A group of phosphorylatable serine residues within the nonhelical domain of NMII-B controls the ability of NMII-B to generate stable migratory front–rear polarity.
Abstract
In this study, we show that the role of nonmuscle myosin II (NMII)-B in front–back migratory cell polarity is controlled by a short stretch of amino acids containing five serines (1935–1941). This motif resides near the junction between the C terminus helical and nonhelical tail domains. Removal of this motif inhibited NMII-B assembly, whereas its insertion into NMII-A endowed an NMII-B–like ability to generate large actomyosin bundles that determine the rear of the cell. Phosphomimetic mutation of the five serines also inhibited NMII-B assembly, rendering it unable to support front–back polarization. Mass spectrometric analysis showed that several of these serines are phosphorylated in live cells. Single-site mutagenesis showed that serine 1935 is a major regulatory site of NMII-B function. These data reveal a novel regulatory mechanism of NMII in polarized migrating cells by identifying a key molecular determinant that confers NMII isoform functional specificity.
Introduction
Front–back polarity is a key feature of migrating cells. It is often defined as an asymmetric distribution of the microtubule-organizing center, the Golgi apparatus, the nucleus, and the protrusive activity (Etienne-Manneville and Hall, 2001). Asymmetry is controlled by different signals, including local activation of Cdc42 upstream of PKC-ζ (Gomes et al., 2005) as well as other Rho GTPases (Hall, 2012). PKC-ζ controls microtubule-organizing center positioning, and it also localizes to the leading edge, forming a complex with Par6, where they jointly regulate protrusion (Tan et al., 2008). Among other functions, Rho GTPases mediate the asymmetric distribution and activation of nonmuscle myosin II (NMII). NMII cross-links and contracts actin, promoting linear structures of bundled filaments. NMII is a hexamer comprised of two common regulatory light chains, two common essential light chains, and two isoform-specific heavy chains. Vertebrates express three NMII isoforms as defined by the myosin heavy chains, which are encoded in three separate genes: Myh9 (NMII-A), Myh10 (NMII-B), and Myh14 (NMII-C). Previous studies show that the two major isoforms, NMII-A and NMII-B, play fundamentally different roles in the organization of the actin in migrating cells (Lo et al., 2004; Even-Ram et al., 2007; Vicente-Manzanares et al., 2007).
NMII-B determines the rear of migrating cells by localizing asymmetrically and increasing actomyosin bundling in cells on stiff substrates (Vicente-Manzanares et al., 2008) but not soft substrates (Raab et al., 2012). The rearward accumulation of stable actomyosin bundles inhibits the signals that generate protrusions in this region (Vicente-Manzanares et al., 2011). Conversely, NMII-A generates minifilaments at the front of the cell that promote actin bundling and adhesion maturation behind the lamellipodium (Vicente-Manzanares et al., 2007; Choi et al., 2008). NMII-A–generated bundles are thin and dynamic, and they can undergo disassembly. NMII-B recruitment to these bundles increases their thickness and impairs their disassembly, and the adhesions at their ends become elongated and stable (Vicente-Manzanares et al., 2011). These properties are related to the different localization and function of NMII-A and NMII-B (Maupin et al., 1994; Kolega, 2003).
Isoform-specific NMII inhibition causes different migratory alterations. NMII-A depletion inhibits rear retraction and also impairs adhesion maturation at the front, whereas NMII-B depletion inhibits front–back polarization (Lo et al., 2004; Cai et al., 2006; Even-Ram et al., 2007; Vicente-Manzanares et al., 2007). Despite sharing high primary structure homology, the isoforms display exquisite functional specificity. Their differential ability to regulate the component processes of cell migration resides in the C terminus, nonhelical domain of the heavy chains, which mediates oligomerization (Sandquist and Means, 2008; Vicente-Manzanares et al., 2008). Previous studies of NMII-B have revealed that phosphorylations within the coiled-coil domain of the heavy chain (Li et al., 2006; Clark et al., 2008) and the nonhelical chain domain (Rosenberg and Ravid, 2006) regulate filament assembly.
In this study, we focus on the role of a group of phosphorylatable serine (Ser) residues within the nonhelical domain of NMII-B in controlling the functions of this isoform. Phosphomimetic and nonphosphorylatable mutants together with mass spectrometric analysis identify serine 1935 as the major regulatory site within this amino acid stretch. Our data demonstrate that this motif uniquely controls the ability of NMII-B to generate stable front–rear polarity and control adhesion dynamics in protrusions.
Results and discussion
A Ser-rich motif in the nonhelical domain of MHCII-B promotes actomyosin stability and cell polarization
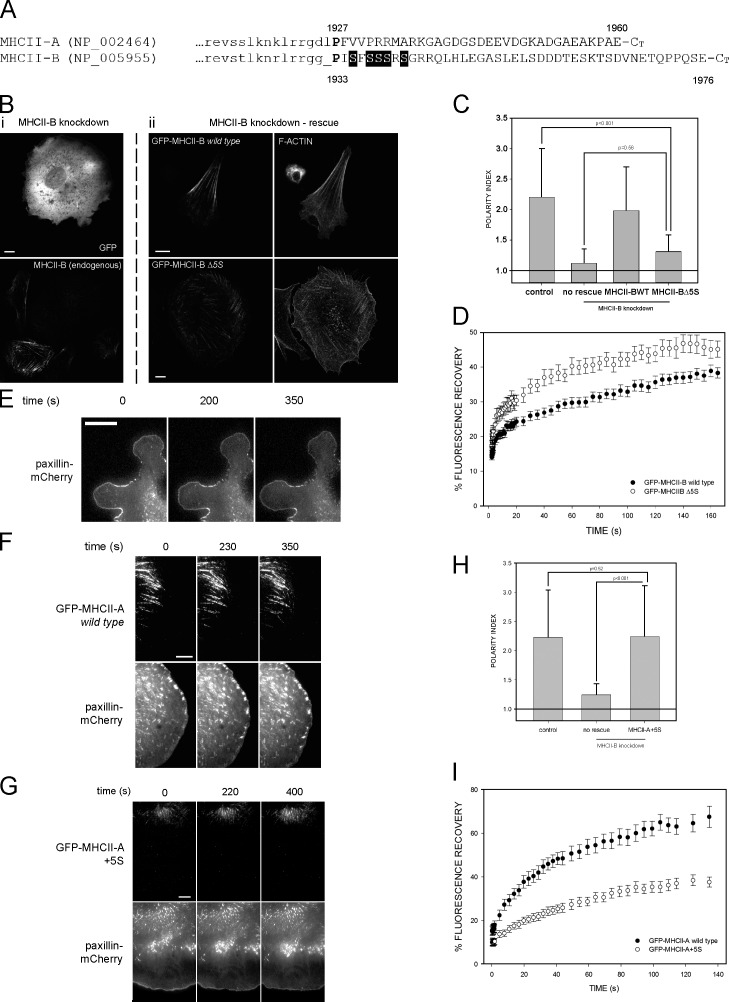
The cellular localization and biochemical properties of NMII-A and NMII-B depend on the C terminus domain of the myosin heavy chain, MHCII (Sandquist and Means, 2008; Vicente-Manzanares et al., 2008). To identify unique motifs that determine this specificity, we aligned the last 200 amino acids of human MHCII-A (NCBI Protein database accession no. NP_002464) and MHCII-B (accession no. NP_005955). Whereas the coiled-coil portion of both molecules displays high homology (Fig. 1 A, last 15 amino acids in lowercase and not depicted), the sequences diverge significantly beyond Pro1927 (MHCII-A)/Pro1933 (MHCII-B; Fig. 1 A, uppercase letters). We hypothesized that these regions contained unique molecular motifs that control the specificity of the cellular function of the isoforms. One such motif, a cluster of five serine residues (SFSSSRS) adjacent to the domain-splitting Pro in MHCII-B (Fig. 1 A, shaded), appeared particularly interesting, as this motif (1935–1941) is absent from the analogous region in MHCII-A.
Figure 1.
The 1935–1941 amino acid region of MHCII-B determines the dynamics of NMII. (A) Alignment of the nonhelical domains of human MHCII-A and MHCII-B. Lowercase, last amino acids of the coiled-coil domain. Uppercase, nonhelical tail domain. P in bold is the domain-breaking proline. Shaded background, 1935–1941 motif. (B, left column [i]) MHCII-B–depleted CHO.K1 cells expressing GFP (top) and stained for endogenous MHCII-B (bottom). (middle and right columns [ii]) MHCII-B–depleted cells expressing shRNA-resistant GFP–MHCII-B (wild type; top row) or GFP–MHCII-B Δ5S (bottom row). (middle column) GFP; (right column) F-actin. Representative examples are shown. (C) Polarity index of MHCII-B–depleted cells rescued or not rescued with wild-type or Δ5S GFP–MHCII-B. Polarity index is calculated as indicated in Materials and methods. Data are the means ± SD of >200 cells from four independent experiments; p-value is the significance according to Mann–Whitney’s test. (D) FRAP curves of wild type and Δ5S GFP–MHCII-B. Data are the means ± SEM of 24 individual measurements per condition in four independent experiments. (E–G) Snapshots of TIRF microscopy time-lapse videos of MHCII-A–depleted CHO.K1 cells transfected with paxillin-mCherry alone (no rescue; E), or cotransfected with paxillin-mCherry and wild-type GFP–MHCII-A (rescue; F) or GFP–MHCII-A+5S (G), plated on fibronectin, and allowed to attach for 45 min. Images were obtained every 5 s. Note the lack of elongated adhesions in protrusions in E and G, and the different distance of the GFP-labeled, wild-type (F), and MHCII-A+5S (G) filaments to the paxillin-decorated leading edge. Images extracted from Videos 1–3. (H) Polarity index of MHCII-B–depleted cells rescued or not rescued with the MHCII-A+5S mutant. Data are the means ± SD of >200 cells from four independent experiments. (I) FRAP curves of wild-type GFP–MHCII-A and GFP–MHCII-A+5S. Data are the means ± SEM of 28 individual measurements per condition in four independent experiments. Bars, 10 µm.
To determine whether the 1935–1941 motif of MHCII-B controls the cellular function of NMII-B, we generated a GFP-coupled deletion mutant (GFP–MHCII-B Δ5S). Wild-type GFP-coupled MHCII-B rescued the polarity defect induced by depletion of endogenous MHCII-B (Fig.1, B and C; Vicente-Manzanares et al., 2007). In contrast, GFP–MHCII-B Δ5S was unable to restore front–rear migratory polarity (Fig. 1, B and C). These cells displayed only small actomyosin bundles and almost no large, rear-defining actomyosin bundles (Fig. 1 B, bottom right). Similar observations were made in MHCII-B–depleted U-2 OS cells, rescued or not with GFP-coupled wild-type or Δ5S MHCII-B (Fig. S1). FRAP analysis of the mutant (as shown in Fig. S2 A) revealed an increased fractional recovery compared with wild-type MHCII-B, consistent with a decreased stability of the mutant in actomyosin bundles (Fig. 1 D). These data indicate that this poly-Ser–containing motif is required for NMII-B to generate large bundles that support front–back polarity in mesenchymal cells.
To address whether this motif is sufficient to define the specific functions of NMII-B in front–rear polarity, we inserted the 1935–1941 motif into the analogous position (P+2) of the nonhelical domain of MHCII-A (GFP–MHCII-A+5S mutant). This insertion produced an NMII-A mutant with properties similar to NMII-B. Unlike wild-type GFP–MHCII-A (Fig. 1 F and Video 2, right), GFP–MHCII-A+5S was not present in protrusions, displaying a central subcellular distribution (Fig. 1 G and Video 3, right). Furthermore, it did not rescue the adhesion maturation defect observed in cells depleted of endogenous NMII-A (Fig. 1 E and Video 1). Adhesion maturation was restored by expression of GFP-coupled wild-type MHCII-A (Fig. 1 F and Video 2, left) but not of wild-type MHCII-B (Vicente-Manzanares et al., 2007). Importantly, GFP–MHCII-A+5S rescued the polarity defect induced by depletion of endogenous MHCII-B, generating large, rear-defining actomyosin bundles (Fig. 1 H and not depicted). FRAP analysis revealed a decreased fractional recovery for this mutant when compared with wild-type MHCII-A (Fig.1 I). These results show that the presence of this motif in the nonhelical tail of MHCII, controls the ability of NMII to support large and stable actomyosin bundles.
Phosphomimetic analysis reveals that phosphorylation of 1935–1941 motif regulates the cellular properties of NMII-B
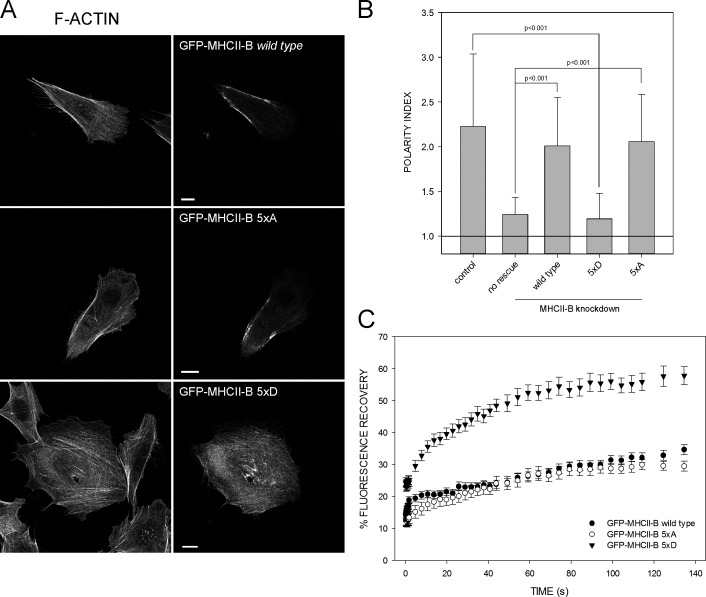
Five of the seven residues of the 1935–1941 motif are serines (Fig. 1 A). We hypothesized that these Ser residues regulate the cellular function of NMII-B by phosphorylation. To test this, we replaced the five Ser with Ala (5×A, nonphosphorylatable) or with Asp (5×D, phosphomimetic). Rescue of endogenous MHCII-B depletion using the 5×A mutant restored front–back polarization to levels comparable to wild-type MHCII-B (Fig. 2, A and B). The mutant localized mainly in large bundles defining the cell sides and its rear (Fig. 2 A). Conversely, the 5×D mutant failed to restore front–back polarity, resulting in poorly defined actomyosin bundles distributed throughout the cell (Fig. 2, A and B). FRAP analysis showed that the fractional recovery of the 5×A mutant was comparable to wild-type MHCII-B, whereas it was increased in the 5×D (Fig. 2 C). These data show that Ser phosphorylation in this amino acid stretch is a negative regulator of NMII-B–dependent actomyosin assembly.
Figure 2.
Phosphomimetic mutation of the five Ser residues adjacent to the nonhelical domain of MHCII-B decreases NMII-B stability and front–back polarization. (A) MHCII-B–depleted CHO.K1 cells expressing the indicated shRNA-resistant GFP–MHCII-B mutants. Column on right depicts GFP. (top) Wild type; (middle) 5×A; (bottom) 5×D. Left column is F-actin. Note the lack of polarity in the cell rescued with the 5×D mutant. Representative examples are shown. Bars, 10 µm. (B) Polarity index of MHCII-B–depleted cells rescued or not rescued with the constructs shown in A. Data are the means ± SD of >200 cells from four independent experiments. (C) FRAP curves of the GFP–MHCII-B mutants used in A and B. Data are the means ± SEM of 24 individual measurements per condition in four independent experiments.
Ser1935 is a central regulatory residue that controls NMII-B assembly and dynamics
We next sought to determine the contribution of individual Ser residues within the 1935–1941 motif to the regulation of NMII-B function. To do this, we analyzed the phosphorylation of endogenous MHCII-B by mass spectrometry (MS). Three Ser (S1935, S1938, and S1939; Table 1 and Fig. S3) were phosphorylated with relative abundances ranging from 1–2% (S1935 and S1939) to 5–10% (S1938). Phosphorylation of S1937 and peptides containing two phosphorylated Ser were detected only in immobilized metal affinity chromatography (IMAC)–enriched preparations (see Materials and methods for details; Table 1). These data suggest that phosphorylation of S1935 and/or S1938 and/or S1939 or a combination of these residues regulates NMII-B. Importantly, S1935 phosphorylation has been reported in HeLa cells (Olsen et al., 2010; Zhou et al., 2013) and liver extracts (Bian et al., 2014).
Table 1.
Phosphorylated residues within the nonhelical tail domain of endogenous MHCII-B identified by MS
| Residues | Sequence | Phosphoresidue | Screening | IMAC |
| 1930–1940 | GGPISFSSSR | 1935 | + | + |
| 1929–1940 | RGGPISFSSSR | 1935 | + | + |
| 1929–1940 | RGGPISFSSSR | 1937 | − | + |
| 1930–1940 | GGPISFSSSR | 1938 | + | + |
| 1929–1940 | RGGPISFSSSR | 1938 | + | + |
| 1930–1940 | GGPISFSSSR | 1939 | + | + |
| 1929–1940 | RGGPISFSSSR | 1939 | + | + |
| 1930–1940 | GGPISFSSSR | * | − | + |
| 1930–1940 | RGGPISFSSSR | * | − | + |
| 1958–1976 | QLHLEGASLELSDDDTESK | 1965 | − | + |
| 1958–1976 | QLHLEGASLELSDDDTESK | 1969 | + | + |
| 1958–1976 | QLHLEGASLELSDDDTESK | 1965 + 1969 | + | + |
Asterisks indicate diphosphorylated forms of the peptide, although the specific combination of phosphorylated sites is not distinguishable from the MS/MS spectra. Plus signs denote positive detection of the indicated phosphorylation; minus signs indicates negative (no) detection of the indicated phosphorylation. Bold letters indicate the phosphorylated residue that has been positively detected or not detected within each peptide.
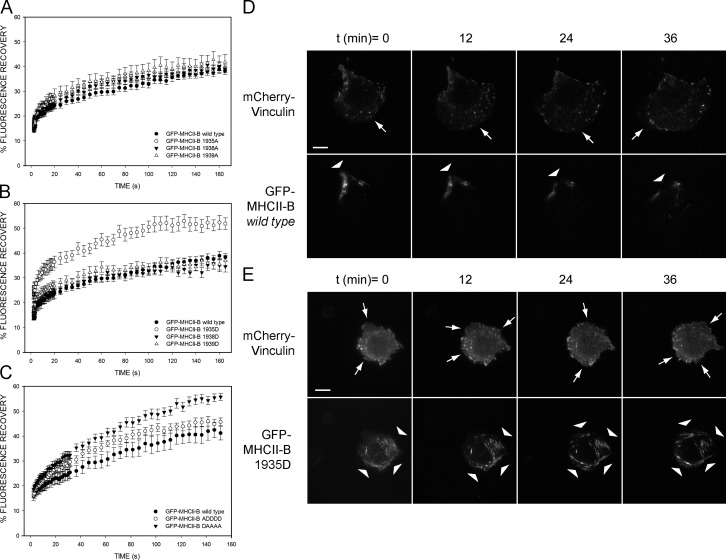
We next focused on the specific role of each Ser of the 1935–1941 motif by generating single phosphomimetic and nonphosphorylatable mutants for S1935, S1938, and S1939. All the single nonphosphorylatable (S to A) mutants displayed FRAP recoveries similar to that of wild-type GFP–MHCII-B (Fig. 3 A). Likewise, mutating S1938 or S1939 to Asp (S to D) had no significant effect. However, the S1935D mutant displayed an elevated fractional recovery (Fig. 3 B). Also, this mutant was unable to rescue the depolarized phenotype caused by NMII-B depletion in CHO.K1 (Table 2) and the organization of large actomyosin filaments in U2OS cells (Fig. S1).
Figure 3.
Ser1935 regulates the stability of NMII-B in actomyosin bundles and controls its ability to locally suppress protrusion during front–back polarization. (A) FRAP curves of wild-type and single mutation S1935A, S1938A, and S1939A GFP–MHCII-B constructs. (B) FRAP curves of wild-type and single mutation S1935D, S1938D, and S1939D GFP–MHCII-B proteins. (C) FRAP curves of wild-type GFP–MHCII-B and DAAAA and ADDDD mutants. (A–C) Data are the means ± SEM of 24 individual measurements per condition in four independent experiments. (D and E) Snapshots of TIRF microscopy time-lapse videos of MHCII-B–depleted CHO.K1 cells cotransfected with shRNA-resistant wild-type GFP–MHCII-B (D) or GFP–1935D-MHCII-B (E) and mCherry-vinculin. Arrows point to protrusive areas of the cell. Arrowheads point to regions decorated with NMII-B filaments. Bars, 10 µm. Images extracted from Videos 4 and 5.
Table 2.
Quantification of the polarity index of the S1935 mutants
| Endogenous MHCII-B depletion | Rescued with | Polarity index ± SD | P-value |
| No (control) | N/A | 2.23 ± 0.81 | <0.01b |
| Yes | No (MHCII-B knockdown) | 1.24 ± 0.19 | <0.01a |
| Yes | GFP–MHCII-B wild type (rescue) | 2.01 ± 0.54 | <0.01b |
| Yes | GFP–MHCII-B 1935A | 2.06 ± 0.65 | <0.01b |
| Yes | GFP–MHCII-B 1935D | 1.35 ± 0.29 | <0.01a |
| Yes | GFP–MHCII-B ADDDD | 1.95 ± 0.73 | <0.01b |
| Yes | GFP–MHCII-B DAAAA | 1.36 ± 0.32 | <0.01a |
Data are the quantification of four experiments (number of cells >200/condition). P-value denotes significance according to the Mann–Whitney test. N/A, not applicable.
Versus MHCII-B–depleted cells rescued with GFP–MHCII-B wild type (row 3).
Versus nonrescued cells (row 2).
To confirm that Ser1935 is the main regulatory residue in this motif, we mutated S1935 to D, the rest of the Ser to A (DAAAA), and vice versa (ADDDD). The DAAAA mutant behaved like the 1935D mutant, displaying high fractional recovery after photobleaching and an inability to restore polarization in MHCII-B–depleted cells (Fig. 3 C and Table 2). In contrast, the ADDDD mutant restored cell polarization (Table 2), and its fractional recovery after photobleaching was slightly higher than wild type or the 1935A mutant (Fig. 3 C). Thus, S1935 acts as an on/off switch: when dephosphorylated, NMII-B is stable and forms large filaments; when phosphorylated, it forms less stable filaments.
Among phosphorylatable residues, S1935 is the closest to P1933. Phosphorylation of S1935 adds negative charge next to the domain-breaking Pro. This may destabilize the coiled-coil tail domain, impairing oligomerization and perturbing actomyosin bundling. Interestingly, phosphomimetic mutation of all of the more distal Ser residues (ADDDD mutant) decreases NMII-B filament stability modestly, but not enough to impair cell polarity, suggesting that additional phosphorylations in this motif may cooperate with S1935. On the other hand, highly dynamic MHCII-A has several positively charged amino acids in this domain. Insertion of the 1935–1941 motif of MHCII-B into its equivalent position (Pro +2) of MHCII-A increases the stability of the filaments (Fig. 1). Conversely, inserting the phosphomimetic (+5D) version of the 1935–1941 motif further decreased the stability of NMII-A compared with wild-type NMII-A (Fig. S2 B), suggesting that net charge in this region is an important regulator of NMII stability.
Mammalian MHCII-B displays a sequence motif (PISFSSSRS) that is well conserved in birds (PITFSSSRS), amphibians (PVSFSSSSS), and fish (Danio rerio, PVSFSSSRS; Callorhinchus milii, GLTFTSSRT). A similar motif appears in zipper, the myosin II equivalent in Drosophila melanogaster (GIGLSSSRLT), but is absent in simple multicellular (Caenorhabditis elegans) and unicellular organisms, e.g., Dictyostelium discoideum and Saccharomyces cerevisiae. This suggests that this motif emerges early in evolution to slow down NMII-related actomyosin turnover, facilitating the appearance of elaborate actin structures required in complex multicellular organisms.
Ser1935 controls the ability of NMII-B to locally suppress protrusion during front–back polarization and regulates its positioning outside the leading edge
To address the role of Ser1935 phosphorylation in establishing migratory front–back polarization, we observed CHO.K1 cells reconstituted with GFP-coupled wild-type MHCII-B or the 1935D mutant and an mCherry-labeled adhesion marker (vinculin) using total internal reflection fluorescence (TIRF) microscopy. Cells reconstituted with the 1935D mutant displayed intermittent protrusions along the cellular perimeter (Fig. 3 E and Video 5), whereas wild type–expressing cells displayed nonprotrusive, MHCII-B–containing stable rears (Fig. 3 D and Video 4). Also, protrusions in 1935D-expressing cells contained many small adhesions and GFP-labeled NMII filaments that turned over frequently (Fig. 3 E and Video 5). Adhesions associated with wild-type MHCII-B–decorated filaments remained stable (Fig. 3 D and Video 4). These experiments suggest that Ser1935 phosphorylation impairs the ability of NMII-B to locally suppress protrusion.
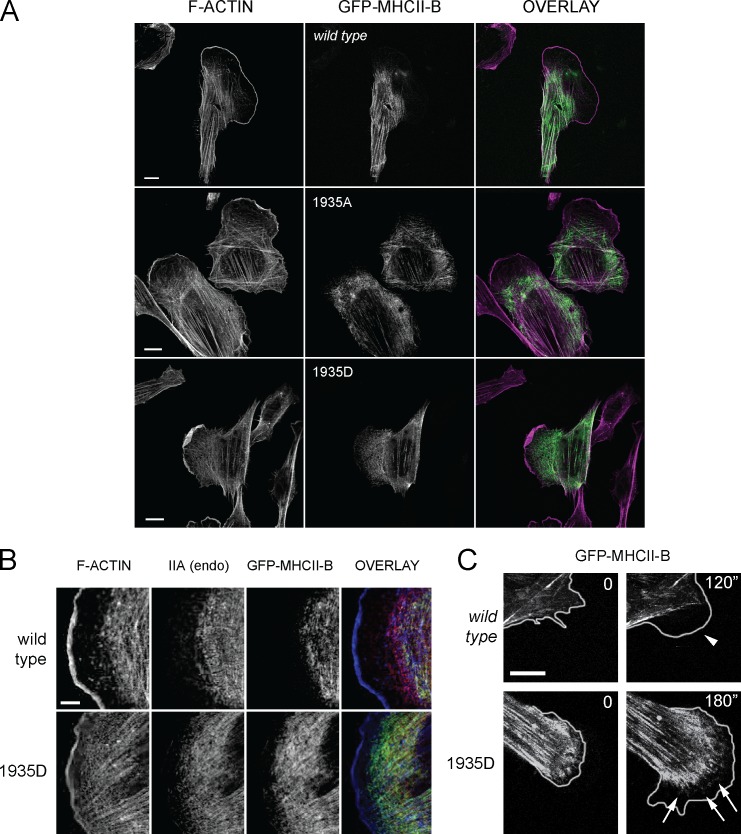
NMII-A forms puncta in protrusive areas, whereas NMII-B seldom appears in these regions (Kolega, 2003; Vicente-Manzanares et al., 2008). Confocal imaging revealed that GFP–MHCII-B 1935D was prominently localized to protrusions in CHO.K1 (Fig. 4 A, bottom row) and U-2 OS cells (Fig. S1, arrows). On the contrary, wild-type NMII-B and the 1935A mutant were not prominent in these regions (Fig. 4 A, top and middle rows; and Fig. S1, arrowheads). Also, whereas wild-type NMII-B appeared behind an NMII-A–only region immediately posterior to the lamellipodium, the 1935D mutant appeared interspersed with NMII-A and close to the leading edge (Fig. 4 B and Fig. S1, arrows). Live-cell confocal imaging experiments showed that the 1935D mutant formed numerous puncta within protrusions, whereas wild-type MHCII-B puncta seldom localized to these regions (Fig. 4 C and Videos 6 and 7). These results suggest that Ser1935 is a crucial regulator of NMII-B assembly and dynamics, and its phosphorylation status controls the proper localization and function of this isoform.
Figure 4.
Ser1935 controls the architecture of the leading edge and the exclusion of NMII-B from this region. (A) CHO.K1 cells transfected with the indicated GFP-MHCII-B constructs (green in overlay) were plated on fibronectin, allowed to attach for 2 h, treated with IGF-1 for 15 min, fixed, and stained for F-actin (magenta in overlay). Representative examples are shown. (B) CHO.K1 cells transfected with the indicated GFP–MHCII-B constructs (green in overlay) were treated as in A and stained for F-actin (blue in overlay) and endogenous NMII-A (IIA; red in overlay). endo, endogenous. (C) Snapshots of confocal time-lapse videos of wild-type GFP–MHCII-B (top) or the 1935D mutant (bottom). The edge of the cell was obtained from corresponding phase-contrast images. Arrowhead in the wild-type image indicates a protrusion devoid of small puncta corresponding to wild-type NMII-B; arrows in the NMII-B 1935D image indicate small puncta corresponding to the mutant appearing within the protrusion. Bars: (A and C) 10 µm; (B) 5 µm. Images extracted from Videos 6 and 7.
Ser1935 regulates the formation of large NMII-B actomyosin bundles and front–rear polarity downstream of PKC-ζ
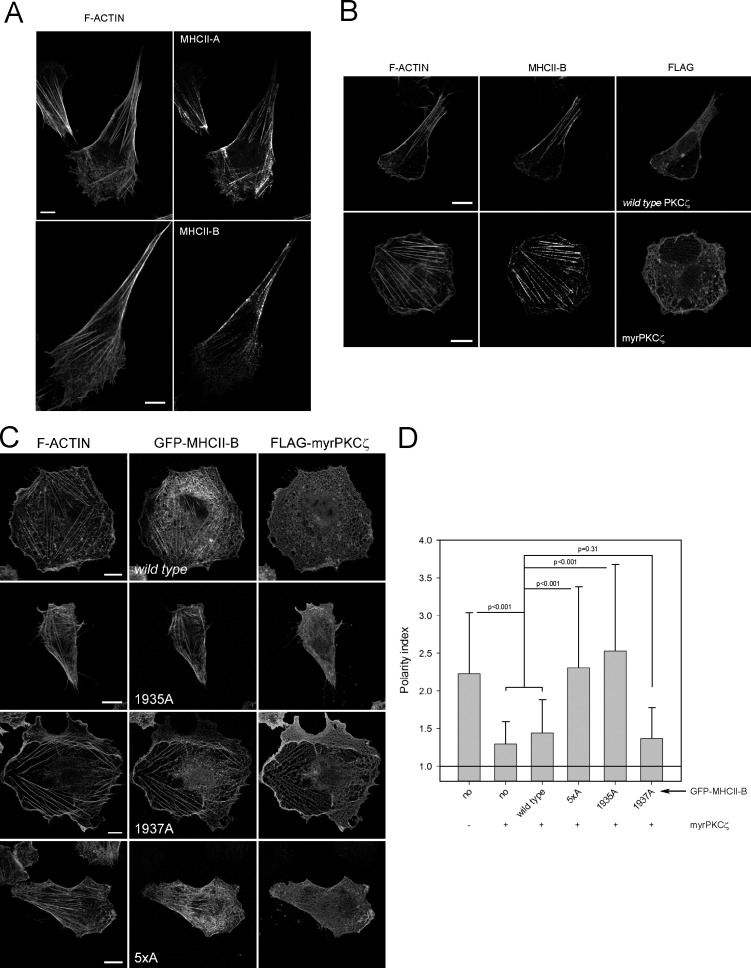
Previous studies showed that PKC-ζ controls cell polarization upstream of NMII (Gomes et al., 2005; Even-Faitelson and Ravid, 2006). To determine whether NMII-B–mediated generation of large, stable bundles is controlled by PKC-ζ, we expressed a constitutively active mutant PKC-ζ (myristoylated [myr]–PKC-ζ). This construct induced isotropic protrusion along the perimeter of the cell and prevented the assembly of large and stable, NMII-B–decorated actomyosin bundles (Fig. 5, A and B).
Figure 5.
PKC-ζ controls cell polarity in a Ser1935 MHCII-B–dependent fashion. (A) Representative images display the localization of endogenous MHCII-A (top right) and MHCII-B (bottom right) and F-actin. Images are z projections of confocal stacks. (B) Effect of myristoylated (myr; constitutively active) PKC-ζ in cell polarity. Cells were transfected with FLAG-tagged wild type (top row) or myr–PKC-ζ (bottom row) and stained for endogenous MHCII-B (middle column) and F-actin (left column). FLAG staining (right column) denotes transfected cells. Representative cells are shown. (C) Reversion of myr–PKC-ζ induced depolarization by NMII-B mutants. Images depict representative cells cotransfected with FLAG-myr–PKC-ζ and the indicated GFP–MHCII-B construct. F-actin is shown in the left column, GFP-MHCII-B mutants are in the middle column, and FLAG is shown in the right column. (D) Quantification of the polarity index of cells as shown in C. Data are the means ± SD of >200 cells from four independent experiments, and p-value is the significance score in the Mann–Whitney test. Bars, 10 µm.
To assess the role of Ser1935 in polarization under these conditions, we coexpressed myr–PKC-ζ with various GFP-coupled MHCII-B constructs. Wild-type MHCII-B did not prevent myr–PKC-ζ–induced loss of polarity. In contrast, the 1935A and 5×A mutant prevented this effect (Fig. 5, C and D). Previous studies have shown that PKC-ζ phosphorylates MHCII-B in Ser1937 downstream of Pak1 (Even-Faitelson et al., 2005; Even-Faitelson and Ravid, 2006), but the nonphosphorylatable 1937A mutant does not overcome the effect of activated PKC-ζ (Fig. 5, C and D). Together, this study shows the existence of at least one crucial regulatory residue that controls the differential ability of NMII-B to form stable actomyosin bundles during cell migration, thereby contributing to the stabilization of migratory polarity in mesenchymal cells.
Materials and methods
Antibodies and reagents
Antibodies against the following molecules were used: MHCII-A and MHCII-B (rabbit IgG; Covance); FLAG (mouse IgG; Sigma-Aldrich). IGF-1, Phalloidin, and secondary antibodies (goat IgG against mouse or rabbit) conjugated to Alexa Fluor dyes (Alexa Fluor 488, 568, or 647 as indicated) were obtained from Invitrogen. Fibronectin and all other reagents were obtained from Sigma-Aldrich unless indicated otherwise.
Plasmids
GFP is in the cytomegalovirus (CMV)-based pEGFP-C1. GFP–MHCII-A and -B are C-terminal fusions of the corresponding human genes in the CMV-based pEGFP-C3 vector. pSUPER–MHCII-A and pSUPER–MHCII-B contain described shRNA sequences against the corresponding rat/hamster (MHCII-A) and rat/hamster/human (MHCII-B) genes (Vicente-Manzanares et al., 2007). Paxillin-mCherry is an N-terminal fusion of chicken paxillin in the CMV-based pEGFP-N3 vectors in which EGFP has been replaced with mCherry (Choi et al., 2008). mCherry-vinculin was generated from chicken vinculin in the CMV-based pEYFP-C1 vector (provided by S. Craig [John Hopkins School of Medicine, Baltimore, MD] through Addgene) by fluorescent probe swap (from pmCherry-C1). The MHCII-A and MHCII-B mutants were constructed from the GFP–MHCII-A or GFP–MHCII-B constructs described in the previous sentences by site-directed mutagenesis using the GeneTailor kit (Invitrogen). These include GFP–MHCII-B Δ5S, GFP–MHCII-A+5S, GFP–MHCII-A+5D, GFP–MHCII-B 5×A, GFP–MHCII-B 5×D, GFP–MHCII-B 1935A, GFP–MHCII-B 1935D, GFP–MHCII-B 1937A, GFP–MHCII-B 1938A, GFP–MHCII-B 1938D, GFP–MHCII-B 1939A, GFP–MHCII-B 1939D, GFP–MHCII-B ADDDD, and GFP–MHCII-B DAAAA. All the human MHCII-B mutants include a silent mutation in the shRNA target sequence (TCA→AGC = Ser→Ser) that renders them shRNA insensitive (Vicente-Manzanares et al., 2007). FLAG-tagged rat PKC-ζ (wild type and myr) constructs in the pCMV6 vector were obtained from A. Toker (Beth Israel Deaconess Medical Center, Boston, MA) through Addgene and have been described previously (Chou et al., 1998).
Cell culture and transfection
Chinese hamster ovary CHO.K1 cells (CCL-61; ATCC) were maintained in DMEM medium with 1 g/dl (low) glucose and supplemented with 10% FBS, 4 mM l-glutamine, 1% nonessential amino acids, and 100 U/ml penicillin/streptomycin. Cells were transfected as described previously (Vicente-Manzanares et al., 2011) using X-tremeGENE HP (Roche) as indicated by the manufacturer. Analysis was performed 24 h (overexpression) or 96 h (depletion and rescue) after transfection. Human osteosarcoma U-2 OS cells (HTB-96; ATCC) were maintained in McCoy’s medium supplemented with 10% FBS, 4 mM l-glutamine, 1% nonessential amino acids, and 100 U/ml penicillin/streptomycin. U-2 OS transfected cells were assayed after 120 h.
Cell plating and immunofluorescence assays
For in vivo as well as staining, cells were allowed to adhere for 1–2 h to 13-mm glass coverslips coated with 2 µg/ml fibronectin in the presence of 100 ng/ml IGF-1 for 15 min where indicated. For staining, cells were fixed using 4% PFA for 10 min at room temperature, permeabilized with 0.2% Triton X-100 for 10 min, and incubated for 2 h with the indicated primary antibodies followed by a 1:500 dilution of the appropriate Alexa Fluor–conjugated antibody and/or Alexa Fluor–conjugated phalloidin for 45 min. Three rinses of 5 min with TBS were performed between incubations and before mounting on glass slides using ProLong antifade medium. Quantification of the polarity index was performed on images acquired using a fluorescence microscope (DMR; Leica). In brief, the polarity index is the result of dividing the length of the main migration axis (which denotes the direction of migration) by the length of the perpendicular axis that passes through the center of the nucleus (Vicente-Manzanares et al., 2007). Polarity index data are always represented as the mean ± SD of >200 cells from four independent experiments. These data did not display a normal distribution (according to Shapiro–Wilk’s test); thus, we used Mann–Whitney’s test to determine significance.
Confocal and TIRF microscopy
FRAP and in vivo imaging was performed using a confocal microscope (SP5; Leica). Images were obtained using a 63× Plan Apochromat objective (NA 1.40) by illumination with light of the appropriate wavelength from a 488-nm Ar laser (fluorochromes used with this laser included GFP or Alexa Fluor 488 as indicated), 561-nm diode-pumped solid-state laser (fluorochromes used with this laser included mCherry or Alexa Fluor 568 as indicated), or a 633-nm He/Ne laser (fluorochrome used with this laser is Alexa Fluor 647) using spectral filtering and hybrid (HyD; Leica) detectors. Images were collected using TCS software (Leica). TIRF images (using GFP- and/or mCherry-tagged probes) corresponding to Fig. 1 and Videos 1–3 were acquired on an inverted microscope (IX70; Olympus) equipped with a 60× Apochromat N objective (NA 1.45). Excitation laser lines were 488 and 543 nm. For dual GFP-mCherry, a dual emission filter (Z488/561) was used. Images were acquired with a charge-coupled device (CCD) camera (Retiga EXi; QImaging) and MetaMorph software (Molecular Devices). TIRF images (using GFP- and/or mCherry-tagged probes) from Fig. 3 and Videos 4 and 5 were acquired on an automated inverted microscope (IX83; Olympus) fitted with a 60× Apochromat N 60× OTIRF (NA 1.49) objective, a cellTIRF Mitico Unit (Olympus), and 491/561-nm laser lines. Dual GFP-mCherry samples were visualized with a dual emission filter (Z488/561). Images were captured using an electron-multiplying CCD camera (ImagEM X2; Hamamatsu Photonics) and Xcellence software (Olympus). In all cases, cells were kept at 37°C in temperature-controlled incubators. Imaging medium was CCM-1 (HyClone) or phenol red–free DMEM (Gibco) + 10% FCS. Images were analyzed using ImageJ (National Institutes of Health).
FRAP
Confocal images for FRAP analysis were acquired using the confocal microscope (SP5) as described in the previous section. Initially, a cellular area (35 µm2) that contained GFP fusion protein-decorated actin filament bundles was scanned three times and then bleached using 30 scans at 100% laser power. To image the recovery of fluorescence intensity after photobleaching, we recorded 15 scans every 0.06 s, 15 scans every 1 s, and subsequent scans every 3–5 s. Background subtraction and normalization were calculated using built-in algorithms, and normalized intensities versus time (seconds) were represented. FRAP data are always represented as the means ± SEM of 24 individual measurements per condition in four independent experiments.
Mass spectrometry—phosphoproteomics
In brief, HEK-293 cells were and maintained in DMEM medium for 24 h. To inhibit phosphatase activity, cells were treated with 1 µM peroxovanadate and 10 nM calyculin A (EMD) 30 min before lysis. Subsequently cells were lysed in a buffer containing 20 mM Tris, 300 mM NaCl, 10 mM MgCl2, and 5 mM ATP with protease and phosphatase inhibitors and immunoprecipitated for 90 min using 5 µl/sample of the MHCII-B antibody (Covance) and 100 µl/sample protein A–conjugated Sepharose (GE Healthcare). The samples were rinsed three times with lysis buffer and resuspended in 100 µl Laemmli buffer. Samples were separated using 7% SDS-PAGE and stained using a commercial silver staining kit. A single band at 230 kD as determined by comparison to a molecular weight ruler (Bio-Rad Laboratories) was excised. The purified protein was reduced, alkylated, and digested in gel with trypsin, and peptides were extracted and desalted. A portion of the peptide mixture was gradient eluted into on a front-end electron transfer dissociation-enabled mass spectrometer (LTQ-FT; Thermo Fisher Scientific). MS1 spectra were acquired in the high-resolution Fourier transform mass analyzer, and MS2 spectra were acquired in the linear ion trap mass analyzer. Additionally, the peptide mixture was enriched for phosphopeptides using iron-IMAC after conversion of peptide carboxyl groups to methyl esters as described elsewhere (Ficarro et al., 2002). In brief, a 150-µm inner diameter fused-silica column with 6 cm of POROS MC 20 µm metal affinity bulk media (Applied Biosystems) was constructed. The column was rinsed with water, 50 mM EDTA, pH 9, and water again. The column was activated with 100 mM FeCl3 (Sigma-Aldrich) and then rinsed with 0.01% acetic acid at 1 µl/min flow rate. Peptide methyl esters were reconstituted in a 1:1:1 solution of acetonitrile/methanol/0.01% acetic acid and then loaded onto the activated IMAC column at a flow rate of 1 µl/min. The column was washed with 1:1:1 solution and 0.01% acetic acid before being butt connected to a C18 precolumn, as previously described. Phosphopeptides were eluted onto the precolumn with 250 mM ascorbic acid (Sigma-Aldrich) at 1 µl/min; combined columns were washed with 0.1% acetic acid at the same flow rate. The IMAC and precolumn were separated after which the loaded precolumn was rinsed with 0.1% acetic acid before connection to an analytical column. Phosphopeptides were gradient eluted and mass analyzed using the methods described earlier in this section. All MS2 spectra were searched against a MHCII-B protein database and the human, rat, and mouse NR (nonredundant) database using the OMSSA (Open Mass Spectrometry Search Algorithm), and they were manually validated by visual inspection of the spectra.
Online supplemental material
Fig. S1 depicts the morphologies, actin, and MHCII-A/B staining of U-2 OS cells depleted of MHCII-B and rescued with the indicated constructs. Fig. S2 shows a representative FRAP experiment and FRAP of MHCII-A+5D. Fig. S3 shows collisionally activated dissociation MS/MS spectrum of the endogenous MHCII-B tryptic peptide GGPIsFSSSR phosphorylated on Ser-1935 and extracted ion chromatogram of endogenous monophosphorylated MIIB peptide Gly 1931–Arg 1940 with the three different detected phosphorylations. Videos 1–3 show adhesion assembly visualized by TIRF in MHCII-A–depleted cells using paxillin-mCherry (Video 1 and Videos 2 and 3, left) and the dynamics of GFP–MHCII-A wild type (Video 2, right) and the +5S mutant (Video 3, right) used for functional rescue. Videos 4 and 5 show adhesion assembly visualized by TIRF in MHCII-B–depleted cells with mCherry-vinculin (left) and dynamics of GFP–MHCII-B wild type (Video 4, right) and 1935D (Video 5, right) used for functional rescue. Videos 6 and 7 show dynamics of GFP–MHCII-B wild type (Video 6, left) and 1935D (Video 7, left) and the corresponding phase images (right). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407059/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201407059.dv.
Supplementary Material
Acknowledgments
The authors thank Prof. Francisco Sánchez-Madrid, Dr. María N. Navarro, and Alvaro Ortega-Carrión for useful discussions and Vanessa Centeno for technical assistance.
This work is supported by grants SAF2011-24953 from MINECO, FP7 Marie Curie CIG-293719 from the EU, CIVP16A1831 from the Ramon Areces Foundation (M. Vicente-Manzanares), GM 23244 (A.R. Horwitz), GM037537 (D.F. Hunt), and the Cell Migration Consortium U54 GM64346 (A.R. Horwitz and D.F. Hunt). M. Vicente-Manzanares is an investigator from the Ramón y Cajal Program (RYC-2010-06094).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- CMV
- cytomegalovirus
- IMAC
- immobilized metal affinity chromatography
- MS
- mass spectrometry
- myr
- myristoylated
- NMII
- nonmuscle myosin II
- TIRF
- total internal reflection fluorescence
References
- Bian Y., Song C., Cheng K., Dong M., Wang F., Huang J., Sun D., Wang L., Ye M., and Zou H.. 2014. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics. 96:253–262 10.1016/j.jprot.2013.11.014 [DOI] [PubMed] [Google Scholar]
- Cai Y., Biais N., Giannone G., Tanase M., Jiang G., Hofman J.M., Wiggins C.H., Silberzan P., Buguin A., Ladoux B., and Sheetz M.P.. 2006. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys. J. 91:3907–3920 10.1529/biophysj.106.084806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., and Horwitz A.R.. 2008. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.M., Hou W., Johnson J., Graham L.K., Lee M.H., Chen C.S., Newton A.C., Schaffhausen B.S., and Toker A.. 1998. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr. Biol. 8:1069–1077 10.1016/S0960-9822(98)70444-0 [DOI] [PubMed] [Google Scholar]
- Clark K., Middelbeek J., Dorovkov M.V., Figdor C.G., Ryazanov A.G., Lasonder E., and van Leeuwen F.N.. 2008. The α-kinases TRPM6 and TRPM7, but not eEF-2 kinase, phosphorylate the assembly domain of myosin IIA, IIB and IIC. FEBS Lett. 582:2993–2997 10.1016/j.febslet.2008.07.043 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., and Hall A.. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell. 106:489–498 10.1016/S0092-8674(01)00471-8 [DOI] [PubMed] [Google Scholar]
- Even-Faitelson L., and Ravid S.. 2006. PAK1 and aPKCζ regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol. Biol. Cell. 17:2869–2881 10.1091/mbc.E05-11-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Faitelson L., Rosenberg M., and Ravid S.. 2005. PAK1 regulates myosin II-B phosphorylation, filament assembly, localization and cell chemotaxis. Cell. Signal. 17:1137–1148 10.1016/j.cellsig.2004.12.015 [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A.D., Conti M.A., Matsumoto K., Adelstein R.S., and Yamada K.M.. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 9:299–309 10.1038/ncb1540 [DOI] [PubMed] [Google Scholar]
- Ficarro S.B., McCleland M.L., Stukenberg P.T., Burke D.J., Ross M.M., Shabanowitz J., Hunt D.F., and White F.M.. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20:301–305 10.1038/nbt0302-301 [DOI] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Hall A.2012. Rho family GTPases. Biochem. Soc. Trans. 40:1378–1382 10.1042/BST20120103 [DOI] [PubMed] [Google Scholar]
- Kolega J.2003. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol. Biol. Cell. 14:4745–4757 10.1091/mbc.E03-04-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhou L., and Gorodeski G.I.. 2006. Estrogen regulates epithelial cell deformability by modulation of cortical actomyosin through phosphorylation of nonmuscle myosin heavy-chain II-B filaments. Endocrinology. 147:5236–5248 10.1210/en.2006-0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.M., Buxton D.B., Chua G.C., Dembo M., Adelstein R.S., and Wang Y.L.. 2004. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell. 15:982–989 10.1091/mbc.E03-06-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P., Phillips C.L., Adelstein R.S., and Pollard T.D.. 1994. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J. Cell Sci. 107:3077–3090. [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. . 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3. [DOI] [PubMed] [Google Scholar]
- Raab M., Swift J., Dingal P.C.D.P., Shah P., Shin J.-W., and Discher D.E.. 2012. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 199:669–683 10.1083/jcb.201205056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., and Ravid S.. 2006. Protein kinase Cγ regulates myosin IIB phosphorylation, cellular localization, and filament assembly. Mol. Biol. Cell. 17:1364–1374 10.1091/mbc.E05-07-0597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandquist J.C., and Means A.R.. 2008. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol. Biol. Cell. 19:5156–5167 10.1091/mbc.E08-05-0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I., Yong J., Dong J.M., Lim L., and Leung T.. 2008. A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell. 135:123–136 10.1016/j.cell.2008.09.018 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Zareno J., Whitmore L., Choi C.K., and Horwitz A.F.. 2007. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176:573–580 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Koach M.A., Whitmore L., Lamers M.L., and Horwitz A.F.. 2008. Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 183:543–554 10.1083/jcb.200806030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Newell-Litwa K., Bachir A.I., Whitmore L.A., and Horwitz A.R.. 2011. Myosin IIA/IIB restrict adhesive and protrusive signaling to generate front–back polarity in migrating cells. J. Cell Biol. 193:381–396 10.1083/jcb.201012159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Di Palma S., Preisinger C., Peng M., Polat A.N., Heck A.J., and Mohammed S.. 2013. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12:260–271 10.1021/pr300630k [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.