Abstract
The DNA transposon piggyBac is a potential therapeutic agent for multiple genetic diseases such as cystic fibrosis (CF). Recombinant piggyBac transposon and transposase are typically codelivered by plasmid transfection; however, plasmid delivery is inefficient in somatic cells in vivo and is a barrier to the therapeutic application of transposon-based vector systems. Here, we investigate the potential for hybrid piggyBac/viral vectors to transduce cells and support transposase-mediated genomic integration of the transposon. We tested both adenovirus (Ad) and adeno-associated virus (AAV) as transposon delivery vehicles. An Ad vector expressing hyperactive insect piggyBac transposase (iPB7) was codelivered. We show transposase-dependent transposition activity and mapped integrations in mammalian cells in vitro and in vivo from each viral vector platform. We also demonstrate efficient and persistent transgene expression following nasal delivery of piggyBac/viral vectors to mice. Furthermore, using piggyBac/Ad expressing Cystic Fibrosis transmembrane Conductance Regulator (CFTR), we show persistent correction of chloride current in well-differentiated primary cultures of human airway epithelial cells derived from CF patients. Combining the emerging technologies of DNA transposon-based vectors with well-studied adenoviral and AAV delivery provides new tools for in vivo gene transfer and presents an exciting opportunity to increase the delivery efficiency for therapeutic genes such as CFTR.
Introduction
Our goal for gene therapy vector development for life-long genetic diseases such as cystic fibrosis (CF) is to create a vehicle with the ability to efficiently, safely, and persistently express a transgene in the appropriate cell types.1,2 There are multiple viral-based vectors for delivering genes to airway epithelia. Each system has its advantages and disadvantages. Nonviral vectors provide an expanded tool-set for gene transfer to cells. For example, research with the “cut-and-paste” DNA transposon Sleeping Beauty pioneered the use of a recombinant transposon and transposase to achieve genomic integration of a transgene.3 Sleeping Beauty-mediated gene transfer resulted in functional correction of coagulation factor deficiencies,4,5,6 lysosomal storage disease,7 as well as in cancer therapeutics.8
PiggyBac is also a DNA transposon and a promising alternative to Sleeping Beauty. Similar to Sleeping Beauty, piggyBac is highly active when introduced into mammalian cells9,10,11 mediates long-term expression in vivo12,13 and is a potential therapeutic agent for multiple genetic diseases such as CF.14,15 Recombinant piggyBac transposon and transposase are typically codelivered by plasmid transfection; however, the greatest barrier of delivering naked DNA is inefficient delivery to somatic cells in vivo.
Yant et al.16 incorporated the Sleeping Beauty integration machinery into Ad vectors. They observed that the Ad genome needed to circularize before releasing the Sleeping Beauty transposon.17,18 They employed a Flp recombination system to drive circularization of the Ad genome in target cells. Here, we explore the potential utility of adenoviral (Ad)- or adeno associated virus (AAV)-based vectors to deliver piggyBac components to airway epithelia without an additional recombination step. For these studies, a second adenoviral vector expressing hyperactive insect piggyBac transposase (iPB7) is codelivered to achieve transposase-mediated genomic integration of the transposon. These novel hybrid vector systems provide valuable additional tools for in vivo gene transfer.
Results
DNA transposition from a piggyBac/AAV vector into the genome
Hybrid piggyBac/AAV vectors expressing the mCherry reporter and puromycin resistance genes were generated (shown schematically, Figure 1a; Supplementary Figure S1). For these studies, iPB7 was codelivered with an Ad5 vector (Ad-iPB7). To determine if iPB7 mobilized the DNA cargo from the piggyBac/AAV vector and catalyze integration into the genome, colony formation assays were performed in a mammalian cell line (Figure 1b). Colony formation assays are a widely used indirect measure of transposition efficiency. PiggyBac/AAV was delivered to HeLa cells at multiplicity of infections (MOIs) ranging from 100 to 100,000. Ad-iPB7 was simultaneously delivered at MOIs ranging from 0 to 100. Following vector transduction, cells were selected for puromycin resistance for ~2 weeks and colonies counted. Puromycin-resistant colony formation is indicative of a successful integration event. In the absence of Ad-iPB7, we observed a dose-dependent increase in puromycin resistant colonies, suggesting that there is baseline level of integration from an AAV vector alone, consistent with previous observations.19 However, in the presence of Ad-iPB7, the colony numbers increased dramatically.
Figure 1.
piggyBac/AAV transduction and colony formation assays. (a) Schematic representation of the piggyBac transposon expressing mCherry and the puromycin resistance gene, puromycin N-acetyl-transferase (Puror), driven by an RSV promoter and delivered by AAV2/5. Hyperactive piggyBac transposase (iPB7) is delivered in trans by an Ad5 vector. ITR, inverted terminal repeat of AAV; TR, piggyBac terminal repeat. (b) HeLa cells were cotransduced with increasing vector multiplicity of infections as indicated. Cells were selected for 10–14 days with 0.5 µg/ml puromycin, stained with methylene blue, and the drug resistant colonies were counted.
As an additional control to confirm that the presence of Ad vector does not enhance piggyBac/AAV conferred colony formation in our assay, we substituted Ad-iPB7 with Ad-GFP (Figure 1b). In the presence of Ad-GFP, the number of puromycin-resistant colonies closely resembled “no transposase” group. These data suggest that iPB7 is necessary to achieve an increase in puromycin resistant colony formation.
In general, the levels of transposition were dose-dependent; however, the piggyBac/AAV (MOI 10,000)/Ad-iPB7 (MOI 100) condition (ratio 100:1) resulted in fewer colonies than the piggyBac/AAV (MOI 10,000)/Ad-PB7 (MOI 10) condition (ratio 1000:1). This result is perhaps counterintuitive because one would expect that if the transposon is constant and the transposase is increased, increased transposition should occur. Yet, we observed that a 1,000:1 ratio consistently resulted in the best fold-increase of colonies over background. This may be the result, in part, of cellular toxicity associated with high MOI delivery of adenoviral vector or overexpression inhibition.
To determine if high MOIs of adenoviral vector might lead to decreased colony counts, we delivered Ad-GFP to HeLa cells at MOIs of 0, 1, 2, 10, 20, 100, and 200 and used flow cytometry to quantify live cells 24 hours later. There was very little variation in the percentage of live cells in all groups. The range was from 94.8 to 97.5% (Supplementary Figure S2). Interestingly, the percentage of GFP-positive cells plateaued at an MOI of 20 (91.1%) and dropped off at MOI 200 (78.7%). These data suggest that the highest nontoxic MOI of adenoviral vector may not be the optimal dose for maximal gene expression.
To confirm transposase-mediated genomic integration, cellular DNA was purified and libraries were generated using LAM-PCR as described in Materials and Methods. The libraries were shotgun-cloned and Sanger sequenced. Of the 54 sequences, ~2/3 (37/54) were bona fide transposase-mediated genomic integrations and the remaining ~1/3 (17/54) were recovered vector. A transposase-mediated integration event is defined by the tell-tale sign of a precise junction between the transposon terminal repeat and genomic DNA occurring at a TTAA. A “no transposase control” was not included because not enough puromycin-resistant colonies could be recovered to yield enough DNA to generate the libraries. Based on the sequencing data, nontransposase-mediated genomic integrations cannot be distinguished from episomal vectors. All but three of the genomic integrations occurred at canonical TTAA sites. The remaining three integrations occurred at either CTAA (1 integration) or TTAG (2 integrations) sites. Low-level NTAA or TTAN integration is consistent with plasmid delivered piggyBac transposon.15,20 These data strongly suggest that delivery with AAV allows for transposase-mediated integration of the piggyBac transposon.
Next, we deep sequenced the libraries using the Illumina HiSeq 2000 platform and recovered 19,059 reads from piggyBac/AAV-transduced HeLa cells and compared the results to 31,078 reads recovered from HeLa cells that were transfected with standard piggyBac transposon and iPB7 expression plasmids. For these analyses, we only included verified genomic integrations that included the TTAA junction between the piggyBac terminal repeat and mappable human genomic sequence. Multiple metrics were used to determine if the piggyBac integration patterns were altered following delivery by an AAV vector. Metrics included distance from transcription start sites, oncogenes, DNase hypersensitive sites, CpG islands, gene density, gene expression, and GC content (Supplementary Figure S3). We observed only subtle differences in the integration profile between AAV- and plasmid-delivered piggyBac transposons or computationally selected matched random controls. There was a very modest increase in integrations near gene dense regions or CpG islands and piggyBac/AAV had a modest decrease in integrations near transcriptional start sites.
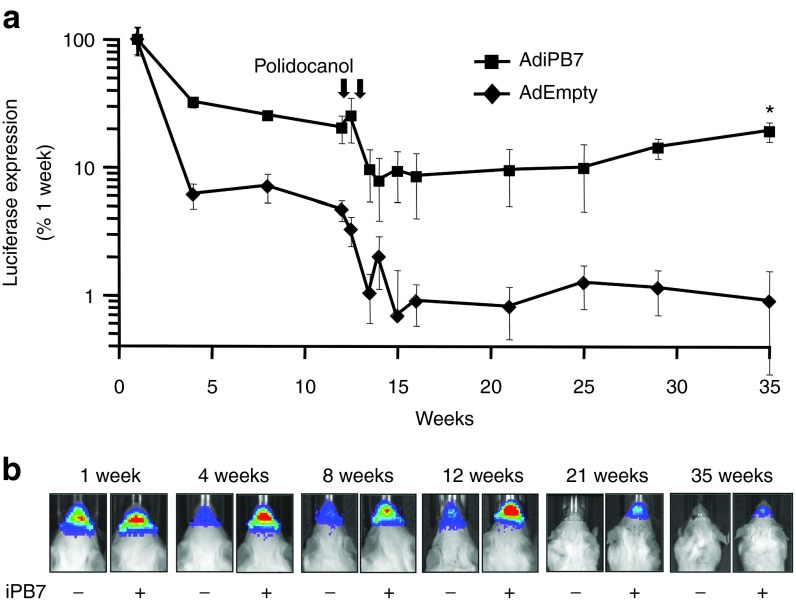
To further test the utility of this hybrid system in vivo, piggyBac/AAV vector expressing firefly luciferase was evaluated for its ability to deliver and transpose into the genome of murine conducting airways. A dose of 9 × 1010 vector genomes (vg) formulated with 1% methylcellulose21 was delivered via nasal instillation to immunocompetent Balb/c mice. Ad-iPB7 was codelivered at a dose of 9 × 107 pfu, achieving the 1,000:1 ratio as determined in vitro. These doses were chosen as a maximal titer that could be delivered in a 50 µl volume. Control mice were untreated (naive) or received Ad-Empty in place of Ad-iPB7. Beginning 4 days post-transduction, luciferase expression was measured at the indicated intervals using a Xenogen CCD camera imaging system (Figure 2a,b).
Figure 2.
AAV delivered piggyBac transposon mediates persistent transgene expression in mouse airways. (a) piggyBac/AAV2/5 expressing firefly luciferase was co-delivered to the nasal airways with Ad-iPB7 or Ad-Empty formulated with 2% methylcellulose. At the indicated time points, mice were given luciferin via i.p. injection and luciferase expression was quantified by bioluminescent imaging. At 12 weeks postdelivery, mice received two doses of polidocanol via nasal delivery, as indicated by arrows and described in Materials and Methods. Luciferase expression was measured as photons/sec/cm2 and is reported as a percentage of levels from week one. *P = 0.002, n = 10. (b) Representative in vivo imaging system images at selected time points are shown.
Both in the presence and absence of iPB7, a decline in nasal bioluminescence was observed between 1 week and 4 weeks postdelivery. The subsequent luciferase expression in animals that received iPB7 stabilized at ~30% initial levels, whereas, expression in animals without iPB7 stabilized at ~6% initial expression. After 12 weeks, the surface epithelial cells were ablated by two consecutive treatments with the detergent 2% polidocanol.22 Monitoring of the bioluminescent expression resumed 2 days following the second polidocanol treatment. Ablation of the surface epithelia is followed by a rapid proliferation and repopulation phase. This procedure helps distinguish between stable integration events in progenitor cells from episomal expression. The luciferase expression in animals that received iPB7 restabilized at ~10% of starting levels and the expression in animals without iPB7 dropped to ~1% of the first timepoint (Figure 2a). These data suggest that the transposase catalyzed piggyBac/AAV integration into a progenitor population of nasal airway cells in mice.
We isolated nasal septa from mice 12 months after being transduced with piggyBac/AAV and Ad-iPB7. Using a similar protocol as described for our in vitro integration library generation, we performed genomic DNA isolation followed by LAM-PCR. Using shot-gun cloning and Sanger sequencing, we mapped 47 piggyBac genomic integrations (Supplementary Figure S4, red arrows). Interestingly, we observed a much lower frequency of recovered vector (1 out of 48). This change in ratios may point to an in vitro sequencing artifact that may not be relevant in vivo.
DNA transposition from a piggyBac/Ad vector into the genome
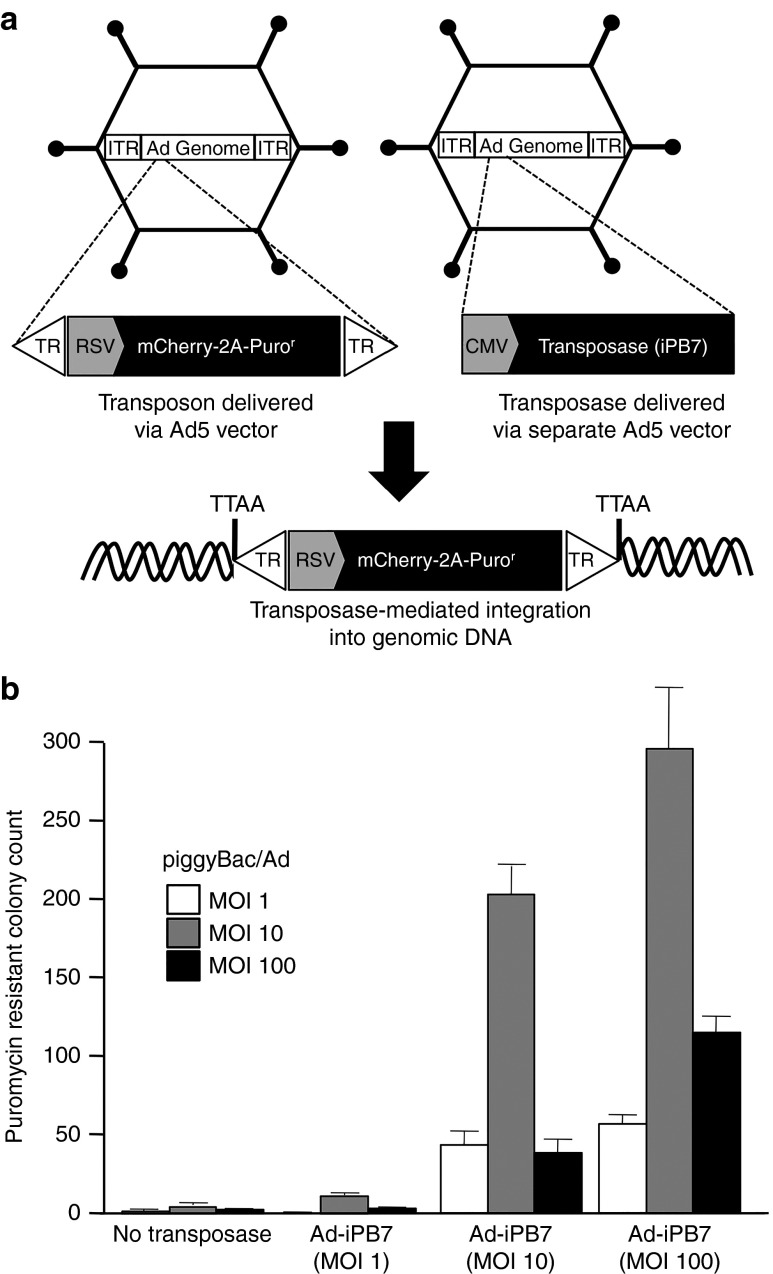
To determine if an Ad5-based viral vector is a suitable delivery vehicle for piggyBac transposon, we inserted the transposon sequence into the E1 region of a first-generation Ad5 vector. Similar to the AAV vector, the transposon expressed mCherry and puromycin resistance genes separated by a T2A element (Figure 3a; Supplementary Figure S1). Transposase-mediated genomic integration was determined by colony formation assays (Figure 3b). Unlike piggyBac/AAV, very few puromycin-resistant colonies were observed for piggyBac/Ad in the absence of iPB7. Interestingly, a piggyBac/Ad MOI of 10 was optimal for each increasing dose of Ad-iPB7. Addition of Ad-iPB7 (MOI = 100) resulted in ~150-fold increase in colony count as compared to piggyBac/Ad without Ad-iPB7. The decreased colony formation counts in the high-dose groups may be the result, in part, of cellular toxicity associated with high MOI delivery of adenoviral vector.
Figure 3.
piggyBac/Ad transduction and colony formation assays. (a) Schematic representation of the piggyBac transposon expressing mCherry and the puromycin resistance gene, puromycin N-acetyl-transferase (Puror), driven by an RSV promoter and delivered by Ad5 vector. iPB7 is delivered in trans by an Ad5 vector. (b) HeLa cells were cotransduced with increasing multiplicity of infections of vector as indicated. Cells were selected for 10–14 days with 0.5 µg/ml puromycin, stained with methylene blue, and the drug resistant colonies were counted.
Transposase-mediated genomic integration in HeLa cells was again verified by LAM-PCR, shotgun cloning, and Sanger sequencing. Of the 65 informative sequences, 23.1% (15/65) were recovered vector and 76.9% (50/65) were confirmed genomic integrations. All but four of the genomic integrations occurred at canonical TTAA sites. The remaining four integrations occurred at TTAG, CTAA (two integrations), or TCAA sites. While the sample size is too small to draw conclusions concerning integration patterns, these data suggest that, like AAV, Ad is a functional delivery vehicle for piggyBac transposon.
To quantify gene transfer efficiencies, piggyBac/Ad expressing firefly luciferase was delivered to SCID mouse airways via nasal instillation. The bioluminescent signal was quantified using a CCD camera 5 minutes following intraperitoneal (i.p.) luciferin delivery. As shown (Figure 4a), in the presence of iPB7, expression stabilized by 8 weeks postdelivery at ~20–30% of the initial time point. In the absence of iPB7, expression continued to decline to ~2% of the initial time point. These results suggest that iPB7 confers persistent expression from a piggyBac/Ad vector in the airways of immunodeficient mice in vivo. Replicate studies in immunocompetent Balb/c mice resulted in loss of expression to naive levels, regardless of the presence or absence of transposase, by 4 weeks postdelivery (Supplementary Figure S5).
Figure 4.

Ad delivered piggyBac transposon mediates persistent transgene expression in mouse airways. (a) piggyBac/Ad expressing firefly luciferase was codelivered to the nasal airways with Ad-iPB7 or Ad-Empty formulated with 2% methylcellulose. At the indicated time points, mice were given luciferin via i.p. injection and luciferase expression was quantified by bioluminescent imaging. Luciferase expression was measured as photons/sec/cm2 and is reported as a percentage of levels from week one. *P < 0.001, n = 10. (b–i) Histological analysis of the airways was performed on SCID mice that received nasal delivery of piggyBac/Ad expressing mCherry. (b) Fluorescent expression and (c) hematoxylin and eosin (H&E) images of naïve mouse lung sections of age matched control mice. (d,f) Representative fluorescent expression and (e,g) H&E images of mice 1 week postdelivery of piggyBac/Ad-mCherry and Ad-iPB7. (f,g) Higher power images of boxed areas in d and e, respectively. (h) Mouse airways 21 weeks postdelivery of piggyBac/Ad-mCherry and Ad-iPB7. (i) Mouse airways 21 weeks postdelivery of piggyBac/Ad-mCherry and Ad-Empty. (j) Fluorescent expression and (k) hematoxylin and eosin (H&E) images of airways of mice 1 year postdelivery of piggyBac/Ad-mCherry and Ad-iPB7. Arrow indicates rare mCherry positive cell. Scale bar = 500 µm. Asterisks indicate large airways. A DAPI stain was used to label nuclei. Representative images from three mice/condition.
To discern the cell types transduced in vivo, similar experiments were repeated using a piggyBac/Ad vector expressing the visual reporter gene mCherry. As before, piggyBac/Ad was delivered via nasal instillation to SCID mice with either Ad-iPB7 or Ad-Empty. Reporter gene expression in fixed frozen tissue was examined at 1 week and 21 weeks postdelivery. At 1 week postdelivery of piggyBac/Ad and Ad-iPB7, we observed abundant mCherry expression that was restricted to the surface epithelia of the conducting airways (Figure 4d–g). At the 1 week time point, the pattern of mCherry expression was indistinguishable between the group receiving iPB7 and the group without iPB7 (data not shown). Furthermore, this pattern of expression is consistent with previous observations using Ad5 vector expressing β-galactosidase and formulated with methylcellulose.21 Consistent with the luciferase reporter gene results (Figure 4a), abundant expression was observed in the conducting airways of mice collected at the 21-week time point from the Ad-iPB7 cohort (Figure 4h) but not the Ad-empty cohort (Figure 4i). As anticipated, no mCherry expression (Figure 4b) or evidence of inflammation (Figure 4c) was observed in control naive SCID mice.
We isolated lungs from SCID mice 12 months after being transduced with piggyBac/Ad and Ad-iPB7. Importantly, abundant mCherry expression was still observed in large airways (Figure 4j,k) at levels similar to 21 weeks postdelivery (Figure 4h). We performed genomic DNA isolation followed by LAM-PCR, using a protocol similar to discussed for in vitro transduced HeLa cells. Using shot-gun cloning and Sanger sequencing, we mapped 49 piggyBac genomic integrations (Supplementary Figure S3, blue arrows). Similar to the piggyBac/AAV mice, but unlike the in vitro shot-gun cloning, we observed a low frequency of recovered vector (2 out of 51). These data strongly support the notion that the observed in vivo persistence of transgene expression in the absence of selection is the result of transposase mediated piggyBac transposition from the Ad genome into the host genome.
piggyBac/Ad-mediated CFTR correction in CF primary airway cells
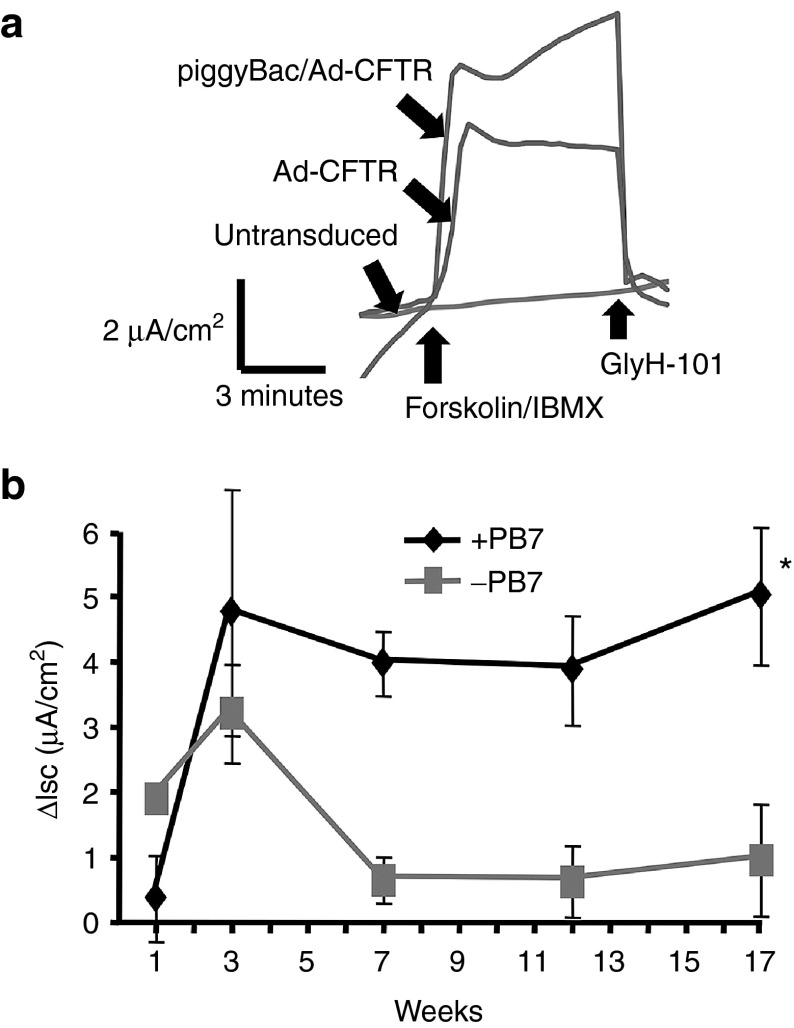
Potential advantages of the piggyBac/Ad vector include a large packaging capacity and preparations with high titers. Because the CFTR cassette flanked by piggyBac terminal repeats exceeded the packaging limit of AAV, we focused on piggyBac/Ad as a delivery vehicle. Specifically, we examined if a piggyBac/Ad-delivered CFTR cDNA can persistently rescue the anion transport defect in CF airway epithelial cells. Well-differentiated human primary CF tracheal epithelia were cultured at air-liquid interface23 and co-transduced with basolateral application of piggyBac/Ad-CFTR (MOI = 50) with Ad-iPB7 or Ad-Empty. At progressive time points, chloride currents were measured in modified Ussing chambers as previously reported.24 Upon addition of forskolin and 3-isobutyl-1-methylxanthine (IBMX), a significant increase in cAMP-stimulated chloride current was observed in epithelia treated with piggyBac/Ad-CFTR (Figure 5a). Changes in short circuit current are not observed in naive CF cultures (Figure 5a). Chloride current (I) increased by ~4 μA/cm2 in these cultures. This increase in cAMP-stimulated chloride current was significant compared to the Ad-empty control-treated cultures (Figure 5b; P < 0.001). Further, these levels of correction are consistent with our previous experience using Ad5-CFTR.22 This observation indicates that sufficient expression was achieved to functionally correct CF airway epithelia.
Figure 5.
piggyBac/Ad-CFTR restores Cl− transport in CF tracheal epithelia. (a) Ad-CFTR or piggyBac/Ad-CFTR were delivered to well-differentiated primary cultures of airway epithelia from CF human donors. Anion channel correction was measured by Cl− current responses to Forskolin/IBMX and GlyH-101 in Ussing chambers. (b) Well-differentiated tracheal epithelia cultures were cotransduced with piggyBac/Ad-CFTR (multiplicity of infection (MOI) = 50) with Ad-iPB7 (MOI = 50) or Ad-Empty (MOI = 50). At the indicated time points, transepithelial Cl− currents were measured in Ussing chambers. Bars represent means of change in Cl− current upon addition of Forskolin (10 μmol/l) and IBMX (100 μmol/l). Error bars represent SEM. n = 3 epithelial cultures/treatment. *P < 0.001.
Discussion
The choice of a vehicle to deliver therapeutic genes to specific target tissues is a vital consideration. Important features for a CFTR gene transfer vector for airway delivery include a large packaging capacity, efficient transduction, persistent expression, and the capacity to be concentrated and purified. There are multiple viral-based vectors for delivering genes to the airways, but none possess all of these attributes. Here, we codelivered piggyBac/Ad or piggyBac/AAV with Ad-transposase to cells in vitro and mouse airways in vivo and demonstrated efficient transduction, transposase-mediated integration, and persistent expression. Hybrid piggyBac/Ad and piggyBac/AAV vectors are valuable new tools for in vivo gene transfer.
Our results indicate the piggyBac/Ad and piggyBac/AAV vectors transpose and express their transgenes in the host genome in vitro and in vivo. We observed transposase-dependent transposition activity in HeLa cells and in the airways of mice. These data suggest that these viral vectors can support piggyBac transposition. This important observation contrasts with previous reports using the Sleeping Beauty based DNA transposon and Ad vectors.16 Yant et al. observed that the Ad genome required Flp-mediated recombination that resulted in circularization before releasing the Sleeping Beauty transposon. It is unclear why piggyBac does not require this circularization requirement. The unmethylated Ad and AAV genomes may be more suitable for piggyBac transposition as compared to Sleeping Beauty.25 The ability of the piggyBac-based viral vectors to integrate without an intermediate recombination step greatly simplifies in vivo delivery and potentially increases integration efficiency.
A prerequisite to life-long expression from a gene therapy vector is genomic integration into progenitor cells; therefore, integrating vector systems may have the greatest potential for treating genetic diseases. There is inherent risk when introducing a transgene with integrating vectors. Insertional mutagenesis may disrupt normal cell functions by inactivating an essential host gene or inappropriately causing expression of an undesirable gene. However, the risk will vary depending on the vector used, the therapeutic gene, and the cell type targeted. In many cases, enhancer effects pose the greatest danger. Using deep sequencing, we mapped viral vector-delivered piggyBac integrations in HeLa cells. Previous reports show that piggyBac preferentially integrates near transcription start sites at a frequency of 16–20%, in a manner similar to murine moloney leukemia virus.11,26,27,28 However, we did not observe this pattern of integration near transcription start sites in our studies (Supplementary Figure S3). In fact, we observed a slightly disfavoured integration pattern at transcription start sites as compared to a matched random control consistent with our previously published mapping data using a plasmid-based delivery system.20 The reason for this discrepancy is unclear but may result from the transposon delivery method, the cell types transduced, or the library generation method. The integration profile of a high capacity Ad-delivered Sleeping Beauty vector was also recently evaluated using adapter mediated-PCR and a near random pattern was observed.29 This finding was consistent with a lentiviral-Sleeping Beauty hybrid vector system or plasmid-based delivery.30,31,32
Several features of adenoviral vectors make them attractive vehicles for delivering therapeutic genes such as CFTR, including their large carrying capacity, efficient gene transfer capabilities, ability to transduce nondividing cells, the ability to be grown to high titer, and ease of purification. Indeed, Ad-based viral vectors were the first to be used for gene therapy trials in CF patients.33 Therapeutic levels of CFTR mRNA were achieved in the airway epithelium of CF patients, but expression quickly waned and subsequent administrations were limited by humoral immunity against the vector.34,35 Thus, the two greatest impediments to adenoviral vectors are robust immune responses and transient expression resulting from episomal expression. As demonstrated, the use of piggyBac/Ad can overcome the limitation of episomal expression; however, immune-deficient SCID mice were required to observe the effect.
Consistent with previous observations using first-generation Ad based vectors, we observed that piggyBac/Ad-mediated transgene expression was transient in immunocompetent mice.36,37,38 This short-lived transgene expression is attributed to the induction of a cytotoxic T lymphocyte immune response against adenoviral antigens.38 Immunogenic responses to adenoviral vectors preclude the potential for readministration. In addition, there are increased levels of inflammation and cytotoxicity associated with adenoviral infections.39 In these studies, when both Ad-iPB7 and piggyBac/Ad were codelivered, the total viral load delivered was 7.5 × 108 pfu. This amount was chosen based on the following three considerations: (i) the optimal ratios determined in vitro, (ii) the titer of the vector preparations, and (iii) a maximal dose that could be delivered in a 25 µl volume. We chose Ad to deliver iPB7 because it efficiently and transiently produces a transgene product. Interestingly, in our Ad-iPB7 and piggyBac/AAV experiments, we only delivered a total dose of 9 × 107 pfu of Ad-based vector and observed persistent transposon expression in immunocompetent Balb/c mice. As before, this amount was chosen based on the same three considerations. A potential explanation for persistent expression in immunocompetent mice from the Ad/AAV combination and not the Ad/Ad combination could be a threshold of Ad (~108 pfu) that triggers a cytotoxic T lymphocyte immune response when delivered intranasally. Perhaps, de-escalation studies would reveal a dose that results in stable expression of the Ad/Ad combination in immunocompetent mice. Pulmonary immune responses in mice may not be predictive of humans. Our data serve as a proof of principle that piggyBac-mediated transgene delivery can lead to long-term transgene persistence in vivo but further testing in a large animal model is necessary.
As we turn our attention to the future, there are at least four strategies to consider when addressing the immune response. (i) Helper dependant (HD)-Ad vectors lack any viral encoded genes and have precedence for long-term expression in vivo.40,41,42 Thus, helper-dependent (HD) piggyBac/Ad and HD-Ad-transposase vectors would likely attenuate a vector-mediated immune response. (ii) Transient immune suppression at the time of delivery could be considered. (iii) The vector load could be reduced by delivering both piggyBac and a self-inactivating transposase from the same Ad vector. Alternatively, the transposase could be supplied as RNA or with an AAV-based vector. (iv) Alternate vector delivery systems could also be considered. Indeed, integrase-deficient lentiviral vectors have been successfully used to deliver Sleeping Beauty.30,32
Our goal for CF gene therapy is to provide a life-long gene replacement strategy for the airways that would be efficacious regardless of the CFTR disease-causing mutation. Primary cultures of airway epithelia derived from humans with CF manifest defective CFTR-dependent anion transport. We determined that the hybrid piggyBac/Ad vector conferred CFTR expression in transduced cells and functionally corrected primary cultures of CF epithelial cells in vitro. We measured the persistence of cAMP-stimulated Cl- dependent short circuit current across the epithelia for 17 weeks.43,44 The demonstration of functional correction of CFTR in primary CF epithelia is a notable benchmark that provides an important foundation for future in vivo studies.
A gene transfer vector with the capacity to persistently express CFTR in airways in vivo may prevent or significantly slow CF disease progression. A high priority will be to demonstrate persistence and stable restoration of CFTR function in large-animal models, such as the CF pigs or ferrets.45,46,47,48 Long-term expression following a single vector dose will require vector delivery to airway cells with progenitor capacity without causing toxicity. Access to potential airway progenitor cells, such as keratin 5-positive basal cells, will be assessed by screening in vivo vector delivery techniques. Transient disruption of tight junctions may help assure vector access to the appropriate cellular compartments. Attaining long-term expression of CFTR and prevention of the progression of changes associated with lung disease in vivo would provide a powerful proof-of-principle for translational gene therapy.
Materials and Methods
Constructs. The piggyBac transposon constructs in these studies expressed either an mCherry-T2A-puromycin N-acetyl-transferase (Puror) cassette or firefly luciferase for in vitro and in vivo studies, respectively. The mCherry-T2A-Puror cassette flanked by the piggyBac terminal repeats (TRs) was designed in silico and in vitro synthesized (GenScript, Piscataway, NJ). Gene cassettes within the transposons were driven by an RSV promoter and cloned from pUC57 into pAAV2 vector to create piggyBac/AAV. For these studies the RSV promoter was chosen because in our experience the RSV promoter is sufficient to drive life-long expression in mice. The AAV2 vector was pseudotyped with the AAV5 serotype capsid. The transposon inserts were separately cloned into the AAV vector using the restriction enzymes XhoI and NotI. The piggyBac/Ad construct was cloned with the same restriction enzymes and ligation protocols from pUC57 into the Ad5 vector. The Ad-iPB7 transposase was cloned by cutting the hyperactive insect piggyBac transposase15 from pcDNA3.1/myc-HisA (Invitrogen, Grand Island, NY) to the Ad5 vector by restriction enzymes EcoRI and NotI. All clones were sequence confirmed. Adenoviral and Adenoviral-Associated Vector production was performed as a fee for service at the University of Iowa Viral Vector Core (http://www.uiowa.edu/~gene).
Colony formation assay. HeLa cells were transduced in a 24-well plate (5 × 104 cells/well) with piggyBac/AAV alone, piggyBac/Ad alone or in combination with Ad-iPB7 at the indicated MOIs in quadruplicate for 4 hours At 24 hours post-transduction, each well was trypsinized and expanded into a 100 mm plate and placed under puromycin selection (0.5 μg/ml). The cells were cultured under puromycin selection for two weeks and the selection media was changed three times per week. Following selection, puromycin-resistant colonies were fixed with 4% paraformaldehyde, stained with methylene blue and counted. Each assay was repeated at least three independent times.
Integration site recovery for Illumina HiSeq2000 sequencing. Integration sites were recovered as described previously.15 Briefly, HeLa cells (5 × 106) were transduced with piggyBac/AAV (MOI = 10,000; 5 × 109 vector genomes (vg)) or piggyBac/Ad (MOI = 10; 5 × 107 plaque forming units (pfu)) in combination with Ad-iPB7 (MOI = 10) for 4 hours. Integrants were selected with puromycin (0.5 μg/ml) for 3 weeks. Genomic DNA from three separate transfections was extracted from the integration library using the DNeasy tissue kit (Qiagen, Valencia, CA). Pooled DNA (2 μg) was digested overnight with ApoI or BstYI at 50 and 60 °C, respectively; DNA was purified with the QIAquick PCR purification kit (Qiagen) and ligated to ApoI and BstYI linkers overnight at 16 °C. Nested PCR was carried out under stringent conditions using transposon end-specific primers AAACCTCGATATACAGACCGATAAAACACATGCGTCAATTTTACGC (primary) and AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTXXXXCGTACGTCACAATATGATTATCTTTC (secondary; XXXX denotes bar code, underlined sequence indicates Illumina cluster-generation sequence) and linker-specific primers CGTAGGGAGCAAGCAGAAGACGG (primary) and CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT (secondary). DNA barcodes were included in the second-round PCR primers in order to track sample origin. The PCR products were gel purified, pooled, and sequenced using the Illumina HiSeq2000 sequencing platform.
Ethics statement. All animal procedures were previously approved (Animal Protocol #1304995) by the Institutional Animal Care and Use Committee (IACUC) and in accordance with National Institutes of Health guidelines.
In vivo delivery and bioluminescence. All mice for this study were housed at the University of Iowa Animal Care Facilities. Female mice (6–8-week-old) were transduced intranasally with piggyBac/Ad or piggyBac/AAV vector plus Ad-iPB7 or Ad-empty formulated 1:1 with 2% methylcellulose (50 μl total volume) as previously described.21 We observed no signs of visible toxicity or mortality as a result of viral delivery. Animals were imaged at indicated time points using the in vivo imaging system (Caliper Life Sciences, Hopkinton, MA). Luciferin substrate (200 μl, 15 mg/ml; Caliper Life Sciences) was administered via intraperitoneal injection and the mice were imaged for 5 minutes. Data were analyzed using Living Image software (Caliper Life Sciences). Polidocanol treatments were performed in mice that received piggyBac/AAV-luciferase. At 3 months postdelivery, 25 µl of 2% polidocanol (Sigma, St Louis, MO) was administered to mice intranasally. A second dose was delivered 48 hours later. Luciferase expression was quantified by bioluminescent imaging at indicated time points before and after polidocanol treatment.22
Primary epithelial cultures and electrophysiology studies. Tracheal epithelial cells from human CF lungs were isolated by enzymatic digestion, seeded onto permeable filters, and grown at air-liquid interface as previously described.23 Cystic Fibrosis transmembrane Conductance Regulator (CFTR)-null porcine tracheal epithelia cultures were studied in modified Ussing chambers as previously described.24 Briefly, epithelia were bathed on both surfaces with solution containing: 135 mmol/l NaCl, 2.4 mmol/l K2HPO4, 0.6 mmol/l KH2PO4, 1.2 mmol/l CaCl2, 1.2 mmol/l MgCl2, 10 mmol/l dextrose, 5 mmol/l HEPES (pH = 7.4) at 37 °C and gassed with compressed air. Baseline transepithelial currents were measured. After apical addition of 100 μmol/l amiloride (Amil) and 100 μmol/l 4,4′-diisothiocyanoto-stilbene-2,2′-disulfonic acid (DIDS), currents were allowed to stabilize and the apical solution was replaced with a 4.8 mmol/l Cl− solution containing 135 mmol/l D-Gluconic Acid, 2.4 mmol/l K2HPO4, 0.6 mmol/l KH2PO4, 1.2 mmol/l CaCl2, 1.2 mmol/l MgCl2, 10 mmol/l dextrose, 5 mmol/l HEPES (pH = 7.4) at 37 °C and gassed with compressed air. cAMP-dependent Cl− current was stimulated by apical addition of 10 μmol/l forskolin and 100 μmol/l 3-isobutyl-1-methylxanthine (IBMX), and CFTR-specific Cl− transport was inhibited with 100 μmol/l GlyH-101. Transepithelial voltage (Vt) was maintained at 0 mV to measure transepithelial current (I). Transepithelial electrical conductance (Gt) was measured by intermittently clamping Vt to +5 and/or −5 mV. Spontaneous values of Vt were measured by transiently removing the voltage clamp.
Immunohistochemistry. Approximately 2.5 × 108 pfu of piggyBac/Ad plus Ad-iPB7 or Ad-empty vector in a 50 µl volume with 1% methylcellulose (1:1) was delivered via nasal instillation to the airways of 3-month-old male SCID/NCr mice (NCI, MD, Balb/c background, 01S11) mice under ketamine/xylazine (87.5 + 2.5 mg/kg) anesthesia. Fluorescence analyses of in vivo mCherry expression in mice lungs were examined as described in ref. 46. Briefly, mice lungs were harvested from a subset of mice and fixed in 4% paraformaldehyde in PBS at 4 °C overnight. Prior to fixation, lungs were gently inflated with 15% sucrose via the trachea to maintain lung architecture. After fixation, lungs were submerged in 15% sucrose for 8 hours and then in 30% sucrose overnight. All steps were performed at 4 °C. Lungs were embedded in OCT media, frozen in liquid nitrogen, and 10 μm sections were obtained using a microtome at −26 °C. The serial sections were then stained with hematoxylin and eosin using standard techniques. Images were captured using an Olympus BX60 fluorescence microscope (Leeds Precision Instrument, Minneapolis, MN).
Statistics. All numerical data are represented as mean ± standard error. Standard one and two sample t-tests were performed using the statistical computer program R (www.r-project.org) with the lme4 package. Analysis of variance was performed using Prism software (GraphPad Software, San Diego, CA).
SUPPLEMENTARY MATERIAL Figure S1. Detailed schematic of vector constructs used in this study. Figure S2. Dose escalation of adenovirus expressing GFP was performed in HeLa cells. Figure S3. The heat maps summarize piggyBac distributions to genomic features. Figure S4. Mouse genomic DNA was isolated from mice 12 months following delivery of piggyBac/AAV or piggyBac/Ad with Ad-iPB7 to the mouse airways. Figure S5. piggyBac/Ad expressing firefly luciferase was co-delivered to the nasal airways with Ad-iPB7 or Ad-Empty formulated with 2% methylcellulose.
Acknowledgments
We thank Paul McCray, Jr., Adam Dupuy, Nancy Craig, Janice Staber, and Erin Burnight for their many insightful discussions. Samantha Osterhaus provided technical assistance and Thomas Bair facilitated the analysis of the deep sequencing results. We also acknowledge the support of the University of Iowa DNA Sequencing Core, In Vitro Models and Cell Culture Core, Viral Vector Core, and Cell Morphology Core. This work was supported by the National Institutes of Health R01 HL-105821 (P.L.S.) and the Cystic Fibrosis Foundation SINN14G0 (P.L.S.). Core facilities were partially supported by the National Institutes of Health: P01 HL-51670, P01 HL-091842 and the Center for Gene Therapy for Cystic Fibrosis P30 DK-54759.
Supplementary Material
Detailed schematic of vector constructs used in this study.
Dose escalation of adenovirus expressing GFP was performed in HeLa cells.
The heat maps summarize piggyBac distributions to genomic features.
Mouse genomic DNA was isolated from mice 12 months following delivery of piggyBac/AAV or piggyBac/Ad with Ad-iPB7 to the mouse airways.
piggyBac/Ad expressing firefly luciferase was co-delivered to the nasal airways with Ad-iPB7 or Ad-Empty formulated with 2% methylcellulose.
References
- Driskell RA, Engelhardt JF. Current status of gene therapy for inherited lung diseases. Annu Rev Physiol. 2003;65:585–612. doi: 10.1146/annurev.physiol.65.092101.142426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dea S, Harrison DJ. CFTR gene transfer to lung epithelium–on the trail of a target cell. Curr Gene Ther. 2002;2:173–181. doi: 10.2174/1566523024605546. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Liu L, Mah C, Fletcher BS. Sustained FVIII expression and phenotypic correction of hemophilia A in neonatal mice using an endothelial-targeted sleeping beauty transposon. Mol Ther. 2006;13:1006–1015. doi: 10.1016/j.ymthe.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ.et al. (2005Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system Blood 1052691–2698. [DOI] [PubMed] [Google Scholar]
- Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- Aronovich EL, Bell JB, Belur LR, Gunther R, Koniar B, Erickson DC.et al. (2007Prolonged expression of a lysosomal enzyme in mouse liver after Sleeping Beauty transposon-mediated gene delivery: implications for non-viral gene therapy of mucopolysaccharidoses J Gene Med 9403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Oh S, Ericson K, Demorest ZL, Vengco I, Gharagozlou S.et al. (2007Transposon-based interferon gamma gene transfer overcomes limitations of episomal plasmid for immunogene therapy of glioblastoma Cancer Gene Ther 14550–560. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S.et al. (2006piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells Proc Natl Acad Sci USA 10315008–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Higuchi Y, Kawakami S, Yamashita F, Hashida M. piggyBac transposon-mediated long-term gene expression in mice. Mol Ther. 2010;18:707–714. doi: 10.1038/mt.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridey SK, Liu L, Doherty JE, Kaja A, Galvan DL, Fletcher BS.et al. (2009PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer Mol Ther 172115–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty JE, Huye LE, Yusa K, Zhou L, Craig NL, Wilson MH. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum Gene Ther. 2012;23:311–320. doi: 10.1089/hum.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnight ER, Staber JM, Korsakov P, Li X, Brett BT, Scheetz TE.et al. (2012A Hyperactive Transposase Promotes Persistent Gene Transfer of a piggyBac DNA Transposon Mol Ther Nucleic Acids 1e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T, Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- Hausl MA, Zhang W, Müther N, Rauschhuber C, Franck HG, Merricks EP.et al. (2010Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B Mol Ther 181896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müther N, Noske N, Ehrhardt A. Viral hybrid vectors for somatic integration - are they the better solution. Viruses. 2009;1:1295–1324. doi: 10.3390/v1031295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Young SM, Jr, Samulski RJ. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet. 2004;38:819–845. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- Li X, Burnight ER, Cooney AL, Malani N, Brady T, Sander JD.et al. (2013piggyBac transposase tools for genome engineering Proc Natl Acad Sci USA 110E2279–E2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Shah AJ, Donovan MD, McCray PB., Jr Viscoelastic gel formulations enhance airway epithelial gene transfer with viral vectors. Am J Respir Cell Mol Biol. 2005;32:404–410. doi: 10.1165/rcmb.2004-0410OC. [DOI] [PubMed] [Google Scholar]
- Burnight ER, Wang G, McCray PB, Jr, Sinn PL. Transcriptional targeting in the airway using novel gene regulatory elements. Am J Respir Cell Mol Biol. 2012;47:227–233. doi: 10.1165/rcmb.2011-0444OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J.et al. (2002An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures Methods Mol Biol 188115–137. [DOI] [PubMed] [Google Scholar]
- Chen JH, Stoltz DA, Karp PH, Ernst SE, Pezzulo AA, Moninger TO.et al. (2010Loss of anion transport without increased sodium absorption characterizes newborn porcine cystic fibrosis airway epithelia Cell 143911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jursch T, Miskey C, Izsvák Z, Ivics Z. Regulation of DNA transposition by CpG methylation and chromatin structure in human cells. Mob DNA. 2013;4:15. doi: 10.1186/1759-8753-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J, Akhtar W, Badhai J, Rust AG, Rad R, Hilkens J.et al. (2014Chromatin landscapes of retroviral and transposon integration profiles PLoS Genet 10e1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan DL, Nakazawa Y, Kaja A, Kettlun C, Cooper LJ, Rooney CM.et al. (2009Genome-wide mapping of PiggyBac transposon integrations in primary human T cells J Immunother 32837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P.et al. (2010Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells Mol Ther 181803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Muck-Hausl M, Wang J, Sun C, Gebbing M, Miskey C.et al. (2013Integration profile and safety of an adenovirus hybrid-vector utilizing hyperactive sleeping beauty transposase for somatic integration PLoS One 8e75344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink CA, Gaspar HB, Gabriel R, Schmidt M, McIvor RS, Thrasher AJ.et al. (2009Sleeping beauty transposition from nonintegrating lentivirus Mol Ther 171197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunstrup NH, Moldt B, Mátés L, Villesen P, Jakobsen M, Ivics Z.et al. (2009Hybrid lentivirus-transposon vectors with a random integration profile in human cells Mol Ther 171205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal RG, McElvaney NG, Rosenfeld MA, Chu CS, Mastrangeli A, Hay JG.et al. (1994Administration of an adenovirus containing the human CFTR cDNA to the respiratory tract of individuals with cystic fibrosis Nat Genet 842–51. [DOI] [PubMed] [Google Scholar]
- Harvey BG, Leopold PL, Hackett NR, Grasso TM, Williams PM, Tucker AL.et al. (1999Airway epithelial CFTR mRNA expression in cystic fibrosis patients after repetitive administration of a recombinant adenovirus J Clin Invest 1041245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal RG. Adenovirus: the first effective in vivo gene delivery vector. Hum Gene Ther. 2014;25:3–11. doi: 10.1089/hum.2013.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakland M, Maury W, McCray PB, Jr, Sinn PL. Intrapulmonary Versus Nasal Transduction of Murine Airways With GP64-pseudotyped Viral Vectors. Mol Ther Nucleic Acids. 2013;2:e69. doi: 10.1038/mtna.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Arias AC, Brogden KA, McCray PB., Jr Lentivirus vector can be readministered to nasal epithelia without blocking immune responses. J Virol. 2008;82:10684–10692. doi: 10.1128/JVI.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michou AI, Santoro L, Christ M, Julliard V, Pavirani A, Mehtali M. Adenovirus-mediated gene transfer: influence of transgene, mouse strain and type of immune response on persistence of transgene expression. Gene Ther. 1997;4:473–482. doi: 10.1038/sj.gt.3300412. [DOI] [PubMed] [Google Scholar]
- Mueller C, Flotte TR. Gene therapy for cystic fibrosis. Clin Rev Allergy Immunol. 2008;35:164–178. doi: 10.1007/s12016-008-8080-3. [DOI] [PubMed] [Google Scholar]
- Cao H, Yang T, Li XF, Wu J, Duan C, Coates AL.et al. (2011Readministration of helper-dependent adenoviral vectors to mouse airway mediated via transient immunosuppression Gene Ther 18173–181. [DOI] [PubMed] [Google Scholar]
- Teramoto S, Bartlett JS, McCarty D, Xiao X, Samulski RJ, Boucher RC. Factors influencing adeno-associated virus-mediated gene transfer to human cystic fibrosis airway epithelial cells: comparison with adenovirus vectors. J Virol. 1998;72:8904–8912. doi: 10.1128/jvi.72.11.8904-8912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad AK, Xiong W, Puntel M, Farrokhi C, Kroeger KM, Salem A.et al. (2012Safety profile of gutless adenovirus vectors delivered into the normal brain parenchyma: implications for a glioma phase 1 clinical trial Hum Gene Ther Methods 23271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Davidson BL, Melchert P, Slepushkin VA, van Es HH, Bodner M.et al. (1998Influence of cell polarity on retrovirus-mediated gene transfer to differentiated human airway epithelia J Virol 729818–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Slepushkin V, Zabner J, Keshavjee S, Johnston JC, Sauter SL.et al. (1999Feline immunodeficiency virus vectors persistently transduce nondividing airway epithelia and correct the cystic fibrosis defect J Clin Invest 104R55–R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB., Jret al. (2008The porcine lung as a potential model for cystic fibrosis Am J Physiol Lung Cell Mol Physiol 295L240–L263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn PL, Cooney AL, Oakland M, Dylla DE, Wallen TJ, Pezzulo AA.et al. (2012Lentiviral vector gene transfer to porcine airways Mol Ther Nucleic Acids 1e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz DA, Rokhlina T, Ernst SE, Pezzulo AA, Ostedgaard LS, Karp PH.et al. (2013Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs J Clin Invest 1232685–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Sui H, Fisher JT, Yan Z, Liu X, Cho HJ.et al. (2010Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis J Clin Invest 1203149–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady T, Lee YN, Ronen K, Malani N, Berry CC, Bieniasz PD.et al. (2009Integration target site selection by a resurrected human endogenous retrovirus Genes Dev 23633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed schematic of vector constructs used in this study.
Dose escalation of adenovirus expressing GFP was performed in HeLa cells.
The heat maps summarize piggyBac distributions to genomic features.
Mouse genomic DNA was isolated from mice 12 months following delivery of piggyBac/AAV or piggyBac/Ad with Ad-iPB7 to the mouse airways.
piggyBac/Ad expressing firefly luciferase was co-delivered to the nasal airways with Ad-iPB7 or Ad-Empty formulated with 2% methylcellulose.