even over short distances, than at other loci (Kawecki and Ebert 2004; Gagnaire et al. 2012).
The present work takes place in the Kerguelen archipelago, isolated in the southern Indian Ocean 4100 km southeast of South Africa and 4000 km west of Australia. The Kerguelen plateau is an obstacle to the eastward flow of the Antarctic Circumpolar Current and creates a large wake zone where water masses strongly mix (Park et al. 2008a,b). The cold superficial Antarctic Waters reach the west coast of the archipelago and separate into two parts drifting along south- and northward (Murail et al. 1977; Edgar 1987; Blain et al. 2001). The morphology of the Kerguelen Archipelago is the result of the volcanic activity combined with glacial erosion that led to a carved coast with protected bays and fjords, and a large enclosed bay with particular environmental conditions, the Gulf of Morbihan. In the very coastal perimeter, the salinity decreases drastically due to the important hydrographical network, and decreases even stronger in the shallow waters of the gulf, or deep inside the bays and fjords like at the Fjord des Portes Noires and the Fjord Henri Bossière (Fig.1) (Arnaud 1974; Murail et al. 1977). This archipelago is also characterized by a high level of endemism (Briggs 1966; McDowall 1968; Poulin and Féral 1995; Hennion and Walton 1997; Brandt et al. 1999; Frenot et al. 2001; Emerson 2002). Given the discrete geographical nature (Emerson 2002) and spatial heterogeneity of the environment, Kerguelen Islands seem particularly suited to investigate the association of the environment and the population differentiation in a marine species with a long planktonic larval stage (thereby a putatively high dispersal potential), such as smooth-shelled Mytilus (L.) mussels.
Figure 1.
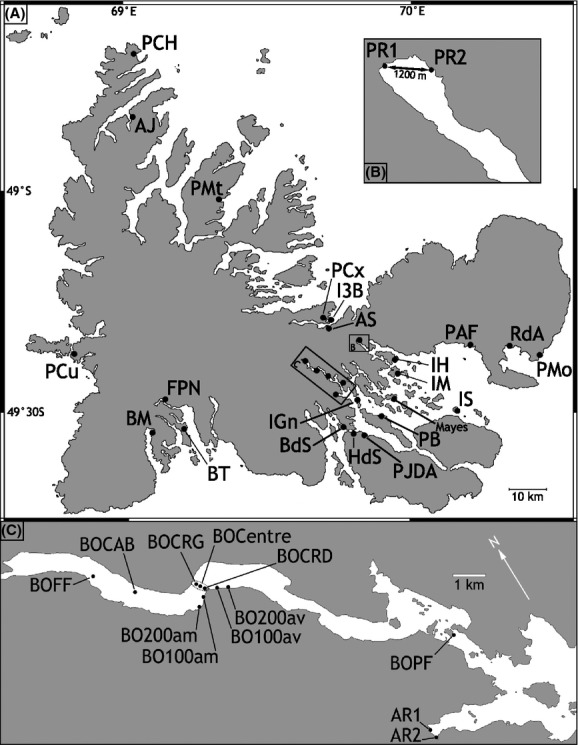
Sampling sites of Kerguelen blue mussels. Sampling details in Table1; (A) Main map. (B) Detail of locations for samples PR1 and PR2. (C) Enlarged map of Henri Bossière Fjord with locations of samples AR and BO. Mayes is the location of the sample ‘KER’ in Borsa et al. (2007).
The blue mussels from Kerguelen are mainly distributed in the intertidal zone from 0 to 2 m depth (Arnaud 1971, 1974), where the environmental conditions are the most variable. Kerguelen blue mussels have been described as M. desolationis (Lamy, 1936). However, their current taxonomic status, determined from morphology, allozymes, and nuclear and mitochondrial DNA sequences, is M. edulis platensis, the Southern-Hemisphere subspecies of M. edulis L. (McDonald et al. 1991; Borsa et al. 2012). Genetic differentiation between smooth-shelled Mytilus spp. mussels (M. edulis L., M. galloprovincialis Lmk., M. trossulus Gould) and the phylogeography of these species has been studied extensively (e.g., McDonald et al. 1991; Sanjuan et al. 1997; Daguin 2000; Daguin and Borsa 2000; Hilbish et al. 2000; Gérard et al. 2008), with particular focus on areas of hybridization (Skibinski et al. 1978; Skibinski 1983; Väinölä and Hvilsom 1991; Viard et al. 1994; Gardner 1996; Bierne et al. 2002a,b, 2003). The most famous example of genetic differentiation linked to the environment in blue mussels is the gradient at the lap locus correlated with salinity gradient along the eastern coast of North America (Koehn 1978). The physiological and selective roles of the lap locus have been highlighted (Hilbish et al. 1982; Hilbish and Koehn 1985a,b) and further studied in blue mussels from Kerguelen (Blot et al. 1989) and New Zealand (Gardner and Kathiravetpillai 1997; Gardner and Palmer 1998). In Kerguelen blue mussels, genetic differences between populations were apparent at three (lap, pgm, pgd) allozyme loci (Blot et al. 1989), and the structure reported to be related to salinity, wave exposure and, to a lesser extent, to the maximum shell length (as a proxy of fitness). However, no statistical analyses were conducted to support this conclusion. Theoretically, genetic differentiation may be due to physical barriers to gene flow but also to local adaptation under selective constraints (Williams 1966; Kawecki and Ebert 2004; Perrin et al. 2004). In cases of barriers to gene flow, the differentiation will affect a majority of loci, whereas in cases of local adaptation only a few loci are concerned.
To determine whether the genetic polymorphism of the blue mussel population of Kerguelen is driven by neutral and/or adaptive forces, we (i) investigated the influence of the water circulation around Kerguelen, first on the total genetic structure and second within differentiated groups and (ii) tested the influence of the habitat type at a smaller scale. To fill in these objectives, we used two nuclear markers polymorphic in Kerguelen blue mussels: Glu-5′ (Inoue et al. 1995; Rawson et al. 1996) and mac-1 (Ohresser et al. 1997), and we also considered the sequence polymorphism at the mitochondrial DNA locus COI (Gérard et al. 2008). We tested the polymorphism at EFbis (Bierne et al. 2002a) and EFprem's (this study), two introns of the elongation factor 1 alpha gene, which are physically linked. We collected blue mussel samples from all around the Kerguelen Archipelago, from contrasted habitats roughly described by five qualitative environmental variables. At a finer grid, a dense network of sites in the complex of islands of the Gulf of Morbihan was sampled to explore the distribution of the allele frequencies, taking into account the environmental changes over short distances.
Material and Methods
Sampling
Blue mussel samples were collected between 1999 and 2003 from 35 sites in the Kerguelen Archipelago (Fig.1). Sample size ranged from 27 to 130 individuals (Table1). Five samples came from the north coast (PCh, PMt, PCx, I3B, AJ), two from the east coast (PMo, RdA), twenty-two from the Gulf of Morbihan (PAF, PR1, PR2, IH, IM, IGn, PF, BOCRD, BOCentre, BOCRG, BO100am, BO200am, BO100av, BO200av, BOCAB, BOFF, Ar1, Ar2, HdS, PJDA, PB, IS), Four from the south coast (BdS, BM, FPN, BM) and one from the west coast (PCu). Pieces of mantle tissue were preserved in 95% ethanol. DNA was extracted using Chelex100© chelating resin (Walsh et al. 1991).
Table 1.
Sample location of Kerguelen blue mussels
| Site | Abbreviation | Region | GPS coordinates |
|---|---|---|---|
| Port Christmas | PCh | North | 48°40′58″; 69°02′12″ |
| Anse du Jardin | AJ | North | 48°50′14″; 69°00′43″ |
| Port Matha | PMt | North | 48°55′59″; 69°02′35″ |
| Port Couvreux | PCx | North | 49°17′07″; 69°41′50″ |
| Ilot des Trois Bergers | I3B | North | 49°17′17″; 69°42′23″ |
| Anse Sablonneuse | AS | North | 49°18′53″; 69°42′30″ |
| Pointe Morne | PMo | East | 49°22′47″; 70°26′36″ |
| Rivière des Albatros | RdA | East | 49°21′08″; 70°22′47″ |
| Port-aux-Français | PAF | Gulf of M. | 49°21′08″; 70°13′07″ |
| Port Raymond (deep bay) | PR1 | Gulf of M. | 49°20′19″; 69°48′20″ |
| Port Raymond (Hut) | PR2 | Gulf of M. | 49°20′27″; 69°49′22″ |
| Ile haute | IH | Gulf of M. | 49°23′13″; 69°56′20″ |
| Ile aux Moules | IM | Gulf of M. | 49°25′04″; 69°56′54″ |
| Ile Guillou (north) | IGn | Gulf of M. | 49°28′10″; 69°48′23″ |
| Portes de Fer | PF | Gulf of M. | 49°26′34″; 69°45′ |
| Henri Bossière Fjord, 200 m downstream from mb | BO200av | Gulf of M. | See Fig.1 |
| Henri Bossière Fjord, 100 m downstream from mb | BO100av | Gulf of M. | See Fig.1 |
| Henri Bossière Fjord, eastern part of mb | BOCRD | Gulf of M. | See Fig.1 |
| Henri Bossière Fjord, center of mb | BOCentre | Gulf of M. | 49°24′28″; 69°40′06″ |
| Henri Bossière Fjord, western part of mb | BOCRG | Gulf of M. | See Fig.1 |
| Henri Bossière Fjord, 100 m upstream from mb | BO100am | Gulf of M. | See Fig.1 |
| Henri Bossière Fjord, 200 m upstream from mb | BO200am | Gulf of M. | See Fig.1 |
| Henri Bossière Fjord, front of hut | BOCAB | Gulf of M. | 49°24′07″; 69°38′37″ |
| Henri Bossière Fjord, deep part | BOFF | Gulf of M. | 49°23′36″; 69°37′48″ |
| Armor | Ar1 | Gulf of M. | 49°27′50″; 69°43′40″ |
| Armor | Ar2 | Gulf of M. | 49°27′50″; 69°43′40″ |
| Halage des Swains | HdS | Gulf of M. | 49°32′57″; 69°48′05″ |
| Port Jeanne d'Arc | PJDA | Gulf of M. | 49°33′07″; 69°49′26″ |
| Port Bizet | PB | Gulf of M. | 49°31′02″; 69°54′13.″ |
| Ile Suhm | IS | Gulf of M. | 49°30′18″; 70°09′38″ |
| Baie des Swains | BdS | South | 49°33′02″; 69°46′23″ |
| Baie de la Table | BT | South | 49°31′; 69°11′15″ |
| Fjord des Portes Noires | FPN | South | 49°29′; 69°06′06″ |
| Baie de la Mouche | BM | South | 49°32′56″; 69°05′10″ |
| Port Curieuse | PCu | West | 49°21′41″; 68°48′35″ |
mb, mussel bed.
Molecular markers
The locus Glu-5′ (Inoue et al. 1995; Rawson et al. 1996) is located at the 5′ extremity of exon Glu coding for an adhesive foot protein (Waite 1992). This locus contains an insertion/deletion (indel) zone, whose amplification reveals three alleles: (T, E and G) that, respectively distinguish M. trossulus, M. edulis, and M. galloprovincialis in the Northern Hemisphere (Borsa et al. 1999). The locus mac-1 is the first intron of the Mytilus actin protein (Ohresser et al. 1997). Among the 49 size-alleles described in the entire range of Mytilus spp., 22 alleles occur in the Southern Hemisphere, 8 of which have not yet been sampled in the Northern Hemisphere (Daguin and Borsa 2000). The four alleles encountered in the Kerguelen blue mussel population in Kerguelen are all shared with Northern-Hemisphere populations (Daguin and Borsa 2000; Borsa et al. 2007). The polymorphism at locus EFbis has been tested in one location in Kerguelen (Mayes Island in the Gulf of Morbihan) and was low, with two alleles detected (160: frequency 0.01; 161: 0.99) (Daguin 2000). We scored locus EFBis in other samples from around the archipelago. A new EPIC marker (EFprem's) in the second intron of EF1α was also scored. Both EFBis and EFprem's loci showed sample monomorphism (see Results). Table S1 summarizes primer names, sequences, and annealing temperatures required for the amplification of nuclear and mitochondrial DNA loci. The genotypes at Glu-5′ and mac-1 were determined from fragment-length variation on, respectively, 2 and 3% agarose gels. The amplification of the Glu-5′ exon by primers Me-15 and Me-17 produced 210-bp (allele E) and 160-bp (allele G) fragments, typical of, respectively, M. edulis and M. galloprovincialis from the Northern Hemisphere (Fig.2A). At locus mac-1, fragments of 400 and 370 bp were revealed. According to Daguin (2000) and Borsa et al. (2007), the 400-bp fragment at locus mac-1 corresponds to allele c4, whereas the 370-bp fragment corresponds to either allele a2 or a3, which differ from one another by one base pair and cannot be distinguished on agarose gels (Fig.2B). Consequently, the 370-bp fragment is here noted ‘a’. The denomination of COI haplotypes (KERF1 to KERF16) follows Gérard et al. (2008).
Figure 2.
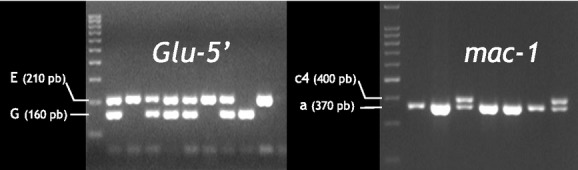
Kerguelen blue mussels. Individual phenotypes scored on agarose gels at nuclear loci. The left lane is a 100-bp DNA ladder.
Genetic analysis
Heterozygosity was estimated by Nei's (1978) nonbiased heterozygosity index (Hn.b). FIS and FST values were estimated according to Weir and Cockerham (1984) using the FSTAT procedure in the program GENETIX 4.02 (Belkhir et al. 2000). The significance of FIS (and respectively FST) values was assessed after 5000 permutations of alleles (resp. individuals) within (resp. between) samples, thus obtaining the distribution of FIS (resp. FST) pseudovalues under the null hypothesis of panmixia (resp. a nonstructured population). The probability (P) values associated to FIS or FST estimates was the proportion of pseudovalues generated by 5000 random permutations larger than, or equal to the observed value. Mantel tests were used to assess the correlation of pairwise FST values and geographical distances (by coastal line) between samples computing the association statistics Z (Mantel 1967). The P-value of Z was the proportion of pseudovalues generated by permutations under the null hypothesis of independence of genetic and geographical distances, larger than, or equal to the observed value of Z. Mantel tests and permutations were computed by the program GENETIX 4.02.
The false discovery rate correction for multiple comparisons was used to adjust levels of statistical significance (Benjamini and Hochberg 1995).
Analyses of molecular variance (AMOVA; Excoffier et al. 1992) were carried out on Glu-5′ and mac-1 genotype data, as well as on COI sequences (conventional FST based on frequencies) for comparative purposes using ARLEQUIN 3.0 (Excoffier et al. 2005). Following the results of pairwise FST (see Results), we defined and tested a geographical structure of three groups based on the origin of the samples (North + East, Gulf, South + West; Table1). In the AMOVA grouping, the unique sample from the west coast (PCu) was lumped with the South group, from which it was not differentiated (see pairwise FST results; Table4). The exact test of sample differentiation (Raymond and Rousset 1995) was run (with 20,000 Markov chain, 1000 dememorization steps) using this same software based on mitochondrial haplotype frequency in the three groups. So-called neutrality tests (Fu's Fs and Tajima's D) were run to check whether the double hypothesis of demographic stability and selective neutrality of the COI marker could be rejected.
Table 4.
Kerguelen blue mussels. Pairwise FST (Weir and Cockerham 1984) values at Glu-5′ (above diagonal) and mac-1 (below diagonal) loci
| Region | Sample | North | Gulf of Morbihan | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCh | AJ | PMt | PCx | I3B | AS | PMo | RdA | PAF | PR1 | PR2 | IH | IM | PJDA | HdS | PB | ||
| North | PCh | – | 0.036* | 0 | 0.004 | 0.016 | 0 | 0 | 0.091 | 0.063** | 0.119** | 0.063 | 0.057** | 0.053 | 0.082 | 0.084 | 0.072** |
| AJ | 0 | – | 0.051** | 0.09 | 0.111 | 0.073 | 0.052 | 0.202 | 0 | 0.019* | 0 | 0 | 0 | 0.006 | 0.005 | 0.003 | |
| PMt | 0 | 0.001 | – | 0 | 0.003 | 0 | 0 | 0.068* | 0.082** | 0.144 | 0.083** | 0.075** | 0.071 | 0.103** | 0.106** | 0.092 | |
| PCx | 0.014 | 0.004 | 0.035* | – | 0 | 0 | 0 | 0.036* | 0.133 | 0.205 | 0.134 | 0.117 | 0.114 | 0.152 | 0.158 | 0.138 | |
| I3B | 0 | 0 | 0.013 | 0 | – | 0 | 0.004 | 0.016 | 0.158 | 0.23 | 0.159 | 0.139 | 0.135 | 0.173 | 0.181 | 0.159 | |
| AS | 0 | 0 | 0.015 | 0 | 0 | – | 0 | 0.048* | 0.111 | 0.177 | 0.112** | 0.099 | 0.095 | 0.123 | 0.135 | 0.118 | |
| PMo | 0 | 0 | 0.002 | 0.003 | 0 | 0 | – | 0.07** | 0.083** | 0.145 | 0.084** | 0.075 | 0.071 | 0.103 | 0.106 | 0.092 | |
| RdA | 0.007 | 0.024 | 0 | 0.071** | 0.044* | 0.046* | 0.025* | – | 0.274 | 0.351 | 0.275 | 0.232 | 0.228 | 0.269 | 0.289 | 0.253 | |
| Gulf of M. | PAF | 0 | 0 | 0.006 | 0 | 0 | 0 | 0 | 0.032* | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PR1 | 0 | 0 | 0.007 | 0 | 0 | 0 | 0 | 0.033* | 0 | – | 0 | 0.006 | 0.006 | 0 | 0 | 0 | |
| PR2 | 0.021 | 0.042* | 0.008 | 0.096** | 0.065** | 0.068** | 0.044* | 0 | 0.051* | 0.053* | – | 0 | 0 | 0 | 0 | 0 | |
| IH | 0.010 | 0 | 0.030* | 0 | 0 | 0 | 0 | 0.069** | 0 | 0 | 0.095 | – | 0 | 0 | 0 | 0 | |
| IM | 0 | 0 | 0.01554 | 0 | 0 | 0 | 0 | 0.049** | 0 | 0 | 0.072** | 0 | – | 0 | 0 | 0 | |
| PJDA | 0.001 | 0 | 0.01636 | 0 | 0 | 0 | 0 | 0.050** | 0 | 0 | 0.074** | 0 | 0 | – | 0 | 0 | |
| HdS | 0.009 | 0 | 0.023* | 0 | 0 | 0 | 0 | 0.066** | 0 | 0 | 0.091** | 0 | 0 | 0 | – | 0 | |
| PB | 0 | 0 | 0 | 0.009 | 0 | 0 | 0 | 0.022 | 0 | 0 | 0.04** | 0.005 | 0 | 0 | 0.005 | – | |
| IGn | 0 | 0 | 0.012 | 0 | 0 | 0 | 0 | 0.042* | 0 | 0 | 0.064 | 0 | 0 | 0 | 0 | 0 | |
| Ar1 | 0 | 0 | 0.006 | 0 | 0 | 0 | 0 | 0.032* | 0 | 0 | 0.051* | 0 | 0 | 0 | 0 | 0 | |
| Ar2 | 0 | 0.001 | 0 | 0.043* | 0.016 | 0.017 | 0.002 | 0 | 0.007 | 0.008 | 0 | 0.037* | 0.019 | 0.020 | 0.035* | 0 | |
| BOCRD | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| BOCentre | 0 | 0 | 0.015 | 0 | 0 | 0 | 0 | 0.045* | 0 | 0 | 0.066** | 0 | 0 | 0 | 0 | 0 | |
| BO100av | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| BO200av | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| BO100am | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| BO200am | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| BOCRG | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| BOFF | 0.006 | 0 | 0.025* | 0 | 0 | 0 | 0 | 0.058* | 0 | 0 | 0.081** | 0 | 0 | 0 | 0 | 0.003 | |
| BOCab | 0.015 | 0.004 | 0.037* | 0 | 0 | 0 | 0.003 | 0.074* | 0 | 0 | 0.099** | 0 | 0 | 0 | 0 | 0.01 | |
| PF | 0 | 0 | 0.012 | 0 | 0 | 0 | 0 | 0.037 | 0 | 0 | 0.057* | 0 | 0 | 0 | 0 | 0 | |
| IS | 0 | 0 | 0 | 0.034 | 0.008 | 0.009 | 0 | 0 | 0.001 | 0.002 | 0.006 | 0.027 | 0.011 | 0.012 | 0.025 | 0 | |
| South | BdS | 0 | 0 | 0 | 0.025* | 0.008 | 0.009 | 0 | 0.005 | 0.002 | 0.002 | 0.018 | 0.020* | 0.01 | 0.01 | 0.012* | 0 |
| BT | 0 | 0.012 | 0 | 0.063** | 0.031 | 0.033 | 0.014 | 0 | 0.020 | 0.022 | 0 | 0.057** | 0.035* | 0.04* | 0.054** | 0.01 | |
| FPN | 0 | 0 | 0.016 | 0 | 0 | 0 | 0 | 0.047* | 0 | 0 | 0.069** | 0 | 0 | 0 | 0 | 0 | |
| BM | 0 | 0 | 0.002 | 0.004 | 0 | 0 | 0 | 0.027* | 0 | 0 | 0.045* | 0 | 0 | 0 | 0 | 0 | |
| West | PCu | 0.009 | 0.001 | 0.029 | 0 | 0 | 0 | 0 | 0.061* | 0 | 0 | 0.084** | 0 | 0 | 0 | 0 | 0.006 |
| IGn | Ar1 | Ar2 | CRD | Centre | 100av | 200av | 100am | 200am | CRG | FF | Cab | PF | IS | South | West | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BdS | BT | FPN | BM | PCu | ||||||||||||||
| 0.193 | 0.338 | 0.136** | 0.149 | 0.198 | 0.077** | 0.08* | 0.167** | 0.111** | 0.138** | 0.072** | 0.063** | 0.044* | 0.023 | 0.016 | 0 | 0.019* | 0 | 0.033* |
| 0.066 | 0.172 | 0.024 | 0.035** | 0.066 | 0.001 | 0 | 0.0461** | 0.015 | 0.029* | 0 | 0 | 0 | 0 | 0 | 0.009 | 0 | 0.031** | 0 |
| 0.220 | 0.369 | 0.163** | 0.175 | 0.226 | 0.099** | 0.103* | 0.194 | 0.135 | 0.164 | 0.092** | 0.083* | 0.062* | 0.037* | 0.03* | 0.006 | 0.032* | 0 | 0.049* |
| 0.283 | 0.432 | 0.229 | 0.238 | 0.293 | 0.153 | 0.162 | 0.26 | 0.198 | 0.227 | 0.144 | 0.133 | 0.112** | 0.078 | 0.063 | 0.030* | 0.064 | 0.011 | 0.092** |
| 0.306 | 0.463 | 0.26 | 0.264 | 0.319 | 0.178 | 0.197 | 0.285 | 0.22 | 0.252 | 0.17 | 0.159 | 0.140 | 0.102** | 0.084** | 0.049* | 0.084 | 0.023 | 0.118** |
| 0.254 | 0.406 | 0.201 | 0.21 | 0.263 | 0.129 | 0.139** | 0.23 | 0.168 | 0.198 | 0.122 | 0.112 | 0.092** | 0.061** | 0.048** | 0.021* | 0.050** | 0.004 | 0.074** |
| 0.221 | 0.369 | 0.164 | 0.176 | 0.227 | 0.1** | 0.105** | 0.195 | 0.136 | 0.165 | 0.093 | 0.084* | 0.064* | 0.038* | 0.029* | 0.007 | 0.032** | 0 | 0.05* |
| 0.415 | 0.579 | 0.403 | 0.386 | 0.441 | 0.3 | 0.354 | 0.407 | 0.341 | 0.373 | 0.287 | 0.275 | 0.276 | 0.22 | 0.179 | 0.142 | 0.167 | 0.091 | 0.239 |
| 0.034* | 0.13** | 0 | 0.01 | 0.033 | 0 | 0 | 0.017 | 0 | 0.005 | 0 | 0 | 0 | 0 | 0.003 | 0.025 | 0.005 | 0.057** | 0 |
| 0.003 | 0.072** | 0 | 0 | 0.002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.022 | 0.035* | 0.069 | 0.036* | 0.113 | 0.015 |
| 0.035* | 0.131 | 0.001 | 0.011 | 0.034* | 0 | 0 | 0.018 | 0 | 0.006 | 0 | 0 | 0 | 0 | 0.004 | 0.026* | 0.005 | 0.057** | 0 |
| 0.044 | 0.14 | 0.01 | 0.02* | 0.044** | 0 | 0 | 0.027* | 0.003 | 0.014 | 0 | 0 | 0 | 0 | 0.005 | 0.023* | 0.005 | 0.051 | 0 |
| 0.045 | 0.141 | 0.008 | 0.02 | 0.045** | 0 | 0 | 0.028* | 0.003 | 0.014 | 0 | 0 | 0 | 0 | 0.003 | 0.021* | 0.004 | 0.049 | 0 |
| 0.022* | 0.104 | 0 | 0.003 | 0.021 | 0 | 0 | 0.009 | 0 | 0 | 0 | 0 | 0 | 0.007 | 0.017 | 0.042** | 0.018* | 0.077 | 0.001 |
| 0.021 | 0.105 | 0 | 0.002 | 0.020* | 0 | 0 | 0.007 | 0 | 0 | 0 | 0 | 0 | 0.006 | 0.016 | 0.043* | 0.017* | 0.079 | 0.001 |
| 0.023* | 0.115 | 0 | 0.008 | 0.029* | 0 | 0 | 0.014 | 0 | 0.004 | 0 | 0 | 0 | 0.003 | 0.012 | 0.035** | 0.013* | 0.067 | 0 |
| – | 0.024* | 0 | 0 | 0 | 0.024 | 0.014 | 0 | 0.006 | 0 | 0.03* | 0.036* | 0.044* | 0.075 | 0.092 | 0.136 | 0.091 | 0.186 | 0.064** |
| 0 | – | 0.06** | 0.05* | 0.022 | 0.113 | 0.109** | 0.038 | 0.079** | 0.06** | 0.123 | 0.133 | 0.153 | 0.193 | 0.211 | 0.27 | 0.206 | 0.323 | 0.179 |
| 0.014 | 0.007 | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.002 | 0.003 | 0.027 | 0.042* | 0.080** | 0.043* | 0.127** | 0.02 |
| – | – | – | – | 0 | 0.003 | 0 | 0 | 0 | 0 | 0.008 | 0.012 | 0.017 | 0.041* | 0.056** | 0.095 | 0.056 | 0.141 | 0.033* |
| 0 | 0 | 0.018 | – | 0.023 | 0.013 | 0 | 0.005 | 0 | 0.029* | 0.035 | 0.043 | 0.074* | 0.092** | 0.138** | 0.092 | 0.19 | 0.064** | |
| – | – | – | – | – | – | 0 | 0.009 | 0 | 0 | 0 | 0 | 0 | 0 | 0.011 | 0.036* | 0.012 | 0.071** | 0 |
| – | – | – | – | – | – | – | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.004 | 0.033 | 0.006 | 0.07* | 0 |
| – | – | – | – | – | – | – | – | 0 | 0 | 0.015 | 0.019 | 0.025 | 0.052* | 0.07** | 0.11 | 0.069** | 0.16 | 0.043* |
| – | – | – | – | – | – | – | – | – | 0 | 0 | 0 | 0 | 0.017 | 0.03* | 0.062* | 0.031* | 0.1042 | 0.011 |
| – | – | – | – | – | – | – | – | – | – | 0.004 | 0.007 | 0.01 | 0.033 | 0.05* | 0.085** | 0.049* | 0.131 | 0.025 |
| 0 | 0 | 0.03* | – | 0 | – | – | – | – | – | – | 0 | 0 | 0 | 0.008 | 0.032* | 0.009 | 0.065 | 0 |
| 0 | 0 | 0.045 | – | 0 | – | – | – | – | – | 0 | – | 0 | 0 | 0.004 | 0.026* | 0.005 | 0.057** | 0 |
| 0 | 0 | 0.012 | – | 0 | – | – | – | – | – | 0 | 0 | – | 0 | 0 | 0.009 | 0 | 0.038 | 0 |
| 0.006 | 0.001 | 0 | – | 0.011 | – | – | – | – | – | 0.021* | 0.034 | 0.007 | – | 0 | 0 | 0 | 0.019 | 0 |
| 0.006 | 0.002 | 0 | – | 0.008 | – | – | – | – | – | 0.016 | 0.0262* | 0.006 | 0 | – | 0 | 0 | 0.013 | 0 |
| 0.03 | 0.02 | 0 | – | 0.033* | – | – | – | – | – | 0.047 | 0.065** | 0.028* | 0 | 0 | – | 0 | 0 | 0.004 |
| 0 | 0 | 0.02 | – | 0 | – | – | – | – | – | 0 | 0 | 0 | 0.012 | 0.01 | 0.035* | – | 0.016* | 0 |
| 0 | 0 | 0.003 | – | 0 | – | – | – | – | – | 0 | 0.004 | 0 | 0 | 0 | 0.014 | 0 | – | 0.027* |
| 0 | 0 | 0.035 | – | 0 | – | – | – | – | – | 0 | 0 | 0 | 0.027 | 0.02 | 0.053* | 0 | 0.001 | – |
In bold: significant values after FDR correction for multiple tests (Benjamini and Hochberg 1995). Bold values without asterisk have P ≤ 0,001.
*P ≤ 0.05; **P ≤ 0.01.
We reanalyzed gene frequency data of nine allozyme loci published in Blot (1989) and Blot et al. (1989). Genotypes were not available, but we could compute FST from gene frequency data using the relationship FST = 1 – Ho/He, the exact tests of differentiation based on Jost's differentiations (see below and Table2), and the contingency tables of the numbers of each allele in each population (Table S2).
Table 2.
Kerguelen blue mussels. Jost's D differentiation estimates and its confidence interval (CI) calculated using a set seven populations common to the allozyme study of Blot (1989) and Blot et al. (1989) and present study. Populations were PMt and PCx from the North region, PAF, IS, HdS, and BOS from the Gulf of Morbihan (all samples within the Bossière Fjord were pooled), and BT (Glu-5′ and mac-1) or “Larose” (allozymes) from the South region. Bold values have confidence intervals that do not include zero
| Locus | D | CI |
|---|---|---|
| Glu-5′ | 0.049 | 0.023–0.075 |
| Mac-1 | 0.002 | 0.000–0.008 |
| LAP1 | 0.014 | 0.000–0.032 |
| LAP2 | 0.011 | 0.000–0.025 |
| PGD | 0.032 | 0.017–0.047 |
| PGI | 0.009 | 0.000–0.027 |
| PGM | 0.007 | 0.000–0.018 |
| GOT1 | 0.000 | 0.000–0.000 |
| GOT2 | 0.001 | 0.000–0.002 |
| MDH1 | 0.001 | 0.000–0.003 |
| MDH2 | 0.001 | 0.000–0.003 |
Two approaches were used to compare differentiation levels among markers or data sets with contrasted samples sizes and polymorphism levels. (i) Jost's differentiation parameters (D) and confidence intervals were computed using the program SPADE (Jost 2008; Chao and Shen 2010). This was carried out for the common subset of seven populations which were analyzed in the present study (for Glu-5′ and mac-1), in Blot (1989) and Blot et al. (1989) for nine allozyme markers. The interest in this approach is that Jost's D is much less affected by polymorphism level than FST and provides confidence intervals. (ii) The POWSIM application (Ryman and Palm 2006) was used to compare results among markers which did not display similar sample sizes and allele frequency distributions. This application uses simulated data sets corresponding to a model of diverging populations (no migration): a given FST level is chosen by the user, by selecting an appropriate pair of values for effective size and divergence time (in number of generations). We thus checked whether the small sample sizes or the reduced polymorphism for the COI and mac-1 data sets, respectively, relative to those of Glu-5′ may affect our results in the finding of significant differentiation among regions. The simulations were run at the differentiation level found with the Glu-5′ genotypic data (we used a value similar to both the overall FST among all populations and the global FCT among regions), using the global frequency distributions of COI haplotypes and mac-1 alleles and their respective sample sizes in the three groups of populations (North + East, Gulf, South + West). The output of the program provides the proportion of cases in which significant differentiation is found.
A median-joining parsimony network (Bandelt et al. 1999) of COI haplotypes was built using NETWORK 4.1.0.7 (available at www.fluxus-technology.com/).
Environmental factors
The habitat at each sampling site was described by five qualitative variables: (i) Substrate (rock, blocks, gravels, or sand); (ii) Wave Exposure (sheltered or exposed); (iii) Slope (flat, steep or hangover); (iv) Salinity (oceanic or influenced by freshwater); (v) Macrocystis (presence or absence). The Region (North, South, Gulf, and West) was also considered as a factor in the following statistical analyses. Correlation among environmental factors and frequency of allele G was assessed by pairwise Spearman's ρ values (Spearman 1904).
We also used AMOVAs on Glu-5′ and mac-1 genotypic data and COI haplotype data, within each geographical region. For each environmental factor, we grouped samples according to the modality of the variable in order to test the effects of environment within regions (FCT, Va), within groups independently of the effects of population differentiation between regions. The AMOVA, although it is restricted to investigate nested factors, has two important advantages over parametric analyses (ANCOVAs were performed using the proportion of the G allele at Glu-5′ as the variable to explain, but not shown): (i) it does not rely upon statistical conditions on the distribution of the data, as the P-value is assessed via permutations (Excoffier et al. 2005) and (ii) it takes into account the statistically important information of the number of individuals in each population.
Results
Polymorphism
The number of individuals characterized was 2248, 1511, and 83 for Glu-5′, mac-1, and COI (Table3), respectively, and 20 for each EFbis and EFprem's.
Table 3.
Allelic frequencies at loci Glu-5′, mac-1, and haplotype composition at locus COI for each sample
| Region | Sample | Locus | Environmental factors | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glu-5′ | mac-1 | COI | Substrate | Slope | Exposure | Salinity | Macro-cystis | |||||||||
| N | Frequency of allele G | Hn.b. | f | N | Frequency of allele a | Hn.b. | f | N | Haplotype | |||||||
| North | PCh | 51 | 0.26 | 0.393 | −0.149 ns | 49 | 0.9 | 0.185 | 0.346 ns | 4 | F1, F3 (3) | R | H | Sh | OW | P |
| North | AJ | 106 | 0.41 | 0.485 | −0.091 ns | 49 | 0.92 | 0.152 | −0.079 ns | – | G | St | Sh | OW | P | |
| North | PMt | 50 | 0.24 | 0.369 | 0.133 ns | 48 | 0.88 | 0.221 | −0.134 ns | 2 | F1, F5 | G | St | Sh | OW | P |
| North | PCx | 109 | 0.2 | 0.324 | 0.150 ns | 48 | 0.96 | 0.081 | −0.033 ns | 4 | F4, F5 (2), F12 | B | H | E | OW | P |
| North | I3B | 50 | 0.17 | 0.285 | 0.230 ns | 49 | 0.94 | 0.116 | 0.303 ns | 4 | F4 (2), F5, F12 | R | H | E | OW | P |
| North | AS | 54 | 0.21 | 0.338 | −0.151 ns | 50 | 0.94 | 0.114 | 0.304 ns | – | G | F | Sh | OW | A | |
| North | PMo | 50 | 0.24 | 0.369 | −0.086 ns | 50 | 0.92 | 0.149 | −0.078 ns | 2 | F2, F16 | B | St | E | OW | P |
| North | RdA | 50 | 0.09 | 0.166 | 0.155 ns | 50 | 0.83 | 0.285 | 0.233 ns | 3 | F1 (3) | S | F | Sh | LSW | A |
| Gulf of M. | PAF | 50 | 0.45 | 0.500 | 0.161 ns | 48 | 0.93 | 0.137 | −0.069 ns | 2 | F2, F4 | B | St | E | OW | P |
| Gulf of M. | PR1 | 50 | 0.52 | 0.504 | 0.129 ns | 49 | 0.93 | 0.134 | −0.067 ns | 4 | F4, F5 (4) | S | F | Sh | LSW | A |
| Gulf of M. | PR2 | 50 | 0.45 | 0.500 | 0.000 ns | 49 | 0.81 | 0.316 | 0.031 ns | – | G | F | Sh | LSW | P | |
| Gulf of M. | IH | 116 | 0.44 | 0.495 | −0.186* | 79 | 0.95 | 0.097 | −0.047 ns | 5 | F1, F5, F13, F14, F15 | R | H | E | OW | P |
| Gulf of M. | IM | 118 | 0.44 | 0.494 | 0.091 ns | 78 | 0.94 | 0.121 | −0.062 ns | 1 | F5 | R | St | E | OW | P |
| Gulf of M. | PJDA | 115 | 0.48 | 0.501 | 0.168 ns | 79 | 0.94 | 0.119 | −0.062 ns | – | B | St | E | OW | P | |
| Gulf of M. | HdS | 71 | 0.48 | 0.503 | 0.048 ns | 69 | 0.95 | 0.097 | −0.046 ns | 3 | F3, F5 (2) | B | F | Sh | OW | A |
| Gulf of M. | PB | 127 | 0.46 | 0.500 | 0.086 ns | 79 | 0.91 | 0.163 | −0.091 ns | 3 | F1, F2 (2) | R | H | E | OW | P |
| Gulf of M. | IGn | 77 | 0.6 | 0.484 | 0.035 ns | 61 | 0.93 | 0.124 | 0.207 ns | 3 | F4, F5 (2) | B | F | Sh | OW | A |
| Gulf of M. | Ar1 | 50 | 0.72 | 0.407 | −0.081 ns | 48 | 0.93 | 0.137 | −0.069 ns | 2 | F1, F2 | R | F | Sh | LSW | A |
| Gulf of M. | Ar2 | 28 | 0.54 | 0.506 | 0.013 ns | 30 | 0.87 | 0.235 | 0.151 ns | – | R | H | Sh | OW | A | |
| Gulf of M. | BO200av | 19 | 0.47 | 0.512 | −0.029 ns | – | – | – | – | – | G | F | Sh | LSW | A | |
| Gulf of M. | BO100av | 49 | 0.47 | 0.503 | 0.109 ns | – | – | – | – | – | G | F | Sh | LSW | A | |
| Gulf of M. | BOCRD | 50 | 0.55 | 0.500 | −0.243 ns | – | – | – | – | – | S | F | Sh | LSW | A | |
| Gulf of M. | BOCentre | 50 | 0.6 | 0.485 | 0.093 ns | 49 | 0.94 | 0.116 | −0.055 ns | 5 | F2, F3, F5, F9, F10 | S | F | Sh | LSW | A |
| Gulf of M. | BOCRG | 50 | 0.54 | 0.502 | 0.044 ns | – | – | – | – | – | S | F | Sh | LSW | A | |
| Gulf of M. | BO100am | 50 | 0.57 | 0.495 | 0.072 ns | – | – | – | – | – | G | F | Sh | LSW | A | |
| Gulf of M. | BO200am | 49 | 0.51 | 0.505 | 0.112 ns | – | – | – | – | – | G | F | Sh | LSW | A | |
| Gulf of M. | BoFF | 50 | 0.46 | 0.502 | −0.198 ns | 49 | 0.95 | 0.098 | −0.044 ns | 3 | F2, F4, F5 | S | F | Sh | LSW | A |
| Gulf of M. | BoCAB | 49 | 0.45 | 0.500 | −0.145 ns | 50 | 0.96 | 0.078 | 0.494 ns | – | B | F | Sh | LSW | A | |
| Gulf of M. | PF | 27 | 0.43 | 0.498 | 0.335 ns | 26 | 0.92 | 0.145 | −0.065 ns | 2 | F1, F4 | R | H | E | LSW | A |
| Gulf of M. | IS | 40 | 0.39 | 0.481 | 0.222 ns | 29 | 0.88 | 0.216 | −0.122 ns | 4 | F2, F3, F4, F5 | B | St | E | OW | P |
| South | BdS | 69 | 0.37 | 0.469 | 0.167 ns | 75 | 0.88 | 0.212 | −0.129 ns | 3 | F1, F4, F5 | B | F | Sh | OW | P |
| South | BT | 50 | 0.32 | 0.440 | −0.093 ns | 30 | 0.85 | 0.259 | −0.150 ns | 3 | F5 (2), F11 | R | H | E | OW | P |
| South | FPN | 130 | 0.37 | 0.470 | 0.001 ns | 50 | 0.94 | 0.114 | −0.054 ns | 3 | F1, F4, F12 | S | F | Sh | LSW | P |
| South | BM | 108 | 0.28 | 0.403 | 0.035 ns | 56 | 0.92 | 0.149 | −0.079 ns | 2 | F5, F8 | R | H | E | OW | P |
| West | PCu | 36 | 0.4 | 0.488 | −0.083 ns | 35 | 0.96 | 0.083 | −0.031 ns | 16 | F1 (3), F2 (3), F3 (2), F4 (1), F5 (3), F6, F7, F12 (2) | R | H | E | OW | P |
N, sample size; Hn.b., nonbiased estimate of genetic diversity; f, Weir and Cockerham's (1984) estimate of Wright's FIS; *, significant values at the level of 5%; ns, non-significant. Substrate: rock (R), blocks (B), gravels (G), sand (S). Slope: flat (F), steep (St), hangover (H). Wave exposure: sheltered (Sh), exposed (E). Salinity: oceanic water (OW), low-salinity water (LSW). Macrocystis: presence (P)/absence (A).
We detected no polymorphism at loci EFbis and EFprem's scored on agarose gels. At locus Glu-5′, G and E allelic frequencies were of 41.6 and 58.4% in the total sample. In the Gulf of Morbihan, the G allele occurred in higher frequencies, sometimes over 50% (samples PR1, IGn, Ar1, Ar2, BoCRD, Bo100am, Bo200am, BoCentre; Table3, Fig.3). At mac-1, the allele a had a frequency of 91.9% in the total sample. Allele c4 had the lowest frequency in all samples (from 4 to 19.7%; Table3, Fig.3). Average Hn.b. values were of 0.453 and 0.152, respectively, for Glu-5′ and mac-1. FIS values at the two nuclear loci were generally nonsignificantly different from 0 (Table3), except for the sample IH (northern part of the Gulf) which shows a heterozygote excess (FIS = −0. 186; P = 0.0362), but this significance level did not pass, by far, the correction for multiple tests. At locus COI, 16 haplotypes were found, haplotype diversity was about Hn.b. = 0.86 within region (it was not computed within population due to a small sample sizes). Neutrality tests (Tajima's D and Fu's Fs) within each region were nonsignificant, and the haplotype network appeared balanced (Fig.4).
Figure 3.
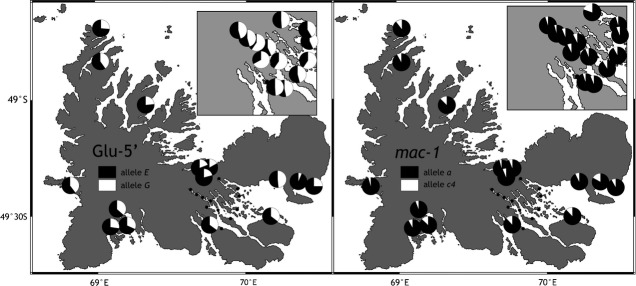
Kerguelen blue mussels. Geographic distribution of allele frequencies at the nuclear loci Glu-5′ and mac-1. Insets: samples from the fjords and islands in the western part of the Gulf of Morbihan.
Figure 4.
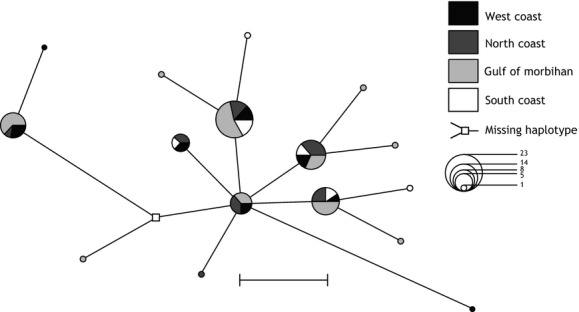
Median-joining parsimony network of COI haplotypes sampled in Kerguelen blue mussels. Scale bar represents one mutational step.
Differentiation among populations
Overall FST values at both nuclear DNA loci were significant (Glu-5′: FST = 0.0627 ± 0.0176, P ≤ 0.0001; mac-1: FST = 0.00945 ± 0.00592; P ≤ 0.004) establishing the presence of significant genetic structure for these loci in Kerguelen. By contrast, the exact test of global differentiation at the COI locus did not appear significant (P = 0.59856 ± 0.08481), and the single non-negative estimate of FST value among regions (pooling individuals from different populations) was 0.007 (nonsignificant).
At Glu-5′, pairwise FST values revealed a differentiation between samples depending on the region to which they belong. Three groups of genetically differentiated samples may be identified (Table1 and 3): (i) the northern group presented the highest frequencies of the E allele, (ii) the Gulf group presented the highest frequencies of the G allele; (iii) the southern group was intermediate. The unique sample from the west coast (PCu) was less differentiated from the southern group than from the northern group and the Gulf. At locus Glu-5′ after the FDR correction, only P-values lower than 0.021 subsisted, mainly those concerning pairwise FST between samples from the north coast and the Gulf of Morbihan. At locus mac-1, fewer pairwise FST values were significant (Table4). However, the samples RdA (East Coast) and PR2 (Gulf) are significantly differentiated from the majority of the other samples, due to the high frequency of allele c4 (17 and 19.4%, respectively). Only PR2 remained differentiated from the remaining samples after FDR correction.
AMOVAs among regions
The differentiation between the three groups of samples (1: north + east coasts, 2: Gulf, 3: south + west coasts) was significant at Glu-5′ (FCT: 0.0773; P < 0.000001), but neither at locus mac-1 (FCT: −0.0003; P = 0.394), nor at COI. (FCT: −0.01244, P = 0.67).
Within-region differentiation
Genetic structure was also evidenced within the three groups of samples genetically differentiated at Glu-5′ (North, Gulf, South). In the northern group, sample AJ was significantly differentiated to all other northern samples (due to its higher frequency of allele G at Glu-5′). In this group, the sample RdA was also differentiated from all northern samples (except I3B) and had the highest frequency of allele E of the whole data set. In the Gulf group, the sample Ar1 was differentiated from all samples of the Gulf except BoCentre, Bo100am, and IGn. Consequently, a significant differentiation was highlighted between samples separated by no more than 500 m: Ar1 and Ar2. At locus mac-1, similarly, samples PR1 and PR2 appeared differentiated but only before FDR correction (Fig.1, Table4).
At the scale of the archipelago, no correlation was detected between genetic differentiation and genetic distance, at any locus. Except along the north coast, at locus Glu-5′, samples PCh to RdA (from northwest to east) (P ≤ 0.04).
Confidence intervals of differentiations and power analyses
Jost's D values were computed for a set of seven populations from the North, Gulf, and the South regions, for Glu-5′, mac-1 and nine allozymes. The maximum value (D = 0.049) was obtained at Glu-5′, and its confidence interval only overlapped that of the enzyme PGD (D = 0.032) which also appeared particularly differentiated (Table2). FST values were all lower than those at Glu-5′. Additionally, whereas Jost's D confidence intervals of all loci except Glu-5′ and PGD included 0.000, the P-values of the exact tests of overall differentiation were generally significant or highly significant (Table S2).
For mac-1 and COI, we used POWSIM to simulate three populations with sample sizes and global allele frequencies corresponding to the three regions for these markers with an FST of 0.07 because Glu-5′ displayed an overall FST of 0.067 in Kerguelen and a FCT of 0.077 in the AMOVA with regional groups. This value of FST was obtained by simulating a fission of three populations of Ne = 1000 each, that occurred 145 generations ago, parameters which allowed maintaining the observed polymorphism. Sample sizes corresponded to samples sizes for mac-1 and COI in each region: 181, 725, 195 and 27, 19, 37, respectively. The allele frequencies used were 0.919 and 0.081 for mac-1, and 0.012, 0.012, 0.012, 0.012, 0.012, 0.012, 0.012, 0.012, 0.012, 0.012, 0.100, 0.144, 0.170, 0.060, 0.130, and 0.276, for COI simulations.
POWSIM simulations indicated that an overall FST of 0.07 would generate significant differences in more than 99.5% of the cases for the COI data set and in more than 92.9% of the cases for the mac-1 data set. When there is polymorphism within populations the maximum value of fixation indices such as FST does not reach one even when no allele is shared among populations (Jost 2008; Meirmans and Hedrick 2011). Glu-5′ and mac-1 both have only two alleles, thus when comparing only two populations, FST values can in theory reach one and the level of intrapopulation diversity should not affect the range of possible FST values. For COI, however, the maximum possible value of FST was higher than at Glu-5′, thus a given FST value corresponds to less differentiation in Glu-5′ than in COI (Jost 2008; Meirmans and Hedrick 2011). Thus, the value of 99.5% of significant FST given by POWSIM for COI is an overestimate.
Statistical analyses with environmental variables
For each environmental variable, the genetic differentiation between samples grouped by category was assessed by pairwise FST for each nuclear marker (Table5). None was significant at mac-1, but many were significant at Glu-5′. For the variable “Substrate”, samples collected on rocks, blocks, gravels, or sand were not significantly differentiated between each other. Regarding “Slope”, only samples collected on flat shores and hangovers were differentiated (P ≤ 0.0001) but steep shore samples appeared significantly differentiated neither from flat nor from hangover locations. For the three remaining factors “Macrocystis”, “Wave exposure” and “Salinity” the samples grouped by category were highly differentiated (P ≤ 0.0001). Concerning the variable “Region”, the single western sample was differentiated from northern samples only (P ≤ 0.004).
Table 5.
Values of FST (Weir and Cockerham 1984), FCT (AMOVA) calculated between samples of Kerguelen blue mussels grouped by categories for each environmental variable at the three loci considering either the whole archipelago or the Gulf of Morbihan. Bold values are significant after FDR correction for multiple tests per column
| Variable | Locus | |||||
|---|---|---|---|---|---|---|
| Glu-5′ | mac-1 | COI | ||||
| Archipelago (35 pop.) | Gulf (22 pop.) | Archipelago (29 pop.) | Gulf (16 pop.) | Archipelago (23 pop.) | Gulf (12 pop.) | |
| Wave | FST: 0.0108 | FST: 0.1315 | FST: NS | FST: NS | – | – |
| Exposure | FCT: 0.0064 | FCT: 0.0123 | FCT: 0.0007 | FCT: −0.0023 | FCT: −0.0036 | FCT: −0.005 |
| Macro-cystis | FST: 0.0223 | FST: 0.0096 | FST: NS | FST: NS | – | – |
| FCT: 0.0239 | FCT: 0.0125 | FCT: −0.0013 | FCT: −0.0025 | FCT: −0.0196 | FCT: −0.0168 | |
| Salinity | FST: 0.0124 | FST: 0.0039 | FST: NS | FST: NS | – | – |
| FCT: 0.0092 | FCT: 0.0033 | FCT: −0.0007 | FCT: −0.0023 | FCT: 0.0272 | FCT: 0.0135 | |
| Slope | FST: NS | FST: NS | FST: NS | FST: NS | – | – |
| FCT: 0.0107 | FCT: 0.0075 | FCT: −0.0021 | FCT: −0.0048 | FCT: −0.0222 | FCT: −0.0114 | |
| Substrate | FST: NS | FST: NS | FST: NS | FST: NS | – | – |
| FCT: −0.0103 | FCT: −0.0025 | FCT: 0.0009 | FCT: 0.0168 | FCT: −0.0349 | FCT: −0.0148 | |
At Glu-5′, AMOVAs realized at the scale of the archipelago for each environmental variable, grouping the 35 samples by categories (Table5) revealed a significant differentiation between the presence and absence of Macrocystis (FCT: 0.0239, P = 0.00684). The groupings were not significantly differentiated, neither for the other factors (at Glu-5′), nor at mac-1 and COI loci. As we tested five environmental variables, the P-values should be corrected taking into account multiple tests. The effect of “Macrocystis” presence remains significant after correction for multiple tests.
Environmental variables “Substrate”, “Wave exposure”, “Slope”, Salinity” and “Macrocystis” were significantly correlated to one another, except the pair “Substrate/Macrocystis”. The frequency of allele G was correlated to “Macrocystis” and “Region” only, whereas the variable “Region” was correlated to none of the other variables (Table6).
Table 6.
Correlation among environmental variables and frequency of allele G at locus Glu-5′ in Kerguelen blue mussels: Spearman's ρ values and levels of significance
| Variable | Variable | |||||
|---|---|---|---|---|---|---|
| Substrate | Slope | Wave exposure | Salinity | Macrocystis | Region | |
| G | 0.0629 | 0.3130 | −0.2487 | 0.2075 | 0.4036* | 0.3785* |
| Substrate | −0.6829*** | 0.5281** | −0.4322* | −0.2192 | 0.2233 | |
| Slope | −0.7176*** | 0.4929** | 0.5210** | 0.0206 | ||
| Wave exposure | −0.4012* | −0.5617** | 0.1474 | |||
| Salinity | 0.6305*** | 0.1025 | ||||
| Macrocystis | −0.0708 | |||||
*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
AMOVAs by environmental variables, restricted to the 22 Gulf samples revealed a significant effect of “Wave Exposure” (accounting for 1.23% of the molecular variance between groups; P = 0.0088) and also of the presence of “Macrocystis” (FCT = 0.0125, P = 0.0489), but they did not overcome the FDR correction for five tests. The presence of Macrocystis in a population was correlated with wave exposure in the Gulf, as most populations where Macrocystis occur, are exposed to waves (except PR2 which is sheltered). The effect of slope was nearly significant (P = 0.058 ± 0.007). At locus mac-1, samples from the Gulf were differentiated (FCT = 0.0168, P = 0.0289) only when grouped by “Substrate” category but this significance level did not pass the FDR correction. In the regions North (eight samples) and South (four samples), the environmental groupings did not reveal any significant differentiation at Glu-5′ or mac-1. At locus COI, none of the environmental grouping of samples was significant.
Discussion
Polymorphism at the three loci and possible departures from neutral expectations
The locus Glu-5′ has traditionally been considered as diagnostic between smooth-shell Mytilus species in the Northern Hemisphere (Inoue et al. 1995; Rawson et al. 1996; Borsa et al. 1999; Daguin and Borsa 2000; Daguin et al. 2001; Luttikhuizen et al. 2002; Gilg and Hilbish 2003a,b; Hilbish et al. 2003), although low frequencies of heterospecific alleles have been reported (Hamer et al. 2012). In Kerguelen, Glu-5′ is polymorphic for heterospecific alleles and at Hardy–Weinberg equilibrium, which was unexpected in a genetic context other than the M. edulis/M. galloprovincialis hybrid zone in the Northern Hemisphere (Borsa et al. 2007). Mitochondria of Kerguelen blue mussels belong to the S1 clade which is endemic to the Southern Ocean (Gérard et al. 2008). The Kerguelen archipelago thus shelters the only wild and stable population (i.e., outside a hybrid zone) of Mytilus known so far, whose polymorphism at Glu-5′ is not in linkage disequilibrium with any of the typical genomes of northern M. edulis, M. galloprovincialis, or M. trossulus.
Unexpected genetic structure was here revealed at Glu-5′ not only at the scale of the archipelago, but also at a much smaller geographic scale, down to a few hundred meters. There was a clear break in allelic frequency at Glu-5′ between samples from the Gulf and the north coast. The highest frequencies of the allele G occurred in the western part of the Gulf, far from the influence of outer marine waters, and reached 60% near Mayes Island (Fig.1) (Daguin 2000; Borsa et al. 2007).
Some of COI haplotypes in Kerguelen blue mussels also occur in southern South America (Gérard et al. 2008). Here, we confirm the homogeneity of COI haplotype frequencies across the four regions of the archipelago. The shape of the haplotype network is compatible with a stable effective size of Kerguelen blue mussel population and with selective neutrality at this locus.
To summarize, in Kerguelen the polymorphism at Glu-5′ is higher than everywhere else, whereas the polymorphism at all other nuclear loci tested (mac-1, EFbis and EFprem's) is lower (Bierne et al. 2002b; this study). The haplotype diversity at the mitochondrial locus COI is also lower in Kerguelen than in Patagonia (Gérard et al. 2008), and allozyme loci are also less polymorphic in Kerguelen than in Northern-Hemisphere populations of M. edulis (Blot et al. 1988). The smaller size of the Kerguelen metapopulation, compared to other less isolated populations worldwide, may explain its lower polymorphism (except at locus Glu-5′). Local adaptation appears as a plausible cause for the maintenance of alleles at balanced frequencies at Glu-5′ in the heterogeneous environment of the Kerguelen archipelago.
The three markers revealed distinct patterns of differentiation between samples
The level of differentiation is much higher at Glu-5′ than at mac-1, COI and eight allozyme loci out of nine. One can hypothesize that allele differences at locus mac-1 may have escaped detection because of the low resolution of agarose gels and that the power to detect possible differences at locus CO1 may have been hampered by insufficient sample sizes. However, these hypotheses were ruled out by analyses of Jost's D and their confidence intervals, as well as the POWSIM analyses suggesting that Glu-5′ was subjected to different constraints.
Thus, Glu-5′ actually reveals highly significant genetic differentiation at all levels, among and within-region, and between environments. Three possible explanations arise: (i) the power analyses might be unreliable, because POWSIM uses a model of fission which may not well represent the actual situation (but Jost's D and confidence intervals are not subject to such doubts); (ii) larvae may preferentially settle (by habitat choice) in certain environments according to their genotype at Glu-5′ or other physically linked loci; (iii) mortality or fecundity may vary among locations according to genotype at Glu-5′ or physically linked genes (i.e., differential selection). Marine species may be subjected to high variance of reproductive success (Hedgecock's sweepstake reproduction hypothesis) which together with collective dispersal of related individuals can generate complex patterns of genetic structure known as chaotic genetic patchiness (Broquet et al. 2013). A skewed offspring distribution also generates departure from the standard Kingman's coalescent and an increased heterogeneity in differentiation levels (Eldon and Wakeley 2009).
Glu-5′ therefore seems to be an outlier displaying particularly high genetic differentiation among Kerguelen populations. However, this observation alone is not sufficient to support hypotheses of natural selection. We will thus use an additional prediction that is not well explained by purely neutral processes which is an association between genetic differentiation and environmental distance (Coop et al. 2010).
Geographic pattern of genetic differentiation associated to ocean circulation
Patterns of genetic differentiation among Kerguelen blue mussels from different groups (North + East, South + West, and Gulf of Morbihan) similar to those here revealed at locus Glu-5′ have been previously reported at allozyme loci (Blot et al. 1989).
After the FDR correction for multiple tests, the most significant differentiations were observed between the north coast and the Gulf. Indeed, the frontier between these regions displays the strongest break of allelic frequencies at Glu-5′, located between samples RdA and PAF. This sample RdA is also differentiated from all others at mac-1, suggesting restricted gene flow toward the most eastern point of Kerguelen. As did Blot et al. (1989), we relate the restricted gene flow to the hydrology and water masses circulation around the archipelago (Murail et al. 1977). All samples are located in the ‘Coastal Hydrological Region’ which has the most changing physical parameters even at fine scale and globally a lower salinity compared to offshore oceanic waters. However, at wider scale, the south coast and northern point of the archipelago receive the same water mass coming from the west (the ACC), but they remain isolated, thus driving to a genetic differentiation among samples from these two regions. The water masses flowing along the north and south coasts only mix far offshore in the northeastern wake zone of the archipelago (Murail et al. 1977). The presence of eddies retaining larvae on the shelf and then dragging them from a site to another on relatively short distance may explain the pattern of isolation by distance observed along the north coast at Glu-5′. Koubbi et al. (2000) have suggested that Lepidonothoten squamifrons larvae are retained by a costal gyre in the Golfe des Baleiniers (the open area off Port Couvreux (PCx), north coast) and also noted the lability of this gyre and the consequent mixing of coastal and oceanic waters during the winter period when winds are the strongest (Razouls et al. 1996; Koubbi et al. 2000). Thus, at the inter-regional scale, hydrological characteristics are able to account for the main genetic differentiation observed, by their effect on migration (i.e., without necessity to invoke selection).
Very fine scale differentiation does not support selective neutrality
In the Gulf, a particular enclosure, genetic differentiations at very fine scale were observed: between samples from the Armor locality, Ar1 and Ar2, which are separated by very short distances (500 m) considering the dispersal potential attributed to the Mytilus mussels. No such differentiation is observed at the locus mac-1. In Armor (Ar), a marked difference in habitat occurs between samples (1 and 2). Ar1 is located near an important freshwater source, where Macrocystis are lacking, and has higher frequency in allele G than its neighbor Ar2. Out of the Gulf, in FPN a comparable habitat (Fjord with freshwater source), we also observed the same trend: a higher frequency of allele G compared to other south coast samples (see Table3). This trend suggests the influence of these protected, low-salinity, sandy habitat on the blue mussels that is expressed by a higher frequency of allele G. However, a third sample with comparable habitat shows the opposite trend: RdA has the lowest frequency of allele G of the whole data set (9%; see Table3).
Genetic differentiation caused by selective pressure from environment?
At Glu-5′, at the scale of the archipelago, the differentiation between groups and between categories of samples with and without Macrocystis, were significant. Typical habitats of protected areas with flat sandy bottoms and low-salinity waters, which are more frequent in the Gulf of Morbihan, lack Macrocystis kelp beds. Conversely, the open coasts are mostly exposed rocky shores, bordered by Macrocystis beds. Consequently, searching for differentiation between samples from the Gulf and those from the south and north coasts, leads to searching the differentiation between samples located in habitats, respectively, without and with Macrocystis kelp beds. Finally, the genetic differentiation among the three main geographic regions may mask the environmental effect (or the reciprocal) on the genetic data. Then, analyzing environmental effect within group would avoid the ‘regions’ effect. At the within-group scale, the results were distinct, mainly due to the contrasting samplings. More precisely, the absence of significant effect of all environmental factors on Glu-5′ data in the north and south Coasts may be due to the low number of samples (8 and 5, respectively) and/or a lower power of Glu-5′ in these regions compared to the Gulf. In the Gulf, the Hn.b. is the highest and both alleles have similar frequencies, thus allowing better detection of small differences. Indeed, within the Gulf, the substantial effect of presence/absence of Macrocystis beds on the sample differentiation was recovered, and the effect of the wave exposure was also revealed (see AMOVAs results). A significant result after the FDR correction cannot be considered an artifact of the number of AMOVAs that were carried out. The environmental effects found by the AMOVAs (even within the Gulf of Morbihan) do not necessarily reflect habitat choice or differential selection linked to Glu-5′ genotypes: geographically close populations tend to share environmental characteristics even within region (for instance, the numerous samples from the Henri Bossière Fjord are all similar) thus if there is fine scale structure due to any other factor, by indirect correlation, a statistical effect of environment may arise even in the absence of causal relationship.
To conclude, three independent lines of evidence suggest that Glu-5′ is affected by selection (or habitat choice): (i) the high polymorphism at this locus in Kerguelen, (ii) highest and more significant FST and FCT at Glu-5′ compared to other loci, and (iii) the significant effects of environmental factors on AMOVAs even within region. However, none is a sufficient proof of selection by itself.
Acknowledgments
We thank the French Polar Institute (IPEV) for the logistic support. Sampling was realized during scientific program IPEV-MACROBENTHOS n° 195 (1999-2003) by the technical volunteers (VATs) from IPEV missions 49–53 in Kerguelen, with a special thanks to Thomas Abiven. We are grateful to Erwan Bronnec for the brave navigation through the Fjord des Portes Noires, to Denis, Jean-Marc and the whole crew of ‘La Curieuse’ ship for great sampling moments in open waters of the archipelago sometimes during very bad weather, and finally to all helping peoples from the 52nd, 53rd missions in Kerguelen. Abigail E Cahill corrected the English of newly written parts of the revised version.
Conflict of interest
None.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1.Molecular markers: locus name, source, primer sequence, annealing temperature (T°C) and fragment length (L) in base pairs.
Table S2. Kerguelen blue mussels.
References
- Arnaud P. Les moulières à Mytilus et Aulacomya des îles Kerguelen (sud de l'Océan Indien). Les “moulières de seuil” et leur intérêt possible pour l'aquaculture des pélécypodes. C. R. Acad. Sci. III. 1971;272:1423–1425. [Google Scholar]
- Arnaud PM. Contribution à la bionomie marine benthique des régions antarctiques et subantarctiques. Théthys. 1974;6:465–656. [Google Scholar]
- Bandelt H-J, Forster P. Röhl A. Median-Joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barton NH. Hewitt GM. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N. Bonhomme F. 2000. and Genetix version 4.02, logiciel sous Windows™ pour la génétique des populations. CNRS UMR 5000, Université Montpellier 2, Montpellier, 2000. Available at: http://www.genetix.univ-montp2.fr/genetix/
- Benjamini Y. Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Bierne N, David P, Boudry P. Bonhomme F. Assortative fertilization and selection at larval stage in the mussels Mytilus edulis and M. galloprovincialis. Evolution. 2002a;56:292–298. doi: 10.1111/j.0014-3820.2002.tb01339.x. [DOI] [PubMed] [Google Scholar]
- Bierne N, David P, Langlade A. Bonhomme F. Can habitat specialisation maintain a mosaic hybrid zone in marine bivalves ? Mar. Ecol. Prog. Ser. 2002b;245:157–170. [Google Scholar]
- Bierne N, Borsa P, Daguin C, et al. Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol. Ecol. 2003;12:447–461. doi: 10.1046/j.1365-294x.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- Blain S, Tréguer P, Belviso S, Bucciarelli E, Denis M, Desabre S, et al. A biogeochemical study of the island mass effect in the context of the iron hypothesis: Kerguelen Islands, Southern Ocean. Deep-Sea Res. II. 2001;48:163–187. [Google Scholar]
- Blot M. 1989. Les populations de Mytilidae de Kerguelen (oce´an austral): ge´ne´tique et adaptation à l'environnement de Mytilus desolationis, Universite´ Paris 7.
- Blot M, Thiriot-Quievreux C. Soyer J. Genetic relationships among population of Mytilus desolationis from Kerguelen, M. edulis from the North Atlantic and M. galloprovincialis from the Mediterranean. Mar. Ecol. Prog. Ser. 1988;44:239–247. [Google Scholar]
- Blot M, Thiriot-Quievreux C. Soyer J. Genetic differences and environments of mussel populations in Kerguelen Islands. Polar Biol. 1989;10:167–174. [Google Scholar]
- Borsa P, Daguin C, Ramos Caetano S. Bonhomme F. Nuclear-DNA evidence that northeasten Atlantic Mytilus trossulus mussels carry M. edulis genes. J. Molluscan Stud. 1999;35:524–527. [Google Scholar]
- Borsa P, Daguin C. Bierne N. Genomic reticulation indicates mixed ancestry in Southern-Hemisphere Mytilus spp. mussels. Biol. J. Linn. Soc. 2007;92:747–754. [Google Scholar]
- Borsa P, Rolland V. Daguin-Thiébaut C. Genetics and taxonomy of Chilean smooth-shelled mussels, Mytilus spp. (Bivalvia: Mytilidae) C. R. Biol. 2012;335:51–61. doi: 10.1016/j.crvi.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Brandt A, Linse K. Mühlenhardt-Siegel U. Biogeography of Crustacea and Mollusca of the Subantarctic and Antarctic regions. Sci. Mar. 1999;63:383–389. [Google Scholar]
- Briggs JC. Oceanic islands, endemism and marine paleotemperatures. Syst. Zool. 1966;15:153–163. [Google Scholar]
- Broquet T, Viard F. Yearsley JM. Genetic drift and collective dispersal can result in chaotic genetic patchiness. Evolution. 2013;67:1660–1675. doi: 10.1111/j.1558-5646.2012.01826.x. [DOI] [PubMed] [Google Scholar]
- Chao A. Shen TJ. 2010. , and Program SPADE (Species Prediction And Diversity Estimation). Program and User's Guide published at http://chao.stat.nthu.edu.tw.
- Coop G, Witonsky D, Di Rienzo A. Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daguin C. 2000. Phylogéographie des moules du complexe d'espèces Mytilus edulis, Université Montpellier II.
- Daguin C. Borsa P. London: Special Publications; 2000. Genetic relationships of Mytilus galloprovincialis Lamarck populations worldwide: evidence for nuclear DNA markers, and. Geological Society. 177:389–397. [Google Scholar]
- Daguin C, Bonhomme F. Borsa P. The zone of sympatry and hybridization of Mytilus edulis and M. galloprovincialis, as described by intron length polymorphism at locus mac-1. Heredity. 2001;86:342–354. doi: 10.1046/j.1365-2540.2001.00832.x. [DOI] [PubMed] [Google Scholar]
- Edgar GJ. Dispersal of fauna and floral propagules associated with drifting Macrocystis pyrifera plants. Mar. Biol. 1987;95:599–610. [Google Scholar]
- Eldon B. Wakeley J. Coalescence times and FST under a skewed offspring distribution among individuals in a population. Genetics. 2009;181:615–629. doi: 10.1534/genetics.108.094342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson BC. Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Mol. Ecol. 2002;11:951–966. doi: 10.1046/j.1365-294x.2002.01507.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P. Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G. Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinf. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R. Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- Frenot Y, Gloaguen JC, Masse L. Lebouvier M. Human activities, ecosystem disturbance and plant invasions in subantarctic Crozet, Kerguelen and Amsterdam Islands. Biol. Conserv. 2001;101:33–50. [Google Scholar]
- Gagnaire P-A, Normandeau E, Côté C, Hansen M. Bernatchez L. The genetic consequences of spatially varying selection in the panmictic American eel (Anguilla rostrata. Genetics. 2012;190:725–736. doi: 10.1534/genetics.111.134825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JPA. The Mytilus edulis species complex in southwest England: effects of hybridization and introgression upon interlocus associations and morphometric variation. Mar. Biol. 1996;125:385–399. [Google Scholar]
- Gardner JPA. Kathiravetpillai G. Biochemical genetic variation at a leucine aminopeptidase (LAP) locus in blue (Mytilus galloprovincialis) and greenshell (Perna canaliculus) mussel populations along a salinity gradient. Mar. Biol. 1997;128:619–625. [Google Scholar]
- Gardner JPA. Palmer NL. Size dependant, spatial and temporal genetic variation at a leucine aminopeptidase (LAP) locus among blue mussel (Mytilus galloprovincialis. Mar. Biol. 1998;132:275–281. [Google Scholar]
- Gérard K, Bierne N, Borsa P, Chenuil A. Féral J-P. Pleistocene separation of mitochondrial lineages of Mytilus spp. mussels from Northern and Southern Hemispheres and strong genetic differentiation among southern populations. Mol. Phylogenet. Evol. 2008;49:84–91. doi: 10.1016/j.ympev.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Gilg M. Hilbish TJ. Patterns of larval dispersal and their effect on the maintenance of a blue mussel hybrid zone in Southwestern England. Evolution. 2003a;57:1061–1077. doi: 10.1111/j.0014-3820.2003.tb00316.x. [DOI] [PubMed] [Google Scholar]
- Gilg MR. Hilbish TJ. The geography of marine larval dispersal: coupling genetics with fine-scale physical oceanography. Ecology. 2003b;84:2989–2998. [Google Scholar]
- Hamer B, Korlevic M, Durmisi E, Nerlovic V. Bierne N. Nuclear marker Me15-Me16 analyses of Mytilus galloprovincialis populations along the eastern Adriatic coast. Cah. Biol. Mar. 2012;53:35–44. [Google Scholar]
- Hedgecock D. Temporal and spatial genetic structure of marine animal populations in the California Current. CalCOFl Rep. 1994;35:75–81. [Google Scholar]
- Hedgecock D. Pudovkin AI. Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 2011;87:971–1002. [Google Scholar]
- Hennion F. Walton DWH. Seed germination of endemic species from Kerguelen phytogeographic zone. Polar Biol. 1997;17:180–187. [Google Scholar]
- Hilbish TJ. Koehn RK. The physiological basis of selection at the LAP locus. Evolution. 1985a;39:1302–1317. doi: 10.1111/j.1558-5646.1985.tb05696.x. [DOI] [PubMed] [Google Scholar]
- Hilbish TJ. Koehn RK. Exclusion of the role of secondary contact in an allele frequency cline in the mussel Mytilus edulis. Evolution. 1985b;39:432–443. doi: 10.1111/j.1558-5646.1985.tb05679.x. [DOI] [PubMed] [Google Scholar]
- Hilbish TJ, Deaton LE. Koehn RK. Effect of an allozyme polymorphism on regulation of cell volume. Nature. 1982;298:688–689. doi: 10.1038/298688a0. [DOI] [PubMed] [Google Scholar]
- Hilbish TJ, Mullinax A, Dolven SI, et al. Origin of the antitropical distribution pattern in the marine mussels (Mytilus spp.): routes and timing of transequatorial migration. Mar. Biol. 2000;136:69–77. [Google Scholar]
- Hilbish TJ, Timmons J, Agrawal V, Schneider KR. Gilg MR. Estuarine habitats protect hybrids mussels from selection. J. Exp. Mar. Biol. Ecol. 2003;292:177–186. [Google Scholar]
- Inoue K, Waite JH, Matsuoka M, Odo S. Harayama S. Interspecific variations in adhesive protein sequences of Mytilus edulis, M. galloprovincialis, and M. trossulus. Biol. Bull. 1995;189:370–375. doi: 10.2307/1542155. [DOI] [PubMed] [Google Scholar]
- Johnson MS. Black R. Pattern beneath the chaos: the effect of recruitment on genetic patchiness in an intertidal limpet. Evolution. 1984;38:1371–1383. doi: 10.1111/j.1558-5646.1984.tb05658.x. [DOI] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol. Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ. Ebert D. Conceptual issues in local adaptation. Ecol. Lett. 2004;7:1225–1241. [Google Scholar]
- Koehn RK. Biochemical aspects of genetic variation at the LAP locus in Mytilus edulis. In: Beardmore JA, editor; Battaglia B, editor. Marine Organisms. New York and London: Plenum Press; 1978. [Google Scholar]
- Koubbi P, Duhamel G. Hebert C. Role of bay, fjord and seamount on the early life history of Lepidonotothen squamifrons from the Kerguelen Islands. Polar Biol. 2000;23:459–465. [Google Scholar]
- Launey S, Ledu C, Boudry P, Bonhomme F. Naciri-Graven Y. Geographic structure in the European Flat Oyster (Ostrea edulis L.) as revealed by microsatellite polymorphism. J. Hered. 2002;93:331–338. doi: 10.1093/jhered/93.5.331. [DOI] [PubMed] [Google Scholar]
- Luttikhuizen PC, Koolhas A, Bol A. Piersma T. Mytilus galloprovincialis-type foot-protein-1 alleles occur at low frequency among mussels in the Dutch Wadden sea. J. Sea Res. 2002;48:241–245. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Maynard Smith J. Sympatric speciation. Am. Nat. 1966;100:637–650. [Google Scholar]
- McDonald JH, Seed R. Koehn RK. Allozymes and morphometric characters of three species of Mytilus in the Northern and Southern Hemispheres. Mar. Biol. 1991;111:323–333. [Google Scholar]
- McDowall RM. Oceanic Islands and Endemism. Syst. Zool. 1968;17:346–350. [Google Scholar]
- Meirmans PG. Hedrick PW. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Murail JF, David P. Panouse M. Hydrologie du plateau continental des îles Kerguelen. CNFRA. 1977;42:41–64. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohresser M, Borsa P. Delsert C. Intron-length polymorphism at the actin gene locus mac-1: a genetic marker for population studies in the marine mussels Mytulis galloprovincialis Lmk and M. edulis L. Mol. Mar. Biol. Biotech. 1997;6:123–130. [PubMed] [Google Scholar]
- Park YH, Gasco N. Duhamel G. Slope currents around the Kerguelen Islands from demersal longline fishing records. Geophys. Res. Lett. 2008a;35:L09604. , and. doi: 10.1029/2008GL033660. [Google Scholar]
- Park YH, Roquet F, Durand I. Fuda J-L. Large-scale circulation over and around the Northern Kerguelen Plateau. Deep-Sea Res. II. 2008b;55:566–581. [Google Scholar]
- Perrin C, Wing SR. Roy MS. Effects of hydrographic barriers on population genetic structure of the sea star Coscinasterias muricata (Echinodermata, Asteroidea) in the New Zealand fiords. Mol. Ecol. 2004;13:2183–2195. doi: 10.1111/j.1365-294X.2004.02241.x. [DOI] [PubMed] [Google Scholar]
- Poulin E. Féral J-P. Pattern of spatial distribution of a brood-protecting schizasterid echinoid, Abatus cordatus, endemic to the Kerguelen Islands. Mar. Ecol. Prog. Ser. 1995;118:179–186. [Google Scholar]
- Rawson PD, Joyner KL, Meetze K. Hilbish TJ. Evidence for intragenic recombination within a novel genetic marker that distinguishes mussels in the Mytilus edulis species complex. Heredity. 1996;77:599–607. doi: 10.1038/hdy.1996.187. [DOI] [PubMed] [Google Scholar]
- Raymond M. Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Razouls S, Koubbi P. Mayraud P. Spatio-temporal distribution of mesozooplankton in a sub-antarctic coastal basin of the Kerguelen archipelago (Southern Indian Ocean) Polar Biol. 1996;16:581–587. [Google Scholar]
- Ryman N. Palm S. POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Mol. Ecol. 2006;6:600–602. doi: 10.1046/j.0962-1083.2001.01345.x. [DOI] [PubMed] [Google Scholar]
- Sanford E. Kelly MW. Local adaptation in marine invertebrates. Annu. Rev. Mar. Sci. 2011;3:509–535. doi: 10.1146/annurev-marine-120709-142756. [DOI] [PubMed] [Google Scholar]
- Sanjuan A, Zapata C. Alvarez G. Genetic differentiation in Mytilus galloprovincialis Lmk. throughout the world. Ophelia. 1997;47:13–31. [Google Scholar]
- Skibinski DOF. Natural selection in hybrid mussel populations. In: Oxford GS, Rollinson D, editors. Protein polymorphisms adaptive and taxonomic significance. New York, NY: Academic Press; 1983. pp. 283–298. [Google Scholar]
- Skibinski DOF, Ahmad M. Beardmore JA. Genetic evidence for naturally occurring hybrids between Mytilus edulis and Mytilus galloprovincialis. Evolution. 1978;32:354–364. doi: 10.1111/j.1558-5646.1978.tb00651.x. [DOI] [PubMed] [Google Scholar]
- Spearman C. The proof and measurement of association between two things. Am. J. Psychol. 1904;15:72–101. [PubMed] [Google Scholar]
- Väinölä R. Hvilsom MM. Genetic divergence and a hybrid zone between Baltic and North Sea Mytilus populations. Biol. J. Linn. Soc. 1991;43:127–148. [Google Scholar]
- Viard F, Delay B, Coustau C. Renaud F. Evolution of the genetic structure of bivalve cohorts at hybridization sites of the Mytilus edulis-M. galloprovincialis complex. Mar. Biol. 1994;119:535–539. [Google Scholar]
- Waite JH. The formation of mussel byssus: anatomy of a natural manufacturing process. In: Case ST, editor. Structure, cellular synthesis and assembly, of biopolymers. Results and problems in cell differentiation. Berlin: Springer-Verlag; 1992. pp. 27–54. , ed. [DOI] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA. Higushi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- Weir BS. Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams GC. Adaptation and natural selection. Princeton: Princeton Univ. Press; 1966. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.Molecular markers: locus name, source, primer sequence, annealing temperature (T°C) and fragment length (L) in base pairs.
Table S2. Kerguelen blue mussels.


