Abstract
Peters anomaly is a rare form of anterior segment ocular dysgenesis, which can also be associated with additional systemic defects. At this time, the majority of cases of Peters anomaly lack a genetic diagnosis. We performed whole exome sequencing of 27 patients with syndromic or isolated Peters anomaly to search for pathogenic mutations in currently known ocular genes. Among the eight previously recognized Peters anomaly genes, we identified a de novo missense mutation in PAX6, c.155G>A, p.(Cys52Tyr), in one patient. Analysis of 691 additional genes currently associated with a different ocular phenotype identified a heterozygous splicing mutation c.1025+2T>A in TFAP2A, a de novo heterozygous nonsense mutation c.715C>T, p.(Gln239*) in HCCS, a hemizygous mutation c.385G>A, p.(Glu129Lys) in NDP, a hemizygous mutation c.3446C>T, p.(Pro1149Leu) in FLNA, and compound heterozygous mutations c.1422T>A, p.(Tyr474*) and c.2544G>A, p.(Met848Ile) in SLC4A11; all mutations, except for the FLNA and SLC4A11 c.2544G>A alleles, are novel. This is the frst study to use whole exome sequencing to discern the genetic etiology of a large cohort of patients with syndromic or isolated Peters anomaly. We report five new genes associated with this condition and suggest screening of TFAP2A and FLNA in patients with Peters anomaly and relevant syndromic features and HCCS, NDP and SLC4A11 in patients with isolated Peters anomaly.
Introduction
Peters anomaly (PA) is a rare form of anterior segment dys-genesis characterized by corneal opacity with or without iridocorneal and/or corneolenticular adhesions with associated defects in the posterior layers of the cornea. These abnormalities may obstruct the visual axis leading to visual impairment. Peters anomaly is often associated with glaucoma due to maldevelopment of the trabecular mesh-work (Reis and Semina 2011; Bhandari et al. 2011). Peters anomaly occurs between the 4th and 7th week of embryonic development as a result of faulty separation of the lens from the surface ectoderm or aberrant reattachment of the lens/iris to the cornea during development of the anterior chamber (Matsubara et al. 2001). Peters anomaly can be found in isolation or in association with systemic features (Ito and Walter 2014). Systemic features are highly variable and may include craniofacial defects (cleft lip/palate, low-set ears, micrognathia, dental defects, abnormal upper lip), central nervous system anomalies (developmental delay, intracranial calcifications, agenesis of the corpus callosum), skeletal defects (brachydactyly, clinodactyly, short limbs, vertebral anomalies, short stature), congenital heart disease, or renal, genital and other anomalies (Ozeki et al. 2000; Weh et al. 2014).
A majority of cases of Peters anomaly lack a genetic diagnosis. To date, mutations in PAX6, PITX2, PITX3, FOXC1, FOXE3, CYP1B1, B3GLCT (formerly B3GALTL) and COL4A1 have been associated with this disorder (Hanson et al. 1994; Reis et al. 2012; Semina et al. 1998; Honkanen et al. 2003; Ormestad et al. 2002; Vincent et al. 2006; Deml et al. 2014). Although mutations in these genes have been shown to occasionally cause Peters anomaly, they all are predominantly responsible for other phenotypes such as aniridia (PAX6), Axenfeld–Rieger spectrum (FOXC1 and PITX2), cataract and microphthalmia (PITX3), primary aphakia (FOXE3), primary congenital/ infantile glaucoma (CYP1B1), and brain small vessel disease (COL4A1) (Reis and Semina 2011). Mutations in B3GLCT have been shown to explain 100 % of classic Peters plus syndrome (PPS) (Lesnik Oberstein et al. 2006; Reis et al. 2008; Weh et al. 2014). This syndrome includes anterior segment abnormalities (Peters anomaly in 85 % of cases) in combination with short stature, brachydactyly, dysmorphic facial features, and mental retardation (Weh et al. 2014; Maillette de Buy Wenniger-Prick and Hennekam 2002). Mutations in B3GLCT have not been found in cases of isolated Peters anomaly or atypical PPS (Reis et al. 2008; Weh et al. 2014).
A recent study by Prokudin and coauthors utilized whole exome sequencing to determine the genetic cause of developmental eye diseases including isolated PA in three patients; causative mutations were identified in two of the patients with PA including a novel PAX6 mutation and compound heterozygous mutations in CYP1B1 (Prokudin et al. 2014). In this study, we have analyzed whole exomes of 27 patients with syndromic or isolated Peters anomaly. We utilized a candidate gene list to identify mutations in genes which are known to result in Peters anomaly when mutated as well as genes currently associated with other ocular phenotypes. Our data expand the list of genes associated with Peters anomaly to include TFAP2A, HCCS, NDP, FLNA and SLC4A11.
Materials and methods
Human patients
The human study was approved by the Institutional Review Board of the Children’s Hospital of Wisconsin with written informed consent obtained from each participant and/or their legal representative. DNA was extracted from blood or buccal samples using standard protocols. The quantity/ quality of DNA was assessed using a NanoDrop 1000 spectrophotometer (Thermo Fisher, USA) and by agarose gel electrophoresis. The study included 20 patients with syndromic Peters anomaly (two or more non-ocular features) and 7 patients with isolated Peters anomaly; three probands had a family history of an ocular condition, two had a family history of non-ocular features, and the remaining cases were sporadic (Supplementary Table 1). Of note, most of the probands were previously examined for B3GLCT, CYP1B1, FOXC1, FOXE3, PITX2 and PITX3 variants by Sanger sequencing with no pathogenic mutations identified (Weh et al. 2014; Reis et al. 2012; data not shown).
Whole exome sequencing and data analysis
Genomic DNA was processed for whole exome sequencing by either Axeq (Rockville, MD) or Perkin Elmer, Inc (Branford, CT). Exome capture was performed with the Agilent Sure Select v4 or v4+ UTR platforms (Santa Clara CA) and 100 base pair paired end sequencing was executed using the Illumina HiSeq 2000 platform (San Diego, CA). The raw reads were aligned by the sequencing company using the Burrows–Wheeler Aligner (BWA) and variants were called using the Genome Analysis Toolkit (GATK v2.20) pipeline available through Perkin Elmer (Branford, CT) or the Sequence Alignment/Map (SAMtools) pipeline through Axeq (Rockville, MD).
The whole exome data were analyzed for mutations in 8 genes previously associated with Peters anomaly (Table 1) and other ocular genes (635 genes from the NEIBank list of Human Eye Disease Genes (http://neibank.nei.nih.gov) and 56 additional genes which have recently been reported in association with ocular phenotypes; Supplementary Table 2) using the SNP & Variation Suite (SVS; Golden Helix, Bozeman, MT). Polymorphic variants that showed more than 1 % allele frequency in general populations (based on the Database of Single Nucleotide Variants (dbSNP, http://www.ncbi.nlm.nih.gov/snp/), 1000 genomes sequencing project (http://www.1000genomes.org/) or the National Heart Lung and Blood Institute Exome Variant Server database (EVS, http://evs.gs.washington.edu/EVS/) were excluded from further analysis. Variants were then classified as either coding or non-coding. Coding variants were further characterized by their predicted impact on the transcript and protein product (insertion, deletion, frameshift indels, nonsense, missense and synonymous). Among non-coding variants, splicing site mutations that alter canonical sequences were considered. The MaxEntScan program (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) was utilized to predict the functional consequences of identified splicing variants. The SIFT, PolyPhen, MutationTaster, Mutation-Assessor, and FATHMM prediction algorithms as well as GERP++ and PhyloP conservation scores were accessed through SVS utilizing data from dbNSFP (Liu et al. 2013) to assist in predicting the functional impact of identified missense variants. Nonsense, splicing, frameshift, or missense variants that were predicted to be damaging by at least 4/5 programs and demonstrated high conservation scores (GERP++ higher than 4 and PhyloP greater than 1.5) were considered to be likely pathogenic mutations. For recessive genes, if one likely pathogenic mutation was identified by these criteria, the criteria to be considered pathogenic were relaxed for the second allele (predicted to be damaging by at least 2/5, GERP ++ score higher than 3 and PhyloP greater than 1).
Table 1.
Summary of genes associated with Peters anomaly (PA)
| Gene | Location | Function | Main phenotype | Reference for PA |
|---|---|---|---|---|
| B3GLCT | 13q12.3 | Beta-1,3-glucosyltransferase | Peters Plus syndrome | Lesnik Oberstein et al. (2006) |
| COL4A1 | 13q34 | Basement membrane collagen | Brain small vessel disease | Deml et al. (2014) |
| CYP1B1 | 2p22.2 | Cytochrome P450 monooxygenase | Primary congenital/infantile glaucoma | Vincent et al. (2006) |
| FLNA | Xq28 | Actin-binding protein | Otopalatodigital spectrum | This manuscript |
| FOXC1 | 6p25 | Transcription factor | Axenfeld–Rieger spectrum | Honkanen et al. (2003) |
| FOXE3 | 1p33 | Transcription factor | Microphthalmia, cataract | Ormestad et al. (2002) |
| HCCS | Xp22.2 | Holocytochrome C-type synthetase | Microphthalmia | This manuscript |
| NDP | Xp11.3 | Cystine knot growth factor | Norrie disease/familial exudative vitreoretinopathy | This manuscript |
| PAX6 | 11p13 | Transcription factor | Aniridia | Hanson et al. (1994) |
| PITX2 | 4q25 | Transcription factor | Axenfeld–Rieger spectrum | Reis et al. (2012) |
| PITX3 | 10q24.32 | Transcription factor | Cataract, multiple types | Semina et al. (1998) |
| SLC4A11 | 20p13 | SLC4 bicarbonate transporter | Corneal endothelial dystrophy 2 | This manuscript |
| TFAP2 | 6p24.3 | Transcription factor | Branchiooculofacial syndrome | This manuscript |
Information about protein structure was obtained using UniProtKB (http://www.uniprot.org) or published reports as specified; records of previously identified mutations in the genes of interest were obtained from publicly available mutation databases [ClinVar (http://www.ncbi.nlm.nih.gov/clinvar); HGMD (http://www.hgmd.cf.ac.uk/ac); LOVD (http://lsdb.hgu.mrc.ac.uk/home.php)] and published reports as indicated.
Variant confirmation
Oligonucleotides flanking the genomic locations of identified variants were designed using the Primer3Plus browser (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi). PCR was performed using genomic DNA from probands and available family members to confirm the variant and assess segregation within the family. Where possible, available family members carrying a variant were also examined clinically. The primers and conditions utilized for the identified mutations are summarized in Supplementary Table 3. PCR products were sequenced bidirectionally using Big Dye Terminator chemistry v3.1 and sequenced using an ABI 3730XL sequencer (Applied Biosystems/Life Technologies, Carlsbad, CA, USA). Sequences were reviewed manually and using Mutation Surveyor (SoftGenetics, State College, PA) and compared to the following reference sequences: NM_000280.3 (PAX6), NM_001032280.2 (TFAP2A), NM_001122608.2 (HCCS), NM_000266.3 (NDP), NM_001456.3 (FLNA), NM_032034.3 (SLC4A11).
Results and discussion
The whole exomes of 27 probands affected with either syndromic (20) or isolated (7) Peters anomaly were analyzed for causative mutations. The average read depth was 54.7 ± 10.8 and on average 94.7 ± 1.8 % of targets were covered by greater than 10× and 99 ± 0.4 % of target nucleotides were covered by 1× or greater. Our initial analysis of the exome data included eight genes known to cause isolated or syndromic Peters anomaly: B3GLCT, CYP1B1, COL4A1, FOXC1, FOXE3, PAX6, PITX2, and PITX3 (Table 1). Using the known gene list for Peters anomaly, a causative mutation in PAX6 was discovered in one case (Table 2). We then expanded our search to include all other genes which have been associated with human ocular phenotypes (NEIBank list of Human Eye Disease Genes and Supplementary Table 2). We identified five additional pathogenic and likely pathogenic variants in genes from this list (Table 2). No pathogenic/likely pathogenic variants were identified in the remaining 21 patients [69 alleles, all heterozygous, were identified based on criteria outlined above; the variants were concluded to be of unknown significance because of their presence in control databases, unclear functional consequences, inconsistency with the known mode of inheritance (heterozygous variants for recessive conditions) and/or a distinctly different phenotype known to be associated with the corresponding genes; Supplementary Table 4].
Table 2.
Summary of identified pathogenic mutations
| Gene | Nucleotide change |
Protein change | Status | Eye |
Short stature/IUGR |
Limb |
DD | CL/P | CHD | Renal | Genital | Other systems | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peters | Other ASD | Other | BD | Other | |||||||||||
| PAX6 | c.155G>A | p.(Cys52Tyr) | Heterozygous, de novo | + | − | − | − | − | − | − | − | − | − | − | Broad nasal bridge |
| TFAP2A | c.1025+2T>A | ND | Heterozygous | + | + | − | + | − | − | − | + | − | + | − | Thin upper lip, accessory nipple (L) |
| HCCS | c.715C>T | p.(Gln239*) | Heterozygous, de novo | + | + | + | − | − | − | − | − | − | − | − | − |
| NDP | c.385G>A | p.(Glu129Lys) | Hemizygous | + | + | − | − | − | − | − | − | − | − | − | − |
| FLNA | c.3446C>T | p.(Pro1149Leu) | Hemizygous | + | + | + | + | − | + | U | + | + | + | + | Multiple congenital defects; neonatal death |
| SLC4A11 | c.1422T>A; c.2544G>A |
p.(Tyr474*) p.(Met848Ile) |
Compound heterozygous |
+ | − | + | − | − | − | M | − | − | − | − | Umbilical hernia |
ASD Anterior segment dysgenesis, IUGR intrauterine growth restriction, BD brachydactyly, DD developmental delay, CL/P cleft lip and/or palate, CHD congenital heart defect, U unknown, M mild delay in gross-motor skills, ND not determined
PAX6 mutation
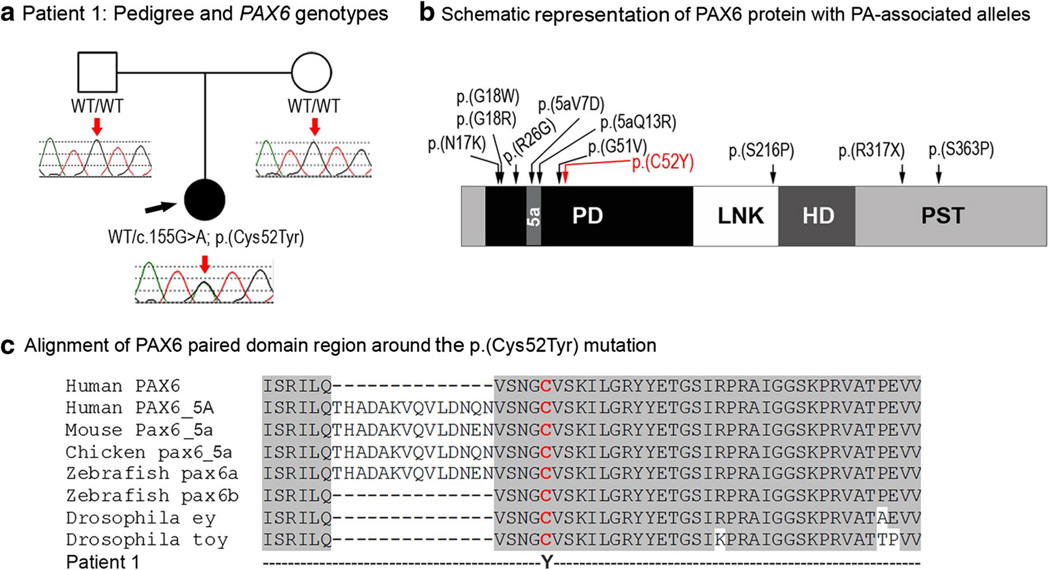
Patient 1, a 9-month-old Caucasian (Serbian/Italian/Irish), had isolated bilateral Peters anomaly. This patient was found to be heterozygous for a novel missense mutation, c.155G>A; p.(Cys52Tyr) that affects the paired domain of the PAX6 gene (Fig. 1). This variant was not found in 13,002 alleles (8,598 European Americans, 4,404 African Americans; mean coverage depth 88 reads) in EVS and not reported in dbSNP or 1,000 genomes, and was predicted to be damaging by 4 out of 5 functional effect prediction programs. In addition, this nucleotide was found to be well conserved with a GERP score of 5.35 and a PhyloP score of 2.49. As neither of the unaffected parents was found to carry this mutation, it is likely a de novo change; however, low-level/gonadal mosaicism in one of the parents cannot be ruled out. Interestingly, a mutation involving the same amino acid has been previously reported in a case of aniridia with cataracts and her mother was affected with juvenile cataracts and glaucoma (Chao et al. 2003), however, that missense variant changed the conserved cysteine to an arginine. The cysteine 52 has been shown to directly contact the phosphate backbone of DNA (Xu et al. 1999), thus changes to this cysteine may affect the ability of PAX6 to properly bind DNA.
Fig. 1.
a Patient 1: Pedigree and PAX6 genotypes. DNA chromatograms of the proband’s and parental PAX6 sequences are shown with a position of c.155G>A, p.(Cys52Tyr) mutation indicated with red arrows; proband is designated with a black arrow. b Schematic representation of the PAX6 protein with PA-associated alleles. PD Paired domain, LNK linker region, HD homeodomain, PST transactivation domain rich of proline, serine, and threonine, 5a- 14-a.a. encoded by the alternatively spliced exon 5a. Positions of all previously reported PAX6 mutations associated with Peters anomaly are shown with black arrows; the cysteine residue at position 52 is shown in red font. c Alignment of PAX6 paired domain region around the p.(Cys52Tyr) mutation. Human PAX6 (NP_000271.1), human PAX6-5A (NP_001245391), mouse Pax6-5a (NP_001231127), chicken pax6-5a (NP_990397), zebrafish pax6a (NP_571379), zebrafish pax6b (NP_571716), Drosophila ey (NP_001014693) and Drosophila toy (NP_524638) sequences were used for the alignment; the position of p.(C52Y) is shown with red arrow
Mutations in PAX6 are associated with autosomal dominant forms of anterior segment dysgenesis, most often aniridia (Reis and Semina 2011), and occasionally other phenotypes including Peters anomaly (Hanson et al. 1994; Zhang et al. 2011). Approximately, one-third of heterozygous Pax6 mutant mice showed Peters anomaly with incomplete separation of the lens from the cornea in one study (Baulmann et al. 2002). Analysis of the previously reported PAX6 mutations identified ten changes associated with Peters anomaly: seven missense mutations in the paired domain, one missense change in the linker domain and two mutations, nonsense and missense, affecting the C-terminal transactivation domain (Fig. 1). Including patient 1 reported here, eight out of the ten missense mutations (80 %) associated with Peters anomaly occur in the paired domain region of PAX6 which appears to be only slightly higher than the overall distribution (68 % of all missense mutations in the PAX6 gene are found in this domain). In contrast, 10 out of the 11 (91 %) Peters anomaly-associated alleles are missense mutations, while among all PAX6 mutations only 28 % are missense.
TFAP2A mutation
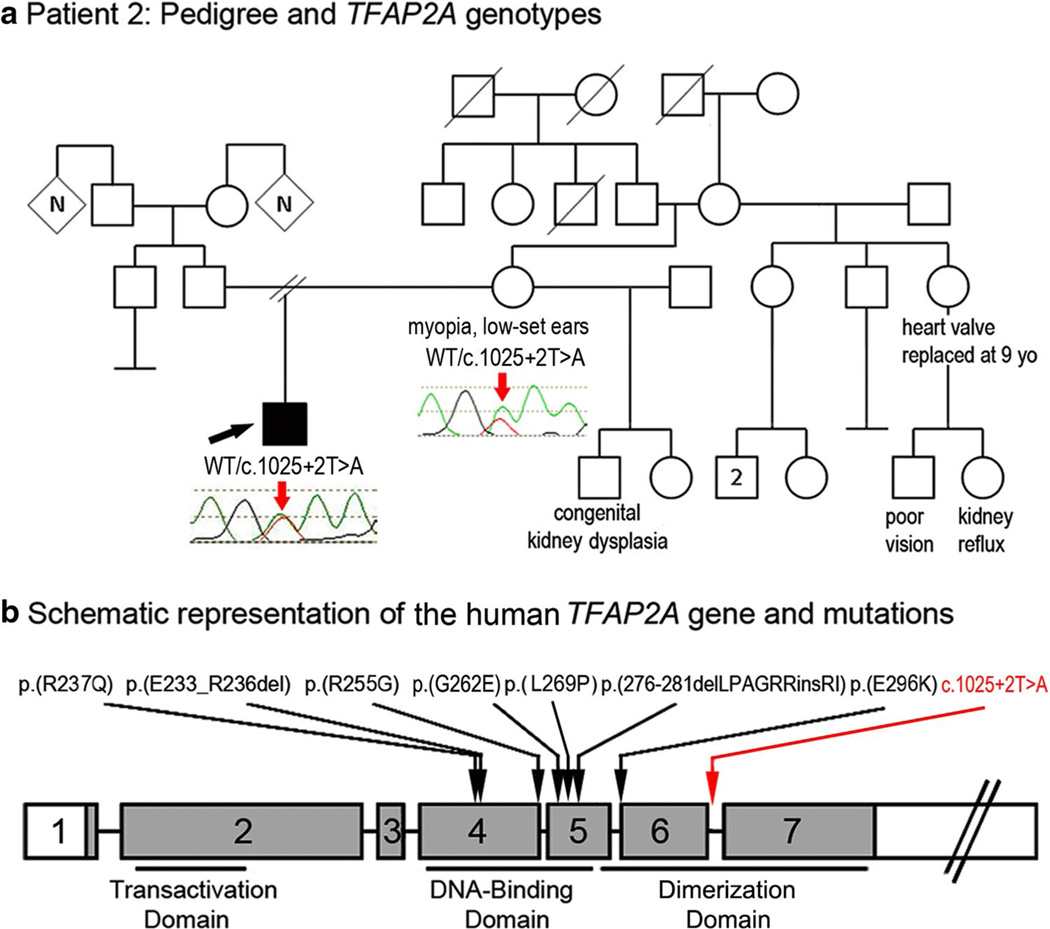
Patient 2, a 9.5-year-old Caucasian (mixed European) male, presented with unilateral Peters anomaly and infantile glaucoma in the right eye; he underwent numerous ocular surgeries including corneal transplant but ultimately the right eye had to be enucleated at 18 months of age. His left eye was normal. This patient also had systemic defects including low-set ears with a right preauricular pit and mild mixed hearing loss with the right ear more severely affected, thin upper lip, and velopharyngeal insufficiency. The patient is of mildly short stature (127 cm, 5–10th percentile) and exhibits global parenchymal hypoplasia of the right kidney. The patient also had an accessory left nipple and showed Bell’s palsy diagnosed at 6 months of age which resolved shortly thereafter. He was found to be heterozygous for a mutation affecting the donor splice site of exon 6 of TFAP2A, c.1025+2T>A. The nucleotide affected by this mutation is well conserved with a GERP score of 5.15 and a PhyloP score of 1.93. The change was absent in 13,006 (8,600 European Americans, 4,406 African Americans; mean coverage depth 131 reads) alleles in EVS and was not present in other control databases. A maternal sample was available and tested positive for the mutation (Fig. 2). The proband’s mother has myopia and minimally low-set ears; her facial features are reported to resemble the patient. Further examination of family history revealed the following congenital/pediatric conditions: a 3-year-old maternal half-brother with congenital multicystic dysplastic left kidney and a maternal aunt who required replacement of a heart valve at age 9 and has a daughter with kidney reflux and poor vision in one eye (Fig. 2). The family was lost to follow-up and no other family members were available for testing or examination.
Fig. 2.
a Patient 2: Pedigree and TFAP2A genotypes. DNA chromatograms of the proband’s and his mother’s TFAP2A sequences are shown with position of the c.1025+2T>A mutation indicated with red arrows; the proband is designated with a black arrow; phenotypes observed in other members of the pedigree are indicated under corresponding symbols. b Schematic representation of the human TFAP2A gene and mutations. Previously reported mutations (Reiber et al. 2010; Gestri et al. 2009; Tekin et al. 2009) are shown with black arrows; the position of the c.1025+2T>A mutation is shown with a red arrow
TFAP2A is located on chromosome 6 and contains seven exons with an alternatively spliced exon 5 and three unique transcription start sites; exons 2–7 are shared between the three transcripts (Gestri et al. 2009). Therefore, the c.1025+2T>A mutation affecting the donor site of exon 6 is likely to disrupt normal splicing of all known TFAP2A isoforms. The mutation was analyzed using MaxEntScan to predict the effect on both 5′ and 3′ splice sites (Yeo and Burge 2004; Samuels et al. 2013; Pauws et al. 2013). This program calculates a numerical score for the probability that a given 5′ (or 3′) splice site sequence is likely to be recognized by the splicing machinery as a splice site with a larger score representing a higher probability. The reference 5′ splice sequence of exon/intron 6 (AAAgtaagg) was given a score of 6.60 while the variant sequence identified in this patient (AAAgaaagg) was given a score of −1.59, thus predicting that this nucleotide change is likely to eliminate the donor splice site of intron 6. The abnormal splicing is likely to disrupt the DNA-binding domain of this transcription factor, which is encoded by exons 6 and 7 (Fig. 2).
TFAP2A is a transcription factor, which is responsive to retinoic acid and essential for normal development of ocular structures (lens and optic cup) and the craniofacial region (Schorle et al. 1996; Pontoriero et al. 2008; Bassett et al. 2010). Heterozygous mutations in TFAP2A have been shown to cause branchio-oculo-facial syndrome (BOFS) (Gestri et al. 2009). Most mutations are missense alleles (usually affecting its DNA-binding domain) or, infrequently, whole or partial gene deletions (Milunsky et al. 2011; Gestri et al. 2009). Only one frameshift mutation has been reported to date (Milunsky et al. 2011) and no splice mutations have been observed. The phenotype of BOFS is variable: major features include thinned erythematous cutaneous defects affecting the cervical or infra/supra-auricular regions, ocular defects (most commonly microphthalmia and lacrimal duct obstruction, less commonly coloboma, strabismus, cataract, and ptosis), and characteristic faces (cleft lip/palate, dolichocephaly, thick nasal tip, upslanted eyes, “pseudo” cleft lip). Patients may also present with short stature, hearing loss, renal anomalies, ectodermal anomalies, supernumerary nipples, and facial weakness (Milunsky et al. 2011). The features of Patient 2 are consistent with an atypical presentation of BOFS. Peters anomaly has not previously been reported in BOFS. The mother, who also carries the mutation, displays only mild features and has another son with congenital kidney defects (who could not be tested for the mutation). The observed phenotypic variability may be explained by the variable expressivity of this mutation; however, the possibility of maternal mosaicism also cannot be excluded.
The complete knockout of Tfap2a in mouse results in perinatal lethality due to incomplete closure of the neural tube, skeletal defects and facial clefting (Schorle et al. 1996). Bassett and colleagues performed a thorough analysis of the ocular anomalies in Tfap2a null mice and found defects of optic cup formation, coloboma and other anomalies. Pontoriero et al. (2008) generated a conditional knockout of Tfap2a in the lens of developing mouse embryos. They found that homozygous conditional knockout mice had Peters anomaly due to improper separation of the lens from the overlying surface ectoderm, which is consistent with the human phenotype observed in our study.
HCCS mutation
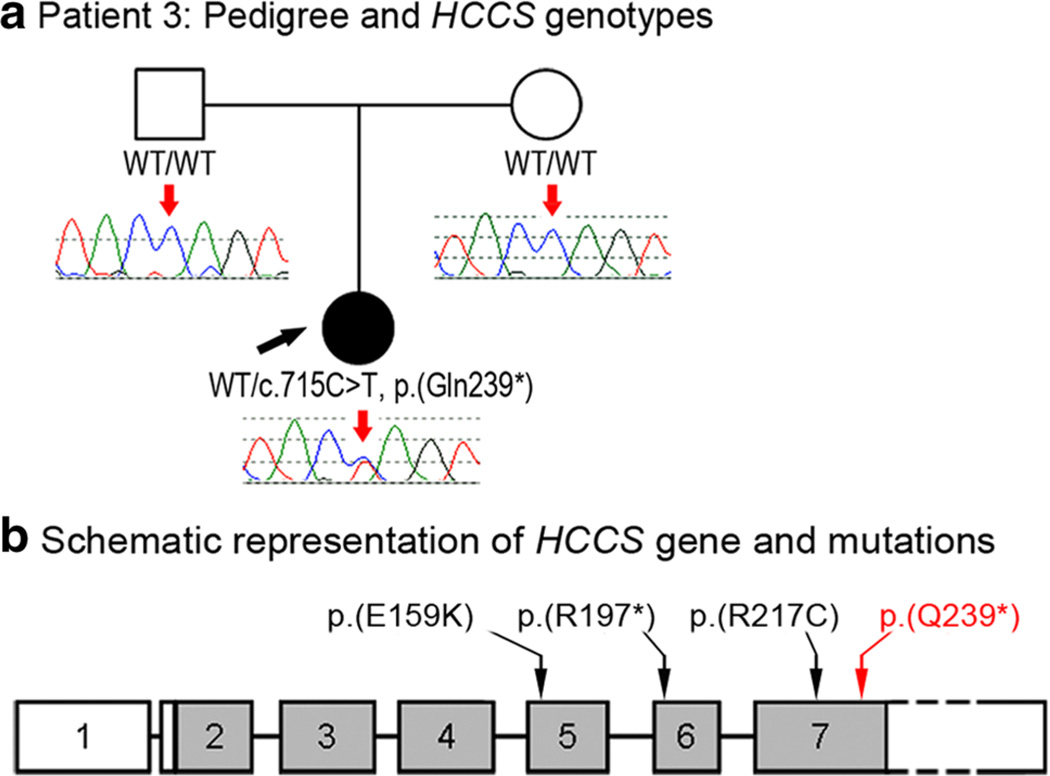
Patient 3, a 5-month-old Caucasian (mixed European) female, demonstrated bilateral Peters anomaly and mild microphthalmia (at 1 week of age, axial length measured 16.2 mm OD and 18.9 mm OS) with an absent/ small residual lens in the left eye. No systemic anomalies were present. Both parents are unaffected. Follow-up at 8 years of age confirmed normal development and lack of skin defects. Due to severe microphthalmia, bilateral sclera shells had been placed. A novel nonsense mutation, c.715C>T, p.(Gln239*), in the HCCS gene was identified. This mutation was not detected in 10,384 alleles (6,606 European Americans, 3,778 African Americans; mean coverage depth 173 reads) available in EVS and not reported in other control databases. This nucleotide is highly conserved with a GERP score of 6.05 and a PhyloP score of 2.57 and the mutation was absent in both unaffected parents (Fig. 3).
Fig. 3.
a Patient 3: pedigree and HCCS genotypes. DNA chromatograms of the proband’s and parental HCCS sequences are shown with the position of the c.715C>T, p.(Gln239*) mutation indicated with red arrows; the proband is designated with a black arrow. b Schematic representation of the human HCCS gene and mutations. HCCS mutations recorded in ClinVar are indicated with black arrows; the position of the p.(Q239*) mutation is shown with a red arrow
Mutations in HCCS have been associated with microphthalmia with linear skin defects (MLS) (Wimplinger et al. 2006). MLS is an X-linked dominant, male lethal phenotype characterized by microphthalmia or other ocular features, including corneal opacities in approximately 35 % and lesions of the skin that heal with age. MLS has also been associated with other variable systemic defects including short stature, congenital heart disease, central nervous system defects including developmental delay, genitourinary anomalies and hearing defects (Sharma et al. 2008). The HCCS gene has 7 exons with exons 2–7 encoding for the protein holocytochrome c synthase (HCCS). HCCS is a conserved heme-binding protein that has been shown to catalyze the attachment of a heme group to apocyctochrome c (Moore et al. 2011). Mutations that affect the C-terminal part of HCCS have been shown to abolish its function while amino acid changes in the N-terminus do not appear to be damaging, consistent with the locations of pathogenic mutations identified in humans (Moore et al. 2011). The mutation identified in patient 3 of this study is the most C-terminal change identified to date. The mutation occurs in the final exon of the gene and is not predicted to be subject to nonsense mediated decay. The translated protein product is predicted to lack 30 highly conserved C-terminal amino acids. Deletions of HCCS are common in MLS, however, missense and a nonsense variant have also been reported (Wimplinger et al. 2006). Involvement of HCCS in the Peters anomaly phenotype may relate to its role in promoting apoptosis (Indrieri et al. 2013) which is thought to play a role in the separation of the lens vesicle from the overlying surface ectoderm.
NDP mutation
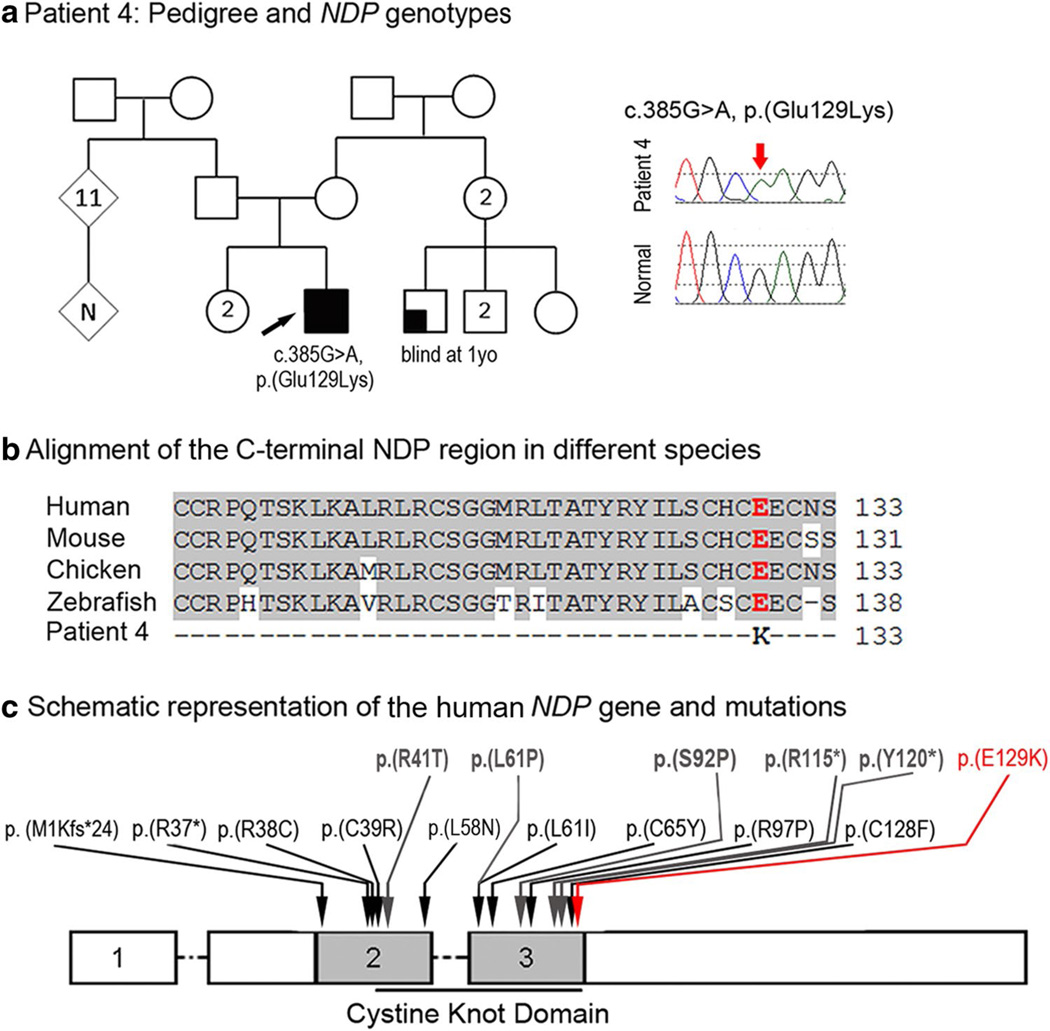
Patient 4, a 5-month-old Hispanic (El Salvador) male, had isolated bilateral Peters anomaly with no other ocular or systemic anomalies reported. The right eye showed central corneal opacity with iridocorneal strands, an irregular pupil edge, and superior corneal opacification. The central anterior chamber was mildly shallow suggesting anterior displacement of the lens. The left eye had more extensive corneal opacity extending from the inferior to superior limbus with iridocorneal adhesions and some central shallowing of the anterior chamber. There was no evidence of cataract or glaucoma in either eye. Corneal diameters and axial lengths were mildly small for age (10.5 mm OD, 9.75 OS and 19.52 mm OD, 18.86 OS). Cornea removed for transplantation showed central thinning due to absence of Descemet’s membrane and endothelial consistent with Peters anomaly as well as some mild stromal corneal edema. The retina in each eye was viewed to the midperiphery after corneal transplant and showed no vascular abnormalities or macular drag; the periphery has not been viewed. The patient also presented with nystagmus and alternating esotropia. Intravenous fluorescein angiography and audiology evaluation have not been performed. This patient was found to be hemizygous for a novel missense mutation, c.385G>A, p.(Glu129Lys), in the NDP gene (Fig. 4); this change has not been seen in 10,511 alleles (6,693 European American, 3,818 African American; mean depth coverage 26 reads) in EVS and was not reported in other control databases. The mutation is predicted to be damaging by 4 of 5 functional effect prediction programs. The nucleotide is well conserved with a GERP score of 5.96 and a PhyloP score of 2.52. Using the 46 species alignment in the UCSC genome browser, the glutamic acid at position 129 in NDP is 100 % conserved. Family history includes an unaffected sister and a male cousin (son of one of his mother’s sisters) who was blind from 1 year of age from unknown cause. Unfortunately, no family members were available for testing.
Fig. 4.
a Patient 4: pedigree and NDP genotypes. Pedigree is shown on the left, proband is indicated with a black arrow; DNA chromatograms of NDP c.385G>A, p.(Glu129Lys) mutant and normal alleles are shown on the right. b Alignment of the C-terminal NDP region in different species. Human (NP_000257), mouse (NP_035013), chicken (NP_001265015) and zebrafish (UniProt ID: E7F073) sequences were used for the alignment; glutamic acid residue at position 129 is shown in red font. c Schematic representation of the human NDP gene and mutations. Positions of some previously reported NDP mutations associated with Norrie disease or EVR2 are shown with black arrows; positions of mutations previously associated with unspecified corneal opacities are shown at the top and indicated with grey arrows; the p.(E129 K) mutation identified in patient 4 is noted with a red arrow
NDP has been identified as the cause of Norrie Disease, an X-linked recessive disorder characterized by retinal dysplasia with hearing loss in most patients and cognitive impairment and/or behavioral/psychiatric abnormalities in some patients (Smith et al. 2012). Mutations in NDP have also been identified in X-linked familial exudative vitreoretinopathy (EVR2), characterized by variable abnormal vascularization of the peripheral retina (Nikopoulos et al. 2010). NDP is divided into three exons, with the coding region contained in the second and third exon. The gene encodes for the protein, Norrin, a 133 amino acid protein that is a member of the canonical Wnt signaling pathway, which functions as a growth factor in an autocrine and/or paracrine fashion. Norrin has a cysteine-rich domain with a predicted tertiary structure of a cysteine knot motif similar to members of the TFGβ family. It has been shown to be important in the development of the retinal vasculature as well as the hyaloid vasculature which supplies nutrients to the developing lens (Ohlmann and Tamm 2012). A knockout of Ndp in the mouse showed overt vascular defects in the developing retina as well as defects in the structures of the inner ear; no defects in the cornea have been noted in Ndp knockout mice (Ohlmann and Tamm 2012).
At the same time, a careful examination of published records revealed that corneal opacities have been reported in suspected (Warburg 1966; Phillips et al. 1986) and NDP-positive (Rehm et al. 1997; Riveiro-Alvarez et al. 2005; Kondo et al. 2007; Liu et al. 2010, Pelcastre et al. 2010; Nikopoulos et al. 2010) cases of Norrie disease. The study by Pelcastre et al. (2010) examined four families with Norrie disease of Mexican descent and found corneal opacities in one proband and posterior polar cataract in several individuals. In the study by Rehm et al. (1997), partially or totally obliterated anterior chambers and corneal opacities were observed in 10 out of 15 affected males from one kindred with Norrie disease and a missense mutation in NDP. Iris atrophy, cataracts and posterior/anterior synechiae were detected in some cases when further examination of the anterior segment was possible. Therefore, the bilateral Peters anomaly observed in Patient 4 further supports the role of NDP in corneal development and also suggests a role in the early events of the lens–cornea separation. Interestingly, five out of six (83 %) NDP alleles associated with corneal phenotypes were found in the last exon of this gene with three of the mutations (50 %) being located within the 18 most C-terminal amino acids (Fig. 4), which may indicate a possible phenotype–genotype correlation. With respect to the other systemic features associated with NDP mutations, no clinical signs of hearing deficiency or the psychiatric signs of Norrie disease were reported in our patient, but these features usually manifest later in childhood (Smith et al. 2012). Alternatively, this mutation may result in a primarily ocular phenotype as was previously shown for one patient (Zhang et al. 2013).
FLNA mutation
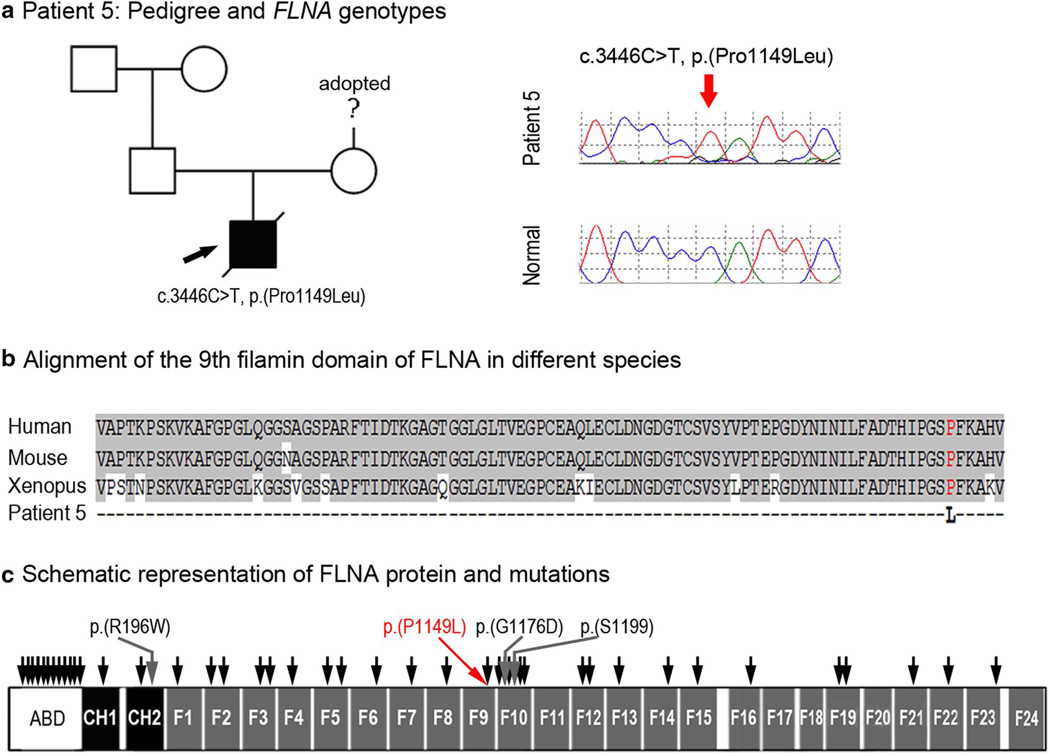
Patient 5, a Caucasian (mixed European) male, was diagnosed prenatally with Tetralogy of Fallot and multiple skeletal anomalies. He was delivered at 35–36 weeks gestational age following placental abruption with multiple congenital anomalies including Peters anomaly, dysmorphic facial features (low-set ears, downslanting palpebral fissures, ‘beak-like’ nose, micrognathia), cleft lip and palate, ambiguous genitalia, small dysplastic kidneys with hydronephrosis and renal failure, and a cyst-like structure between the bladder and rectum. The patient was also small for gestational age with microcephaly (height, weight, and head circumference all <3rd percentile). The patient died at 7 days of age. Postmortem ocular examination confirmed Peters anomaly with corneal opacity, absence of Bowman’s and Descemet’s membranes, iridocorneal adhesion, peripheral anterior synechia with angle closure and cataract. Hypoplasia of the thumbs and great toes with proximal placement and camptodactyly of the left 2nd finger was noted along with tracheal stenosis, abnormally hyperplastic erector pili in the skin, unusual pattern of fibrous septa in the spleen, single umbilical artery, and neuroglial heterotopia in the cerebellum. Complex cardiac anomalies were confirmed, including Tetralogy of Fallot along with absence of the ductus arteriosus, redundancy of pulmonary and aortic valves, and hypertrophy of both ventricles. Skeletal defects included malrotation of the lower extremities with genu recurvatum and posterior knees, femoral shortening, and wide anterior and posterior fontanelles. Skeletal radiographs demonstrated a decrease in cranial ossification, dysmorphic base of skull, dysmorphic middle and inner ears, micrognathia with decreased tooth buds, fusion of C2 and C3 posterior elements and C5 and C6 posterior elements, platyspondyly, squaring of lateral margins of iliac wings with medial spikes at the caudal portion of the iliac bones, dislocation of hips, broad medial ends of the clavicles with caudal hooking, bilateral dysplasia of the glenoid, dysplasia of scapulae, dislocation of proximal radius, posterior bowing of proximal ulna, absence of ossification within the distal phalanx of the thumb, shortening of femora and bones of upper arms, dislocation of hips and knees, and proximal placement of the great toes. Cytogenetic studies showed normal 46, XY karyotype with normal hybridization of subtelomeric probes; no other clinical genetic studies were performed.
This patient was found to have a hemizygous missense mutation in FLNA, c.3446C>T, p.(Pro1149Leu) (Fig. 5), which was predicted to be damaging by all five functional effect prediction programs, was not seen in 10,355 alleles in EVS (6,618 European Americans and 3,737 African Americans; mean depth coverage 73 reads), was not reported in other control databases, and has been previously reported in a female with frontometaphyseal dysplasia (FMD) (Robertson et al. 2006). The nucleotide is well conserved with a GERP score of 4.91 and a PhyloP score of 2.04. The patient is the first child born to these parents. Maternal family history is unknown due to adoption; paternal family history is unremarkable. Parental samples were not available for testing. The FLNA gene encompasses 48 exons and encodes Filamin A, an actin-binding protein that interacts with integrins, transmembrane receptors and other proteins to regulate remodeling of the actin cytoskeleton; the protein contains the actin-binding domain, two calponin homology domains and 24 filamin repeats. The remodeling of the cytoskeleton plays an important role in many processes including cell shape changes, cell migration and signaling (Adams et al. 2012). The c.3446C>T nucleotide transition is located in exon 22 and affects the 9th filamin repeat domain of the protein. This domain was shown to be involved in the interaction of FLNA with supervillin, which regulates cell spreading and cytokinesis (Smith et al. 2010).
Fig. 5.
a Patient 5: Pedigree and FLNA genotypes. Pedigree is shown on the left, proband is indicated with a black arrow; DNA chromatograms of FLNA c.3446C>T, p.(Pro1149Leu) mutant and normal alleles are shown on the right. b Alignment of the 9th filamin domain of FLNA in different species. Human (NP_001104026), mouse (NP_001277350), and Xenopus (UniProt ID:F6R7N1) sequences were used for the alignment; proline residue at position 1149 is shown in red font. c Schematic representation of the human FLNA protein and mutations. FLNA domains are indicated according to UniProtKB (P21333): ABD Actin-binding domain, CH1, CH2 calponin homology domains 1 and 2, F1–F24 filamin repeats 1 through 24. The approximate positions of the previously reported FLNA mutations associated with various non-ocular phenotypes are shown by black arrows; mutations associated with ocular phenotypes (please see text) are shown with grey arrows and specified; the p.(P1149L) mutation identified in this study is indicated with red arrow/font
FLNA mutations are linked to a broad range of phenotypes, including frontometaphyseal dysplasia (FMD), periventricular nodular heterotopia, otopalatodigital syndrome type 1 (OPD1), otopalatodigital syndrome type 2 (OPD2), and Melnick–Needles syndrome (MNS) (Robertson 2005). The majority of mutations are found in the actin-binding domain (at least 35 mutations) and the remaining mutations are somewhat evenly distributed throughout the protein with a small cluster (at least 9 pathogenic alleles) in the 10th filamin repeat. Overall, the patient’s phenotype is consistent with FMD, but ocular anomalies have not been previously linked with this condition. Careful examination of all published reports revealed several additional cases of eye anomalies in association with FLNA mutations. Corneal opacity has been reported in one patient with a missense mutation in FLNA [c.514C>G, p.(Leu172Val)] and features consistent with OPD2 (Murphy-Ryan et al. 2011). Congenital glaucoma and cataracts were reported in another patient with OPD2 and a missense mutation in FLNA [c.586C>T, p.(Arg196Trp)] (Kondoh et al. 2007). And finally, two unrelated patients with MNS and missense mutations in FLNA (c.3776_3777delinsAT, p.(Gly1176Asp); c.3596C>T, p.(Ser1199Leu) were reported with sclerocornea, exophthalmos, and congenital cataracts (Santos et al. 2010). Interestingly, including patient 5 reported in this study, three out of the four (75 %) FLNA mutations associated with ocular phenotypes occurred in the adjacent 9th and 10th filamin repeats (Fig. 5); further studies are needed to determine whether this finding reveals a potential genotype–phenotype correlation. Congenital glaucoma, cataracts, and/or corneal clouding have been reported in other patients with a clinical diagnosis of OPD2 or MNS without FLNA testing (Murphy-Ryan et al. 2011). To further support a role of FLNA in ocular development, some female mice with heterozygous Flna mutations were reported to show a ‘dilated pupil’ phenotype (Hart et al. 2006). Also, FLNA has been shown to interact with the protein encoded by FOXC1, which is known to play an important role in anterior segment ocular development (Berry et al. 2005).
SLC4A11 mutations
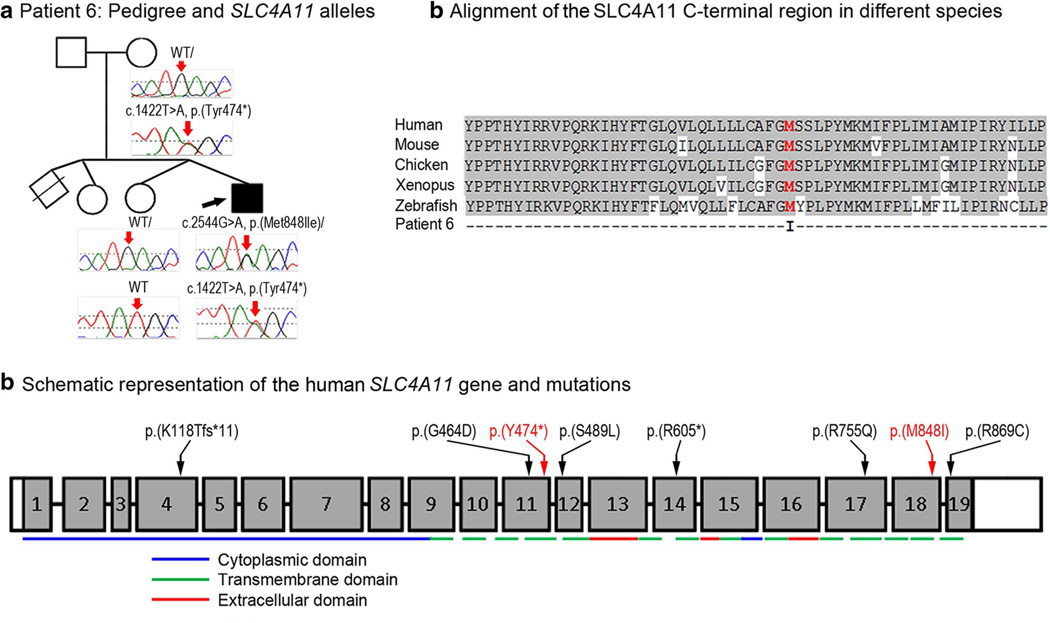
Patient 6, a 2.5-month-old Jamaican male, presented with bilateral Peters anomaly with microphthalmia. The corneas were small (8 and 9 mm) and axial length was 13.5 mm. There was fusion of the iris and rudimentary lens to the posterior cornea in the right eye and no pupil was present. In the left eye, filamentous strands extended from the iris and rudimentary lens to a central defect in the posterior cornea. Abnormal signals in the vitreous cavities by B-scan ultrasonography were possibly indicative of maldeveloped and malpositioned retinal tissue. The only systemic anomaly reported was umbilical hernia. Follow-up at 13 months of age showed normal growth (10–25 percentile), mild delay in gross-motor skills, and placement of eye prostheses bilaterally. The patient was found to be compound heterozygous for mutations in SLC4A11: a novel nonsense mutation, c.1422T>A, p.(Tyr474*), and a missense change, c.2544G>A, p.(Met848Ile) (Fig. 6). The missense mutation is predicted to be damaging by 3 of 5 programs and the nucleotide is conserved with a GERP score of 4.41 and a PhyloP score of 1.24. Analysis of samples from the unaffected mother and unaffected twin sibling identified the c.1422T>A, p.(Tyr474*) allele in the mother and wild-type sequence for both alleles in the unaffected twin. The unaffected father was not available for testing (Fig. 6). The c.1422T>A, p.(Tyr474*) mutation has not been detected in 13,006 alleles in EVS (8,600 European American, 4,406 African American; mean coverage depth 107 reads) or other control populations, while the c.2544G>A, p.(Met848Ile) allele has been reported in 121/13,006 (0.9 %) controls in EVS (1/8,600 European Americans and 120/4,406 African American; mean depth coverage 98 reads). Neither allele has been previously reported in association with ocular phenotypes (Kodaganur et al. 2013).
Fig. 6.
a Patient 6: Pedigree and SLC4A11 genotypes. DNA chromatograms of SLC4A11 alleles are shown with positions of c.1422T>A, p.(Tyr474*) and c.2544G>A, p.(Met848Ile) mutations indicated with red arrows; proband is designated with a black arrow. b Alignment of the SLC4A11 C-terminal region in different species. Human (NP_114423), mouse (XP_006499668), chicken (XP_004936340), Xenopus (XM_002936363) and zebrafsh (NM_001159828) sequences were used for the alignment; methionine residue at position 848 is shown in red font. c Schematic representation of the human SLC4A11 gene and mutations. The SLC4A11 exons are shown as numbered grey boxes; positions of cytoplasmic, transmembrane, and extracellular domains encoded by corresponding exonic sequences are indicated at the bottom with blue, green and red lines, correspondingly; positions of the previously described SLC4A11 mutations are indicated with black arrows/font while alleles reported in this study are shown with red arrows/font. SLC4A11 domains and mutations are shown as presented in Vilas et al. (2011) and Vithana et al. (2006)
SLC4A11 encodes for a transmembrane sodium/hydroxide transporter (Jalimarada et al. 2013). The gene is divided into 19 coding exons; the protein product is 891 amino acids in length. SLC4A11 appears to have a broad expression pattern in adult human tissues (Parker et al. 2001). The nonsense mutation in the 11th coding exon in patient 6 is predicted to result in nonsense mediated decay, but if the mutant transcript were translated it would result in a protein with C-terminal truncation, missing 417 amino acids. This deletion removes 11 transmembrane regions and the entire extracellular and C-terminal cytoplasmic domains (Vilas et al. 2011). The transmembrane domains and extracellular domain of this protein family are thought to be essential for their function (Vilas et al. 2011). The missense mutation changes a highly conserved methionine at position 848, which is the last amino acid of the 13th transmembrane domain, to isoleucine. Out of the 46 species aligned using the UCSC Genome Browser, only 4 orthologs have an amino acid other than methionine and none have isoleucine at that position.
Mutations in SLC4A11 have been linked to autosomal recessive congenital hereditary endothelial dystrophy (CHED2) (Vithana et al. 2006), autosomal recessive corneal dystrophy and perceptive deafness (CDPD, also known as Harboyan syndrome) (Desir et al. 2007), and autosomal dominant Fuchs endothelial corneal dystrophy (FECD4) (Vithana et al. 2008). Each of these phenotypes is associated with diffuse corneal clouding that often leads to vision loss; disrupted homeostasis of the inner layers of the cornea (Baratz et al. 2010), corneal edema associated with a reduced number of endothelial cells (Desir et al. 2007) and disruption of the endothelium and Descemet’s membrane (Vithana et al. 2006; Han et al. 2013; Vithana et al. 2008) have been reported as possible underlying mechanisms. Developmental delay has not been reported in other patients with SLC4A11 mutations; given the young age of the patient, the mild gross-motor delays may be due to the severe visual impairment. Several mouse models lacking functional Slc4a11 have been generated, each with a slightly variable phenotype. The model generated by Han et al. (2013) shows a progressive corneal dystrophy with an increasing thickness of the cornea and a progressive loss of endothelial cells with age. Their data suggest that Slc4a11 functions to maintain the homeostasis of the endothelium and that loss of this protein results in osmotic imbalance leading to death of corneal endothelial cells.
Conclusion
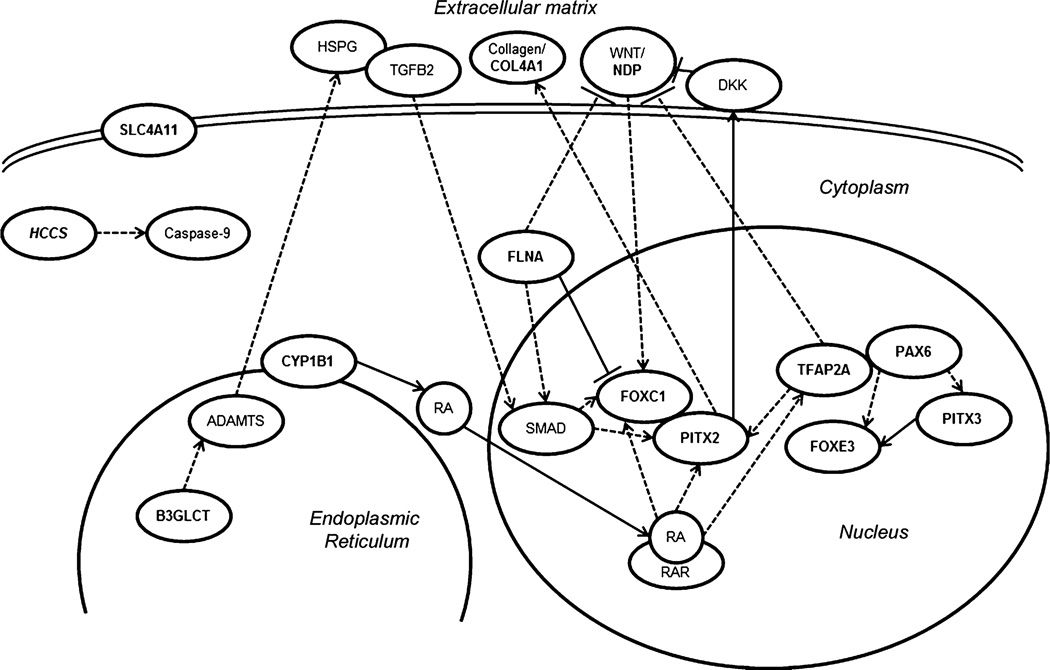
Peters anomaly is a highly heterogeneous disorder that has been previously associated with mutations in 8 different genes, each explaining only a small portion of the overall spectrum. We present evidence for the possible involvement of five additional factors, TFAP2A, HCCS, NDP, FLNA and SLC4A11, in the etiology of this condition. The processes involved in embryonic lens–cornea separation as well as subsequent differentiation of these ocular tissues are currently poorly understood. Identification of genetic factors controlling these early events in humans through studies of relevant ocular disorders provides valuable insight into mechanisms of anterior segment development. Analysis of the genetic pathways linked with the 13 factors associated with Peters anomaly revealed several possible connections that may begin to elucidate the processes which are affected in this disorder (Fig. 7). Additional genetic studies are needed to better define the contribution of these and other factors to the Peters anomaly spectrum. Our data suggest that TFAP2A and FLNA screening should be considered in patients with Peters anomaly and systemic malformations while HCCS, NDP and SLC4A11 should be considered for testing in cases of isolated Peters anomaly.
Fig. 7.
Schematic representation of reported interactions between factors associated with Peters anomaly. The genes implicated in PA based on previous reports as well as this study are marked in bold. PITX2 and FOXC1 have been shown to physically bind and modulate the activity of each other (Acharya et al. 2011) as well as to be regulated by TGFβ (Silla et al. 2014; Iwao et al. 2009), WNT and retinoic acid signaling pathways (Gage and Zacharias 2009; Kumar and Duester 2010; Hatou et al. 2013); in connection with the WNT pathway, PITX2 has been reported to physically bind the DKK2 promoter (Gage and Zacharias 2009). Also, PITX2 has been shown to regulate the deposition of collagens by regulating the procollagen lysyl hydroxylase gene (Hjalt et al. 2001). B3GLCT is a glucosyltransferase which catalyzes the addition of glucose to O-linked fucose via a β-1,3 linkage onto trombospondin type 1 repeats which have been found in the ADAMTS family of proteins (Ricketts et al. 2007; Wang et al. 2009); some ADAMTS proteins have been shown to cleave heparan sulfate proteoglycans (HSPGs) (Barbouri et al. 2014; Kuno and Matsuhima 1998) and thus potentially affect TGFβ signaling (Lin 2004; Iwao et al. 2009). NDP represents a WNT ligand (Ohlmann and Tamm 2012). TFAP2A has been shown to be responsive to retinoic acid, to inhibit the WNT pathway (Li and Dashwood 2004) and be involved in PITX2 regulation (Bamforth et al. 2004). PAX6 deficiency has been shown to negatively affect FOXE3 (Blixt et al. 2007) and PITX3 expression (Chauhan et al. 2002); PITX3 has been reported to directly regulate FOXE3 expression (Shi et al. 2006; Ahmad et al. 2013). PAX6 and TFAP2A have been demonstrated to physically interact and promote transcription of some corneal genes (Sivak et al. 2004). CYP1B1 participates in the synthesis of retinoic acid (Chambers et al. 2007). FLNA appears to negatively regulate FOXC1 by physical binding and sequestering it to heterochromatin and is important for SMAD translocation into the nucleus (Berry et al. 2005; Huang et al. 2009). FLNA may also be important in maintaining appropriate levels of WNT signaling (Adams et al. 2012)
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the patients and their families for their participation in research studies. This work was supported by the National Institutes of Health awards R01EY015518 and funds provided by the Children’s Hospital of Wisconsin (EVS) and 1UL1RR031973 from the Clinical and Translational Science Award (CTSA) program.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-014-1481-x) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no relevant financial conflict of interest.
Ethical standards All experiments described comply with the current laws of the United States of America and the Declaration of Helsinki.
Contributor Information
Eric Weh, Department of Pediatrics and Children’s Research Institute, Medical College of Wisconsin, Milwaukee, WI 53226, USA; Cell Biology, Neurobiology and Anatomy Department, Medical, College of Wisconsin, Milwaukee, WI 53226, USA.
Linda M. Reis, Department of Pediatrics and Children’s Research Institute, Medical College of Wisconsin, Milwaukee, WI 53226, USA
Hannah C. Happ, Department of Pediatrics and Children’s Research Institute, Medical College of Wisconsin, Milwaukee, WI 53226, USA
Alex V. Levin, Pediatric Ophthalmology and Ocular Genetics, Wills Eye, Hospital, Philadelphia, PA 19107, USA
Patricia G. Wheeler, Nemours Children’s Clinic, Orlando, FL 32806, USA
Karen L. David, Division of Genetics, Department of Medicine, New York, Methodist Hospital, Brooklyn, NY 11215, USA
Erin Carney, Division of Genetics, Department of Medicine, New York, Methodist Hospital, Brooklyn, NY 11215, USA.
Brad Angle, Department of Pediatrics, Northwestern University Feinberg School of Medicine and Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL 60611, USA.
Natalie Hauser, Center for Human Genetics, UT Southwestern Medical Center, Dallas, TX 75390, USA.
Elena V. Semina, Department of Pediatrics and Children’s Research Institute, Medical College of Wisconsin, Milwaukee, WI 53226, USA esemina@mcw.edu Cell Biology, Neurobiology and Anatomy Department, Medical, College of Wisconsin, Milwaukee, WI 53226, USA.
References
- Acharya M, Huang L, Fleisch VC, Allison WT, Walter MA. A complex regulatory network of transcription factors critical for ocular development and disease. Hum Mol Genet. 2011;20:1610–1624. doi: 10.1093/hmg/ddr038. [DOI] [PubMed] [Google Scholar]
- Adams M, Simms RJ, Abdelhamed Z, Dawe HR, Szymanska K, Logan CV, Wheway G, Pitt E, Gull K, Knowles MA, Blair E, Cross SH, Sayer JA, Johnson CA. A meckelin-filamin A interaction mediates ciliogenesis. Hum Mol Genet. 2012;21:1272–1286. doi: 10.1093/hmg/ddr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad N, Aslam M, Muenster D, Horsch M, Khan MA, Carlsson P, Beckers J, Graw J. Pitx3 directly regulates Foxe3 during early lens development. Int J Dev Biol. 2013;57:741–751. doi: 10.1387/ijdb.130193jg. [DOI] [PubMed] [Google Scholar]
- Bamforth SD, Bragança J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA, Anderson RH, Bhattacharya S. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- Baratz KH, Tosakulwong N, Ryu E, Brown WL, Branham K, Chen W, Tran KD, Schmid-Kubista KE, Heckenlively JR, Swaroop A, Abecasis G, Bailey KR, Edwards AO. E2-2 protein and Fuchs’s corneal dystrophy. N Engl J Med. 2010;363:1016–1024. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- Barbouri D, Afratis N, Gialeli C, Vynios DH, Theocharis AD, Karamanos NK. Syndecans as modulators and potential pharmacological targets in cancer progression. Front Oncol. 2014;4:4. doi: 10.3389/fonc.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Williams T, Zacharias AL, Gage PJ, Fuhrmann S, West-Mays JA. AP-2alpha knockout mice exhibit optic cup patterning defects and failure of optic stalk morphogenesis. Hum Mol Genet. 2010;19:1791–1804. doi: 10.1093/hmg/ddq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulmann DC, Ohlmann A, Flügel-Koch C, Goswami S, Cvekl A, Tamm ER. Pax6 heterozygous eyes show defects in chamber angle differentiation that are associated with a wide spectrum of other anterior eye segment abnormalities. Mech Dev. 2002;118:3–17. doi: 10.1016/s0925-4773(02)00260-5. [DOI] [PubMed] [Google Scholar]
- Berry FB, O’Neill MA, Coca-Prados M, Walter MA. FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A-mediated manner. Mol Cell Biol. 2005;25:1415–1424. doi: 10.1128/MCB.25.4.1415-1424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Ferri S, Whittaker B, Liu M, Lazzaro DR. Peters anomaly: review of the literature. Cornea. 2011;30:939–944. doi: 10.1097/ICO.0b013e31820156a9. [DOI] [PubMed] [Google Scholar]
- Blixt A, Landgren H, Johansson BR, Carlsson P. Foxe3 is required for morphogenesis and differentiation of the anterior segment of the eye and is sensitive to Pax6 gene dosage. Dev Biol. 2007;302:218–229. doi: 10.1016/j.ydbio.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Chambers D, Wilson L, Maden M, Lumsden A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development. 2007;134:1369–1383. doi: 10.1242/dev.02815. [DOI] [PubMed] [Google Scholar]
- Chao LY, Mishra R, Strong LC, Saunders GF. Missense mutations in the DNA-binding region and termination codon in PAX6. Hum Mutat. 2003;21:138–145. doi: 10.1002/humu.10163. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Zhang W, Cveklova K, Kantorow M, Cvekl A. Identification of differentially expressed genes in mouse Pax6 heterozygous lenses. Invest Ophthalmol Vis Sci. 2002;43:1884–1890. [PMC free article] [PubMed] [Google Scholar]
- Deml B, Reis LM, Maheshwari M, Griffis C, Bick D, Semina EV. Whole exome analysis identifies dominant COL4A1 mutations in patients with complex ocular phenotypes involving microphthalmia. Clin Genet (Epub ahead of print) 2014 doi: 10.1111/cge.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desir J, Moya G, Reish O, Van Regemorter N, Deconinck H, David KL, Meire FM, Abramowicz MJ. Borate transporter SLC4A11 mutations cause both Harboyan syndrome and non-syndromic corneal endothelial dystrophy. J Med Genet. 2007;44:322–326. doi: 10.1136/jmg.2006.046904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Zacharias AL. Signaling “cross-talk” is integrated by transcription factors in the development of the anterior segment in the eye. Dev Dyn. 2009;238:2149–2162. doi: 10.1002/dvdy.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestri G, Osborne RJ, Wyatt AW, Gerrelli D, Gribble S, Stewart H, Fryer A, Bunyan DJ, Prescott K, Collin JR, Fitzgerald T, Robinson D, Carter NP, Wilson SW, Ragge NK. Reduced TFAP2A function causes variable optic fissure closure and retinal defects and sensitizes eye development to mutations in other morphogenetic regulators. Hum Genet. 2009;126:791–803. doi: 10.1007/s00439-009-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SB, Ang HP, Poh R, Chaurasia SS, Peh G, Liu J, Tan DT, Vithana EN, Mehta JS. Mice with a targeted disruption of Slc4a11 model the progressive corneal changes of congenital hereditary endothelial dystrophy. Invest Ophthalmol Vis Sci. 2013;54:6179–6189. doi: 10.1167/iovs.13-12089. [DOI] [PubMed] [Google Scholar]
- Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters’ anomaly. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- Hatou S, Yoshida S, Higa K, Miyashita H, Inagaki E, Okano H, Tsubota K, Shimmura S. Functional corneal endothelium derived from corneal stroma stem cells of neural crest origin by retinoic acid and Wnt/β-catenin signaling. Stem Cells Dev. 2013;22:828–839. doi: 10.1089/scd.2012.0286. [DOI] [PubMed] [Google Scholar]
- Hjalt TA, Amendt BA, Murray JC. PITX2 regulates procollagen lysyl hydroxylase (PLOD) gene expression: implications for the pathology of Rieger syndrome. J Cell Biol. 2001;152:545–552. doi: 10.1083/jcb.152.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkanen RA, Nishimura DY, Swiderski RE, Bennett SR, Hong S, Kwon YH, Stone EM, Sheffield VC, Alward WL. A family with Axenfeld-Rieger syndrome and Peters anomaly caused by a point mutation (Phe112Ser) in the FOXC1 gene. Am J Ophthalmol. 2003;135:368–375. doi: 10.1016/s0002-9394(02)02061-5. [DOI] [PubMed] [Google Scholar]
- Huang J, Dattilo LK, Rajagopal R, Liu Y, Kaartinen V, Mishina Y, Deng CX, Umans L, Zwijsen A, Roberts AB, Beebe DC. FGF-regulated BMP signaling is required for eyelid closure and to specify conjunctival epithelial cell fate. Development. 2009;136:1741–1750. doi: 10.1242/dev.034082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrieri A, Conte I, Chesi G, Romano A, Quartararo J, Tatè R, Ghezzi D, Zeviani M, Goffrini P, Ferrero I, Bovolenta P, Franco B. The impairment of HCCS leads to MLS syndrome by activating a non-canonical cell death pathway in the brain and eyes. EMBO Mol Med. 2013;5:280–293. doi: 10.1002/emmm.201201739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito YA, Walter MA. Genomics and anterior segment dysgenesis: a review. Clin Exp Ophthalmol. 2014;42:13–24. doi: 10.1111/ceo.12152. [DOI] [PubMed] [Google Scholar]
- Iwao K, Inatani M, Matsumoto Y, Ogata-Iwao M, Takihara Y, Irie F, Yamaguchi Y, Okinami S, Tanihara H. Heparan sulfate deficiency leads to Peters anomaly in mice by disturbing neural crest TGF-beta2 signaling. J Clin Invest. 2009;119:1997–2008. doi: 10.1172/JCI38519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalimarada SS, Ogando DG, Vithana EN, Bonanno JA. Ion transport function of SLC4A11 in corneal endothelium. Invest Ophthalmol Vis Sci. 2013;54:4330–4340. doi: 10.1167/iovs.13-11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaganur SG, Kapoor S, Veerappa AM, Tontanahal SJ, Sarda A, Yathish S, Prakash DR, Kumar A. Mutation analysis of the SLC4A11 gene in Indian families with congenital hereditary endothelial dystrophy 2 and a review of the literature. Mol Vis. 2013;19:1694–1706. [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Qin M, Kusaka S, Tahira T, Hasebe H, Hayashi H, Uchio E, Hayashi K. Novel mutations in Norrie disease gene in Japanese patients with Norrie disease and familial exudative vitreoretinopathy. Invest Ophthal Vis Sci. 2007;48:1276–1282. doi: 10.1167/iovs.06-1042. [DOI] [PubMed] [Google Scholar]
- Kondoh T, Okamoto N, Norimatsu N, Uetani M, Nishimura G, Moriuchi H. A Japanese case of oto-palato-digital syndrome type II: an apparent lack of phenotype-genotype correlation. J Hum Genet. 2007;52:370–373. doi: 10.1007/s10038-007-0108-7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Duester G. Retinoic acid signaling in perioptic mesenchyme represses Wnt signaling via induction of Pitx2 and Dkk2. Dev Biol. 2010;340:67–74. doi: 10.1016/j.ydbio.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno K, Matsushima K. ADAMTS-1 protein anchors at the extracellular matrix through the thrombospondin type I motifs and its spacing region. J Biol Chem. 1998;273:13912–13917. doi: 10.1074/jbc.273.22.13912. [DOI] [PubMed] [Google Scholar]
- Lesnik Oberstein SA, Kriek M, White SJ, Kalf ME, Szuhai K, den Dunnen JT, Breuning MH, Hennekam RC. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet. 2006;79:562–566. doi: 10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Dashwood RH. Activator protein 2alpha associates with adenomatous polyposis coli/beta-catenin and Inhibits beta-catenin/T-cell factor transcriptional activity in colorectal cancer cells. J Biol Chem. 2004;279:45669–45675. doi: 10.1074/jbc.M405025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Liu D, Hu Z, Peng Y, Yu C, Liu Y, Mo X, Li X, Lu L, Xu X, Su W, Pan Q, Xia K. A novel nonsense mutation in the NDP gene in a Chinese family with Norrie disease. Mol Vis. 2010;16:2653–2658. [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillette de Buy Wenniger-Prick LJ, Hennekam RC. The Peters’ plus syndrome: a review. Ann Genet. 2002;45:97–103. doi: 10.1016/s0003-3995(02)01120-6. [DOI] [PubMed] [Google Scholar]
- Matsubara A, Ozeki H, Matsunaga N, Nozaki M, Ashikari M, Shirai S, Ogura Y. Histopathological examination of two cases of anterior staphyloma associated with Peters’ anomaly and persistent hyperplastic primary vitreous. Br J Ophthalmol. 2001;85:1421–1425. doi: 10.1136/bjo.85.12.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky JM, Maher TM, Zhao G, Wang Z, Mulliken JB, Chitayat D, Clemens M, Stalker HJ, Bauer M, Burch M, Chénier S, Cunningham ML, Drack AV, Janssens S, Karlea A, Klatt R, Kini U, Klein O, Lachmeijer AM, Megarbane A, Mendelsohn NJ, Meschino WS, Mortier GR, Parkash S, Ray CR, Roberts A, Roberts A, Reardon W, Schnur RE, Smith R, Splitt M, Tezcan K, Whiteford ML, Wong DA, Zori R, Lin AE. Genotype–phenotype analysis of the branchio-oculo-facial syndrome. Am J Med Genet A. 2011;155A:22–32. doi: 10.1002/ajmg.a.33783. [DOI] [PubMed] [Google Scholar]
- Moore RL, Stevens JM, Ferguson SJ. Mitochondrial cytochrome c synthase: CP motifs are not necessary for heme attachment to apocytochrome c. FEBS Lett. 2011;585:3415–3419. doi: 10.1016/j.febslet.2011.08.042. [DOI] [PubMed] [Google Scholar]
- Murphy-Ryan M, Babovic-Vuksanovic D, Lindor N. Bifid tongue, corneal clouding, and Dandy–Walker malformation in a male infant with otopalatodigital syndrome type 2. Am J Med Genet A. 2011;155A:855–859. doi: 10.1002/ajmg.a.33901. [DOI] [PubMed] [Google Scholar]
- Nikopoulos K, Venselaar H, Collin RW, Riveiro-Alvarez R, Boonstra FN, Hooymans JM, Mukhopadhyay A, Shears D, van Bers M, de Wijs IJ, van Essen AJ, Sijmons RH, Tilanus MA, van Nouhuys CE, Ayuso C, Hoefsloot LH, Cremers FP. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat. 2010;31:656–666. doi: 10.1002/humu.21250. [DOI] [PubMed] [Google Scholar]
- Ohlmann A, Tamm ER. Norrin: molecular and functional properties of an angiogenic and neuroprotective growth factor. Prog Retin Eye Res. 2012;31:243–257. doi: 10.1016/j.preteyeres.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Ormestad M, Blixt A, Churchill A, Martinsson T, Enerbäck S, Carlsson P. Foxe3 haploinsufficiency in mice: a model for Peters’ anomaly. Invest Ophthalmol Vis Sci. 2002;43:1350–1357. [PubMed] [Google Scholar]
- Ozeki H, Shirai S, Nozaki M, Sakurai E, Mizuno S, Ashikari M, Matsunaga N, Ogura Y. Ocular and systemic features of Peters’ anomaly. Graefes Arch Clin Exp Ophthalmol. 2000;238:833–839. doi: 10.1007/s004170000177. [DOI] [PubMed] [Google Scholar]
- Parker MD, Ourmozdi EP, Tanner MJ. Human BTR1, a new bicarbonate transporter superfamily member and human AE4 from kidney. Biochem Biophys Res Commun. 2001;282:1103–1109. doi: 10.1006/bbrc.2001.4692. [DOI] [PubMed] [Google Scholar]
- Pauws E, Peskett E, Boissin C, Hoshino A, Mengrelis K, Carta E, Abruzzo MA, Lees M, Moore GE, Erickson RP, Stanier P. X-linked CHARGE-like Abruzzo–Erickson syndrome and classic cleft palate with ankyloglossia result from TBX22 splicing mutations. Clin Genet. 2013;83:352–358. doi: 10.1111/j.1399-0004.2012.01930.x. [DOI] [PubMed] [Google Scholar]
- Pelcastre EL, Villanueva-Mendoza C, Zenteno JC. Novel and recurrent NDP gene mutations in familial cases of Norrie disease and X-linked exudative vitreoretinopathy. Clin Exp Ophthalmol. 2010;38:367–374. doi: 10.1111/j.1442-9071.2010.02245.x. [DOI] [PubMed] [Google Scholar]
- Phillips CI, Newton M, Duvall J, Holloway S, Levy AM. Probably Norrie’s disease due to mutation. Two sporadic sibships of two males each, a necropsy of one case, and, given Norrie’s disease, a calculation of the gene mutation frequency. Br J Ophthalmol. 1986;70:305–313. doi: 10.1136/bjo.70.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoriero GF, Deschamps P, Ashery-Padan R, Wong R, Yang Y, Zavadil J, Cvekl A, Sullivan S, Williams T, West-Mays JA. Cell autonomous roles for AP-2alpha in lens vesicle separation and maintenance of the lens epithelial cell phenotype. Dev Dyn. 2008;237:602–617. doi: 10.1002/dvdy.21445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokudin I, Simons C, Grigg JR, Storen R, Kumar V, Phua ZY, Smith J, Flaherty M, Davila S, Jamieson RV. Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur J Hum Genet. 2014;22(7):907–915. doi: 10.1038/ejhg.2013.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Gutiérrez-Espeleta GA, Garcia R, Jiménez G, Khetarpal U, Priest JM, Sims KB, Keats BJ, Morton CC. Norrie disease gene mutation in a large Costa Rican kindred with a novel phenotype including venous insufficiency. Hum Mutat. 1997;9:402–408. doi: 10.1002/(SICI)1098-1004(1997)9:5<402::AID-HUMU4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Reiber J, Sznajer Y, Posteguillo EG, Müller D, Lyonnet S, Baumann C, Just W. Additional clinical and molecular analyses of TFAP2A in patients with the branchio-oculo-facial syndrome. Am J Med Genet A. 2010;152A:994–999. doi: 10.1002/ajmg.a.33331. [DOI] [PubMed] [Google Scholar]
- Reis LM, Semina EV. Genetics of anterior segment dysgenesis disorders. Curr Opin Ophthalmol. 2011;22:314–324. doi: 10.1097/ICU.0b013e328349412b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis LM, Tyler RC, Abdul-Rahman O, Trapane P, Wallerstein R, Broome D, Hoffman J, Khan A, Paradiso C, Ron N, Bergner A, Semina EV. Mutation analysis of B3GALTL in Peters Plus syndrome. Am J Med Genet A. 2008;146A:2603–2610. doi: 10.1002/ajmg.a.32498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis LM, Tyler RC, Volkmann Kloss BA, Schilter KF, Levin AV, Lowry RB, Zwijnenburg PJ, Stroh E, Broeckel U, Murray JC, Semina EV. PITX2 and FOXC1 spectrum of mutations in ocular syndromes. Eur J Hum Genet. 2012;20:1224–1233. doi: 10.1038/ejhg.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts LM, Dlugosz M, Luther KB, Haltiwanger RS, Majerus EM. O-fucosylation is required for ADAMTS13 secretion. J Biol Chem. 2007;282:17014–17023. doi: 10.1074/jbc.M700317200. [DOI] [PubMed] [Google Scholar]
- Riveiro-Alvarez R, Trujillo-Tiebas MJ, Gimenez-Pardo A, Garcia-Hoyos M, Cantalapiedra D, Lorda-Sanchez I, Rodriguez de Alba M, Ramos C, Ayuso C. Genotype-phenotype variations in five Spanish families with Norrie disease or X-linked FEVR. Mol Vis. 2005;11:705–712. [PubMed] [Google Scholar]
- Robertson SP. Filamin A: phenotypic diversity. Curr Opin Genet Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Robertson SP, Jenkins ZA, Morgan T, Adès L, Aftimos S, Boute O, Fiskerstrand T, Garcia-Miñaur S, Grix A, Green A, Der Kaloustian V, Lewkonia R, McInnes B, van Haelst MM, Mancini G, Illés T, Mortier G, Newbury-Ecob R, Nicholson L, Scott CI, Ochman K, Brozek I, Shears DJ, Superti-Furga A, Suri M, Whiteford M, Wilkie AO, Krakow D. Frontometaphyseal dysplasia: mutations in FLNA and phenotypic diversity. Am J Med Genet A. 2006;140:1726–1736. doi: 10.1002/ajmg.a.31322. [DOI] [PubMed] [Google Scholar]
- Samuels ME, Majewski J, Alirezaie N, Fernandez I, Casals F, Patey N, Decaluwe H, Gosselin I, Haddad E, Hodgkinson A, Idaghdour Y, Marchand V, Michaud JL, Rodrigue MA, Desjardins S, Dubois S, Le Deist F, Awadalla P, Raymond V, Maranda B. Exome sequencing identifies mutations in the gene TTC7A in French–Canadian cases with hereditary multiple intestinal atresia. J Med Genet. 2013;50:324–329. doi: 10.1136/jmedgenet-2012-101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos HH, Garcia PP, Pereira L, Leão LL, Aguiar RA, Lana AM, Carvalho MR, Aguiar MJ. Mutational analysis of two boys with the severe perinatally lethal Melnick–Needles syndrome. Am J Med Genet A. 2010;152A:726–731. doi: 10.1002/ajmg.a.33260. [DOI] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–170. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- Sharma VM, Ruiz de Luzuriaga AM, Waggoner D, Greenwald M, Stein SL. Microphthalmia with linear skin defects: a case report and review. Pediatr Dermatol. 2008;25:548–552. doi: 10.1111/j.1525-1470.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- Shi X, Luo Y, Howley S, Dzialo A, Foley S, Hyde DR, Vihtelic TS. Zebrafsh foxe3: roles in ocular lens morphogenesis through interaction with pitx3. Mech Dev. 2006;123:761–782. doi: 10.1016/j.mod.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Silla ZT, Naidoo J, Kidson SH, Sommer P. Signals from the lens and Foxc1 regulate the expression of key genes during the onset of corneal endothelial development. Exp Cell Res. 2014 doi: 10.1016/j.yexcr.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Sivak JM, West-Mays JA, Yee A, Williams T, Fini ME. Transcription factors Pax6 and AP-2alpha interact to coordinate corneal epithelial repair by controlling expression of matrix metalloproteinase Gelatinase B. Mol Cell Biol. 2004;24:245–257. doi: 10.1128/MCB.24.1.245-257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Fang Z, Luna EJ. Novel interactors and a role for supervillin in early cytokinesis. Cytoskeleton (Hoboken) 2010;67:346–364. doi: 10.1002/cm.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Mullen TE, Graham D, Sims KB, Rehm HL. Norrie disease: extraocular clinical manifestations in 56 patients. Am J Med Genet A. 2012;158A:1909–1917. doi: 10.1002/ajmg.a.35469. [DOI] [PubMed] [Google Scholar]
- Tekin M, Sirmaci A, Yüksel-Konuk B, Fitoz S, Sennaroğlu L. A complex TFAP2A allele is associated with branchio-oculo-facial syndrome and inner ear malformation in a deaf child. Am J Med Genet A. 2009;149A:427–430. doi: 10.1002/ajmg.a.32619. [DOI] [PubMed] [Google Scholar]
- Vilas GL, Morgan PE, Loganathan SK, Quon A, Casey JR. A biochemical framework for SLC4A11, the plasma membrane protein defective in corneal dystrophies. Biochemistry. 2011;50:2157–2169. doi: 10.1021/bi101887z. [DOI] [PubMed] [Google Scholar]
- Vincent A, Billingsley G, Priston M, Glaser T, Oliver E, Walter M, Ritch R, Levin A, Heon E. Further support of the role of CYP1B1 in patients with Peters anomaly. Mol Vis. 2006;12:506–510. [PubMed] [Google Scholar]
- Vithana EN, Morgan P, Sundaresan P, Ebenezer ND, Tan DT, Mohamed MD, Anand S, Khine KO, Venkataraman D, Yong VH, Salto-Tellez M, Venkatraman A, Guo K, Hemadevi B, Srinivasan M, Prajna V, Khine M, Casey JR, Inglehearn CF, Aung T. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2) Nat Genet. 2006;38:755–757. doi: 10.1038/ng1824. [DOI] [PubMed] [Google Scholar]
- Vithana EN, Morgan PE, Ramprasad V, Tan DT, Yong VH, Venkataraman D, Venkatraman A, Yam GH, Nagasamy S, Law RW, Rajagopal R, Pang CP, Kumaramanickevel G, Casey JR, Aung T. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17:656–666. doi: 10.1093/hmg/ddm337. [DOI] [PubMed] [Google Scholar]
- Wang LW, Leonhard-Melief C, Haltiwanger RS, Apte SS. Post-translational modification of thrombospondin type-1 repeats in ADAMTS-like 1/punctin-1 by C-mannosylation of tryptophan. J Biol Chem. 2009;284:30004–30015. doi: 10.1074/jbc.M109.038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg M. Norrie’s disease. A congenital progressive oculoa-coustico-cerebral degeneration. Acta Ophth Suppl. 1966;89:1–47. [PubMed] [Google Scholar]
- Weh E, Reis LM, Tyler RC, Bick D, Rhead WJ, Wallace S, McGregor TL, Dills SK, Chao MC, Murray JC, Semina EV. Novel B3GALTL mutations in classic Peters plus syndrome and lack of mutations in a large cohort of patients with similar phenotypes. Clin Genet. 2014;86:142–148. doi: 10.1111/cge.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimplinger I, Morleo M, Rosenberger G, Iaconis D, Orth U, Meinecke P, Lerer I, Ballabio A, Gal A, Franco B, Kutsche K. Mutations of the mitochondrial holocytochrome c-type synthase in X-linked dominant microphthalmia with linear skin defects syndrome. Am J Hum Genet. 2006;79:878–889. doi: 10.1086/508474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zhang X, Tong Y, Xu W, Dong B, Yang H, Xu L, Li Y. Two novel mutations of the PAX6 gene causing different phenotype in a cohort of Chinese patients. Eye (London) 2011;25:1581–1589. doi: 10.1038/eye.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Jiang WY, Chen LM, Chen SQ. A novel Norrie disease pseudoglioma gene mutation, c.-1_2delAAT, responsible for Norrie disease in a Chinese family. Int J Ophthalmol. 2013;6:739–743. doi: 10.3980/j.issn.2222-3959.2013.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.