Abstract
Several cationic antimicrobial peptides have been investigated as potential anti-cancer drugs due to their demonstrated selective toxicity towards cancer cells relative to normal cells. For example, intracellular delivery of KLA, a pro-apoptotic peptide, results in toxicity against a variety of cancer cell lines; however, the relatively low activity and small size leads to rapid renal excretion when applied in vivo, limiting its therapeutic potential. In this work, apoptotic peptide-polymer hybrid materials were developed to increase apoptotic peptide activity via multivalent display. Multivalent peptide materials were prepared with comb-like structure by RAFT copolymerization of peptide macromonomers with N-(2-hydroxypropyl) methacrylamide (HPMA). Polymers displayed a GKRK peptide sequence for targeting p32, a protein often overexpressed on the surface of cancer cells, either fused with or as a comonomer to a KLA macromonomer. In three tested cancer cell lines, apoptotic polymers were significantly more cytotoxic than free peptides as evidenced by an order of magnitude decrease in IC50 values for the polymers compared to free peptide. The uptake efficiency and intracellular trafficking of one polymer construct was determined by radiolabeling and subcellular fractionation. Despite their more potent cytotoxic profile, polymeric KLA constructs have poor cellular uptake efficiency (<1%). A significant fraction (20%) of internalized constructs localize with intact mitochondrial fractions. In an effort to increase cellular uptake, polymer amines were converted to guanidines by reaction with O-methylisourea. Guanidinylated polymers disrupted function of isolated mitochondria more than their lysine-based analogs, but overall toxicity was decreased, likely due to inefficient mitochondrial trafficking. Thus, while multivalent KLA polymers are more potent than KLA peptides, these materials can be substantially improved by designing next generation materials with improved cellular internalization and mitochondrial targeting efficiency.
Keywords: RAFT polymerization, peptide delivery, multivalency, apoptotic peptides
1 Introduction
One class of potential anti-cancer agents draws inspiration from cationic antimicrobial peptides (CAP), natural host defense mechanisms widely conserved in diverse species [1, 2]. These peptides eliminate a wide range of bacteria, fungi, viruses, and protozoa [3, 4] by disrupting negatively-charged membranes through electrostatic interactions, leading to pore formation, cellular depolarization, and cell death [5]. Minimal bacterial resistance was developed against antimicrobial amphiphilic polymers over several hundred cellular divisions compared to rapid development of antibiotic resistance, suggesting that drug strategies based on membrane-disruption are less susceptible to drug resistance [6]. Most CAPs have low cytotoxicity towards healthy eukaryotic cells, whose cellular membranes contain high levels of zwitterionic phosphatidylcholine resulting in minimal CAP interaction. Cancer cells, however, frequently overexpress anionic phospholipids, such as phosphatidylserine and O-glycosylated mucins, resulting in net-negative membranes that interact with CAPs [2, 7]. Therefore, many CAPs show selective toxicity towards cancer cells relative to normal cells.
Additionally, intracellular delivery of these CAPs can induce mitochondrial dysfunction [8]. Mitochondrial membranes resemble bacterial membranes and are disrupted upon exposure to CAPs, inducing cellular apoptosis through the release of cytochrome c [8]. The peptide sequence (KLAKLAK)2, or “KLA”, has been shown to permeabilize mitochondrial membranes in a local peptide concentration-dependent manner [4, 8]. KLA has therefore been investigated as a pro-apoptotic agent in fusion peptide [8-11], polymer conjugate [12], and nanoparticle conjugate [13] form. These materials have been studied in several cancer cell lines and animal models both in vitro and in vivo, showing promising cancer cell killing [10, 12, 13]. However, the requirement for high intracellular concentrations pose a significant barrier to clinical translation.
Multivalent polymeric display can significantly increase the activity of functional peptides and drugs. Dendrimeric display of folate, for example, has been shown to increase binding avidity up to 5 orders of magnitude [14]. Likewise, multivalent display of apoptotic peptides increased activity by over an order of magnitude [12, 15]. Multivalent strategies to increase peptide bioactivity can allow for rational design and optimization of materials for cancer applications.
In this work, peptide copolymers were synthesized via reverse addition-fragmentation chain transfer (RAFT) polymerization of N-(2-hydroxypropyl) methacrylamide (HPMA) with methacrylamido-functionalized peptide macromonomers and evaluated in several cancer lines. Two peptide sequences were used, the KLA apoptotic sequence and a GKRK targeting ligand for p32, a mitochondrial protein frequently overexpressed on the surface of tumor cells [16], isolated from phage display. Two peptide-HPMA copolymers with differing display of the peptides were evaluated: (i) pHGcK, a copolymer of GKRK, KLA, and HPMA, and (ii) pHGfK, a copolymer of GKRK-KLA fusion peptide and HPMA. These polymers were evaluated for in vitro cellular toxicity, plasma membrane disruption, intracellular trafficking, and inhibition of mitochondrial respiration.
2 Materials and Methods
2.1 Materials
N-(2-hydroxypropyl)methacrylamide (HPMA) was purchased from Polysciences (Warrington, PA). The initiator VA-044 was purchased from Wako Chemicals (Richmond, VA). Fmoc-protected amino acids and HBTU were purchased from AAPPTec (Louisville, KY), N-succinimidyl methacrylate from TCI America (Portland, Oregon), and Rink Amide Resin from EMD Biosciences (Darmstad, Germany). All other materials were reagent grade or better and were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise stated.
2.2 Material synthesis
2.2.1. Peptide monomers
Three peptides were synthesized using (d) and (l) amino acids and 6-aminohexanoic acid (Ahx): Ahx(d)[KLAKLAK]2 (composed of only (d) amino acids); AhxGKRK(d)[KLAKLAK]2 (composed of (l) amino acid uptake sequence GKRK with (d)-amino acid KLA); and AhxGKRK (composed of only (l) amino acids). Peptides were synthesized on solid support with Rink amide linker following standard Fmoc chemistry on an automated PS3 peptide synthesizer (Protein Technologies, Phoenix, AZ). Prior to peptide cleavage from the resin, the amino termini of the peptides were deprotected and coupled with N-succinimidyl methacrylate. These functionalized peptide monomers are respectively called MaAhxKLA, MaAhxGKRK-KLA, and MaAhxGKRK. Synthesized peptides were cleaved from resin by treatment of solid support with a solution of TFA/H2O/triisopropylsilane (TIPS)/1,3-dimethoxybenzene (90:2.5:2.5:5, v/v/v) for 2.5 hours under gentle mixing. Cleaved peptide monomers were precipitated in cold ether, dissolved in methanol and re-precipitated twice in cold ether. Each peptide monomer was purified to > 95% purity using RP-HPLC and analyzed by MALDI-TOF MS.
2.2.2. Polymers
Four polymers were synthesized: HPMA-co-(MaAhxGKRK-KLA) (pHGfK), HPMA-co-MaAhxKLA-co-MaAhxGKRK (pHGcK), and two HPMA-co-MaAhxGKRK copolymers (pHG35k, pHG64k). pHGfK, pHGcK, and pHG35k were synthesized with a monomer to chain transfer agent ratio of 142 and pHG64k with a ratio of 226; all polymers had 10% peptide mole feed. Monomers were dissolved in 9:1 acetate buffer (1 M, pH 5.1):ethanol (v/v) such that the final monomer concentration of the solution was 0.7 M. The RAFT chain transfer agent (CTA) used was ethyl cyanovaleric trithiocarbonate (ECT, molecular weight 263.4 g/mol) and the initiator (I) used was VA-044. The molar ratios of total monomer:CTA:I at the start of polymerization were 142:1:0.1 and 226:1:0.1, respectively. The reaction solutions were transferred to round bottom flasks, capped with a rubber septum, purged with argon for 10 min, and the submerged in a 44 °C oil bath to initiate polymerization. The polymerization was allowed to proceed for 24 hrs. The flasks were removed from the oil bath and polymers dialyzed against distilled H2O to removed unreacted monomers and buffer salts. The dialyzed polymers were lyophilized dry.
2.2.3. Guanidinylation of peptide and polymers
15 mg of the pHGcK and pHGfK copolymers and 12 mg of AcAhxKLA were dissolved in 1 mL of half-saturated NaHCO3. 60 mg of o-methylisourea was dissolved in 1 mL of half-saturated NaHCO3 and added to each solution. Guanidinylation reaction was allowed to proceed at room temperature under stirring for 3 days. After 3 days, polymer reactions were dialyzed again distilled H2O to purify. Guanidinylated peptide was purified by RP-HPLC and analyzed by MALDI-TOF MS.
2.2.4 Size exclusion chromatography
Molecular weight analysis was carried out by size exclusion chromatography. The copolymers were dissolved at 2 mg/mL in running buffer (150 mM acetate buffer, pH 4.4) for analysis by size exclusion chromatography-multiangle laser light scattering (SEC-MALLS). Analysis was carried out on an OHpak SB-804 HQ column (Shodex, New York, NY) in line with a miniDAWN TREOS multiangle light scattering detector (Wyatt, Santa Barbara, CA) and an OptiLab rEX refractive index detector (Wyatt). Absolute molecular weight averages (Mn, Mw) were calculated using ASTRA software (Wyatt).
2.2.5 Amino acid analysis
The polymer composition was determined through modified amino acid analysis following the method of Bidlingmeyer and coworkers [17]. Briefly, hydrolyzed polymer samples were run on a ZORBAX Eclipse Plus C18 (Agilent Technologies, Santa Clara, CA) HPLC column with pre-column derivatization using o-phthalaldehyde/β-mercaptopropionic acid to label hydrolyzed amino acids and 1-amino-2-propanol (hydrolyzed HPMA). Calibration curves were generated using serial dilutions of (l)-lysine, (l)-arginine, and reagent grade 1-amino-2-propanol.
2.2.6 3H-pHGfK radiolabeling
pHGfK was 3H-labeled using 3H-acetic anhydride. 5 mg of polymer was dissolved in 500 μL of 5% triethylamine in N,N-dimethylformamide. 2.5 μL of H3-acetic anhydride was added and reaction allowed to proceed under mixing for 2 hrs. Polymer was precipitated in ice-cold ether, dissolved in methanol and reprecipitated twice in ice-cold ether.
2.3 Polymer cytotoxicity
The cytotoxicity of the polymers was evaluated in vitro using the MTS assay. GL261 (murine glioma), SNB-19 (human glioblastoma), and HeLa (human cervical cancer) cells were plated overnight in 96-well plates at a density of 3000, 1500, and 2500 cells per well per 0.1 mL growth media, respectively. Polymers of various concentrations were prepared in phosphate buffered saline (PBS) and then diluted 10-fold in complete growth media. The cells were rinsed once with PBS and incubated with 100 μL of the polymer solution for 48 hrs at 37 °C, 5% CO2. At 48 hrs, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) (Promega, Madison, WI) was added to each well. Cells were then incubated for 3 hrs and absorbance measured at 1.5 hrs and 3 hrs at 490 nm using a plate reader (Tecan Safire2, Männerdorf, Switzerland). IC50 values were determined using a nonlinear fit (four-parameter variable slope) in GraphPad Prism v.6 (San Diego, CA).
2.4 Hemolysis assay
A hemolysis assay was used to evaluate the membrane-lytic activity of the synthesized materials following the procedure described by Hoffman and co-workers [18]. Briefly, plasma from freshly isolated human blood was removed by centrifugation. The cell layer containing the erythrocytes was washed three times with 150 mM NaCl and resuspended into phosphate buffer at pH 7.4. 16 μL of polymer at various concentrations and 1% Triton X-100 as control were added to 184 μL of erythrocyte suspensions in a 96-well conical plate and incubated for 1 h at 37 °C. The plate was then centrifuged, pelleting intact erythrocytes, and 100 μL of supernatant transferred to a 96-well flat bottom plate. Released hemoglobin within the supernatant was measured at 541 nm absorbance and percent hemolysis was calculated relative to the Triton X-100 control. Experiments were performed in triplicate.
2.5 Cellular uptake and subcellular fractionation
Subcellular fractionation experiments were completed as previously described by Shi et al with minor modifications [19]. HeLa cells were seeded in 150 mm2 dishes at 5 × 106 cells per 20 mL media per dish 24 h prior to the start of the experiment. Radiolabeled pHGfK was added to cells for a final concentration of 1 mM polymer and incubated for 6 hrs at 37 °C, 5% CO2. After a 6 h incubation, media was collected. Cells will be washed once with PBS, incubated with CellScrub (Genlantis, San Diego, CA) for 15 min at room temperature, washed twice in DPBS (no MgCl2, CaCl2), lifted off the plates in PBS, and then transferred to conical tubes. To remove dead/compromised cells, cells were washed twice with PBS, pelleting cells at 500g for 5 min after each wash. The cells were washed once with homogenization buffer (HB) (250 mM sucrose, 10 mM HEPES-NaOH, 1 mM EDTA, pH 7.4), pelleting the cells at 1000g for 6 min. The resulting pellet was then resuspended in 2.5× the wet pellet mass of HB (containing 1× protease inhibitors). Cells were homogenized with a 25-gauge needle until greater than 90% cell lysis was achieved.
Fractionation into a nuclear (N), heavy mitochondrial (HM), light mitochondrial (LM), microsomal (MF), and cytosolic (C) fractions was completed via differential centrifugation as previously described [19]. Briefly, the cell lysate was centrifuged at 1000g for 10 min. The resulting pellet (N) was resuspended in HB and centrifuged again. The remaining post-nuclear supernatant (PNS) was combined from both washes, and centrifuged at 3000g, 15,000g, and 100,000g, each with a wash step, to yield the HM, LM, MF pellets and C supernatant. For radioactivity analysis, samples were mixed with Ultima Gold XR scintillation fluid (Perkin Elmer, Waltham, MA), and then analyzed for radioactivity using a Beckman LS-6500 scintillation counter (Beckman Coulter Inc, Pasadena, CA).
2.6 Mitochondrial respiration assay
Mitochondria were isolated using established protocols with minor modifications [20]. Four confluent T225 flasks of SNB-19 cells were trypsinized, collected, and pelleted at 400g for 5 min at 4 °C. The cell pellet was washed twice with ice-cold Isolation Buffer (70 mM sucrose, 220 mM mannitol, 5 mM HEPES, 1 mM EGTA, pH 7.2, 0.5% (w/v) fatty-acid free BSA). The resulting pellet was resuspended in 3× the wet pellet volume in Isolation Buffer containing 1× Roche Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). The cells were homogenized by 15 passes through a 26-gauge needle and the homogenate was centrifuged twice at 600g for 10 min at 4 °C, discarding the pellet (unbroken cells and nuclei) each time. The supernatant was centrifuged at 7000g for 10 min at 4 °C, the supernatant (lysosomes and microsomes) discarded, and the pellet resuspended in Isolation Buffer and centrifuged again at 7000g for 10 min at 4 °C. The supernatant was removed, leaving a concentrated crude mitochondrial pellet. The pellet was resuspended in 120 μL of Measurement Buffer (250 mM sucrose, 15 mM KCl, 1 mM EGTA, 5 mM MgCl2, 30 mM K2HPO4, pH 7.4).
Mitochondrial respiration was studied using the Mito-ID® O2 Extracellular Sensor Kit (Enzo Life Sciences, Farmingdale, NY) with minor modifications to manufacturer’s protocol. For each condition tested, 3 μL of mitochondria suspension was diluted to 30 μL with Measurement Buffer in a 96 well plate. 25 μL of 0.53 mM peptide or equivalent polymer concentration was added to the mitochondria solutions and incubated at room temperature for 20 min. After 20 min, 100 μL of O2 sensor probe and 50 μL of 6.6 mM ADP/100 mM succinate in Measurement Buffer were added to each well following manufacturer’s protocol. 100 μL of oil was added on top of each well and then the plate was incubated at 30 °C for 10 min prior to beginning reading fluorescence. Fluorescence at ex/em 380/650 nm was read every 1.5 min for 30 min. Materials were tested in triplicate.
3 Results and discussion
3.1 Polymer characterization
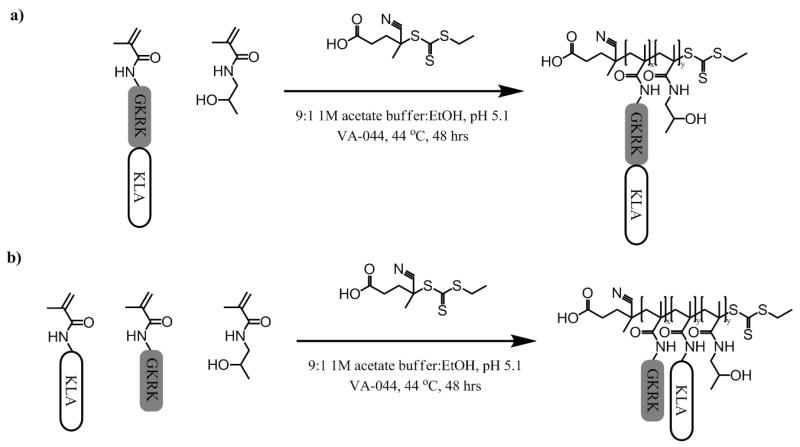
Two KLA-containing HPMA copolymers and two control HPMA-co-GKRK copolymers were synthesized via RAFT polymerization of methacrylamido-functionalized peptide monomers with HPMA to investigate the effects of multivalent display on cellular toxicity of KLA (Scheme 1). Polymers contained the GKRK p32 targeting sequence either as a separate comonomer (pHGcK) or fused with the KLA pro-apoptotic sequence (pHGfK). Degree of polymerization was chosen to target polymers around 50 kDa in size in order to be below the renal filtration threshold. HPMA was copolymerized to provide an inert, hydrophilic backbone.
Scheme 1.
Synthetic scheme of (a) pHGfK and (b) pHGcK.
The molecular weight and composition of the synthesized copolymers are summarized in Table 1. Copolymers were synthesized with narrow polydispersities (≤ 1.2). KLA-containing copolymers were around 50 kDa and control polymers lacking the KLA sequence (pHG35k and pHG64k) about 35.5 kDa and 63.6 kDa, respectively. Polymers contained 7-12% peptide, near quantitative incorporation of peptides (~10%) based on molar feed, as determined by amino acid analysis. pHG35k and pHG64k polymers served as cationic polymer controls lacking the KLA sequence.
Table 1. Properties of HPMA-Peptide Copolymers.
| Peptide-HPMA Copolymer | Abbrev. | Mn (kDa)a | Mw/Mna | % GKRKb | % KLAb | % GK-KLAb |
|---|---|---|---|---|---|---|
| HPMA-co-MaAhxGKRK-co-MaAhxKLA | pHGcK | 49.3 | 1.1 | 7.86 | 7 | --- |
| HPMA-co-MaAhxGKRK-KLA | pHGfK | 55.7 | 1.18 | --- | --- | 12.13 |
| HPMA-co-MaAhxGKRK | pHG35k | 35.5 | 1.15 | 9.1 | --- | --- |
| HPMA-co-MaAhxGKRK | pHG64k | 63.6 | 1.2 | 10.2 | --- | --- |
Determined by SEC-MALS.
Determined by amino acid analysis.
3.2 Polymer cytotoxicity
The IC50 values (concentration of polymer for 50% growth inhibition) of acetylated KLA peptide, acetylated GK-KLA peptide (GKRK targeting sequence fused with KLA), and KLA polymers were determined in three cancer cell lines - HeLa (human adenocarcinoma), SNB-19 (human glioblastoma), and GL261 (murine glioma) - with results summarized in Table 2. Cell viability following incubation with a range of material concentrations was determined by MTS assay, a measure of metabolic activity. The SNB-19 cell line was most sensitive to KLA toxicity (IC50 = 60 μM) while the HeLa cell line was least sensitive to KLA toxicity (IC50 > 250 μM). The fusion peptide GK-KLA, which includes the GKRK targeting peptide to increase cell uptake, showed ~2-5-fold lower IC50 compared to KLA. This result confirms previous reports where conjugation of GKRK to KLA was shown to enhance mitochrondrial localization and cellular apoptosis [13, 16]. Administration of the two KLA copolymers resulted in a 20-70 fold decrease in IC50 values compared to KLA, indicating enhanced cytotoxicity (Table 2); this result is consistent with prior reports of increased toxicity due to multivalent display of pro-apoptotic peptides, such as BH3 [15], KLA [12, 13], and antimicrobial peptides [21], relative to monomeric peptides. Interestingly, the architectural display of the two peptide sequences does not affect the cytotoxicity of the polymers – pHGfK and pHGcK show nearly-identical IC50 values for all cell types tested despite differences in peptide display as either individual or fused sequences. This suggests that targeting ligands can be fused to KLA sequences in polymeric constructs without compromising KLA activity, simplifying material synthesis.
Table 2. Peptide and Polymer IC50 Values.
| GL261 | SNB-19 | HeLa | ||||
|---|---|---|---|---|---|---|
| Peptide or Polymer | IC50 (μg/mL) | IC50 (μM KLA) | IC50 (μg/mL) | IC50 (μM KLA) | IC50 (μg/mL) | IC50 (μM KLA) |
| KLA1 | 314.3 | 165.8 | 115.1 | 60.7 | > 500 | > 250 |
| GK-KLA2 | 182.1 | 73.6 | 65.1 | 26.3 | 84.9 | 34.3 |
| pHGcK | 25.7 | 5.4 | 11.7 | 2.5 | 22.0 | 4.4 |
| pHGfK | 20.1 | 5.4 | 11.2 | 3 | 18.8 | 5.0 |
Full peptide sequence: AcAhx-d[KLAKLAK]2.
Full peptide sequence: AcAhxGKRK-d[KLAKLAK]2
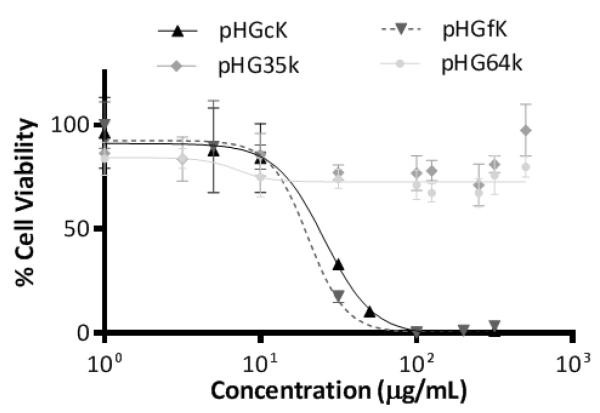
Cytotoxic cationic polymers can exert their toxicity through plasma and mitochondrial membrane disruption [22]. To evaluate whether the increased toxicity observed with pHGcK and pHGfK copolymers was due to the cationic nature of the polymers resulting from multivalent display of the GKRK targeting ligand, two copolymers of HPMA and GKRK were synthesized as controls. pHG35k was synthesized with the same monomer:CTA ratio as the KLA copolymers and pHG64k was synthesized with target molecular weight of 50 kDa. Neither pHG copolymer demonstrated dose-dependent cytotoxicity at mass concentrations up to 100-fold higher than the IC50 values of the two KLA copolymers (Figure 1). KLA copolymer cytotoxicity is therefore likely due to the KLA sequence and not the cationic targeting sequence.
Figure 1.
Cytotoxicity curves for various peptide HPMA copolymers in GL261 cells.
3.3 Hemolysis assay
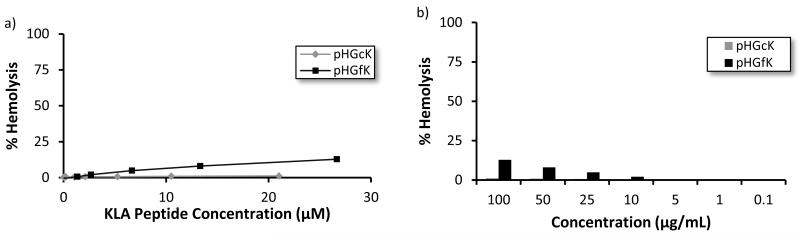
To investigate the route of cellular toxicity, the ability of the pHGcK and pHGfK copolymers to lyse plasma membranes was determined via hemolysis assay. Polymers were incubated with freshly isolated human erythrocytes and tested for plasma membrane disruption by detecting hemoglobin release (Figure 2). Minimal hemolysis was observed for both copolymers at concentrations an order of magnitude higher than the IC50 values for the cancer lines. KLA peptide was previously reported to have hemolytic activity at concentrations > 750 μM [23]. Observed cellular toxicity is therefore likely not due to direct plasma membrane disruption but rather through an intracellular route. Cationic polymers containing membrane-active domains have shown hemolysis at concentrations as low as 1 μg/mL [24]. High cationic charge density is correlated with plasma membrane disruption [25]; KLA materials possibly differ in membrane lytic characteristics due to lower charge density.
Figure 2.
Percent hemolysis of pHGck and pHGfK polymers relative to Triton X-100 control in (a) KLA molar concentration and (b) polymer mass concentration.
Interestingly, the manner of GKRK display affected the membrane-lytic behavior of the KLA-containing copolymers. pHGfK, with the GKRK sequence fused to KLA, shows significantly higher membrane-lytic behavior; at 100 μg/mL, pHGfK induces 13% hemoglobin release when incubated with erythrocytes, whereas pHGcK induces < 2% hemoglobin release at the same mass concentration (Figure 2b). However, despite differences in hemolytic activity, the IC50 values are similar. pHGcK has more charge delocalization due to spatial separation of the GKRK and KLA sequences along the polymer backbone; comparatively, the two sequences are fused together in pHGfK. This could lead to differences in membrane specificity and lytic activity. We previously have shown that 60 kDa HPMA-oligolysine copolymers demonstrated differences in cytotoxicity based on pendant oligolysine chain length despite keeping the overall charge/mass ratio constant [26]. Therefore, molecular architecture may play a significant role in cytotoxicity.
3.4 Cellular uptake and intracellular trafficking
The cellular uptake and intracellular localization of pHGfK were investigated through subcellular fractionation of radiolabeled polymer. HeLa cells were chosen as a model cell line to study intracellular trafficking due to (1) well-established fractionation protocols [19] and (2) previous demonstration of p32-targeted delivery of nanoparticles to HeLa cells [27]. 3H-labeled pHGfK was incubated with HeLa cells for 6 hrs at 37 °C and then cells were washed, lysed, and relative radioactivity measured in the various fractions. Less than 3% of polymer was found to be cell associated, with about 2% surface bound and less than 1 % internalized after 6 hrs (Figure 3a). Low cellular association and poor internalization therefore pose significant barriers towards the efficacy of these polymers. Similarly, low uptake was seen with the cationic polymer PEI, where 5% cellular association was observed in HeLa cells after 4 hrs, suggesting cationic polymers may not be efficiently internalized [19].
Figure 3.
(a) [ 3H]-labeled pHGfK copolymer uptake in HeLa cells after 6 hr incubation at 37 ° C. (b) Subcellular fractionation and localization of polymer in HeLa cell lysates after 6 hr exposure to polymers. NP = nuclear pellet; HM = heavy mitochondria; LM = light mitochondria; MF = microsomes; C = cytosol.
The cellular lysate was fractionated via differential centrifugation (Figure 3b) to determine intracellular distribution of the polymer after a 6 hr incubation with cells. Of the internalized polymer, 20% was found in the heavy mitochondrial fraction that contains intact mitochondria while 11% and 7% were found respectively in the light mitochondrial and microsomal fractions, which contain some mitochondria with other intracellular membrane vesicles including lysosomes, Golgi membranes, and endosomes [28]. Only 5% remained in the cytosol. In comparison, a cationic HPMA-peptide polymer with similar structure to pHGfK except displaying oligolysine instead of the GK-KLA peptide, showed lower distribution in the HM fraction (12%) [29]. The GKRK sequence has been shown to bind to p32 overexpressed on many cancer lines, including glioblastoma [16]; GKRK-functionalized nanoworms have previously been shown to traffic to mitochondria in several glioblastoma cell lines [13]. This sequence may contribute towards increased trafficking of the pHGfK polymer to mitochondria compared to the HPMA-oligolysine polymers.
3.5 Guanidinylation of KLA polymers
Due to the poor efficiency of cell uptake (Figure 3), pHGcK and pHGfK were guanidinylated in an attempt to increase cell internalization. Guandinylation of chitosan [30] and aminoglycosides [31] has previously been shown to significantly increase cellular uptake. In addition, a fusion peptide of oligoarginine (R7) with KLA was shown to have more potent cytotoxic properties and to enhance permeabilitization and aggregation of mitochondria [9, 32]. We therefore hypothesized that conversion of the primary lysine amines of pHGcK and pHGfK to guanidines would increase cellular uptake and therefore cytotoxicity. The lysine residues on pHGcK and pHGfK were converted to homoarginine via reaction with o-methylisourea to yield guanidinylated polymer analogs pHGchR and pHGfhR, respectively [33]. Amino acid analysis confirmed complete lysine conversion as noted by the disappearance of the lysine peak and the concurrent emergence of a distinct homoarginine peak (data not shown). The toxicity of these constructs was evaluated in vitro in HeLa and SNB-19 cells. For both cell lines, guanidinylated polymers demonstrated 2-4 fold decrease in cytotoxicity compared to the original KLA polymers (Table 3). This was in contrast to guanidinylated KLA peptide (hRLA) which had significantly higher cytotoxicity (over 10-fold decrease in IC50) than KLA in SNB-19 cells (Table 3). Additionally, HPMA-KLA copolymers lacking the GKRK sequence showed very low cytotoxicity with IC50 > 300 μg/mL in HeLa cells (data not shown). Receptor-mediated endocytosis is expected to be important for efficient delivery and polymer activity, as knockdown of p32 in several glioblastoma tumors have shown > 60% reduction in GKRK phage binding [16]. Therefore, the lower cytotoxicity of the guanidinylated polymers could be due to the guanidinylation of the GKRK sequence which negatively affects uptake, trafficking, and cytotoxicity, or due to reduced mitochondrial disruption due to guanidinylation of the KLA sequence.
Table 3. Guanidinylated Peptide and Polymer IC50 Values.
| SNB-19 | HeLa | |||
|---|---|---|---|---|
| Peptide or Polymer | IC50 (μg/mL) | IC50 (μM KLA) | IC50 (μg/mL) | IC50 (μM KLA) |
| hRLA | 10.64 | 4.95 | --------- | --------- |
| pHGchR | 41.4 | 10.78 | 26.78 | 6.98 |
| pHGfhR | 44.7 | 10.72 | 26.33 | 6.38 |
3.6 Effect of polymers on mitochondrial respiration.
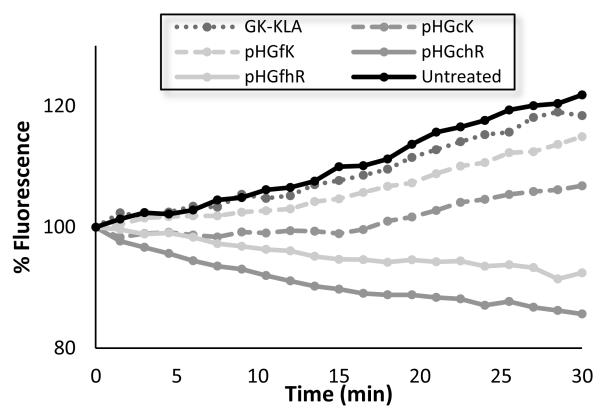
The effect of pHGcK and pHGfK and guanidinylated analogs pHGchR and pHGfhR on mitochondrial activity was therefore determined using an assay for oxygen consumption from isolated mitochondria. The function of isolated mitochondria was monitored for 30 minutes following incubation with polymers or peptide using an oxygen-sensitive, phosphorescent probe (Figure 4). Oxygen consumption, which correlates directly with mitochondrial respiration, was decreased by 17% when treated with GK-KLA peptide, and by 69% and 32% when treated with pHGcK and pHGfK polymers, respectively. Therefore, the observed increased cytotoxicity of the KLA copolymers relative to KLA peptide could be due in part to differences in membrane activity independent of cellular uptake. Guanidinylated polymers have a much greater effect on mitochondrial function; no oxygen consumption was observed and slightly decreased signal, attributed to probe photobleaching, was seen. These results suggest that guanidinylation of KLA-containing polymers increases mitochondrial disruption activity but that overall cytotoxicity may be reduced due to altered intracellular trafficking. Lipophilicity and charge distribution were shown to affect cellular uptake and intracellular trafficking of cationic materials [34]. Additionally, guanidine groups bind more strongly to sulfates than primary amines, which may result in greater binding to membrane proteins such as heparan sulfates and therefore reduced trafficking to mitochondria [35].
Figure 4.
Relative fluorescence of an O2-sensitive fluorescence probe as a measure of mitochondrial function in isolated mitochondria treated with the various polymers.
4 Conclusions
In this work, p32-targeted polymers displaying multiple pendant pro-apoptotic KLA peptides were synthesized and tested for their cytotoxicity. Targeting sequences were presented in the polymers either as fusion sequences with KLA or as separate monomers. Differences in display of the targeting peptide did not affect overall polymer toxicity; polymeric constructs are at least 10-fold more potent against cancer cell lines compared to KLA peptide. The internalization efficiency and intracellular trafficking of one polymeric KLA construct was determined by radiolabeling with subcellular fractionation analysis. Cellular uptake and intracellular localization studies show < 1% of dosed polymer is internalized within 6 hrs but that ~20% of internalized polymer is localized to fractions containing intact mitochondria. Guanidinylation of the copolymers was investigated to improve cellular uptake but despite improved ability to disrupt the function of isolated mitochondria, cytotoxicity of the guanidinylated polymers decreased relative to the original polymers. Thus, while polymeric display of pro-apoptotic peptides improves potency, there is significant room for improving uptake and mitochondrial targeting efficiency.
Acknowledgements
This was work was supported by NIH 1R01CA177272. David Chu was supported by a NIH T32 training grant (NIH CA138312). Julie Shi was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-0718124. We thank Profs. Anthony Convertine and Patrick Stayton for their generous donation of the ECT chain transfer agent.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PubMed] [Google Scholar]
- [2].Mader JS, Hoskin DW. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert Opin. Inv. Drug. 2006;15:933–946. doi: 10.1517/13543784.15.8.933. [DOI] [PubMed] [Google Scholar]
- [3].Nicolas P, Mor A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 1995;49:277–304. doi: 10.1146/annurev.mi.49.100195.001425. [DOI] [PubMed] [Google Scholar]
- [4].Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. BBA-Biomembranes. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- [6].Sovadinova I, Palermo EF, Urban M, Mpiga P, Caputo GA, Kuroda K. Activity and mechanism of antimicrobial peptide-mimetic amphiphilic polymethacrylate derivatives. Polymers. 2011;3:1512–1532. [Google Scholar]
- [7].Leuschner C, Hansel W. Membrane disrupting lytic peptides for cancer treatments. Curr. Pharm. Des. 2004;10:2299–2310. doi: 10.2174/1381612043383971. [DOI] [PubMed] [Google Scholar]
- [8].Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Del Rio G, Krajewski S, Lombardo CR, Rao R, Ruoslahti E, Bredesen DE, Pasqualini R. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- [9].Law B, Quinti L, Choi Y, Weissleder R, Tung C-H. A mitochondrial targeted fusion peptide exhibits remarkable cytotoxicity. Mol. Cancer Ther. 2006;5:1944–1949. doi: 10.1158/1535-7163.MCT-05-0509. [DOI] [PubMed] [Google Scholar]
- [10].Mai JC, Mi Z, Kim S-H, Ng B, Robbins PD. A proapoptotic peptide for the treatment of solid tumors. Cancer Res. 2001;61:7709–7712. [PubMed] [Google Scholar]
- [11].Alves ID, Carré M, Montero M-P, Castano S, Lecomte S, Marquant R, Lecorché P, Burlina F, Schatz C, Sagan S, Chassaing G, Braguer D, Lavielle S. A proapoptotic peptide conjugated to penetratin selectively inhibits tumor cell growth. BBA-Biomembranes. 2014;1838:2087–2098. doi: 10.1016/j.bbamem.2014.04.025. [DOI] [PubMed] [Google Scholar]
- [12].Adar L, Shamay Y, Journo G, David A. Pro-apoptotic peptide-polymer conjugates to induce mitochondrial-dependent cell death. Polym. Advan. Technol. 2011;22:199–208. [Google Scholar]
- [13].Agemy L, Friedmann-Morvinski D, Kotamraju VR, Roth L, Sugahara KN, Girard OM, Mattrey RF, Verma IM, Ruoslahti E. Targeted nanoparticle enhanced proapoptotic peptide as potential therapy for glioblastoma. P. Natl. Acad. Sci. USA. 2011;108:17450–17455. doi: 10.1073/pnas.1114518108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery. Chem. Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- [15].Richter M, Chakrabarti A, Ruttekolk IR, Wiesner B, Beyermann M, Brock R, Rademann J. Multivalent design of apoptosis-inducing Bid-BH3 peptide–oligosaccharides boosts the intracellular activity at identical overall peptide concentrations. Chem-Eur J. 2012;18:16708–16715. doi: 10.1002/chem.201202276. [DOI] [PubMed] [Google Scholar]
- [16].Agemy L, Kotamraju VR, Friedmann-Morvinski D, Sharma S, Sugahara KN, Ruoslahti E. Proapoptotic Peptide-mediated cancer therapy targeted to cell surface p32. Mol. Ther. 2013;21:2195–2204. doi: 10.1038/mt.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J. Chromatogr. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- [18].Bulmus V, Woodward M, Lin L, Murthy N, Stayton P, Hoffman A. A new pH-responsive and glutathione-reactive, endosomal membrane-disruptive polymeric carrier for intracellular delivery of biomolecular drugs. J. Control. Release. 2003;93:105–120. doi: 10.1016/j.jconrel.2003.06.001. [DOI] [PubMed] [Google Scholar]
- [19].Shi J, Chou B, Choi JL, Ta AL, Pun SH. Investigation of polyethylenimine/DNA polyplex transfection to cultured cells using radiolabeling and subcellular fractionation methods. Mol. Pharm. 2013;10:2145–2156. doi: 10.1021/mp300651q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protocols. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- [21].Chamorro C, Boerman MA, Arnusch CJ, Breukink E, Pieters RJ. Enhancing membrane disruption by targeting and multivalent presentation of antimicrobial peptides. BBA-Biomembranes. 2012;1818:2171–2174. doi: 10.1016/j.bbamem.2012.04.004. [DOI] [PubMed] [Google Scholar]
- [22].Chu DSH, Schellinger JG, Shi J, Convertine AJ, Stayton PS, Pun SH. Application of living free radical polymerization for nucleic acid delivery. Accounts Chem. Res. 2012;45:1089–1099. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Javadpour MM, Juban MM, Lo W-CJ, Bishop SM, Alberty JB, Cowell SM, Becker CL, McLaughlin ML. De novo antimicrobial peptides with low mammalian cell toxicity. J. Med. Chem. 1996;39:3107–3113. doi: 10.1021/jm9509410. [DOI] [PubMed] [Google Scholar]
- [24].Schellinger JG, Pahang JA, Johnson RN, Chu DS, Sellers DL, Maris DO, Convertine AJ, Stayton PS, Horner PJ, Pun SH. Melittin-grafted HPMA-oligolysine based copolymers for gene delivery. Biomaterials. 2013;34:2318–2326. doi: 10.1016/j.biomaterials.2012.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomedicine. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Johnson RN, Chu DS, Shi J, Schellinger JG, Carlson PM, Pun SH. HPMA-oligolysine copolymers for gene delivery: optimization of peptide length and polymer molecular weight. J. Control. Release. 2011;155:303–311. doi: 10.1016/j.jconrel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF, Hirsch MS, Mesirov JP, Drapkin R, Root DE, Lo J, Fogal V, Ruoslahti E, Hahn WC, Bhatia SN. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer target ID4. Sci. Transl. Med. 2012;4:147ra112–147ra112. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Graham JM. Current Protocols in Cell Biology. John Wiley & Sons, Inc.; 2001. Purification of a crude mitochondrial fraction by density-gradient centrifugation. [DOI] [PubMed] [Google Scholar]
- [29].Shi J, Choi JL, Chou B, Johnson RN, Schellinger JG, Pun SH. Effect of polyplex morphology on cellular uptake, intracellular trafficking, and transgene expression. ACS Nano. 2013;7:10612–10620. doi: 10.1021/nn403069n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhai X, Sun P, Luo Y, Ma C, Xu J, Liu W. Guanidinylation: A simple way to fabricate cell penetrating peptide analogue-modified chitosan vector for enhanced gene delivery. J. Appl. Polym. Sci. 2011;121:3569–3578. [Google Scholar]
- [31].Luedtke NW, Carmichael P, Tor Y. Cellular uptake of aminoglycosides, guanidinoglycosides, and polyarginine. J. Am. Chem. Soc. 2003;125:12374–12375. doi: 10.1021/ja0360135. [DOI] [PubMed] [Google Scholar]
- [32].Lemeshko VV. Potential-dependent membrane permeabilization and mitochondrial aggregation caused by anticancer polyarginine-KLA peptides. Arch. Biochem. Biophys. 2010;493:213–220. doi: 10.1016/j.abb.2009.11.004. [DOI] [PubMed] [Google Scholar]
- [33].Carlson PM, Schellinger JG, Pahang JA, Johnson RN, Pun SH. Comparative study of guanidinebased and lysine-based brush copolymers for plasmid delivery. Biomater. Sci. 2013;1:736–744. doi: 10.1039/C3BM60079C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yousif LF, Stewart KM, Horton KL, Kelley SO. Mitochondria-penetrating peptides: sequence effects and model cargo transport. Chembiochem. 2009;10:2081–2088. doi: 10.1002/cbic.200900017. [DOI] [PubMed] [Google Scholar]
- [35].Fromm JR, Hileman RE, Caldwell EEO, Weiler JM, Linhardt RJ. Differences in the interaction of heparin with arginine and lysine and the importance of these basic amino acids in the binding of heparin to acidic fibroblast growth factor. Arch. Biochem. Biophys. 1995;323:279–287. doi: 10.1006/abbi.1995.9963. [DOI] [PubMed] [Google Scholar]