Abstract
Mucopolysaccharidosis (MPS) IIIA is a neuropathic lysosomal storage disease caused by deficiency in N-sulfoglucosamine sulfohydrolase (SGSH). Genome-wide gene expression microarrays in MPS IIIA mice detected broad molecular abnormalities (greater than or equal to twofold, false discovery rate ≤10) in numerous transcripts (314) in the brain and blood (397). Importantly, 22 dysregulated blood transcripts are known to be enriched in the brain and linked to broad neuronal functions. To target the root cause, we used a self-complementary AAVrh74 vector to deliver the human SGSH gene into 4–6 weeks old MPS IIIA mice by an intravenous injection. The treatment resulted in global central nervous system (CNS) and widespread somatic restoration of SGSH activity, clearance of CNS and somatic glycosaminoglycan storage, improved behavior performance, and significantly extended survival. The scAAVrh74-hSGSH treatment also led to the correction of the majority of the transcriptional abnormalities in the brain (95.9%) and blood (97.7%), of which 182 and 290 transcripts were normalized in the brain and blood, respectively. These results demonstrate that a single systemic scAAVrh74-hSGSH delivery mediated efficient restoration of SGSH activity and resulted in a near complete correction of MPS IIIA molecular pathology. This study also demonstrates that blood transcriptional profiles reflect the biopathological status of MPS IIIA, and also respond well to effective treatments.
Introduction
Mucopolysaccharidosis (MPS) IIIA, is a neuropathic lysosomal storage disease (LSD) caused by autosomal recessive deficiency in N-sulfoglucosamine sulfohydrolase (SGSH). SGSH is essential for the stepwise degradation of heparan sulfate glycosaminoglycans (GAG) in lysosomes.1 The SGSH defect results in lysosomal accumulation of heparan sulfate-GAGs in cells of virtually all organs, especially in cells throughout the central nervous system (CNS). The lysosomal GAG storage leads to complex secondary pathologies, which remain poorly understood.2,3,4,5,6,7,8 Infants with MPS IIIA appear normal at birth. Progressive clinical disorders usually emerge between 2 and 6 years of age, predominantly neurological, presenting as developmental delay, behavior abnormalities, reduced cognitive capacity, and seizures.1,9 Somatic manifestations have been well documented in patients with MPS IIIA, such as splenohepatomegaly, skeletal deformations, and gastrointestinal tissues.1,10 Death usually occurs prior to age 20. There are currently no effective treatments for MPS IIIA.
The most critical challenge in developing therapies for MPS III has been the presence of the blood-brain-barrier (BBB). Significant therapeutic advancements have been made for treating LSDs, such as hematopoietic stem cell transplantation, recombinant enzyme replacement therapy, and gene therapy.11 The currently approved systemic enzyme replacement therapy or hematopoietic stem cell transplantation have shown somatic benefits; however, MPS III (A-D) disorders are not amenable to these approaches because the BBB prevents effective CNS access of either recombinant enzyme or enzyme produced by transplanted hematopoietic stem cells. Alternative intrathecal enzyme replacement therapy has been shown to correct neuropathology in animal models,12,13,14 leading to the ongoing clinical trials that target the CNS disorders in patients with MPS I, II, and IIIA (http://clinicaltrials.gov), but will require life-long repetitive administration. In contrast, gene therapy has the potential for long-term treatment of the LSDs that can benefit from bystander effects from a relatively small number of transduced cells. Numerous gene therapy studies, mostly designed to restore the missing enzyme activity, have shown various degrees of success in the correction of lysosomal storage in vitro and in vivo in LSD animal models, using various viral vectors.15 Recombinant adeno-associated virus (rAAV) has been a favored gene delivery vector because of demonstrated long-term expression in the CNS and periphery.15,16 Previous studies showed functional correction of CNS lesions in MPS IIIA mice by intracerebral SGSH and SUMF1 gene delivery using rAAV vectors.17 The trans-BBB neurotropism of emerging new AAV serotypes offers an effective solution for CNS gene therapy, showing a great potential for the treatment of LSDs and other neurological diseases.18,19,20,21,22,23 A single systemic delivery of rAAV9 vector can lead to global CNS and widespread somatic restoration of enzyme activity, correction of lysosomal storage pathology, and functional neurological benefits in mice with MPS IIIB or IIIA mice.18,19 A recent study reported whole body correction of MPS IIIA in a mouse model following intracerebrospinal fluid (CNS) rAAV9 gene delivery.24 The therapeutic potential of hematopoietic stem cell–lentiviral gene therapy and myeloid-driven autologous hematopoietic-lentiviral gene therapy have recently been reported in MPS IIIA mice.25,26
In order to develop alternate vectors to AAV9 for potential use in patients with preexisting Ab, or for repeat administration, we used a self-complementary (sc) AAVrh74-hSGSH vector that also has the ability to cross the BBB. We have further used this study as a platform to test gene expression arrays as a potential surrogate marker for correction of MPS IIIA pathogenesis.
A single systemic delivery of scAAVrh74-hSGSH vector in MPS IIIA mice led to effective restoration of SGSH activity in the CNS and periphery. Using genome-wide gene expression microarrays, we detected broad molecular abnormalities in the brain and peripheral blood in MPS IIIA mice, and demonstrate a strong blood–brain association between anomalously expressed genes. Importantly, both the blood and brain transcriptional alterations responded well to the scAAVrh74-hSGSH gene delivery treatment, strongly supporting the potential of blood transcriptional signatures as surrogate outcome measures for MPS IIIA.
Results
To assess the therapeutic impacts of scAAVrh74 gene delivery, we treated 4–6 weeks old MPS IIIA mice with an intravenous (IV) injection of scAAVrh74-U1a-hSGSH (5 × 1012 vector genome/kg, n = 13), using wild-type (wt) and nontreated MPS IIIA littermates (n = 15) as controls. The animals were tested for behavioral performance in a hidden task in Morris water maze (n = 13) and observed for longevity (n = 9). At 7 months postinjection, blood and tissue were collected from groups of male mice (n = 4/per group) for analyses to assess therapeutic impact. We focused on males to minimize individual variation in the gene array analysis. Total RNA was isolated from the brain and peripheral blood of individual wt, nontreated MPS IIIA, and scAAVrh74-treated mice (all male, n = 4/group) and then analyzed separately (n = 4/group) using genome-wide gene expression microarrays of >60,000 transcripts, to assess the potential blood–brain molecular association in MPS IIIA and their response to the vector treatment.
Restoration of SGSH activity in the CNS and somatic tissues
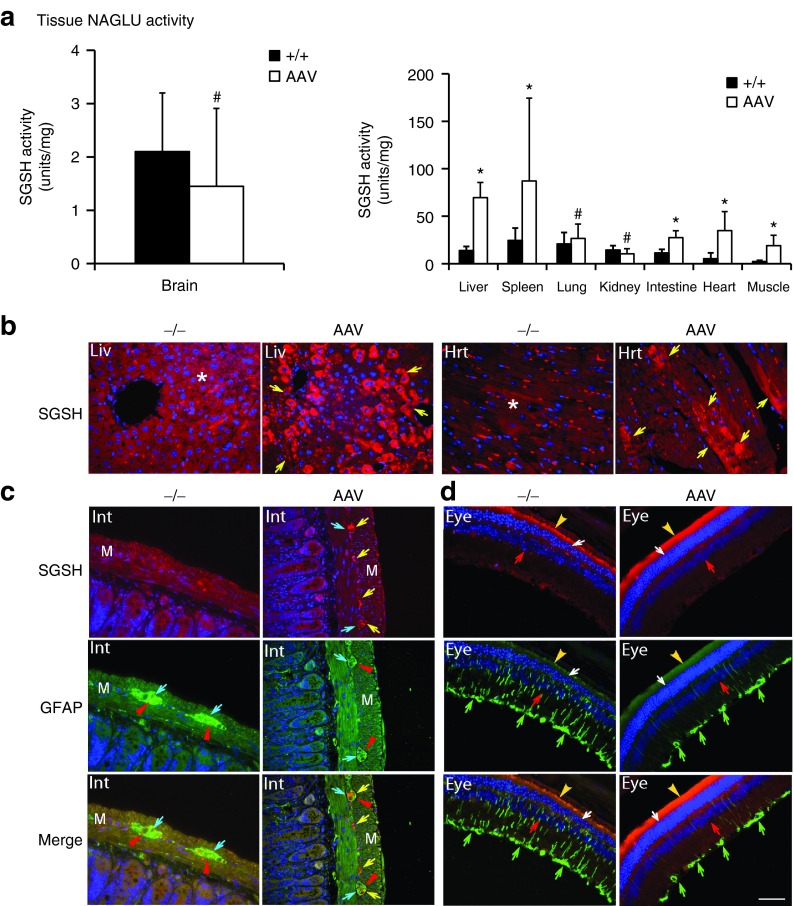
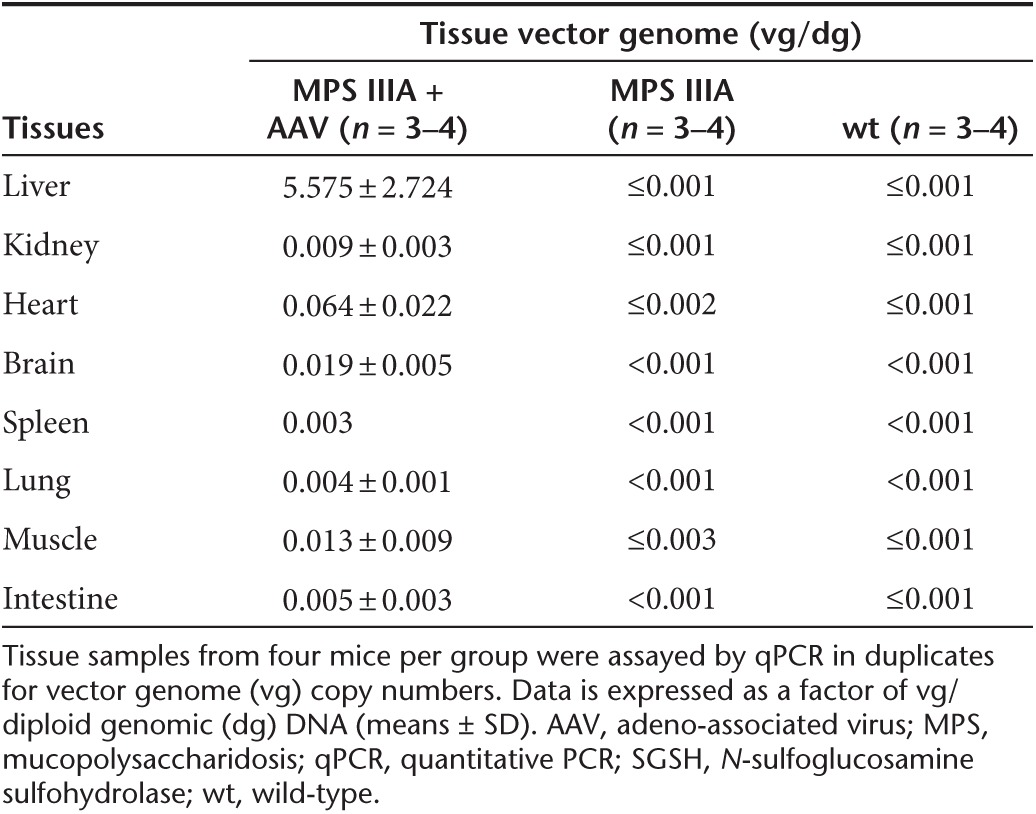
Tissues were assayed for SGSH enzymatic activity at 7 months postinjection to quantify the expression and functionality of the recombinant SGSH (rSGSH). We detected SGSH activity at or near wt levels in the brain, lung, and kidney, and supraphysiological levels in the liver, heart, skeletal muscle, intestine, and spleen (Figure 1a) in the scAAVrh74-treated MPS IIIA mice. In nontreated MPS IIIA control mice, very low levels of residual SGSH activity was observed in the majority of somatic tissues (<0.1% of wt levels) and no detectable SGSH activity was observed in the brain, heart, and muscle. Using immunofluorescence (IF) staining, we detected rSGSH-positive cells and signals in widespread somatic tissues (Figure 1b, Supplementary Figure S1). The CNS rSGSH expression was diffuse (Supplementary Figure S1a), with the IF staining signals lower than in the liver where >40% of hepatocytes were rSGSH-positive (Figure 1b). In heart, ~10% of cardiomyocytes were rSGSH-positive (Figure 1b). In the intestine, the discernible rSGSH signals were seen only in neurons of myenteric plexus and submucosal plexus (Figure 1c). These data indicate that the IV-delivered scAAVrh74 vector mediated efficient expression of enzymatically functional rSGSH in the CNS, peripheral nervous system (PNS), and broad somatic tissues in MPS IIIA mice. Using qPCR, we further confirmed the broad biodistribution of AAVrh74-hSGSH vector (Table 1).
Figure 1.
Restoration of functional SGSH in MPS IIIA mice following a single systemic scAAVrh74-hSGSH gene delivery. MPS IIIA mice (4–6 weeks old) were treated with an intravenous injection of scAAVrh74-U1a-hSGSH vector (5 × 1012 vg/kg). At 7 months postinjection, tissues were assayed by (a) SGSH activity assay and immunofluorescence for (b, c) hSGSH and (c, d) GFAP. SGSH activity is expressed as units/mg protein, and 1 unit = mmol of 4MU released/17 hour. +/+: wt; −/−: nontreated MPS IIIA mice; AAV: scAAVrh74-treated MPS IIIA mice. #P ≥ 0.05 versus wt; *P ≤ 0.05 versus wt; Liv: liver, Hrt: heart; Int: small intestine; M: muscularis externa. Eye: retina. Red fluorescence: hSGSH-positive cells/signals; green fluorescence: GFAP-positive cells/signals; blue fluorescence: DAPI-stained nuclei. *Red signals in nontreated liver and heat tissues are autofluorescence. Yellow arrows: hSGSH-positive cells and signals; blue arrows: neurons of myenteric plexus; red arrowheads: GFAP-positive staining surrounding myenteric neurons; green arrows: GFAP-positive Müller cells; red arrows: inner nuclear layer; white arrows: outer nuclear layer; yellow arrowheads: pigment layer. Scale bar = 50 μm. AAV, adeno-associated virus; DAPI, 4′,6-diamidino-2-phenylindole; GFAP, glial fibrillary acidic protein; MPS, mucopolysaccharidosis; SGSH, N-sulfoglucosamine sulfohydrolase; vg, vector genome; wt, wild-type.
Table 1. Estimated tissue vector genome in MPS IIIA mice treated with scAAVrh74-U1a-hSGSH vector.
Clearance of CNS and somatic tissue GAG accumulation and correction of gliosis in the CNS and PNS
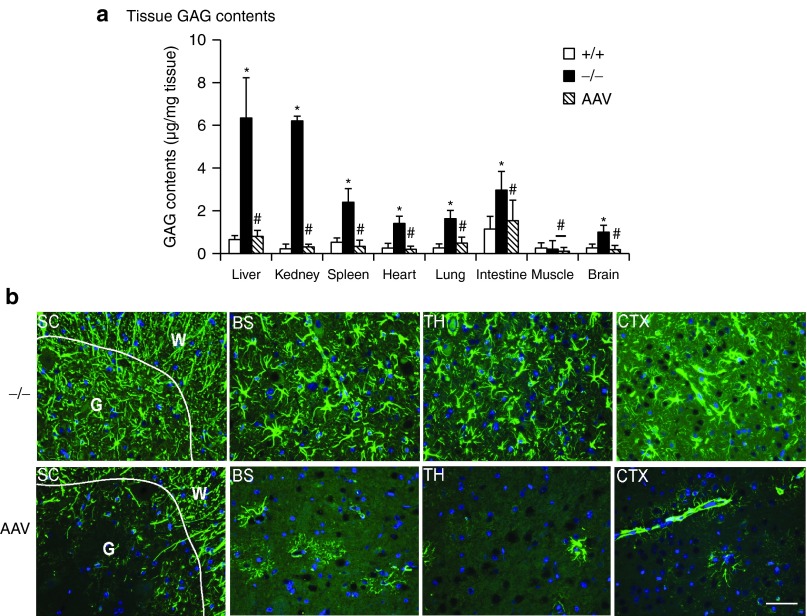
To further assess the functionality of the scAAVrh74-mediated rSGSH, tissues were assayed for GAG content to quantify the lysosomal GAG storage. The results showed a significant reduction of GAG to wt levels in virtually all tested tissues in scAAVrh74-treated MPS IIIA mice, including brain, liver, spleen, heart, lung, intestine, and kidney (Figure 2a). Using IF staining for glial fibrillary acidic protein (GFAP), we observed the correction of astrogliosis throughout the brain and spinal cord (Supplementary Figure S1a, Figure 2b). Importantly, our data also showed a significant decrease in GFAP signals in the intestine (Figure 1c) and retina (Figure 1d). These results further suggest that the AAVrh74-mediated rSGSH was functional, leading to the clearance of GAG storage in both the CNS and periphery, as well as correction of neuroinflammation in both the CNS and PNS.
Figure 2.
Intravenous (IV) administration of AAVRh74-hSGSH corrects GAG storage and astrocytosis. MPS IIIA mice (4–6 weeks old) were treated with an IV injection of scAAVrh74-u1a-hSGSH vector (5 × 1012 vg/kg). At 7 months postinjection, (a) tissues were assayed for GAG contents and (b) immunofluorescence was performed for GFAP. GAG contents are expressed as μg/mg wet tissue. #P ≥ 0.05 versus wt; *P ≤ 0.05 versus wt; green fluorescence: GFAP-positive cells/signals; blue fluorescence: DAPI-stained nuclei. +/+: wt; −/−: nontreated MPS IIIA mice; AAV: scAAVrh74-treated MPS IIIA mice. SC: spinal cord; G: gray matter; W: white matter; BS: brain stem; TH: thalamus; CTX: cerebral cortex. Scale bar = 50 μm. AAV, adeno-associated virus; DAPI, 4′,6-diamidino-2-phenylindole; GAG, glycosaminoglycan; GFAP, glial fibrillary acidic protein; MPS, mucopolysaccharidosis; SGSH, N-sulfoglucosamine sulfohydrolase; vg, vector genome; wt, wild-type.
Interestingly, the scAAVrh74-hSGSH vector treatment led to the complete clearance of GAGs in the kidney (Figure 2a), which was not observed previously in similar studies in MPS IIIB mice using rAAV9-CMV-hNAGLU18 or in our ongoing studies in MPS IIIA mice using scAAV9-U1a-hSGSH (unpublished data). Using IF staining, we observed rSGSH-positive signals in the kidney in glomerulus, proximal tubules, and collective tubule (Supplementary Figure S1b). This added tissue tropism suggests broader therapeutic potential of AAVrh74 as gene delivery vector.
Significant behavioral improvements and increase in survival
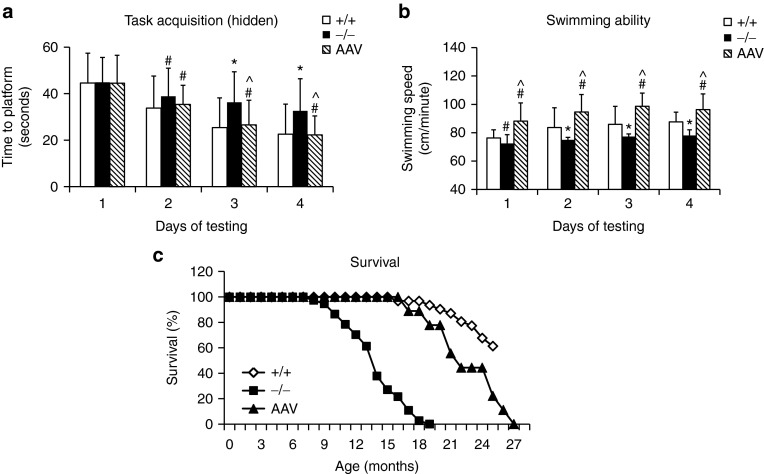
To assess neurological impact, MPS IIIA mice treated with an IV injection of 5 × 1012 vector genome/kg scAAVrh74-hSGSH vector (n = 13) were tested for behavior performance at 7–7.5 months of age in a hidden task in the Morris water maze. The vector-treated MPS IIIA mice exhibited a significant decrease in latency to find the hidden platform (Figure 3a) and an increase in swimming speed (Figure 3b) over 4 days of testing, compared with wt and nontreated MPS IIIA mice (n = 15/group), indicating the correction of learning and swimming ability.
Figure 3.
Restoration of SGSH activity corrects behavior abnormalities and extends lifespan. MPS IIIA mice (4–6 weeks old) were treated with an intravenous injection of scAAVrh74-U1a-hSGSH vector (5 × 1012 vg/kg). (a,b) Mice were tested for behavior performance at age 7–7.7 months, in a hidden task in water maze: +/+: wt (n = 41); −/−: nontreated MPS IIIA mice (n = 33); AAV: scAAVrh74-treated MPS IIIA mice (n = 13). (c) Longevity: wt: n = 42; IIIA: n = 31; AAVrh74: n = 7. #P > 0.05 versus wt; *P ≤ 0.05 versus wt; ^P ≤ 0.05 versus nontreated IIIA. AAV, adeno-associated virus; MPS, mucopolysaccharidosis; SGSH, N-sulfoglucosamine sulfohydrolase; vg, vector genome; wt, wild-type.
Nine scAAVrh74-treated MPS IIIA mice were observed for longevity and survived >15.6–25.4 months (21.0 ± 3.2 months), while all nontreated MPS IIIA mice died at 7.4–17.3 months (13.2 ± 2.7 months) of age (P < 0.001) (Figure 3c). These data demonstrate that a single IV scAAVrh vector injection alone is functionally beneficial in treating the CNS disorders and increasing longevity in MPS IIIA mice.
Extensive correction of molecular abnormalities in the brain and blood in MPS IIIA mice
Total RNA samples from the brain and peripheral blood of 8-month-old male wt, nontreated MPS IIIA, and scAAVrh74-treated MPS IIIA mice (n = 4/group) were analyzed using genome-wide gene expression microarrays with >60,000 probes, to assess the comprehensive molecular pathological changes in MPS IIIA and their responsiveness to the rAAVrh74-hSGSH gene delivery.
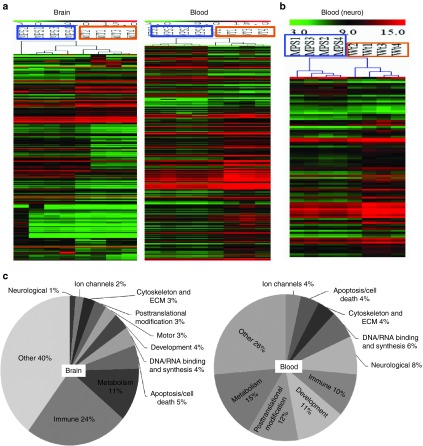
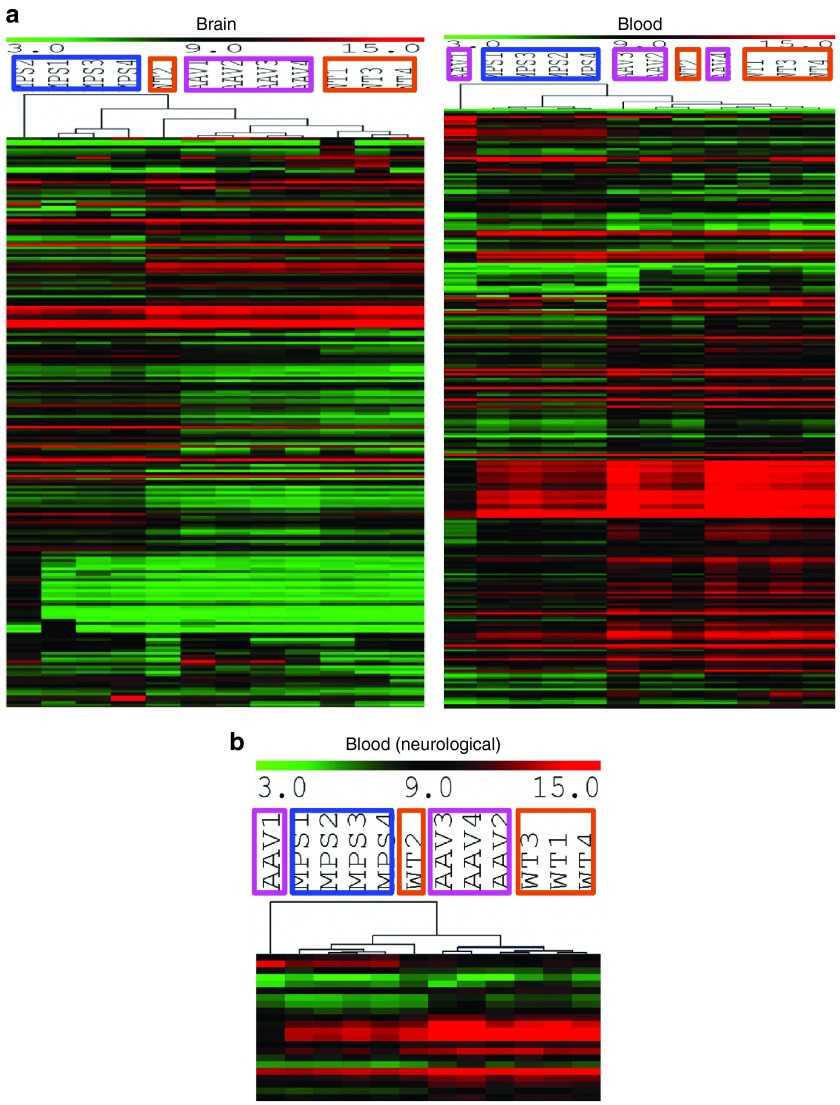
We detected significant alterations in numerous transcripts (greater than or equal to twofold, false discovery rate (FDR) <10%) in both the brain and blood in MPS IIIA mice (Table 2), compared with wt mice. These significant transcriptional changes involved 314 transcripts in the brain including 218 known genes and 96 unidentified genes, and 397 transcripts in the blood including 261 known genes and 136 unidentified genes (Table 2). In addition, >1,000 transcripts in the blood and >1,000 transcripts in the brain were significantly altered ≥1.5-fold (FDR <10%). Further array data analyses presented below were focused on genes dysregulated greater than or equal to twofold (FDR <10%). Of these significantly changed genes, only 20 were changed in both the brain and blood, of which 19 were altered in the same direction, while the astrocyte marker Gfap was downregulated in the blood but upregulated in the brain (Table 3). An additional 10 transcripts with unknown functions were also altered in the same direction in the brain and blood (data not shown). Further, heat map analyses of array data showed clear separation of both blood and brain gene expression profiles between MPS IIIA and wt mice (Figure 4a,b). Functional analyses showed that the gene dysregulations detected in both the brain and blood were associated with numerous biological functions (Figure 4b,c, Supplementary Table S1), suggesting broad functional impairments, including changes linked to major components of MPS IIIA neuropathology. The blood gene alterations reflect the major secondary neuropathology of MPS IIIA, including inflammation and metabolic impairments (Figure 4c). Importantly, 22 altered genes in the blood are known to be enriched in the brain and essential for neurological functions (Figure 4b,c, Supplementary Table S1). These results suggest very complex molecular abnormalities in MPS IIIA.
Table 2. Broad transcriptional abnormalities in the brain and blood of MPS IIIA mice.
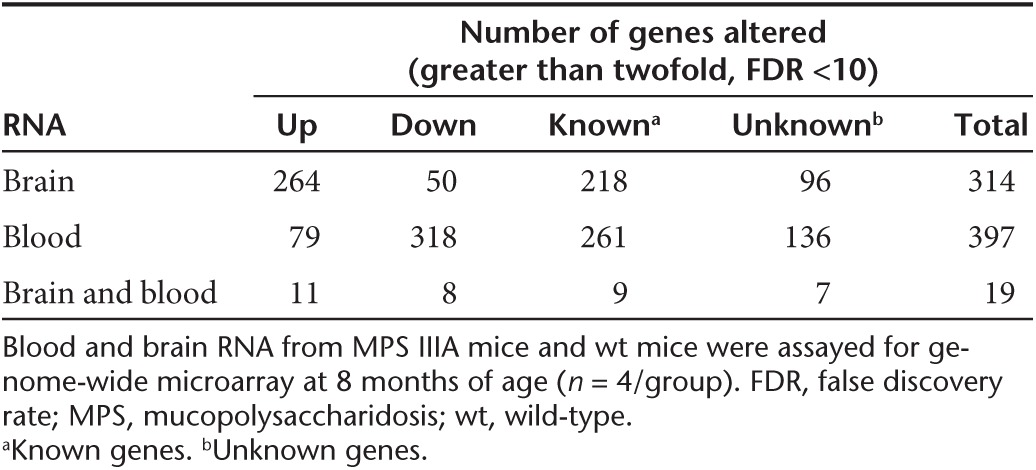
Table 3. Genes significantly altered in both the brain and blood.
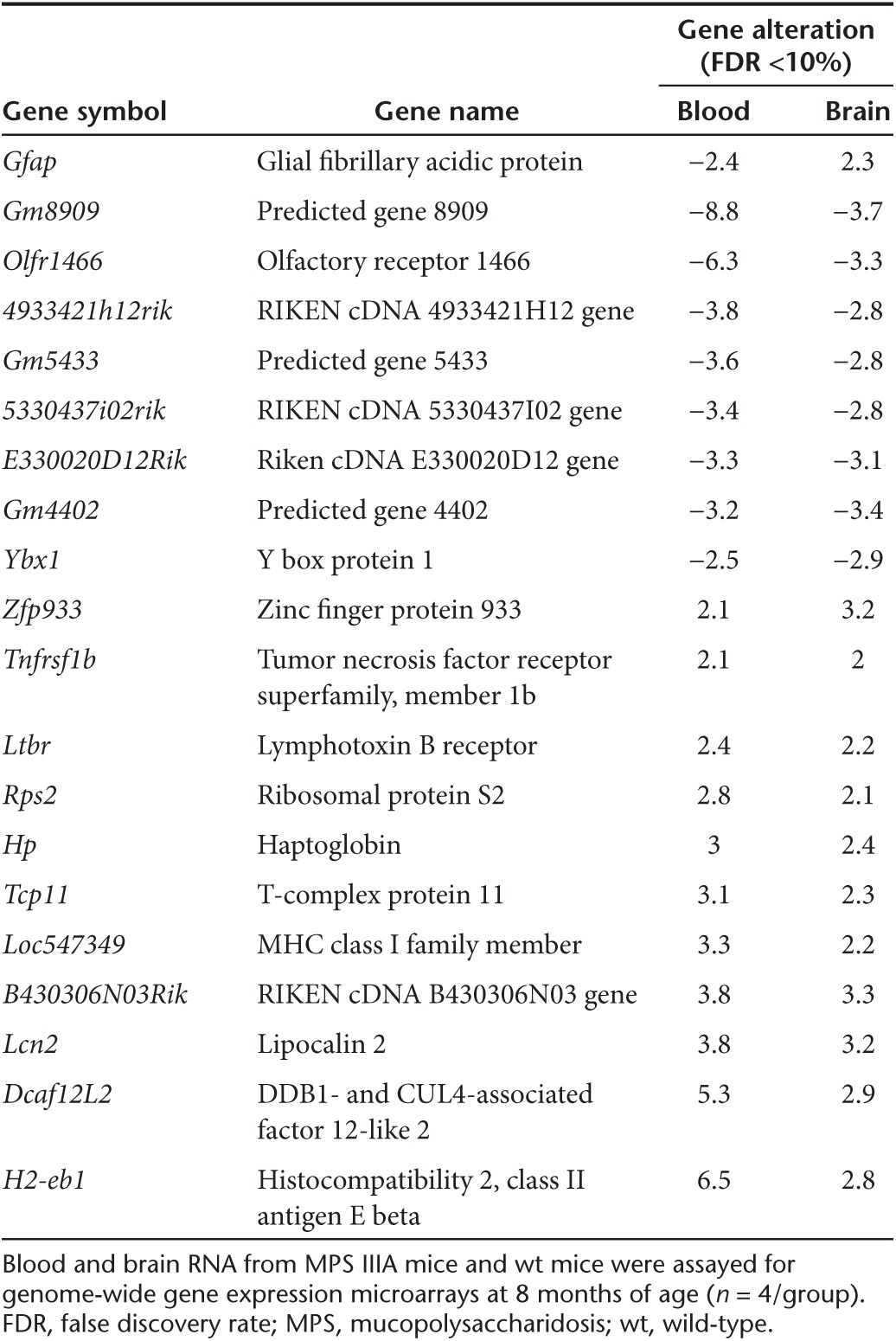
Figure 4.
Brain and blood transcriptional abnormalities affect broad biological functions in MPS IIIA mice. Brain and blood RNAs from 8-month-old wt and nontreated MPS IIIA mice (n = 4/group) were assayed by genome-wide gene expression microarrays. (a, b) Significantly altered transcripts in MPS IIIA mice (greater than or equal to twofold, FDR <10%) were analyzed to generate heat maps using MeV analysis software. MPS 1–4: MPS IIIA mice (in brown frames); WT 1–4: wt mice (in blue frames); red: upregulated genes in AD; green: downregulated genes in AD; color intensity: degrees of changes. (c) These transcriptional dysregulations of known genes were further analyzed by IPA for functional associations and used to generate pie charts. ECM, extracellular matrix; FDR, false discovery rate; IPA, Ingenuity Pathway Analysis; MPS, mucopolysaccharidosis; wt, wild-type.
In MPS IIIA mice treated with scAAVrh74-hSGSH vector, our data showed extensive correction of transcriptional abnormalities (greater than or equal to twofold, FDR <10%) in both the brain (95.9% of transcripts) and blood (97.7% of transcripts) (Table 4, Supplementary Table S1). We observed complete normalization in 182 transcripts (58%), partial correction in 83 transcripts (26.4%), and over correction in 36 transcripts (16.5%) in the brains of MPS IIIA mice. Similarly, in the blood, the scAAVrh74-hSGSH treatment led to normalization in 290 transcripts (73%), partial correction in 79 transcripts (19.9%), and over correction in 19 transcripts (4.8%). No correction was observed in only 13 and 9 dysregulated transcripts in the brain and blood, respectively (Supplementary Table S1). Heat map analyses showed that both the blood and brain transcripts in AAVrh74-treated MPS IIIA mice largely clustered with those in wt mice, but not with those of nontreated MPS IIIA mice (Figure 5).
Table 4. Extensive correction of brain and blood gene dysregulations in MPS IIIA mice by a systemic rAAVrh74-hSGSH gene delivery.
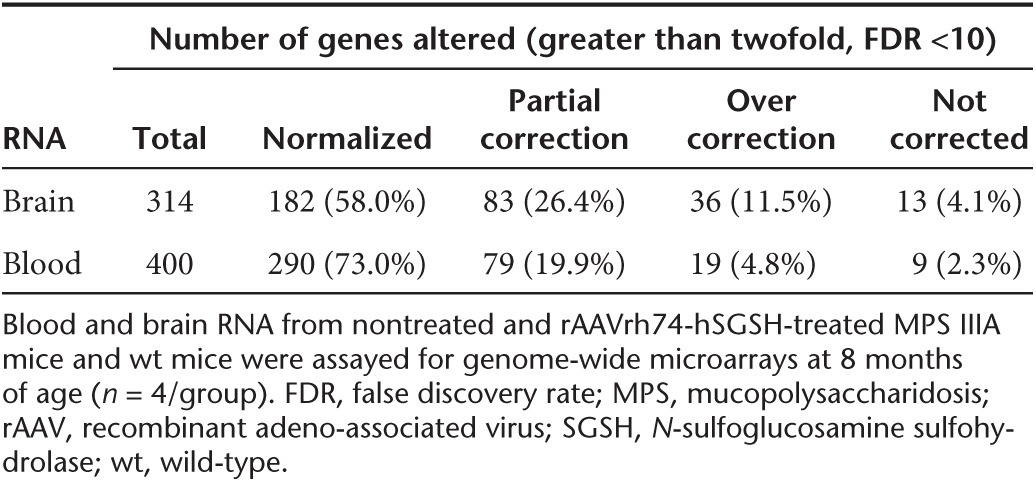
Figure 5.
Brain and blood transcriptional profiles responded well to a single systemic scAAVrh74-hSGSH gene delivery. Brain and blood RNAs from 8 months old wt, nontreated MPS IIIA, and AAVrh74-treated MPS IIIA mice (n = 4/group) were assayed by genome-wide gene expression microarrays. (a, b) Significantly altered transcripts in MPS IIIA mice (≥2-fold, FDR <10%) were analyzed to generate heat maps using MeV analysis software. MPS 1–4: MPS IIIA mice (in brown frames); WT 1–4: wt mice (in blue frames); AAV 1–4: scAAVrh74-treated MPS IIIA mice (in pink frames); red: upregulated genes in AD; green: downregulated genes in AD; color intensity: degrees of changes. AAV, adeno-associated virus; FDR, false discovery rate; MPS, mucopolysaccharidosis; SGSH, N-sulfoglucosamine sulfohydrolase; wt, wild-type.
Discussion
This study demonstrates the therapeutic potential of systemic human SGSH (hSGSH) gene delivery for treating MPS IIIA, using a novel scAAVrh74 vector, an AAV serotype that was initially isolated from nonhuman primates and has the ability to cross the BBB. A single IV injection of rAAVrh74-hSGSH vector mediated the expression of functional rSGSH diffusely throughout the CNS, and also in the PNS and widespread somatic tissues, demonstrating the trans-BBB neurotropism and broad tissue affinity of AAVrh74. Furthermore, the restoration of functional SGSH led to the complete clearance of accumulated GAG in the brain and all seven tested somatic tissues. Additionally, this treatment corrected astrocytosis in both the CNS and PNS of MPS IIIA animals, demonstrating correction of not only the primary pathology (lysosomal GAG accumulation) but also neuroinflammation, a hallmark neuropathology in MPS IIIA. Most importantly, this systemic scAAVrh74-hSGSH gene delivery approach is functionally beneficial, resulted in the behavioral correction and significantly extended survival in MPS IIIA mice. A previously published study showed similar therapeutic effect in MPS IIIA mice, using a conventional single-stranded rAAV9 vector to deliver murine SGSH gene controlled by a stronger promoter, at a vector dose ~10-fold higher than that used in this study.19 This further supports the previous observations that a scAAV vector system offers higher transduction efficiency at lower vector doses than ssAAV vector. We believe that we have developed an effective and minimally invasive gene therapy approach for treating MPS IIIA. The use of the hSGSH gene in the present study makes this approach readily translatable for treating human disease. We also demonstrate the potential of AAVrh74 as an alternative vector of AAV9 for trans-BBB gene delivery, in consideration of preexisting AAV antibodies and the possible need for readministration in patients.
One of the important findings in this study is that genome-wide blood transcriptional profiles reflect the complex pathophysiological status in MPS IIIA and may have potential as effective surrogate outcome measures for therapeutic assessments. Our data showed significant dysregulation in numerous genes involving a broad range of biological functions in both the blood and brain, reflecting the complexity of this profound disease pathology. While there is very little overlap in between them, both the blood and brain transcriptional signatures represent complex molecular abnormalities. These blood and brain gene dysregulations reflect the major components of MPS IIIA pathology, particularly neuroinflammatory processes such as astrocytosis and metabolic impairments, as well as abnormalities associated with DNA/RNA binding and syntheses, apoptosis/cell death and repair, posttranslational modification, cytoskeletal and extracellular matrix functions. Most importantly, we demonstrate for the first time that the transcriptional abnormalities in both the brain and blood responded well to a single systemic scAAVrh74-hSGSH gene delivery in MPS IIIA mice. The treatment led to correction of 95.9 and 97.7% of the transcriptional alterations in the brain and blood respectively, the majority of which were normalized, further supporting the therapeutic potential of our approach in treating both the neurological and somatic disorders of MPS IIIA.
Our data showed more than 20 significantly dysregulated blood genes in MPS IIIA mice that are known to be either enriched in the brain, or are essential to neurological function. These altered blood genes include Gfap, prion protein (Prnp), dedicator of cytokinesis (Dock3), ermin, ERM-like protein (Ermn), gamma-aminobutyric acid (Gaba) A receptor, rho 2 (Gabrr2), huntingtin-associated protein 1 (Hap1), teneurin transmembrane protein 4 (Tenm4), opioid receptor, mu 1 (Oprm1), retinol binding protein 3, interstitial (Rbp3), synaptosomal-associated protein, 25 kDa (Snap25), and synaptophysin (Syp) (http://www.genecards.org). Of these neurological-associated genes, only Gfap was altered inversely in the brain and blood (increased in brain and decreased in blood). None of these genes have a known role in blood cells, suggesting that changes in these transcript levels may be a response to conditions in the CNS, either by altered transcriptional activity in blood or by as yet unknown mechanisms. We hypothesize that the abnormal blood transcriptional profiles reflect biopathophysiological status, including neuropathology in MPS IIIA. It is possible that blood transcriptional signatures have the potential for tracking disease-stage specific pathology, especially changes associated with molecular neuropathology, during disease progression, which may be used as CNS-specific biomarkers for MPS IIIA. This is further supported by our recently published findings of strong blood–brain links in MPS IIIB mice.27 This is particularly important as therapeutic development advances and therapies for MPS IIIA become available, because no specific outcome measures are currently available for MPS corresponding to neurological disease severity or therapeutic responsiveness. This lack of outcome measures will pose a critical challenge for therapeutic assessment.
Finally, our data demonstrate a surprisingly efficient scAAVrh74-mediated transduction and functional impact in the kidney. While showing broad tissue affinity similar to that of AAV9, the IV injection of AAVrh74-hSGSH mediated near wt levels of rSGSH expression and the complete clearance of GAG accumulation in the kidney of MPS IIIA mice. The lysosomal storage pathology manifests predominantly the distal tubule cells in the kidney of MPS IIIA mice.28 The kidney has been a difficult target for AAV-mediated gene delivery. We have observed little impact in previous studies in MPS IIIB mice using rAAV9 or rAAV2 vector,18,29,30 or our ongoing study in MPS IIIA mice using a scAAV9-hSGSH vector (unpublished data). This added tissue tropism suggests broader therapeutic potential of AAVrh74 as gene delivery vector.
In summary, we have developed an efficient gene therapy approach that is functionally beneficial for the treatment of both the neuropathic and somatic manifestations of MPS IIIA. We demonstrate that rAAVrh74 has the ability to cross the BBB, and a single IV injection of scAAVrh74-U1a-hSGSH vector led to efficient restoration of functional SGSH and the correction of lysosomal storage pathology throughout the CNS in MPS IIIA mice. We also report that the SGSH deficiency triggers profound transcriptional impairments in the brain and blood in MPS IIIB mice. Importantly, the blood and brain transcriptional abnormalities were largely corrected in response to systemic scAAVrh74-hSGSH gene delivery, suggesting the therapeutic surrogate outcome measure potential of blood transcriptional signatures.
Materials and Methods
Animals. A spontaneous MPS IIIA mouse colony28 was kindly provided by Dr. Steve U Walkley (Albert Einstein College of Medicine), and maintained on an inbred background (C57BL/6) of backcrosses from heterozygotes in the vivarium at the Research Institute at Nationwide Children's Hospital. The genotypes of progeny mice were identified by PCR with restriction digest.31,32 All animal care and procedures were performed strictly following the approved protocol, in accordance with the Guide for the Care and Use of Laboratory Animals (8th Edition, 2011).
MPS IIIA mice and their age-matched wt littermates were used in the experiments.
rAAV viral vector. An scAAV vector plasmid was constructed to produce scAAVrh74-U1a-hSGSH viral vector. The vector genome contains AAV2 terminal repeats, a murine small nuclear RNA promoter U1a,33 hSGSH coding sequence, and polyadenylation signal from the bovine growth hormone gene. The viral vectors were produced in HEK293 cells using three-plasmid cotransfection including AAV helper plasmid encoding AAV serotype rh74 capsid, and purified by banding on a CsCl step gradient,34 and followed by dialysis into Tris-buffered saline, pH 8.0. The titer of the vector was determined using dot blotting.
Systemic vector delivery. MPS IIIA mice (4–6 weeks old, n = 13), were treated with an IV injection of scAAVrh74-U1a-hSGSH vector (5 × 1012 vector genome/kg, in 150–200 μl Tris-buffered saline) via tail vein. The animals were briefly anesthetized by isoflurane inhalation for vector injection accuracy. Age- and sex-matched wt and nontreated MPS IIIA littermates were used as controls (n = 15/group).
Behavioral tests: hidden task in the Morris water maze. The scAAVrh74-U1a-hSGSH-treated MPS IIIA mice (n = 13, M:F = 7:6) and controls (n = 15/group) were tested for behavioral performance at ~7–7.5 months of age in a hidden task in Morris water maze.35 The water maze consisted of a large circular pool (diameter = 122 cm) filled with water (45-cm deep, 24–26 °C) containing 1% white TEMPERA paint, located in a room with numerous visual cues. Mice were tested for their ability to find a hidden escape platform (20 × 20 cm) 0.5 cm under the water surface. Each animal was given four trials per day, across 4 days. For each trial, the mouse was placed in the pool at one of four randomly ordered locations, and then given 60 seconds to swim to the hidden platform. If the mouse found the platform, the trial ended, and the animal was allowed to remain 10 seconds on the platform before the next trial began. If the platform was not found, the mouse was placed on the platform for 10 seconds, and then given the next trial. Measures were taken of latency to find the platform, swimming distance (cm), and swimming speed (cm/minute) through an automated tracking system (San Diego Instruments, San Diego, CA).
Longevity observation. Following the scAAVrh74-U1a-hSGSH vector injection, mice were continuously observed for the development of humane endpoint criteria, or until death occurred. The endpoint was when the symptoms of late-stage MPS IIIA clinical manifestation (urine retention, rectal prolapse, protruding penis) became irreversible, or when mice showed significant weight loss, dehydration, or morbidity.
Blood and tissue analyses. Blood and tissue analyses were carried out when mice were 8–8.5 months old (n = 4). After the mice were anesthetized with an intraperitoneal injection of Ketamine/Xylazine Cocktail, blood samples were harvested in Paxgene blood tubes for RNA extraction. The animals were then perfused transcardially with cold phosphate-buffered saline (pH 7.4), and brain and multiple somatic tissue samples were collected on dry ice and stored at −80 °C before being processed for analyses.
SGSH activity assay. Tissue samples were assayed for SGSH enzyme activity following previously published procedures.36 The assay measures 4-methylumbelliferone, a fluorescent product formed by hydrolysis of the substrate 4-Methylumbelliferyl-alpha-D-N-sulphoglucosaminide. The SGSH activity is expressed as unit/mg protein. One unit is equal to 1 nmol 4-methylumbelliferone released/17 hour at 37 °C.
GAG content measurement. GAGs were extracted from wet tissues following published procedures with modification.31,37 Dimethylmethylene blue assay was used to measure GAG content.38 The GAG samples (from 0.5 to 1.0 mg tissue) were mixed with H2O to 40 μl before adding 35 nmol/l DMB (Polysciences, Warrington, PA) in 0.2 mmol/l sodium formate buffer (pH 3.5). The product was measured using a spectrophotometer (OD535). The GAG content was expressed as μg/mg tissue.
Total RNA isolation. Total RNA was isolated from brain tissue samples using Qiagen RNeasy Kit, and from whole blood using the Paxgene Blood RNA Kit (Qiagen, Valencia, CA), following the protocols recommended by the manufacturer. To ensure the RNA quality, each extracted total RNA sample was cleaned using Qiagen RNeasy column. Blood RNA were then further processed using GLOBINclear Kit (Ambion, Grand Island, NY), to rapidly deplete α and β-globin messenger RNA for more sensitive gene expression data. RNA concentration was determined using the NanoDrop spectrophotometer (NanoDrop products, Wilmington, DE).
Genome-wide gene expression microarray. RNA samples from study subjects were assayed for transcriptional profiles using the Sureprint G3 Mouse GE 8x60k v2 microarrays (Agilent Technologies, Santa Clara, CA), following manufacturer-provided procedures. This array provides full coverage of murine genes and transcripts, including large intergenic noncoding RNAs. In brief, RNA samples were labeled with Cy3, purified using Qiagen columns, and checked for labeling efficiency using the Nanodrop. The labeled samples were then fragmented and hybridized to the array overnight. Microarray slides were then washed and scanned with Agilent G2505C Microarray Scanner, at 2-μm resolution. Images were analyzed with Feature Extraction 10.10 (Agilent). Median foreground intensities were then be obtained for each spot and imported into the mathematical software package “R.” Using the negative controls on the arrays, the background threshold was determined and all values less than this value were flagged. The data were then normalized by the print tip loess method using the LIMMA (Linear models for microarray data) (Supplementary Table S2) package in “R” as previously described.39 The median value for the replicate probes was used as the expression measurement for that given open reading frame. Complete statistical analysis were performed in “R” using both the LIMMA and Siggenes Bioconductor packages (Significance Analysis of Microarrays) (http://www-stat.stanford.edu/~tibs/SAM/), to determine the FDR and q value for each gene/transcript and combined with a fold change cutoff (e.g., ≤1.5-fold downregulation or ≥1.5-fold upregulation) to identify the final list of differentially expressed transcripts.
Functional analysis of gene expression. Functional analyses were performed to identify the biological functions and/or disease associations of the most significant gene alterations (greater than or equal to twofold, FDR ≤10%) from array data, using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood, CA). Using this same cutoff, transcripts were also identified as known genes and assigned tissue expression categories by the Database for Annotation, Visualization and Integrated Discovery (DAVID) (NIAID, Bethesda, MD)40 in conjunction with Gene Cards (http://www.genecards.org/). Heat maps were generated by MultiExperiment Viewer software (MeV, MA), using the average linkage clustering algorithm and the Pearson correlation as a measure of similarity.
Immunofluorescence. Tissues were processed for thin paraffin sections (4 μm) and IF. The IF staining was performed to identify cells expressing rSGSH, or GFAP for astrocytes, using antibodies against hSGSH (Abcam, Cambridge, MA) or GFAP (Millipore, Billerica, MA), and corresponding secondary antibody conjugated with AlexaFluor568 or AlexaFluor488 (Invitrogen, Grand Island, NY), following procedures recommended by the manufacturers. The sections were imaged under a fluorescence microscope.
Statistics. Data were analyzed using Student's t-test and/or separate one-way analysis of variance to examine group differences. Statistical analyses of microarray data were performed as described above. For all comparisons, significance was set at P < 0.05.
SUPPLEMENTARY MATERIAL Figure S1. AAV-mediated rSGSH expression in the brain and kidney and the correction of astrocytosis. Table S1. Transcriptional abnormalities in the (a) brains and the (b) blood of MPSIIIA mice. Table S2. Confirmation of microarray data by qRT-PCR.
Acknowledgments
We thank Steve U Walkley at Albert Einstein College of Medicine for providing us the MPS IIIA mouse model. We also thank Sanfilippo community for supporting this study through Elisa's Life - Sanfilippo Children's Research Foundation (Canada), Team Sanfilippo, Ben's Dream - Sanfilippo Research Foundation, and LivLife. Both H.F. and D.M.M. are scientific cofounders of Abeona Therapeutics.
Supplementary Material
References
- Neufeld EF, Muenzer J.2001The mucopolysaccharidoses. Scriver CR, Beaudet AL, Sly WS, Valle D.eds.). The Metabolic & Molecular Basis of Inherited Disease8th ed. McGraw-Hill; New York; St Louis; San Francisco; pp. 3421–3452. [Google Scholar]
- McGlynn R, Dobrenis K, Walkley SU. Differential subcellular localization of cholesterol, gangliosides, and glycosaminoglycans in murine models of mucopolysaccharide storage disorders. J Comp Neurol. 2004;480:415–426. doi: 10.1002/cne.20355. [DOI] [PubMed] [Google Scholar]
- Crawley AC, Gliddon BL, Auclair D, Brodie SL, Hirte C, King BM, et al. Characterization of a C57BL/6 congenic mouse strain of mucopolysaccharidosis type IIIA. Brain Res. 2006;1104:1–17. doi: 10.1016/j.brainres.2006.05.079. [DOI] [PubMed] [Google Scholar]
- Wilkinson FL, Holley RJ, Langford-Smith KJ, Badrinath S, Liao A, Langford-Smith A, et al. Neuropathology in mouse models of mucopolysaccharidosis type I, IIIA and IIIB. PLoS One. 2012;7:e35787. doi: 10.1371/journal.pone.0035787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraldi A, Hemsley K, Crawley A, Lombardi A, Lau A, Sutherland L, et al. Functional correction of CNS lesions in an MPS-IIIA mouse model by intracerebral AAV-mediated delivery of sulfamidase and SUMF1 genes. Hum Mol Genet. 2007;16:2693–2702. doi: 10.1093/hmg/ddm223. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Galvin JE, Lee VM, Rorke LB, Dickson DW, Wolfe JH, et al. Accumulation of intracellular amyloid-beta peptide (A beta 1-40) in mucopolysaccharidosis brains. J Neuropathol Exp Neurol. 1999;58:815–824. doi: 10.1097/00005072-199908000-00004. [DOI] [PubMed] [Google Scholar]
- Winder-Rhodes SE, Garcia-Reitböck P, Ban M, Evans JR, Jacques TS, Kemppinen A, et al. Genetic and pathological links between Parkinson's disease and the lysosomal disorder Sanfilippo syndrome. Mov Disord. 2012;27:312–315. doi: 10.1002/mds.24029. [DOI] [PubMed] [Google Scholar]
- Yogalingam G, Hopwood JJ. Molecular genetics of mucopolysaccharidosis type IIIA and IIIB: diagnostic, clinical, and biological implications. Hum Mutat. 2001;18:264–281. doi: 10.1002/humu.1189. [DOI] [PubMed] [Google Scholar]
- Cleary MA, Wraith JE. Management of mucopolysaccharidosis type III. Arch Dis Child. 1993;69:403–406. doi: 10.1136/adc.69.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valayannopoulos V, Wijburg FA. Therapy for the mucopolysaccharidoses. Rheumatology (Oxford) 2011;50 suppl. 5:v49–v59. doi: 10.1093/rheumatology/ker396. [DOI] [PubMed] [Google Scholar]
- Dickson P, McEntee M, Vogler C, Le S, Levy B, Peinovich M, et al. Intrathecal enzyme replacement therapy: successful treatment of brain disease via the cerebrospinal fluid. Mol Genet Metab. 2007;91:61–68. doi: 10.1016/j.ymgme.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley KM, King B, Hopwood JJ. Injection of recombinant human sulfamidase into the CSF via the cerebellomedullary cistern in MPS IIIA mice. Mol Genet Metab. 2007;90:313–328. doi: 10.1016/j.ymgme.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Crawley AC, Marshall N, Beard H, Hassiotis S, Walsh V, King B, et al. Enzyme replacement reduces neuropathology in MPS IIIA dogs. Neurobiol Dis. 2011;43:422–434. doi: 10.1016/j.nbd.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Byrne BJ, Falk DJ, Clément N, Mah CS. Gene therapy approaches for lysosomal storage disease: next-generation treatment. Hum Gene Ther. 2012;23:808–815. doi: 10.1089/hum.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High KA, Aubourg P. rAAV human trial experience. Methods Mol Biol. 2011;807:429–457. doi: 10.1007/978-1-61779-370-7_18. [DOI] [PubMed] [Google Scholar]
- Fu H, McCarty DM. Treating lysosomal storage diseases that affect the central nervous system: overcoming the blood-brain-barrier. Curr Med Lit. 2013;11:33. [Google Scholar]
- Fu H, Dirosario J, Killedar S, Zaraspe K, McCarty DM. Correction of neurological disease of mucopolysaccharidosis IIIB in adult mice by rAAV9 trans-blood-brain barrier gene delivery. Mol Ther. 2011;19:1025–1033. doi: 10.1038/mt.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzo A, Marcó S, García M, Villacampa P, Ribera A, Ayuso E, et al. Correction of pathological accumulation of glycosaminoglycans in central nervous system and peripheral tissues of MPSIIIA mice through systemic AAV9 gene transfer. Hum Gene Ther. 2012;23:1237–1246. doi: 10.1089/hum.2012.029. [DOI] [PubMed] [Google Scholar]
- Duque S, Joussemet B, Riviere C, Marais T, Dubreil L, Douar AM, et al. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:3254–3271. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang H, Yang B, Mu X, Ahmed SS, Su Q, He R, et al. Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol Ther. 2011;19:1440–1448. doi: 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurigot V, Marcó S, Ribera A, Garcia M, Ruzo A, Villacampa P, et al. Whole body correction of mucopolysaccharidosis IIIA by intracerebrospinal fluid gene therapy. J Clin Invest. 2013;123:3254–3271. doi: 10.1172/JCI66778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford-Smith A, Wilkinson FL, Langford-Smith KJ, Holley RJ, Sergijenko A, Howe SJ, et al. Hematopoietic stem cell and gene therapy corrects primary neuropathology and behavior in mucopolysaccharidosis IIIA mice. Mol Ther. 2012;20:1610–1621. doi: 10.1038/mt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergijenko A, Langford-Smith A, Liao AY, Pickford CE, McDermott J, Nowinski G, et al. Myeloid/microglial driven autologous hematopoietic stem cell gene therapy corrects a neuronopathic lysosomal disease. Mol Ther. 2013;21:1938–1949. doi: 10.1038/mt.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton BJ, Duncan FJ, Murrey D, Ware T, Meadows A, McCarty DM, et al. Amyloidosis, synucleinopathy, and prion encephalopathy in a neuropathic lysosomal storage disease: the CNS-biomarker potential of peripheral blood. PLoS One. 2013;8:e80142. doi: 10.1371/journal.pone.0080142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik M, Muller VJ, Rozaklis T, Johnson L, Dobrenis K, Bhattacharyya R, et al. A mouse model for mucopolysaccharidosis type III A (Sanfilippo syndrome) Glycobiology. 1999;9:1389–1396. doi: 10.1093/glycob/9.12.1389. [DOI] [PubMed] [Google Scholar]
- McCarty DM, DiRosario J, Gulaid K, Muenzer J, Fu H. Mannitol-facilitated CNS entry of rAAV2 vector significantly delayed the neurological disease progression in MPS IIIB mice. Gene Ther. 2009;16:1340–1352. doi: 10.1038/gt.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Kang L, Jennings JS, Moy SS, Perez A, Dirosario J, et al. Significantly increased lifespan and improved behavioral performances by rAAV gene delivery in adult mucopolysaccharidosis IIIB mice. Gene Ther. 2007;14:1065–1077. doi: 10.1038/sj.gt.3302961. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya R, Gliddon B, Beccari T, Hopwood JJ, Stanley P. A novel missense mutation in lysosomal sulfamidase is the basis of MPS III A in a spontaneous mouse mutant. Glycobiology. 2001;11:99–103. doi: 10.1093/glycob/11.1.99. [DOI] [PubMed] [Google Scholar]
- Chicoine LG, Rodino-Klapac LR, Shao G, Xu R, Bremer WG, Camboni M, et al. Vascular delivery of rAAVrh74.MCK.GALGT2 to the gastrocnemius muscle of the rhesus macaque stimulates the expression of dystrophin and laminin α2 surrogates. Mol Ther. 2014;22:713–724. doi: 10.1038/mt.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JS, Sethna M, Ramamurthy L, Gowen SA, Samulski RJ, Marzluff WF. Efficient expression of protein coding genes from the murine U1 small nuclear RNA promoters. Proc Natl Acad Sci USA. 1996;93:8852–8857. doi: 10.1073/pnas.93.17.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino-Klapac LR, Montgomery CL, Bremer WG, Shontz KM, Malik V, Davis N, et al. Persistent expression of FLAG-tagged micro dystrophin in nonhuman primates following intramuscular and vascular delivery. Mol Ther. 2010;18:109–117. doi: 10.1038/mt.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton EC, Baird A, Morgan A, Muir JL, Aggleton JP. The conjoint importance of the hippocampus and anterior thalamic nuclei for allocentric spatial learning: evidence from a disconnection study in the rat. J Neurosci. 2001;21:7323–7330. doi: 10.1523/JNEUROSCI.21-18-07323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova EA, Voznyi YaV, Keulemans JL, Hoogeveen AT, Winchester B, Tsvetkova IV, et al. A fluorimetric enzyme assay for the diagnosis of Sanfilippo disease type A (MPS IIIA) J Inherit Metab Dis. 1996;19:278–285. doi: 10.1007/BF01799255. [DOI] [PubMed] [Google Scholar]
- van de Lest CH, Versteeg EM, Veerkamp JH, van Kuppevelt TH. Quantification and characterization of glycosaminoglycans at the nanogram level by a combined azure A-silver staining in agarose gels. Anal Biochem. 1994;221:356–361. doi: 10.1006/abio.1994.1425. [DOI] [PubMed] [Google Scholar]
- de Jong JG, Wevers RA, Laarakkers C, Poorthuis BJ. Dimethylmethylene blue-based spectrophotometry of glycosaminoglycans in untreated urine: a rapid screening procedure for mucopolysaccharidoses. Clin Chem. 1989;35:1472–1477. [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.