An Msh2/Msh6-dependent DNA repair mechanism mitigates the mutagenicity of photolesions and induces cell cycle responses by excising incorrect nucleotides incorporated by postreplicative translesion synthesis.
Abstract
In addition to correcting mispaired nucleotides, DNA mismatch repair (MMR) proteins have been implicated in mutagenic, cell cycle, and apoptotic responses to agents that induce structurally aberrant nucleotide lesions. Here, we investigated the mechanistic basis for these responses by exposing cell lines with single or combined genetic defects in nucleotide excision repair (NER), postreplicative translesion synthesis (TLS), and MMR to low-dose ultraviolet light during S phase. Our data reveal that the MMR heterodimer Msh2/Msh6 mediates the excision of incorrect nucleotides that are incorporated by TLS opposite helix-distorting, noninstructive DNA photolesions. The resulting single-stranded DNA patches induce canonical Rpa–Atr–Chk1-mediated checkpoints and, in the next cell cycle, collapse to double-stranded DNA breaks that trigger apoptosis. In conclusion, a novel MMR-related DNA excision repair pathway controls TLS a posteriori, while initiating cellular responses to environmentally relevant densities of genotoxic lesions. These results may provide a rationale for the colorectal cancer tropism in Lynch syndrome, which is caused by inherited MMR gene defects.
Introduction
The integrity of mammalian genomes incessantly is compromised by damage to nucleotides, resulting either from their spontaneous decay or from exposure to endogenous or exogenous genotoxic agents. Specifically, structurally aberrant nucleotides, when not removed by NER, entail the risk of inducing nucleotide substitutions or genomic rearrangements. To prevent carcinogenic derailment, cells can transiently arrest their cycle or, ultimately, induce senescence or apoptosis. Single-stranded DNA (ssDNA) patches represent key intermediates in the induction of these protective responses, as these activate the canonical RPA–ATR–CHK1 DNA damage signaling pathway, upon cooperative binding of the ssDNA-binding protein replication protein A (RPA; Jackson and Bartek, 2009; Ciccia and Elledge, 2010). ssDNA patches in response to nucleotide lesions may be induced by various DNA transactions, during different stages of the cell cycle. Thus, in nonreplicating cells, small NER-induced ssDNA patches can be extended by the EXO1 nuclease, generating the structures to trigger DNA damage signaling (Novarina et al., 2011; Sertic et al., 2011). During S phase, small lesion-containing ssDNA patches can originate from stalling of the replicative DNA polymerases at unrepaired lesions, followed by downstream repriming of processive replication or by convergence of replication from an adjacent replicon, both at leading and lagging strands (Lopes et al., 2006; Elvers et al., 2011; Novarina et al., 2011). These ssDNA patches can rapidly be filled by DNA damage tolerance mechanisms, notably by TLS, comprising the replicative bypass of the lesions by specialized DNA polymerases (Lopes et al., 2006; Chang and Cimprich, 2009; Sale et al., 2012). Although TLS averts DNA damage responses and gross genomic instability, the associated polymerases frequently incorporate incorrect nucleotides opposite poorly instructive or noninstructive nucleotide lesions. For this reason, TLS is responsible for DNA damage–induced substitution mutations. The mutagenicity of TLS is controlled at multiple levels, including the regulation of the recruitment of TLS polymerases to chromatin, by their intrinsically distributive activities and, possibly, by proofreading in trans provided by replicative polymerases (Bebenek et al., 2001; Stelter and Ulrich, 2003; Avkin et al., 2006).
DNA mismatch repair is an excision repair pathway that corrects mispaired, undamaged or slightly aberrant nucleotides (Zlatanou et al., 2011; Peña-Diaz et al., 2012; Rodriguez et al., 2012; Jiricny, 2013). Cells with defects in components of the MMR machinery are characterized by spontaneous mutator phenotypes. Surprisingly, MMR proteins are also involved in cell cycle or apoptotic responses to agents that induce structurally aberrant, poorly instructive or noninstructive nucleotide lesions (Wu et al., 2003; Shin-Darlak et al., 2005; van Oosten et al., 2005; Borgdorff et al., 2006; Smith-Roe et al., 2006; Seifert et al., 2008). In addition, we have reported that the mutagenicity of short-wave ultraviolet light (UVC), which induces only helix-distorting nucleotide lesions, is significantly increased in mouse embryonic stem (ES) cells deficient for the heterodimeric mismatch-binding protein Msh2/Msh6, but not its paralog Msh2/Msh3 (Borgdorff et al., 2006). To investigate the mechanistic basis for the involvement of MMR proteins in these responses, we have generated a set of isogenic ES cell lines with single or combined disruptions in core components of NER, TLS, and Msh2/Msh6. These cell lines were exposed to low, physiologically relevant, doses of short-wave UVC light. By cross-linking dipyrimidines, UVC induces only two well-defined mutagenic nucleotide lesion types: moderately distorting cyclobutane pyrimidine dimers (CPD), as well as severely distorting, noninstructive, and highly genotoxic (6–4)pyrimidine-pyrimidone dimers [(6–4)PP]. Although CPD in mouse cells are refractory to repair, (6–4)PP is a good substrate for NER, specifically outside of S phase (Van Sloun et al., 1999; Beukers et al., 2008).
Our data unveil an unanticipated Msh2/Msh6-dependent DNA repair mechanism, termed post-TLS repair, that excises nontemplated incorporations by TLS opposite both CPD and (6–4)PP. Post-TLS repair mitigates the mutagenicity of these photolesions, while persisting excision tracts opposite (6–4)PP are strong inducers of canonical RPA–ATR–CHK1-dependent cell cycle checkpoints. When the resulting ssDNA patches, containing an embedded (6–4)PP (henceforth called ss[6–4]PP patches), are transferred to the subsequent cell cycle, they are converted to double-stranded DNA (dsDNA) breaks, and these are associated with the delayed induction of apoptosis. Based on these results, we hypothesize that post-TLS repair plays a significant role in the suppression of cancer induced by environmentally relevant doses of genotoxic agents.
Results
Suppression of UVC-induced mutations by Msh2/Msh6 is not caused by known canonical or noncanonical MMR subpathways
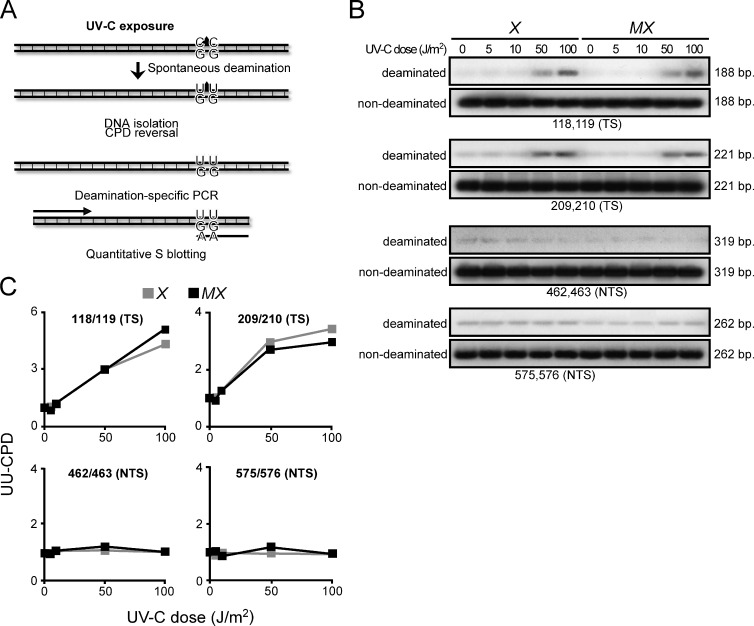
Recently, a noncanonical MMR pathway has been described that, outside of S phase, processes U-G mismatches derived from the enzymatic or spontaneous deamination of cytidine in C-G base pairs (Peña-Diaz et al., 2012). Also C-containing CPD can efficiently undergo spontaneous deamination to U-containing CPD, which, during replication, encodes the incorporation of A by TLS. This process is responsible for a significant fraction of UVC-induced C-G to T-A transitions (Fig. 1 A; Pfeifer et al., 2005). We argued that this noncanonical MMR pathway might be capable of excising U-containing CPD that is mispaired with G, thereby preventing UVC-induced C to T transitions (Shin-Darlak et al., 2005). This hypothesis was investigated by measuring levels of di-U-containing CPD (UU-CPD), resulting from the spontaneous double-deamination of CC-CPD, at four positions within an ectopic Hprt minigene at 16 h after exposure of (NER-deficient) Xpa−/− and derived Msh6−/−Xpa−/− ES cells. Indeed, UU-CPD were detected specifically at the transcribed DNA strand of Hprt, confirming previous observations (Fig. 1 B; Hendriks et al., 2010). However, the levels of these UU-CPD were not increased in the absence of Msh2/Msh6 (Fig. 1 C). This result excludes the possibility that noncanonical MMR to any significant extent excises deaminated CC-CPD.
Figure 1.
Msh2/Msh6-dependent suppression of UVC-induced mutations is not related to ncMMR. (A) C’s within CPD lesions can deaminate spontaneously to U’s that instruct the mutagenic incorporation of adenines during replication. Excision of U-containing CPD by ncMMR might preclude their mutagenicity. Levels of site-specific UU-CPD at an ectopic Hprt minigene can be quantified, after photoreversal of the CPD in vitro, by PCR using primers with a 3′ AA dinucleotide and quantitative Southern blotting. Primers with a 3′ GG dinucleotide are used as standards (see Hendriks et al., 2010 for primer sequences). (B) Autoradiographs of Southern blots of PCR products, specific for the four tested UU-CPD. Numerals refer to the position in the Hprt cDNA. TS, UU-CPD at the transcribed strand; NTS, UU-CPD at the nontranscribed strand; X, Xpa−/−; MX, Msh6−/−Xpa−/−. (C) Quantification of deaminated CC-CPD from the autoradiographs shown in B, corrected for the standards.
Msh2/Msh6 acts independently of NER to suppress the mutagenicity of photolesions
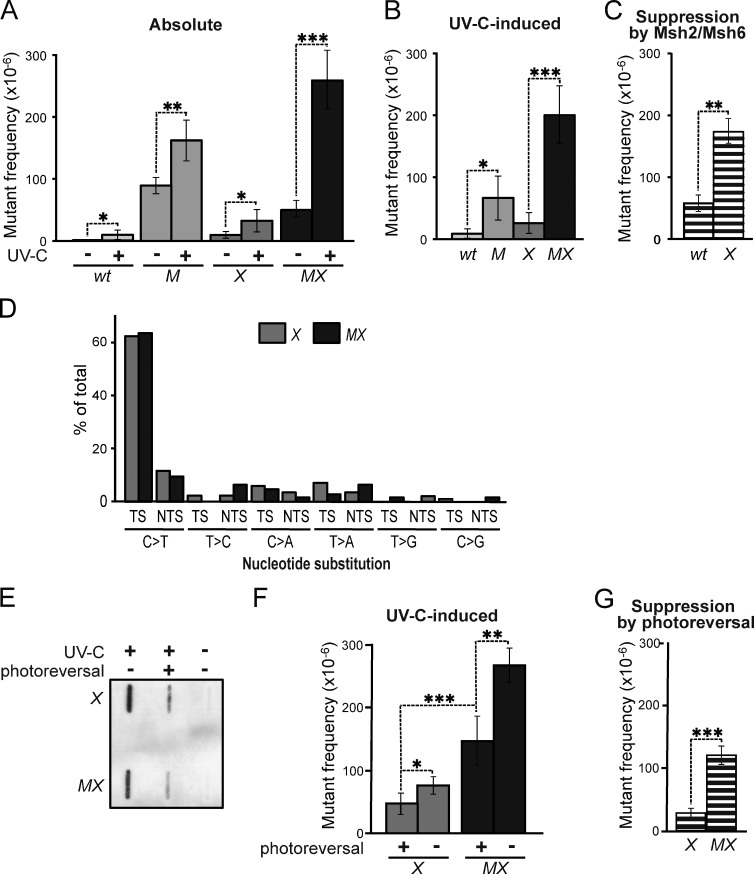
We asked whether the Msh2/Msh6-dependent suppression of UVC-induced mutations reflects an involvement of MMR components in NER (Lee et al., 2004). This possibility predicts that the disruption of Msh2/Msh6 affects UVC-induced mutagenesis in NER-proficient cells, but not in NER-deficient cells. This was investigated by measuring spontaneous and UVC-induced mutant frequencies in isogenic WT, Msh6−/−, Xpa−/−, and two independently derived Msh6−/−Xpa−/− ES cell lines (lines 4 and 30; Fig. 2 A and Fig. S1 A). We then calculated UVC-induced mutant frequencies by subtracting mock treatment–induced frequencies from mutant frequencies upon UVC exposure (Fig. 2 B). Msh2/Msh6 provided protection against low-dose UVC-induced mutagenesis in NER-proficient cells, confirming our previous results (Fig. 1 B and Fig. S1 B; Borgdorff et al., 2006). Importantly, in NER-deficient cells, Msh2/Msh6 conferred even better protection against UVC-induced mutagenesis (Fig. 2 C and Fig. S1 C), demonstrating that Msh2/Msh6 operates independently of NER to mitigate UVC-induced mutagenesis. Of note, the protection against induced mutant frequencies provided by Msh2/Msh6 was even greater than that provided by NER (compare both M and X with WT in Fig. 2 B and Fig. S1 B). This result attests to the physiological relevance of Msh2/Msh6 in determining the mutagenic consequences of UVC exposure.
Figure 2.
Msh2/Msh6 acts independently of NER to suppress the mutagenicity of photolesions. (A) Frequencies of mutants at the genomic Hprt gene in isogenic WT, Msh6−/− (M), Xpa−/− (X), and Msh6−/−Xpa−/− (MX, line 4) ES cells after mock or low-dose UVC (0.75 J/m2), treatment. Bars represent averages of at least three independent experiments. See Fig. S1 for results using an independently derived Msh6−/−Xpa−/− ES cell line (line 30). (B) UVC-induced mutant frequencies in isogenic ES cell lines, derived from data in A. (C) Msh2/Msh6-dependent suppression of UVC-induced mutagenesis in NER-proficient (WT) and in NER-deficient (X) backgrounds. Frequencies were derived by subtraction of induced mutant frequencies in Msh6-proficient cells from those in Msh6-deficient cells (B) in each individual experiment, followed by averaging. (D) Relative frequencies of UVC (0.75 J/m2)-induced nucleotide substitutions at dipyrimidines in Xpa−/− (X) and Msh6−/−Xpa−/− (MX) ES cells, derived from Table S1. TS, dipyrimidine (photolesion site) located at the transcribed DNA strand; NTS, dipyrimidine at the nontranscribed DNA strand. (E) Slot blot illustrating photoreversal of the majority of CPD in UVC-treated (0.75 J/m2) ES cell lines. (F) UVC (0.75 J/m2)-induced mutant frequencies in Xpa−/− (X) and in Msh6−/−Xpa−/− (MX) ES cells, with or without CPD photoreversal. All frequencies were corrected for spontaneous mutant frequencies. Bars represent averages of three independent experiments. (G) Effect of CPD photoreversal on UVC-induced mutant frequencies in Xpa−/− (X) and in Msh6−/−Xpa−/− (MX) cells. Frequencies were calculated by subtraction of induced mutant frequencies in Msh6-proficient cells from those in Msh6-deficient cells (F) in each individual experiment, followed by averaging.
We considered two different Msh2/Msh6-dependent mechanisms for the suppression of mutations induced by UVC: (i) canonical MMR, either at misincorporations provoked by UVC-induced nucleotide pool imbalances (Newman and Miller, 1985), or caused by misincorporations by TLS while replicating (undamaged) nucleotides adjacent to the lesions; (ii) a novel Msh2/Msh6-dependent pathway that operates at incorrect insertions opposite poorly instructive or noninstructive, nucleotide lesions. Comparison of UVC-induced mutation spectra at Hprt in Xpa−/− and Msh6−/−Xpa−/− cells demonstrated that Msh2/Msh6 suppresses mutations at dipyrimidines, the fingerprints of photolesions. Moreover, we did not observe significant differences between both genotypes with respect to (i) the strong inclination of substitutions toward the 3′ (non- or poorly instructive) nucleotide of dipyrimidines, (ii) the nature of these substitutions, (iii) their location within the Hprt gene, or (iv) their distribution at the transcribed versus nontranscribed DNA strands (Fig. 2 D and Table S1). Thus, rather than by repairing inadvertent misincorporations caused by nucleotide pool imbalances, or by error-prone replication by TLS, both at undamaged templates, Msh2/Msh6 presumably suppresses mutations that are targeted by photolesions at dipyrimidines.
To investigate whether Msh2/Msh6 suppresses the mutagenicity of CPD lesions that, although moderately helix-distorting, have retained basepairing capacity we stably introduced the marsupial CPD photolyase gene in Xpa−/− and Msh6−/−Xpa−/− ES cells. This enables the reversal of CPD at will by exposing UVC-treated cells to visible light (Fig. 2 E; Jans et al., 2005; Beukers et al., 2008). CPD reversal prevented UVC-induced mutagenesis, which attests to the mutagenicity of these prevalent and persistent photolesions, and this suppression was observed both in Msh2/Msh6-proficient cells and in Msh2/Msh6-deficient cells (Fig. 2 F). However, this prevention was more pronounced in Msh2/Msh6-deficient cells than in Msh2/Msh6-proficient cells (Fig. 2 G). Thus, CPD reversal and Msh2/Msh6 represent independent pathways that each reduce the mutagenicity of CPD. Nevertheless, even after removal of almost all CPD by photoreversal, the mutagenicity of UVC was significantly reduced in an Msh2/Msh6-dependent fashion (Fig. 2 F). This result surprisingly suggests that Msh2/Msh6, in addition to mutations induced by CPD, also suppresses mutations induced by strongly distorting, noninstructive (6–4)PP.
Msh2/Msh6-dependent induction of persistent gaps opposite (6–4)PP
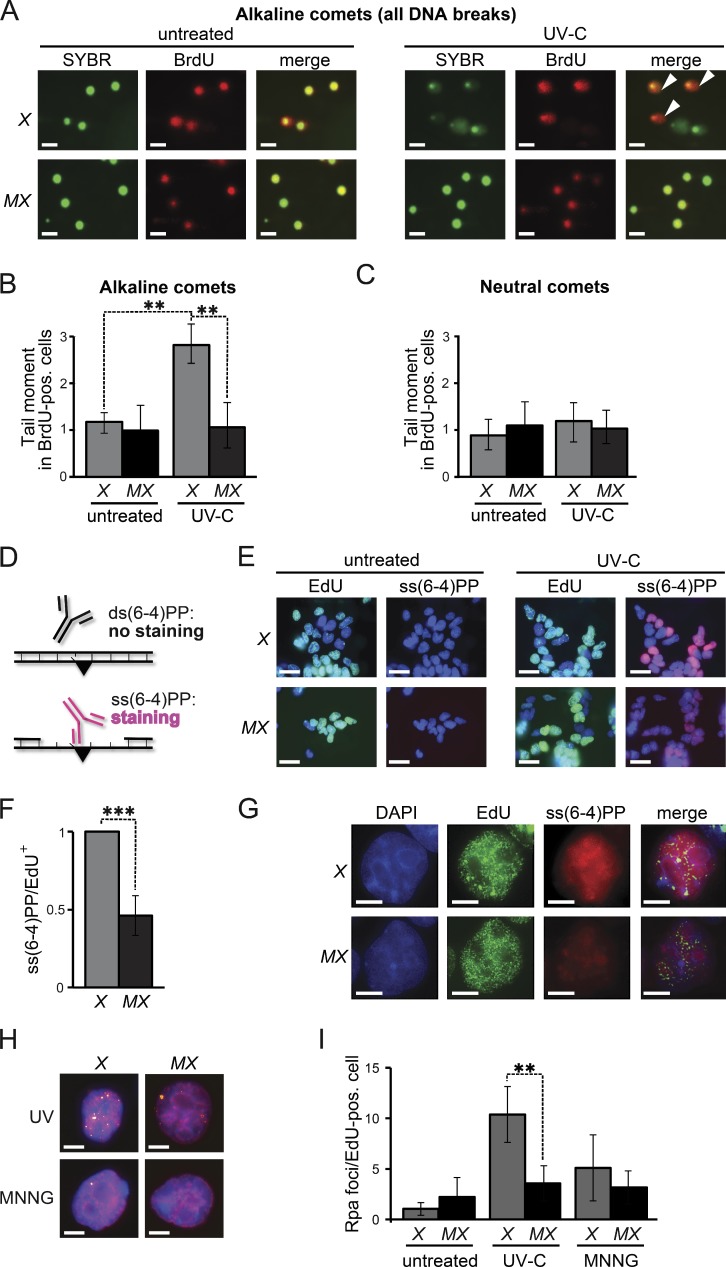
We investigated the possibility that Msh2/Msh6 is involved in an excision repair pathway in response to UVC treatment by investigating the Msh2/Msh6-dependent induction of DNA strand interruptions during the cell cycle of low-dose UVC exposure. Immediately upon exposure, replicating DNA was pulse-labeled with BrdU, and cell cycle progression was measured by bivariate flow cytometry. Both Xpa−/− and Msh6−/−Xpa−/− ES cells progressed through the cell cycle and most of the cells had reached the second S phase after 16 h, despite the persistence of both CPD and (6–4)PP (Fig. S2). When the spindle poison nocodazole was added immediately upon exposure, virtually all cells of both genotypes had arrested at mitosis after 16 h (Fig. S2). We then used single-cell electrophoresis (comet assays) to investigate the presence of ssDNA or dsDNA discontinuities in these mitotic cells, using Xpa−/− cells to avoid possible interference from NER-mediated incisions. Comets apparent after electrophoresis under alkaline conditions represent all (ss and ds) DNA strand discontinuities, whereas comets apparent after neutral electrophoresis specifically represent dsDNA breaks. No comets were detected in BrdU-positive, mitotic, Msh6−/−Xpa−/− cells under either electrophoresis condition, indicating that efficient DNA damage tolerance does not depend on Msh2/Msh6 (Fig. 3, A–C). In contrast, in BrdU-positive Xpa−/− cells, comets were detected under alkaline, but not under neutral, electrophoresis conditions. Thus, low-dose UVC exposure during S phase specifically induces Msh2/Msh6-dependent ssDNA discontinuities, and these persist at least to mitosis.
Figure 3.
Msh2/Msh6-dependent, S-phase-associated, induction of ss(6–4)PP. (A) Alkaline comet assays of nocodazole-treated (mitotic) cells 16 h after UVC-treatment (0.75 J/m2). Nuclei are stained with SYBR green. BrdU-positive nuclei were replicating during exposure. Comets (white arrowheads) are representative of ssDNA breaks. X, Xpa−/− ES cells, MX, Msh6−/−Xpa−/− line 4. Bars, 10 µM. (B) Quantification of tail moments, representing ssDNA breaks, in BrdU-positive mitotic cells after alkaline electrophoresis. Bars represent averages of three independent experiments. (C) Quantification of tail moments, representing dsDNA breaks, in BrdU-positive mitotic cells after neutral electrophoresis. Bars represent averages of three independent experiments. (D) Immunostaining to detect ss(6–4)PP (black triangle). (E) Staining for ss(6–4)PP (magenta) in ES cell lines, UVC treated (2 J/m2) during replication (EdU-positive nuclei, green), at 4 h after exposure. X, Xpa−/− ES cells; MX, Msh6−/−Xpa−/− line 4. Blue, nuclear stain (DAPI). Bars, 10 µM. (F) Relative levels of ss(6–4)PP in EdU-positive nuclei at 4 h after UVC exposure. Values were corrected for the 0 h time point. The ss(6–4)PP level in Xpa−/− ES cells was set to 1. Bars represent averages of three independent experiments. (G) Partial co-localization of Msh2/Msh6-dependent ss(6–4)PP with EdU (replication foci; pulse-labeled immediately after UVC exposure). Cells were stained at 4 h after UVC exposure. In the merged panels, the brightness of the EdU channel was reduced slightly. Bars, 2.5 µM. (H) Chromatin-associated Rpa foci (yellow) at 8 h after treatment of replicating (EdU positive, magenta) cells with UVC (0.75 J/m2), or with MNNG (4 µM) to induce canonical MMR. Blue, nuclear stain (DAPI). Bars, 2.5 µM. (I) Quantification of the numbers of Rpa foci in EdU-positive nuclei, at 8 h after UVC or MNNG treatment. One representative experiment of in total three experiments is shown.
We argued that these ssDNA discontinuities might reflect Msh2/Msh6-dependent excision at the DNA strand opposite noninstructive (6–4)PP. This possibility predicts that, by excising the opposing strand, (6–4)PP embedded in dsDNA will be converted to ss(6–4)PP, in an Msh2/Msh6-dependent fashion. This was investigated using a highly sensitive staining procedure, based on a monoclonal antibody that specifically detects ss(6–4)PP (Fig. 3 D; Jansen et al., 2009). To avoid any interference with NER-dependent excision, we used Xpa−/− and Msh6−/−Xpa−/− ES cells. In addition, the cells were pulse-labeled with the replication marker 5-ethynyl-2′-deoxyuridine (EdU) immediately upon low-dose UVC exposure, enabling us to identify cells at S phase during UVC treatment. At 4 h after treatment, pronounced Msh2/Msh6-dependent staining of ss(6–4)PP was visible, surrounding the EdU-positive replication foci (Fig. 3, E–G). These results suggest that Msh2/Msh6 either inhibits post-replicative TLS, resulting in the persistence of unreplicated ss(6–4)PP, or mediates excision at the strand opposite (6–4)PP, resulting in the de novo induction of ss(6–4)PP patches.
We then investigated whether these ss(6–4)PP patches are large and persistent enough to be visualized as chromatin-bound Rpa foci. Indeed, heterogeneously-sized Rpa foci were induced by low-dose UVC treatment during S phase, in an Msh2/Msh6-dependent fashion (Fig. 3, H and I). Consistent with previous data, the methylating drug methyl-N′-nitro-N-nitrosoguanidine (MNNG), which is known to induce canonical MMR, did not induce Rpa foci, presumably reflecting the transient nature of these ssDNA patches (Stojic et al., 2004; York and Modrich, 2006; Mojas et al., 2007). This result provides additional evidence that Msh2/Msh6-dependent ss(6–4)PP patches do not result from canonical MMR.
Dependence of the UVC-induced intra-S checkpoint on Msh2/Msh6
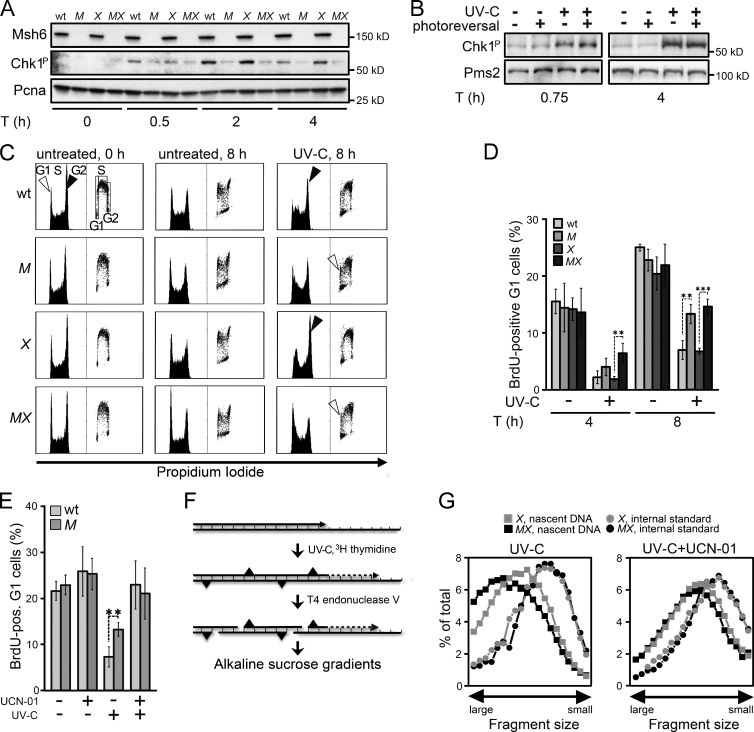
We hypothesized that the Rpa-covered ss(6–4)PP patches contribute to canonical Rpa–Atr–Chk1-mediated DNA damage signaling. Surprisingly, in NER-deficient ES cells treated with low-dose UVC, phosphorylation of Chk1 largely depended on Msh2/Msh6 proficiency, suggesting that the Msh2/Msh6-dependent ss(6–4)PP patches are dominant inducers of DNA damage signaling (Fig. 4 A and Fig. S3 A). Consistently, CPD photoreversal did not reduce the Msh2/Msh6-dependent Chk1 phosphorylation, suggesting a relation between the helical distortion conferred by the lesion and its propensity to induce DNA damage signaling via Msh2/Msh6-dependent excision (Fig. 4 B). Of note, although Msh2/Msh6-dependent Chk1 phosphorylation was most pronounced in Xpa−/− cells, Msh2/Msh6 appeared to be required for optimal DNA damage signaling also in NER-proficient cells (Fig. 4 A).
Figure 4.
Requirement of Msh2/Msh6 for optimal checkpoint induction. (A) Immunoblot displaying Chk1 phosphorylation upon UVC (0.75 J/m2) exposure of ES cell lines. WT, WT ES cells; M, Msh6−/−; X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. Pcna, internal standard. Shown is a representative experiment of three independent experiments. See Fig. S3 A for one independent experiment. (B) Chk1 phosphorylation in UVC (0.75 J/m2)-treated Xpa−/− ES cells is not affected by photoreversal of CPD. The 45-min time point represents cells immediately after photoreversal. Pms2, internal standard. Shown is a representative experiment of three independent experiments. (C) Bivariate cytometry displaying cell cycle progression upon UVC exposure. NER-proficient ES cells were treated with 2 J/m2 UVC, NER-deficient cells with 0.75 J/m2 UVC. Exposure was immediately followed by pulse labeling of replicating cells with BrdU. Vertical axis, cell numbers (left) or BrdU staining intensity (right). M, Msh6−/−; X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. G1, G1 phase; S, S phase;, G2, G2/M phase. Filled arrowheads, accumulation at late S/G2/M phase. Open arrowheads, appearance of BrdU-positive cells at the G1 phase after UVC-exposure. A representative experiment of three independent experiments is shown. (D) Quantification of UVC-exposed BrdU-positive cells at G1 at 4 and 8 h after treatment from three independent experiments. M, Msh6−/−; X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. (E) Quantification of UVC-exposed BrdU-positive ES cells at G1, cultured for 8 h in the presence of the Chk1 inhibitor UCN-01. Bars represent averages from three independent experiments. M, Msh6−/−. (F) Assessment of maturation of nascent DNA strands at 4 h after UVC treatment. [14C]-labeled intra-CPD fragments in fractions from the gradients are derived from parental DNA, serving as internal standard. [3H]-labeled fragments in fractions from the gradients represent nascent DNA. (G) Maturation of nascent DNA strands of ES cell lines, cultured for 4 h after UVC (5 J/m2) treatment in the absence (left) or presence (right) of the Chk1 inhibitor UCN-01. X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. Depicted is a representative experiment from two independent experiments. See Fig. S3 E for a similar experiment, using the Atr inhibitor Caffeine.
It was shown previously that Msh2/Msh6-deficient cells fail to efficiently accumulate at late S and G2 phases of the cell cycle upon low-dose UV treatment, suggesting a defect in checkpoint activation (van Oosten et al., 2005; Seifert et al., 2008). This was confirmed and extended here using both NER-proficient and NER-deficient ES cells and their Msh2/Msh6-deficient derivatives (Fig. 4 C and Fig. S3, B and C). Corroborating the checkpoint defect, Msh2/Msh6-deficient ES cells displayed accelerated progression to the subsequent G1 phase upon low-dose UVC treatment, irrespective of their NER status (Fig. 4 D and Fig. S3 D). The difference in cell cycle progression was abolished by culturing the cells in the presence of the Chk1 inhibitor UCN-01 after exposure (Fig. 4 E). Cumulatively, these data demonstrate that Msh2/Msh6-dependent ss(6–4)PP patches are dominant inducers of Chk1-dependent cell cycle arrests in response to low-dose UVC treatment.
We asked whether the Msh2/Msh6-dependent cell cycle delay reflects the intra-S checkpoint that slows the initiation and progression of replication during late S phase after low-dose UV exposure (Heffernan et al., 2002; Seiler et al., 2007). To address this, we used a sensitive alkaline sucrose gradient–based sedimentation assay that measures the maturation of daughter DNA strands beyond an internal standard (Fig. 4 F). This analysis revealed that the delay in the maturation of daughter strands, induced by UVC exposure, was abolished by loss of Msh2/Msh6 (Fig. 4 G). The difference in replicon maturation between WT and Msh2/Msh6-deficient cells was abolished when UVC-exposed cells were cultured in the presence of UCN-01 or the Atr inhibitor caffeine (Figs. 4 G and S3 E). Thus, irrespective of the NER status, Msh2/Msh6 mediates a bona fide, Rpa–Atr–Chk1-mediated, intra-S checkpoint upon UVC exposure, most likely via the induction of persistent ss(6–4)PP patches.
We entertained the possibility that the intra-S checkpoint might induce the selective elimination of heavily damaged cells that carry the highest risk of mutations, which would explain the Msh2/Msh6-dependent reduction in UVC-induced mutagenesis. This possibility was investigated by measuring UVC-induced mutant frequencies in WT and Msh6−/− ES cells, cultured in the presence of UCN-01 to abolish the checkpoint (Fig. S3 F). This treatment did not alter the Msh2/Msh6-dependent suppression of induced mutant frequencies (Fig. S3G). We conclude that the Msh2/Msh6-dependent intra-S checkpoint itself is not responsible for the suppression of UVC-induced mutations.
Delayed cytotoxicity of (6–4)PP depends on Msh2/Msh6
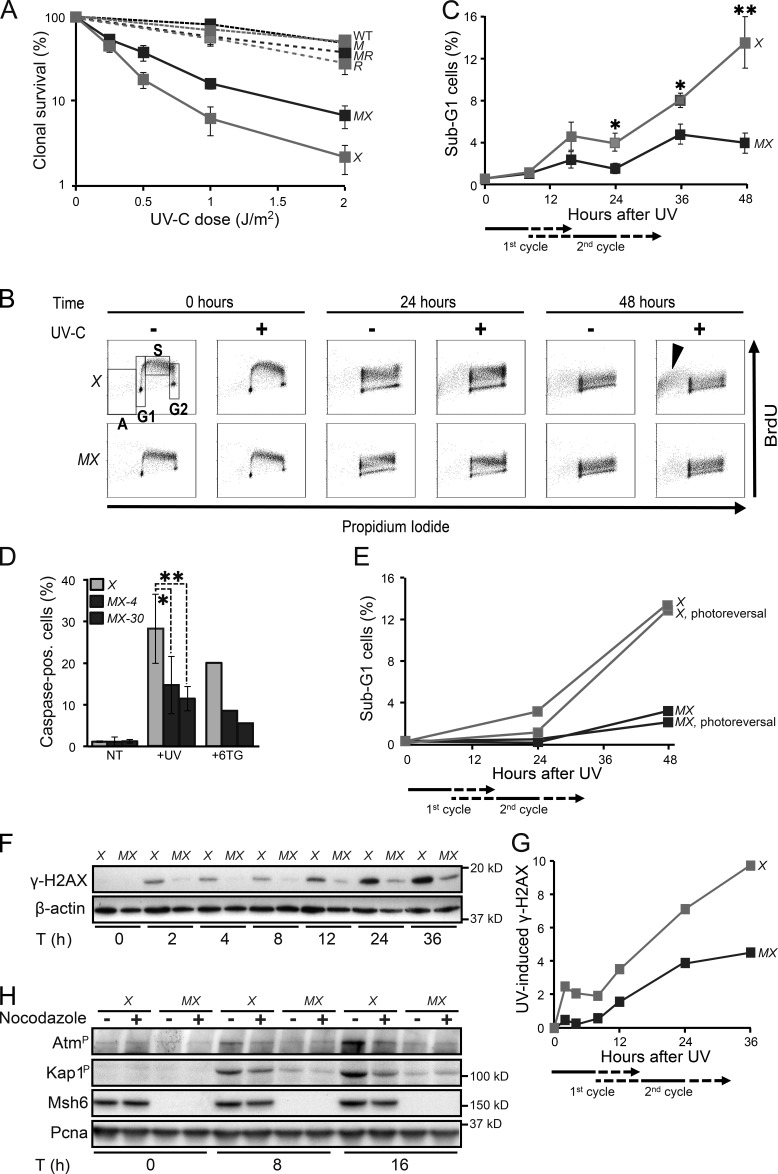
Previously, we have found that disruption of Msh2/Msh6 does not affect the UVC sensitivity of NER-proficient ES cells (Borgdorff et al., 2006). However, Msh2/Msh6 deficiency resulted in a significant reduction of the toxicity of UVC in Xpa−/− cells, which suggests that the cytotoxicity of UVC in part is mediated by Msh2/Msh6-dependent ss(6–4)PP patches that are expected to be more abundant in the absence of NER (Fig. 5 A). To investigate the kinetics of the Msh2/Msh6-dependent cell death we labeled replicating DNA with BrdU, immediately upon low-dose UVC exposure. In Xpa−/− ES cells, a BrdU-positive sub-G1 population became apparent only beyond 24 h post-exposure, during or after the second cell cycle after exposure (Fig. 5, B and C; and Fig. S4). Caspase staining confirmed that the delayed sub-G1 population represents apoptotic cells (Fig. 5 D). Such a delayed response resembles the apoptotic response that is induced during the second S phase after exposure, after the collapse of transient, but iterative, ssDNA tracts induced by canonical MMR at (near-normal) 6-methylthioguanine.T mispairs (Fig. 5 D; Mojas et al., 2007). Photoreversal of CPD did not quench the Msh2/Msh6-dependent induction of apoptosis in Xpa−/− cells, in further support of the causal involvement of ss(6–4)PP patches (Fig. 5 E).
Figure 5.
Msh2/Msh6-dependent delayed apoptotic responses to UVC. (A) Clonal survival of ES cell lines used in this study in response to UVC. M, Msh6−/−; X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4; R, Rev1B/B; MR, Msh6−/−Rev1B/B. Lines represent averages from three independent experiments. (B) Bivariate cytometry of cell cycle progression in isogenic ES cell lines after exposure to 0.75 J/m2 UVC or mock treatment, followed by pulse labeling with BrdU, at later time points. Cells were analyzed for DNA content (propidium iodide staining) and for cell cycle progression (BrdU staining). A, sub-G1 fraction; G1, G1 phase; S, early S phase; G2, late S/G2/M phase. Arrowhead, late-appearing sub-G1 population. See Fig. S4 for an independent experiment. X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. 10,000 cells were analyzed per data point. (C) Quantification of sub-G1 fractions (fractions A; Fig. 5 B). The progression of cells to the second cell cycle after treatment is deduced from the dilution of the BrdU signal (see also Figs. S2 and S4). X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. Lines represent averages from three independent experiments. (D) Identification of apoptotic cells by staining for activated caspases. Two independent Msh6−/−Xpa−/− ES cell lines (4 and 30) were tested. 6-thioguanine (6TG) was used as a positive control for the induction of delayed apoptosis by canonical MMR (Mojas et al., 2007). X, Xpa−/−; MX-4 and MX-30, Msh6−/−Xpa−/− lines 4 and 30, respectively. One experiment is shown from three independent experiments. (E) UVC (0.75 J/m2)-induced apoptosis is not mitigated by photoreversal of CPD. X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. One representative experiment is shown from three independent experiments. (F) Immunoblot displaying γ-H2AX levels in adherent cells upon UVC treatment (0.75 J/m2). β-actin, internal standard. X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. One representative experiment is shown from three independent experiments. G. Quantification of the Immunoblot depicted in Fig. 5F. The signals for γ-H2AX were normalized with respect to the corresponding signals for β-actin. X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. H. Msh2/Msh6-dependent induction of the dsDNA breaks markers phospho-Atm and phospho-Kap1 during the second cell cycle after low-dose (0.75 J/m2) UVC treatment. PCNA, internal standard. X, Xpa−/−; MX, Msh6−/−Xpa−/− line 4. One representative experiment is shown from three independent experiments.
To better understand the origin of the Msh2/Msh6-dependent apoptosis, we stained cells treated with low-dose UVC for the ss and dsDNA breaks marker γ (phosphorylated) H2AX (de Feraudy et al., 2010). In Xpa−/− ES cells, the induction of γ-H2AX upon UVC treatment appeared biphasic, and both phases largely depended on Msh2/Msh6 (Fig. 5, F and G). The first phase coincided with the induction of ss(6–4)PP patches, and with the phosphorylation of Chk1, during the cell cycle of exposure (Fig. 3; Fig. 4, A and B; and Fig. S3 A). However, the second phase of γ-H2AX appeared only during the second cell cycle upon exposure, preceding the induction of apoptosis (Fig. 5 C). To investigate whether dsDNA breaks are induced during the second S phase, we measured autophosphorylation of the dsDNA break signaling kinase Atm, as well as the Atm-dependent phosphorylation of Kap1 (Shiloh and Ziv, 2013). Both Atm and Kap1 phosphorylation were induced in an Msh2/Msh6-dependent fashion, most strongly at 16 h after low-dose UVC treatment, when most cells reside in the second cycle after exposure (Fig. 5 H). Indeed, Atm and Kap1 phosphorylation were significantly reduced when progression of the exposed cells to the second cycle was blocked using nocodazole (Fig. 5 H). Collectively, these results demonstrate the Msh2/Msh6-dependent induction of dsDNA breaks, but only during the second cell cycle upon exposure.
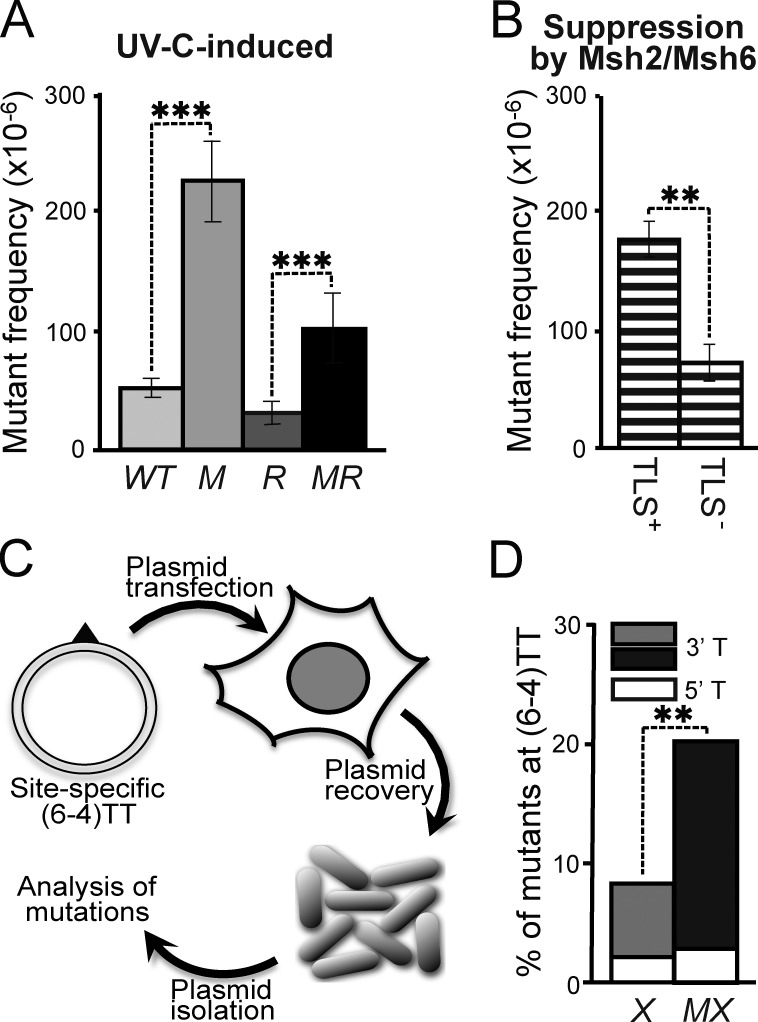
Msh2/Msh6-dependent correction of mutagenic TLS at (6–4)PP
Because ss(6–4)PP are noninstructive, any incorporation at this lesion (even the correct one) cannot be instructed by basepairing. Nevertheless, the Msh2/Msh6-dependent reduction of the mutagenicity of (6–4)PP, combined with the induction of (6–4)PP patches, suggests the selective excision of nucleotides that are ‘incorrectly’ incorporated by mutagenic TLS opposite these photolesions. This hypothesis predicts that Msh2/Msh6 should only suppress UVC-induced nucleotide substitutions at (6–4)PP, in case mutagenic TLS is operational. To investigate this, we used ES cells with a targeted deletion of the N-terminal BRCT domain of Rev1 (called Rev1B/B cells) that display a reduction in mutagenic TLS at (6–4)PP (Fig. 6 A; Jansen et al., 2005; Jansen et al., 2009; Diamant et al., 2012). Rev1B/B cells were only slightly sensitized to UVC, whereas the additional disruption of Msh6 did not alter this sensitivity (Fig. 5 A). Next, we determined frequencies of UVC-induced Hprt mutants in the WT, Msh6−/−, Rev1B/B, and Rev1B/BMsh6−/− ES cells. This revealed that the suppression of UVC-induced mutations by Msh2/Msh6 was significantly attenuated in Rev1B/B cells, compared with WT cells (Fig. 6 B). This epistasis between Rev1 and Msh6 provides strong genetic evidence for the Msh2/Msh6-dependent, selective repair of Rev1 BRCT domain–dependent incorrect incorporations at noninstructive photolesions. The observation that disruption of Msh2/Msh6 resulted in a residual increase of UVC-induced mutant frequencies also in the Rev1B/B cells (Fig. 6 B) indicates that Msh2/Msh6 also mediates excision of incorrect nucleotides incorporated by Rev1 BRCT domain-independent mutagenic TLS.
Figure 6.
Epistasis of mutagenic TLS and Msh2/Msh6. (A) UVC (2 J/m2)-induced mutant frequencies in isogenic WT, Msh6−/− (M), Rev1B/B (R), and Msh6−/−Rev1B/B (MR) ES cells. Bars represent averages from three independent experiments. (B) Msh2/Msh6-dependent suppression of UVC-induced mutant frequencies in Rev1-proficient (TLS+) and Rev1B/B (TLS−) ES cells, derived from A. Frequencies were calculated by subtraction of induced mutant frequencies in Rev1B/B cells from those in Rev1B/BMsh6−/− cells in each individual experiment, followed by averaging. (C) In vivo assay to quantify mutagenic TLS at a site-specific (6–4)TT (triangle) on a transfected replicating plasmid. (D) Mutant frequencies at the 5′ and the 3′ thymidines of a site-specific (6–4)TT in NER-deficient (Xpc−/−) mouse embryonic fibroblast lines in the presence (X) or absence (MX) of Msh2/Msh6. Bars represent averages from two experiments (see Fig. S5).
We next set out to provide direct evidence for such a novel post-TLS repair pathway. This was done by measuring the mutagenicity of TLS at a site-specific (6–4)TT within a replicating plasmid, transfected into gene-targeted mouse embryonic fibroblast (MEF) lines (Figs. 6 C and S5 A). In this setting, TLS of the lesion and its mutagenicity cannot be affected by DNA damage responses of the cell. In this experiment, Msh2/Msh6 deficiency did not significantly affect the overall TLS efficiency at the site-specific lesion (Fig. S5 B), providing additional evidence that Msh2/Msh6 is not required for TLS itself. However, the mutagenicity of TLS at the most distorted and noninstructive 3′ thymidine of the (6–4)TT was significantly increased in the absence of Msh2/Msh6 (Fig. 6 D; Fig. S5 C; and Table S2). These results are fully consistent with the increase of mutations at the 3′ nucleotide of dipyrimidines within the genome in the absence of Msh2/Msh6 (Fig. 2, B and D; Fig. S1 B; and Table S1). For these reasons this experiment provides direct evidence of an Msh2/Msh6-dependent repair pathway that excises nucleotides incorrectly incorporated by TLS opposite (6–4)PP.
Discussion
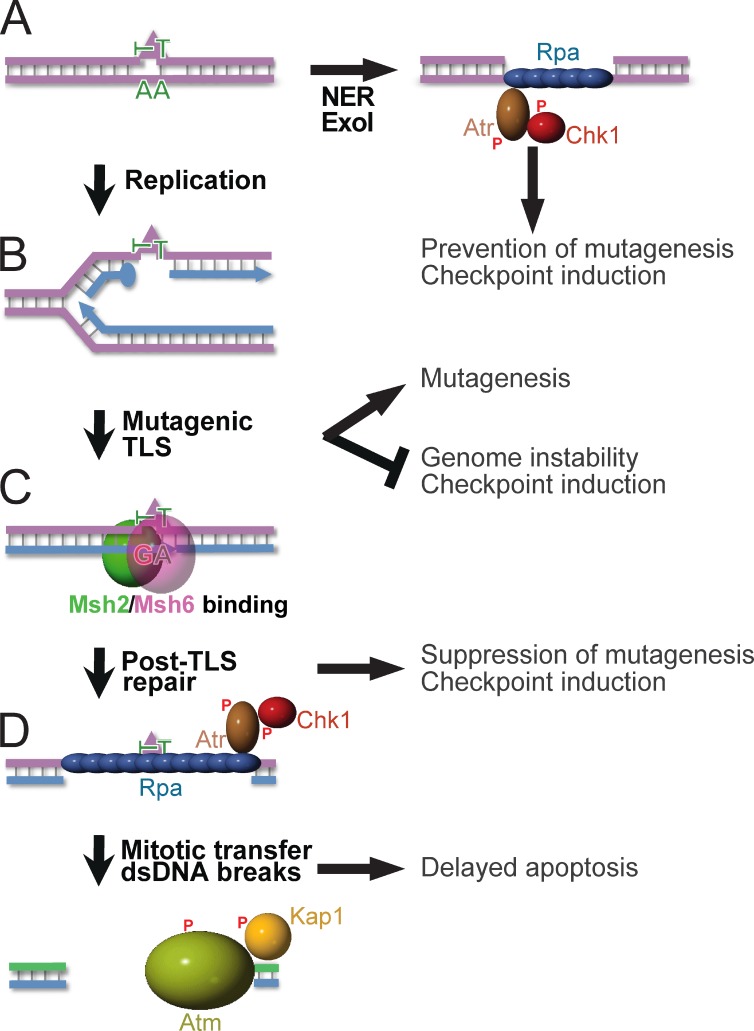
Cellular responses to genotoxic, carcinogenic agents are highly diverse, including nucleotide substitution mutagenesis, gross-genomic instability, cell cycle arrests, and apoptosis. Here, we wanted to resolve the relative roles of Msh2/Msh6, NER, and TLS in these responses, and to provide a mechanistic basis for Msh2/Msh6 in the orchestration of these pleiotropic responses to physiologically relevant densities of lesions. This work has unveiled a novel excision repair pathway, called post-TLS repair, that excises misincorporations by TLS at structurally aberrant, even noninstructive, nucleotide lesions. This results in suppression of their mutagenicity, in the induction of the canonical Rpa–Atr–Chk1-dependent, intra-S checkpoint, and when ss(6–6)PP patches are transferred through mitosis to the subsequent cell cycle in dsDNA breaks and apoptosis (Fig. 7). Providing biophysical support for this Msh2/Msh6-dependent repair pathway, purified Msh2/Msh6 specifically binds to mismatched, but not to matched CPD and (6–4)PP in vitro (Wang et al., 1999, 2006). Our data demonstrate that TLS itself is not measurably affected by loss of Msh2/Msh6 (Figs. 3 and S5). Ubiquitination of PCNA upon UVC exposure, a prerequisite for error-free TLS (Chang and Cimprich, 2009; Sale et al., 2012), largely depends on Msh2/Msh6 and it has been suggested that the recruitment of TLS polymerases to photolesions depends on Msh2 (Lv et al., 2013). However, our data suggest that TLS is normal in the absence of Msh2/Msh6 and that PCNA ubiquitination and the Msh2/Msh6-dependent recruitment of TLS polymerases may reflect the requirement of TLS to perform filling of ss(6–4)PP and ssCPD gaps only after Msh2/Msh6-dependent post-TLS repair.
Figure 7.
Orchestration of diverse responses to low-dose UVC by post-TLS repair. P, Phosphate moieties A. NER repairs structurally aberrant DNA lesions (triangle). In nonreplicating cells, Exo1 can lengthen NER-induced excision tracts, provoking Rpa–Atr–Chk1-dependent checkpoint responses (Novarina et al., 2011; Sertic et al., 2011). (B) DNA lesions that escape NER arrest processive DNA polymerases δ or ε, necessitating postreplicative TLS for lesion bypass. By incorporating a nucleotide opposite the lesions, TLS precludes gross-genomic instability and the induction of checkpoints. The frequent incorporation of an incorrect nucleotide at poor or noninstructive lesions causes the inherent mutagenicity of TLS. (C) Msh2/Msh6 specifically recognizes incorrect nucleotides incorporated by TLS. This initiates their excision, which prevents mutagenesis. Persistent excision tracts by post-TLS repair opposite noninstructive lesions underlie the Rpa–Atr–Chk1-mediated intra-S checkpoint. (D) Excision tracts that are transferred to the subsequent cell cycle collapse to dsDNA breaks, reflected by autophosphorylation of Atm and by Atm-dependent phosphorylation of Kap1. We hypothesize that these dsDNA breaks originate from replicative runoff at the gap within the template, and are causative of the delayed Msh2/Msh6-dependent apoptosis.
Although downstream components of post-TLS repair remain to be identified and may be similar or even identical to those of canonical MMR, our data imply that this repair pathway is distinct from canonical and noncanonical MMR in several important respects. Thus, canonical and noncanonical MMR act at mispaired, non-, or slightly damaged nucleotides (such as uridine and methylated or oxidized guanines), without inducing rapid cell cycle checkpoints (Zlatanou et al., 2011; Peña-Diaz et al., 2012; Rodriguez et al., 2012; Jiricny, 2013).
Post-TLS repair, the intra-S checkpoint, and delayed apoptosis
One hypothesis explaining the involvement of MMR proteins in DNA damage signaling entails the direct recruitment of DNA damage signaling proteins, including Atr, by MMR proteins tethered to damaged DNA, in the absence of excision (Yoshioka et al., 2006; Bai et al., 2010; Liu et al., 2010; Pabla et al., 2011). In contrast, our data rather reveal that canonical Rpa–Atr–Chk1-dependent checkpoint responses are induced by persistent ss(6–6)PP patches, derived from post-TLS repair opposite noninstructive lesions, at least at low lesion densities (Figs. 3, 4, and S3). The persistence of these ss(6–6)PP patches may be related to the requirement of PCNA ubiquitination and of both replicative and TLS polymerases for their filling (see above), and it may be pertinent to their propensity to induce DNA damage signaling. Most likely, excision tracts generated by canonical MMR at unmodified or slightly aberrant (such as methylated) nucleotides are filled rapidly, precluding the induction of DNA damage signaling (Stojic et al., 2004; Mojas et al., 2007). Furthermore, assuming that post-TLS repair-induced excision tracts have the same length as those induced by canonical MMR (up to 3 kg bps; Kadyrov et al., 2006), the cooperative nature of Rpa binding to long ssDNA patches (MacDougall et al., 2007) might further exacerbate DNA damage signaling. The poor checkpoint activation in the absence of Msh2/Msh6 (Figs. 4 and S3) indicates that, in contrast to previously assumed (Novarina et al., 2011), postreplicative gaps at arrested replication forks may not be able to strongly induce DNA damage responses. This possibly is related to the small size of these postreplicative gaps and to their rapid filling by TLS (Lopes et al., 2006; Elvers et al., 2011).
Our data, furthermore, strongly suggest that post-TLS repair-induced ss(6–4)PP patches that persist beyond mitosis induce apoptosis following the second S phase. Since accumulation of the dsDNA breaks markers phosphorylated Atm, Kap1 and H2AX precedes apoptosis (Figs. 5 and S3) it is conceivable that replicative runoff at the ss(6–4)PP patches, resulting in dsDNA breaks, is the direct trigger of the delayed apoptosis. This mechanistically is reminiscent of the apoptotic response to canonical MMR-induced transient but iterative excision tracts, during the second S phase upon exposure to methylating drugs or to 6-thioguanine (Stojic et al., 2004; Mojas et al., 2007).
Post-TLS repair and cancer
Remarkably, post-TLS repair is more efficacious than NER in mitigating the mutagenicity of UVC (Fig. 2 B). Moreover, after a low UVC dose (0.75 J/m2), the frequency of induced mutations prevented by post-TLS repair is already equal or greater than either the frequency of spontaneous mutations prevented by canonical MMR at normal misincorporations or the frequency of mutations prevented by NER at photolesions (Fig. 2, A and B; and Fig. S1 B). For these reasons, and also because post-TLS repair induces protective cell cycle and apoptotic responses, it is conceivable that post-TLS repair has a tumor-suppressive activity that matches or exceeds that of either canonical MMR or of NER. Indeed, acquired loss of MMR is associated with sunlight-induced skin cancer in mice and humans (Ponti et al., 2006; Young et al., 2008). Post-TLS repair likely is not confined to photolesions because mutagenic or apoptotic responses to intestinal genotoxins also largely depend on proficiency for MMR genes, irrespective of the NER status (Wu et al., 2003; Smith-Roe et al., 2006). Inherited defects in MMR genes cause Lynch syndrome, a common predisposition to, predominantly, colorectal cancer, whereas epigenetic silencing of the MMR gene hMLH1 characterizes a significant fraction of sporadic colorectal cancers (Boland and Goel, 2010). We hypothesize that loss of pleiotropic tumor-suppressing activities of post-TLS repair, in combination with continuous exposure to intestinal genotoxins, provides a rationale to understand the tissue tropism of MMR gene-deficient colorectal cancer.
Materials and methods
Cell lines and cell culture
WT primary diploid 129/OLA mouse-derived ES cell line E14 (Wakayama et al., 1999) was parental to all cell lines used in this study. Msh2−/− ES cell line sMSH2-9 carries a targeted insertion of a Pgk promoter-driven hygromycin-resistance cassette in exon 4 at both Msh2 alleles (de Wind et al., 1995). An ES cell line with disruptions of exons 3 and 4 of each Xpa allele by a Pgk promoter-hygromycin and a Pgk promoter-neomycin resistance marker, respectively, and a Tet-regulatable Hprt minigene at the Rosa26 locus was previously described (Hendriks et al., 2010). Rev1B/B ES cells were generated by first replacing exons 2 and 3 of one Rev1 allele by a Pgk promoter-hygromycin marker (Jansen et al., 2005). This was followed by selection of loss of heterozygosity using a very high (1 mg/ml) hygromycin concentration, resulting in a biallelic Rev1 mutation. In all of these ES cell lines, Msh6 was additionally disrupted by electroporation with a gene-targeting construct that replaces part of exon 4 of Msh6 with a Pgk promoter-puromycin marker (de Wind et al., 1999). This was followed by selection of cells that have become Msh6−/−, resulting from loss of heterozygosity, using 6-thioguanine (Borgdorff et al., 2005). Loss of Msh6 expression was verified using immunoblotting (e.g., Fig. 5 H). Xpc−/− and Xpc−/−Msh2−/− MEFs were derived from 13.5-d embryos carrying a homozygous disruption of exon 10 of Xpc by a neomycin-resistance cassette (Cheo et al., 1997), crossed with Msh2+/− mice. MEF lines were immortalized using a 3T3 protocol.
Stable CPD photolyase-expressing ES cell lines were generated by electroporation with an human elongation factor-1α promoter-driven expression construct (Chung et al., 2002), expressing the marsupial CPD photolyase gene (Beukers et al., 2008). Photolyase expression was analyzed by semi-quantitative reverse-transcriptase PCR, and cell lines with similar expression were used for experiments. Photoreversal of CPD was performed by exposing the cells for 45 min to cool-white fluorescent light in a 30°C incubator, immediately upon UVC treatment. Photoreversal was verified using the MINIFOLD I slot-blot system (Schleicher & Schuell) and anti-CPD mouse monoclonal antibody TDM-2 (Cosmobio).
UVC treatment and analysis of cell cycle responses, cytotoxicity, mutant frequencies and mutational spectra
The exposure to 1 J/m2 short-wave UVC induces density of photolesions at the genome of ES cells that is similar to the induction of photolesions in the basal layer of the unprimed epidermis after exposure to bright sunlight for 5 min (Kuluncsics et al., 1999). Unless stated otherwise, we have used UVC, at doses of 2 J/m2 (for NER-proficient cells; roughly equaling a 10-min sunlight exposure of the skin) or of 0.75 J/m2 (for NER-deficient cells).
Treatment of ES cells with UVC, measurement of mutant frequencies and generation and analysis of mutation spectra were performed as described before (Borgdorff et al., 2006). In brief, asynchronically growing ES cells were washed with PBS and exposed to UVC (245 nm wavelength) light using a Philips TUV lamp at a dose rate of 0.06 J/s. To determine mutant frequencies, ES cells were exposed to UVC or mock treated, and propagated for 6 d on feeder cells in ES complete medium to allow loss of WT Hprt mRNA and protein. Subsequently, ES cells were seeded in medium containing 30 µM 6-thioguanine to select for Hprt-deficient clones. In parallel, cloning efficiencies were determined by seeding 250 cells of each population. After 9 d, clones were stained with Methylene blue solution [0.15% (wt/vol) in methanol] and counted. The frequencies of Hprt mutants were determined by correcting the number of 6TG-resistant colonies in the mock-treated and UVC-treated cell populations for the cloning efficiencies. Mutant frequencies and mutation spectra from Msh6−/− cell lines, obtained after UVC treatment, were corrected for spontaneous mutants in the same cell line, obtained after mock treatment. Msh2/Msh6-dependent suppression of UVC-induced mutant frequencies was calculated for each independent experiment, followed by averaging. Occasional interexperimental variation in mutant frequencies, presumably caused by fluctuations in the flux of the UVC lamps, was corrected for by using a WT internal control, included in all experiments.
Spectra of mutations at the genomic Hprt gene were determined of 6TG-resistant ES cell clones from independently UVC-treated populations. After RNA isolation, cDNA synthesis was performed using an Hprt-specific primer (5′-GCAGCAACTGACATTTCTAAA-3′). Hprt cDNA was amplified by PCR using primers hprt-mus1 (5′-TTTTTGCCGCGAGCCGACC-3′) and san2m13 (5′-CGACGTTGTAAAACGACGGCCAGTGCAGATTCAACTTGCCGCTC-3′). Automated DNA sequence analysis of the PCR products was performed using the M13 primer (5′-TTGTAAAACGACGGCCAGT-3′) on Applied Biosystems equipment.
To measure the induction of apoptosis, cells were treated with UVC or with 6-thioguanine (2 h, 40 µM) as a positive control for canonical MMR-induced delayed apoptosis. At 48 h after treatment, cells were collected by trypsinization, merged with detached cells from the culture supernatant and analyzed using the fluorescent Apofluor Green Caspase activity assay (ICN Biomedicals, Inc.).
Quantification of deaminated dicytosine-CPD
Spontaneous deamination of dicytidine-CPD to diuridine-CPD at the Hprt gene was determined as described in Fig. 2 A (Hendriks et al., 2010). Upon UVC exposure, cells were incubated at 37°C for 16 h in medium to allow cytosine deamination. UVC-induced CPDs were removed from the DNA with CPD photolyase enzyme. Equal amounts of DNA were used for PCR analysis with primers containing either 3′ GG (to amplify nondeaminated dicytosines) or AA (to amplify deaminated dicytosines), at either transcribed or nontranscribed DNA strands (Hendriks et al., 2010). PCR products were separated on agarose gels, transferred to Zeta-Probe membranes (Bio-Rad Laborattories), and hybridized with an [α32P]dATP-labeled Hprt probe. The intensity of the deamination-specific products was quantified on scanned x-ray films, using Photoshop 5.5 software (Adobe).
Maturation of nascent strands and single-cell gelelectrophoresis (Comet assays)
Maturation of nascent strands after exposure to 5 J/m2 UVC was quantified using an alkaline sucrose gradient assay (Fig. 4 E; Van Zeeland and Filon, 1982). In brief, cells were prelabeled with [14C]thymidine for ∼24 h to label parental DNA. After exposure to 5 J/m2 of UVC, cells were with [3H]thymidine for 4 h to label nascent DNA. A small aliquot of a concentrated cell suspension was permeabilized by freeze-thawing twice. Subsequently, the cell lysate was treated with T4 endonuclease V, which cuts specifically at CPDs and subjected to alkaline sucrose gradient sedimentation using a linear alkaline sucrose gradient that was centrifuged at 40,000 g for 123 min (ω2t = 1.3 × 1011 rads2/s) at 20°C. The gradients were fractionated and each fraction was assayed for radioactivity. This protocol is more sensitive than classical alkaline sucrose gradient assays.
Persistent ssDNA and dsDNA in mitotic cells that were UVC-exposed during replication was investigated using single-cell electrophoresis (’BrdU Comet assays’; McGlynn et al., 1999), under alkaline or neutral conditions. In brief, cells were pulse-labeled for 10 min with 5 µM BrdU, immediately after treatment with 0.75 J/m2 UVC. Next, nocodazole (300 ng/ml) was added to arrest the cells at mitosis. At 16 h after UVC treatment cells were subjected to single-cell (Comet) gelelectrophoresis under alkaline of neutral conditions. After electrophoresis nuclei were stained with SYBR green and for BrdU. Tail moments of at least 50 BrdU-positive cells were measured in each experiment.
Immunoblotting and immunocytochemistry
The following antibodies were used for immunoblotting: γ-H2AX (mouse; Millipore), P.Chk1Ser317 or P.Chk1Ser345 (rabbit; Bethyl Labs), P.Kap1Ser824 (rabbit; Bethyl Labs), P.AtmSer1981 (mouse; Cell Signaling Technology), PCNA (mouse PC10; Santa Cruz Biotechnology, Inc.), β-actin (rabbit; EMD Millipore), Pms2 and Msh6 (both mouse; BD). After incubation with secondary peroxidase-conjugated antibodies (goat; Bio-Rad Laboratories) proteins were visualized by enhanced chemiluminescence detection. For quantification, the signal for γ-H2AX was calibrated with respect to the β-actin signal derived from hybridization of the same membrane.
To investigate the induction of ssDNA opposite photolesions, we used a modification of an immunocytochemical assay that identifies photolesions, specifically when embedded within ssDNA (Jansen et al., 2009; Temviriyanukul et al., 2012). In brief, cells were treated with UVC (2 J/m2), followed by labeling of replicating cells with EdU (10 µM). At 4 h after exposure, cells were permeabilized in CSK buffer (10 mM HEPES-KOH, pH 7.4, 300 mM sucrose, 100 mM NaCl, and 3 mM MgCl2) + 1% Triton X-100) and fixed in 3.7% formaldehyde for 20 min. Cells were stained for incorporated EdU using Alexa Fluor 488–conjugated azide according to the manufacturer’s (Invitrogen) recommendations and then for ss(6–4)PP using mouse monoclonal antibody 64M-2 (Cosmobio). The sensitivity of the latter staining was increased by using Tyramide signal amplification (TSA-biotin conjugate; Perkin Elmer) and visualized using Streptavidin-Cy3 conjugate (Jackson ImmunoResearch Laboratories). The intensity of ss(6–4)PP staining in 30–50 EdU-positive cells was measured in each experimental sample.
Staining for chromatin-associated Rpa was performed as follows: ES cells were irradiated with 0.75 J/m2 UVC or treated with 4 µM MNNG (1 h, in serum-free medium, in the presence of 10 µM of the alkyltransferase inhibitor O6−Benzylguanine). Then, serum and EdU (10 µM) were added for 1 h and medium was replaced. At 8 h after exposure, cells were harvested, permeabilized, and fixed as above. Next, the cells were centrifuged onto poly(L)lysine-coated coverslips and fixed again, followed by washing in PBS. Incorporated EdU was visualized with Alexa Fluor 647–conjugated azide, as described above. Chromatin-associated Rpa foci were stained by subsequent incubation with a rat Rpa32 antibody (Cell Signaling Technology) and an anti–rat Cy3 secondary antibody (Jackson ImmunoResearch Laboratories), according to established protocols. Nuclei were counterstained with DAPI and mounted in Vectashield mounting medium (Vector laboratories). For each sample, Rpa foci in 70–100 EdU-positive cells were quantified.
Immunofluorescent stainings were viewed using a Zeiss Axioplan 2 or M2 microscope equipped with Zeiss plan-Apochromat 63×/1,4 oil or Zeiss plan-Apochromat 40×/1,3 oil objectives, at room temperature. Microscopy images were acquired an AxiopCam MRm camera using Zen 2011 software (Zeiss). Contrast, saturation and exposure of images were adjusted using Photoshop 5.5 software (Adobe), using the Focus Magic 3 plugin for sharpening of subnuclear foci. The same settings were applied for all images within an experiment, unless indicated otherwise.
Bivariate flow cytometry analysis
Exponentially growing ES cells were exposed to UVC or mock treated and immediately pulse-labeled with 10 µM BrdU (Millipore) for 30 min at 37°C in the dark. Next, culture medium containing BrdU was replaced by medium containing 5 µM Thymidine (Sigma-Aldrich). At different time points, cells were trypsinized and fixed with 70% ethanol. Before staining, cells were permeabilized by 2 M HCl/0.05% Triton-X for 35 min and subsequently neutralized by 1 M Tris-HCl, pH 7.5. Neutralized cells were incubated overnight with a mouse monoclonal antibody against BrdU (BD) at 4°C, followed by incubation with a FITC-conjugated rat anti–mouse antibody (BD) for 1 h at room temperature in the dark. Next, genomic DNA of the cells was stained with 10 µg/ml propidium iodide. BrdU incorporation and DNA content were determined using a FACSCalibur apparatus (BD).
Site-specific TLS assay
To produce a substrate for TLS carrying a site-specific (6–4)PP, a damaged oligonucleotide (5′-CTCGCTAGCATCTTCATCATACAGTCAGTG-3′; the position of the (6−4)TT is indicated in bold) was prepared as previously described (Iwai et al., 1996). The oligonucleotide was annealed to single-stranded pMTEX6 (top strand in Fig. S5 A) that contains a Polyoma virus replication origin and the large T antigen gene (a gift from M. Moriya and K. Hashimoto, Stony Brook University, NY), and then extended with T4 DNA polymerase (TaKaRa) and ligated with T4 DNA ligase (TaKaRa). The resulting covalently closed circular DNA was purified by CsCl-ethidium bromide density gradient centrifugation. Site-specific TLS assays in vivo were essentially performed as previously described (Yang et al., 2009). In brief, 1 µg of the substrate DNA was transfected into MEFs (2.5 × 105 cells in a 25-cm2 flask) by FuGENE6 transfection reagent (Roche) according to the manufacturer’s protocol. The MEFs were incubated for 24 h, and then detached by treating with trypsin-EDTA, seeded in a 75-cm2 flask, and cultured for an additional 24 h. Progeny plasmids were recovered by Hirt extraction and then treated with DpnI (New England Biolabs) for 1 h to remove nonreplicated input DNA. After phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation, the recovered plasmids were treated with or without EcoRV-HF (New England Biolabs). E. coli strain NEB10β (New England Biolabs) was transformed with the mixture of above plasmids and 0.08 ng of pZeo, which serves as an internal control for transformation efficiency, and then plated onto 1xYT plates with carbenicillin/blasticidin S and 1xYT plates with zeocin, respectively. The carbenicillin/blasticidin S-resistant colonies carrying EcoRV-digested plasmids were subjected to colony PCR to amplify the sequence surrounding the (6–4)TT oligonucleotide, followed by direct sequencing. Ratios of modified strand/unmodified strand, correct TLS, mutagenic TLS, and deletions were calculated as described in Fig. S5 C.
Statistical analysis
Unpaired two-tailed t tests were used to analyze data on mutant frequencies, apoptotic fractions, comets, and Rpa foci. All experiments were performed at least three times. Unless specified otherwise, error bars represent standard deviations in all figures. *, P < 0.05; **, P < 0.01; ***, P < 0.001. A χ2 test was used to analyze mutant frequencies in the TLS assay.
Online supplemental material
Table S1 lists full mutation spectra at Hprt of Xpa−/− Msh6−/−Xpa−/− (line 4) ES cells, after mock treatment or treatment with 0.75 J/m2 UVC. T2 lists the full mutation spectrum induced by TLS at a site-specific (6–4)TT at a replicating plasmid, after rescue from Xpc−/− Msh2−/−Xpc−/− MEFs (Fig. 6 C). Fig. S1 depicts spontaneous and UVC-induced mutant frequencies at Hprt in Xpa−/− and Msh6−/−Xpa−/− (line 30) ES cells. Fig. S2 displays cell cycle profiles of Xpa−/− and Msh6−/−Xpa−/− (line 4) ES cells, untreated or treated with low-dose UVC and nocodazole. Fig. S3 shows the role of the Msh2/Msh6 heterodimeric protein in UVC-induced DNA damage signaling and in intra-S checkpoint induction. Fig. S4 shows the delayed appearance of a sub-G1 fraction specifically in Msh2/Msh6-proficient ES cells. Fig. S5 displays additional data on TLS on a site-specific (6–4)TT at a replicating plasmid, after rescue from Xpc−/− Msh2−/−Xpc−/− MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408017/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201408017.dv.
Supplementary Material
Acknowledgments
We thank Mark Drost for critical comments on the manuscript. We would like to dedicate this work to the late John B. Hays (1937-2014) whose pioneering work and rigid scientific standards have been a source of inspiration.
This work was supported by grants from the EU (FP7) and from the Dutch Cancer Foundation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- (6-4)PP
- (6-4)pyrimidine-pyrimidone dimers
- 6TG
- 6-thioguanine
- CPD
- cyclobutane pyrimidine dimer
- dsDNA
- double-stranded DNA
- EdU
- 5-ethynyl-2’-deoxyuridine
- ES
- embryonic stem
- MEF
- mouse embryonic fibroblast
- MMR
- DNA mismatch repair
- MNNG
- methyl-N’-nitro-N-nitrosoguanidine
- NER
- nucleotide excision repair
- ss(6-4)PP
- single-stranded (6-4)pyrimidine-pyrimidone dimers
- ssDNA
- single-stranded DNA
- TLS
- DNA translesion synthesis
- UVC
- short-wave ultraviolet light
- X
- Xpa-deficient cells
- XM
- Xpa,Msh6 double-deficient cells
References
- Avkin S., Sevilya Z., Toube L., Geacintov N., Chaney S.G., Oren M., and Livneh Z.. 2006. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 22:407–413 10.1016/j.molcel.2006.03.022 [DOI] [PubMed] [Google Scholar]
- Bai H., Madabushi A., Guan X., and Lu A.-L.. 2010. Interaction between human mismatch repair recognition proteins and checkpoint sensor Rad9-Rad1-Hus1. DNA Repair (Amst.). 9:478–487 10.1016/j.dnarep.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K., Matsuda T., Masutani C., Hanaoka F., and Kunkel T.A.. 2001. Proofreading of DNA polymerase eta-dependent replication errors. J. Biol. Chem. 276:2317–2320 10.1074/jbc.C000690200 [DOI] [PubMed] [Google Scholar]
- Beukers R., Eker A.P.M., and Lohman P.H.M.. 2008. 50 years thymine dimer. DNA Repair (Amst.). 7:530–543 10.1016/j.dnarep.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Boland C.R., and Goel A.. 2010. Microsatellite instability in colorectal cancer. Gastroenterology. 138:2073–2087: e3 10.1053/j.gastro.2009.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgdorff V., van Hees-Stuivenberg S., Meijers C.M., and de Wind N.. 2005. Spontaneous and mutagen-induced loss of DNA mismatch repair in Msh2-heterozygous mammalian cells. Mutat. Res. 574:50–57 10.1016/j.mrfmmm.2005.01.021 [DOI] [PubMed] [Google Scholar]
- Borgdorff V., Pauw B., van Hees-Stuivenberg S., and de Wind N.. 2006. DNA mismatch repair mediates protection from mutagenesis induced by short-wave ultraviolet light. DNA Repair (Amst.). 5:1364–1372 10.1016/j.dnarep.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Chang D.J., and Cimprich K.A.. 2009. DNA damage tolerance: when it’s OK to make mistakes. Nat. Chem. Biol. 5:82–90 10.1038/nchembio.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheo D.L., Ruven H.J., Meira L.B., Hammer R.E., Burns D.K., Tappe N.J., van Zeeland A.A., Mullenders L.H., and Friedberg E.C.. 1997. Characterization of defective nucleotide excision repair in XPC mutant mice. Mutat. Res. 374:1–9 10.1016/S0027-5107(97)00046-8 [DOI] [PubMed] [Google Scholar]
- Chung S., Andersson T., Sonntag K.-C., Björklund L., Isacson O., and Kim K.-S.. 2002. Analysis of different promoter systems for efficient transgene expression in mouse embryonic stem cell lines. Stem Cells. 20:139–145 10.1634/stemcells.20-2-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., and Elledge S.J.. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Feraudy S., Revet I., Bezrookove V., Feeney L., and Cleaver J.E.. 2010. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 107:6870–6875 10.1073/pnas.1002175107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wind N., Dekker M., Berns A., Radman M., and te Riele H.. 1995. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 82:321–330 10.1016/0092-8674(95)90319-4 [DOI] [PubMed] [Google Scholar]
- de Wind N., Dekker M., Claij N., Jansen L., van Klink Y., Radman M., Riggins G., van der Valk M., van’t Wout K., and te Riele H.. 1999. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat. Genet. 23:359–362 10.1038/15544 [DOI] [PubMed] [Google Scholar]
- Diamant N., Hendel A., Vered I., Carell T., Reissner T., de Wind N., Geacinov N., and Livneh Z.. 2012. DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity. Nucleic Acids Res. 40:170–180 10.1093/nar/gkr596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvers I., Johansson F., Groth P., Erixon K., and Helleday T.. 2011. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 39:7049–7057 10.1093/nar/gkr420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan T.P., Simpson D.A., Frank A.R., Heinloth A.N., Paules R.S., Cordeiro-Stone M., and Kaufmann W.K.. 2002. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 22:8552–8561 10.1128/MCB.22.24.8552-8561.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks G., Calléja F., Besaratinia A., Vrieling H., Pfeifer G.P., Mullenders L.H.F., Jansen J.G., and de Wind N.. 2010. Transcription-dependent cytosine deamination is a novel mechanism in ultraviolet light-induced mutagenesis. Curr. Biol. 20:170–175 10.1016/j.cub.2009.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S., Shimizu M., Kamiya H., and Ohtsuka E.. 1996. Synthesis of a Phosphoramidite Coupling Unit of the Pyrimidine (6-4) Pyrimidone Photoproduct and Its Incorporation into Oligodeoxynucleotides. J. Am. Chem. Soc. 118:7642–7643 10.1021/ja9603158 [DOI] [Google Scholar]
- Jackson S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J., Schul W., Sert Y.-G., Rijksen Y., Rebel H., Eker A.P.M., Nakajima S., van Steeg H., de Gruijl F.R., Yasui A., et al. . 2005. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 15:105–115 10.1016/j.cub.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Jansen J.G., Tsaalbi-Shtylik A., Langerak P., Calléja F., Meijers C.M., Jacobs H., and de Wind N.. 2005. The BRCT domain of mammalian Rev1 is involved in regulating DNA translesion synthesis. Nucleic Acids Res. 33:356–365 10.1093/nar/gki189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J.G., Tsaalbi-Shtylik A., Hendriks G., Gali H., Hendel A., Johansson F., Erixon K., Livneh Z., Mullenders L.H.F., Haracska L., and de Wind N.. 2009. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol. Cell. Biol. 29:3113–3123 10.1128/MCB.00071-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J.2013. Postreplicative mismatch repair. Cold Spring Harb. Perspect. Biol. 5:a012633 10.1101/cshperspect.a012633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov F.A., Dzantiev L., Constantin N., and Modrich P.. 2006. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 126:297–308 10.1016/j.cell.2006.05.039 [DOI] [PubMed] [Google Scholar]
- Kuluncsics Z., Perdiz D., Brulay E., Muel B., and Sage E.. 1999. Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts. J. Photochem. Photobiol. B. 49:71–80 10.1016/S1011-1344(99)00034-2 [DOI] [PubMed] [Google Scholar]
- Lee D.F., Drouin R., Pitsikas P., and Rainbow A.J.. 2004. Detection of an involvement of the human mismatch repair genes hMLH1 and hMSH2 in nucleotide excision repair is dependent on UVC fluence to cells. Cancer Res. 64:3865–3870 10.1158/0008-5472.CAN-03-3193 [DOI] [PubMed] [Google Scholar]
- Liu Y., Fang Y., Shao H., Lindsey-Boltz L., Sancar A., and Modrich P.. 2010. Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway. J. Biol. Chem. 285:5974–5982 10.1074/jbc.M109.076109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Foiani M., and Sogo J.M.. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 21:15–27 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Lv L., Wang F., Ma X., Yang Y., Wang Z., Liu H., Li X., Liu Z., Zhang T., Huang M., et al. . 2013. Mismatch repair protein MSH2 regulates translesion DNA synthesis following exposure of cells to UV radiation. Nucleic Acids Res. 41:10312–10322 10.1093/nar/gkt793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall C.A., Byun T.S., Van C., Yee M.-C., and Cimprich K.A.. 2007. The structural determinants of checkpoint activation. Genes Dev. 21:898–903 10.1101/gad.1522607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn A.P., Wasson G., O’Connor J., McKerr G., McKelvey-Martin V.J., and Downes C.S.. 1999. The bromodeoxyuridine comet assay: detection of maturation of recently replicated DNA in individual cells. Cancer Res. 59:5912–5916. [PubMed] [Google Scholar]
- Mojas N., Lopes M., and Jiricny J.. 2007. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 21:3342–3355 10.1101/gad.455407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman C.N., and Miller J.H.. 1985. Mechanism of UV-induced deoxynucleoside triphosphate pool imbalance in CHO-K1 cells. Mutat. Res. 145:95–101. [DOI] [PubMed] [Google Scholar]
- Novarina D., Amara F., Lazzaro F., Plevani P., and Muzi-Falconi M.. 2011. Mind the gap: keeping UV lesions in check. DNA Repair (Amst.). 10:751–759 10.1016/j.dnarep.2011.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabla N., Ma Z., McIlhatton M.A., Fishel R., and Dong Z.. 2011. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J. Biol. Chem. 286:10411–10418 10.1074/jbc.M110.210989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Diaz J., Bregenhorn S., Ghodgaonkar M., Follonier C., Artola-Borán M., Castor D., Lopes M., Sartori A.A., and Jiricny J.. 2012. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 47:669–680 10.1016/j.molcel.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Pfeifer G.P., You Y.-H., and Besaratinia A.. 2005. Mutations induced by ultraviolet light. Mutat. Res. 571:19–31 10.1016/j.mrfmmm.2004.06.057 [DOI] [PubMed] [Google Scholar]
- Ponti G., Losi L., Pedroni M., Lucci-Cordisco E., Di Gregorio C., Pellacani G., and Seidenari S.. 2006. Value of MLH1 and MSH2 mutations in the appearance of Muir-Torre syndrome phenotype in HNPCC patients presenting sebaceous gland tumors or keratoacanthomas. J. Invest. Dermatol. 126:2302–2307 10.1038/sj.jid.5700475 [DOI] [PubMed] [Google Scholar]
- Rodriguez G.P., Romanova N.V., Bao G., Rouf N.C., Kow Y.W., and Crouse G.F.. 2012. Mismatch repair-dependent mutagenesis in nondividing cells. Proc. Natl. Acad. Sci. USA. 109:6153–6158 10.1073/pnas.1115361109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E., Lehmann A.R., and Woodgate R.. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13:141–152 10.1038/nrm3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert M., Scherer S.J., Edelmann W., Böhm M., Meineke V., Löbrich M., Tilgen W., and Reichrath J.. 2008. The DNA-mismatch repair enzyme hMSH2 modulates UV-B-induced cell cycle arrest and apoptosis in melanoma cells. J. Invest. Dermatol. 128:203–213 10.1038/sj.jid.5700941 [DOI] [PubMed] [Google Scholar]
- Seiler J.A., Conti C., Syed A., Aladjem M.I., and Pommier Y.. 2007. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol. 27:5806–5818 10.1128/MCB.02278-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertic S., Pizzi S., Cloney R., Lehmann A.R., Marini F., Plevani P., and Muzi-Falconi M.. 2011. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc. Natl. Acad. Sci. USA. 108:13647–13652 10.1073/pnas.1108547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y., and Ziv Y.. 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14:197–210 10.1038/nrm3546 [DOI] [PubMed] [Google Scholar]
- Shin-Darlak C.Y., Skinner A.M., and Turker M.S.. 2005. A role for Pms2 in the prevention of tandem CC —> TT substitutions induced by ultraviolet radiation and oxidative stress. DNA Repair (Amst.). 4:51–57 10.1016/j.dnarep.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Smith-Roe S.L., Hegan D.C., Glazer P.M., and Buermeyer A.B.. 2006. Mlh1-dependent suppression of specific mutations induced in vivo by the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine (PhIP). Mutat. Res. 594:101–112 10.1016/j.mrfmmm.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Stelter P., and Ulrich H.D.. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 425:188–191 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- Stojic L., Mojas N., Cejka P., Di Pietro M., Ferrari S., Marra G., and Jiricny J.. 2004. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 18:1331–1344 10.1101/gad.294404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temviriyanukul P., van Hees-Stuivenberg S., Delbos F., Jacobs H., de Wind N., and Jansen J.G.. 2012. Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome. DNA Repair (Amst.). 11:550–558 10.1016/j.dnarep.2012.03.007 [DOI] [PubMed] [Google Scholar]
- van Oosten M., Stout G.J., Backendorf C., Rebel H., de Wind N., Darroudi F., van Kranen H.J., de Gruijl F.R., and Mullenders L.H.. 2005. Mismatch repair protein Msh2 contributes to UVB-induced cell cycle arrest in epidermal and cultured mouse keratinocytes. DNA Repair (Amst.). 4:81–89 10.1016/j.dnarep.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Van Sloun P.P.H., Jansen J.G., Weeda G., Mullenders L.H., van Zeeland A.A., Lohman P.H., and Vrieling H.. 1999. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res. 27:3276–3282 10.1093/nar/27.16.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zeeland A.A., and Filon A.R.. 1982. Post-replication repair: elongation of daughter strand DNA in UV-irradiated mammalian cells in culture. Progress in Mutation Research. 4:375–384. [Google Scholar]
- Wakayama T., Rodriguez I., Perry A.C., Yanagimachi R., and Mombaerts P.. 1999. Mice cloned from embryonic stem cells. Proc. Natl. Acad. Sci. USA. 96:14984–14989 10.1073/pnas.96.26.14984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lawrence C.W., Li G.-M., and Hays J.B.. 1999. Specific binding of human MSH2.MSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J. Biol. Chem. 274:16894–16900 10.1074/jbc.274.24.16894 [DOI] [PubMed] [Google Scholar]
- Wang H., Hoffman P.D., Lawrence C., and Hays J.B.. 2006. Testing excision models for responses of mismatch-repair systems to UV photoproducts in DNA. Environ. Mol. Mutagen. 47:296–306 10.1002/em.20206 [DOI] [PubMed] [Google Scholar]
- Wu J., Zhu B.-B., Yu J., Zhu H., Qiu L., Kindy M.S., Gu L., Seidel A., and Li G.-M.. 2003. In vitro and in vivo modulations of benzo[c]phenanthrene-DNA adducts by DNA mismatch repair system. Nucleic Acids Res. 31:6428–6434 10.1093/nar/gkg875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I.-Y., Hashimoto K., de Wind N., Blair I.A., and Moriya M.. 2009. Two distinct translesion synthesis pathways across a lipid peroxidation-derived DNA adduct in mammalian cells. J. Biol. Chem. 284:191–198 10.1074/jbc.M806414200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York S.J., and Modrich P.. 2006. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. J. Biol. Chem. 281:22674–22683 10.1074/jbc.M603667200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K., Yoshioka Y., and Hsieh P.. 2006. ATR kinase activation mediated by MutSα and MutLα in response to cytotoxic O6-methylguanine adducts. Mol. Cell. 22:501–510 10.1016/j.molcel.2006.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L.C., Listgarten J., Trotter M.J., Andrew S.E., and Tron V.A.. 2008. Evidence that dysregulated DNA mismatch repair characterizes human nonmelanoma skin cancer. Br. J. Dermatol. 158:59–69. [DOI] [PubMed] [Google Scholar]
- Zlatanou A., Despras E., Braz-Petta T., Boubakour-Azzouz I., Pouvelle C., Stewart G.S., Nakajima S., Yasui A., Ishchenko A.A., and Kannouche P.L.. 2011. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol η in response to oxidative DNA damage in human cells. Mol. Cell. 43:649–662 10.1016/j.molcel.2011.06.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.