ADAPT is an ImageJ plug-in that can be used for rapid whole-cell analysis of time-lapse videos, thereby providing data on cell morphology, membrane velocity, and temporal changes in any fluorescent protein of interest at the cell periphery, as exemplified by the morphological characterization of cellular blebs.
Abstract
Cell migration is frequently accompanied by changes in cell morphology (morphodynamics) on a range of spatial and temporal scales. Despite recent advances in imaging techniques, the application of unbiased computational image analysis methods for morphodynamic quantification is rare. For example, manual analysis using kymographs is still commonplace, often caused by lack of access to user-friendly, automated tools. We now describe software designed for the automated quantification of cell migration and morphodynamics. Implemented as a plug-in for the open-source platform, ImageJ, ADAPT is capable of rapid, automated analysis of migration and membrane protrusions, together with associated fluorescently labeled proteins, across multiple cells. We demonstrate the ability of the software by quantifying variations in cell population migration rates on different extracellular matrices. We also show that ADAPT can detect and morphologically profile filopodia. Finally, we have used ADAPT to compile an unbiased description of a “typical” bleb formed at the plasma membrane and quantify the effect of Arp2/3 complex inhibition on bleb retraction.
Introduction
Cell motility is a central process in the maintenance and development of multicellular organisms. For example, highly coordinated cell migration is essential for tissue morphogenesis and wound healing (Ridley et al., 2003). However, motility can also play an important role in disease progression, as in the migration of tumor cells through complex environments to effect metastasis (Sahai and Marshall, 2003). Perhaps the most well-characterized effectors of morphological change and migration are lamellipodia and filopodia, localized protrusions at the cell membrane driven by actin polymerization (Mattila and Lappalainen, 2008; Krause and Gautreau, 2014). Another form of protrusion is the cellular bleb, observed during cytokinesis and amoeboid cell motility, the latter of which has been observed in development and tumor cell invasion (Sahai and Marshall, 2003; Fackler and Grosse, 2008; Paluch and Raz, 2013). These protrusions occur at regions where the plasma membrane separates from the underlying actin cortex or the cortex itself ruptures, driven by increased hydrostatic pressure within the cell (Paluch et al., 2005; Charras and Paluch, 2008; Tinevez et al., 2009). The analysis of cell blebbing has the potential to not only provide insights into the mechanism of bleb retraction, and, therefore, amoeboid cell motility, but also presents an opportunity to interrogate factors involved in the reformation and regulation of the actin cortex. Furthermore, unbiased analysis of bleb morphologies and dynamics can aid the development of mathematical models aimed at furthering our understanding of cell migration in complex environments (Tozluoğlu et al., 2013).
The sophistication of imaging techniques available to cell biologists has increased rapidly in recent years, from advances in digital camera technology to new labeling methods and microscope designs. However, the development of computational algorithms to analyze the vast amounts of data produced is lagging behind (Myers, 2012). The application of automated, unbiased, computational methods for morphodynamic quantification is rare, with the use of kymographs, for example, still popular (Suraneni et al., 2012; Ura et al., 2012; Wiggan et al., 2012; Dang et al., 2013). Such analyses are time consuming, subject to individual bias, and extract relatively low levels of information. Software has been described to enable quantitative analysis of cell dynamics (Dormann et al., 2002; Bosgraaf et al., 2009; Machacek et al., 2009; Biro et al., 2013; Tsygankov et al., 2014), but shortcomings include the requirement for proprietary software, the unavailability of source code, and/or limited functionality (Table 1). The need for specialist, proprietary software (such as MATLAB) possibly limits availability to cell biologists, whereas the withholding of source code impedes customization to specific problems, such as the analysis of spatially and temporally localized morphodynamic events. In cases in which such functionality has been incorporated, analysis is restricted to a limited number of features or correlation with temporal changes in protein localization is not possible (Biro et al., 2013; Tsygankov et al., 2014).
Table 1.
Comparison of ADAPT with analysis software described in other publications
| Software | Open source | Proprietary software required | Correlation with fluorescence intensity | Analysis of individual protrusions | References |
| QuimP | No | No | Yes | No | Dormann et al., 2002; Bosgraaf et al., 2009; Bosgraaf and Van Haastert, 2010; Zatulovskiy et al., 2014 |
| KoreTechs | Yes | Yes | Yes | Limited to one | Biro et al., 2013 |
| CellGeo | Yes | Yes | No | Yes | Tsygankov et al., 2014 |
| ADAPT | Yes | No | Yes | Yes |
Here, we present a plug-in for the popular open source platform ImageJ (Schneider et al., 2012), which we believe addresses the shortcomings of other software designed for the analysis of cellular morphodynamics (Table 1). ADAPT (automated detection and analysis of protrusions) permits rapid whole-cell analysis of time-lapse videos, providing data on cell morphology, membrane velocity, and temporal changes in any fluorescent protein of interest at the cell periphery (Videos 1 and 2). Such data facilitate the quantification of, for example, intercellular variations in morphodynamics and protein localization over time (Figs. 1 and 2).
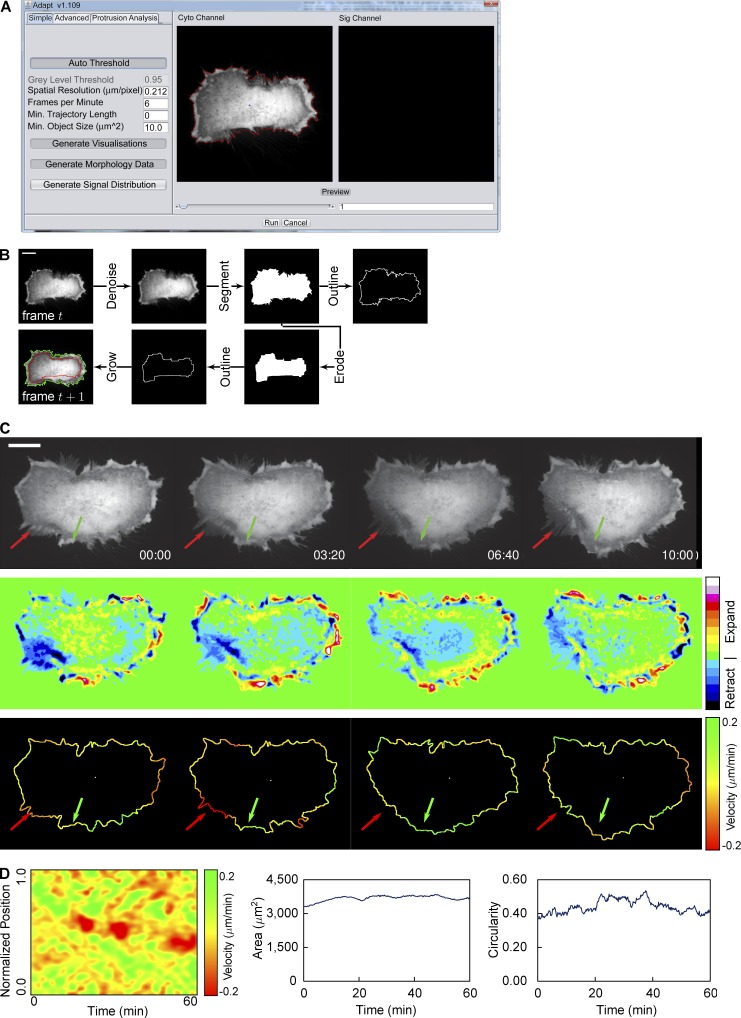
Figure 1.
Overview of whole cell analysis. (A) The ADAPT user interface, illustrating the preview segmentation (red line around a HeLa cell expressing mCherry). (B) Illustration of the segmentation process. Images are Gaussian filtered to suppress noise and then a gray-level threshold is applied to create a binary image. The cell boundary is taken as pixels bordering segmented regions. The resulting segmentation is used as the seed for the region-growing algorithm in the next frame. The red line denotes the eroded segmentation of the cell in frame t, whereas the green line represents the segmentation of the cell in frame t + 1. (C) Velocity is calculated at each point on the cell boundary based on the change in gray level between frames: expansion results in an increase in gray level at a particular spatial coordinate over time and retraction a decrease, as shown in the second row of images. This change in gray level can be used to calculate the membrane velocity at each point, as shown in the bottom row. The green and red arrows indicate regions undergoing expansion and retraction, respectively. (D) Resulting velocity map (left) and plots showing changes in area (center) and circularity (right) over time for a single cell (shown in Video 1). Bars, 20 µM.
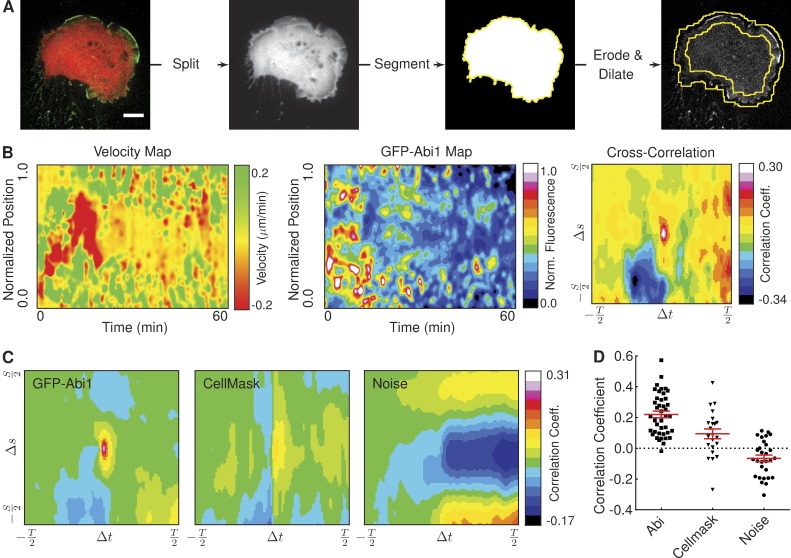
Figure 2.
Correlation of protein recruitment with plasma membrane protrusion velocity. (A) The image shows an HT1080 cell stably expressing GFP-Abi1 and mCherry. The image is split into constituent channels, and the mCherry signal is segmented to construct a cell mask image. Eroded and dilated versions of this mask image are used to construct the region of interest (denoted by the yellow lines) in the GFP-Abi1 image. Bar, 10 µM. (B) Velocity and GFP-Abi1 intensity maps for the cell in A, together with the result of a cross-correlation. (C) Mean cross-correlations of velocity and fluorescence intensity maps reveal a strong correlation between membrane velocity and protein localization in HT1080 cells expressing GFP-Abi1. Correlations between velocity and CellMask or noise were considerably weaker (22 ≥ ncell ≤ 41). (D) Peak values (at Δs = 0 and Δt = 0) in cross-correlations of velocity and fluorescence intensity maps for individual cells (each dot represents a single cell). A one-way analysis of variance (ANOVA) test shows the differences in mean values to be highly significant (P < 0.0001). Error bars represent standard error of the mean.
ADAPT also permits the tracking of cell migration and the automated detection of individual membrane protrusions, outputting data on local membrane velocity, change in protrusion size, and local recruitment of proteins to the cell membrane. We have applied our software to the quantification of cell migration rates on different concentrations of fibronectin. We have also quantified the effect of a formin inhibitor, SMIFH2 (Rizvi et al., 2009), on filopodia formation. Finally, we used ADAPT to characterize vaccinia virus-induced cell blebbing in the early stages of infection. By analyzing videos of blebbing cells stably expressing GFP- or RFP-tagged lifeact (a marker for F-actin), MLC2 (myosin light chain 2), and ezrin, we have compiled an unbiased description of the lifetime of a typical bleb. We also demonstrate the utility of the software in investigating system perturbations by quantifying the reduction in bleb dynamics affected by the Arp2/3 complex inhibitor CK-869 (Nolen et al., 2009).
Results
Quantification of cell morphodynamics
An overview of the ADAPT analysis pipeline for whole-cell analysis is presented in Fig. 1. The first tab on the graphical user interface allows the user to preview the segmentation of their selected dataset (Fig. 1 A). In the example shown, cells were expressing mCherry as a cytosolic volume marker, and so, gray-level segmentation and boundary localization are easily achieved (Fig. 1 B). This preview segmentation is influenced by the choice of input parameters, most significantly by the gray-level threshold, which is either specified by the user (“Auto Threshold” disabled) or calculated automatically (“Auto Threshold” enabled). Segmentation is achieved using a region-growing approach, with the initial seed point either specified by the user or calculated using a Euclidean distance map. An eroded form of this segmented cell is then used to seed the segmentation of the next frame in the dataset (Fig. 1 B). The velocity at each point on the cell boundary may then be estimated using a method similar to that described by Döbereiner et al. (2006; Fig. 1 C). This calculation, based on the concept of optical flow, makes use of the observation that, as a cell extends a protrusion, the gray level in the vicinity of that protrusion increases with time. Conversely, as the protrusion retracts, the gray level decreases. These changes in gray level may be scaled according to the specified spatial and temporal resolution to provide estimates of membrane velocity at every point on the cell boundary for each video frame. Given this velocity data, the construction of a “velocity map” is possible, illustrating membrane velocities for all points on the cell boundary at all time points (Fig. 1 D). Morphological parameters, such as area and circularity, are also calculated for each time point, based on an analysis of the segmented cell boundary (Fig. 1 D). This data may be considered an end result in itself, or it can be used as the basis for a more extensive analysis of protrusions and/or protein localization.
Correlating protein localization with membrane velocity
Correlations can be sought between membrane dynamics and the localization of a fluorescent protein of interest by constructing a “map” of fluorescence intensity around the cell periphery (Fig. 2, A and B). In the example shown, the HT1080 cell is stably expressing GFP-Abi1, a subunit of the WAVE (Wiskott–Aldrich syndrome protein family verprolin homologous proteins) regulatory complex (Chen et al., 2014). The cell has also been transfected with mCherry, to facilitate segmentation. A region of interest is constructed based on this segmentation, within which the intensity of GFP-Abi1 at the cell periphery is analyzed (Fig. 2 A). Performing such an analysis on subsequent frames in a video permits the construction of a map of fluorescence intensities for all points on the cell boundary over time (Fig. 2 B).
Visual inspection of the velocity and GFP-Abi1 maps in Fig. 2 B suggests a relationship between the intensity of GFP-Abi1 at the cell membrane and velocity: green regions in the velocity map tend to coincide with white/red regions in the GFP-Abi1 map, indicating that Abi localizes where lamellipodia extend. This is particularly apparent in the first 20–30 min of the video—it is possible that beyond this point, bleaching of the GFP-Abi1 signal has become significant. Performing a cross-correlation analysis of the two maps confirms this relationship (Fig. 2 B). A correlation coefficient taken in isolation is relatively meaningless. However, extending this analysis to populations of cells can demonstrate whether this correlation is significant relative to negative controls.
To determine whether the localization of Abi at the cell periphery during lamellipodia outgrowth was indeed significant, we applied our cross-correlation analysis to three populations of HT1080 cells. All three were expressing either GFP or mCherry as cytosolic markers. In addition, one population was expressing GFP-Abi1 and another was incubated with CellMask plasma membrane stain (Video 2). The mean cross-correlation maps show a marked difference between correlations of velocity with GFP-Abi1 and noise or CellMask (Fig. 2 C). A comparison of the peak correlation values at the center of individual maps showed a statistically significant difference (Fig. 2 D). Seeking correlations between membrane velocity and fluorescence intensity in this manner represents a powerful, unbiased tool for investigating relationships between protein localization and changes in cell morphology.
Automated quantification of cell migration
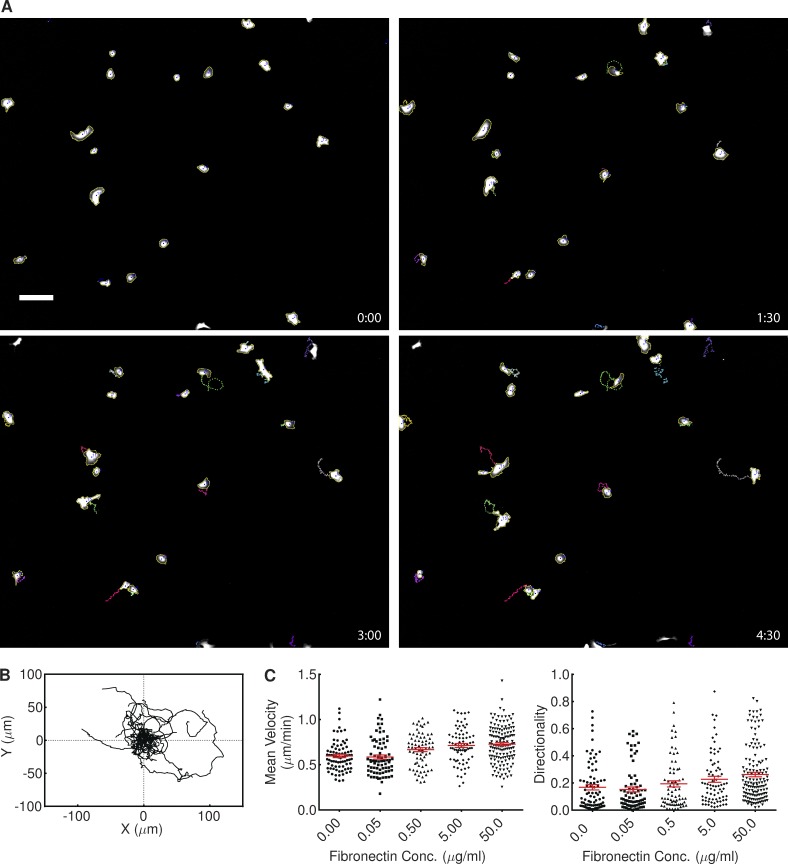
As cell segmentation is based on an iterative erosion and region growing procedure (Fig. 1 B), this naturally lends itself to the tracking of moving cells, provided the rate of cell migration is low relative to the frame rate (Fig. 3). The number of cells that may be tracked is limited only by available memory. The centroid of each cell is calculated in each frame, and this allows the construction of a map of cell trajectories, illustrating the range of migration in a population (Fig. 3 B). We applied such an analysis to populations of HT1080 cells expressing GFP plated on different concentrations of fibronectin. We found that an increase in fibronectin concentration resulted in a statistically significant increase in both mean velocity and the mean directionality of the cells (70 ≤ ncell ≤ 146; Fig. 3 C). The 95% confidence interval of mean velocity increased from 0.560–0.633 µm/min in the absence of fibronectin to 0.695–0.761 µm/min on 50.0 µg/ml fibronectin, representing a statistically significant increase. Directionality also increased, from 0.129–0.206 to 0.229–0.295. This demonstrates the ability of ADAPT to automatically identify, in an unbiased fashion, small but statistically significant differences in cell migration across populations.
Figure 3.
Application of ADAPT to tracking cell migration. (A) Still images of HT1080 cells stably expressing GFP, taken from Video 3. Image dimensions are 1,705 × 1,368 µm, with labels representing elapsed time in hours and minutes. Bar, 150 µM. (B) Trajectories of the cells shown in Video 3, relative to the starting position of each cell (ncell = 20). (C) The influence of fibronectin concentration on the velocity and directionality of HT1080 cells. Plots show the combined data from two independent experiments. A one-way ANOVA test showed fibronectin concentration (Conc.) to have a significant impact on both measures (P < 0.0001). Error bars represent standard error of the mean.
Automated detection and tracking of filopodia
Filopodia are thin cellular protrusions, ∼200 nm in diameter, consisting of 10–30 actin filaments in a bundle (Chhabra and Higgs, 2007). Given their involvement in several cellular processes, including migration, adhesion, and embryonic development (Mattila and Lappalainen, 2008), we have incorporated the ability to automatically detect and track filopodia within ADAPT.
Filopodia are initially identified by segmentation from the cell body. This is achieved by first performing ne erosion operations on a binary representation of a cell, followed by ne dilation operations (Fig. 4 A), in which ne is a number specified by the user. This has the effect of removing any filopodia-like structures from the periphery of the cell, with just the cell “core” remaining. This core can then be subtracted from the original binary image of the cell, resulting in an image consisting solely of filopodia. Automated tracking of the filopodia is then achieved by inputting the sequence of filopodia images that result into the same algorithm used to track cell migration (Videos 4 and 5).
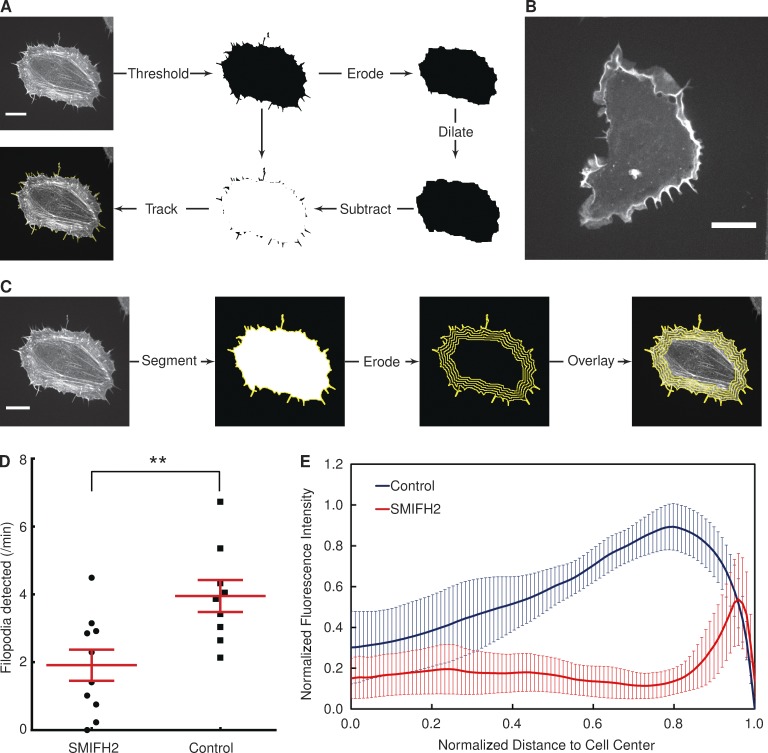
Figure 4.
Influence of formin inhibition on filopodia formation and actin distribution. (A) Illustration of the filopodia detection scheme, with a HeLa cell expressing lifeact-GFP. (B) An example of a HeLa cell expressing lifeact-GFP treated with 40 µM SMIFH2 for 1 h. (C) Using a scheme similar to that shown in Fig. 2 A, the distribution of a fluorescent signal of interest (in this case, lifeact-GFP) can be quantified. The segmented image of a cell is iteratively eroded, and at each step, the fluorescence signal along the image boundary is summed. This permits the plotting of fluorescence intensity as a function of distance to the cell center. (D) The number of filopodia detected per minute by ADAPT in the presence or absence of a formin inhibitor (**, P < 0.01). Each dot represents one cell (ncell = 10 in each population). Error bars represent standard error of the mean. (E) Cellular distribution of actin in control and SMIFH2-treated cells. Error bars represent 95% confidence intervals. Bars, 20 µM.
Using this approach, we tested whether we could detect differences in the number of filopodia produced by two different populations of cells. Given the demonstrated role of formins in filopodia formation (Yang et al., 2007), we used a formin inhibitor, SMIFH2 (Rizvi et al., 2009), to suppress filopodia formation in HeLa cells. An example of a SMIFH2-treated HeLa cell expressing lifeact-GFP is shown in Fig. 4 B. Performing a quantitative analysis of SMIFH2-treated and nontreated cells reveals that the number of filopodia produced per minute is reduced by approximately twofold in the presence of the inhibitor (Fig. 4 D).
Apart from the apparent reduction in the number of filopodia relative to control cells, there is also a striking difference in the distribution of actin within the cell in the presence of SMIFH2, with fluorescence most intense around the cell edge. We have equipped ADAPT with the means to characterize such differences in fluorescence distribution within cells (Fig. 4 C). This is achieved using an approach similar to that illustrated in Fig. 2 A. But, rather than combining an eroded and dilated mask to form a single region of interest, fluorescence distribution is mapped by successively eroding a segmentation and summing the fluorescence intensities along the edge of that segmentation after each erosion iteration (Fig. 4 C). This allows the plotting of total fluorescence intensity against distance to/from the cell center/periphery (Fig. 4 E).
Using these combined approaches, we found that SMIFH2 radically alters actin distribution in HeLa cells. In the presence of the inhibitor, actin is predominantly concentrated in a rim around the cell edge (Fig. 4 E). This is in contrast to the study by Rotty et al. (2015), who reported that SMIFH2 had no effect on the actin distribution in mouse embryonic fibroblasts, although the inhibitor was used at a lower concentration (15 µM), and their analysis was qualitative in nature. Although these data confirm that formins are needed for filopodia formation, it also lends weight to the theory that they play a key role in the formation of stress fibers (Hotulainen and Lappalainen, 2006).
Automated detection and tracking of blebs
Lamellipodial and filopodial protrusions are generally characteristic of two-dimensional cell migration. However, in recent years, as improved imaging technologies have permitted superior observation of three-dimensional environments, there has been increased interest in amoeboid or bleb-based migration (Bergert et al., 2012; Paluch and Raz, 2013; Tozluoğlu et al., 2013; Zatulovskiy et al., 2014). Various mechanisms have been proposed to promote bleb-based motility in complex three-dimensional environments, but experimental evidence is still quite rare (Paluch and Raz, 2013). Cell blebbing is also frequently observed as a viral cytopathic effect. For example, ∼3 h after infection with vaccinia virus, cells begin to transiently contract and bleb (Schramm et al., 2006). We took advantage of this effect and used vaccinia-infected HeLa cells as our model system for the purpose of developing software to automatically detect and track blebs.
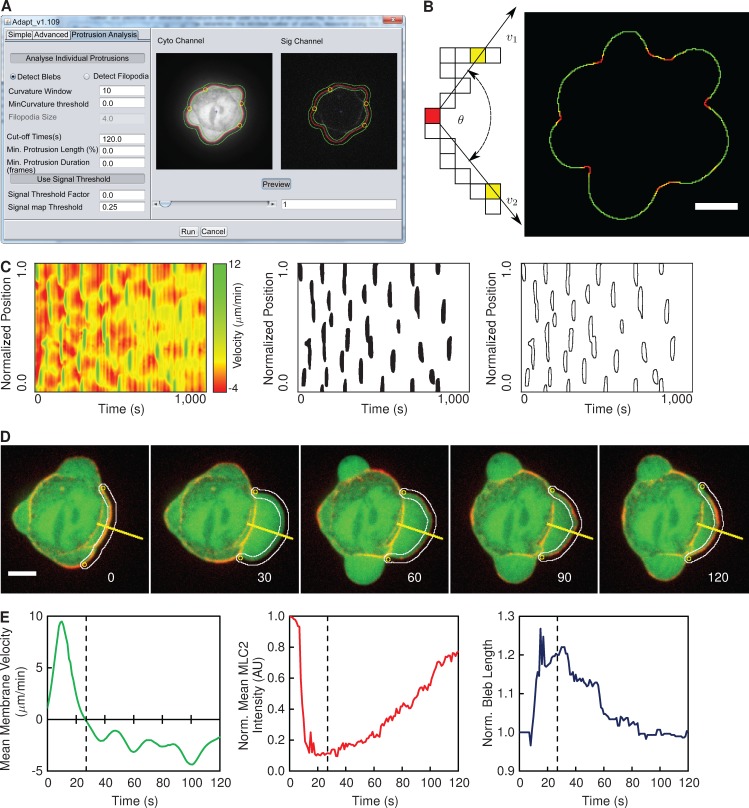
The initial analysis for the detection of blebs is identical to that described in Fig. 1. ADAPT then uses localized changes in cell boundary curvature to track blebs (Fig. 5, A and B), after their initial detection as local maxima in velocity maps (Fig. 5 C). The number and position of detected curvature extrema used to track blebs may be controlled by the user, by varying the values of “Curvature Window” and “Curvature Threshold” in the graphical user interface (Fig. 5 A). The curvature window determines the minimum number of pixels, measured along the cell boundary, between adjacent curvature extremes, whereas the curvature threshold represents the minimum curvature value required for a point to qualify as an anchor point.
Figure 5.
Overview of bleb detection and analysis. (A) The ADAPT user interface, illustrating the preview segmentation of a vaccinia-infected HeLa cell expressing GFP (Cyto Channel) and MLC2-RFP (Sig Channel). The green lines delineate the region over which fluorescence intensities will be quantified. The yellow circles indicate detected curvature extrema. The red line outlines the segmented cell. (B) Curvature is evaluated at each point on the cell boundary as the angle (θ) subtended by vectors (v1 and v2) to two points n pixels away in each direction. When all curvature values are superimposed on the cell boundary, bleb necks are identifiable as sharp concavities (in red) on a generally convex curve (green). (C) Blebs are detected as local maxima in the velocity map. (D) Blebs are tracked using local extrema in curvature as “anchor points.” The yellow lines denote the reference for the kymograph in Fig. 7 A. Image labels show seconds after bleb initiation. The white lines delineate the region over which fluorescence intensities will be quantified. (E) The mean membrane velocity, mean fluorescence intensity of the cortical component of interest (MLC2-RFP), and the length of boundary between the two anchor points for a single bleb (shown in D). The dotted line in the plots indicates the onset of retraction, and image labels correspond to seconds after bleb initiation. AU, arbitrary unit. Bars, 5 µM.
Using these curvature extrema as “anchor points,” each bleb is individually tracked for a user-specified period of time or until a new bleb forms in the same location (Fig. 5 D). In the presence of a second marker, such as MLC2-RFP (Fig. 5 A, Sig Channel), velocity may be related to dynamic changes in protein localization at the membrane. For each frame in which a bleb is monitored, a region of specified cortical width is defined along the cell boundary, within which the fluorescence intensity of a protein of interest (MLC2-RFP, for example) is analyzed (Fig. 5 A). The total fluorescence intensity corresponds to the sum of all pixel values within the defined cortical region above a threshold value. The bleb length corresponds to the number of contour pixels between the anchor points. The velocity values along the contour between the two anchor points are summed and divided by the bleb length to give the mean velocity for that time point. The mean fluorescence intensity is calculated by dividing the total fluorescence intensity by the bleb length. A unique dataset may thus be compiled for each individual bleb, detailing changes in mean membrane velocity, mean fluorescence intensity of a localized protein of interest at the bleb membrane, and the length of the bleb membrane over time (Fig. 5 E).
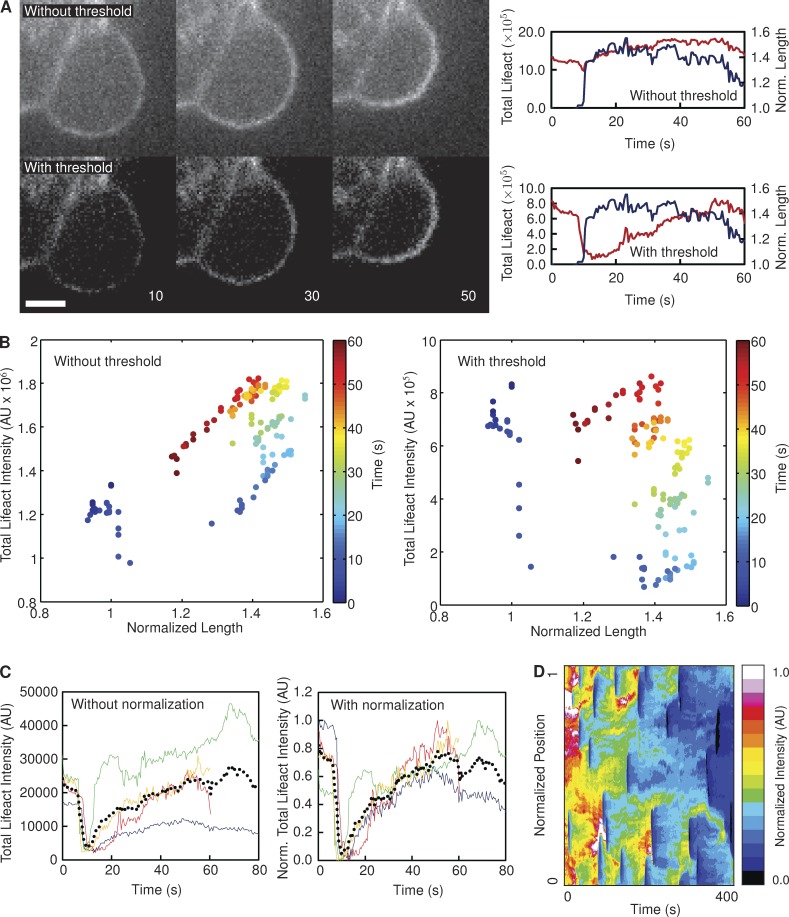
Raw images of fluorescent proteins in live cells often exhibit a low signal-to-noise ratio (Fig. 6 A), particularly when camera exposure times are short. In our analysis of cell blebbing, this can result in the total fluorescence intensity of a protein of interest at the membrane correlating strongly with bleb length (Fig. 6, A and B). This is a consequence of relatively low image contrast, where the background pixel values are similar in magnitude to the pixel values corresponding to the fluorescent protein of interest. As such, the total fluorescence intensity at the bleb membrane (Fig. 6 A, red lines) closely follows the number of pixels summed to calculate the fluorescence value, which is obviously directly proportional to bleb length (Fig. 6 A, blue lines). This correlation is particularly evident when the series of total fluorescence intensities calculated for an individual bleb over its lifetime are plotted directly against the corresponding series of length measurements for that bleb—a strong underlying linear trend is evident (Fig. 6 B).
Figure 6.
Justification for postprocessing of data. (A) Images of a bleb on a HeLa cell expressing lifeact-GFP, with and without the application of a threshold to remove noise. In the absence of a threshold, the total fluorescence intensity (red lines) closely follows the length of the bleb (blue lines). Applying a threshold disrupts this correlation. Image labels correspond to seconds after bleb initiation. Data represent the analysis of a single bleb. Bar, 2.5 µM. (B) The change in total lifeact-GFP intensity along the periphery of a single bleb (shown in A) over time, with (right) and without (left) the use of a threshold value to exclude noise. (C) Each colored line in the plots represents a single bleb on the same cell (nbleb = 4), with (right) and without (left) normalization. The black dotted line represents the mean in each case. (D) The heat map represents the lifeact intensity recorded at every boundary point over the course of a video in one particular cell. It is evident that the signal intensity diminishes over extended time periods. AU, arbitrary unit.
The influence of low signal-to-noise ratio can be alleviated by setting pixels below a certain threshold value to zero. This threshold value should be high enough to exclude most of the noisy pixels but low enough to retain the pixels corresponding to protein localization (Fig. 6 A). An appropriate threshold value disrupts the linear relationship between fluorescence intensity and bleb length and also results in a significant increase in the dynamic range of the measured fluorescent signal (Fig. 6 B). However, even with this noise threshold in place, a large degree of variation in total fluorescence intensity is evident between individual blebs within the same cell (Fig. 6 C). As such, a curve representing the cell mean (Fig. 6 C, dotted lines) is not representative of all the individual blebs. Photobleaching was found to be a major contributor to this intracellular variation. The effect of bleaching is illustrated in Fig. 6 D—it is evident that normalized fluorescence intensity decreases with time. Normalizing each fluorescence profile to its own maximum largely accounted for this variation, permitting the compilation of a representative mean fluorescence profile (Fig. 6 C).
Comparison of ADAPT with manual analysis of blebs
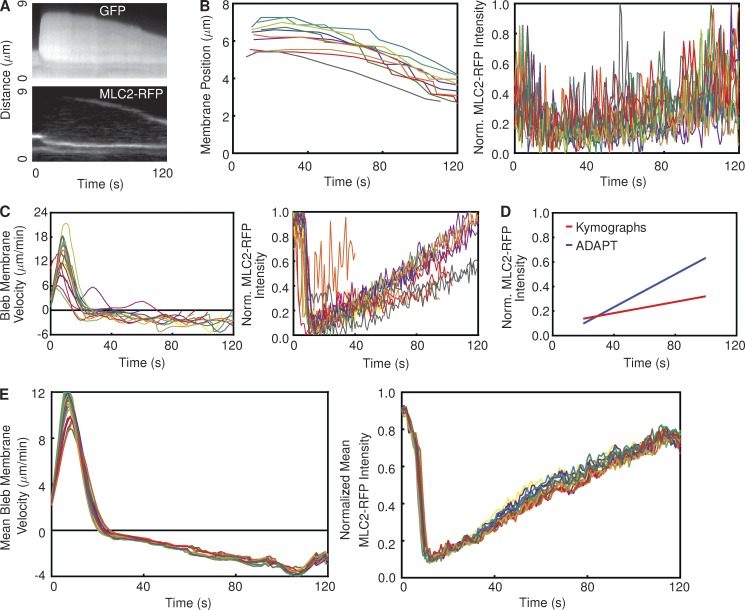
To compare the performance of ADAPT to more frequently used approaches, we manually quantified the mean rates of bleb retraction and MLC2-RFP increase at bleb membranes in a single cell using kymographs (Fig. 7 A). Using cytosolic GFP as the reference for kymograph construction, the mean rate of bleb retraction was calculated as ∼1.7 µm/min (n = 10; Fig. 7 B). The change in MLC2-RFP intensity at the bleb membrane over time was also derived using kymographs (Fig. 7 B). Although a substantial level of noise is evident, we calculated a rate of increase in MLC2-RFP intensity of ∼0.002 s−1.
Figure 7.
Comparison between ADAPT and kymograph analysis of blebs. (A) Kymograph of a bleb on a vaccinia-infected HeLa cell expressing GFP and MLC2-RFP, corresponding to the yellow line in Fig. 5 D. (B) Each line represents the membrane displacement (left) and MLC2-RFP intensity (right) derived from one kymograph (representing one bleb; nbleb = 10). (left) The retraction speed was calculated as 1.74 ± 0.08 µm/min by fitting a straight line to the linear portions of the plots representing membrane position over time. The rate of increase in MLC2-RFP intensity was measured as 2.29 × 10−3 ± 1.74 × 10−4 s−1. (C) Analysis of the same cell using ADAPT. Each line represents the mean membrane velocity (left) and MLC2-RFP intensity (right) over time on a single bleb (nbleb = 10). (D) Comparison of mean MLC2-RFP recruitment rates derived from the data in B and C. The rates of increase are 1.95–2.63 × 10−3 and 6.44–6.97 × 10−3 per second for the kymograph- and ADAPT-derived data, respectively. (E) The results of 20 analyses of the same cell as that in B and C using ADAPT, with a different, random set of input parameters for each analysis.
We repeated the analysis of the same number of blebs on the same cell using ADAPT (Fig. 7 C). The data provided by ADAPT on bleb membrane velocity is not directly comparable with that provided by kymographs. This is because kymographs can only provide a single estimate of retraction velocity for each bleb over its entire lifetime (Fig. 7 B). ADAPT, on the other hand, provides an estimate of bleb membrane velocity, averaged over the entire bleb length, at every time point. The change in MLC2-RFP intensity, as calculated by ADAPT, is in stark contrast to that provided by kymographs: a consistent, linear trend is evident for most blebs (Fig. 7 C). The mean rate of increase derived from these data was approximately 6.7 × 10−3 s−1, roughly three times higher than the rate calculated using kymographs (Fig. 7 B). The difference between the two mean trends is shown in Fig. 7 D.
The greater spread of data points in the case of the kymograph analysis (Fig. 7 B) suggests that the measured recruitment rate of MLC2-RFP is sensitive to the position on the bleb where the kymograph is generated. This may be explained by the heterogeneous nature of myosin recruitment at the membrane (Biro et al., 2013). ADAPT averages the recruitment rate along the length of the entire bleb membrane, removing any influence of heterogeneous protein distribution and reducing data variability.
It could be argued that this large difference in measured MLC2-RFP recruitment rates is a product of a specific set of input parameters used by ADAPT for this particular analysis. To test this hypothesis, we analyzed the same cell 20 times with different, randomly generated values for the input parameters (Table S1). The resultant mean plots for bleb membrane velocity and MLC2-RFP fluorescence intensity are extremely consistent (Fig. 7 E). The mean rate of MLC2-RFP fluorescence increase was calculated as 6.85 × 10−3 ± 6.12 × 10−5 s−1, which is in good agreement with that shown in Fig. 7 D. This demonstrates that the data produced by ADAPT are relatively insensitive to small changes in input parameters.
However, it is apparent in Fig. 7 C that the mean velocity and profile of MLC2-RFP recruitment of some blebs deviates substantially from the means in Fig. 7 E. It is likely that such data result from inaccurate tracking or false detections of blebs. For example, considering the detections shown in Video 6, it is apparent that detection number 6 is a false detection, 4 and 14 could each be considered two blebs detected as one, and one of the anchor points associated with 0 does not really coincide with the bleb neck. Calculating the mean bleb membrane velocity and the mean MLC2-RFP intensity over time with or without these erroneous detections, however, results in very similar trends (Fig. S2), illustrating that small errors will not have a significant influence on the mean data output by ADAPT. It should of course be noted that the frequency and severity of errors will increase as image quality (signal to noise ratio, spatial, and temporal resolution) is decreased.
Characterization of an “average” bleb
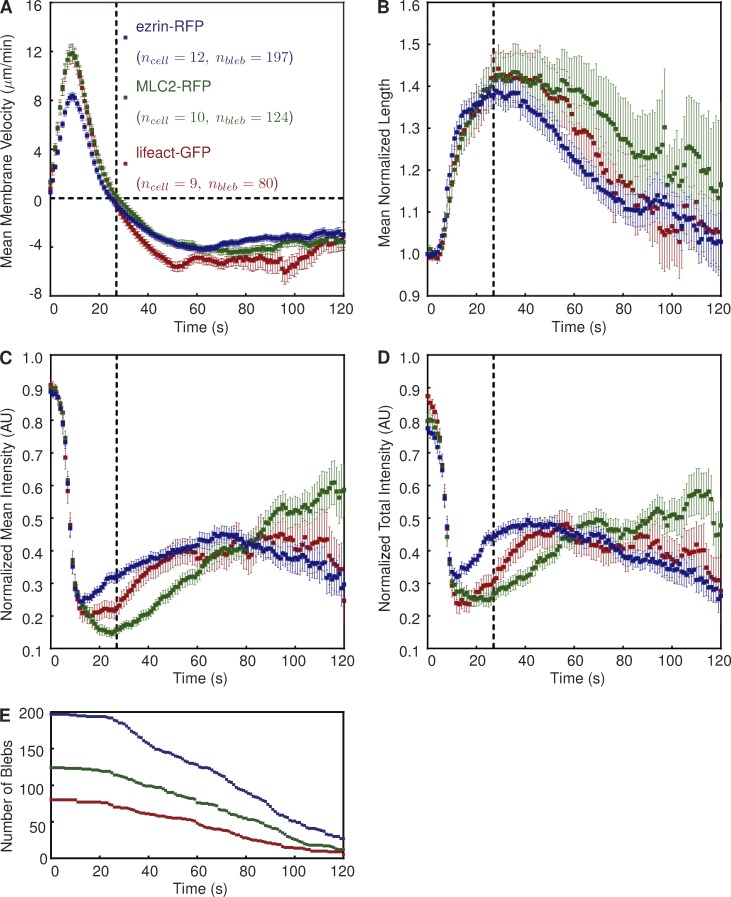
Having developed ADAPT, we applied the software to the characterization of cortical proteins involved in cortex reformation during bleb retraction. We used ADAPT to characterize three populations of blebbing vaccinia-infected HeLa cells, each expressing a different fluorescently tagged protein (lifeact-GFP, MLC2-RFP, and ezrin-RFP). The mean morphodynamic data for a large number of blebs (n = 401) illustrate that their retraction consistently occurs ∼27 s after bleb initiation (Fig. 8, A and B). This is similar to a previously reported value of 30 s in filamin-deficient M2 cells (Charras et al., 2006). Complete bleb retraction requires ∼90 s. Small variations in velocity and normalized length profiles are evident between the three populations, possibly caused by the presence of different tagged cortical components in each population.
Figure 8.
Description of a “typical” bleb in vaccinia-infected, blebbing HeLa cells. (A–D) Graphs show mean bleb velocity (A), mean normalized bleb perimeter length (B), normalized mean fluorescence intensity (C), and normalized mean fluorescence intensity (D) of the indicated protein. Positive and negative velocity values are indicative of extension and retraction, respectively. Fluorescence intensities are normalized to signal strength in the cortex before bleb formation. The dotted vertical line at t = 27 s indicates the onset of bleb retraction. Error bars represent standard error of the mean. The number of blebs (nbleb) indicates the population size at t = 0. (E) The number of blebs used to derive the mean curves depicted in A–D over time. AU, arbitrary unit.
The average description of lifeact-GFP, MLC2-RFP, and ezrin-RFP recruitment (Fig. 8, C and D) is consistent with the canonical description of bleb retraction, whereby actin–membrane linker proteins such as ezrin are first recruited to the membrane, followed by actin and then myosin motors (Charras et al., 2006; Fackler and Grosse, 2008). However, it is evident that ezrin recruitment begins before the onset of retraction (Fig. 8, C and D). This is in contrast to previous suggestions that ezrin is not present in the expanding bleb, although this was based on the manual analysis of a relatively small number of blebs (n = 23; Charras et al., 2006). It has also been previously stated, again based on a limited number of blebs, that total actin intensity remains constant during bleb retraction (Charras et al., 2008). However, we found that the intensity of normalized lifeact-GFP continues to increase long after the onset of retraction (Fig. 8 D). It is also apparent that the commencement of MLC2 recruitment coincides with the onset of bleb retraction (Fig. 8, A–D, denoted by the dotted lines).
The profiles of MLC2 and lifeact (actin) recovery are broadly similar to those reported for laser-induced blebs in HeLa cells blocked in metaphase (Biro et al., 2013). However, our data have been compiled from a wide variety of blebs, illustrating the utility of ADAPT in reliably extracting data from a relatively heterologous population in an unbiased fashion. A previous study has also attempted to estimate the point at which different components of the cytoskeleton first appear at the membrane (Charras et al., 2006). However, this requires the specification of arbitrary signal-to-noise thresholds, above which the recruitment of the protein of interest is considered to have commenced. The estimated time at which recruitment commences is thus heavily influenced by the choice of threshold. Although our study also makes use of noise thresholds, their use is restricted to the enhancement of trends corrupted by noise (Fig. 6 B), rather than the estimation of absolute values.
The apparent increase in data spread (as indicated by wider error bars) over time in Fig. 8 (A–D) may be attributed to differences in bleb lifetimes (Fig. 8 E). That is, not all blebs are tracked from formation to complete retraction. It is possible that, in some cases, this is a result of a loss of anchor point detection, leading to a breakdown in bleb tracking. However, the most common explanation for the termination of tracking is the appearance of a new bleb at the site of a retracting bleb.
Influence of Arp2/3 inhibition on bleb retraction
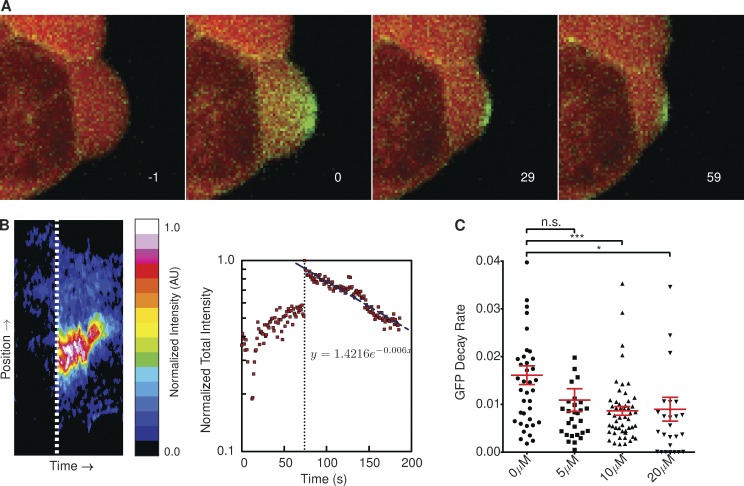
Given our description of a “typical” bleb, we proceeded to investigate whether ADAPT was capable of detecting and quantifying perturbations to the system. It has been reported that Arp2/3, together with mDia1, plays an important role in nucleating cortical actin (Bovellan et al., 2014). We found that increasing concentrations of the Arp2/3 inhibitor, CK-869, resulted in lower bleb retraction speeds (Fig. 9 A). We also found the dynamics of MLC2 localization were affected by the inhibitor, with increasing concentrations resulting in a reduced rate of MLC2-RFP recruitment to the bleb membrane (Fig. 9 B). Given the observed reduction in myosin recruitment rates in the presence of CK-869, we investigated whether we could identify any differences in the nature of the actin cortex forming at the bleb membrane during retraction in the presence of the inhibitor. To this end, we used cells stably expressing photoactivatable Cherry-GFPPA–β-actin to examine the rate of actin turnover in retracting blebs (Fig. 10 A). The photoactivated GFP-actin signal at the bleb membrane can be well represented by a decaying exponential function (Fig. 10 B). We found the rate of decay of the photoactivated Cherry-GFPPA–β-actin signal decreased as the concentration of CK-869 was increased (Fig. 10 C), indicative of reduced actin turnover.
Figure 9.
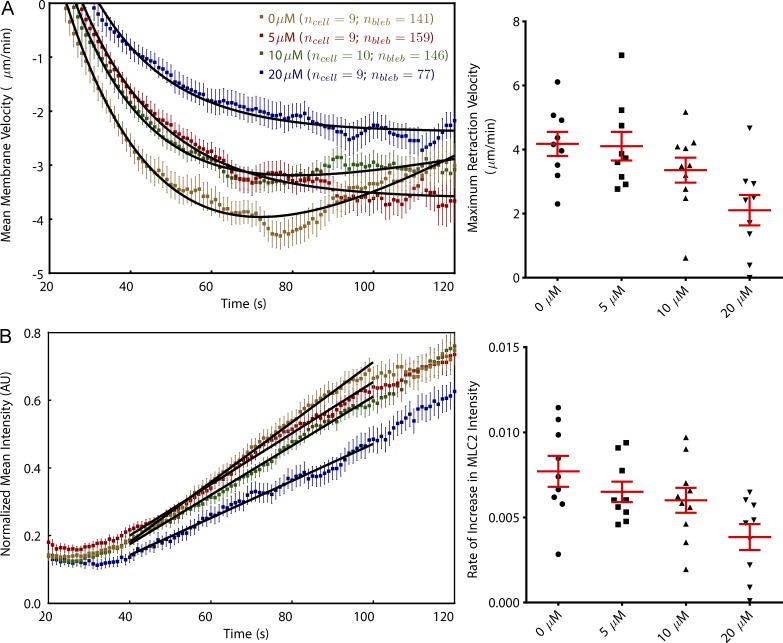
Inhibition of Arp2/3 complex retards bleb retraction and MLC2 recruitment. (A) Fitting curves on a per-cell basis shows that the mean maximum bleb retraction velocity decreases with increasing CK-869 dose (P < 0.001 for post-ANOVA test for linear trend). (B) MLC2 recruitment is slowed in the presence of CK-869, with the rate of recovery approximately dependent on dose (P < 0.05). Colors correspond to the concentrations shown in A. Error bars represent standard error of the mean. AU, arbitrary unit.
Figure 10.
Inhibition of Arp2/3 complex slows actin turnover at bleb membrane. (A) Decay of photoactivated Cherry-GFPPA–β-actin shows actin turnover at the plasma membrane during bleb retraction. Image labels correspond to seconds post-GFP activation. (B) The heat map represents the GFP intensity at all points on the boundary of a single photoactivated bleb (shown in A) over its lifetime. Dotted lines represent the point at which the Cherry-GFPPA–β-actin is activated. The decay rate of the activated GFP signal is approximately exponential. Data represent the analysis of a single bleb. (C) Rate of actin turnover, measured as the exponential decrease in photoactivated Cherry-GFPPA–β-actin intensity, was significantly reduced in the presence of CK-869 (P < 0.05). Annotation above columns indicates results of Student’s t tests: n.s., not significant; *, P < 0.05; ***, P < 0.001. Error bars represent standard error of the mean. AU, arbitrary unit.
CK-869 inhibits Arp2/3 by locking the complex in an inactive conformation, preventing it from nucleating new actin filaments (Hetrick et al., 2013). Rotty et al. (2015) suggest that in the absence of Arp2/3, the polymerization of actin within a cell is dominated by profilin-I in conjunction with Ena/vasodilator-stimulated phosphoprotein (VASP). This would favor the formation of thick bundles of F-actin over the branched meshwork associated with Arp2/3-mediated polymerization. Given this, our data suggest that inhibition of Arp2/3 results in the formation of a more stable actin cortex beneath the bleb membrane, which is less favorable for actomyosin contraction.
Discussion
In this study, we have introduced ADAPT, an ImageJ plug-in designed for rapid, whole-cell analysis, providing data on both global and local morphodynamic changes. We have demonstrated the utility of ADAPT in quantifying cell protrusions and migration, while also extracting metrics of comparative value at the population level. We provide evidence that ADAPT is sensitive enough to detect small differences in cell migration on different concentrations of fibronectin. We also show that ADAPT can quantify subtle drug-induced variations in the dynamics of actin and myosin within retracting blebs, along with perturbations to the speed of bleb retraction. To the best of our knowledge, this is the first study describing an analysis of hundreds of blebs, providing detailed data on bleb retraction velocity and actin turnover, as well as recruitment of ezrin, actin, and myosin, in a typical bleb (Fig. 8).
Bovellan et al. (2014) recently reported an effect of Arp2/3 inhibition on actin dynamics in blebbing HeLa cells. They suggest that laser-induced blebs on metaphase HeLa cells accumulate actin more rapidly in the presence of the Arp2/3 inhibitor CK-666. However, the biphasic profile of actin recovery they reported under control conditions is markedly different from that shown here (Fig. 8 C). We found, in the absence of Arp2/3 inhibition, that actin begins to recover rapidly at the onset of bleb retraction before becoming relatively constant at ∼60 s after initiation. Bovellan et al. (2014) report that actin recovers rapidly for ∼20 s, before slowing to a lower, constant rate of increase. Nonetheless, the general idea of a new cortex forming more rapidly in the presence of Arp2/3 inhibition does not contradict our own suggestion that Arp2/3 inhibition results in a more stable, less plastic cortex. Certainly, the detection of all seven Arp2/3 subunits in blebs (isolated from M2 cells) by Bovellan et al. (2014) strongly suggests a role for the complex in cortical actin regrowth and bleb retraction. Our own attempts to show localization of Arp2/3 to bleb membranes were inconclusive. Live-cell imaging of HeLa cells expressing ArpC2-GFP with a spinning-disc confocal microscope showed that Arp2/3 is certainly present in the bleb cytosol, but it is not clear whether it strongly localizes to the membrane (Video 7).
It has recently been reported that profilin acts as a regulator of F-actin network homeostasis, controlling the distribution of actin assembly between Arp2/3-, formin-, and Ena/VASP-mediated pathways (Rotty et al., 2015; Suarez et al., 2015). In mouse embryonic fibroblasts lacking Arp2/3, actin assembly was dependent on profilin and Ena/VASP (Rotty et al., 2015). Fission yeast expressing a loss-of-function profilin mutant had formin-mediated actin assembly restored by the addition of CK-666, reducing competition for G-actin monomers from Arp2/3 (Suarez et al., 2015). In the presence/absence of profilin, Suarez et al. (2015) also demonstrated a link between lower actin turnover and inhibition of Arp2/3 branch formation in vitro. There is also evidence that the rate of actin turnover influences the recruitment of myosin II. Myosin IIA and IIB recruitment to epithelial cell junctions was impaired in the absence of Arp2/3, although the precise explanation was unclear (Verma et al., 2012). More recently, a mathematical model of epithelial cells that showed a dependence of actomyosin pattern formation and dynamics on F-actin turnover (Moore et al., 2014). These studies support our observations, suggesting that inhibition of Arp2/3 results in a more rigid cortex forming at the membrane of retracting blebs, exhibiting a lower actin turnover and a geometry less favorable for myosin recruitment. However, variation in the overall level of F-actin within the cell in the presence of CK-869 may also be a factor. Bovellan et al. (2014) observed a significant reduction in F-actin levels when M2 cells were treated with CK-666. On the other hand, Rotty et al. (2015) reported no reduction in F-actin levels in mouse embryonic fibroblasts lacking Arp2/3. A reduction in the total level of F-actin within the cell should result in impaired actomyosin contractility, which would lower turgor pressure and favor faster bleb retraction. It is clear that a more detailed spatiotemporal analysis of actin and myosin dynamics at the bleb membrane is required to elucidate the precise molecular details.
We have also demonstrated that ADAPT is suitable for the analysis not only of blebs but also of other protrusions, such as lamellipodia (Fig. 2) and filopodia (Fig. 4). However, unlike blebs, lamellipodia do not have distinct start and end points but are rather more wavelike in nature. As such, determining where a lamellipodium begins and ends is difficult, if not impossible. It is our view that the analysis of cells producing such structures should be performed at the whole-cell level, treating the entire cell boundary as single protruding/retracting structure, as per Fig. 2. Filopodia, on the other hand, are more similar to blebs in the sense that they are relatively distinct structures with an identifiable start/end point (Video 5). However, they are obviously much smaller structures, and their accurate detection and tracking with ADAPT will depend heavily on the input images being of sufficiently high spatial and temporal resolution. Furthermore, distinguishing between “true” filopodia and other fingerlike structures, such as protrusions that form as the trailing cell edge retracts, represents a significant challenge. It is possible that the use of a suitable fluorescent protein associated with filopodia, such as fascin or VASP (Mattila and Lappalainen, 2008), could be used for more accurate identification. However, in its current implementation, it is likely that analysis with ADAPT will include false filopodia detections, but the associated data for such false detections can be manually removed by the user upon examination of the output (Videos 4 and 5).
ADAPT is also suitable for the tracking of cell migration (Fig. 3). Accurate tracking is dependent on a high frame rate, relative to the speed of migration, and a relatively dispersed cell population. A degree of cell–cell contact will not disrupt the tracking procedure, but clusters of cells in close proximity are unlikely to be successfully tracked. Although the algorithm underlying the ADAPT tracking procedure could be improved (at the cost of additional processing time), it is questionable as to whether this would deliver a significant improvement in performance. Tracking objects in fluorescence microscopy datasets still represents a significant challenge, and, as yet, there is no single approach that will prove superior in all scenarios (Chenouard et al., 2014).
Because ADAPT is written in Java as a plug-in for ImageJ, it may be run on any PC or Mac on which Java is installed, making it accessible to virtually the entire cell biology community. This is in contrast to other recently described software, which require expensive proprietary packages as prerequisites (Biro et al., 2013; Tsygankov et al., 2014). ADAPT is also completely open source, the source code being provided with this publication. Access to source code permits others to view exactly how mathematical functions and algorithms were implemented, increasing reproducibility. Furthermore, the hosting of the source code in an open online repository permits continuous improvement and extension of the software as well as customization to other experimental scenarios through the incorporation of additional code modules. Combined with a user-friendly interface and the provision of test data for validation purposes, ADAPT may thus be considered to have a high degree of “usability” (user friendly, developer friendly, modular, validated, etc.), as described by Carpenter et al. (2012).
For example, in its current form, ADAPT is designed to analyze 2D representations of cells. In theory, the analysis could be extended to 3D stacks. This could involve rendering the segmented cell boundary as a 3D mesh, detecting local velocity maxima in that mesh and anchoring bleb tracking by detecting curvature extrema as curves rather than points. Alternatively, individual 2D slices could be analyzed as described in this article and then assembled into a 3D representation. However, acquiring 3D stacks of blebbing cells at sufficiently high spatial and temporal resolution to conduct such an analysis represents a significant challenge.
Although the current implementation of ADAPT is capable of analyzing multiple cells simultaneously, this capability will be limited by available memory and the capacity of ImageJ to handle large datasets (more than 1–2 gigabytes). However, the potential exists to implement the same algorithms underlying the operation of ADAPT in a massively parallel framework, permitting its application to, for example, high-throughput screening. Software for live-cell high-throughput trials has previously been reported, but this was specific to the screening of cell cycle perturbations (Held et al., 2010). A high-throughput implementation of ADAPT, on the other hand, would provide a more generally applicable screening tool.
More generally, we assert that the use of such automated methods for the analysis of cell dynamics represents an essential tool for rapid, unbiased data acquisition. An absence of quantitative analysis limits the potential of the sophisticated image acquisition tools available to modern cell biologists. Furthermore, the sheer scale of data produced using modern imaging modalities will necessitate the real-time analysis of images as they are produced, alleviating the need for expenditure on high levels of data storage. Ultimately, it should be the aim of cell biologists to move away from the use of “representative images” in their publications in favor of truly representative, unbiased data. This should result in the conception of more accurate models of biological processes, furthering our molecular understanding of cellular processes.
Materials and methods
The files necessary to run ADAPT are provided as supplemental software, together with Java source code and test data. These resources, along with a wiki containing instructions for using ADAPT, are also freely available online (https://bitbucket.org/djpbarry/adapt).
Generation of stable cell lines
A pEL-GFPPACherry–β-actin expression vector was previously created by PCR using pAREK1 GPAC (variant1)–β-actin as a template (Welman et al., 2010). The PCR product was cloned into NotI–BamHI sites of the pELGFP vector in place of GFP (Humphries et al., 2012). The pLVX-Cherry-GFPPA–β-actin lentiviral expression vector was then created by digesting pEL-Cherry-GFPPA–β-actin with BglII–XbaI and ligating this into the pLVX-puro vector (Takara Bio Inc.) digested with BamHI–XbaI. Lentivirus was produced in HEK293FT cells by cotransfection of pLVX-Cherry-GFPPA–β-actin with psPAX2 and pMD2.g vectors at a ratio of 10:7:3 µg DNA, respectively (Trono laboratory second generation packaging system; Addgene). Supernatants from transfected HEK293FT cells containing the lentivirus were collected 24 and 48 h after transfection, pooled, and filtered. HeLa cells were then infected with lentivirus for 48 h, and cells stably expressing Cherry-GFPPA–β-actin were selected by 1 µg/ml puromycin selection. MLC2-RFP, lifeact-GFP, and ezrin-RFP constructs were created by subcloning relevant genes into a pLVX puro vector (Takara Bio Inc.), and stable HeLa cell lines were generated using the lentivirus system, as previously described (Humphries et al., 2012). Human Abi1 was cloned into the Not1–EcoRI site of a modified pLVX-puro-GFP vector and used to generate lentiviruses. HT1080 fibrosarcoma cell lines stably expressing GFP or the GFP-tagged Abi1 were then selected with puromycin and FACS sorting (Chen et al., 2014).
Cell culture and live-cell imaging
Cells were seeded onto fibronectin-coated 35-mm MatTek dishes, transfected using FuGENE 6 (Promega) with pcDNA-GFP or pLVX-mCherry ∼6 h later (if required), and cultured overnight. For the characterization of vaccinia virus-induced blebbing, cells were then infected with a WR strain of vaccinia virus in serum-free MEM (Arakawa et al., 2007). This medium was replaced with phenol red–free MEM with 10% FCS 45 min after infection. Where indicated, Arp2/3 inhibitor (CK-869; EMD Millipore), dissolved in DMSO, was also added 45 min after infection. Live-cell imaging was commenced ∼3 h after infection using either a Plan Apochromat 63×/1.40 NA or an α Plan Apochromat 100×/1.46 NA oil objective (Carl Zeiss) in a temperature-controlled chamber at 37°C. Images were captured on a camera (Evolve 512; Photometrics) mounted on an inverted microscope (AxioObserver.Z1; Carl Zeiss), constituents of a custom-built spinning-disc confocal system (Intelligent Imaging Innovations). All hardware was controlled with SlideBook software (Intelligent Imaging Innovations). Images were collected at 0.03–2.4 Hz, with each acquisition consisting of excitation with a 488-nm, followed by a 568-nm, laser with a maximum switching time of 200 ms. For experiments involving Cherry-GFPPA–β-actin, photoactivation was achieved using a focused 405-nm laser. For quantification of filopodia formation, the formin inhibitor, SMIFH2 (Sigma-Aldrich) was dissolved in DMSO and added to cells ∼1 h before imaging. Cell migration assays were conducted by seeding HT1080 cells on 96-well plates (Costar) in FluoroBrite DMEM (Gibco) supplemented with 1.25% vol/vol penicillin streptomycin, 2.4 nM glutamine, and 10% vol/vol FCS. Cells were then imaged for 8–12 h in an IncuCyte FLR (Essen Bioscience) at 37°C.
Image analysis
Image segmentation.
Before segmentation, images are Gaussian filtered to suppress noise (Fig. 1 B). Kalman filtering was avoided so as not to introduce motion blurring artifacts, which may influence the calculation of membrane velocity. ADAPT then segments each individual frame of the cytosolic channel by way of a region-growing algorithm, with a gray-level threshold, user specified, or automatically calculated using any one of the thresholding algorithms available in ImageJ (specified by the user; National Institutes of Health), constituting the termination condition. The seed point for the first frame is determined automatically if not specified by the user. The final segmentation of one frame is subjected to a series of erosion operations and then used as the seed for the segmentation of the subsequent frame (Fig. 1 B). The cell boundary contour is taken as the set of foreground points bordering background in each segmented frame, resulting in a time-varying contour (S(t)). The resulting segmentation is used to calculate various morphological parameters, such as perimeter length (P), projected area (Ap), and circularity (C = 4πAp/P2).
Quantification of membrane velocity.
Estimation of membrane velocity is achieved using a method similar to that previously described (Döbereiner et al., 2006). Given a contour S(t), the normal velocity, vn, may be estimated at each point (x, y, t)∈S(t) according to
where I(x, y, t) is the gray level at (x, y) in frame t, h(t) is the gray-level threshold for frame t, Δ(x, y, t) is the local image gradient, and δt is an integer number of image frames. The set of values constituting vn(x, y, t) are then translated into a two-dimensional velocity map (Fig. 1 C).
Quantification of fluorescence intensity.
Before running ADAPT, the video channel containing a fluorescent signal of interest (if present) is Kalman filtered to suppress noise. For the purposes of analyzing fluorescence intensity at the cell periphery, a region of interest, Rc, is defined in each frame, the width of which is determined by the user (Cortex Depth; Fig. 1 A). The outer and inner edges of this region are based on the cell segmentation (Fig. 2 A). A binary mask image is created from this segmentation and Cortex Depth/Spatial Resolution erosion/dilation operations are performed, yielding the inner/outer segmentations shown in Fig. 1 A.
A threshold fluorescence value, Tf, is calculated for each video sequence, defined as
where μf is the mean intensity of all pixels in every Rc, δσf is the user-specified Signal Threshold Factor (Fig. 5 A), and σf is the standard deviation of all pixel intensities in all Rc. In the case of analyzing the distribution of fluorescence intensities within a cell, the cell segmentation is subjected to a series of erosion operations (Fig. 4 C). After each erosion, representing a step to the cell center, the segmentation boundary is identified, and the mean fluorescence intensity along this boundary is calculated—no threshold is taken into consideration.
The cross-correlation value, c(x,y), at coordinate (x, y) within the cross-correlation map, C, of two images, I1 and I2, is calculated as follows:
where m × n are the dimensions of I1 and I2, σ1 and σ2 are the standard deviations of I1 and I2, μ1 and μ2 are the means of I1 and I2, t = x − m/2, and s = y − n/2.
Quantification of cell migration.
The mean velocity of a cell is calculated as the total distance traveled, L, over the course of a video, divided by the duration (in minutes) of that video. Directionality, D, is calculated as follows:
where (x0, y0) and (xf, yf) are the starting and final positions of the cell, respectively.
Detection and tracking of filopodia.
Filopodia are considered as any object that may be eroded from the periphery of a cell (Fig. 4 A). Objects smaller than a user-specified minimum area are excluded. The objects are then treated as individual “cells” and tracked from frame to frame in the same manner that cells are. New filopodia-like objects are searched for in each frame by searching for objects, larger than the minimum area, that are not currently being tracked. A minimum number of frames over which an object must be tracked in order for it to be counted as a filopodia can also be specified by the user.
Detection and tracking of blebs.
Blebs are initially identified as local maxima in velocity maps by calculating a minimum velocity threshold. The portion of the boundary contour constituting a bleb is tracked in time using estimates of curvature, similar to a previously described approach for detecting single blebs (Fig. 5 B; Biro et al., 2013). Given S(t), the local curvature at a point (x, y) may be estimated by calculating the angle subtended by two lines passing through (x, y), each intersecting another point n pixels along the contour in the clockwise and counter-clockwise directions (Fig. 5 B). The lower the value of n, the more sensitive the calculation is to segmentation and pixilation error. Bleb necks are manifested as local extrema in contour curvature and may be used as “anchor” points to demarcate the portion of the cell boundary that is undergoing blebbing (Fig. 5 D). However, as a result of noise, it is not always possible to detect the bleb neck in each frame, and this can cause the tracking of a bleb to destabilize. Constructing trajectories for each curvature extrema can reduce this potential instability, as “gaps” in detections can be “bridged” (Fig. S1). Each bleb may then be monitored for a predetermined number of frames (Fig. 5 A, Cut-off Time) or until a new bleb forms at the same point on the cell boundary.
For the analysis of fluorescent protein recruitment at the bleb membrane, the anchor points define the left and right extremes of subregions, Rb∈Rc, for each bleb. The total fluorescence intensity of the protein of interest is calculated in each frame by summing all pixels values above Tf within Rb. The mean fluorescence intensity is calculated by dividing this value by the bleb length, defined as the length of cell boundary between the two bleb anchor points.
Data postprocessing
Accurate tracking of individual blebs may be validated by the user by examining an image stack output showing each bleb tracked (Video 6). All plots of velocity, perimeter length, and cortical signal extracted for each individual bleb are averaged together across all cells to provide a description of a typical bleb within the population. Individual length plots are first normalized to initial length before averaging. To combat differences in expression levels, together with the effects of residual noise and bleaching (Fig. 6), plots of total and mean signal recovery are minimum subtracted and then normalized to their respective maximum values before averaging. For experiments involving cells expressing Cherry-GFPPA–β-actin, fluorescent signals were manually analyzed for each bleb, and a curve was fit to the portion of the plot corresponding to exponential decay.
Parameterizing velocity and fluorescence intensity profiles
To derive parameters describing individual cells, mean plots of velocity and increase in MLC2-RFP fluorescence intensity for individual cells were parameterized. Bleb retraction velocity could be modeled as follows:
where a, b, c, and k are constants. The minimum of such curves was taken as the maximum retraction velocity attained. All curve fitting was performed with Prism (GraphPad Software). Increase in MLC2-RFP fluorescence intensity, I(t), was generally well described by a straight line I(t) = mt + c1, where m is the rate of MLC2-RFP recruitment and c1 is a constant.
Online supplemental material
Fig. S1 shows the algorithm used to track anchor points. Fig. S2 shows the influence of manually excluding inaccurate bleb detections on data means. Table S1 shows the input parameters used to generate the data in Fig. 7 E. Video 1 illustrates whole-cell analysis with ADAPT. Video 2 illustrates the correlation of H1080 cell morphodynamics with fluorescence intensities. Video 3 shows tracking of HT1080 cell migration with ADAPT. Videos 4 and 5 illustrate the tracking of filopodia on a HeLa cell. Video 6 illustrates the analysis of a blebbing, vaccinia-infected HeLa cell with ADAPT. Video 7 shows a blebbing HeLa cell expressing ArpC2-GFP. A ZIP file is also provided that includes the JAR files necessary to install and run ADAPT, the source code is packaged within the JAR files, and test data are also included within the ZIP file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501081/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201501081.dv.
Supplementary Material
Acknowledgments
The authors would like to thank Paul Bates and Erik Sahai for their comments on the manuscript.
This work has been supported by postdoctoral fellowship awards from Cancer Research UK and the Canadian Institutes of Health Research (number 234201).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- VASP
- vasodilator-stimulated phosphoprotein
References
- Arakawa Y., Cordeiro J.V., and Way M.. 2007. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host Microbe. 1:213–226 10.1016/j.chom.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Bergert M., Chandradoss S.D., Desai R.A., and Paluch E.. 2012. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc. Natl. Acad. Sci. USA. 109:14434–14439 10.1073/pnas.1207968109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biro M., Romeo Y., Kroschwald S., Bovellan M., Boden A., Tcherkezian J., Roux P.P., Charras G., and Paluch E.K.. 2013. Cell cortex composition and homeostasis resolved by integrating proteomics and quantitative imaging. Cytoskeleton (Hoboken). 70:741–754 10.1002/cm.21142 [DOI] [PubMed] [Google Scholar]
- Bosgraaf L., and Van Haastert P.J.. 2010. Quimp3, an automated pseudopod-tracking algorithm. Cell Adhes. Migr. 4:46–55 10.4161/cam.4.1.9953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L., van Haastert P.J., and Bretschneider T.. 2009. Analysis of cell movement by simultaneous quantification of local membrane displacement and fluorescent intensities using Quimp2. Cell Motil. Cytoskeleton. 66:156–165 10.1002/cm.20338 [DOI] [PubMed] [Google Scholar]
- Bovellan M., Romeo Y., Biro M., Boden A., Chugh P., Yonis A., Vaghela M., Fritzsche M., Moulding D., Thorogate R., et al. 2014. Cellular control of cortical actin nucleation. Curr. Biol. 24:1628–1635 10.1016/j.cub.2014.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Kamentsky L., and Eliceiri K.W.. 2012. A call for bioimaging software usability. Nat. Methods. 9:666–670 10.1038/nmeth.2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G., and Paluch E.. 2008. Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 9:730–736 10.1038/nrm2453 [DOI] [PubMed] [Google Scholar]
- Charras G.T., Hu C.-K., Coughlin M., and Mitchison T.J.. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175:477–490 10.1083/jcb.200602085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G.T., Coughlin M., Mitchison T.J., and Mahadevan L.. 2008. Life and times of a cellular bleb. Biophys. J. 94:1836–1853 10.1529/biophysj.107.113605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.J., Squarr A.J., Stephan R., Chen B., Higgins T.E., Barry D.J., Martin M.C., Rosen M.K., Bogdan S., and Way M.. 2014. Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell. 30:569–584 10.1016/j.devcel.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenouard N., Smal I., de Chaumont F., Maška M., Sbalzarini I.F., Gong Y., Cardinale J., Carthel C., Coraluppi S., Winter M., et al. 2014. Objective comparison of particle tracking methods. Nat. Methods. 11:281–289 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E.S., and Higgs H.N.. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9:1110–1121 10.1038/ncb1007-1110 [DOI] [PubMed] [Google Scholar]
- Dang I., Gorelik R., Sousa-Blin C., Derivery E., Guérin C., Linkner J., Nemethova M., Dumortier J.G., Giger F.A., Chipysheva T.A., et al. 2013. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 503:281–284. [DOI] [PubMed] [Google Scholar]
- Döbereiner H.-G., Dubin-Thaler B.J., Hofman J.M., Xenias H.S., Sims T.N., Giannone G., Dustin M.L., Wiggins C.H., and Sheetz M.P.. 2006. Lateral membrane waves constitute a universal dynamic pattern of motile cells. Phys. Rev. Lett. 97:038102 10.1103/PhysRevLett.97.038102 [DOI] [PubMed] [Google Scholar]
- Dormann D., Libotte T., Weijer C.J., and Bretschneider T.. 2002. Simultaneous quantification of cell motility and protein-membrane-association using active contours. Cell Motil. Cytoskeleton. 52:221–230 10.1002/cm.10048 [DOI] [PubMed] [Google Scholar]
- Fackler O.T., and Grosse R.. 2008. Cell motility through plasma membrane blebbing. J. Cell Biol. 181:879–884 10.1083/jcb.200802081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M., Schmitz M.H.A., Fischer B., Walter T., Neumann B., Olma M.H., Peter M., Ellenberg J., and Gerlich D.W.. 2010. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods. 7:747–754 10.1038/nmeth.1486 [DOI] [PubMed] [Google Scholar]
- Hetrick B., Han M.S., Helgeson L.A., and Nolen B.J.. 2013. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20:701–712 10.1016/j.chembiol.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P., and Lappalainen P.. 2006. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173:383–394 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries A.C., Dodding M.P., Barry D.J., Collinson L.M., Durkin C.H., and Way M.. 2012. Clathrin potentiates vaccinia-induced actin polymerization to facilitate viral spread. Cell Host Microbe. 12:346–359 10.1016/j.chom.2012.08.002 [DOI] [PubMed] [Google Scholar]
- Krause M., and Gautreau A.. 2014. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15:577–590 10.1038/nrm3861 [DOI] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., and Danuser G.. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- Moore T., Wu S.K., Michael M., Yap A.S., Gomez G.A., and Neufeld Z.. 2014. Self-organizing actomyosin patterns on the cell cortex at epithelial cell-cell junctions. Biophys. J. 107:2652–2661 10.1016/j.bpj.2014.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G.2012. Why bioimage informatics matters. Nat. Methods. 9:659–660 10.1038/nmeth.2024 [DOI] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch E.K., and Raz E.. 2013. The role and regulation of blebs in cell migration. Curr. Opin. Cell Biol. 25:582–590 10.1016/j.ceb.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch E., Piel M., Prost J., Bornens M., and Sykes C.. 2005. Cortical actomyosin breakage triggers shape oscillations in cells and cell fragments. Biophys. J. 89:724–733 10.1529/biophysj.105.060590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., Wu C., Haynes E.M., Suarez C., Winkelman J.D., Johnson H.E., Haugh J.M., Kovar D.R., and Bear J.E.. 2015. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev. Cell. 32:54–67 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., and Marshall C.J.. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719 10.1038/ncb1019 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm B., de Haan C.A., Young J., Doglio L., Schleich S., Reese C., Popov A.V., Steffen W., Schroer T., and Locker J.K.. 2006. Vaccinia-virus-induced cellular contractility facilitates the subcellular localization of the viral replication sites. Traffic. 7:1352–1367 10.1111/j.1600-0854.2006.00470.x [DOI] [PubMed] [Google Scholar]
- Suarez C., Carroll R.T., Burke T.A., Christensen J.R., Bestul A.J., Sees J.A., James M.L., Sirotkin V., and Kovar D.R.. 2015. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell. 32:43–53 10.1016/j.devcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Rubinstein B., Unruh J.R., Durnin M., Hanein D., and Li R.. 2012. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197:239–251 10.1083/jcb.201112113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.-Y., Schulze U., Salbreux G., Roensch J., Joanny J.-F., and Paluch E.. 2009. Role of cortical tension in bleb growth. Proc. Natl. Acad. Sci. USA. 106:18581–18586 10.1073/pnas.0903353106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoğlu M., Tournier A.L., Jenkins R.P., Hooper S., Bates P.A., and Sahai E.. 2013. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat. Cell Biol. 15:751–762 10.1038/ncb2775 [DOI] [PubMed] [Google Scholar]
- Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., and Elston T.C.. 2014. CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204:443–460 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura S., Pollitt A.Y., Veltman D.M., Morrice N.A., Machesky L.M., and Insall R.H.. 2012. Pseudopod growth and evolution during cell movement is controlled through SCAR/WAVE dephosphorylation. Curr. Biol. 22:553–561 10.1016/j.cub.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Han S.P., Michael M., Gomez G.A., Yang Z., Teasdale R.D., Ratheesh A., Kovacs E.M., Ali R.G., and Yap A.S.. 2012. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 23:4601–4610 10.1091/mbc.E12-08-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welman A., Serrels A., Brunton V.G., Ditzel M., and Frame M.C.. 2010. Two-color photoactivatable probe for selective tracking of proteins and cells. J. Biol. Chem. 285:11607–11616 10.1074/jbc.M110.102392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggan O., Shaw A.E., DeLuca J.G., and Bamburg J.R.. 2012. ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev. Cell. 22:530–543 10.1016/j.devcel.2011.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Czech L., Gerboth S., Kojima S., Scita G., and Svitkina T.. 2007. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5:e317 10.1371/journal.pbio.0050317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatulovskiy E., Tyson R., Bretschneider T., and Kay R.R.. 2014. Bleb-driven chemotaxis of Dictyostelium cells. J. Cell Biol. 204:1027–1044 10.1083/jcb.201306147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.