Abstract
The majority of bacteria live not planktonically, but as residents of sessile biofilm communities. Such populations have been defined as ‘matrix-enclosed microbial accretions, which adhere to both biological and nonbiological surfaces’. Bacterial formation of biofilm is implicated in many chronic disease states. Growth in this mode promotes survival by increasing community recalcitrance to clearance by host immune effectors and therapeutic antimicrobials. The human gastrointestinal (GI) tract encompasses a plethora of nutritional and physicochemical environments, many of which are ideal for biofilm formation and survival. However, little is known of the nature, function, and clinical relevance of these communities. This review summarizes current knowledge of the composition and association with health and disease of biofilm communities in the GI tract.
Keywords: biofilm, microbiota, gastrointestinal disease, gastrointestinal tract
Introduction
The human gastrointestinal (GI) tract extends from the esophagus through the stomach, small intestine, and large intestine (colon) and terminates in the rectum (Fig. 1). The small intestine is divided proximally-to-distally into the duodenum, jejunum, and ileum. This collection of interconnected organs harbors a diversity of microhabitats that are colonized by microorganisms to varying degrees, depending on local environmental conditions. For the purposes of this article, the oral and nasal cavities will not be regarded as being part of the GI tract, although these anatomical spaces also contain great microbiological complexity (Ledder et al., 2007).
Fig. 1.
The human gastrointestinal tract.
There exists in the GI tract a gradient of colonization, from the relatively sparsely populated esophagus and stomach to the much more heavily colonized colon, which can contain up to 1012 culturable bacteria per gram luminal contents (Hopkins et al., 2002). Evolution has dictated that the GI tract possess a large surface area to facilitate efficient nutrient uptake, its primary physiological role in the body. This coupled to high nutrient availability and a constant influx of microorganisms, together with stable autochthonous populations, makes the GI tract an ideal site for the development of sessile microbial biofilm communities. The microbiome of the gut has recently been determined in 124 subjects, and the microbial diversity indicates that the entire cohort harbors only between 1000 and 1150 prevalent bacterial species and each individual at least 160 such species (Qin et al., 2010). In addition, there were common microbial flora in subjects tested with 75 species common to > 50% of individuals and 57 species common to > 90%.
Those microorganisms in closest proximity to host tissues have the most opportunity for interaction with host physiology, immunity, and metabolism; thus, mucosal populations are arguably the most important component of any host–microbiota interaction, whether beneficial or detrimental. The GI tract microbiota has been implicated in disease states such as inflammatory bowel disease (IBD; Macpherson et al., 1996), colon cancer (Horie et al., 1999a, b), gastric cancer (Björkholm et al., 2003), and irritable bowel syndrome (IBS; Swidsinski et al., 2005). In addition, recent microbiome studies have uncovered a relationship between diet, microbiota, and health status, particularly in older subjects (Claesson et al., 2012).
The GI tract is anatomically divided into ‘upper’ and ‘lower’ sections by the ligament of Treitz; however, from a microbial perspective, this division applies to the GI tract poorly. The colonization gradient in the GI tract, and particularly the large and rapid (relative to the length of the GI tract) increase in microbial population density from the terminal ileum to the cecum, renders possible a convenient – if somewhat artificial given their connectedness – microbial distinction between the ‘upper’ and ‘lower’ GI tracts at the level of the ileocecal valve. We will consider first the nature and influence of microbial biofilms in the upper GI tract, that is to say the esophagus, stomach and small intestine. Following this, we shall venture forth into the lower GI tract.
The upper GI tract
In quantitative terms, the esophagus and stomach carry the lightest bacterial load in the entire digestive system. In comparison with the lower GI tract, comparatively few microbiological investigations have been made on this part of the gut; this is due in part to difficulties in obtaining representative samples. In contradistinction, fecal effluent provides a ready supply of material for investigations of lower gut microbiology. Studies of the upper GI tract that have been carried out indicate that it is sparsely colonized in terms of microbial population density, but exhibits considerable diversity. Culturable bacteria in the healthy esophagus are mainly Gram-positive facultatively anaerobic species such as lactobacilli and streptococci. These are thought to originate primarily in the oral cavity (Macfarlane & Dillon, 2007). While traditionally the stomach has been considered inhospitable for bacteria due to its acidity, using sensitive molecular techniques Bik et al. (2006) identified a surprisingly diverse bacterial population in gastric mucosal biopsies.
Barrett’s esophagus
Barrett’s esophagus (BE) arises in individuals suffering from long-term gastroesophageal reflux disease. In this condition, squamous epithelial cells lining the distal esophagus undergo metaplastic changes, forming a columnar mucosa (Winters et al., 1987). Estimates of BE prevalence vary markedly; indeed, the two largest recent studies gave prevalences of 1.6% and 6.8%, in the general community (Ronkainen et al., 2005) and individuals undergoing endoscopic examination (Rex et al., 2003), respectively. Patients diagnosed with BE have a markedly higher risk of esophageal dysplasia and subsequent adenocarcinoma (Spechler et al., 2001).
To date, there have been three investigations of esophageal mucosal bacterial populations in BE patients. One such retrospective analysis of stored esophageal tissue (Osias et al., 2004) reported increased microbial colonization (mainly Gram-positive cocci) in patients with BE. However, no significant difference was found when aerobic cultures of fresh esophageal biopsy specimens were analyzed. In another investigation, a molecular cloning, and thus nonquantitative, approach was used to identify the bacteria on a mucosal sample from a single BE patient. Twenty-one bacterial species were detected, of which circa 50% were categorized as ‘unidentified’ rumen and oral isolates (Pei et al., 2005).
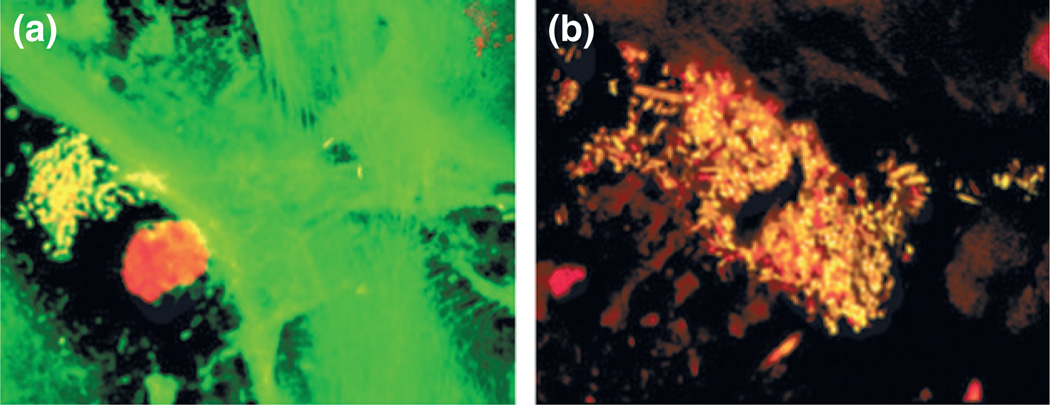
The third, and more detailed, study by Macfarlane et al. (2007) involved analysis of esophageal biopsy and aspirate specimens taken from (1) seven individuals with confirmed BE; and (2) seven controls. Controls, for the purposes of this study, were defined as those persons attending the GI clinic for upper GI tract endoscopy procedures, but who had no evidence of BE by either endoscopic or histologic examination. Each specimen was subjected to analysis by culturing techniques on a variety of solid media under aerobic, anaerobic, and microaerophilic conditions, and bacterial isolates were identified by 16S rRNA gene sequencing. The spatial location of bacterial biofilms on mucosal samples was determined by fluorescence microscopy. A total of 46 bacterial species were detected; interestingly, high levels of Campylobacter concisus and Campylobacter rectus were detected in four of the seven (57.1%) patients with BE, but none of those without. Examination of biopsy material using fluorescence microscopy revealed distinct microcolonies existing within the mucosal layer (Fig. 2).
Fig. 2.
Fluorescence microscopy image of mucosal biopsies from BE patients showing distinct microcolonies existing within the mucosal layer. Original magnification, × 60 (Macfarlane et al., 2007).
Nitrate in the human body is concentrated in the saliva. Some is reduced by bacterial nitrate reductase in the mouth, but the rest is washed into the esophagus and stomach. The finding that the esophagus in some Barrett’s patients was colonized heavily by nitrate-reducing campylobacters raises the possibility that some of the cellular damage observed in the esophagi of BE patients is caused by nitrate and nitric oxide formation. Under low pH conditions, chemical reduction of nitrate can lead to the generation of carcinogenic N-nitroso compounds and nitric oxide (Suzuki et al., 2005). Nitric oxide is capable of inhibiting DNA repair enzymes and can also be mutagenic at high concentrations (Liu et al., 2002). Interestingly, the principal area of nitrite production has been shown to occur at the gastroesophageal junction (Iijima et al., 2002), lending support to the notion of bacterial involvement in mutagenic events associated with BE. Increased numbers of nitrate-reducing veillonellas were also found in patients with BE (Macfarlane et al., 2007) compared with control subjects, and these organisms have been reported to be present in higher levels in oral squamous cell carcinomas (Nagy et al., 1998).
Thus, the role of microorganisms and specifically sessile biofilm bacteria in the pathogenesis of BE is intriguing. However, more work is needed to ascertain what, if any, affect the unique bacterial communities identified in BE patients exert on the host.
The stomach
Historically, the stomach was thought to be a sterile environment; the discovery of Helicobacter pylori colonization dramatically altered this belief. More recently, sensitive molecular techniques have identified the presence of a diverse population of bacteria, including 128 phylotypes from eight bacterial phyla in a study of gastric mucosal biopsies taken from 23 adult subjects (Bik et al., 2006). Not surprisingly, 67% of the identified phylotypes had previously been identified in oral specimens. Sampling contamination or passage of transient microorganisms, either from ingested food or from swallowed oropharyngeal bacteria that are not resident in the stomach, is certainly also present, but their importance is unknown.
Helicobacter pylori
In a significant proportion of the population, the gastric mucosa is colonized by H. pylori (Lehours & Yilmaz, 2007), a phenomenon associated with peptic ulcer disease, achlorhydria (Graham et al., 1988), corpus-predominant gastritis (Harford et al., 2000), and gastric (Peek & Blaser, 2002), and possibly also esophageal (Ye et al., 2004), adenocarcinomas.
Biofilm formation by H. pylori has been observed in vitro at air/liquid interfaces in media with a high carbon/nitrogen ratio (Stark et al., 1999). The capacity to form biofilm does not appear related to cell surface hydrophobicity, motility, or auto-aggregation (Yonezawa et al., 2010), but is strain-dependent (Yonezawa et al., 2009). Furthermore, attachment of H. pylori to glass surfaces and biofilm formation has been reported (Cole et al., 2004). Surface properties affected H. pylori morphology; the highly infectious spiral form was associated with attachment to nonpolymeric substances. Presence of serum in the medium inhibits attachment (Williams et al., 2008). Interestingly, addition of mucin (10% w/v type III porcine) resulted in an increase in planktonic, but not biofilm, H. pylori numbers; thus, the proportion of adherent cells declined upon addition of mucin (Cole et al., 2004). This may be due to mucin-mediated inhibition of H. pylori binding (Simon et al., 1997). However, the significance of this finding is uncertain as the actual number of adherent H. pylori cells remained unchanged. Helicobacter pylori strain TK1402 was able to produce biofilms with greater biomass than other strains; such biofilms contained abundant outer membrane vesicles (Yonezawa et al., 2009).
Helicobacter pylori biofilms have also been directly visualized within the gastric mucosa (Carron et al., 2006; Coticchia et al., 2006; Cellini et al., 2008; Cammarota et al., 2010). Indeed, in subjects with peptic ulcer disease, biofilm covered c. 97% of the surface of urease-positive biopsies compared to c. 1.5% of urease-negative controls (Coticchia et al., 2006). Within 3 days of initial colonization of the gastric mucosa, H. pylori induces profound hypochlorhydria and activates pro-inflammatory pathways that are involved in further development of mucosal pathology (Zavros et al., 2005). Although the precise mechanism of pathogenesis remains unclear, production of IL-1beta by monocytes and neutrophils, themselves recruited through H. pylori-induced IL-8 production by mucosal epithelial cells (Bimczok et al., 2010), inhibits H+, K+-ATPase (proton pump) α-subunit expression (Göõz et al., 2000; Saha et al., 2007). In addition, these infections often demonstrate in vitro and in vivo recalcitrance to even quadruple antimicrobial therapy using antibiotics to which the strains are supposedly sensitive (Megraud et al., 1991; Gisbert, 2008; Cammarota et al., 2010).
Helicobacter pylori possesses a number of virulence factors that assist in gastric mucosal colonization and persistence. Recent evidence has suggested that H. pylori heat shock protein 60 (Hsp60) may be involved in angiogenesis (Lin et al., 2010), itself vital for tumor development. Helicobacter pylori vacuolating toxin (VacA) disrupts actin interaction with parietal cell apical membranes, preventing recruitment and fusion of H, K-ATPase-containing tubulovesicles and causing hypochlorhydria (Wang et al., 2008). Perhaps the best-known H. pylori virulence factor is urease (Mobley et al., 1988), which assists colonization and persistence by modulating the highly acidic conditions in the immediate environment of H. pylori cells. Urease may act either within the bacterial cytoplasm (Weeks et al., 2000), on the cell surface (Baik et al., 2004), or extracellularly (Gobert et al., 2002). Urease-mediated increases in gastric pH may be useful not only for survival of H. pylori; recent evidence suggests that the viscoelasticity of gastric mucus increases as pH rises, facilitating movement of H. pylori through the mucus layer (Celli et al., 2009).
Recently, a study of the biofilm-disrupting compound N-acetylcysteine (NAC) has demonstrated the importance of the biofilm phenotype in human H. pylori infection (Cammarota et al., 2010). In this study of 40 patients, all with a history of multiple failed attempts at H. pylori eradication, SEM documented biofilm in all patients (100%). Patients were randomized to receive 1-week treatment with NAC or placebo prior to culture-guided antibiotic therapy. Thirteen of the 20 patients (65%) who received NAC cleared their infection while only four of the 20 patients (20%) who received placebo did so (P < 0.01). Ten of those who successfully eradicated their H. pylori infection agreed to a follow-up upper endoscopy, and in these patients, SEM showed disappearance of biofilm in all. While these exciting findings should be confirmed in larger studies, they suggest that the biofilm phenotype plays an important role in human GI infection and provides the first evidence that biofilm-directed therapy can be successful for GI diseases.
The small intestine
After being expelled from the stomach through the pyloric sphincter, digestive material is in a highly liquid state due to the addition of gastric juices in the stomach, bile, mucus, and other secretions present in the duodenum itself. The end result is a high flow rate through the small intestine, with average transit times being in the region of 2–4 h. This washing-through of gut contents contributes to the low bacterial load of the duodenum, jejunum, and ileum; bacteria passing through these organs have little opportunity to attach to the mucosa and form biofilm. Bacterial population density increases along the length of the small intestine until a colonic-like community structure is established in the vicinity of the ileo-cecal valve, where numbers of microorganisms present can reach 108–109 CFU per gram contents. A variety of disease states can result in larger numbers of bacteria in the small bowel, for example, achlorhydria (Williams & McColl, 2006).
Enteral nutrition
Patients who are unable to masticate or swallow normally, typically due to cerebrovascular disease, oropharyngeal or esophageal carcinoma, or craniofacial trauma, require nutritional support via an enteral tube. Enteral nutrition (EN) is typically preferred to parenteral nutrition as both animal and human studies have shown it to be safer and more physiological in that it preserves gut barrier and absorptive functions, and immune mechanisms. The 2011 American Society for Gastrointestinal Endoscopy guidelines on the role of endoscopy in enteral feeding recommends nasoenteric feeding as the preferred approach to feeding patients who are expected to resume peroral nutrition within 30 days (Jain et al., 2011). In patients not predicted to resume peroral nutrition within 30 days, they suggest that nutrition be provided by a percutaneous endoscopic gastrostomy (PEG) feeding tube, after first addressing factors such as patient preferences, quality of life, and overall prognosis with the patient and their family. Alternatives to PEG include surgically placed or interventional radiology–placed gastrostomy tubes. Patients with severe gastroesophageal reflux, delayed gastric emptying, or repeated tube feeding-related aspiration pneumonia may benefit from direct or trans-gastric jejunal feeding.
Low gastric pH is generally considered to be a major factor suppressing microbial colonization of the stomach; however, some enteric bacteria possess one or more acid resistance mechanism(s) (Castanie-Cornet et al., 1999) that can confer protection from the bactericidal effects of acid during passage through the stomach. Many innate defense mechanisms break down in EN patients, where a lack of sensory stimuli associated with food intake inhibits saliva production and peristalsis, while reduced swallowing may result in lower gastric acid production and reduce nitrite concentrations. The net effect is greater susceptibility to microbial overgrowth in the stomach and small intestine, at times resulting in diarrhea, although more serious complications such as malabsorption and sepsis also occur (Cabré & Gassull, 1993). The formation of microbial biofilms on EN tubes is an unavoidable consequence of bacterial overgrowth. These structures are difficult to eradicate with antimicrobial agents (Walters et al., 2003) and can harbor pathogens (Bauer et al., 2002) and/or microorganisms carrying antibiotic resistance genes (Ohlsen & Lorenz, 2010).
Nasogastric feeding
During passage through the nasal cavity and esophagus, the NG tube is exposed to nasopharyngeal and esophageal microbiotas. Additionally, the exterior environment and the feeding formula itself, which may be contaminated (Mathus-Vliegen et al., 2006), are other sources of tube contamination. The location of NG tubes in the nasopharynx, esophagus, and stomach ensures a regular supply of nutrients, together with the presence of large numbers of bacteria. Under such conditions, biofilm formation is inevitable. It should also be noted that the NG tube passes close to the larynx, raising the possibility of respiratory tract colonization.
Marrie et al. (1990) undertook microbiological assessments of the external surfaces of the gastric portion of NG tubes recovered from hospitalized patients. They reported that the majority of such tubes were covered in an amorphous biofilm, composed primarily of microcolonies within which bacterial cells were enclosed by an extracellular matrix. These microcolonies were composed both of bacteria of varying morphotypes and yeast cells. Interestingly, a proportion of the observed microcolonies were found to be composed of dead cells and empty cell walls. NG tubes that had been in situ for as little as 24 h were colonized extensively.
A further study evaluated colonization of the oropharynx of elderly patients by Pseudomonas aeruginosa (Leibovitz et al., 2003). Pseudomonas aeruginosa was detected in 18 of 53 (34%) patients receiving NG and none of the controls, while other Gram-negative bacteria were detected in 34 (64%) of NG patients and four (8%) of the controls. Additionally, SEM revealed P. aeruginosa biofilm on tube surfaces. Pulsed-field gel electrophoresis analysis suggested that the oropharynx was the source of tube contamination.
A further study used first-ever introduced NG tubes that had been self-removed by patients between one and 7 days after placement; these tubes were examined by SEM and confocal laser scanning microscopy (Leibovitz et al., 2005). The surfaces of the majority of tubes were covered by biofilm. No quantitative data on the extent, morphology, or composition of NG biofilm was provided in this study.
Segal et al. (2006) investigated the microbiological composition of gastric juices and the nasal cavities of 107 subjects undergoing NG feeding. Potentially pathogenic microorganisms (defined in this study as Gram-negative bacteria or Staphylococcus aureus) were isolated from 74% and 69% of gastric and nasopharyngeal samples, respectively. The most common organisms isolated from gastric juice were Proteus spp. (26%) and Escherichia coli (22%), while Proteus spp. (24%) and Pseudomonas spp. (21%) were the species isolated most frequently from the oropharynx. This study also noted high gastric pH (4.57 ± 0.65 after 3 h NG feeding, and 4.2 ± 0.9 after 12 h). High pH correlated strongly with isolation of pathogenic bacteria, underlining the importance of gastric acid in host defense. The authors hypothesized that the colonized stomach may act as a reservoir of pathogens, leading to aspiration pneumonia in some cases.
Due to the presence of this array of pathogenic biofilm populations on NG tubes, it is not surprising that they can act as a microbial reservoir for a number of diseases associated with NG tubes including nasogastric tube syndrome, microbial pneumonia, sinusitis, middle ear effusion, acute necrotizing esophagitis, and even death (Goldenberg et al., 1990; Le Moal et al.,1999; Apostolakis et al., 2001; Bullock et al., 2004; Lin et al., 2006). As with all mature biofilms forming on indwelling medical devices, the NG tube should be removed and antimicrobial chemotherapy applied to resolve the infection.
Gastrostomy feeding
PEG has the advantage of reduced nasal and oropharyngeal irritation and is typically easier to manage in the home or other community setting, and PEG insertion can facilitate discharge from hospital. PEG tubes can be left in situ for extended periods, but often they require replacement due to either deterioration of the PEG tube itself or its accidental removal by patients.
Candida spp. readily colonize PEG tubes, a phenomenon that may lead to tube deterioration (Gottlieb et al., 1992). Dautle et al. undertook a comprehensive analysis of PEG tube microbiotas using molecular techniques. Random amplified polymorphic DNA (RAPD) analysis was used on material obtained from biofilms that had formed on 18 gastrostomy devices taken from pediatric patients whose age ranged from 6 months to 17 years. These devices had remained in place for a mean time of 20 months (range, 3–47 months). Data indicated that PEG tube biofilms in pediatric patients were compositionally diverse, containing enterococci, staphylococci, E. coli, lactobacilli, candidas, pseudomonads, and bacilli (Dautle et al., 2003).
The gastric and duodenal microbiotas of PEG patients and populations on PEG tube surfaces themselves were evaluated by culturing methods. Interestingly, those individuals who received antibiotics prior to PEG tube placement had both an increased prevalence of some types of infection and decreased mortality rates. The organisms isolated were mainly candidas, enterobacteria, streptococci, staphylococci, and lactobacilli (Table 1; O’May et al., 2005a, b). Data suggested that gastric pH had no significant effect on the density of colonization in the stomachs and duodena of EN patients, although it did affect microbiota composition: Bifidobacterium, Klebsiella, and Staphylococcus spp. were detected only in aspirates with a pH of greater than three. Significantly, E. coli, staphylococci, and candidas were detected only in aspirates from patients who had received antibiotic treatment during their stay in hospital. This was supported by the work of Smith et al. (2011) who used real-time PCR and FISH to investigate microbial colonization of the gastric mucosa of eight PEG patients. Mean levels of enterobacteria and staphylococci were significantly higher in PEG patients than in controls; however, levels of the pro-inflammatory cytokines IL-1α, IL-6, and TNF-α were lower in PEG patients. As with NG tubes, PEG tubes contaminated with a variety of pathogenic microbial biofilms can produce a number of infections, most importantly peristomal infection and the potential for sepsis (Blomberg et al., 2012). Resolution of infection, and prevention of re-infection, may require removal of the PEG because antibiotics alone will not clear biofilm pathogens from a contaminated tube.
Table 1.
Characterization of microorganisms detected in gastric and duodenal aspirates obtained from patients undergoing a PEG placement procedure (O’May et al., 2005a, b)
| Genus | Population size* | |
|---|---|---|
| Gastric aspirates | Duodenal aspirates | |
| Streptococcus | 5.2 ± 0.6 (5) | 4.8 ± 0.5 (11) |
| Staphylococcus | 5.8 ± 0.7 (4) | 4.7 ± 0.8 (6) |
| Proprionibacterium | 3.8 ± 0.4 (3) | ND |
| Peptostreptococcus | 3.8 ± 0.4 (3) | 5.7 ± 0.9 (4) |
| Lactobacillus | 4.0 ± 0.2 (6) | 4.0 ± 0.3 (6) |
| Klebsiella | ND | 4.7 ± 0.6 (5) |
| Gemella | 3.7 (1) | 4.5 ± 1.2 (2) |
| Eubacterium | 3.6 ± 0.1 (3) | 4.6 ± 0.4 (3) |
| Escherichia | 5.4 ± 0.4 (5) | 4.5 ± 0.6 (6) |
| Corynebacterium | 4.4 ± 1.1 (3) | 4.4 ± 0.6 (5) |
| Clostridium | 3.5 ± 0.4 (2) | 4.7 ± 0.4 (2) |
| Bifidobacterium | 4.7 ± 0.3 (3) | 4.8 ± 0.4 (6) |
| Actinomyces | 3.9 ± 0.1 (2) | 5.5 ± 0.6 (3) |
| Candida | 4.6 ± 0.5 (5) | 3.7 ± 0.2 (5) |
ND, Not detected.
Data are expressed as mean log10 CFU ml−1 ± standard deviation (N); Ntotal = 20.
In general, data obtained by in vitro modeling using a chemostat-based system mirrored those of human studies (O’May et al., 2005a, b). Lowering of pH from six to three had no significant effect on the density of planktonic or biofilm communities; indeed, a significant (circa 107 CFU ml−1) microbiota was detected at pH 3. It is important to note that because of the continuous culture methods employed in this study, these recovery data must represent cells actively multiplying in such low pH values. Low pH altered markedly microbiota composition: candidas and lactobacilli were aciduric while numbers of E. coli and Klebsiella pneumoniae decreased concomitantly with pH. Visualization of PEG tube surface-associated biofilm using BacLight™ showed microcolonies composed of both living and dead cells; in many cases, yeast pseudohyphae were found to be invading the interior of microcolonies. Where this occurred, bacterial cells surrounding the pseudohyphae were red-stained. More recent work has established the existence of an interaction between S. aureus and Candida albicans pseudohyphae during biofilm growth (Peters et al., 2010). Differential in-gel electrophoresis demonstrated differential expression of 27 proteins during co-culture biofilm growth. Variation in expression of the virulence-related factors such as α-lactate dehydrogenase 1 (upregulated; Richardson et al., 2008) and CodY (downregulated by contact with C. albicans hyphae; Levdikov et al., 2006) suggests synergistic pathogenesis. CodY has been shown to repress polysaccharide intercellular adhesion-dependent biofilm formation, and production of hemolysins alpha and delta and proteins involved in the agr-dependent quorum-sensing system, a global regulator of virulence (Majerczyk et al., 2010). Thus, downregulation of CodY expression may enable enhanced toxin-mediated virulence and increased biofilm formation in S. aureus. This phenomenon is potentially highly significant and merits further study.
The frequent use of EN makes understanding the mechanisms behind and consequences of microbial colonization in such patients increasingly important. Biofilm formation is inevitable when the upper GI tract becomes overgrown, and a stable nonshedding surface, the tube itself, is in situ for long periods. Early data suggest that the use of antibiotics in such patients may actually increase the probability of colonization by potentially pathogenic microorganisms such as S. aureus and C. albicans. Dosing with pro-, pre-, and synbiotics either before or after tubes are placed may represent a novel method of altering biofilm composition toward a more commensal-type structure.
The lower GI tract
Epithelial surfaces in the GI tract are covered by a layer of mucus, which prevents most microorganisms reaching and persisting on the mucosal surface. This viscoelastic gel is protective against adhesion and invasion by many pathogenic microorganisms, bacterial toxins, end products of metabolism, pancreatic endopeptidases, microbial antigens, and other damaging agents present in the lumen of the bowel. Mucus consists primarily of water (c. 95%) and glycoproteins that give mucus its viscosity and ability to form gel structures.
Mucins are chemically and structurally diverse molecules; however, they always are comprised, to some extent, of galactose and hexosamines, with smaller quantities of fucose (Quigley & Kelly, 1995). The carbohydrate groups exist as both linear and branched oligosaccharides; these can comprise as much as 85% of the molecule (Smith & Podolsky, 1986). Mucin oligosaccharides are attached to a protein core via serine or threonine residues. The attachment of sulfate and sialic acids to terminal mucin oligosaccharides confers resistance to digestion by microbial glycosidases (Corfield et al., 2001). To survive, bacteria resident in the colon must produce a number of hydrolytic enzymes, for example, polysaccharidases, glycosidases, proteases, peptidases. Mucins are important sources of carbohydrate for saccharolytic bacteria, particularly populations in the distal colon, where the supply of fermentable carbohydrate is usually limiting (Macfarlane et al., 1992).
Some bacteria can invade the mucus layer, and many intestinal microorganisms use these molecules as carbon, nitrogen, and energy sources (McCormick et al., 1988). The removal of carbohydrates and other components, such as sulfate, from the glycoprotein compromises its protective function (Schrager & Oates, 1978), particularly when the rate of mucus breakdown exceeds that of its synthesis and secretion.
Pure and mixed culture studies have established that in many gut bacteria, synthesis of degradative enzymes, particularly β-galactosidase, N-acetyl β-glucosaminidase, and neuraminidase, is catabolite regulated (Macfarlane et al., 1989, 1997; Macfarlane & Gibson, 1991) and therefore dependent on local concentrations of mucin and other carbohydrates. While some colonic microorganisms can produce several different glycosidases (Macfarlane et al., 1990), the majority of experimental data suggest that the breakdown of mucin is a cooperative activity (Macfarlane et al., 1999). Studies on biofilm communities in the gut have demonstrated the presence of bacterial microcolonies on mucosal surfaces in healthy people (Fig. 3; Macfarlane & Macfarlane, 2004). Despite its undoubted significance, few studies have focused on mucosal bacterial communities. However, there is evidence to suggest that mucosal populations are distinct from those in the gut lumen (Macfarlane & Macfarlane, 2004), and these are thought to play an important role in IBD (see below). Despite this, little is known about bacterial growth in the mucus layer, the organisms that colonize this microcosm, or their role in disease processes.
Fig. 3.
Confocal laser scanning microscopy of a bacterial microcolony on healthy rectal mucosa stained with a live/dead stain. The microcolony was sectioned in 1.5 µm slices from the lumen (a) to the mucosal surface (i). Original magnification, × 60 (Macfarlane & Macfarlane, 2004).
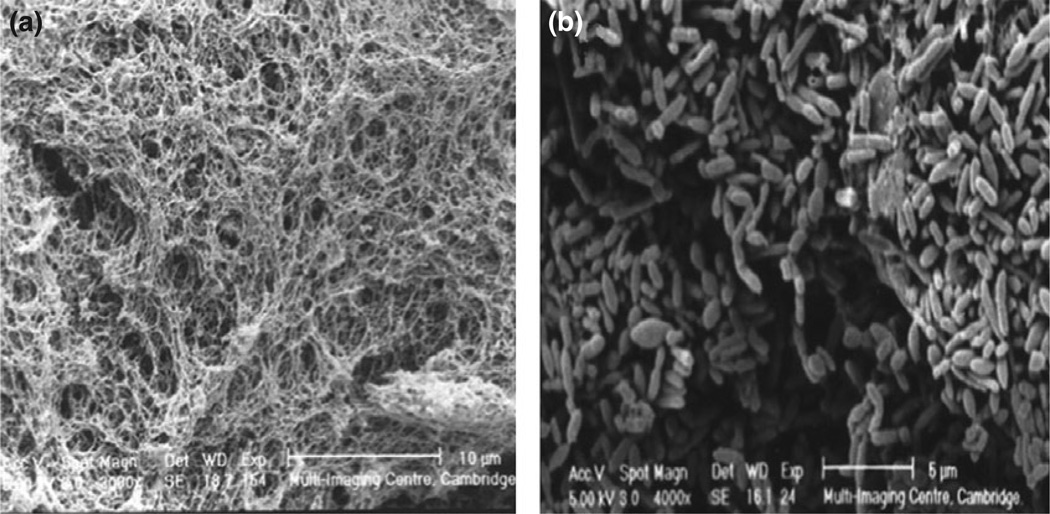
Chemostat-based modeling studies (Macfarlane et al., 2005) have shown differential colonization of artificial mucin gels by fecal bacteria in a two-stage continuous culture system, simulating the nutrient availability of the proximal (vessel 1) and distal (vessel 2) colon. The establishment of bacterial communities in mucin gels was investigated by selective culture methods, SEM, and confocal laser scanning microscopy, in association with fluorescently labeled 16S rRNA gene oligonucleotide probes. Mucin gels were rapidly colonized by heterogeneous bacterial populations (Fig. 4), particularly members of the Bacteroides fragilis group, enterobacteria, and clostridia. Intestinal bacterial populations growing on mucin surfaces were found to be phylogenetically and metabolically distinct from their planktonic counterparts.
Fig. 4.
SEM image of chemostat-housed mucin gels showing rapid colonization by heterogeneous bacterial populations, particularly members of the Bacteroides fragilis group, enterobacteria, and clostridia (Macfarlane et al., 2005).
Inflammatory bowel disease
The two most common forms of idiopathic IBD are UC and Crohn’s disease (CD). It is estimated that more than one million Americans suffer from IBD. UC affects only the mucosal surfaces in the large intestine and rectum. CD can occur anywhere in the digestive tract, often with inflammatory lesions spreading deep into the layers of affected tissues. UC, CD, and acute self-limited colitis (ASLC) all cause diarrhea, with or without accompanying bleeding. However, UC and CD are chronic inflammatory diseases, as opposed to ASLC (mainly infectious agents) and IBS, which is not accompanied by overt inflammation (Steed et al., 2008).
Recent studies of the gut microbiota of patients with IBD have in general terms found a decline in microbial flora diversity (Frank et al., 2007) and methanogens (Scanlan et al., 2008), and an increase in fungal diversity (Ott et al., 2008). Furthermore, despite strenuous efforts to identify microbial community compositions unique to IBD states, none have as yet been elucidated (Reiff & Kelly, 2010). Frank et al. (2007) performed an rRNA sequence analysis of diverse intestinal biopsies from both diseased and normal tissues of patients with IBD and healthy controls. Data suggested depletion of the commensal phyla Firmicutes and Bacteroidetes. The authors suggest treatment of at least some forms of IBD by targeted antimicrobial chemotherapy. More recently, Qin et al. (2009) utilized Illumina-based bacterial profiling to determine the microbiome differences between the healthy individuals and those suffering from IBD. Patients’ microbial profiles clearly separated patients with IBD from healthy individuals and the patients with UC from the patients with CD.
Other authors have echoed this view. Notably, Greenberg suggested that although a cursory examination of available clinical trials would lead to the conclusion that the use of antibiotics in Crohn’s is – at best – ineffective, a more in-depth examination of both clinical and laboratory evidence may lead to the opposite conclusion (Greenberg, 2004). As it is likely that IBD represents a number of disease states, the symptoms of which are often indistinguishable, it follows that microbial community composition will be similarly diverse. Thus, any attempt at treating such a diversity of disease states with a single strategy is likely to fail.
Ulcerative colitis
UC is a chronic relapsing form of IBD, and the precise etiology of which is unknown. In UC, the inflammatory response is located principally within the colonic mucosa. The distal colon is always affected, and the disease usually progresses from its initiation site in the distal bowel toward the proximal large intestine. UC, depending on the severity of the condition, can severely affect the quality of life, and if medical treatments are not effective, surgical removal of all or most of the colon is necessary.
Involvement of commensal gut bacteria in both the initiation and maintenance of UC has been suggested since the early 1970s (Hill et al., 1971). Antimicrobial agents specifically active against obligate anaerobes have been shown to prevent ulceration in guinea pigs (Onderdonk & Bartlett, 1979), while experiments using germ-free animals show that they only develop colitis when repopulated with fecal bacteria (Sadlack et al., 1993). A variety of species including Fusobacterium spp., Shigella spp. (Onderdonk et al., 1983) and adhesive E. coli (Dickinson et al., 1980) isolated from the colitic bowel have been implicated in disease etiology; however, no specific microorganisms have been found in all individuals suffering from UC, and Koch’s postulates cannot be demonstrated. The luminal microbiota of patients with UC has been examined extensively (Swidsinski et al., 2005, 2008a, b; Macfarlane et al., 2009; Swidsinski et al., 2009; Ott et al., 2008; Reiff & Kelly, 2010). There is good evidence that bacteria growing on the gut wall play an important role in UC, because they exist in close juxtaposition to host tissues, and can interact with the host immune and neuroendocrine systems. This is particularly so given that FISH imaging has suggested that mucosal bacterial populations are in contact with the mucosal epithelium in UC and Crohn’s patients, but not in healthy individuals (Swidsinski et al., 2009).
Bacterial populations compositionally distinct from those in the gut lumen are known to exist on the mucosal surface, and in the mucus layer in the large gut (Poxton et al., 1997), where Bacteroides and fusobacteria appear to predominate, but other groups such as eubacteria, clostridia, and anaerobic Gram-positive cocci are also present as either heterogeneous populations or microcolonies (Croucher et al., 1983). Until relatively recently, there have been comparatively few studies on bacteria that inhabit the colonic mucosa, largely due to two factors: Firstly, feces and other types of material from the gut lumen are easier to obtain than tissue samples from the gut wall, and secondly, in most studies individuals taking part have been treated prophylactically with antibiotics and other types of drug (e.g. anti-inflammatory drugs and steroids), or the bowel has been purged before colonoscopy. As a consequence, the metabolic and health-related significance of bacteria growing as biofilms on the colonic mucosa is only now beginning to be elucidated.
The notion that biofilm growth in the mucus layer is important in the pathogenesis of UC is considered likely given that (1) mucosal bacteria have been visualized colonizing the colonic mucosa in patients with UC (Macfarlane et al., 2004); and (2) the condition’s intractability to antibiotic treatment. Antimicrobial agents are still used in treating patients with IBD, mostly in people with severe disease, as in patients with fistulae or other septic-type complications, and occasionally as a first-line therapy. The employment of antibiotic therapy seems mainly to be based on reported benefits observed in individual patients, that is, on small numbers of or individual case studies (Greenberg, 2004; Thompson-Chagoyán et al., 2005). Also, in a recent meta-analysis, Wang et al. (2012) found that antimicrobial therapy improved clinical outcomes of patients with IBD. However, the long-term improvement may be limited due to the ‘rebound effect’ following cessation of antibiotic treatment described by Swidsinski et al. (2008a, b). This study suggested that while mucosal bacterial populations are suppressed during antibiotic treatment, those communities re-establish to at least their previous level after therapy is stopped. In this study, the ‘rebound effect’ was observed when bacterial populations in antibiotic-treated individuals were measured 4 weeks after cessation of treatment. Bacterial numbers were circa 25 times higher than in those who had not been treated. This rebound effect was found to diminish over time, but was still present up to 36 weeks after cessation of antimicrobial therapy. The ‘rebound effect’ seemed to cause increases in the very types of bacteria that were the targets of antibiotic therapy, for example, Bacteroides (targeted by metronidazole) and enterobacteria (targeted by ciprofloxacin). The data collected in this study also suggested, although inconclusively, that the organisms detected were less metabolically active than in nontreated individuals. Bacteria in antibiotic-treated samples were visualized by DAPI staining, but not by fluorescence in situ hybridization (FISH). The authors postulate that this may have been due to reduced rRNA levels within the bacteria, reflecting a lower level of protein synthesis and so reduced metabolic activity and possibly also lower viability.
Results from this study may provide some insight as to why IBD does not seem to respond to antibiotic treatment, despite the widely held belief that gut mucosa-associated bacteria are involved in disease pathogenesis. The mechanism behind the ‘rebound effect’ remains unclear, although it seems likely that survivor bacteria in the mucus layer are able to utilize nutrients that are not assimilated by microbial communities killed by the antibiotic. Further work is needed to confirm this, however. Of wider importance is the question of whether this ‘rebound effect’ is a general property of biofilm, either in the body or more universally. If so, it represents a potentially important new area of inquiry.
A promising new therapy for IBD involves the oral administration of probiotics, prebiotics, or synbiotics. Probiotics are defined as live microorganisms with a demonstrable health benefit when ingested by or otherwise administered to the human host; prebiotics are food ingredients that selectively stimulate the growth and/or the activity of intestinal bacteria that have health-promoting properties (Steed et al., 2008). At the present time, the overwhelming preponderance of prebiotics are nondigestible oligosaccharides (NDO), of which galacto-oligosaccharides (GOS), lactulose, inulins, and their fructooligosaccharide (FOS) derivatives have been by far the most extensively investigated (Macfarlane et al., 2006, 2008). It is important to note that the term nondigestible refers only to the host; bacteria resident in the gut are capable of utilizing prebiotic polysaccharides as energy sources. One key difference between pro- and prebiotics is that probiotics are allochthonous microorganisms, whereas prebiotics can only influence those bacteria already resident with the gut of the patient. Therefore, incoming probiotic bacteria have to overcome the colonization resistance offered by the bacteria in the resident microbiota who have already established themselves within the metabolic and spatial microenvironments close to or on the gut wall. A synbiotic is the combination of a pro- and prebiotic in one; the terms comes from the idea that the two, when used together, will (1) be more likely to be able to overcome colonization resistance; and (2) may have a synergistic effect on the host.
Furrie et al. (2005) reported on a double-blinded randomized controlled trial in which a synbiotic was fed to patients with UC for a period of 1 month. Eighteen patients took part in this study; those selected to receive the synbiotic were provided with six grams of synergy 1 (oligofructose-enriched inulin) and 2 × 1011 live Bifidobacterium longum per day, which they were asked to take twice daily. Results showed that bifidobacterial numbers on the rectal mucosa increased by > 40-fold in those subjects who had received the synbiotic compared with a fourfold increase in the control group. This was accompanied by significant reductions in mucosal pro-inflammatory cytokines (TNF-α, IL-1β) together with inducible human β-defensins 2, 3, and 4. β-Defensins are antimicrobial short-chain peptides produced by gut epithelial cells during inflammation. However, unlike other immune system mediators such as TNF-α and IL-1β, β-defensins are not formed by immune inflammatory cells infiltrating the mucosa. For this reason, β-defensins are useful markers of epithelial surface healing. Histologic assessments indicated marked, although not significant, reductions in inflammatory cells and crypt abscesses in patients receiving the synbiotic, together with regeneration of normal tissue, while sigmoidoscopy scores and clinical activity indices in these individuals also improved. This short-term pilot study provided preliminary data supporting the notion that synbiotic administration has the potential to be developed into acceptable therapies for patients suffering from active UC, but further work is needed to investigate the long-term efficacy of synbiotics in inducing and maintaining remission.
Crohn’s disease
Compared to UC, the evidence for sessile mucosal bacterial involvement in the pathogenesis and maintenance of CD is sparse. Concentrations of mucosal bacteria in patients with CD were found to be two logs higher than in healthy controls or patients with IBS. Of these, Bacteroides spp. predominated in patients with CD, in some individuals comprising c. 80% of total mucosal bacteria, compared with c. 15% in IBS (Swidsinski et al., 2005). Furthermore, these populations were found to be directly adjacent to the epithelium in patients with CD but not healthy controls (Swidsinski et al., 2009). The stability of bacterial diversity over time, particularly during active CD episodes and relapses, in patients with CD is lower than that in healthy controls (Scanlan et al., 2006). Therefore, the constantly changing microbial populations on the colonic mucosa of patients with CD may account – at least in part – for the aberrant immune responses characteristic of the condition. Alternatively, these alterations in the microbiome may themselves be caused by changes in disease activity.
In contrast, an rRNA sequence analysis of the microbial communities of colonic biopsies from patients with CD and healthy controls suggested depletion of normal commensals, such as Bacteroides spp. Furthermore, stratification of patients into a number of microbiota groupings suggests that CD represents a number of disease states (Frank et al., 2007). However, another study suggested that the dominant mucosal-associated bacteria in inflamed and noninflamed tissue in patients with CD did not differ (Vasquez et al., 2007).
Interest in a role for adherent-invasive E. coli (AIEC) in CD (Darfeuille-Michaud, 2002) is increasing because this microorganism is more prevalent in patients with CD than in healthy individuals in a number of countries, for example, the UK (Martin et al., 2004), France (Darfeuille-Michaud et al., 2004), and the United States (Baumgart et al., 2007). AIEC strains are adherent to and can invade colonic epithelial cells in vitro, as well as survive and multiply inside macrophages. Furthermore, intracellular growth of AIEC does not induce apoptosis or tumor necrosis factor (TNF) production. AIEC does not appear to be genetically unique, but does possess genes associated with the virulence of extra-intestinal pathogenic E. coli (Martinez-Medina et al., 2009a). The biofilm-producing capacity of AIEC strains from the colonic mucosa was compared to that of non-AIEC strains by Martinez-Medina et al. Specific biofilm formation indices were significantly higher among AIEC strains compared to other colonic E. coli isolates (Martinez-Medina et al., 2009b). Moreover, AIEC strains also exhibited greater adherence and invasion indices. Biofilm-producing AIEC strains were more frequently motile and positive for the S fimbriae-encoding sfa/focDE virulence genes. Thus, the extant data on the role of AIEC in CD warrants further investigation into the nature and pathogenic mechanisms of this bacterium.
Patients with CD have higher levels of serum IgG specific to a number of microbial antigens. IgG levels to the ASCA epitope of Saccharomyces cerevisiae are elevated in many patients with CD (McKenzie et al., 1990). This is particularly interesting given (1) the increased incidence of S. cerevisiae in patients with CD has been reported (Ott et al., 2008); and (2) that this epitope is also expressed by both C. albicans and Mycobacterium paratuberculosis (Mpofu et al., 2007). Levels of flagellin-specific serum IgG, for example, CBir1, are higher in CD populations, but not in either those suffering from UC or in healthy controls (Lodes et al., 2004). An intestinal E. coli strain, O83:H1, has been found to adhere to and invade colonic epithelial cells in vitro when flagellated, but not in the absence of a flagellum (Eaves-Pyles et al., 2008). The serum IgG response to OmpC, gASCA, AMPCA, ALCA, and ACCA in patients with CD has been linked to both the complicated disease phenotype and the need for surgery (Papp et al., 2008). However, it is also possible that the increases in serum IgG levels reported in the aforementioned studies are merely reflective of a more general increase in IgG levels to multiple microbial antigens in patients with CD. Indeed, Adams and co-authors reported that levels of IgG specific to mannan and flagellin were no more effective for diagnosis of CD than IgG levels against complex mixtures of antigens from gut commensal bacteria such as Bacteroides vulgates (Adams et al., 2008).
The link between biofilms and disease
As described in Table 2, there have been a number of studies that have shown the simultaneous inflammation, a disease process, and microbial biofilm communities in the affected GI location. A set of criteria were previously proposed by Parsek & Singh (2005) to demonstrate a link between biofilm formation and human disease. These criteria include direct examination of an infected tissue revealing pathogenic bacteria in communities attached to a surface where there is a localized infection and evidence of recalcitrance to antibiotic treatment despite the antibiotic sensitivity demonstrated by planktonic forms.
Table 2.
Evidence of microbial populations existing as biofilms in the GI tract
| Biofilm location | Disease process | Biofilm evidence | References |
|---|---|---|---|
| Esophagus mucosa of acid reflux patients | BE | FISH on biopsy samples | Macfarlane et al. (2007) |
| Stomach | Helicobacter pylori –induced ulcers | Culture, SEM | Megraud et al. (1991); Carron et al. (2006); Coticchia et al. (2006); Cellini et al. (2008); Gisbert (2008); Cammarota et al. (2010) |
| Nasogastric tubes | Pseudomonas aeruginosa, Enterobacteriaceae, biofilms on tubes | Culture, SEM | Goldenberg et al. (1990); Le Moal et al. (1999); Apostolakis et al. (2001); Leibovitz et al. (2003, 2005); Bullock et al. (2004); Lin et al. (2006); Hurrell et al. (2009) |
| PEG | Contamination of tubing with Candida spp., lactobacilli, E. coli and Klebsiella pneumoniae biofilms | Culture, fluorescence microscopy | O’May et al. (2005a, b); Blomberg et al. (2012) |
| Large intestines | IBD (UC and Crohn’s) | FISH imaging showing mucosal bacterial populations in contact with the mucosal epithelium in patients with IBD, not in healthy individuals | Macfarlane & Macfarlane (2004); Swidsinski et al. (2009) |
| Large intestines | Biofilms in healthy colons with normal flora | Culture, fluorescence microscopy | Macfarlane & Macfarlane (2004) |
GI biofilm diseases that may fulfill these criteria include H. pylori infection, BE, IBD including Crohn’s and ulcerative colitis (UC), and nasogastric (NG)/PEG tubes. In the case of H. pylori biofilms in GI diseases, the causal link between localized biofilms and host disease, as well as recalcitrance to antimicrobial therapy, is well documented. Helicobacter pylori biofilms have been directly visualized within the gastric mucosa, and the resistance of these microbial populations to eradication by antimicrobials can make treatment difficult (Megraud et al., 1991; Carron et al., 2006; Coticchia et al., 2006; Cellini et al., 2008; Gisbert, 2008; Cammarota et al., 2010). Another GI disease, BE, is correlated with the local nitrate reduction demonstrated by the biofilm communities of campylobacters and veillonellas that may contribute to the metaplastic changes seen in the squamous epithelial cells of the esophagus in BE patients (Macfarlane et al., 2007). Although intriguing, designing a prospective study to demonstrate a causal relationship between the presence of these bacteria and progression to BE represents a significant challenge. The microbial communities associated with IBD have been described as well as the positive effects on antibiotic treatment in these diseases (Macfarlane & Macfarlane, 2004; Wang et al., 2012). However, like other biofilm diseases, once antibiotic therapy is withdrawn, patients can suffer from a ‘rebound effect’ in which the biofilm bacteria not eliminated by the antimicrobial agents are able to reseed the GI tract and restore the symptoms associated with IBD, whether Crohn’s or UC (Swidsinski et al., 2009). Biofilms have also been well documented in the contamination of indwelling medical devices on neonatal and elderly nasogastric tubes and PEGs (Goldenberg et al., 1990; Le Moal et al.,1999; Apostolakis et al., 2001; Leibovitz et al., 2003; Bullock et al., 2004; Leibovitz et al., 2005; O’May et al., 2005a, b; Lin et al., 2006; Hurrell et al., 2009; Blomberg et al., 2012). The microbial species includes Enterobacteriaceae, S. aureus, lactobacilli, and Candida spp., all having well-described recalcitrance to antimicrobial agents when grown as a biofilm compared to their planktonic counterparts. Therefore, in the plethora of diseases associated with these tubes, removal of the device may be the only way to resolve the infection.
Conclusions
The GI tract contains the highest concentration of bacteria anywhere within the human body. It is constantly exposed to materials originating from the external environment, which help to maintain a constant supply of nutrients for its resident microbiotas. A more conducive environment for biofilm formation is difficult to imagine. Information available at the present time suggests that microorganisms residing in the GI tract do indeed form biofilms on any available surface, including those introduced as part of a medical intervention. Despite this ubiquity, the number of studies on these unique microbial communities is small when compared to other sites in the human body. These communities will, in future, no doubt be found to be involved in the pathogenesis of many human diseases.
Footnotes
This timely review on the significance of microbial biofilms and gastrointestinal disease will stimulate research in this field.
References
- Adams RJ, Heazlewood SP, Gilshenan KS, O’Brien M, McGuckin MA, Florin TH. IgG antibodies against common gut bacteria are more diagnostic for Crohn’s disease than IgG against mannan or flagellin. Am J Gastroenterol. 2008;103:386–396. doi: 10.1111/j.1572-0241.2007.01577.x. [DOI] [PubMed] [Google Scholar]
- Apostolakis LW, Funk GF, Urdaneta LF, McCulloch TM, Jeyapalan MM. The nasogastric tube syndrome: two case reports and review of the literature. Head Neck. 2001;23:59–63. doi: 10.1002/1097-0347(200101)23:1<59::aid-hed9>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Baik SC, Kim KM, Song SM, et al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J Bacteriol. 2004;186:949–955. doi: 10.1128/JB.186.4.949-955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer TT, Torres A, Ferrer R, Heyer CM, Schultze-Werninghaus G, Rasche K. Biofilm formation in endotracheal tubes. Association between pneumonia and the persistence of pathogens. Monaldi Arch Chest Dis. 2002;57:84–87. [PubMed] [Google Scholar]
- Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. P Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimczok D, Clements RH, Waites KB, Novak L, Eckhoff DE, Mannon PJ, Smith PD, Smythies LE. Human primary gastric dendritic cells induce a Th1 response to H. pylori. Mucosal Immunol. 2010;3:260–269. doi: 10.1038/mi.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm B, Falk P, Engstrand L, Nyrén O. Helicobacter pylori: resurrection of the cancer link. J Intern Med. 2003;253:102–119. doi: 10.1046/j.1365-2796.2003.01119.x. [DOI] [PubMed] [Google Scholar]
- Blomberg J, Lagergren J, Martin L, Mattsson F, Lagergren P. Complications after percutaneous endoscopic gastrostomy in a prospective study. Scand J Gastroenterol. 2012;47:737–742. doi: 10.3109/00365521.2012.654404. [DOI] [PubMed] [Google Scholar]
- Bullock TK, Waltrip TJ, Price SA, Galandiuk S. A retrospective study of nosocomial pneumonia in postoperative patients shows a higher mortality rate in patients receiving nasogastric tube feeding. Am Surg. 2004;70:822–826. [PubMed] [Google Scholar]
- Cabré E, Gassull MA. Complications of enteral feeding. Nutrition. 1993;9:1–9. [PubMed] [Google Scholar]
- Cammarota G, Branca G, Ardito F, et al. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol. 2010;8:817–820.e813. doi: 10.1016/j.cgh.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Carron MA, Tran VR, Sugawa C, Coticchia JM. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg. 2006;10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, Bansil R. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. P Natl Acad Sci USA. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellini L, Grande R, Di Campli E, Traini T, Giulio MD, Lannutti SN, Lattanzio R. Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand J Gastroenterol. 2008;43:178–185. doi: 10.1080/00365520701675965. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. 2004;186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield AP, Carroll D, Myerscough N, Probert CS. Mucins in the gastrointestinal tract in health and disease. Front Biosci. 2001;6:D1321–D1357. doi: 10.2741/corfield. [DOI] [PubMed] [Google Scholar]
- Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg. 2006;10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Croucher SC, Houston AP, Bayliss CE, Turner RJ. Bacterial populations associated with different regions of the human colon wall. Appl Environ Microbiol. 1983;45:1025–1033. doi: 10.1128/aem.45.3.1025-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn’s disease. Int J Med Microbiol. 2002;292:185–193. doi: 10.1078/1438-4221-00201. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Dautle MP, Wilkinson TR, Gauderer MW. Isolation and identification of biofilm microorganisms from silicone gastrostomy devices. J Pediatr Surg. 2003;38:216–220. doi: 10.1053/jpsu.2003.50046. [DOI] [PubMed] [Google Scholar]
- Dickinson RJ, Varian SA, Axon AT, Cooke EM. Increased incidence of faecal coliforms with in vitro adhesive and invasive properties in patients with ulcerative colitis. Gut. 1980;21:787–792. doi: 10.1136/gut.21.9.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Jezek GE, Islas-Islas M, Torres AG. Escherichia coli isolated from a Crohn’s disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397–409. doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. P Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O’neil DA, Macfarlane GT. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut. 2005;54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisbert JP. “Rescue” regimens after Helicobacter pylori treatment failure. World J Gastroenterol. 2008;14:5385–5402. doi: 10.3748/wjg.14.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, Mersey BD, Cheng Y, Blumberg DR, Newton JC, Wilson KT. Cutting edge: urease release by Helicobacter pylori stimulates macrophage inducible nitric oxide synthase. J Immunol. 2002;168:6002–6006. doi: 10.4049/jimmunol.168.12.6002. [DOI] [PubMed] [Google Scholar]
- Goldenberg SP, Wain SL, Marignani P. Acute necrotizing esophagitis. Gastroenterology. 1990;98:493–496. doi: 10.1016/0016-5085(90)90844-q. [DOI] [PubMed] [Google Scholar]
- Göõz M, Hammond CE, Larsen K, Mukhin YV, Smolka AJ. Inhibition of human gastric H(+)-K(+)-ATPase alpha-subunit gene expression by Helicobacter pylori. Am J Physiol Gastrointest Liver Physiol. 2000;278:G981–G991. doi: 10.1152/ajpgi.2000.278.6.G981. [DOI] [PubMed] [Google Scholar]
- Gottlieb K, DeMeo M, Borton P, Mobarhan S. Gastrostomy tube deterioration and fungal colonization. Am J Gastroenterol. 1992;87:1683. [PubMed] [Google Scholar]
- Graham DY, Alpert LC, Smith JL, Yoshimura HH. Iatrogenic Campylobacter pylori infection is a cause of epidemic achlorhydria. Am J Gastroenterol. 1988;83:974–980. [PubMed] [Google Scholar]
- Greenberg GR. Antibiotics should be used as first-line therapy for Crohn’s disease. Inflamm Bowel Dis. 2004;10:318–320. doi: 10.1097/00054725-200405000-00021. [DOI] [PubMed] [Google Scholar]
- Harford WV, Barnett C, Lee E, Perez-Perez G, Blaser MJ, Peterson WL. Acute gastritis with hypochlorhydria: report of 35 cases with long term follow up. Gut. 2000;47:467–472. doi: 10.1136/gut.47.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Drasar BS, Hawksworth G, Aries V, Crowther JS, Williams RE. Bacteria and aetiology of cancer of large bowel. Lancet. 1971;1:95–100. doi: 10.1016/s0140-6736(71)90837-3. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34(suppl 2):S12–S18. doi: 10.1016/s1590-8658(02)80157-8. [DOI] [PubMed] [Google Scholar]
- Horie H, Kanazawa K, Okada M, Narushima S, Itoh K, Terada A. Effects of intestinal bacteria on the development of colonic neoplasm: an experimental study. Eur J Cancer Prev. 1999a;8:237–245. doi: 10.1097/00008469-199906000-00012. [DOI] [PubMed] [Google Scholar]
- Horie H, Kanazawa K, Kobayashi E, Okada M, Fujimura A, Yamagiwa S, Abo T. Effects of intestinal bacteria on the development of colonic neoplasm II. Changes in the immunological environment. Eur J Cancer Prev. 1999b;8:533–537. doi: 10.1097/00008469-199912000-00009. [DOI] [PubMed] [Google Scholar]
- Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Hilton A, Armstrong R, Smith C, Grant J, Shoo S, Forsythe S. Neonatal enteral feeding tubes as loci for colonisation by members of the Enterobacteriaceae. BMC Infect Dis. 2009;9:146. doi: 10.1186/1471-2334-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima K, Henry E, Moriya A, Wirz A, Kelman AW, McColl KE. Dietary nitrate generates potentially mutagenic concentrations of nitric oxide at the gastroesophageal junction. Gastroenterology. 2002;122:1248–1257. doi: 10.1053/gast.2002.32963. [DOI] [PubMed] [Google Scholar]
- Jain R, Maple JT, Anderson MA, et al. The role of endoscopy in enteral feeding. Gastrointest Endosc. 2011;74:7–12. doi: 10.1016/j.gie.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Le Moal G, Lemerre D, Grollier G, Desmont C, Klossek JM, Robert R. Nosocomial sinusitis with isolation of anaerobic bacteria in ICU patients. Intensive Care Med. 1999;25:1066–1071. doi: 10.1007/s001340051013. [DOI] [PubMed] [Google Scholar]
- Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, McBain AJ. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73:516–523. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehours P, Yilmaz O. Epidemiology ofHelicobacter pylori infection. Helicobacter. 2007;12(suppl 1):1–3. doi: 10.1111/j.1523-5378.2007.00541.x. [DOI] [PubMed] [Google Scholar]
- Leibovitz A, Dan M, Zinger J, Carmeli Y, Habot B, Segal R. Pseudomonas aeruginosa and the oropharyngeal ecosystem of tube-fed patients. Emerg Infect Dis. 2003;9:956–959. doi: 10.3201/eid0908.030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitz A, Baumoehl Y, Steinberg D, Segal R. Biodynamics of biofilm formation on nasogastric tubes in elderly patients. Isr Med Assoc J. 2005;7:428–430. [PubMed] [Google Scholar]
- Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J Biol Chem. 2006;281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- Lin CC, Lin CD, Cheng YK, Tsai MH, Chang CS. Middle ear effusion in intensive care unit patients with prolonged endotracheal intubation. Am J Otolaryngol. 2006;27:109–111. doi: 10.1016/j.amjoto.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Lin CS, He PJ, Hsu WT, Wu MS, Wu CJ, Shen HW, Hwang CH, Lai YK, Tsai NM, Liao KW. Helicobacter pylori-derived Heat shock protein 60 enhances angiogenesis via a CXCR2-mediated signaling pathway. Biochem Biophys Res Commun. 2010;397:283–289. doi: 10.1016/j.bbrc.2010.05.101. [DOI] [PubMed] [Google Scholar]
- Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O(6)-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer Res. 2002;62:3037–3043. [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn’s disease. J Clin Invest. 2004;113 doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR. Formation of glycoprotein degrading enzymes by Bacteroides fragilis. FEMS Microbiol Lett. 1991;61:289–293. doi: 10.1016/0378-1097(91)90567-t. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Bacterial diversity in the human gut. Adv Appl Microbiol. 2004;54:261–289. doi: 10.1016/S0065-2164(04)54010-8. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Cummings JH, Macfarlane S, Gibson GR. Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol. 1989;67:520–527. [PubMed] [Google Scholar]
- Macfarlane GT, Hay S, Macfarlane S, Gibson GR. Effect of different carbohydrates on growth, polysaccharidase and glycosidase production by Bacteroides ovatus in batch and continuous culture. J Appl Bacteriol. 1990;68:179–187. doi: 10.1111/j.1365-2672.1990.tb02564.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR, Cummings JH. Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol. 1992;72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, McBain AJ, Macfarlane GT. Consequences of biofilm and sessile growth in the large intestine. Adv Dent Res. 1997;11:59–68. doi: 10.1177/08959374970110011801. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, JH C, Macfarlane G. Bacterial colonisation of surfaces in the large intestine. In: Gibson G, Roberfroid M, editors. Colonic Microflora, Nutrition and Health. London: Chapman & Hall; 1999. pp. 71–87. [Google Scholar]
- Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis. 2004;38:1690–1699. doi: 10.1086/420823. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Woodmansey EJ, Macfarlane GT. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol. 2005;71:7483–7492. doi: 10.1128/AEM.71.11.7483-7492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT, Cummings JH. Review article: prebiotics in the gastrointestinal tract. Aliment Pharmacol Ther. 2006;24:701–714. doi: 10.1111/j.1365-2036.2006.03042.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Furrie E, Macfarlane GT, Dillon JF. Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin Infect Dis. 2007;45:29–38. doi: 10.1086/518578. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Steed H, Macfarlane S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol. 2008;104:305–344. doi: 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. The gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15:1528–1536. doi: 10.2174/138161209788168146. [DOI] [PubMed] [Google Scholar]
- Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–375. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. Direct targets of CodY in Staphylococcus aureus . J Bacteriol. 2010;192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrie TJ, Sung JY, Costerton JW. Bacterial biofilm formation on nasogastric tubes. J Gastroenterol Hepatol. 1990;5:503–506. doi: 10.1111/j.1440-1746.1990.tb01431.x. [DOI] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009a;15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC) BMC Microbiol. 2009b;9:202. doi: 10.1186/1471-2180-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathus-Vliegen EM, Bredius MW, Binnekade JM. Analysis of sites of bacterial contamination in an enteral feeding system. JPEN J Parenter Enteral Nutr. 2006;30:519–525. doi: 10.1177/0148607106030006519. [DOI] [PubMed] [Google Scholar]
- McCormick BA, Stocker BA, Laux DC, Cohen PS. Roles of motility, chemotaxis, and penetration through and growth in intestinal mucus in the ability of an avirulent strain of Salmonella typhimurium to colonize the large intestine of streptomycin-treated mice. Infect Immun. 1988;56:2209–2217. doi: 10.1128/iai.56.9.2209-2217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie H, Main J, Pennington CR, Parratt D. Antibody to selected strains of Saccharomyces cerevisiae (baker’s and brewer’s yeast) and Candida albicans in Crohn’s disease. Gut. 1990;31:536–538. doi: 10.1136/gut.31.5.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraud F, Trimoulet P, Lamouliatte H, Boyanova L. Bactericidal effect of amoxicillin on Helicobacter pylori in an in vitro model using epithelial cells. Antimicrob Agents Chemother. 1991;35:869–872. doi: 10.1128/aac.35.5.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley HL, Cortesia MJ, Rosenthal LE, Jones BD. Characterization of urease from Campylobacter pylori. J Clin Microbiol. 1988;26:831–836. doi: 10.1128/jcm.26.5.831-836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpofu CM, Campbell BJ, Subramanian S, Marshall-Clarke S, Hart CA, Cross A, Roberts CL, McGoldrick A, Edwards SW, Rhodes JM. Microbial mannan inhibits bacterial killing by macrophages: a possible pathogenic mechanism for Crohn’s disease. Gastroenterology. 2007;133:1487–1498. doi: 10.1053/j.gastro.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Nagy KN, Sonkodi I, Szöke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34:304–308. [PubMed] [Google Scholar]
- Ohlsen K, Lorenz U. Immunotherapeutic strategies to combat staphylococcal infections. Int J Med Microbiol. 2010;300:402–410. doi: 10.1016/j.ijmm.2010.04.015. [DOI] [PubMed] [Google Scholar]
- O’May GA, Reynolds N, Macfarlane GT. Effect of pH on an in vitro model of gastric microbiota in enteral nutrition patients. Appl Environ Microbiol. 2005a;71:4777–4783. doi: 10.1128/AEM.71.8.4777-4783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’May GA, Reynolds N, Smith AR, Kennedy A, Macfarlane GT. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J Clin Microbiol. 2005b;43:3059–3065. doi: 10.1128/JCM.43.7.3059-3065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk AB, Bartlett JG. Bacteriological studies of experimental ulcerative colitis. Am J Clin Nutr. 1979;32:258–265. doi: 10.1093/ajcn/32.1.258. [DOI] [PubMed] [Google Scholar]
- Onderdonk AB, Cisneros RL, Bronson RT. Enhancement of experimental ulcerative colitis by immunization with Bacteroides vulgatus. Infect Immun. 1983;42:783–788. doi: 10.1128/iai.42.2.783-788.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osias GL, Bromer MQ, Thomas RM, Friedel D, Miller LS, Suh B, Lorber B, Parkman HP, Fisher RS. Esophageal bacteria and Barrett’s esophagus: a preliminary report. Dig Dis Sci. 2004;49:228–236. doi: 10.1023/b:ddas.0000017443.44802.4b. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- Papp M, Altorjay I, Dotan N, et al. New serological markers for inflammatory bowel disease are associated with earlier age at onset, complicated disease behavior, risk for surgery, and NOD2/CARD15 genotype in a Hungarian IBD cohort. Am J Gastroenterol. 2008;103:665–681. doi: 10.1111/j.1572-0241.2007.01652.x. [DOI] [PubMed] [Google Scholar]
- Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2005;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Pei Z, Yang L, Peek RM, Jr, Levine SM, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett’s esophagus. World J Gastroenterol. 2005;11:7277–7283. doi: 10.3748/wjg.v11.i46.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. Microbial interactions and differential protein expression in Staphylococcus aureus -Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol. 2010;59:493–503. doi: 10.1111/j.1574-695X.2010.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poxton IR, Brown R, Sawyerr A, Ferguson A. Mucosa-associated bacterial flora of the human colon. J Med Microbiol. 1997;46:85–91. doi: 10.1099/00222615-46-1-85. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M, Kelly S. Structure, Function and Metabolism of Host Mucus Glycoproteins. Boca Raton FL: CRC Press; 1995. pp. 175–199. [Google Scholar]
- Reiff C, Kelly D. Inflammatory bowel disease, gut bacteria and probiotic therapy. Int J Med Microbiol. 2010;300:25–33. doi: 10.1016/j.ijmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Rex DK, Cummings OW, Shaw M, Cumings MD, Wong RK, Vasudeva RS, Dunne D, Rahmani EY, Helper DJ. Screening for Barrett’s esophagus in colonoscopy patients with and without heartburn. Gastroenterology. 2003;125:1670–1677. doi: 10.1053/j.gastro.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- Ronkainen J, Aro P, Storskrubb T, Johansson SE, Lind T, Bolling-Sternevald E, Vieth M, Stolte M, Talley NJ, Agréus L. Prevalence of Barrett’s esophagus in the general population: an endoscopic study. Gastroenterology. 2005;129:1825–1831. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- Saha A, Hammond CE, Gooz M, Smolka AJ. IL-1beta modulation of H, K-ATPase alpha-subunit gene transcription in Helicobacter pylori infection. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1055–G1061. doi: 10.1152/ajpgi.00338.2006. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8:79. doi: 10.1186/1471-2180-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrager J, Oates MD. Relation of human gastrointestinal mucus to disease states. Br Med Bull. 1978;34:79–82. doi: 10.1093/oxfordjournals.bmb.a071463. [DOI] [PubMed] [Google Scholar]
- Segal R, Pogoreliuk I, Dan M, Baumoehl Y, Leibovitz A. Gastric microbiota in elderly patients fed via nasogastric tubes for prolonged periods. J Hosp Infect. 2006;63:79–83. doi: 10.1016/j.jhin.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Simon PM, Goode PL, Mobasseri A, Zopf D. Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect Immun. 1997;65:750–757. doi: 10.1128/iai.65.2.750-757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Podolsky DK. Colonic mucin glycoproteins in health and disease. Clin Gastroenterol. 1986;15:815–837. [PubMed] [Google Scholar]
- Smith AR, Macfarlane S, Furrie E, Ahmed S, Bahrami B, Reynolds N, Macfarlane GT. Microbiological and immunological effects of enteral feeding on the upper gastrointestinal tract. J Med Microbiol. 2011;60:359–365. doi: 10.1099/jmm.0.026401-0. [DOI] [PubMed] [Google Scholar]
- Spechler SJ, Lee E, Ahnen D, et al. Long-term outcome of medical and surgical therapies for gastroesophageal reflux disease: follow-up of a randomized controlled trial. JAMA. 2001;285:2331–2338. doi: 10.1001/jama.285.18.2331. [DOI] [PubMed] [Google Scholar]
- Stark RM, Gerwig GJ, Pitman RS, et al. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol. 1999;28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- Steed H, Macfarlane GT, Macfarlane S. Prebiotics, synbiotics and inflammatory bowel disease. Mol Nutr Food Res. 2008;52:898–905. doi: 10.1002/mnfr.200700139. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Iijima K, Scobie G, Fyfe V, McColl KE. Nitrate and nitrosative chemistry within Barrett’s oesophagus during acid reflux. Gut. 2005;54:1527–1535. doi: 10.1136/gut.2005.066043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008a;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Bengmark S, Scholze J, Doerffel Y. Bacterial biofilm suppression with antibiotics for ulcerative and indeterminate colitis: consequences of aggressive treatment. Arch Med Res. 2008b;39:198–204. doi: 10.1016/j.arcmed.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn’s disease and ulcerative colitis – an overview. J Physiol Pharmacol. 2009;60(suppl 6):61–71. [PubMed] [Google Scholar]
- Thompson-Chagoyán OC, Maldonado J, Gil A. Aetiology of inflammatory bowel disease (IBD): role of intestinal microbiota and gut-associated lymphoid tissue immune response. Clin Nutr. 2005;24:339–352. doi: 10.1016/j.clnu.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Vasquez N, Mangin I, Lepage P, et al. Patchy distribution of mucosal lesions in ileal Crohn’s disease is not linked to differences in the dominant mucosa-associated bacteria: a study using fluorescence in situ hybridization and temporal temperature gradient gel electrophoresis. Inflamm Bowel Dis. 2007;13:684–692. doi: 10.1002/ibd.20084. [DOI] [PubMed] [Google Scholar]
- Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xia P, Wu F, et al. Helicobacter pylori VacA disrupts apical membrane-cytoskeletal interactions in gastric parietal cells. J Biol Chem. 2008;283:26714–26725. doi: 10.1074/jbc.M800527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SL, Wang ZR, Yang CQ. Meta-analysis of broad-spectrum antibiotic therapy in patients with active inflammatory bowel disease. Exp Ther Med. 2012;4:1051–1056. doi: 10.3892/etm.2012.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- Williams C, McColl KE. Review article: proton pump inhibitors and bacterial overgrowth. Aliment Pharmacol Ther. 2006;23:3–10. doi: 10.1111/j.1365-2036.2006.02707.x. [DOI] [PubMed] [Google Scholar]
- Williams JC, McInnis KA, Testerman TL. Adherence of Helicobacter pylori to abiotic surfaces is influenced by serum. Appl Environ Microbiol. 2008;74:1255–1258. doi: 10.1128/AEM.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters C, Spurling TJ, Chobanian SJ, et al. Barrett’s esophagus. A prevalent, occult complication of gastroesophageal reflux disease. Gastroenterology. 1987;92:118–124. [PubMed] [Google Scholar]
- Ye W, Held M, Lagergren J, Engstrand L, Blot WJ, McLaughlin JK, Nyrén O. Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. J Natl Cancer Inst. 2004;96:388–396. doi: 10.1093/jnci/djh057. [DOI] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009;9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, Kamiya S. Assessment of in vitro biofilm formation by Helicobacter pylori. J Gastroenterol Hepatol. 2010;25(suppl 1):S90–S94. doi: 10.1111/j.1440-1746.2009.06213.x. [DOI] [PubMed] [Google Scholar]
- Zavros Y, Eaton KA, Kang W, Rathinavelu S, Katukuri V, Kao JY, Samuelson LC, Merchant JL. Chronic gastritis in the hypochlorhydric gastrin-deficient mouse progresses to adenocarcinoma. Oncogene. 2005;24:2354–2366. doi: 10.1038/sj.onc.1208407. [DOI] [PubMed] [Google Scholar]