Abstract
Environmental toxicants such as toxic metals can alter epigenetic regulatory features such as DNA methylation, histone modification, and non-coding RNA expression. Heavy metals influence gene expression by epigenetic mechanisms and by directly binding to various metal response elements in the target gene promoters. Given the role of epigenetic alterations in regulating genes, there is potential for the integration of toxic metal-induced epigenetic alterations as informative factors in the risk assessment process. Here, we focus on recent advances in understanding epigenetic changes, gene expression, and biological effects induced by toxic metals.
Keywords: Epigenetics, Gene expression, DNA methylation, Histone modification, Noncoding RNA, Heavy metal, Environmental exposure
INTRODUCTION
Epigenetics describes the study of mitotically and meiotically heritable changes in gene expression without mutating the DNA sequence. Epigenetic alterations regulate key events in cellular homeostasis, including transcriptional and translational regulation of gene expression. Epigenetic modifications include three commonly studied alterations: (1) DNA methylation; (2) covalent, post-translational modifications (PTMs) of histones; and (3) expression of non-coding RNAs (ncRNA). Epigenetic regulation of gene expression can be influenced by a variety of environmental factors, and their dysregulations has been implicated in various diseases (1,2).
DNA methylation is the most well-studied epigenetic alteration and plays a key role in development such as genomic imprinting and X chromosome inactivation (3). Also, DNA methylation is associated with cancer, Alzheimer’s disease, diabetes, atherosclerosis, Friedrich’s ataxia, immunodeficiency, rheumatoid arthritis, multiple sclerosis, and systemic lupus erythematosus (4). The transfer of a methyl group to cytosine forms 5-methyl cytosine (5-mC) in CpG dinucleotides. The DNA methyltransferase (DNMT) family consists of DNMT1, DNMT2, DNMT3a and DNMT3b. DNMT1 maintains the methylation status once it is established by DNMT3a and DNMT3b through de novo methylation (3). These DNMTs cooperate to maintain a precise DNA methylation pattern. Methyl binding domain (MBD) proteins including MeCP2, MBD1, MBD2 and MBD4 are readers that recognize and bind to methylated CpG sites and presumably mediate transcription repression or silencing. However the mechanism by which this occurs has yet to be uncovered. So far several proteins have been introduced as DNA methylation erasers including TET (teneleventranslocation) and AID (activation-induced cytidine deaminase) (5). These erasers seem to have a distinct function in different cellular contexts.
Histone modifications provide another important mechanism of epigenetic regulation. Histones package and order DNA into basic structural units called nucleosomes. The Nterminal tails of histones can undergo various post-translational modifications (PTMs) including acetylation, methylation, phosphorylation, ubiquitylation and sumoylation (6). There are now at least 16 classes of histone modifications on over 60 different residues of histones, which regulate important cellular processes such as transcription, replication and repair. Histone modifications are relatively labile compared to DNA methylation. Unlikely DNA methylation, histone modifications can lead to both activation and repression of gene expression depending on the type of modification at a specific residue (7,8). Whereas lysine acetylation of histones is broadly linked with gene activation, lysine methylation of histones creates specific and unique signals depending on the residue modified. For example, active promoter regions are enriched in trimethylated histone H3 at lysine 4 (H3K4me3), whereas inactive promoters are enriched in trimethylated histone H3 at lysine 27 and trimethylated histone H3 at lysine 9 (H3K27me3 and H3K9me3), and regulatory enhancers are enriched in monomethylated histone H3 at lysine 4 (H3K4me1) and/or acetylation histone H3 at lysine 27 (H3K27Ac) (7). PTMs on histone proteins function in elaborate combinations to regulate the many activities associated with chromatin. Thus, altered histone modifications have the potential to impact differential gene and subsequent protein expression. Differential histone modifications have been implicated in several diseases such as cancers and Parkinson’s and Huntington’s diseases (9). The diseases associated with differential histone modifications are also associated with aberrant DNA methylation.
Another epigenetic modulator of gene expression is noncoding RNA. ncRNA is composed of long ncRNAs and small ncRNAs. Small ncRNAs regulate gene expression by two mechanisms including translational repression (by microRNA, miRNA) and mRNA degradation (by small interfering RNA, siRNA). A complete sequence match between miRNA and target mRNA results in translational repression (miRNA pathway), whereas a base mismatch triggers the mRNA degradation pathway (siRNA pathway) and the mRNA is degraded (10). At present, over 17,000 distinct mature miRNAs have been identified in over 140 species (11). Aberrant expression of miRNAs has been associated with various human diseases such as cancer, cardiovascular disease, and genetic disorders (12). Circulating miRNAs have been shown to be correlated with cardiovascular disease and tumors. Therefore, miRNAs could be readily employed as biomarkers of exposure and disease due to the availability of circulating extracellular miRNAs found in body fluids, such as amniotic fluid, saliva, serum, and plasma.
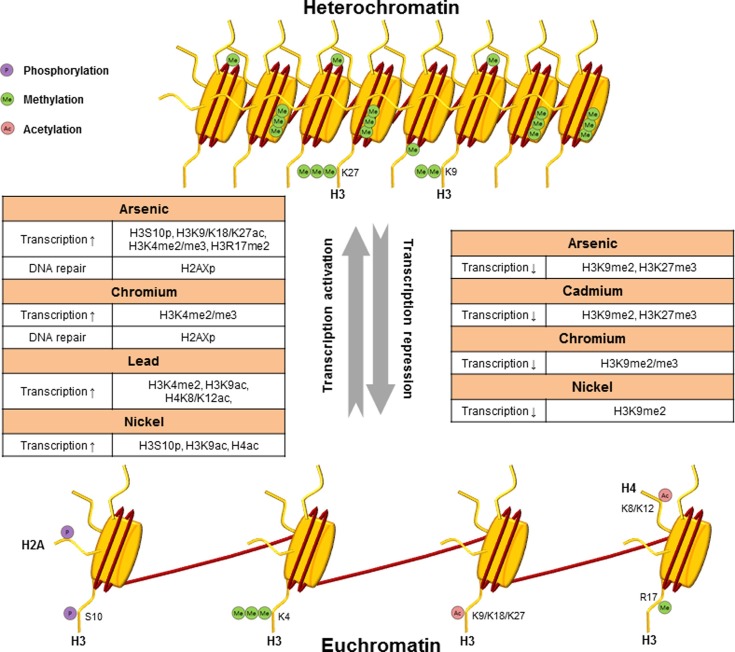
The increasing evidence that various environmental factors affect epigenetic alterations suggests that the influence of the environment on an organism at the molecular level can extend well beyond interactions with the DNA sequence. To date, emerging work has focused to address the effect of environmental exposures on histone modifications and miRNA expression as well as DNA methylation. This review highlights the current evidence that epigenetic alterations are associated with environmental exposure to toxic metals and considers the potential mechanisms behind environmentally related epigenetic alterations. A summary of the data discussed is presented in Table 1 and Fig. 1.
Table 1. An overview of the epigenetic effects of metal exposure.
| DNA methylation | Histone modifications | ncRNA expression | |
|---|---|---|---|
|
| |||
| Arsenic | +/− | H3S10p, H2AXp, H3K9/K27/K16/K18ac, H3K4me2/me3, H3K9me2, H3K27me3, H3R3me2, H3R17me2 | Let-7 family, miR-16, miR-17, miR-19, miR-20, miR-22, miR-24, miR-26b, miR-29a, miR-30, miR-34a, miR-96, miR-98, miR-107, miR-126, miR-195, miR-200b, miR-210, miR-221, miR-222, miR-454 |
| Cadmium | +/− | H3p, H3ac, H3K4me3, H3K9me3, H3K27me3 | Let-7 family, miR-15b, miR-23b, miR-101, miR-144, miR-130a, miR-361-5p, miR-455-3p, miR-1233, miR-1275 |
| Chromium | +/− | H2AXp, H3K4me2/me3, H3K9me2/me3 | miR-143, miR-222. miR-3940-5p |
| Copper | ND | H3ac, H4ac | miR-398 |
| Lead | − | H3K9ac, H4K8/K12ac, H3K4me2 | miR-146a, miR-222 |
| Mercury | +/− | H3ac, H3K27me3 | miR-141, miR-196b, miR-302b, miR-367, miR-382 |
| Nickel | + | H3S10p, H3K9ac, H4ac, H2Aub, H2Bub, H3K9me2 | miR-222 |
| Organotin | − | H3ac, H4ac | ND |
(+), hypermethylation; (−), hypomethylation. Ac, acetylation; Me, methylation; P, phosphorylation; Ub, ubiquitination. ND, no data.
Fig. 1. Metals can activate or silence transcription through epigenetic modifications. Ac, acetylation; Me, methylation; P, phosphorylation.
METAL EXPOSURE AND ASSOCIATED EPIGENETIC ALTERATIONS
Arsenic. Inorganic arsenic (As) compounds (pentavalent arsenate [As(V)] and trivalent arsenite [As(III)]) are important environmental carcinogens that affect DNA methylation status in the cell (13). The toxicity of As is most often linked to the arsenite owing to its ability to bind thiol groups in various cellular components (14). Because As is biotransformed into a methylated form using S-adenosyl methionine (SAM) as the methyl donor, As metabolism may deplete intracellular methyl group stores, which may result in hypomethylation of genes. However, the effect of As-induced SAM depletion on DNA and histone methylation is controversial; both hypo- and hypermethylation of different genes can be found following As exposure (15), and alteration of histone methylation is also observed, such as increased H3K9me2 and decreased H3K27me3 (16-18). Smeester et al. identified 183 differentially methylated promoters associated with As exposure in adult subjects from Zimapan, Hildago, Mexico and out of this group were 17 tumor suppressor or tumor suppressor-associated genes with hypermethylated promoters (19).
In individuals exposed to inorganic As in Bangladesh, total urinary As was inversely correlated with global H3K9ac and positively correlated with H3K9me2 (20). A positive correlation was reported between H3K4me2 and H3K9ac and cumulative exposure to As and nickel-containing particulate matter (PM) in peripheral blood lymphocytes (PBL) (21). As is a potent inducer of H3S10 phosphorylation (H3S10p) and this modification has been associated with up-regulation of genes involved in tumorigenesis (22). Recently, it was reported that As induces the expression of the anti-oxidant gene ferritin through methylation of H4 arginine 3 (H4R3) and H3 arginine 17 (H3R17) in a dose dependent manner in human keratinocytes (23). Several studies have demonstrated the mechanisms of As-induced histone modifications, focusing on the expression and activity of histone modifying enzymes. In hepatocellular carcinoma cells, the increased level of H3K9 acetylation by As was correlated with inhibition of HDAC activity (24). As was shown to increase H3K9me2 and H3K9me3 through increased expression of the H3K9 methyltransferase G9a/KMT1C without change in expression of lysine (K)-specific demethylase 3A (JHDM2A/KDM3A) (25). This suggests that these genes represent possible biomarkers of inorganic As-induced disease.
Exposure to As also causes changes in miRNA expression (26). Sodium arsenite up-regulated the expression of four miRNAs (miR-22, miR-34a, miR-221 and miR-222) and strongly down-regulated the expression of miR-210 in human lymphoblasts. Malignant transformation of TP53−/− cells treated with As was correlated with decreased levels of miR-200b (27). Decreased expression of miR-200b caused methylation of the miR-200 promoter, suggesting that As may transcriptionally repress miRNA expression through DNA methylation. The alteration pattern of these miRNAs could potentially serve as a biomarker for assessing As toxicity and As-induced disease risk.
Cadmium. Cadmium (Cd) is an environmental contaminant and is classified as a Group I carcinogen associated with cancers of the liver, bone, kidney, and pancreas as well as a known risk factor for cardiovascular disease (28). Cd exposure occurs primarily through cigarette smoke and dietary sources. Cd is present in almost all foods and its levels depend on the type of food and source contamination (29). Cd exposure results in DNA hypermethylation and hypomethylation. Acute Cd exposure has been shown to decrease DNA methylation via noncompetitive inhibition of DNMT activity. Conversely, chronic Cd exposure results in global DNA hypermethylation and enhanced DNMT activity. This increased methylation was correlated with increased cell proliferation followed by transformation (30). Consistent with this study, long-term Cd treatment resulted in increased DNMT3b transcript levels leading to increased DNMT activity, increased global DNA methylation, promoter hypermethylation and loss of expression of tumor suppressor genes RASSF1A and p16. Importantly, this data suggests that genome-wide hypermethylation along with DNMT3b overexpression may be a useful biomarker to specifically identify Cd-induced human prostate cancers (31).
Studies of Cd-induced histone modifications are mainly limited to in vitro studies. In Cd-transformed urothelial cells, levels of H3K4me3, H3K27me3 and H3K9me3 occupancy at the metallothionein 3 (MT3) promoter were increased compared to untransformed cells (32). In addition to altering methylation of histone H3 lysine residues, Cd was shown to decrease H3 phosphorylation via inhibition of the human vaccinia-related kinase VRK1/2 (33). This demonstrates that Cd may block histone modifications through inhibition of histone modifying enzymes.
Like histone modification, Cd has been shown to alter miRNA levels in vitro. Cd upregulates the expression of miR-101 and miR-144, which target the cystic fibrosis transmembrane conductance regulator (CFTR) (34). Indeed, Cd decreased expression of CFTR in human bronchial epithelial cells, suggesting that Cd, through miRNA induction, may be involved in the pathogenesis of cystic fibrosis.
Chromium. Chromium (Cr) can exist in several valence states, such as +6, +3 and 0 oxidative states. Cr(III) is the final oxidative form in all biological systems (13). Exposure to Cr(VI) is mainly through drinking water contaminated through industrial Cr use and inhalation in occupations dealing with plating, production and welding using chromate. Cr is highly mutagenic and associated with many types of cancers including prostate, bone, leukemia, lymphoma, renal, gastrointestinal, brain, nasal and lung cancer (35,36). The mutagenic and carcinogenic nature of Cr may be explained in part by the variety of genotoxic lesions that it produces; Cr forms adducts with DNA, induces DNA strand breaks, DNA intra- and interstrand crosslinks, and DNA-protein crosslinking (37,38). Cr also interferes with DNA damage response and repair. The risk of developing Cr(VI)-associated neoplasms is the highest for lung, followed by nasal cancers (16). For instance, 33% of chromate-induced lung cancers exhibited methylation of the p16 promoter, while the same applied to 26% of non-chromate lung cancers (39). In another study, two other tumor suppressor genes, adenomatosis polyposis coli (APC) and human mut-L homologue 1 (hMLH1) were hypermethylated in chromate-induced lung cancers (86 and 28%, respectively), compared with 44% and 0% in nonchromate lung cancers, respectively (40).
In addition to DNA methylation, Cr can also alter specific histone marks in chromatin. Cr cross-linked HDAC1-DNMT1 complexes to Cyp1a1 promoter-associated chromatin and inhibited histone marks induced by Ah receptormediated gene transactivation, including H3S10p, H3K4me3, and various acetylation marks in histones H3 and H4 (41,42). Also, it has been demonstrated in vitro that Cr interacts with histone arginine and lysine residues (43). Cr increased expression of the H3K9-specific methyltransferase G9a, which was correlated with increased levels of H3K9me3 as well as H3K4 (44). H3K9me occurred not only globally, but also at the promoter of MLH1, a DNA mismatch repair enzyme. Cr exposure decreased expression of MLH1, suggesting that Cr represses expression of this gene through H3K9me and DNA methylation, providing an epigenetic link between Cr and carcinogenesis.
Cr was negatively associated with miR-146a in peripheral blood leukocytes (45). In Cr-transformed human lung epithelial cells, miR-143 is repressed and this repression activates angiogenesis through the IL-8-insulin growth factor receptor (IGFR)-HIF-1 pathway (46). Cr level showed significant association with plasma miR-3940-5p levels. XRCC2 and BRCC3 protein levels, which are DNA repair genes, were positively associated with Cr and micronuclei frequency. These data suggest that Cr-induced genetic damage may be due to the regulation of miRNA on DNA repair genes responsive to high Cr(VI) exposure (47).
Copper. Occupational exposure to copper (Cu) has been reported to result in chromosome aberrations and increased frequency of micronuclei in peripheral blood leukocytes (48). In addition to genomic toxicity, Cu can also alter the epigenome. In human hepatocyte Hep3B cells, Cu treatment induced significant decreases in global acetylation of histone H3 and H4 through direct inhibition of histone acetyltransferase (HAT) activity without affecting histone deacetylase (HDAC) activity (49). Cu can directly bind to with critical histidine residues of histones H2A, H3 and H4 in vitro (50,51). Cu binding to the C-terminal peptide of H2B (H2B94-125) could interfere with the ubiquitination of Lys120, which has been associated with gene silencing (52,53). In addition to histones, Cu interacts with DNA, resulting in induced conformational changes of chromatin and altered gene expression.
Currently, the majority of reports on miRNA expression resulting from Cu exposure have been performed in nonhuman studies. Cu has been shown to alter miRNA level in the plant. miR-398 was the first miRNA identified to be regulated by oxidative stress. Its targets are the Cu/Zn superoxide dismutases (CSD) enzymes and the stress upregulated Nodulin 19 (Nod19) (54). In bean plants under Cu toxicity, miR-398b down-regulation and up-regulation of its target genes, CSD1 and Nod19, were observed (55). CSD1 and Nod19 participate in reactive oxygen species (ROS) detoxification. Down-regulation of miR-398b and up-regulation of its targets was observed in common bean roots during the oxidative stress resulting from short-time exposure to high Cu. Thus, this data suggests that the miR-398bmediated up-regulation of CSD and Nod19 may be relevant for common bean plants to cope with oxidative stress generated in abiotic and biotic stresses.
Lead. Lead (Pb) is a naturally occurring element that is widely known for its human health impacts and is one of the most widely studied environmental toxicants (13). It is a genotoxic agent and has been shown to induce DNA breaks and chromosome aberrations. Exposure to Pb has been primarily associated with adverse effects in a variety of organs, including the neuronal and reproductive systems (56). The relationship between infantile exposure to Pb and adult-onset Alzheimer’s disease (AD) were investigated in primate model systems (57,58). Adult primates demonstrated a significant decrease in DNA methyltransferases, DNMT1 and DNMT3A; as well as a marked reduction in H3K9ac, H4K8ac, H4K12ac and H3K4me2 compared with controls. This epigenetic perturbation caused aberrant upregulation in certain genes important for brain function, such as neuron-derived orphan receptor 1 (NOR1), membrane-associated phospholipase A2 precursor (PLA2) and flavoprotein subunit of complex II. It may play a role in the development of late onset Alzheimer’s disease. Currently, no human studies have investigated histone modification resulting from exposure to environmental Pb.
At present, only one study has addressed changes in miRNA expression upon exposure to Pb. The study found that miR-222 expression was positively associated with Pb exposure, while miR-146a expression was negatively correlated with Pb exposure in peripheral blood leukocytes exposed via inhalation (59). miR-146a, which was downregulated, has been shown to be associated with inflammation in rodent models whereas miR-222, which was up-regulated by Pb exposure, has been shown to regulate the tumor suppressor, Cyclin-dependent kinase inhibitor 1B (p27Kip1). Down-regulation of p26Kip1 has been linked to increased cell proliferation and higher incidences of various cancers (60).
Mercury. Humans are primarily exposed to mercury (Hg) by consuming contaminated seafood and via occupational exposure including metal smelting, gold mining, coal burning, electric industries, and wood production (61,62). Hg has been shown to be genotoxic and can cause damage to neuronal, cardiovascular and renal systems (63). One study found hypermethylation of the promoter region of glutathione S-transferase mu 1 (GSTM1) in women with elevated blood Hg levels (above 2.9 µg/L) without reduced expression levels of GSTM1 (64). Another study showed a correlation between DNA hypomethylation of the selenoprotein P plasma 1 (SEPP1) promoter and increasing Hg levels in male hair samples (65). Currently, the majority of reports on histone modification resulting from exposure to environmental Hg have been performed in non-human studies. In pregnant mouse models, exposure to methylmercury (MeHg) was associated with increased H3K27me3, decreased histone H3 acetylation and increased DNA methylation in the promoter region of brain derived neurotrophic factor (BDNF) (66). Exposure to methyl mercury chloride (MeHgCl) led to increased expression of miR-302b, miR-367, miR-372, miR-196b, and miR-141 in carcinoma pluripotent stem cells (67). This study showed that these miRNAs were associated with developmental processes and cellular responses to stress and pathway analysis on probable mRNA targets revealed possible links to neurological development including learning and memory formation.
Nickel. Both water-soluble (e.g. NiCl2, NiSO4) and water-insoluble (e.g. Ni3S2, NiO) nickel (Ni) compounds are human carcinogens, although it is nonmutagenic or weakly mutagenic in most animals (68). Thus, the mechanism of Ni-induced health effects, including carcinogenicity, is largely unknown. Nickel’s carcinogenic potential is thought to be due to its ability to precipitate epigenetic changes (69). Ni condenses chromatin to a greater extent than Mg, the natural divalent cation of the cell (70). Ni-induced carcinogenesis may be mediated by structural change of chromatin, such as heterochromatinization, which is associated with gene repression. Ni-induced tumorigenesis was associated with promoter hypermethylation of the tumor suppressor gene p16 (71). Ni exposure both in vitro and in vivo was also shown to reduce or silence the expression of another tumor suppressor gene Fhit (72). Ji et al. (2008) investigated the mechanistic link between Ni-induced epigenetic changes and cellular transformation in human bronchial epithelial (16HBE) cells (73). The DNA repair gene, O6-methylguanine DNA methyltransferase (MGMT) exhibits epigenetic changes such as hypermethylation of its promoter, recruitment of MeCP2, MBD2 and DNMT1 to the promoter and transcription-repressive histone modifications including high levels of H3K9me2 and low levels of H4ac and H3K9ac. Ni has been shown to cause a global loss of acetylation at all four core histones and an increase in the repressing methylation maker H3K9me2, but no change in H3K9me3 in human lung A549 cells (74). Niinduced histone hypermethylation is due to the direct inhibition of H3K9 demethylase, a jumonji-domain containing protein 1A (JMJD1A/KDM4A), by replacing the Fe2+ in the catalytic center by Ni2+ (74,75). Ni also induces increased levels of both H2A and 2B ubiquitination (76) and phosphorylation of histone H3S10 in response to stimulation of the JNK-MAPK pathway in A549 cells (77).
Karaczyn et al. reported a different type of effect of Ni on histones (78,79). Like Cu, Ni binds to the C-terminal TESHHKAKGK motif of histone H2A and assists in the hydrolysis of the E-S peptide bond, resulting in the cleavage and release of the terminal octapeptide SHHKAKGK from histone’s C-terminal tail. Similarly, both C- and N-terminal ends of histone H2B are cleaved off in cells exposed to Ni (78). This effect was also induced by cobalt, but not by Cu, Cd and Zn. This data suggests that the truncation of histone H2A and H2B may alter chromatin structure and affect gene expression, which may be involved in mediating Ni toxicity and carcinogenicity.
In Ni-induced tumors, miR-222 was highly expressed and its target genes, CDKN1B and CDKN1C, were down-regulated in Wista rats (80). The same alteration of miR-222 and its target genes was also found in malignant 16HBE cells induced with Ni3S2 compounds. This suggests that miR-222 may promote cell proliferation infinitely during nickelinduced tumorigenesis in part by regulating the expression of its target genes CDKN1B and CDKN1C.
Organotin. The organotin class of compounds includes chemicals with tin covalently bound to hydrocarbon substituents (13). Animal studies using organotin revealed carcinogenicity, neurotoxicity, reproductive and developmental toxicity, immunotoxicity and endocrine activity (81). HAT activity is enhanced by organotins including not only tributylin (TBT), diphenyltin (TPT) and their derivatives but also related compounds such as dibutyltin, diphenyltin, triethyltin and tripropyltin (82). Organotin exposure has also been shown to induce DNA hypomethylation in liver samples from Marble trouts (S. marmoratus) (83). Whether or not the DNA hypomethylation and histone hyperacetylation caused by organotins are correlated with their carcinogenic and tumor-promoting activity in fish or in mammals requires further study, because these two changes can work synergistically, or antagonistically to affect gene transcription (13).
CONCLUSIONS
Epigenetic changes in the form of DNA methylation, histone modification or ncRNA expression have been linked to environmental factors, including heavy metal exposure (Table 1 and Fig. 1) and in some cases to various human diseases such as cancer. Due to the fact that epigenetic changes can occur at any time in an individual’s lifetime and that they can be passed on to future generations, the exact mechanisms by which metals induce these changes are of great significance (84). There remain significant questions regarding the mechanisms through which environmental exposure drives or selects epigenetic alteration. Nevertheless, only a few mechanisms, such as oxidative stress induction, disruption of cell metabolism or epigenetic changes, have been proposed (84-86). Among these mechanisms, the mechanisms that induce epigenetic alterations as well as the changes themselves could be used as biomarkers for the prognosis and the diagnosis of several diseases. Also, the understanding and characterization of heavy metal-induced epigenetic changes will be of great significance in prognosis, diagnosis, and treatment of disease and drug development, with respect to the control of DNA methylation secondary to toxic factor exposure.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2014R1A1A2054026).
References
- 1.Baccarelli A., Bollati V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. (2009);21:243–251. doi: 10.1097/MOP.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koturbash I., Beland F.A., Pogribny I.P. Role of epigenetic events in chemical carcinogenesis--a justification for incorporating epigenetic evaluations in cancer risk assessment. Toxicol. Mech. Methods. (2011);21:289–297. doi: 10.3109/15376516.2011.557881. [DOI] [PubMed] [Google Scholar]
- 3.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. (2002);16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 4.Heyn H., Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat. Rev. Genet. (2012);13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 5.De Carvalho D.D., You J.S., Jones P.A. DNA methylation and cellular reprogramming. Trends Cell Biol. (2010);20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suganuma T., Workman J.L. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. (2011);80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 7.Hon G.C., Hawkins R.D., Ren B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. (2009);18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S., Antosiewicz-Bourget J., Ye Z., Espinoza C., Agarwahl S., Shen L., Ruotti V., Wang W., Stewart R., Thomson J.A., Ecker J.R., Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. (2010);6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyn H., Carmona F.J., Gomez A., Ferreira H.J., Bell J.T., Sayols S., Ward K., Stefansson O.A., Moran S., Sandoval J., Eyfjord J.E., Spector T.D., Esteller M. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis. (2013);34:102–108. doi: 10.1093/carcin/bgs321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhuri S., Cui Y., Klaassen C.D. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol. Appl. Pharmacol. (2010);245:378–393. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. (2011);39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell J.T., Olson E.N. MicroRNAs in stress signaling and human disease. Cell. (2012);148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng T.F., Choudhuri S., Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: a review of the literature. J. Appl. Toxicol. (2012);32:643–653. doi: 10.1002/jat.2717. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta S., McArthur J.M., Sarkar A., Leng M.J., Ravenscroft P., Howarth R.J., Banerjee D.M. Do ponds cause arsenic-pollution of groundwater in the Bengal basin? An answer from West Bengal. Environ. Sci. Technol. (2008);42:5156–5164. doi: 10.1021/es702988m. [DOI] [PubMed] [Google Scholar]
- 15.Zhong C.X., Mass M.J. Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol. Lett. (2001);122:223–234. doi: 10.1016/S0378-4274(01)00365-4. [DOI] [PubMed] [Google Scholar]
- 16.Salnikow K., Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem. Res. Toxicol. (2008);21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J., Ye J., Zhao X., Li A., Zhou J. JWA is required for arsenic trioxide induced apoptosis in HeLa and MCF-7 cells via reactive oxygen species and mitochondria linked signal pathway. Toxicol. Appl. Pharmacol. (2008);230:33–40. doi: 10.1016/j.taap.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L., Hou J., Fu W., Wang D., Yuan Z., Jiang H. Arsenic trioxide and 2-methoxyestradiol reduce betacatenin accumulation after proteasome inhibition and enhance the sensitivity of myeloma cells to Bortezomib. Leuk. Res. (2008);32:1674–1683. doi: 10.1016/j.leukres.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 19.Benton M.A., Rager J.E., Smeester L., Fry R.C. Comparative genomic analyses identify common molecular pathways modulated upon exposure to low doses of arsenic and cadmium. BMC Genomics. (2011);12:173. doi: 10.1186/1471-2164-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chervona Y., Hall M.N., Arita A., Wu F., Sun H., Tseng H.C., Ali E., Uddin M.N., Liu X., Zoroddu M.A., Gamble M.V., Costa M. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol. Biomarkers Prev. (2012);21:2252–2260. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantone L., Nordio F., Hou L., Apostoli P., Bonzin M., Tarantini L., Angelici L., Bollati V., Zanobetti A., Schwartz J., Bertazzi P.A., Baccarelli A. Inhalable metalrich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ. Health Perspect. (2011);119:964–969. doi: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J., Gorospe M., Barnes J., Liu Y. Tumor promoter arsenite stimulates histone H3 phosphoacetylation of proto-oncogenes c-fos and c-jun chromatin in human diploid fibroblasts. J. Biol. Chem. (2003);278:13183–13191. doi: 10.1074/jbc.M300269200. [DOI] [PubMed] [Google Scholar]
- 23.Huang B.W., Ray P.D., Iwasaki K., Tsuji Y. Transcriptional regulation of the human ferritin gene by coordinated regulation of Nrf2 and protein arginine methyltransferases PRMT1 and PRMT4. FASEB J. (2013);27:3763–3774. doi: 10.1096/fj.12-226043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez T., Brocher J., Stopper H., Hock R. Sodium arsenite modulates histone acetylation, histone deacetylase activity and HMGN protein dynamics in human cells. Chromosoma. (2008);117:147–157. doi: 10.1007/s00412-007-0133-5. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X., Sun H., Ellen T.P., Chen H., Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis. (2008);29:1831–1836. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsit C.J., Eddy K., Kelsey K.T. MicroRNA responses to cellular stress. Cancer Res. (2006);66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Zhao Y., Smith E., Goodall G.J., Drew P.A., Brabletz T., Yang C. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol. Sci. (2011);121:110–122. doi: 10.1093/toxsci/kfr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellez-Plaza M., Navas-Acien A., Menke A., Crainiceanu C.M., Pastor-Barriuso R., Guallar E. Cadmium exposure and all-cause and cardiovascular mortality in the U.S. general population. Environ. Health Perspect. (2012);120:1017–1022. doi: 10.1289/ehp.1104352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Järup L., Akesson A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. (2009);238:201–208. doi: 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Takiguchi M., Achanzar W.E., Qu W., Li G., Waalkes M.P. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. (2003);286:355–365. doi: 10.1016/S0014-4827(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 31.Benbrahim-Tallaa L., Waterland R.A., Dill A.L., Webber M.M., Waalkes M.P. Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ. Health Perspect. (2007);115:1454–1459. doi: 10.1289/ehp.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somji S., Garrett S.H., Toni C., Zhou X.D., Zheng Y., Ajjimaporn A., Sens M.A., Sens D.A. Differences in the epigenetic regulation of MT-3 gene expression between parental and Cd+2 or As+3 transformed human urothelial cells. Cancer Cell Int. (2011);11:2. doi: 10.1186/1475-2867-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barcia-Sanjurjo I., Vazquez-Cedeira M., Barcia R., Lazo P.A. Sensitivity of the kinase activity of human vaccinia-related kinase proteins to toxic metals. J. Biol. Inorg. Chem. (2013);18:473–482. doi: 10.1007/s00775-013-0992-6. [DOI] [PubMed] [Google Scholar]
- 34.Hassan F., Nuovo G.J., Crawford M., Boyaka P.N., Kirkby S., Nana-Sinkam S.P., Cormet-Boyaka E. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PloS One. (2012);7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langåvdrd S. One hundred years of chromium and cancer: a review of epidemiological evidence and selected case reports. Am. J. Ind. Med. (1990);17:189–215. doi: 10.1002/ajim.4700170205. [DOI] [PubMed] [Google Scholar]
- 36.Nickens K.P., Patierno S.R., Ceryak S. Chromium genotoxicity: A double-edged sword. Chem. Biol. Interact. (2010);188:276–288. doi: 10.1016/j.cbi.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X., Mao Y., Knapton A.D., Ding M., Rojanasakul Y., Gannett P.M., Dalal N., Liu K. Reaction of Cr(VI) with ascorbate and hydrogen peroxide generates hydroxyl radicals and causes DNA damage: role of a Cr(IV)-mediated Fenton-like reaction. Carcinogenesis. (1994);15:2475–2478. doi: 10.1093/carcin/15.11.2475. [DOI] [PubMed] [Google Scholar]
- 38.Zhitkovich A. Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium(VI). Chem. Res. Toxicol. (2005);18:3–11. doi: 10.1021/tx049774+. [DOI] [PubMed] [Google Scholar]
- 39.Kondo K., Takahashi Y., Hirose Y., Nagao T., Tsuyuguchi M., Hashimoto M., Ochiai A., Monden Y., Tangoku A. The reduced expression and aberrant methylation of p16(INK4a) in chromate workers with lung cancer. Lung Cancer. (2006);53:295–302. doi: 10.1016/j.lungcan.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Ali A.H., Kondo K., Namura T., Senba Y., Takizawa H., Nakagawa Y., Toba H., Kenzaki K., Sakiyama S., Tangoku A. Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol. Carcinog. (2011);50:89–99. doi: 10.1002/mc.20697. [DOI] [PubMed] [Google Scholar]
- 41.Schnekenburger M., Peng L., Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim. Biophys. Acta. (2007);1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnekenburger M., Talaska G., Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol. Cell. Biol. (2007);27:7089–7101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levina A., Harris H.H., Lay P.A. Binding of chromium(VI) to histones: implications for chromium(VI)-induced genotoxicity. J. Biol. Inorg. Chem. (2006);11:225–234. doi: 10.1007/s00775-005-0068-3. [DOI] [PubMed] [Google Scholar]
- 44.Fan J., Sun Y., Wang J., Fan M. An organicreagent-free method for determination of chromium(VI) in steel alloys, sewage sludge and wastewater. Anal. Chim. Acta. (2009);640:58–62. doi: 10.1016/j.aca.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Bollati V., Marinelli B., Apostoli P., Bonzini M., Nordio F., Hoxha M., Pegoraro V., Motta V., Tarantini L., Cantone L., Schwartz J., Bertazzi P.A., Baccarelli A. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ. Health Perspect. (2010);118:763–768. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He J., Qian X., Carpenter R., Xu Q., Wang L., Qi Y., Wang Z.X., Liu L.Z., Jiang B.H. Repression of miR-143 mediates Cr (VI)-induced tumor angiogenesis via IGF-IR/IRS1/ERK/IL-8 pathway. Toxicol. Sci. (2013);134:26–38. doi: 10.1093/toxsci/kft101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Li P., Yu S., Zhang J., Wang T., Jia G. miR-3940-5p associated with genetic damage in workers exposed to hexavalent chromium. Toxicol. Lett. (2014);229:319–326. doi: 10.1016/j.toxlet.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 48.De Olivera J.V., Boufleur L.A., Dos Santos C.E., Dias J.F., Squeff C.H., Silva G.R., Ianistcki M., Benvegnú V.C., Da Silva J. Occupational genotoxicity among copper smelters. Toxicol. Ind. Health. (2012);28:789–795. doi: 10.1177/0748233711422735. [DOI] [PubMed] [Google Scholar]
- 49.Kang J., Lin C., Chen J., Liu Q. Copper induces histone hypoacetylation through directly inhibiting histone acetyltransferase activity. Chem. Biol. Interact. (2004);148:115–123. doi: 10.1016/j.cbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Zoroddu M.A., Kowalik-Jankowska T., Kozlowski H., Molinari H., Salnikow K., Broday L., Costa M. Interaction of Ni(II) and Cu(II) with a metal binding sequence of histone H4: AKRHRK, a model of the H4 tail. Biochim. Biophys. Acta. (2000);1475:163–168. doi: 10.1016/S0304-4165(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 51.Karavelas T., Mylonas M., Malandrinos G., Plakatouras J.C., Hadjiliadis N., Mlynarz P., Kozlowski H. Coordination properties of Cu(II) and Ni(II) ions towards the C-terminal peptide fragment -ELAKHA- of histone H2B. J. Inorg. Biochem. (2005);99:606–615. doi: 10.1016/j.jinorgbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 52.Zavitsanos K., Nunes A.M., Malandrinos G., Hadjiliadis N. Copper effective binding with 32-62 and 94-125 peptide fragments of histone H2B. J. Inorg. Biochem. (2011);105:102–110. doi: 10.1016/j.jinorgbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Nunes A.M., Zavitsanos K., Malandrinos G., Hadjiliadis N. Coordination of Cu(2+) and Ni(2+) with the histone model peptide of H2B N-terminal tail (1-31 residues): A spectroscopic study. Dalton Trans. (2010);39:4369–4381. doi: 10.1039/b927157k. [DOI] [PubMed] [Google Scholar]
- 54.Bonnet E., Wuyts J., Rouze P., Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. U.S.A. (2004);101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naya L., Paul S., Valdés-López O., Mendoza-Soto A.B., Nova-Franco B., Sosa-Valencia G., Reyes J.L., Hernández G. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PloS One. (2014);9:e84416. doi: 10.1371/journal.pone.0084416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasuba V., Rozgaj R., Milić M., Zeljezić D., Kopjar N., Pizent A., Kljaković-Gaspić Z. Evaluation of lead exposure in battery-manufacturing workers with focus on different biomarkers. J. Appl. Toxicol. (2010);30:321–328. doi: 10.1002/jat.1497. [DOI] [PubMed] [Google Scholar]
- 57.Wu J., Basha M.R., Brock B., Cox D.P., Cardozo-Pelaez F., McPherson C.A., Harry J., Rice D.C., Maloney B., Chen D., Lahiri D.K., Zawia N.H. Alzheimer's disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J. Neurosci. (2008);28:3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bihaqi S.W., Huang H., Wu J., Zawia N.H. Infant exposure to lead (Pb) and epigenetic modifications in the aging primate brain: implications for Alzheimer’s disease. J. Alzheimers Dis. (2011);27:819–833. doi: 10.3233/JAD-2011-111013. [DOI] [PubMed] [Google Scholar]
- 59.Wright R.O., Schwartz J., Wright R.J., Bollati V., Tarantini L., Park S.K., Hu H., Sparrow D., Vokonas P., Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ. Health Perspect. (2010);118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.le Sage C., Nagel R., Egan D.A., Schrier M., Mesman E., Mangiola A., Anile C., Mercatelli N., Ciafrè S.A., Farace M.G., Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. (2007);26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tchounwou P.B., Ayensu W.K., Ninashvili N., Sutton D. Environmental exposure to mercury and its toxicopathologic implications for public health. Environ. Toxicol. (2003);18:149–175. doi: 10.1002/tox.10116. [DOI] [PubMed] [Google Scholar]
- 62.Spiegel S.J. Occupational health, mercury exposure, and environmental justice: learning from experiences in Tanzania. Am. J. Public Health. (2009);99(Suppl 3):S550–558. doi: 10.2105/AJPH.2008.148940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grotto D., Valentini J., Fillion M., Passos C.J., Garcia S.C., Mergler D., Barbosa F., Jr. Mercury exposure and oxidative stress in communities of the Brazilian Amazon. Sci. Total Environ. (2010);408:806–811. doi: 10.1016/j.scitotenv.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 64.Hanna C.W., Bloom M.S., Robinson W.P., Kim D., Parsons P.J., vom Saal F.S., Taylor J.A., Steuerwald A.J., Fujimoto V.Y. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum. Reprod. (2012);27:1401–1410. doi: 10.1093/humrep/des038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodrich J.M., Basu N., Franzblau A., Dolinoy D.C. Mercury biomarkers and DNA methylation among Michigan dental professionals. Environ. Mol. Mutagen. (2013);54:195–203. doi: 10.1002/em.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Onishchenko N., Karpova N., Sabri F., Castrén E., Ceccatelli S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J. Neurochem. (2008);106:1378–1387. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- 67.Pallocca G., Fabbri M., Sacco M.G., Gribaldo L., Pamies D., Laurenza I., Bal-Price A. miRNA expression profiling in a human stem cell-based model as a tool for developmental neurotoxicity testing. Cell Biol. Toxicol. (2013);29:239–257. doi: 10.1007/s10565-013-9250-5. [DOI] [PubMed] [Google Scholar]
- 68.Sutherland J.E., Costa M. Epigenetics and the environment. Ann. N. Y. Acad. Sci. (2003);983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 69.Arita A., Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. (2009);1:222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellen T.P., Kluz T., Harder M.E., Xiong J., Costa M. Heterochromatinization as a potential mechanism of nickel-induced carcinogenesis. Biochemistry. (2009);48:4626–4632. doi: 10.1021/bi900246h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Govindarajan B., Klafter R., Miller M.S., Mansur C., Mizesko M., Bai X., LaMontagne K., Jr., Arbiser J.L. Reactive oxygen-induced carcinogenesis causes hypermethylation of p16(Ink4a) and activation of MAP kinase. Mol. Med. (2002);8:1–8. doi: 10.1007/s00894-001-0054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kowara R., Salnikow K., Diwan B.A., Bare R.M., Waalkes M.O., Kasprzak K.S. Reduced Fhit protein expression in nickel-transformed mouse cells and in nickelinduced murine sarcomas. Mol. Cell. Biochem. (2004);255:195–202. doi: 10.1023/B:MCBI.0000007275.22785.91. [DOI] [PubMed] [Google Scholar]
- 73.Ji W., Yang L., Yu L., Yuan J., Hu D., Zhang W., Yang J., Pang Y., Li W., Lu J., Fu J., Chen J., Lin Z., Chen W., Zhuang Z. Epigenetic silencing of O6-methylguanine DNA methyltransferase gene in NiS-transformed cells. Carcinogenesis. (2008);29:1267–1275. doi: 10.1093/carcin/bgn012. [DOI] [PubMed] [Google Scholar]
- 74.Chen H., Ke Q., Kluz T., Yan Y., Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell. Biol. (2006);26:3728–3737. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X., Fan Y., Long X., Sun X. Similar DNA methylation and histone H3 lysine 9 dimethylation patterns in tripronuclear and corrected bipronuclear human zygotes. J. Reprod. Dev. (2010);56:324–329. doi: 10.1262/jrd.09-170A. [DOI] [PubMed] [Google Scholar]
- 76.Ke Q., Ellen T.P., Costa M. Nickel compounds induce histone ubiquitination by inhibiting histone deubiquitinating enzyme activity. Toxicol. Appl. Pharmacol. (2008);228:190–199. doi: 10.1016/j.taap.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ke Q., Li Q., Ellen T.P., Sun H., Costa M. Nickel compounds induce phosphorylation of histone H3 at serine 10 by activating JNK-MAPK pathway. Carcinogenesis. (2008);29:1276–1281. doi: 10.1093/carcin/bgn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karaczyn A.A., Bal W., North S.L., Bare R.M., Hoang V.M., Fisher R.J., Kasprzak K.S. The octapeptidic end of the C-terminal tail of histone H2A is cleaved off in cells exposed to carcinogenic nickel(II). Chem. Res. Toxicol. (2003);16:1555–1559. doi: 10.1021/tx0300277. [DOI] [PubMed] [Google Scholar]
- 79.Karaczyn A.A., Golebiowski F., Kasprzak K.S. Truncation, deamidation, and oxidation of histone H2B in cells cultured with nickel(II). Chem. Res. Toxicol. (2005);18:1934–1942. doi: 10.1021/tx050122a. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J., Zhou Y., Ma L., Huang S., Wang R., Gao R., Wu Y., Shi H., Zhang J. The alteration of miR-222 and its target genes in nickel-induced tumor. Biol. Trace Elem. Res. (2013);152:267–274. doi: 10.1007/s12011-013-9619-6. [DOI] [PubMed] [Google Scholar]
- 81.Boyer I.J. Toxicity of dibutyltin, tributyltin and other organotin compounds to humans and to experimental animals. Toxicology. (1989);55:253–298. doi: 10.1016/0300-483X(89)90018-8. [DOI] [PubMed] [Google Scholar]
- 82.Osada S., Nishikawa J., Nakanishi T., Tanaka K., Nishihara T. Some organotin compounds enhance histone acetyltransferase activity. Toxicol. Lett. (2005);155:329–335. doi: 10.1016/j.toxlet.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y., Wang C., Zhang J., Chen Y., Zuo Z. DNA hypomethylation induced by tributyltin, triphenyltin, and a mixture of these in Sebastiscus marmoratus liver. Aquat. Toxicol. (2009);95:93–98. doi: 10.1016/j.aquatox.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 84.Fragou D., Fragou A., Kouidou S., Njau S., Kovatsi L. Epigenetic mechanisms in metal toxicity. Toxicol. Mech. Methods. (2011);21:343–352. doi: 10.3109/15376516.2011.557878. [DOI] [PubMed] [Google Scholar]
- 85.Coppin J.F., Qu W., Waalkes M.P. Interplay between cellular methyl metabolism and adaptive efflux during oncogenic transformation from chronic arsenic exposure in human cells. J. Biol. Chem. (2008);283:19342–19350. doi: 10.1074/jbc.M802942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franco R., Schoneveld O., Georgakilas A.G., Panayiotidis M.I. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. (2008);266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]