The TDP-43 target G3BP1 is essential for a functional interaction between stress granules and processing bodies.
Abstract
G3BP1, a target of TDP-43, is required for normal stress granule (SG) assembly, but the functional consequences of failed SG assembly remain unknown. Here, using both transformed cell lines and primary neurons, we investigated the functional impact of this disruption in SG dynamics. While stress-induced translational repression and recruitment of key SG proteins was undisturbed, depletion of G3BP1 or its upstream regulator TDP-43 disturbed normal interactions between SGs and processing bodies (PBs). This was concomitant with decreased SG size, reduced SG–PB docking, and impaired preservation of polyadenylated mRNA. Reintroduction of G3BP1 alone was sufficient to rescue all of these phenotypes, indicating that G3BP1 is essential for normal SG–PB interactions and SG function.
Introduction
Organisms have evolved a variety of strategies to overcome exposure to environmental stressors such as thermal stress, oxidative stress, osmotic stress, UV irradiation, or viral infection. One of these mechanisms is the formation of “stress-induced RNA granules” such as stress granules (SGs) and processing bodies (PBs; Kedersha et al., 1999; Sheth and Parker, 2003). Although SGs and PBs share many of the same protein and mRNA components, their exact compositions remain unknown (Lavut and Raveh, 2012; Stoecklin and Kedersha, 2013). SGs are cytoplasmic inclusions (≤5 µm2) that are absent in normal growth conditions but can be induced by stress stimuli. PBs are smaller, dense inclusions that are present in basal conditions, and can be induced by stress. Both structures recruit mRNA during stress, and this correlates with/further contributes to translational repression. The exact functions of SGs and PBs are a matter of some debate in the literature. The prevailing view is that SGs form primarily to protect endogenous mRNA from stress degradation, actively sorting transcripts for this storage versus degradation or reinitiation of translation, although this has never been explicitly demonstrated (Anderson and Kedersha, 2008). However, the chief function of PBs is considered to be mRNA degradation, although there are some reports that mRNA can be returned for reinitiation (Brengues et al., 2005). The stress-induced RNA granule response is conserved throughout evolution and is tightly regulated to facilitate cell survival after exposure to noxious stimuli (Thomas et al., 2011).
There is increasing evidence that SGs may participate in the pathogenesis of the late-onset neurodegenerative disease amyotrophic lateral sclerosis (ALS; Strong, 2010; Li et al., 2013). ALS-affected neurons frequently feature the mislocalization/misaccumulation of two RNA binding proteins (RBPs), TDP-43 and FUS. Both are nuclear RBPs involved in many aspects of RNA metabolism, and are known genetic causes of familial and sporadic ALS, which suggests deregulation of the RNA life cycle as a common pathogenic mechanism (Bentmann et al., 2013). While mutant TDP-43 and FUS demonstrate recruitment to SGs in response to many different stimuli, whether the consequent disruption in SG dynamics is due to a loss or gain of function is still a matter of debate (Andersson et al., 2008; Colombrita et al., 2009; Bosco et al., 2010; Liu-Yesucevitz et al., 2010; Dewey et al., 2011; McDonald et al., 2011; Aulas et al., 2012; Bentmann et al., 2012). We have previously reported that depletion of TDP-43, but not FUS, yields a significant down- and up-regulation of the core SG proteins G3BP1 and TIA-1, respectively (McDonald et al., 2011). Moreover, we have demonstrated that SG size is decreased by TDP-43 depletion via blocked secondary aggregation, which can be restored by exogenous expression of G3BP1 (Aulas et al., 2012). In neuronal-like cells, we have linked cellular vulnerability to oxidative stress to the loss of G3BP1 and its upstream regulator TDP-43 (Aulas et al., 2012).
In this study, we investigated the consequences of disrupted SG dynamics: namely the inability of SGs to achieve secondary aggregation as a consequence of decreased G3BP1 expression. We demonstrate that stress-induced translational arrest is normal despite TDP-43/G3BP1 depletion, but uncover that both molecules influence basal translation rates. In addition, in both transformed cells and primary neurons, TDP-43/G3BP1–mediated regulation of SG size determines the stability of interactions with PBs, another type of RNA granule, which is involved in mRNA decay. The disturbance in SG–PB interactions in stress conditions results in a progressive loss of polyadenylated transcripts. Collectively, these data demonstrate that the TDP-43 target G3BP1 is essential for a functional interaction between SGs and PBs.
Results
SG proteins and polyadenylated mRNA are localized normally to SGs despite impaired secondary aggregation
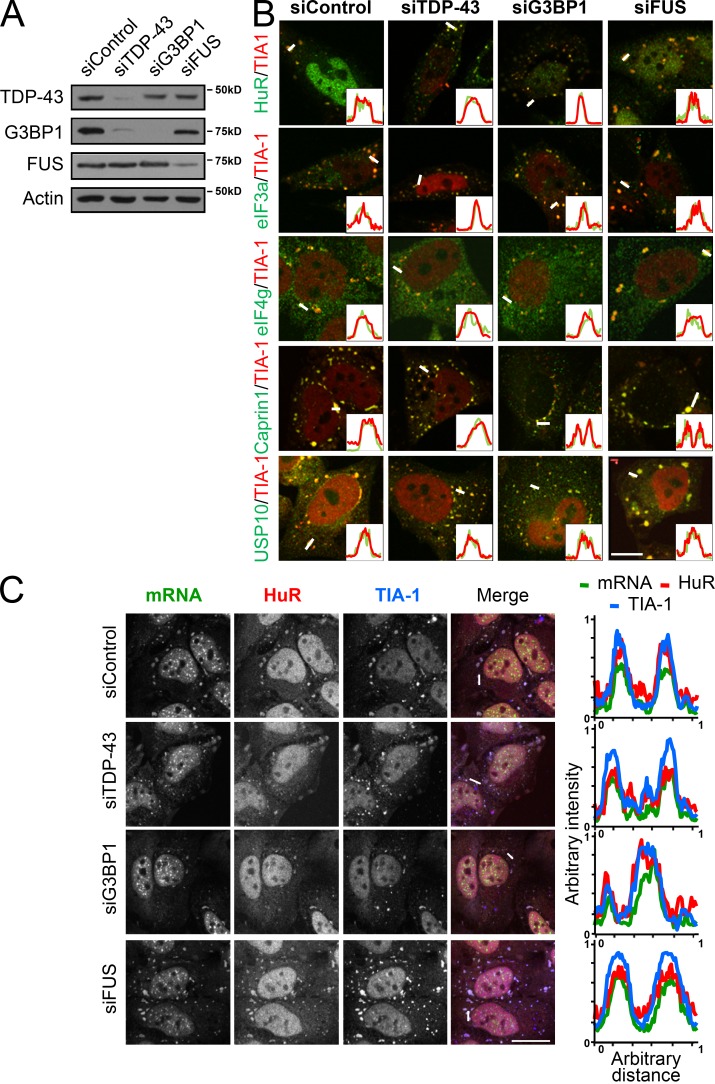
We have previously reported that depletion of TDP-43, or its target G3BP1, impairs the secondary aggregation step of SG assembly in response to oxidative stress, yielding increased numbers of SGs of reduced size. In contrast, SG assembly proceeds normally in FUS-depleted cells (Aulas et al., 2012). Given the reduction in size, we hypothesized that the recruitment of one or more SG components is disrupted by TDP-43/G3BP1 depletion. Because there is no method available to robustly isolate or enrich for SGs, we used a candidate approach to evaluate if the recruitment of certain SG components was altered in these smaller SGs. Specifically, we selected a panel of SG markers that are commonly used in the literature and have been previously shown to localize to SGs in a variety of contexts, namely HuR, eIF3A, and eIF4G (Kedersha and Anderson, 2002; Kimball et al., 2003; Liu-Yesucevitz et al., 2010; Dewey et al., 2011). Inspection of TDP-43– and G3BP1-depleted HeLa cells 60 min after a 30-min exposure to 0.5 mM sodium arsenite (SA; i.e., 90 min, the time at which SG secondary aggregation is complete) demonstrated undisturbed localization of HuR, eIF3A, and eIF4G to SGs marked with TIA-1 (Fig. 1, A and B). In fact, it was comparable to siControl and siFUS cells where SG size is normal. We noted the same localization in response to thermal stress and with a second set of siRNA for each of our target genes (unpublished data). Quantitative analysis also showed no major differences in SG protein localization between the groups (Fig. 1 B, histograms). The localization of Caprin1 and USP10, both known interacting partners of G3BP1 (Solomon et al., 2007; Takahashi et al., 2013), are also not affected by either TDP-43, G3BP1, or FUS depletion (Fig. 1 B).
Figure 1.
SG proteins and polyadenylated mRNA are well recruited to SGs despite impaired SG aggregation. (A–C) HeLa cells were transfected with the indicated siRNAs for 72 h. Coverslips were collected at 90 min after SA treatment (0.5 mM, 30 min). (A) Representative Western blot showing the efficiency of siRNA transfections. (B) Coverslips were labeled for eIF4G, eIF3A, HuR, Caprin1, or USP10 and the SG marker TIA-1. Bar, 25 µm. (C) Polyadenylated mRNA was visualized via fluorescence in situ hybridization with an oligo(dT) probe and HuR and TIA-1 labeling. Bar, 10 µm. Representative micrographs from ≥3 independent experiments are shown. Line scans (from the white lines) were used to assess colocalization (histograms).
SGs are composed of proteins and mRNA (Kedersha et al., 1999). Even though the function of SGs remains to be better defined, the leading hypothesis is that they are assembled to store and/or sort polyadenylated mRNA during a stress exposure (Kedersha et al., 2005). An obligate feature of SGs is the presence of polyadenylated mRNA. Thus, we performed FISH using a fluorescently conjugated oligo(dT) probe followed by HuR and TIA-1 immunostaining to label SGs (Fig. 1 C). In all conditions, mRNA was highly colocalized with TIA-1+HuR+ SA-induced SGs to normal levels. The same result was obtained with a second set of siRNAs and in response to thermal stress (unpublished data). Although we cannot exclude the possibility that depletion of TDP-43 or G3BP1 may influence the localization of specific transcripts or proteins to SGs, from our experiments we conclude that the localization of polyadenylated mRNA and these core SG proteins is normal despite impaired SG assembly.
Normal stress-induced translational repression in TDP-43–, G3BP1-, and FUS-depleted cells
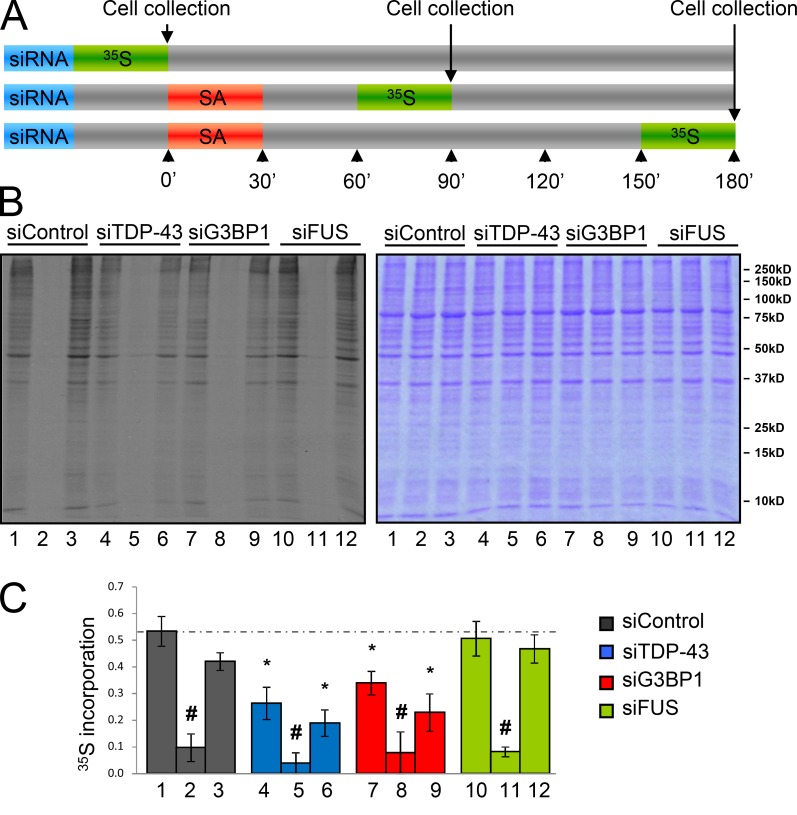
It is our hypothesis that the process of secondary aggregation in SG assembly (i.e., the fusion of numerous small SGs to fewer larger SGs organized perinuclearly) must hold some functional significance. However, there is no assay to test SG function per se. SG formation is concomitant to a translational repression after stress exposure, so as to provide translational priority to stress-responsive genes (Nover et al., 1983, 1989; Kedersha et al., 1999; Kedersha and Anderson, 2002). Although mRNA localization appeared normal, we speculated that the failure of SGs to coalesce may have an impact on the efficiency of this stress-induced translational repression and contribute to the increased cellular vulnerability we have previously reported in TDP-43–depleted cells (McDonald et al., 2011; Aulas et al., 2012). After a 72-h siRNA transfection, whole cell lysates were collected after pulsing with [35S]methionine for 30 min before SG assembly (i.e., unstressed), when SG coalescence is completed (90 min), and when SGs are fully resolved (180 min; McDonald et al., 2011; Aulas et al., 2012; Fig. 2). As expected, siControl cells displayed a robust transient repression of translation after SA treatment (82%) at 90 min with a full recovery to basal levels by 180 min (Fig. 2 B). Stress-induced translational repression occurred normally in TDP-43–, G3BP1-, and FUS-depleted cells, compared with siControl cells (Fig. 2 C; P = 0.421, 0.850, 0.791 for siTDP-43, siG3BP1, and siFUS, respectively), indicating that stress-induced translational repression is independent of SG size.
Figure 2.
TDP-43 and G3BP1 depletion induce a defect in basal translation. (A–C) HeLa cells were transfected with the indicated siRNAs for 72 h. Cells were stressed with 0.5 mM SA for 30 min and collected at 90 (lanes 2, 5, 8, and 11) and 180 min (lanes 3, 6, 9, and 12) and before stress (lanes 1, 4, 7, and 10). All samples were incubated with 50 µM [35S]methionine 30 min before cell lysis. Samples were prepared in Laemmli buffer and subjected to SDS-PAGE. (A) Experimental design. (B) Autoradiography of the cell lysates (left) and same gel stained with Coomassie blue to demonstrate equivalent protein loading (right). (C) The histogram shows densitometric quantification of the entire lane expressed relative to the Coomassie-stained gel. The means ± SEM (error bars) of ≥3 independent experiments are plotted. *, P < 0.05, compared with siControl; #, P < 0.05, compared with the unstressed sample.
Basal translation is diminished by TDP-43 and G3BP1 siRNA
In unstressed conditions, siTDP-43 and siG3BP1 cells exhibited a significant reduction in basal translation compared with siControl cells. Specifically, unstressed siTDP-43 and siG3BP1 cells were metabolically labeled to 49% and 64% (P = 0.003 and P = 0.008, respectively) of that observed in siControl cells, respectively (Fig. 2 B, left: compare lanes 4 and 7 to lane 1). Protein synthesis was not significantly different between siTDP-43 and siG3BP1 cells (P = 0.224). Interestingly, at 180 min after stress, translation returned to the lower pre-stress/basal level in both siTDP-43 and siG3BP1 cells (Fig. 2 B, top: compare lanes 4 and 6, 7 and 9). However, FUS-depleted cells exhibited basal translation levels comparable to siControl cells (P = 0.693). Coomassie blue staining confirms comparable protein loading between lanes (Fig. 2 B, right). These results were confirmed with a complementary ribopuromycylation assay (Fig. S1, A and B). This assay features a short pulse with puromycin, which is intercalated into nascent polypeptides, followed by immunoblotting with an anti-puromycin antibody. With this assay, basal translation levels in siTDP-43– and siG3BP1-treated HeLa cells were reduced 38 ± 7% and 36 ± 2% (P = 0.0053 and P = 0.0009, respectively) compared with siControl cells (Fig. S1, A and B), which is consistent with metabolic labeling.
To investigate further the defect in basal translation, we performed polysome profiling of siRNA-treated cells. Compared with siControl cells, TDP-43–depleted cells demonstrate a decrease in total RNA associated with polysomes, as detected by adsorption at 254 nm. This is accompanied by an increase in the peak corresponding to 80S ribosome-associated RNA, which is consistent with a defect in translation initiation (polysome/monosome ratio: siControl, 3.46 ± 0.31; siTDP-43, 2.8 ± 0.29; P < 0.001; Fig. S1 C, top). siG3BP1 cells also show a trend for reduced formation of polysomes (polysome/monosome ratio: siControl, 3.58 ± 0.11; siG3BP1, 3.39 ± 0.22; P = 0.057; Fig. S1 C, middle). In contrast, and consistent with metabolic labeling, siFUS cells exhibit a normal polysome-to-monosome ratio (polysome/monosome ratio: siControl, 3.42 ± 0.27; siFUS, 3.45 ± 0.09; P = 0.142; Fig. S1 C, bottom). Collectively, the data indicate that TDP-43 and its target G3BP1 are required for normal translation in basal conditions, whereas FUS is dispensable.
Increased levels of polyadenylated mRNA and PBs
To evaluate whether the observed deficits in protein synthesis in basal conditions were due to a reduced steady-state level of polyadenylated mRNA, we performed FISH with a fluorescently conjugated oligo(dT) probe and quantified single plane confocal images for total cellular fluorescence intensity (Fig. 3 A). This approach was chosen so as to provide a global appreciation of polyadenylated mRNA levels, and not focus on individual targets which may be under regulation by these multifunctional RNA-binding proteins. Compared with cells treated with control siRNA, polyadenylated mRNA levels were increased 1.35 ± 0.07-, 1.23 ± 0.06-, and 1.45 ± 0.2-fold in siTDP-43, siG3BP1, and siFUS cells, respectively (Fig. 3 A; P = 0.007, 0.025, and 0.038, respectively). A second independent approach involving spectrophotometric measurement of directly isolated polyadenylated mRNA yielded similar results (Fig. S1 D). These changes are not accounted for by deregulated steady-state levels of the PB-resident mRNA degradation enzymes Dcp1a or Xrn-1 (Fig. 3 B). The effect on polyadenylated mRNA was specific because total RNA levels were unchanged by TDP-43 siRNA (unpublished data). Also, despite an increased amount of polyadenylated mRNA, its nuclear–cytoplasmic distribution was comparable to siControl cells for all conditions (unpublished data).
Figure 3.
Increased mRNA correlates with alterations in PBs in basal conditions. (A–F) HeLa cells were transfected with the indicated siRNAs for 72 h. (A) Polyadenylated mRNA was visualized via FISH with an oligo(dT) probe. Total cellular fluorescence was quantified from at least 10 cells per experiment from ≥3 experiments (35 cells per sample in total). Bar, 10 µm. (B) Whole cell lysates were examined by immunoblotting for PB proteins Dcp1a and Xrn-1. Bands were quantified via densitometry and normalized to Actin. (C–F) Unstressed HeLa cells were labeled for Dcp1a (to visualize PBs) and TIA-1 (to visualize the nucleus). These data are extracted from the kinetics presented in Fig. S2 C. (C) Representative images of ≥3 experiments. Bar, 25 µm. (D) Percentage of cells with PBs in basal conditions. (E) Quantification of the number of individual PBs per cell. (F) Quantification of individual PB size. In all cases, the means ± SEM (error bars) of three independent experiments are plotted. *, P < 0.05.
Decreased levels of translation and increased polyadenylated mRNA levels in basal conditions have been linked to changes in another type of RNA granule known as PBs (Coller and Parker, 2005; Eulalio et al., 2007). PBs are present in basal conditions and are also induced by stress (Sheth and Parker, 2003; Kedersha et al., 2005). In general, PBs contain mRNA decapping and degradative enzymes and thus are considered to be sites of mRNA degradation (Sheth and Parker, 2003; Cougot et al., 2004). Using Dcp1a as a marker for PBs, we counted the number of cells containing PBs in normal culture conditions (Fig. 3, C and D). In siControl cells, 30 ± 4% of HeLa cells contained PBs in basal conditions. However, in siTDP-43, siG3BP1, and siFUS cells, the percentage of cells was increased two- and threefold (62 ± 4%, 60 ± 6%, and 90 ± 3%), respectively, compared with siControl cells (Fig. 3 D; P < 0.005 for all conditions). Collectively, these observations correlate well with the increased levels of polyadenylated transcripts and were confirmed with a second set of siRNAs (Fig. S2 A).
We also examined the total number of PBs per cell and the size of individual PBs in basal conditions. We observed there to be a trend of 1.4 ± 0.1-fold and a significant 1.5 ± 0.1-fold more PBs per individual cell in unstressed siTDP-43 and siFUS cells, but not siG3BP1 (Fig. 3 E; P = 0.079, 0.018, and 0.935, respectively). In addition, individual PB size was not significantly different for siTDP-43 and siG3BP1 cells, but was reduced 36 ± 12% in FUS-depleted cells (Fig. 3 F; P = 0.026). A size distribution analysis revealed there to be an increased proportion of the smallest sized PBs (0.1–0.2 µm2) in siFUS cells and a similar trend that doesn’t reach significance in siTDP-43 cells (Fig. S2 B; P = 0.041 and 0.075, respectively) in basal conditions. Thus, basal PB dynamics are variably disrupted in all siRNA conditions tested.
Stress-induced PB kinetics are normal in all conditions
Like SGs, PBs can also be induced by exogenous stress (Sheth and Parker, 2003; Kedersha et al., 2005; Eulalio et al., 2007). As we observed some PB deregulation in basal conditions, we next investigated whether there are alterations in stress-induced PB dynamics. Inspection of the number of cells forming PBs immediately after a 30-min exposure to SA shows that stress-induced PB formation occurs normally in TDP-43–, G3BP1-, and FUS-depleted cells (Fig. S2 C). In addition, PBs are observed in nearly 100% of SA-stressed HeLa cells at 30 and 90 min, and start to dissolve by 180 min, paralleling our previous observations for SG formation and disassembly (McDonald et al., 2011). Interestingly, we noted that at 180 min, regardless of siRNA condition, the number of cells containing PBs returns to the pre-stress level (Fig. S2 C). While the number of PBs is increased to a maximum of ~20 per cell compared with unstressed conditions for all siRNA conditions, we did not observe a stress-induced increase in the mean size of individual PBs (Fig. S2 D). However, at 180 min, an increased proportion of smaller PBs (0.15–1 µm2) was observed in siTDP-43 and siFUS cells when a size distribution analysis is performed (Fig. S2 B; P = 0.023 and P = 0.008, respectively).
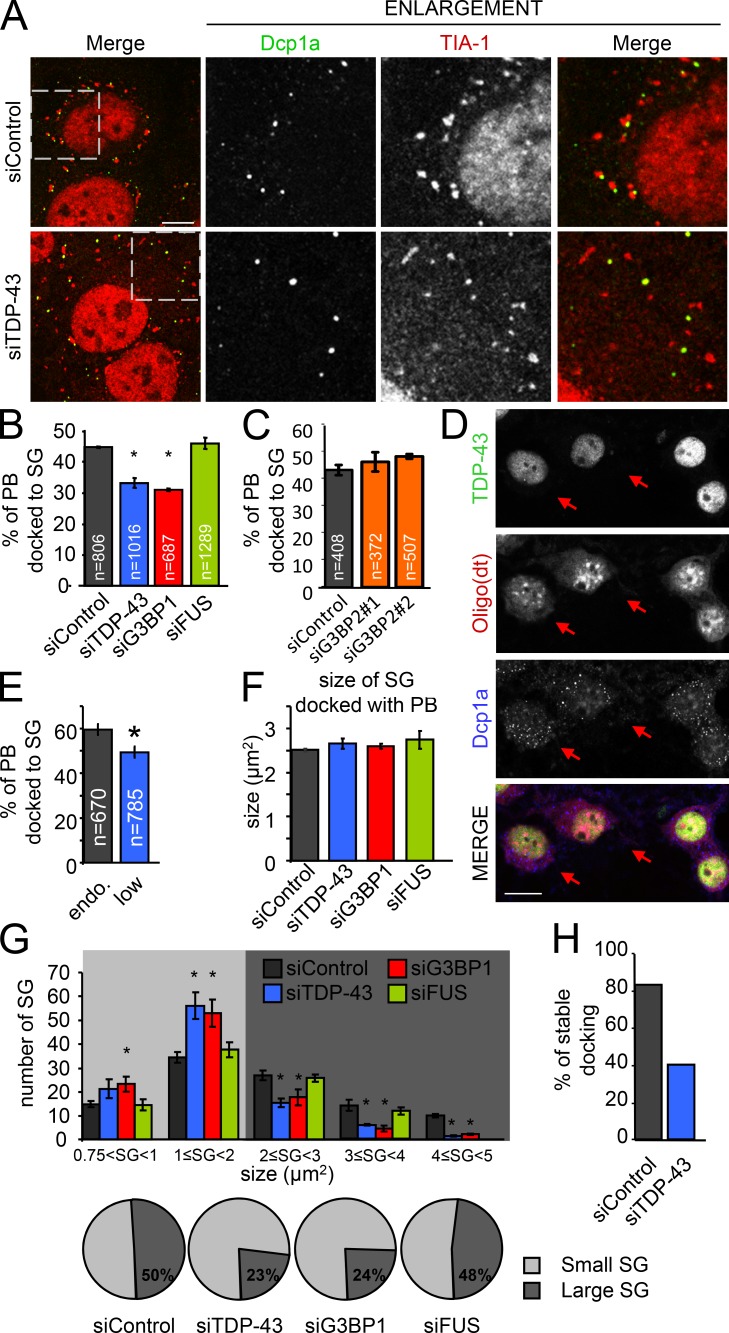
SG–PB docking is compromised in TDP-43– and G3BP1-depleted cells
After SA stress exposure, one or more PBs are frequently observed to make close and transient contact with SGs, referred to as docking (Kedersha et al., 2005; Stoecklin and Kedersha, 2013). The association of these two distinct types of RNA granules has been proposed to permit or facilitate the exchange of mRNA and proteins between the two structures (Kedersha et al., 2005). To determine if SG–PB docking is influenced by the absence of TDP-43, G3BP1, or FUS, we performed immunofluorescent labeling with markers defined for each structure. Although Dcp1a has been well established as being found only in PBs, TIA-1 is recognized as an SG marker that has also been observed in PBs in certain contexts (Dang et al., 2006; Leung et al., 2006; Stoecklin and Kedersha, 2013). However, in our experience with HeLa cells stressed with SA, TIA-1 is never detected in PBs. For these experiments, SGs are defined as TIA-1+ cytoplasmic puncta with a minimum size of 0.75 µm2. Coverslips were fixed at 90 min (including 30 min of SA exposure) so as to permit SGs and stress-induced PBs to fully assemble (Fig. 4 A). SG–PB docking was defined as when a Dcp1a+ puncta and a TIA-1+ puncta were in close proximity to each other such that there were no visible unstained pixels between the structures and/or when there was a partial overlap of the signals (Fig. 4 A). In siControl SA-treated HeLa cells, 45 ± 0.5% of PBs were docked with SGs. In contrast, SG–PB docking was significantly reduced in TDP-43– and G3BP1-depleted conditions (31 ± 2.0% and 31 ± 0.5%, respectively; P = 0.0002 and 0.000004), whereas siFUS exhibited SG–PB docking equivalent to controls (47 ± 1.3%, P = 0.225; Fig. 4 B). A second independent set of siRNAs confirmed these results (Fig. S2 E). Moreover, a second set of markers, namely p54/RCK and an oligo(dT) probe to label PBs and SGs, respectively, yielded similar results (Fig. S2 F). In addition, depletion of G3BP2, a close relative of G3BP1, via two independent siRNAs did not disturb SG–PB docking (Fig. 4 C) or SG size (not depicted), whereas a double knockdown of G3BP1 and G3BP2 yielded a near total block in SG formation, as previously reported (Matsuki et al., 2013; not depicted). Lastly, the observed deficits in SG–PB docking cannot be attributed to PBs because all samples demonstrate a similar PB size and number at this time point (Fig. S2 D).
Figure 4.
SG–PB docking is dependent on SG size. (A–C and F–H) HeLa cells were transfected with the indicated siRNAs for 72 h. (D and E) Cortical neurons treated with TDP-43 siRNA. (A) Representative images of docking in siControl and siTDP-43 cells. Cells were labeled for TIA-1 and Dcp1a to follow SGs and PBs, respectively. The boxed regions are enlarged on the right. Bar, 10 µm. (B and C) Quantification of docking between SGs and PBs in HeLa using TIA-1 and Dcp1a. PBs are considered docked when SGs and PBs were in contact. n = total number of events analyzed. (D) Representative images of unstressed transfected cortical neurons. Arrows indicate TDP-43Low cells. Bar, 10 µm. (E) Quantification of SG–PB docking in cortical neurons. TDP-43Low, n = 45 neurons; TDP-43Endogenous, n = 48 neurons. (F) Size of SGs when docked with PBs. (G) Distribution analysis of SG size. SG size was separated between small (0.75–2 µm2) and large (2–5 µm2) SGs, as shown in the pie charts. (H) The number of stable SG–PB docking events (large SGs only) during a 10-min window, as captured with live stage imaging. All data are representative of ≥3 independent experiments and plotted as the mean ± SEM (error bars). *, P < 0.05.
We also investigated SG–PB docking in primary cortical neurons. Neurons were transfected with siTDP-43 and subsequently stressed with SA. Due to the low transfection efficiency in neurons, cells were selected according to TDP-43 expression, as revealed by immunofluorescence labeling, and classified as either TDP-43Low– or TDP-43Endogenous–expressing cells (Fig. 4 D). Using the PB marker Dcp1a and an oligo(dT) probe to follow SGs, we observed an 11% decrease in SG–PB docking in TDP-43Low neurons compared with neurons exhibiting normal levels of TDP-43 (TDP-43Endogenous; P = 0.0045; Fig. 4 E). A similar decrease in docking was also observed in the neuronal-like cell line SH-SY5Y treated with siTDP-43 and siG3BP1 (P = 0.002 and P > 0.001) but not siFUS (P = 0.803; Fig. S2 G). Thus, using multiple cell types and markers, these data demonstrate that reduced levels of TDP-43 and/or G3BP1 similarly compromise SG–PB docking.
Stable SG–PB interactions are dependent on G3BP1-mediated SG size
Because SG–PB docking was reduced in siTDP-43 and siG3BP1 cells where SG size is diminished (Aulas et al., 2012), we wondered if SG size is a determinant of PB docking. To address this, we measured the size of SGs when they were docked with PBs. In HeLa cells stressed with SA, the mean size of a docked SG is ~2.5 µm2 regardless of siRNA condition (siControl, 2.48 ± 0.16 µm2; siTDP-43, 2.49 ± 0.22 µm2; siG3BP1, 2.54 ± 0.21 µm2; siFUS, 2.7 ± 0.18 µm2; Fig. 4 F). A distribution analysis for SG size in all siRNA-treated cells analyzed at 90 min after stress (including 30 min of SA treatment) reveals that SGs distribute into two broad classes: “small SGs” measuring 0.75 to <2 µm2 and “large SGs” measuring 2–5 µm2. In siControl cells, SGs are divided evenly between the two classes. In the context of siTDP-43 and siG3BP1, the proportion of SG that measure as “large SGs” is similarly reduced by approximately half to 23 ± 2% and 24 ± 4%, respectively (P = 0.002 and 0.035). In contrast, in siFUS cells where SG–PB docking is normal, the distribution of SGs into small and large classes is indistinguishable from siControl (52 ± 4% and 48 ± 4%, respectively; P = 0.600; Fig. 4 G).
To assess the dynamics of SG–PB docking in real time, we performed live cell imaging using transient transfection of HeLa cells with Dcp1a-RFP and TIA-1-GFP plasmids in the context of TDP-43 siRNA. We first verified that transient exogenous expression of the TIA-1-GFP reporter construct at the low amount we used (0.3 µg each) did not induce SG formation and that cells underwent SG formation with normal kinetics after SA treatment (Fig. S3 A). In cells treated with SA and then imaged for a 10-min window at 90 min (the time point where SGs are largest), images were collected every 30 s and individual PBs were monitored for contact with SGs. Although PBs are dynamic, the number of contacts between SGs and PBs was unchanged by TDP-43 depletion (Fig. S3 B). However, the mean duration of PBs docking onto large SGs (>2 µm2) during the 10-min imaging window was reduced by 41% in TDP-43–depleted cells compared with siControl cells (4.95 ± 0.98 min vs. 8.35 ± 0.76 min; P = 0.0104) but was comparable when small SGs (<2 µm2) were analyzed (Fig. S3 C). We further noted that in siTDP-43 cells, only 40% of PBs were continuously docked onto large SGs for the entire 10-min imaging window, in contrast to 83% in siControl cells (Fig. 4 H). Thus, we conclude that SG size, mediated by G3BP1-dependent secondary aggregation of SG, is a determinant of stable SG–PB docking.
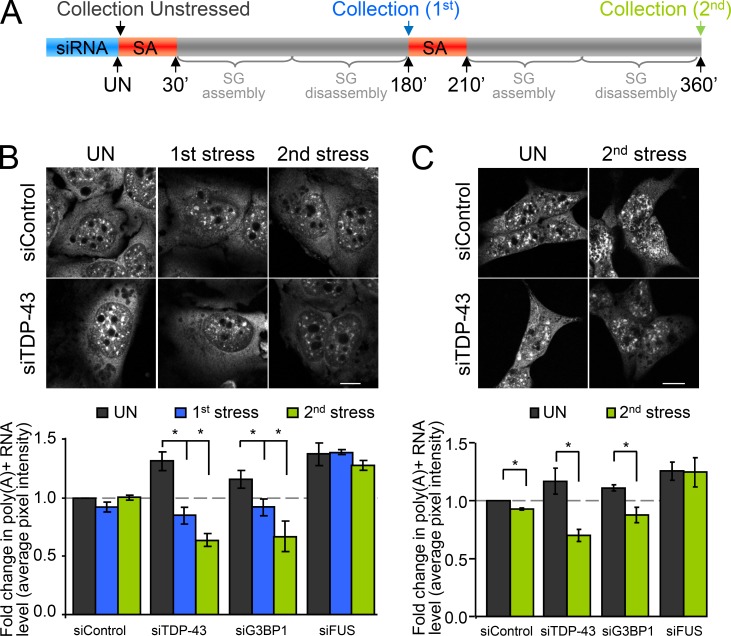
Loss of polyadenylated mRNA correlates with reduced SG–PB docking
The field consensus is that the function of SGs is to sort mRNA for storage, reinitiation, or degradation (Kedersha et al., 2005). It has been proposed that mRNA and RNPs can be exchanged between SGs and PBs via docking and that this may serve to target certain transcripts for degradation in PBs while other transcripts are stored in SGs and thus protected from degradation (Kedersha et al., 2005). The prevailing view in mammalian cells is that mRNAs are selectively transferred from SGs to PBs during docking, with localization of mRNAs in PBs being a one-way ticket to decapping and degradation (Kedersha et al., 2005). However, evidence in yeast suggests that transcripts can be recapped and that PBs also participate in the sorting of transcripts, such that mRNAs are proposed to be transferred from PBs to SGs for storage (Brengues et al., 2005; Parker and Sheth, 2007). To determine if deficient SG–PB docking has a functional consequence on mRNA preservation, we designed an experiment where cells were induced to undergo two full cycles of SG (and stress-induced PB) assembly and disassembly with polyadenylated mRNA levels being measured before the first stress, and then after each SG disassembly (i.e., 180 min after each SA addition; Fig. 5 A). In siControl cells, the levels of polyadenylated mRNA after two consecutive SA treatments, as measured via total cellular pixel intensity after FISH with an oligo(dT) probe, is equivalent to unstressed cells (Fig. 5 B). However, in cells depleted of TDP-43 or G3BP1, although mRNA levels are initially higher in unstressed conditions (as shown in Fig. 3 A), there is a progressive step-wise decrease in polyadenylated mRNA after two consecutive rounds of SA treatment. Specifically, relative to unstressed cells, we observe a decrease in polyadenylated mRNA levels of 35 ± 4% and 52 ± 3% in siTDP-43 cells and 21 ± 2% and 43 ± 7% in siG3BP1 cells (P = 0.013 and 0.002 for siTDP-43, P = 0.048 and 0.031 for siG3BP1). In contrast, siFUS cells, which also exhibit higher levels of polyadenylated mRNA in basal conditions, did not demonstrate a loss of mRNA but rather were constant, comparable to siControl cells (Fig. 5 B). A similar result was obtained with a second set of siRNAs after a single SA exposure (Fig. S4). We further validated this result in SH-SY5Y cells treated with siTDP-43 and siG3BP1 and examined after the second SA exposure (Fig. 5 C). Thus, the functional interaction between SGs and PBs serves to preserve transcripts during stress.
Figure 5.
Decreased mRNA preservation after stress exposure. (A–C) Cells were transfected with the indicated siRNAs for 72 h and then subjected to two consecutive rounds of stress response induced by SA (SG assembly and disassembly). Coverslips were collected before stress, and after the first and the second round of SG assembly/disassembly. Polyadenylated mRNA was visualized via FISH with an oligo(dT) probe. (A) Experimental design. (B) Representative images of HeLa cells labeled with an oligo(dT) probe and quantification of total pixel intensity. (C) Representative images and quantification of a similar experiment performed in SH-SY5Y cells and examined only after the second SA stress exposure. All data are representative of >3 independent experiments and plotted as the mean ± SEM (error bars). *, P < 0.05. Bars, 10 µm.
Exogenous G3BP1 fully rescues SG–PB docking and maintains mRNA levels
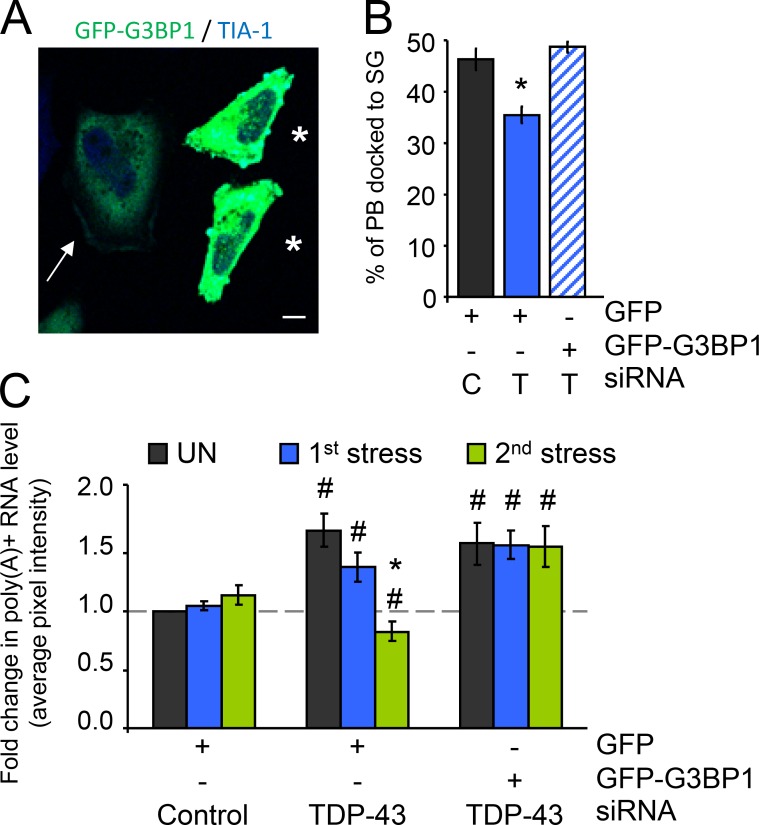
We have previously shown that depletion of TDP-43 induces a down-regulation of G3BP1 transcript and protein (McDonald et al., 2011), and that defective SG assembly induced by TDP-43 depletion is fully restored by exogenous G3BP1 expression (Aulas et al., 2012). In the current manuscript, the linear relationship from TDP-43 to G3BP1 is preserved since nearly every phenotype observed in siTDP-43 cells is also observed in G3BP1-depleted cells. To determine if G3BP1 is the molecule directly mediating these effects, we reintroduced G3BP1 expression into siTDP-43 cells. As we have previously published, in order to avoid artifacts from G3BP1 overexpression, G3BP1 cDNA is limited to 0.3 µg and only cells with low total fluorescence and without spontaneous cytoplasmic aggregates (sometimes referred to as constitutive SGs) are analyzed (Aulas et al., 2012; Fig. 6 A, arrow). SG–PB docking, analyzed as described in Fig. 4 B, was fully restored by exogenous G3BP1 expression (Fig. 6 B). In addition, the progressive loss of polyadenylated mRNA after consecutive SA exposures was fully rescued by G3BP1 expression (Fig. 6 C). Thus, TDP-43 regulation of G3BP1 is central to coordinating the functional interaction between SGs and PBs.
Figure 6.
G3BP1 rescues SG–PB interactions and maintains polyadenylated mRNA levels. (A–C) HeLa cells transfected with control (C) or TDP-43 (T) siRNA for 48 h and transfected with GFP or G3BP1-GFP for 24 h, then treated with SA. (A) Representative confocal micrograph demonstrating low-level transient expression of G3BP1. The arrow indicates a typical example of a low-expressing cell that was quantified. Asterisks indicate cells with high levels of G3BP1 expression that were excluded from the analysis. Bar, 10 µm. (B) Coverslips were collected before and at 90 min after stress and then labeled for Dcp1a and TIA-1 to follow PBs and SGs, respectively. Docking was quantified as before in low fluorescence GFP-expressing cells. (C) Cells were subjected to two consecutive rounds of stress response induced by SA (SG assembly and disassembly). Coverslips were collected before stress, and after the first and second rounds of stress. Polyadenylated mRNA was visualized via FISH with a oligo(dT) probe and quantified as before. All data are representative of ≥3 independent experiments and plotted as the mean ± SEM (error bars). *, P < 0.05 compared with unstressed siControl; #, P < 0.05 compared with unstressed of the same sample.
Discussion
TDP-43 and G3BP1 depletion yields small SGs (Aulas et al., 2012). However, we show here that these smaller SGs contain canonical SG proteins such as eIF3G, eIF4A, HuR, and normal levels of polyadenylated mRNA. In addition, the localization of known G3BP1 binding partners, Caprin1 and USP10, is also undisturbed. Importantly, stress-induced translational arrest proceeds normally despite a defect in secondary SG aggregation. However, by three different assays, we demonstrate that TDP-43/G3BP1 depletion is associated with a defect in basal translation rates in HeLa cells, consistent with a previous study (Higashi et al., 2013), although this remains to be confirmed in additional cell types. In addition, we demonstrate that the defect in basal translation is not due to a loss of polyadenylated mRNA levels, which were increased in all siRNA conditions examined, compared with controls. It has been previously reported that a translational defect exists in TDP-43–depleted HeLa cells with a similar abundance in stalled preinitiation complexes. However, this was attributed to a decrease in total mRNA that could be available for translation (Higashi et al., 2013). Although we did not observe a decrease in total mRNA, in TDP-43– and G3BP1-depleted cells, less RNA was associated with polysomes and more was associated with monosomes, which is further consistent with impaired translation initiation. We speculate that, at least in TDP-43/G3BP1–depleted cells, the increase in basal polyadenylated mRNA could be attributed to reduced G3BP1 expression because G3BP1 has been reported to function as an endoribonuclease (Tourrière et al., 2001). However, FUS does not regulate G3BP1 expression. It remains unknown if FUS regulates posttranslational modifications on G3BP1 (e.g., phosphorylation, methylation) that are known to functionally impact G3BP1 (Tourrière et al., 2003; Kim et al., 2008). Alternatively, increased transcript levels in FUS-depleted cells could be due to some other unknown mechanism. The molecular aspects of translation that are affected in TDP-43– and G3BP1-depleted conditions warrant further study.
We have previously shown that TDP-43–mediated regulation of G3BP1 determines SG size (Aulas et al., 2012). We now conclude that SG size is a determinant of SG–PB docking. TDP-43 localization to SGs is maintained in G3BP1-depleted cells (Fig. S5). However, G3BP1 is absent from SGs in siTDP-43 cells due to down-regulation of its expression (Aulas et al., 2012). While we cannot exclude the possibility that there are other TDP-43–regulated SG proteins not investigated here that could be similarly absent from SGs, the data strongly suggest that G3BP1 is the central determinant of SG size and functional SG–PB interactions, as introduction of exogenous G3BP1 alone was sufficient to completely restore these aspects. Interestingly, the transition from primary to secondary aggregation has been postulated to integrate cellular signaling molecules via piggyback recruitment on core SG proteins, such as G3BP1 (Kedersha et al., 2000; Anderson and Kedersha, 2002; Anderson and Kedersha, 2009; Bartoli et al., 2011; Wolozin, 2012). Our data extends this proposal to include SG–PB interactions. In addition, our data demonstrate that although there is some functional redundancy for G3BP1 and G3BP2 in SG nucleation, consistent with a published study (Matsuki et al., 2013), G3BP2 does not participate in SG secondary aggregation (unpublished data) or SG–PB docking in HeLa cells.
The current model in mammalian cells posits that upon stress, nontranslating mRNAs complexed with RNPs preferentially transit from polysomes to SGs (Brengues et al., 2005). It is widely considered that at SGs, mRNPs are sorted either for degradation (presumably via docking and exchange with PBs), returned to the polysome for reinitiation, or stored until the stress is abated and translation returns to normal (Kedersha et al., 2005). However, in yeast, there is evidence that suggests mRNPs localized to PBs can be remodeled and subsequently accumulated in SGs, effectively assigning the triage point to PBs rather than SGs (Buchan and Parker, 2009). As proposed by Buchan and Parker (2009), the relative rates of exchange of translation factors for PB proteins versus the aggregation of SG components might be the determining factor for mRNA fate (SG or PB) when translation is interrupted. Essentially, the question becomes: what determines the path of nontranslating mRNAs in times of stress? In our attempt to define the fate of mRNA immediately after polysome disassembly in TDP-43/G3BP1–depleted conditions, we developed an assay to determine the levels of polyadenylated mRNA after two consecutive rounds of SG assembly and disassembly. Here, we observed that although polyadenylated mRNA levels remain constant in siControl cells, implying efficient protection/storage of transcripts, there is a progressive and step-wise loss of mRNA when cells are depleted of TDP-43 or G3BP1. However, there is no evidence for decreased RNA synthesis, as steady-state polyadenylated mRNA levels are increased in basal conditions. In our view, as docking is not increased (to exchange mRNA from SGs to PBs), this scenario is only possible if nontranslating mRNAs preferentially transit from polysomes to PBs. At PBs, the transcripts are presumably degraded outright, and/or fail to be transferred to SGs. Moreover, because docking is not completely abolished, this suggests that both models coexist in mammalian cells (i.e., nontranslating mRNA can flow to both SGs and PBs). As TDP-43 localizes to SGs despite G3BP1 depletion, and reintroduction of G3BP1 alone to TDP-43–depleted cells rescues all phenotypes, we conclude that G3BP1 is a critical RBP that directs the preferential flow of mRNA from polysomes to SGs, possibly due to its role in driving the aggregation of SG components. In the absence of G3BP1 (either via direct siRNA or down-regulation as a consequence of TDP-43 siRNA), SG–PB contacts are less stable, resulting in the path to PBs to be used more often, consistent with the Buchan and Parker model. This implies that a functional significance of SG size is the preservation of mRNA transcripts.
Our data also clearly demonstrate an interesting subplot: while FUS does not influence SG–PB interactions, it does influence PB number and size, which suggests that PB dynamics might be a common underlying mechanism in ALS pathogenesis. In addition, translation levels remain comparable between siControl and siFUS cells, but there is an approximately threefold increase in PB formation and twofold increase in mRNA levels in basal conditions. This is consistent with reports that although changes in PB formation, mRNA levels, and translation often correlate, they are not fully interdependent, and thus FUS siRNA may be an important tool in further dissecting this mechanism (Teixeira et al., 2005; Sheth and Parker, 2006).
In the current report, using transformed cells and primary neurons, we establish that G3BP1, a target of TDP-43, is the critical regulator of the interaction between RNA granule subtypes, namely SGs and PBs. The data presented here are the first report that SG size, as determined by the presence of G3BP1, dictates the interactions between SGs and PBs. Moreover, we show that the progressive loss of polyadenylated transcripts is a functional consequence of sequentially disrupting normal SG assembly and subsequent interactions with PBs. We have previously reported that G3BP1 is a downstream target of the ALS-causing gene TDP-43 and that its protein levels are reduced in one of two transformed lymphoblastoid cells bearing TARDBP mutations (McDonald et al., 2011; Aulas et al., 2012). Moreover, TDP-43 misaccumulates into cytoplasmic inclusions concomitantly with nuclear depletion in 97% of all ALS cases (Ling et al., 2013). Thus, we speculate that TDP-43 mislocalization (and concomitant nuclear depletion) or mutation may result in dysregulation of G3BP1, which could be an important intersecting point in ALS pathogenesis.
Materials and methods
Immortalized cell culture and transfection
HeLa cells were grown in Dulbecco’s modified Eagle medium (Invitrogen), supplemented with 10% FBS and 1% glutamine. For transfection of siRNA, 125 pmol of custom siRNAs were transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. siRNA sequences used were: human FUS/TLS #1, 5′-GGCCAAGAUCAAUCCUCCAUGAGUA-3′, and #2, 5′-CGGGACAGCCCAUGAUUAAUUUGUA-3′; human G3BP1 #1, 5′-ACAUUUAGAGGAGCCUGUUGCUGAA-3′, and #2, 5′-GCGCAUUAACAGUGGUGGGAAAUUA-3′; human TDP-43 #1, 5′-AAGCAAAGCCAAGAUGAGCCUUUGA-3′, and #2, 5′-AAGAUGAGAACGAUGAGCCCAUUGA-3′; human G3BP2 #1, 5′-GAGCUAAAGGAAUUCUUCAUGAGUU-3′, and #2, 5′-GGAGCCUUUGGAAGAAUCCUCUCAU-3′; and negative control low GC siRNA (catalog no. 12935-200; Invitrogen). HeLa cells were transfected at 30–50% confluence and collected after 72 h. For cDNA transfection, 0.3 µg G3BP1-GFP (provided by J. Tazi, Institut de Génétique Moléculaire de Montpellier), 0.3 µg Dcp1a-RFP (provided by P. Anderson, Harvard University, Cambridge, MA), and 0.3 µg TIA-1-GFP (provided by J. Goodier, Johns Hopkins University, Baltimore, MD) was transfected with Lipofectamine LTX (Invitrogen) according to the manufacturer’s instructions. Cells were transfected with cDNA 48 h after siRNA transfection and collected after an additional 24 h. SH-SY5Y cells were cultured in DMEM/F12 (Invitrogen), supplemented with 10% FBS and 10% nonessential amino acid, and transfected with 100 pmol siRNA using Nucleofector, as per the manufacturer’s instructions (Lonza). After 8 h, the media were replaced. Cells were collected 48 h after nucleofection. To induce SGs, cells were treated with 0.5 mM SA (30 min, 37°C; Sigma-Aldrich). Recovery was initiated by changing the media.
Mouse cortical neurons
The use of animals and all surgical procedures described in this article were performed according to guidelines set by the Canadian Council on Animal Care. Neurons were prepared from embryonic day 16 mouse embryos (C57BL/6). Cerebral cortices were treated with trypsin (0.025%, 37°C, 20 min). The reaction was stopped with trypsin inhibitor solution containing DNase. Neurons were dissociated by several passages through a Pasteur pipette and then plated either on glass coverslips or on culture dishes coated with polylysine and laminin (Sigma-Aldrich). Neurons were maintained in Neurobasal medium (Invitrogen) supplemented with Glutamax (Invitrogen) and B27 (Invitrogen).
Immunoblot analysis
Cells were collected in ice-cold PBS, lysed in RIPA buffer (150 mM NaCl, 50 mM Tris, pH 7.4, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, and protease inhibitors), incubated for 10 min on ice, then 10 min at room temperature, and centrifuged at 13,800 g. Supernatants were collected and quantified with the BCA Protein Assay kit (Thermo Fisher Scientific). Antibody information is listed in Table S1. Blots were visualized with peroxidase-conjugated secondary antibodies and ECL Western Blotting Substrate (Thermo Fisher Scientific). Densitometry was performed with Photoshop CS4 (Adobe).
Immunofluorescence
Cells grown on coverslips were fixed in 1% formaldehyde in PBS and subsequently permeabilized with 0.1% Triton X-100 in PBS for 15 min. Coverslips were blocked with 0.1% BSA in PBS for 15 min and incubated with primary antibodies (Table S1) diluted in blocking buffer for 1 h at room temperature. Coverslips were subsequently washed once with 0.1% Triton X-100 in PBS and twice with 0.1% BSA in PBS. Labeling was visualized with fluorescently conjugated secondary antibodies. Coverslips were washed tree times in PBS and mounted with ProLong Antifade reagent (Invitrogen). Images were collected on a confocal microscope (SP5; Leica).
Fluorescence in situ hybridization
Cells grown on coverslips were fixed in 4% formaldehyde in PBS and subsequently permeabilized with 0.1% Triton X-100 in 2× saline sodium citrate (SSC) for 15 min, then washed with 1 M Tris, pH 8, for 5 min. Coverslips were blocked with 0.0005% BSA, 10 mg/ml yeast RNA diluted in 2× SSC, washed with 1 M Tris, pH 8, for 5 min, and incubated with hybridization buffer (1.3 ng oligo(dT) probe, 0.005% BSA, 1 mg/ml yeast RNA, 10% dextran sulfate, and 25% formamide, diluted in 2× SSC) for 1 h in a humid chamber. Coverslips were subsequently washed twice with 4× SSC for 5 min and once with 2× SSC for 5 min. Coverslips where incubated with primary antibodies for 1 h (details in the previous section) and then washed twice with 4× SSC for 5 min and once with 2× SSC for 5 min. Labeling was visualized with fluorescently conjugated secondary antibodies, as above. Coverslips were washed twice with 4× SSC, once with 2× SSC, and then mounted with ProLong Antifade reagent (Invitrogen). Images were collected using 40× (1.25 NA) and 63× oil (1.7 NA) objective lenses on a confocal microscope (SP5; Leica) equipped with LAS AF software (Leica) for acquisition. Total cellular fluorescence intensity of the oligo(dT) probe was determined in Photoshop CS4. The Dynabeads mRNA DIRECT kit (Invitrogen) was used to validate this approach.
Quantification of SG and PB size and number
SG and PB parameters were quantified with ImageJ. SGs and PBs were identified by TIA-1 and Dcp1a staining (or oligo(dT) and p54), respectively, and cells were scored as positive when they had at least two foci with a minimum size of 0.75 and 0.15 µm2, respectively. Automatic recognition by ImageJ used the following parameters: SGs ranging from 0.75 to 5 µm2, PBs ranging from 0.15 to 5 µm2 in a minimum of 10 randomly selected cells per condition.
Metabolic labeling
Cells were stressed (0.5 mM SA, 30 min) and collected at 0 (before stress), 90, and 180 min. Cells were pulsed with 50 µM [35S]methionine diluted in normal growth medium lacking methionine 30 min before collection in ice-cold PBS and lysed in RIPA buffer. Equal amounts of protein were loaded on SDS-PAGE gel and then stained with Coomassie blue and dried for 2 h at 80°C. Dried gels were exposed to film overnight. Densitometry of the whole lane was performed using Photoshop CS4.
Polysome profiling
After 72 h of siRNA transfection, polysome profiling was performed as described previously (Gkogkas et al., 2013). In brief, cells were washed with ice-cold PBS containing 100 µg/ml cycloheximide and subsequently lysed in 5 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 100 µg/ml cycloheximide, 2 mM DTT, 0.5% Triton X-100, and 0.5% sodium deoxycholate. Lysates were loaded onto 10–50% sucrose density gradients (20 mM Hepes-KOH, pH 7.6, 100 mM KCl, and 5 mM MgCl2) and centrifuged at 235,000 g in an SW40i rotor for 2 h at 4°C. Absorbance of different fractions at 254 nm was continuously recorded using an ISCO fractionator with a UV detector (Teledyne ISCO). The polysome-to-monosome ratio was calculated as the area under the A254 absorbance curve, using the function describing the absorbance values, processed with the definite integral command in MATLAB (MathWorks).
Live imaging
Live imaging was performed on HeLa cells grown on glass-bottomed dishes (MatTek Corporation) 48 h after cDNA transfection. The environment was kept at 37°C and 5% CO2. Cells were acclimated to the microscope incubator chamber before imaging. Only cells exhibiting low levels of TIA-1-GFP and devoid of spontaneous SGs were selected for subsequent SA treatment and imaging. Time-lapse movies of cells in DMEM phenol red–free media (#31053-028; Gibco) were taken using an inverted microscope (DMI 6000B; Leica) equipped with an HC Plan-Apochromat CS2 63×/1.40 NA oil objective lens and a 1394 ORCA-ERA camera (Hamamatsu Photonics) equipped with an incubation chamber and heated stage set to 37°C. Images were collected every 30 s for 10 min with maximum sample protection using two different channels using Volocity 6.1.1 software for acquisition. Analysis was made manually using the same software following individual PBs between frames. Only PBs and SGs that remained in focus for the 10 min of filming were included in the analysis.
Statistics
Data were compared via two-tailed Student t test or Mann-Whitney, where appropriate.
Online supplemental material
Fig. S1 shows further analysis of impaired basal translation by ribopuromycylation and polysome profiling as well as alternate measure of polyadenylated mRNA levels in siRNA-treated cells. Fig. S2 is an analysis of PBs in basal conditions and examination of SG–PB docking with a second set of siRNAs, additional markers, and in a second cell type. Fig. S3 shows analysis of live-stage imaging experiments. Fig. S4 is confirmation of impaired mRNA preservation in HeLa cells using a second set of siRNAs. Fig. S5 shows normal TDP-43 recruitment to SGs in G3BP1- and FUS-depleted cells. Table S1 includes information for all antibodies used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408092/DC1.
Supplementary Material
Acknowledgments
We thank the members of the Vande Velde laboratory and N. Grandvaux for helpful discussions, and we especially thank S. Peyrard for technical support; A. Prat, N. Arbour, and the University of Montreal Hospital Research Centre (CRCHUM) Cancer Axe for access to equipment; and J. Tazi, P. Anderson, and J. Goodier for reagents and the CRCHUM Imaging platform.
This work was supported by the Canadian Institutes of Health Research (CIHR) Neuromuscular Research Partnership, the Muscular Dystrophy Association (grant MDA238828), the National Science and Engineering Research Council (NSERC), and the Canadian Foundation for Innovation (CFI; all to C. Vande Velde). C. Vande Velde is a Canadian Institutes of Health Research New Investigator. A. Aulas has received support from the Réseau de Médecine Génétique Appliquée.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ALS
- amyotrophic lateral sclerosis
- PB
- processing body
- RBP
- RNA binding protein
- SA
- sodium arsenite
- SG
- stress granule
- SSC
- saline sodium citrate
References
- Anderson P., and Kedersha N.. 2002. Stressful initiations. J. Cell Sci. 115:3227–3234. [DOI] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2009. Stress granules. Curr. Biol. 19:R397–R398 10.1016/j.cub.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Andersson M.K., Ståhlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., Nilsson O., and Aman P.. 2008. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 9:37 10.1186/1471-2121-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A., Stabile S., and Vande Velde C.. 2012. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener. 7:54 10.1186/1750-1326-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli K.M., Bishop D.L., and Saunders W.S.. 2011. The role of molecular microtubule motors and the microtubule cytoskeleton in stress granule dynamics. Int. J. Cell Biol. 2011:939848 10.1155/2011/939848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E., Neumann M., Tahirovic S., Rodde R., Dormann D., and Haass C.. 2012. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43). J. Biol. Chem. 287:23079–23094 10.1074/jbc.M111.328757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E., Haass C., and Dormann D.. 2013. Stress granules in neurodegeneration—lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 280:4348–4370 10.1111/febs.12287 [DOI] [PubMed] [Google Scholar]
- Bosco D.A., Lemay N., Ko H.K., Zhou H., Burke C., Kwiatkowski T.J. Jr, Sapp P., McKenna-Yasek D., Brown R.H. Jr, and Hayward L.J.. 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 19:4160–4175 10.1093/hmg/ddq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., and Parker R.. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 310:486–489 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., and Parker R.. 2005. General translational repression by activators of mRNA decapping. Cell. 122:875–886 10.1016/j.cell.2005.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., and Ratti A.. 2009. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111:1051–1061 10.1111/j.1471-4159.2009.06383.x [DOI] [PubMed] [Google Scholar]
- Cougot N., Babajko S., and Séraphin B.. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.K., Romo D., Gorospe M., Kaufman R., Anderson P., and Liu J.O.. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- Dewey C.M., Cenik B., Sephton C.F., Dries D.R., Mayer P. III, Good S.K., Johnson B.A., Herz J., and Yu G.. 2011. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell. Biol. 31:1098–1108 10.1128/MCB.01279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., and Izaurralde E.. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Gkogkas C.G., Khoutorsky A., Ran I., Rampakakis E., Nevarko T., Weatherill D.B., Vasuta C., Yee S., Truitt M., Dallaire P., et al. 2013. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 493:371–377 10.1038/nature11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S., Kabuta T., Nagai Y., Tsuchiya Y., Akiyama H., and Wada K.. 2013. TDP-43 associates with stalled ribosomes and contributes to cell survival during cellular stress. J. Neurochem. 126:288–300 10.1111/jnc.12194 [DOI] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 10.1042/BST0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., and Anderson P.. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., and Anderson P.. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., and Anderson P.. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.E., Ryu I., Kim W.J., Song O.K., Ryu J., Kwon M.Y., Kim J.H., and Jang S.K.. 2008. Proline-rich transcript in brain protein induces stress granule formation. Mol. Cell. Biol. 28:803–813 10.1128/MCB.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S.R., Horetsky R.L., Ron D., Jefferson L.S., and Harding H.P.. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273–C284 10.1152/ajpcell.00314.2002 [DOI] [PubMed] [Google Scholar]
- Lavut A., and Raveh D.. 2012. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 8:e1002527 10.1371/journal.pgen.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., and Sharp P.A.. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 103:18125–18130 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.R., King O.D., Shorter J., and Gitler A.D.. 2013. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201:361–372 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S.C., Polymenidou M., and Cleveland D.W.. 2013. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 79:416–438 10.1016/j.neuron.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., et al. 2010. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE. 5:e13250 (published erratum appears in PLoS One 2011. 6) 10.1371/journal.pone.0013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., and Fujii M.. 2013. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 18:135–146 10.1111/gtc.12023 [DOI] [PubMed] [Google Scholar]
- McDonald K.K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G.A., and Vande Velde C.. 2011. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20:1400–1410 10.1093/hmg/ddr021 [DOI] [PubMed] [Google Scholar]
- Nover L., Scharf K.D., and Neumann D.. 1983. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 3:1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L., Scharf K.D., and Neumann D.. 1989. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9:1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., and Sheth U.. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Sheth U., and Parker R.. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., and Parker R.. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 125:1095–1109 10.1016/j.cell.2006.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Xu Y., Wang B., David M.D., Schubert P., Kennedy D., and Schrader J.W.. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324–2342 10.1128/MCB.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., and Kedersha N.. 2013. Relationship of GW/P-bodies with stress granules. Adv. Exp. Med. Biol. 768:197–211 10.1007/978-1-4614-5107-5_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M.J.2010. The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS). J. Neurol. Sci. 288:1–12 10.1016/j.jns.2009.09.029 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., and Fujii M.. 2013. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol. 33:815–829 10.1128/MCB.00763-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., and Parker R.. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.G., Loschi M., Desbats M.A., and Boccaccio G.L.. 2011. RNA granules: the good, the bad and the ugly. Cell. Signal. 23:324–334 10.1016/j.cellsig.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Gallouzi I.E., Chebli K., Capony J.P., Mouaikel J., van der Geer P., and Tazi J.. 2001. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 21:7747–7760 10.1128/MCB.21.22.7747-7760.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., and Tazi J.. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wolozin B.2012. Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7:56 10.1186/1750-1326-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.