Abstract
Ubiquitin C-terminal hydrolase L1 (UCH-L1) is a key neuronal deubiquitinating enzyme which is mutated in Parkinson disease (PD) and in childhood-onset neurodegenerative disorder with optic atrophy. Furthermore, reduced UCH-L1 protein levels are associated with a number of neurodegenerative diseases, whereas up-regulation of UCH-L1 protein expression is found in multiple types of cancer. However, very little is known about how UCH-L1 protein level is regulated in cells. Here, we report that UCH-L1 is a novel interactor and substrate of PD-linked E3 ubiquitin-protein ligase parkin. We find that parkin mediates K63-linked polyubiquitination of UCH-L1 in cooperation with the Ubc13/Uev1a E2 ubiquitin-conjugating enzyme complex and promotes UCH-L1 degradation by the autophagy-lysosome pathway. Targeted disruption of parkin gene expression in mice causes a significant decrease in UCH-L1 ubiquitination with a concomitant increase in UCH-L1 protein level in brain, supporting an in vivo role of parkin in regulating UCH-L1 ubiquitination and degradation. Our findings reveal a direct link between parkin-mediated ubiquitin signaling and UCH-L1 regulation, and they have important implications for understanding the roles of these two proteins in health and disease.
Keywords: Parkinsonism, Ubiquitin, Lysine 63-linked polyubiquitination, Autophagy, Lysosome
Introduction
Ubiquitin-dependent signaling is required for myriad cellular functions, and disruptions in this system are associated with human disease, including neurodegenerative disorders and cancer [1–4]. The ubiquitin system participates in diverse cellular signaling events through the addition or removal of ubiquitin moieties to target proteins. Protein ubiquitination involves the coordinated and sequential action of three proteins—E1-activating, E2-conjugating, and E3-ligating enzymes that together mediate isopeptide bond formation between the ubiquitin C-terminal glycine and substrate lysine residue, after which additional ubiquitin moieties can be joined at one of the ubiquitin’s seven internal lysine residues to form a polyubiquitin chain [5–7]. The nature of the ubiquitin linkage dictates downstream signaling. For example, K48-linked polyubiquitination is the canonical signal for proteasomal degradation [8], whereas K63-linked polyubiquitination plays a signaling role in regulation of various proteasome-independent cellular processes, including endocytosis, DNA repair, and protein trafficking for lysosomal degradation [9–11]. Deubiquitinating enzymes (DUBs) remove ubiquitin from ubiquitinated proteins or cleave C-terminal adducts of ubiquitin to regenerate the ubiquitin monomer, and therefore play an important role in regulating ubiquitin-dependent signaling events as well as in ubiquitin recycling [12, 13].
Ubiquitin C-terminal hydrolase L1 (UCH-L1) is a neuronal DUB with a critical role in the control of cellular ubiquitin homeostasis [14–18]. Human genetic studies reveal that UCH-L1 is mutated in a rare form of autosomal dominant Parkinson disease (PD) [19] and in the recently described childhood-onset neurodegenerative disorder with optic atrophy (NDGOA) [20]. The PD-linked I93M mutation reduces UCH-L1 activity by around 50 % [19–21], while the NDGOA-linked E7A mutation reduces activity by over 90 % [20]. Furthermore, we have shown that UCH-L1 is oxidatively damaged and down-regulated in sporadic PD and Alzheimer disease [22]. In mice, loss-of-function mutations in UCH-L1 cause a severe neurodegenerative phenotype dubbed gracile axonal dystrophy which is characterized by progressive ataxia and hindlimb paralysis [23–26]. Although predominantly expressed in neurons [27, 28], UCH-L1 protein is up-regulated in multiple tumors and cancer cells and is likely to have an oncogenic role in tumorigenesis [29–32]. Despite these established links between UCH-L1 and human diseases, little is known about how UCH-L1 is regulated in cells.
Another ubiquitin system component involved in PD pathogenesis is parkin, an E3 ubiquitin-protein ligase whose mutations cause autosomal recessive juvenile Parkinsonism (ARJP) [33–35]. Parkin mutations and down-regulation are also found in several types of cancer, supporting a function of parkin as a tumor suppressor [36–39]. Although UCH-L1 and parkin have been linked to PD and cancer, the relationship between these two proteins remains unknown.
In this study, we investigated the interaction of UCH-L1 with parkin and the role of parkin in UCH-L1 regulation. Our results reveal that parkin binds and facilitates K63-linked polyubiquitination of UCH-L1, and the parkin-mediated ubiquitination promotes UCH-L1 degradation through the autophagy-lysosomal pathway.
Materials and methods
Expression constructs and antibodies
Conventional molecular biological techniques were used to generate the following expression constructs: N-terminal Myc- and His-tagged human wild-type or mutant UCH-L1; N-terminal S-, GFP-, or GST-tagged human wild-type or mutant parkin. Other expression constructs used in this study include N-terminally HA-tagged Ub-WT, Ub-K48, Ub-K63, Ub-K0 (provided by T. Dawson, Johns Hopkins University, Baltimore, MD) and Ub-K48R and Ub-K63R (provided by M. Wooten, Auburn University, Auburn, AL). Polyclonal anti-UCH-L1 antibody against a synthetic peptide (residues 201–219) of human UCH-L1 was generated in rabbit and affinity-purified as described [40]. Other antibodies used in this study include anti-actin (clone C4, Millipore), anti-HA (clone 12CA5), anti-Myc (clone 9E10), anti-S-tag (Abcam), anti-ubiquitin (clone P4G7, Abcam), anti-GST (clone B14, Santa Cruz), anti-p62 (BD Biosciences), anti-LC3 (Sigma), and anti-parkin (Cell Signaling). The rabbit polyclonal anti-PINK1 antibody was generated and described in our previous study [41, 42]. Horseradish peroxidase- and fluorophore-conjugated secondary antibodies were from Jackson ImmunoResearch.
Cell culture and transfection
SH-SY5Y or HeLa cells were cultured in DMEM (Gibco) supplemented with 10 % fetal bovine serum (Atlanta Biologicals) and 1 % penicillin/streptomycin (Fisher). Cell transfections were performed using LipofectAMINE 2000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. Lysates were harvested at 24–72 h post-transfection for subsequent analyses.
Co-immunoprecipitation and S-tag pulldown assays
For co-immunoprecipitation analysis of the interaction between endogenous UCH-L1 and parkin or PINK1, anti-UCH-L1 antibodies or rabbit serum IgG controls were cross-linked to G-Sepharose agarose (Millipore) using 25 mM dimethyl pimelimidate dihydrochloride (Thermo Scientific). Immunoprecipitation was carried out as described [43] by incubation of the cross-linked anti-UCH-L1 or IgG beads with SH-SY5Y cell lysates prepared in Triton lysis buffer (50 mM Tris HCl pH 7.6, 150 mM NaCl, 0.1 % Triton-X-100, 1 % IGEPAL CA630 supplemented with proteinase and phosphatase inhibitors). Immunocomplexes were eluted with 100 mM glycine (pH = 2.8) followed by immunoblotting with anti-parkin, anti-PINK1, and anti-UCH-L1 antibodies. For co-immunoprecipitation analyses of the interaction between Myc-tagged UCH-L1 WT or mutant and GFP-tagged parkin, lysates from transfected HeLa cells were subjected to immunoprecipitation with anti-Myc antibody followed by recovery of protein complexes with protein G-Sepharose and subsequent immunoblotting analyses. S-tag pulldown assays were performed as described [44] by incubation of lysates from HeLa cells coexpressing S-tagged parkin WT or mutant and Myc-UCH-L1 with S-protein-agarose (Novagen), and the pulled down protein complexes were analyzed by immunoblotting with anti-UCH-L1 and anti-S-tag antibodies.
Recombinant protein purification and in vitro binding assays
Recombinant His-UCH-L1, glutathione S-transferase (GST), and GST-tagged parkin proteins were expressed in E. coli BL21 or Arctic Express cells and purified as previously described [45, 46]. In vitro binding assays were performed as described [46, 47] by incubation of GST-parkin and GST proteins immobilized on glutathione agarose with mouse brain lysate or soluble His-UCH-L1 for 2 h. Bound proteins were analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence confocal microscopy
SH-SY5Y cells were fixed in 4 % paraformaldehyde, stained with indicated primary and secondary antibodies, and then processed for indirect immunofluorescence confocal microscopy as previously described [48]. Images were acquired with a Nikon C1 confocal laser-scanning microscope, exported in TIFF format with the Nikon EZ-C1 viewer (Nikon Instruments Inc., Melville, NY) and processed using Adobe Photoshop CS4 (Adobe Systems, Inc.) to adjust contrast and brightness.
In vivo and in vitro ubiquitination assays
In vivo ubiquitination assays were performed as previously described [11, 42]. In brief, lysates from HeLa cells coexpressing S-tagged parkin, Myc-tagged UCH-L1, and HA-tagged wild-type or mutant ubiquitin plasmids as indicated were immunoprecipitated under denaturing conditions with anti-Myc antibody, and ubiquitinated UCH-L1 was detected by immunoblotting with anti-HA antibody. In vitro ubiquitination assays were performed as described [11, 42] by incubation of purified His-UCH-L1 (1 µg) with E1 enzyme (18 nM), E2 enzyme (UbcH7, UbcH8, or UbcH13/Uev1a; 250 nM), ubiquitin (10 µg) and GST or GST-parkin (1 µg) in reaction buffer (50 mM Tris–HCl, pH 7.6, 5 mM MgCl2, 100 mM NaCl, 25 µM ZnCl2, 2 mM dithiothreitol, and 4 mM ATP) for 2 h at 37 °C. Ubiquitin, E1, and E2 enzymes were from Boston Biochem, and the total volume of the reaction was 100 µL. Ubiquitinated UCH-L1 was detected by immunoblotting with anti-ubiquitin antibody.
Analysis of UCH-L1 ubiquitination and protein levels in parkin−/− mice
A breeding colony of parkin knockout (parkin −/−) mice was established from breeding pairs provided by R. Palmiter, University of Washington, Seattle, WA [49, 50]. For the assessment of endogenous UCH-L1 ubiquitination in parkin −/− and parkin +/+ mouse brain, immobilized GST-tagged, tandem ubiquitin binding entities (GST-TUBE2, LifeSensors) were used as described [51] to isolate ubiquitinated proteins from brain extracts from 4-month-old male parkin −/− mice and wild-type controls, followed by immunoblotting with anti-UCH-L1 and anti-GST antibodies. For the analysis of total UCH-L1 protein levels, brains from 3-month-old parkin −/− and parkin +/+ mice were homogenized in 1 % SDS and then subjected to SDS–PAGE and immunoblotting with anti-UCH-L1 and anti-β-actin antibodies.
Treatment of cells with proteasome, lysosome, and autophagy inhibitors
SH-SY5Y cells expressing S-parkin or the S-vector control were subjected to 24 h treatments with the proteasome inhibitor MG132 (20 µM, Sigma), lysosome inhibitor chloroquine (CQ, 100 µM, Sigma), autophagy inhibitor 3-methyl-adenine (3MA, 10 mM, Sigma), or 0.1 % Me2SO (DMSO, Fisher) vehicle control. Equal amounts of whole cell lysates were subjected to SDS-PAGE followed by immunoblotting with anti-UCH-L1 and anti-β-actin antibodies. The relative level of UCH-L1 was determined as described [43] by normalizing the immunoblot intensity of UCH-L1 against that of β-actin using Image J software.
UCH-L1 degradation assays
For the measurement of UCH-L1 degradation rate, SH-SY5Y cells expressing S-parkin or the S-vector control were treated with protein synthesis inhibitor cycloheximide (10 µg/mL) for the indicated lengths of time. Cells were lysed at the indicated time points, and equal amounts of lysates (normalized to cell number) were analyzed by immunoblotting with anti-UCH-L1 and anti-S-tag antibodies. UCH-L1 half-life (t 1/2) was calculated from the equation t 1/2 = ln(2)/λ, where λ represents the decay constant determined after fitting the plotted data points to an exponential curve.
Statistical analyses
Data were analyzed by Student’s t test or ANOVA with a Tukey’s post hoc test, where p < 0.05 was considered statistically significant. Results were expressed as mean ± SEM from three independent experiments.
Results
UCH-L1 interacts with parkin in vitro and in vivo
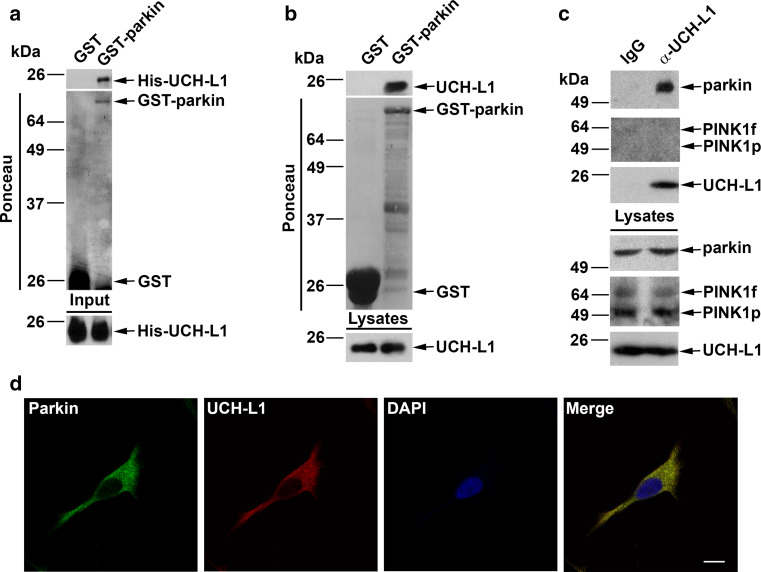
UCH-L1 was identified as a potential interactor of parkin in a recent proteomic screen [52], but this result has not yet been validated and it is unknown if these two proteins interact either in vitro or in vivo. To test if UCH-L1 and parkin can physically interact with each other, we performed in vitro binding assays with purified recombinant proteins. We found that His-tagged UCH-L1 bound selectively to GST-tagged parkin but not to GST alone (Fig. 1a), indicating a direct interaction between UCH-L1 and parkin. To confirm this interaction, we performed GST pulldown assays and found that purified GST-parkin, but not the GST control, was able to pulldown endogenous UCH-L1 from mouse brain homogenates (Fig. 1b), further supporting a specific interaction between these two proteins. We then performed co-immunoprecipitation analysis to examine the association of endogenous UCH-L1 and parkin in dopaminergic SH-SY5Y cells (Fig. 1c). Anti-UCH-L1 antibody, but not the IgG control, was able to co-immunoprecipitate UCH-L1 and parkin from cell lysates (Fig. 1c), indicating that UCH-L1 interacts with parkin in vivo. As a control, we tested the interaction between UCH-L1 and the parkin binding partner PINK1 by co-immunoprecipitation analysis and found no interaction between these two proteins (Fig. 1c). The ability of UCH-L1 to co-immunoprecipitate selectively with parkin but not PINK1 further confirms the specificity of the observed UCH-L1-parkin interaction. Consistent with the biochemical data, our confocal immunofluorescence microscopy analyses revealed colocalization of endogenous UCH-L1 and parkin proteins in SH-SY5Y cells (Fig. 1d), providing additional evidence supporting an in vivo association of UCH-L1 with parkin.
Fig. 1.
Specific interaction of UCH-L1 with parkin. a In vitro binding assays were performed by incubation of soluble His-tagged UCH-L1 (input) with immobilized GST or GST-parkin proteins (shown by Ponceau staining). Analysis of bound UCH-L1 by immunoblotting with anti-UCH-L1 antibody reveals direct binding of UCH-L1 to parkin. b GST pulldown assays were performed by incubation of equal amounts of mouse brain lysates with immobilized GST or GST-parkin (Ponceau staining) followed by immunoblotting with anti-UCH-L1 antibody to detect binding of endogenous UCH-L1 to parkin. c Co-immunoprecipitation analysis reveals a specific interaction of UCH-L1 with parkin but not PINK1. SH-SY5Y cell lysates were subjected to immunoprecipitation with anti-UCH-L1 antibody cross-linked to G-protein agarose or immunoprecipitation with the IgG controls. The immunoprecipitated proteins were eluted and analyzed by immunoblotting with anti-parkin, anti-PINK1, and anti-UCH-L1 antibodies d SH-SY5Y cells were double-immunostained with antibodies against endogenous parkin (green) and UCH-L1 (red). Colocalization is indicated by the yellow color in the merge panel. Scale bar 10 µm
The parkin-UCH-L1 interaction is impaired by PD-linked parkin mutations but not by UCH-L1 mutations
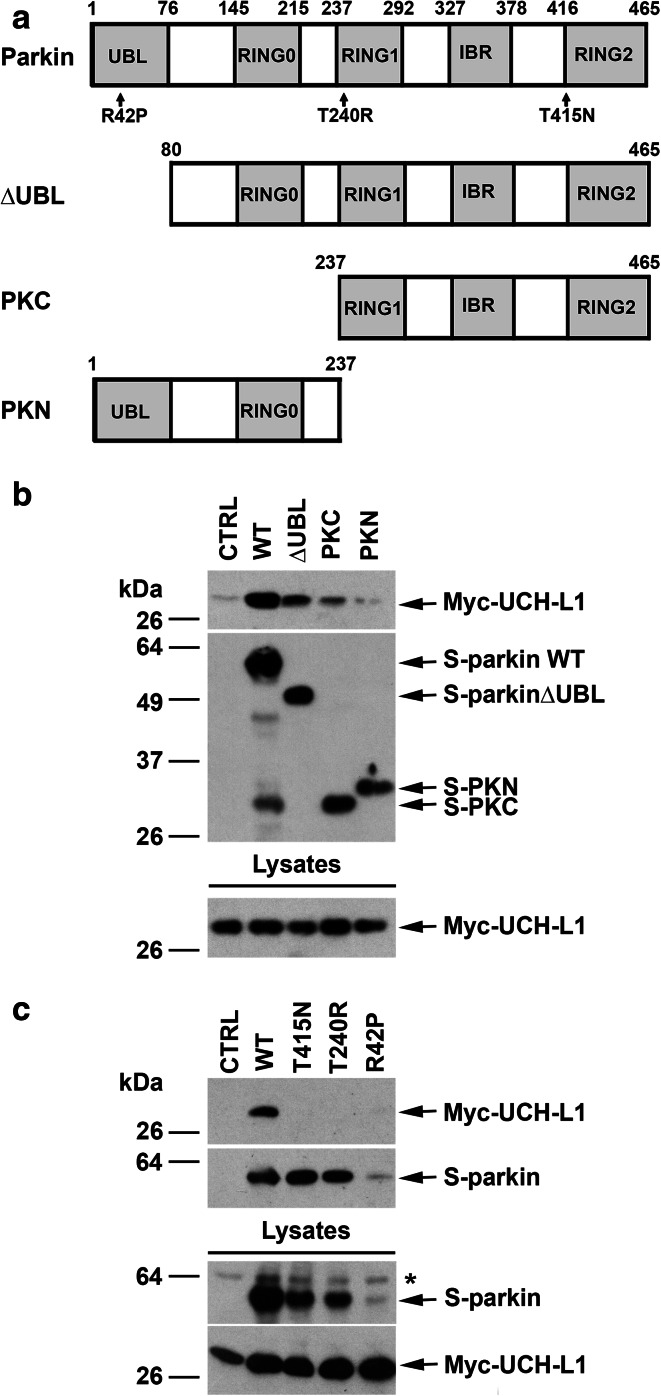
Because UCH-L1 binds monoubiquitin [18], we hypothesized that the parkin-UCH-L1 interaction may be mediated by the ubiquitin-like domain (UBL) at the N-terminus of parkin. To test this hypothesis, we examined the interaction of UCH-L1 with N- or C-terminal truncation mutants of parkin (Fig. 2a). We found that parkin ΔUBL, a truncated form of parkin lacking the UBL domain, retained the ability to bind UCH-L1 (Fig. 2b), arguing against the involvement of the UBL domain in mediating the parkin–UCH-L1 interaction. In addition, the parkin N-terminal region (PKN), which contains the UBL and RING0 domains, was virtually incapable of binding UCH-L1 (Fig. 2b), indicating that parkin UBL and RING0 domains are dispensable for the parkin–UCH-L1 interaction. In contrast, the parkin C-terminal region (PKC), which contains the RING1, RING2, and in-between RING-finger (IBR) domains, was able to bind UCH-L1 (Fig. 2b), indicating that the UCH-L1-binding region is located between amino acid residues 237 and 465 of parkin.
Fig. 2.
Pathogenic mutations and C-terminal truncation of parkin disrupt the interaction with UCH-L1. a Schematic representation of parkin and its mutants used in this study. The location of PD-linked parkin mutations are indicated on the domain structure. UBL, ubiquitin-like domain; IBR, in between RING-finger domain; PKC, C-terminal parkin; PKN, N-terminal parkin. b The interaction of parkin with UCH-L1 occurs within the C-terminal portion of parkin. S-pulldown assays were performed with lysates from HeLa cells coexpressing Myc-UCH-L1 and S-parkin WT or deletion mutant or the S-vector control, followed by immunoblotting with anti-Myc and anti-S-tag antibodies. c Impairment of the parkin-UCH-L1 interaction by PD-linked parkin mutations. S-pulldown assays were performed using lysates from HeLa cells coexpressing Myc-UCH-L1 and S-vector or S-parkin WT or mutant as indicated, followed by immunoblotting with anti-Myc and anti-S-tag antibodies. Asterisk indicates a nonspecific band
We next examined the effects of several familial PD-linked parkin mutations (Fig. 2a) on the parkin–UCH-L1 interaction by performing S-pulldown assays using lysates from HeLa cells co-expressing Myc-tagged UCH-L1 and S-tagged parkin WT or pathogenic mutant. We found that the ability of parkin to bind UCH-L1 is abrogated by parkin T240R and T415 N mutations (Fig. 2c). Consistent with previous reports that parkin R42P mutant is misfolded and rapidly degraded by the proteasome [53], we observed low levels of parkin R42P mutant in cells (Fig. 2c). Our results showed an apparent lack of UCH-L1 binding to parkin R42P mutant (Fig. 2c).
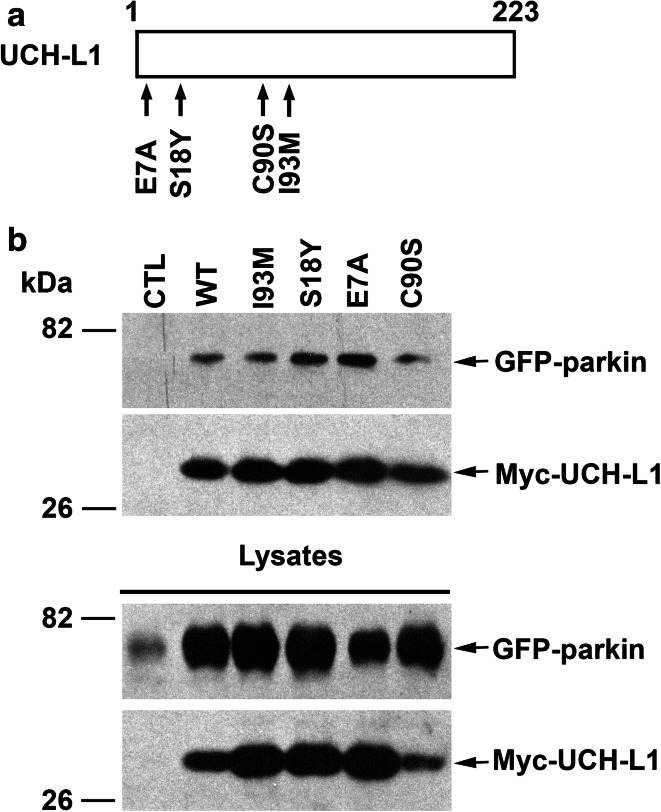
We then assessed the effects of several UCH-L1 mutations (Fig. 3a) on the parkin–UCH-L1 interaction and found that the ability of UCH-L1 to interact with parkin is not impaired by the PD-linked UCH-L1 I93 M mutation or by the NDGOA-associated E7A mutation (Fig. 3b). In addition, UCH-L1 S18Y substitution, a polymorphism in UCH-L1 which is thought to confer protection against sporadic PD [54–57], also had no inhibitory effect on the parkin-UCH-L1 interaction (Fig. 3b). In fact, UCH-L1 S18Y and E7A mutants showed an enhanced interaction with parkin (Fig. 3b), which might be due to the increased hydrophobicity at the N-terminal UCH-L1 protein surface induced by substitution of the polar serine (S18) and acidic glutamic acid (E7) residues with hydrophobic tryptophan and alanine residues. The increased hydrophobicity at UCH-L1 N-terminal region might provide a better binding surface for parkin. In addition, we found that the UCH-L1 catalytic site-specific C90S mutation did not disrupt the ability of UCH-L1 to interact with parkin (Fig. 3b), indicating that the DUB activity of UCH-L1 is not required for its interaction with parkin.
Fig. 3.
Interaction of parkin with wild-type and mutant UCH-L1. a Schematic representation of UCH-L1 and its mutants used in this study. The locations of neurodegenerative disease-linked UCH-L1 mutations (E7A and I93 M), protective polymorphism (S18Y), and catalytically inactive mutation (C90S) are indicated. b Lysates from HeLa cells coexpressing GFP-parkin and the indicated Myc-UCH-L1 WT or mutant were immunoprecipitated with anti-Myc antibodies followed by immunoblotting with anti-Myc and anti-parkin antibodies
Parkin mediates K63-linked polyubiquitination of UCH-L1
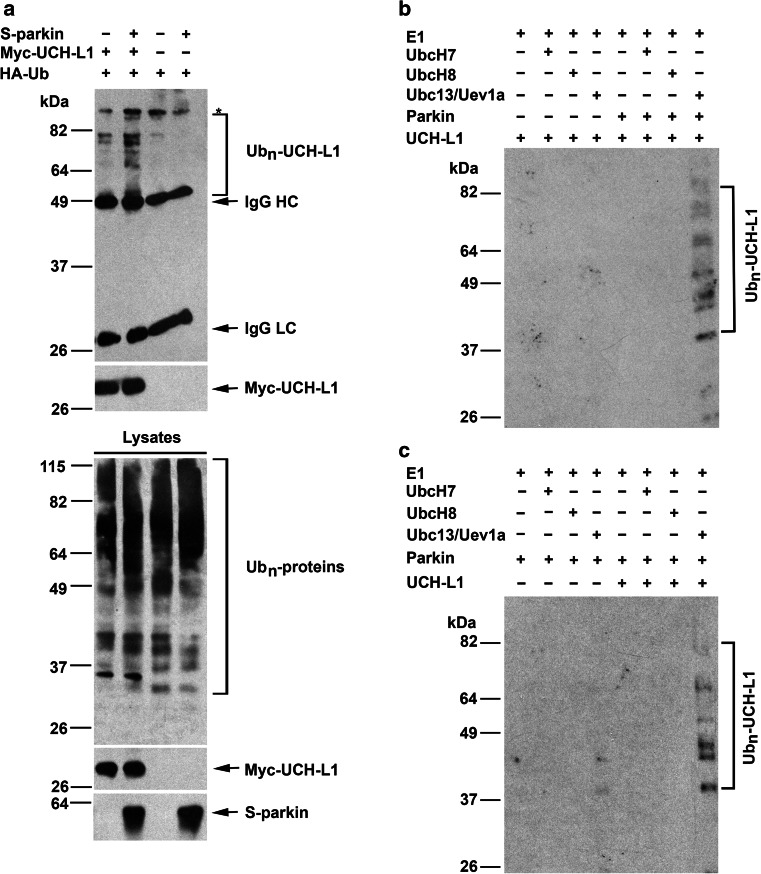
Our finding of an interaction between UCH-L1 and parkin (Fig. 1) raises the possibility that UCH-L1 may be a substrate of parkin E3 ligase. To test this possibility, we assessed the ability of parkin to promote UCH-L1 ubiquitination in cells by using a well-established in vivo ubiquitination assay [58, 59]. We found that ectopic parkin expression in HeLa cells, which lack endogenous parkin, resulted in enhanced ubiquitination of Myc-tagged UCH-L1 compared to the vector-transfected control (Fig. 4a), supporting a role of parkin in facilitating UCH-L1 ubiquitination in vivo. We next performed in vitro ubiquitination analyses with recombinant proteins to test if UCH-L1 is ubiquitinated by parkin in the presence of various E2 ubiquitin-conjugating enzymes (UbcH7, UbcH8, or the Ubc13/Uev1a complex) which are known to facilitate parkin E3 ligase activity [11]. We observed parkin-dependent UCH-L1 ubiquitination in the presence of Ubc13/Uev1a, but not in the presence of either UbcH7 or UbcH8 (Fig. 4b). No ubiquitination signal was detected when UCH-L1 was omitted from the reaction mixture (Fig. 4c). These results, together with our previous finding that Ubc13/Uevla is the cognate E2 enzyme for parkin-mediated K63-linked polyubiquitination, whereas UbcH7 and UbcH8 are the cognate E2 enzymes for parkin-mediated K48-linked polyubiquitination [11], suggest that parkin cooperates with the Ubc13/Uev1a E2 enzyme to catalyze K63-linked polyubiquitination of UCH-L1.
Fig. 4.
UCH-L1 is a substrate of parkin E3 ubiquitin ligase. a Parkin ubiquitinates UCH-L1 in vivo. Parkin-mediated ubiquitination of UCH-L1 was assessed in HeLa cells expressing HA-tagged ubiquitin, Myc-UCH-L1, and S-parkin as indicated. In vivo ubiquitination of UCH-L1 was determined by immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-HA and anti-Myc antibodies. Ubn-UCH-L1, polyubiquitinated UCH-L1; IgG HC, immunoglobulin heavy chain; IgG LC, immunoglobulin light chain. Asterisk indicates a nonspecific band. b Parkin ubiquitinates UCH-L1 in vitro in cooperation with the Ubc13/Uevla E2 enzyme. In vitro ubiquitination assays were performed with recombinant His-tagged UCH-L1 in the presence of E1, E2 (UbcH7, UbcH8, or the Ubc13/Uev1a complex), GST or GST-parkin, and ubiquitin as indicated. c Parkin-mediated ubiquitination in cooperation with Ubc13/Uevla is selective for His-UCH-L1. No ubiquitination signal was detected when UCH-L1 was omitted from the reaction mixture. Ubiquitinated UCH-L1 was detected by immunoblotting with anti-ubiquitin antibody
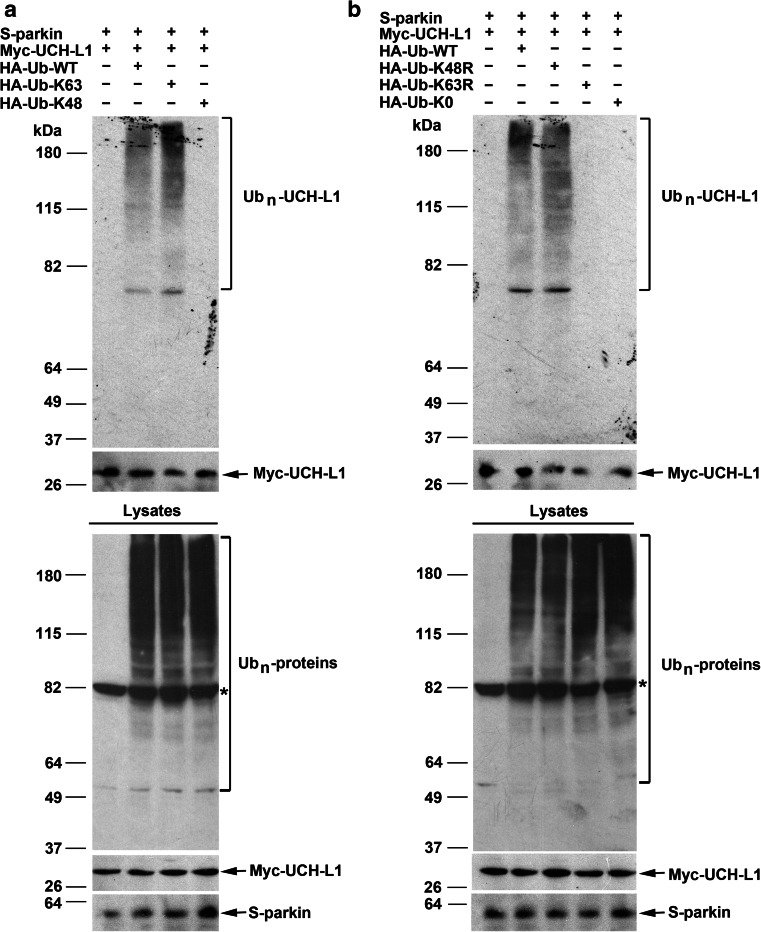
To determine if parkin-mediated K63-linked polyubiquitination of UCH-L1 takes places in cells, we performed in vivo ubiquitination assays using ubiquitin mutants, Ub-K48 and Ub-K63, which permits only the formation of K48-linked and K63-linked polyubiquitin chains, respectively, because all other lysine residues of ubiquitin were mutated to arginine. We detected robust UCH-L1 polyubiquitination by parkin in cells expressing Ub-WT or Ub-K63, but not in cells expressing Ub-K48 (Fig. 5a), supporting that parkin-mediated UCH-L1 polyubiquitination occurs via the K63-linkage. To further confirm this linkage, we used ubiquitin mutants, Ub-K48R and Ub-K63R, which contain a single lysine-to-arginine mutation at the indicated residues and are therefore incapable of forming K48-linked and K63-linked polyubiquitin chains, respectively. We found that parkin-mediated UCH-L1 polyubiquitination was abolished by replacement of Ub-WT with Ub-K63R but not by the replacement with Ub-K48R (Fig. 5b). In addition, parkin-mediated UCH-L1 polyubiquitination was not detected in cells expressing Ub-K0, an ubiquitin mutant which is incapable of forming polyubiquitin chains because all its lysine residues were mutated to arginines (Fig. 5b). Together, these data provide strong support for a function of parkin in facilitating K63-linked polyubiquitination of UCH-L1.
Fig. 5.
Parkin-mediated UCH-L1 polyubiquitination occurs via the K63 linkage. a, b HeLa cells were transfected with S-parkin, Myc-UCH-L1, and HA-tagged wild-type or mutant ubiquitin as indicated. Polyubiquitinated UCH-L1 (Ubn-UCH-L1) was detected by immunoprecipitation with anti-Myc antibody under denaturing conditions followed by immunoblotting with anti-HA and anti-Myc antibodies. The expression of S-parkin, Myc-UCH-L1, and HA-ubiquitin-conjugated proteins (Ubn-proteins) in the cell lysates were confirmed by immunoblotting with anti-parkin, anti-Myc, and anti-HA antibodies, respectively. Asterisk indicates a nonspecific band
Parkin is required for polyubiquitination of endogenous UCH-L1 in the brain
To further assess the role of parkin in the regulation of UCH-L1 ubiquitination in vivo, we examined the ubiquitination status of endogenous UCH-L1 in brains from wild-type (parkin +/+) and parkin knockout (parkin −/−) mice [49, 50] by using the tandem ubiquitin binding entities (TUBEs) approach. Previous studies have shown that GST-tagged TUBEs, such as GST-tagged TUBE2 based on the UBA1 domain from human RAD23A, preferentially capture endogenous poly-ubiquitinated proteins, but bind monoubiquitinated proteins at much lower affinity [51]. Our analysis of UCH-L1 ubiquitination with GST-tagged TUBE2 revealed that endogenous UCH-L1 in mouse brain was polyubiquitinated and the UCH-L1 polyubiquitination was significantly reduced by targeted disruption of parkin expression in mice (Fig. 6a, b). In addition to polyubiquitinated UCH-L1 forms, we also observed a ubiquitinated UCH-L1 species at ~40 kDa, which could represent UCH-L1 diubiquitination or monoubiquitination at two different sites (Fig. 6a). The level of this ubiquitinated UCH-L1 species was not significantly changed in the parkin −/− mouse brain compared to that in the parkin +/+ mouse brain (Fig. 6a, c). These findings indicate that parkin is required for the regulation of UCH-L1 polyubiquitination but not UCH-L1 diubiquitination or monoubiquitination in vivo.
Fig. 6.
UCH-L1 polyubiquitination is reduced in parkin −/− mouse brain. a Brain extracts (500 µg) from two individual parkin +/+ or parkin −/− mice were subjected to pulldown using GST-TUBEs, and ubiquitinated UCH-L1 was detected by immunoblotting with anti-UCH-L1 antibodies. The level of polyubiquitinated UCH-L1 (Ubn-UCH-L1) (b) and the level of diubiquitinated UCH-L1 (Ub2-UCH-L1) (c) were quantified using Image J software and expressed as a percentage of the corresponding control level in the parkin +/+ mice. Data represent mean ± SEM (n = 3 animals per genotype). *p < 0.05 versus the parkin +/+ control, unpaired two-tailed student’s t test
Parkin promotes UCH-L1 degradation through the autophagy-lysosome pathway
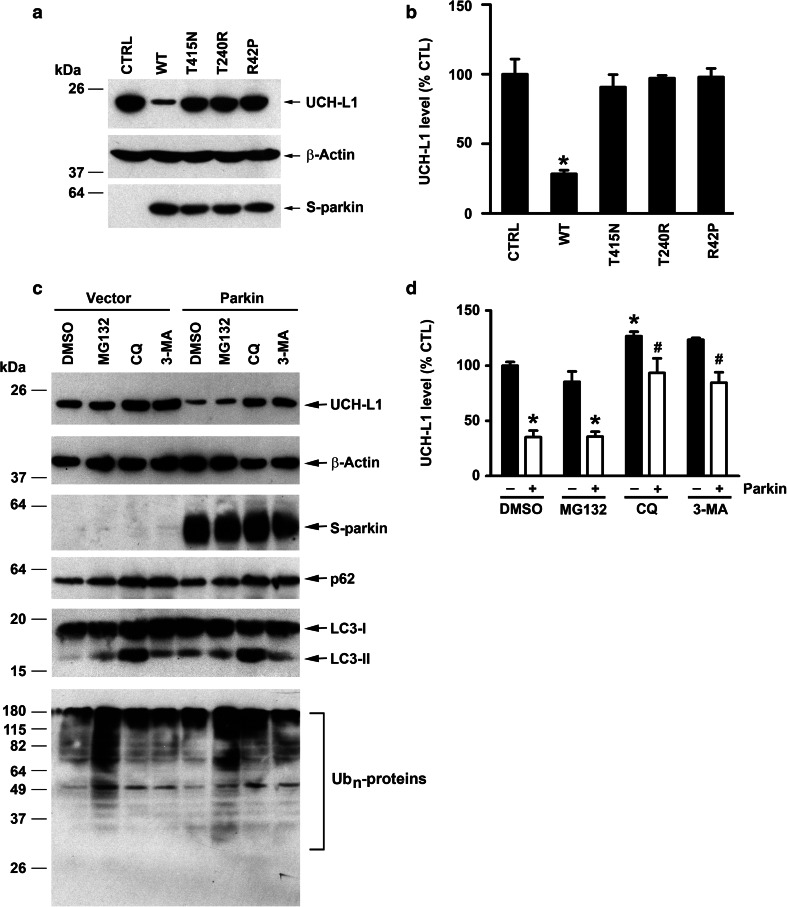
Next, we assessed the effect of targeted parkin deletion on the total level of endogenous UCH-L1 in mouse brain by performing quantitative Western blot analysis. We found that the total UCH-L1 level in the brain was significantly higher in parkin −/− mice than that in the parkin +/+ controls (Fig. 7a, b), suggesting a role for parkin in regulation of UCH-L1 degradation. To test this possibility, we analyzed the effect of parkin overexpression on the turnover rate of endogenous UCH-L1 protein in SH-SY5Y cells (Fig. 7c, d). We found that endogenous UCH-L1 protein is a long-lived protein with a half-life of ~59 h and the UCH-L1 protein half-life was reduced to ~40 h by parkin overexpression, indicating that parkin indeed promotes UCH-L1 degradation. Consistent with the accelerated UCH-L1 turnover rate induced by parkin overexpression, the steady-state level of endogenous UCH-L1 protein was significantly lower in parkin WT-overexpressing SH-SY5Y cells than that in the vector-transfected control cells (Fig. 8a, b). Moreover, we found that the steady-state level of endogenous UCH-L1 protein was unaltered by overexpression of PD-linked parkin R42P, T240R, and T415N mutants (Fig. 8a, b), indicating that these pathogenic mutations abolish the ability of parkin to promote UCH-L1 degradation.
Fig. 7.
Regulation of endogenous UCH-L1 protein degradation by parkin. Total brain UCH-L1 protein level is elevated in parkin −/− mice. Immunoblot analysis of brain homogenates from two individual parkin +/+ or parkin −/− mice with anti-UCH-L1, anti-β-actin and anti-parkin antibodies. a The relative level of total UCH-L1 was normalized to the β-actin level and expressed relative to the normalized UCH-L1 level in the parkin +/+ brain homogenate. b Data represent mean ± SEM (n = 5 animals per genotype). *p < 0.05 versus the parkin +/+ control, unpaired two-tailed student’s t test. c Equal numbers of SH-SY5Y cells expressing S-tag vector or S-parkin were treated with cycloheximide (10 µg/mL) for 0–72 h as indicated. Cells were lysed in equal volumes of SDS at each time point, and equal amounts of lysates (normalized to cell number) were subjected to SDS-PAGE followed by immunoblotting with anti-UCH-L1 and anti-parkin antibodies. d The levels of endogenous UCH-L1 protein at each time point were quantified and plotted relative to the corresponding UCH-L1 at 0 h
Fig. 8.
Parkin targets UCH-L1 for degradation by the autophagy-lysosome pathway. a Cell lysates of SH-SY5Y cells expressing S-tagged parkin WT, indicated S-tagged parkin mutant, or S-tag vector control were analyzed by immunoblotting with anti-UCH-L1, anti-S-tag, and anti-β-actin antibodies. b The level of endogenous UCH-L1 was normalized to the β-actin level and expressed as a percentage of the normalized UCH-L1 level in vector-transfected control cells. Results are shown as mean ± SEM from three independent experiments. *p < 0.05 versus vector-transfected control, one-way ANOVA with a Tukey’s post hoc test. c SH-SY5Y cells expressing S-tag vector or S-parkin were treated for 24 h with the indicated protein degradation inhibitors or vehicle (DMSO), and cell lysates were analyzed by immunoblotting with anti-UCH-L1, anti-S-tag, anti-p62, anti-LC3, anti-ubiquitin, and anti-β-actin antibodies. d The level of endogenous UCH-L1 was normalized to the β-actin level and expressed as a percentage of the normalized UCH-L1 level in the DMSO-treated, vector-transfected control cells. Results are shown as mean ± SEM from three independent experiments. *p < 0.05 versus the DMSO-treated, vector-transfected control; #p < 0.05 versus the DMSO-treated, parkin-transfected control, two-way ANOVA with a Tukey’s post hoc test
There are two major protein degradation pathways in cells: the ubiquitin-proteasome pathway and the autophagy-lysosome pathway [60, 61]. To determine which degradation pathway is involved in mediating the effect of parkin on UCH-L1 protein turnover, we assessed the effects of proteasome, lysosome and autophagy inhibition on the steady-state level of endogenous UCH-L1 in SH-SY5Y cells in the absence or presence of exogenous parkin. We found that parkin overexpression reduced the steady-state level of endogenous UCH-L1 protein (Fig. 8c, d), consistent with a role of parkin in promoting UCH-L1 degradation. The parkin-induced UCH-L1 degradation was blocked by the lysosome inhibitor chloroquine (CQ) or the autophagy inhibitor 3MA but not by the proteasome inhibitor MG132 (Fig. 8c, d). Together, these results support that parkin promotes UCH-L1 degradation through the autophagy-lysosome pathway but not the proteasome pathway.
Discussion
While parkin and UCH-L1 have both been implicated in the pathogenesis of PD and cancer, it is not established whether there is a physical or functional link between these two proteins. Our work described in this study shows that UCH-L1 is a substrate of parkin E3 ligase and reveals a function of parkin as a regulator of UCH-L1 degradation.
Despite ample evidence indicating the importance of UCH-L1 in health and disease, our knowledge of UCH-L1 post-translational modifications and their roles in UCH-L1 regulation is limited. A previous study reported that His-tagged UCH-L1 in transfected COS-7 monkey kidney cells is monoubiquitinated, with a ~33 kDa ubiquitinated UCH-L1 species which corresponds to monoubiquitination at a single site [62]. However, the E3 ligase for mediating UCH-L1 monoubiquitination remains unidentified, and the ubiquitination status of endogenous UCH-L1 is unknown. Our TUBE analysis of endogenous UCH-L1 ubiquitination in mouse brain revealed the presence of multiple polyubiquitinated UCH-L1 forms as well as a ~40 kDa diubiquitinated UCH-L1 species (which could also represent UCH-L1 monoubiquitination at two sites), but no ~33 kDa monoubiquitinated UCH-L1 species. Interestingly, we found that the levels of the polyubiquitinated UCH-L1 forms, but not ~40 kDa diubiquitinated UCH-L1 species, were significantly reduced by targeted parkin deletion in mice, indicating a role for parkin in regulation of endogenous UCH-L1 polyubiquitination but not UCH-L1 diubiquitination or monoubiquitination.
Because ubiquitin has seven internal lysine residues, polyubiquitination can occur via different linkages, leading to distinct outcomes. For example, K48-linked polyubiquitination targets protein to the proteasome for degradation, whereas K63-linked polyubiquitination acts in a proteasome-independent manner to regulate a number of cellular processes, including protein trafficking and autophagosome formation [7, 9]. We and others have previously shown that, through its association with different E2 enzymes, parkin is capable of mediating multiple forms of ubiquitination, including K48-linked polyubiquitination and K63-linked polyubiquitination [11, 63, 64]. Our in vivo ubiquitination studies revealed that, in cells, parkin-mediated UCH-L1 polyubiquitination occurs via the K63 linkage but not the K48 linkage. Furthermore, by using in vitro ubiquitination assays with recombinant proteins, we obtained evidence that parkin cooperates with the UbcH13/Uevla E2 ubiquitin-conjugating complex to facilitate K63-linked polyubiquitination of UCH-L1.
Emerging evidence indicates that K63-linked polyubiquitination could serve as a signal for targeting protein or other cargo to the autophagy machinery for subsequent degradation by the lysosome [64, 65]. The recognition of this targeting signal is thought to be mediated by adaptor proteins, such as p62, which binds simultaneously to K63-polyubiquitinated cargo via its ubiquitin-binding UBA domain and to the autophagy machinery component LC3 via its LC3-interacting region (LIR) to induce autophagosome formation [66, 67]. The autophagosome then fuses with the lysosome, leading to degradation of the cargo by lysosomal hydrolases. Consistent with this view, our findings support that parkin-mediated K63-linked polyubiquitination of UCH-L1 promotes the degradation of UCH-L1 by the autophagy-lysosome pathway. These findings provide new insights into the mechanism that control UCH-L1 degradation in cells.
Protein degradation plays an important role in regulating the expression levels of specific proteins and consequently the cellular processes in which these proteins participate [60]. UCH-L1 has a well-characterized deubiquitinating activity for catalyzing the hydrolysis of C-terminal esters and amides of ubiquitin to generate monomeric ubiquitin [68] and may also possess a dimerization-dependent E3 ligase activity [69]. Although the exact function of UCH-L1 remains unclear, UCH-L1 has been shown to control the cellular pool of free ubiquitin [18, 24], and thereby could affect many ubiquitin-dependent cellular processes.
PD is the second most common neurodegenerative disease characterized by the degeneration of dopaminergic neurons in the substantia nigra and the presence of intraneuronal inclusions known as Lewy bodies. The majority (>90 %) of PD cases are sporadic with unknown etiology, and familial PD with genetic mutations accounts for less than 10 % [70]. Loss-of-function mutations in parkin are a major cause of familial PD, responsible for approximately 50 % of recessively transmitted early-onset PD cases [33–35]. In contrast, only a single mutation (I93 M) in UCH-L1 has been associated with a single family (two siblings) with autosomal dominant familial PD [19], and it remains controversial whether the I93M variant is a pathogenic mutation or a rare polymorphism. Unlike sporadic PD patients, familial PD (ARJP) patients with parkin mutations are typically devoid of Lewy bodies [71, 72]. Consistent with a lack of Lewy bodies in ARJP patients, parkin −/− mice show no protein aggregates in the brain [73, 74]. How parkin mutations trigger neurodegeneration is poorly understood. Our finding that PD-linked parkin mutations abrogated the parkin-UCH-L1 interaction suggests that these mutations cause a loss of parkin-mediated regulation of UCH-L1 degradation, leading to increased levels of UCH-L1 in brains of human ARJP patients with parkin mutations. Although there is no report of UCH-L1 levels in ARJP brains, our observation of elevated UCH-L1 levels in parkin −/− mouse brains supports this notion. Interestingly, Drosophila genetic studies have shown that increased UCH-L1 levels in the eye imaginal discs induces caspase-dependent apoptosis, resulting in a rough eye phenotype [75]. Taken together, these results suggest that the loss of parkin-mediated UCH-L1 regulation may be a pathogenic mechanism contributing to neurodegeneration in human ARJP patients with parkin mutations.
Despite the identification of genetic defects responsible for several rare, familial forms of PD, the etiology of the more common, sporadic forms of PD is poorly understood. Oxidative stress has been implicated in the pathogenesis of sporadic PD [70], and oxidative damage to both parkin and UCH-L1 were found in postmortem brains of sporadic PD patients [22, 76, 77]. According to this study, one would predict that oxidative damage to parkin should cause increased UCH-L1 levels in sporadic PD brains by impairing parkin-mediated regulation of UCH-L1 degradation, but we have previously shown that UCH-L1 levels are reduced in sporadic PD brains as well as in Alzheimer disease brains [22]. This apparent discrepancy could be reconciled by the fact that only the soluble levels of UCH-L1 protein were measured in our previous study [22] and that UCH-L1 protein was also found to aggregate with Lewy bodies in PD brains and with neurofibrillary tangles in AD brains [22, 78], likely as a result of the observed oxidative damage to UCH-L1 protein [22].
In conclusion, our findings obtained from this study uncover a new function of parkin in regulation of UCH-L1 ubiquitination and degradation, and they provide novel insights into roles of these two proteins in health and neurodegenerative disease.
Acknowledgments
We thank Dr. Samuel M. Lee and Soe Thein for assistance with parkin and UCH-L1 protein purifications, Drs. Ted Dawson and Marie Wooten for providing plasmids, and Dr. Richard Palmiter for providing breeding pairs of parkin knockout mice. This work was supported in part by Grants from the National Institutes of Health (NS067950 to J.E.M, GM103613 to L.L., and AG034126 to L.S.C.) and the Emory University Research Committee (SK46673 to L.L.).
Conflict of interest
The authors declare that they have no conflict of interest.
Abbreviations
- UCH-L1
Ubiquitin carboxyl-terminal hydrolase L1
- UPS
Ubiquitin proteasome system
- DUB
Deubiquitinating enzyme
- UBL
Ubiquitin-like domain
- PD
Parkinson disease
- ARJP
Autosomal recessive juvenile parkinsonism
- RING
Really interesting new gene
- IBR
In-between RING-finger domain
- PKC
C-terminal parkin
- PKN
N-terminal parkin
References
- 1.Satija YK, Bhardwaj A, Das S. A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer. Int J Cancer. 2013;133(12):2759–2768. doi: 10.1002/ijc.28129. [DOI] [PubMed] [Google Scholar]
- 2.McKinnon C, Tabrizi SJ. The ubiquitin-proteasome system in neurodegeneration. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5802. [DOI] [PubMed] [Google Scholar]
- 3.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8(11):1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 4.Kowalski JR, Juo P. The role of deubiquitinating enzymes in synaptic function and nervous system diseases. Neural Plast. 2012;2012:892749. doi: 10.1155/2012/892749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Tijn P, Hol EM, van Leeuwen FW, Fischer DF. The neuronal ubiquitin-proteasome system: murine models and their neurological phenotype. Prog Neurobiol. 2008;85(2):176–193. doi: 10.1016/j.pneurobio.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19(1):94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Olzmann JA, Chin LS. Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy. 2008;4(1):85–87. doi: 10.4161/auto.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178(6):1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta . 2014;1843(1):114–128. doi: 10.1016/j.bbamcr.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen CN, Krantz BA, Wilkinson KD. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry. 1998;37(10):3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]
- 15.Case A, Stein RL. Mechanistic studies of ubiquitin C-terminal hydrolase L1. Biochemistry. 2006;45(7):2443–2452. doi: 10.1021/bi052135t. [DOI] [PubMed] [Google Scholar]
- 16.Wang YL, Takeda A, Osaka H, Hara Y, Furuta A, Setsuie R, Sun YJ, Kwon J, Sato Y, Sakurai M, Noda M, Yoshikawa Y, Wada K. Accumulation of beta- and gamma-synucleins in the ubiquitin carboxyl-terminal hydrolase L1-deficient gad mouse. Brain Res. 2004;1019(1–2):1–9. doi: 10.1016/j.brainres.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246(4930):670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 18.Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, Li H, Sato Y, Nishikawa K, Sun YJ, Sakurai M, Harada T, Hara Y, Kimura I, Chiba S, Namikawa K, Kiyama H, Noda M, Aoki S, Wada K. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12(16):1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 19.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 20.Bilguvar K, Tyagi NK, Ozkara C, Tuysuz B, Bakircioglu M, Choi M, Delil S, Caglayan AO, Baranoski JF, Erturk O, Yalcinkaya C, Karacorlu M, Dincer A, Johnson MH, Mane S, Chandra SS, Louvi A, Boggon TJ, Lifton RP, Horwich AL, Gunel M. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc Natl Acad Sci USA. 2013;110(9):3489–3494. doi: 10.1073/pnas.1222732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishikawa K, Li H, Kawamura R, Osaka H, Wang YL, Hara Y, Hirokawa T, Manago Y, Amano T, Noda M, Aoki S, Wada K. Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem Biophys Res Commun. 2003;304(1):176–183. doi: 10.1016/S0006-291X(03)00555-2. [DOI] [PubMed] [Google Scholar]
- 22.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279(13):13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 23.Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23(1):47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 24.Walters BJ, Campbell SL, Chen PC, Taylor AP, Schroeder DG, Dobrunz LE, Artavanis-Tsakonas K, Ploegh HL, Wilson JA, Cox GA, Wilson SM. Differential effects of Usp14 and Uch-L1 on the ubiquitin proteasome system and synaptic activity. Mol Cell Neurosci. 2008;39(4):539–548. doi: 10.1016/j.mcn.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci USA. 2010;107(4):1636–1641. doi: 10.1073/pnas.0911516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamazaki K, Wakasugi N, Tomita T, Kikuchi T, Mukoyama M, Ando K. Gracile axonal dystrophy (GAD), a new neurological mutant in the mouse. Proc Soc Exp Biol Med. 1988;187(2):209–215. doi: 10.3181/00379727-187-42656. [DOI] [PubMed] [Google Scholar]
- 27.Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40(6):1542–1547. doi: 10.1111/j.1471-4159.1983.tb08124.x. [DOI] [PubMed] [Google Scholar]
- 28.Day IN, Thompson RJ. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol. 2010;90(3):327–362. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 29.Hurst-Kennedy J, Chin LS, Li L. Ubiquitin C-terminal hydrolase l1 in tumorigenesis. Biochem Res Int. 2012;2012:123706. doi: 10.1155/2012/123706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mastoraki A, Ioannidis E, Apostolaki A, Patsouris E, Aroni K. PGP 9.5 and cyclin D1 coexpression in cutaneous squamous cell carcinomas. Int J Surg Pathol. 2009;17(6):413–420. doi: 10.1177/1066896909336018. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Gharib TG, Huang CC, Thomas DG, Shedden KA, Taylor JM, Kardia SL, Misek DE, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin Cancer Res. 2002;8(7):2298–2305. [PubMed] [Google Scholar]
- 32.Tezel E, Hibi K, Nagasaka T, Nakao A. PGP9.5 as a prognostic factor in pancreatic cancer. Clin Cancer Res. 2000;6(12):4764–4767. [PubMed] [Google Scholar]
- 33.West AB, Maidment NT. Genetics of parkin-linked disease. Hum Genet. 2004;114(4):327–336. doi: 10.1007/s00439-003-1074-6. [DOI] [PubMed] [Google Scholar]
- 34.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 35.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, Agid Y, Brice A. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med. 2000;342(21):1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 36.Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, D’Andrilli G, Scambia G, Picchio MC, Alder H, Godwin AK, Croce CM. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25–q27. Proc Natl Acad Sci USA. 2003;100(10):5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picchio MC, Martin ES, Cesari R, Calin GA, Yendamuri S, Kuroki T, Pentimalli F, Sarti M, Yoder K, Kaiser LR, Fishel R, Croce CM. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin Cancer Res. 2004;10(8):2720–2724. doi: 10.1158/1078-0432.CCR-03-0086. [DOI] [PubMed] [Google Scholar]
- 38.Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, Arends MJ. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci USA. 2010;107(34):15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, Ladanyi M, Sander C, Heguy A, Holland EC, Paty PB, Mischel PS, Liau L, Cloughesy TF, Mellinghoff IK, Solit DB, Chan TA. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42(1):77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin LS, Nugent RD, Raynor MC, Vavalle JP, Li L. SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J Biol Chem. 2000;275(2):1191–1200. doi: 10.1074/jbc.275.2.1191. [DOI] [PubMed] [Google Scholar]
- 41.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5(7):e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19(2):352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giles LM, Li L, Chin LS. Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis. J Biol Chem. 2009;284(32):21765–21775. doi: 10.1074/jbc.M109.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hackbarth JS, Lee SH, Meng XW, Vroman BT, Kaufmann SH, Karnitz LM. S-peptide epitope tagging for protein purification, expression monitoring, and localization in mammalian cells. Biotechniques. 2004;37(5):835–839. [PubMed] [Google Scholar]
- 45.Lee SM, Chin LS, Li L. Charcot-Marie-Tooth disease-linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. J Cell Biol. 2012;199(5):799–816. doi: 10.1083/jcb.201204137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Chin LS, Weigel C, Li L. Spring, a novel RING finger protein that regulates synaptic vesicle exocytosis. J Biol Chem. 2001;276(44):40824–40833. doi: 10.1074/jbc.M106141200. [DOI] [PubMed] [Google Scholar]
- 47.Webber E, Li L, Chin LS. Hypertonia-associated protein Trak1 is a novel regulator of endosome-to-lysosome trafficking. J Mol Biol. 2008;382(3):638–651. doi: 10.1016/j.jmb.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SM, Olzmann JA, Chin LS, Li L. Mutations associated with Charcot-Marie-Tooth disease cause SIMPLE protein mislocalization and degradation by the proteasome and aggresome-autophagy pathways. J Cell Sci. 2011;124(Pt 19):3319–3331. doi: 10.1242/jcs.087114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102(6):2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez FA, Curtis WR, Palmiter RD. Parkin-deficient mice are not more sensitive to 6-hydroxydopamine or methamphetamine neurotoxicity. BMC Neurosci. 2005;6:71. doi: 10.1186/1471-2202-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10(11):1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davison EJ, Pennington K, Hung CC, Peng J, Rafiq R, Ostareck-Lederer A, Ostareck DH, Ardley HC, Banks RE, Robinson PA. Proteomic analysis of increased Parkin expression and its interactants provides evidence for a role in modulation of mitochondrial function. Proteomics. 2009;9(18):4284–4297. doi: 10.1002/pmic.200900126. [DOI] [PubMed] [Google Scholar]
- 53.Schlehe JS, Lutz AK, Pilsl A, Lammermann K, Grgur K, Henn IH, Tatzelt J, Winklhofer KF. Aberrant folding of pathogenic Parkin mutants: aggregation versus degradation. J Biol Chem. 2008;283(20):13771–13779. doi: 10.1074/jbc.M707494200. [DOI] [PubMed] [Google Scholar]
- 54.Carmine Belin A, Westerlund M, Bergman O, Nissbrandt H, Lind C, Sydow O, Galter D. S18Y in ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) associated with decreased risk of Parkinson’s disease in Sweden. Parkinsonism Relat Disord. 2007;13(5):295–298. doi: 10.1016/j.parkreldis.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Satoh J, Kuroda Y. A polymorphic variation of serine to tyrosine at codon 18 in the ubiquitin C-terminal hydrolase-L1 gene is associated with a reduced risk of sporadic Parkinson’s disease in a Japanese population. J Neurol Sci. 2001;189(1–2):113–117. doi: 10.1016/S0022-510X(01)00555-X. [DOI] [PubMed] [Google Scholar]
- 56.Elbaz A, Levecque C, Clavel J, Vidal JS, Richard F, Correze JR, Delemotte B, Amouyel P, Alperovitch A, Chartier-Harlin MC, Tzourio C. S18Y polymorphism in the UCH-L1 gene and Parkinson’s disease: evidence for an age-dependent relationship. Mov Disord. 2003;18(2):130–137. doi: 10.1002/mds.10326. [DOI] [PubMed] [Google Scholar]
- 57.Maraganore DM, Lesnick TG, Elbaz A, Chartier-Harlin MC, Gasser T, Kruger R, Hattori N, Mellick GD, Quattrone A, Satoh J, Toda T, Wang J, Ioannidis JP, de Andrade M, Rocca WA. UCHL1 is a Parkinson’s disease susceptibility gene. Ann Neurol. 2004;55(4):512–521. doi: 10.1002/ana.20017. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler TC, Chin LS, Li Y, Roudabush FL, Li L. Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J Biol Chem. 2002;277(12):10273–10282. doi: 10.1074/jbc.M107857200. [DOI] [PubMed] [Google Scholar]
- 59.Lee JT, Wheeler TC, Li L, Chin LS. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet. 2008;17(6):906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 60.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12(9):1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 61.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 62.Meray RK, Lansbury PT., Jr Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1. J Biol Chem. 2007;282(14):10567–10575. doi: 10.1074/jbc.M611153200. [DOI] [PubMed] [Google Scholar]
- 63.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord. 2009;25(Suppl 1):9. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chin LS, Olzmann JA, Li L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans. 2010;38(Pt 1):144–149. doi: 10.1042/BST0380144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 66.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 67.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283(11):6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 68.Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35(21):6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 69.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111(2):209–218. doi: 10.1016/S0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 70.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 71.Mori H, Kondo T, Yokochi M, Matsumine H, Nakagawa-Hattori Y, Miyake T, Suda K, Mizuno Y. Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology. 1998;51(3):890–892. doi: 10.1212/WNL.51.3.890. [DOI] [PubMed] [Google Scholar]
- 72.Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, Morita T, Tsuji S, Ikuta F. Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology. 1994;44(3 Pt 1):437–441. doi: 10.1212/WNL.44.3_Part_1.437. [DOI] [PubMed] [Google Scholar]
- 73.Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, Gajendiran M, Roth BL, Chesselet MF, Maidment NT, Levine MS, Shen J. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278(44):43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 74.Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, Laville M, Pratt J, Corti O, Pradier L, Ret G, Joubert C, Periquet M, Araujo F, Negroni J, Casarejos MJ, Canals S, Solano R, Serrano A, Gallego E, Sanchez M, Denefle P, Benavides J, Tremp G, Rooney TA, Brice A, Garcia de Yebenes J. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12(18):2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- 75.Thao DT, An PN, Yamaguchi M, LinhThuoc T. Overexpression of ubiquitin carboxyl terminal hydrolase impairs multiple pathways during eye development in Drosophila melanogaster. Cell Tissue Res. 2012;348(3):453–463. doi: 10.1007/s00441-012-1404-x. [DOI] [PubMed] [Google Scholar]
- 76.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304(5675):1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 77.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101(29):10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lowe J, McDermott H, Landon M, Mayer RJ, Wilkinson KD. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J Pathol. 1990;161(2):153–160. doi: 10.1002/path.1711610210. [DOI] [PubMed] [Google Scholar]