SUMMARY
Metabolic adaptation is essential for cell survival during nutrient deprivation. We report that eukaryotic elongation factor 2 kinase (eEF2K), which is activated by AMP-kinase (AMPK), confers cell survival under acute nutrient depletion by blocking translation elongation. Tumor cells exploit this pathway to adapt to nutrient deprivation by reactivating the AMPK-eEF2K axis. Adaptation of transformed cells to nutrient withdrawal is severely compromised in cells lacking eEF2K. Moreover, eEF2K knockdown restored sensitivity to acute nutrient deprivation in highly resistant human tumor cell lines. In vivo, overexpression of eEF2K rendered murine tumors remarkably resistant to caloric restriction. Expression of eEF2K strongly correlated with overall survival in human medulloblastoma and glioblastoma multiforme. Finally, C. elegans strains deficient in efk-1, the eEF2K ortholog, were severely compromised in their response to nutrient depletion. Our data highlight a conserved role for eEF2K in protecting cells from nutrient deprivation and in conferring tumor cell adaptation to metabolic stress.
INTRODUCTION
Nutrient deprivation (ND) is a severe physiological stress with dire consequences for cell viability. Living organisms have therefore evolved molecular mechanisms to respond to ND, including metabolic reprogramming to preserve energy balance (Caro-Maldonado and Muñoz-Pinedo, 2011). A key mediator is the highly conserved energy sensor AMP-activated protein kinase (AMPK), which is activated when cellular AMP:ATP or ADP:ATP ratios increase (Hardie, 2011). AMPK limits energy-consuming processes such as proliferation and protein synthesis and induces catabolic processes such as glycolysis and fatty acid oxidation to preserve energy (Hardie, 2011). Another critical nutrient sensor is mammalian target of rapamycin complex 1 (mTORC1), which is regulated by ATP and amino acid levels (Zoncu et al., 2011). This complex couples nutrient abundance to control of protein synthesis through phosphorylation of 4EBP1 and p70S6K (Hay and Sonenberg, 2004). When nutrient availability is compromised, mTORC1 is inactivated, in part through AMPK (Inoki et al., 2003), thereby blocking protein synthesis, the most energy-demanding process in the cell (Buttgereit and Brand, 1995).
Pathologic ND occurs along with hypoxia in early stages of tumor development before new blood vessels form or at later stages due to abnormal tumor vasculature (Nagy et al., 2009). While metabolic stress prevents tumor development by inducing growth arrest and necrosis, it may also select for metabolically adapted cells that can form aggressive tumors (Jones and Thompson, 2009). Proto-oncogenes such as MYC, AKT, and mTOR that stimulate anabolic metabolism sensitize cells to ND (Buzzai et al., 2005; Choo et al., 2010; Shim et al., 1998). This argues that, to balance initial oncogenic events driving energy-demanding processes such as proliferation, tumors must develop adaptive responses to protect cells from ND (Jones and Thompson, 2009). Several factors have been linked to such responses, including ATF4, NFκB, and CPT1C, which affect amino acid synthesis, mitochondrial respiration, and fatty acid oxidation, respectively (Mauro et al., 2011; Ye et al., 2010; Zaugg et al., 2011). However, our understanding of this process is incomplete, and uncovering the molecular pathways involved is critical for potential therapeutic targeting in tumors.
In this study, we report that eukaryotic translation elongation factor 2 kinase (eEF2K) is a conserved mediator of the cellular response to ND. EEF2K inhibits activity of translation elongation factor eEF2, which mediates the translocation stage of translation elongation, whereby polypeptidyl-tRNAs move from the A to the P site of the ribosome (Carlberg et al., 1990). Activity of eEF2K is tightly controlled by nutrient availability, notably through direct positive regulation by AMPK and inhibition by mTORC1 and Ras-Erk-p90RSK pathways (Proud, 2007). In the absence of nutrients, eEF2K is activated to phosphorylate and inactivate eEF2 (Ryazanov et al., 1988), thereby blocking energy-demanding messenger RNA (mRNA) translation elongation (Carlberg et al., 1990). Our data demonstrate a critical role for eEF2K in protecting normal tissues from acute ND through inhibition of eEF2 and show that this pathway is exploited by tumor cells in adapting to metabolic stress.
RESULTS
Oncogenic Transformation Sensitizes Fibroblasts to Acute ND in Association with Defective eEF2 Signaling
We first tested effects of oncogenic transformation on responses to acute ND using National Institutes of Health (NIH) 3T3 fibroblasts transformed by activated K-RasV12 (RasV12) or the ETV6-NTRK3 (EN) chimeric tyrosine kinase (Knezevich et al., 1998). Both oncoproteins constitutively activate Ras-Erk and PI3K-Akt (Tognon et al., 2002), allowing us to study whether these pathways impact acute responses to ND. Transformed fibroblasts cultured in media lacking glucose, amino acids, and serum showed massive cell death compared to nontransformed control cells under ND (Figures 1A and S1A available online). Apoptosis was confirmed by Annexin V staining (Figure S1B). Glucose depletion alone induced cell death in transformed cells, whereas amino acid depletion had little effect (Figure S1C). Nevertheless, withdrawal of both glucose and amino acids significantly enhanced cell death in transformed cells (Figure S1C). Increased cell death was not linked to increased proliferation or reactive oxygen species (ROS), as these were similarly reduced or increased, respectively, in control and transformed cells under ND (Figures S1D and S1E). Acute ND precipitously reduced ATP levels in control cells, but, unexpectedly, this was not observed in transformed cells (Figure S1F), arguing against a severe energetic crisis in the latter. Finally, autophagy was similarly induced in all three cell lines based on LC3-II accumulation (data not shown), ruling against differential autophagy flux as the basis of the observed phenotype.
Figure 1. Oncogenic Transformation Sensitizes Cells to ND and Alters eEF2 Signaling Pathways.
(A) Caspase-3 activity assays for NIH 3T3 MSCV, EN, and RasV12 cells grown in complete media or deprived of nutrients (ND) for the indicated times (n = 3).
(B) Schematic representation of nutrient-responsive signaling pathways assessed in this study. Gray arrows and bars indicate release from regulatory effects of upstream pathways.
(C) Immunoblot analysis of NIH 3T3 MSCV, EN, and RasV12 cells grown in complete media or under ND for the indicated times.
(D) Intracellular levels of AMP and ATP in NIH 3T3 MSCV, EN, RasV12, EN-S, and RasV12-S cells grown in complete media (Ctrl) or under ND for 6 hr. Results are expressed as relative fold increases of AMP:ATP ratio over MSCV Ctrl for n = 3.
(E) Immunoblot analysis of NIH 3T3 EN and RasV12 cells deprived of nutrients (1 hr) and treated with Akt inhibitor VII (Akti; 1 μM), rotenone (0.5 μM), 2-deoxyglucose (2-DG; 25 mM), or vehicle.
(F) Scheme for generation of EN-S and RasV12-S cells from NIH 3T3 EN and RasV12 cells (top). Caspase-3 activity assays of NIH 3T3 EN, EN-S, RasV12, and RasV12-S cells grown in complete media or under ND as indicated (n = 3) (bottom).
(G) Immunoblot analysis of NIH 3T3 EN, EN-S, RasV12, and RasV12-S cells grown in complete media or under ND for the indicated times.
(H) Immunoblot analysis of NIH 3T3 EN-S/MSCV, EN-S/DN-AMPK, RasV12-S/MSCV, and RasV12-S/DN-AMPK cells grown in complete media or under ND for the indicated times.
(I) Caspase-3 assays of NIH 3T3 EN-S/MSCV, EN-S/DN-AMPK, RasV12-S/MSCV, and RasV12-S/DN-AMPK cells grown in complete media (Ctrl) or under ND for 48 hr (n = 3).
Where shown, data are reported as means ± SD with indicated significance (*p < 0.05, **p < 0.01, and ***p < 0.005). See also Figures S1 and S2.
We next assessed mTORC1 signaling, but this was rapidly and profoundly inhibited in all three cell lines by ND (Figure 1C). However, activation of another key energy sensor, AMPK, was compromised in transformed cells under ND, as indicated by reduced phosphorylation of AMPK or acetyl CoA-carboxylase (ACC) (Figure 1C). Moreover, phosphorylation of eEF2 at Thr56, a known marker of AMPK activation, was markedly blunted under ND in transformed versus control cells (Figure 1C). EEF2 is essential for translation elongation (Buttgereit and Brand, 1995), and Thr56 is phosphorylated by eEF2 kinase (eEF2K), the only known kinase for eEF2, to block eEF2 activity (Carlberg et al., 1990). This suggested that eEF2 activity was partially retained in transformed cells under ND. In addition, phosphorylation of eEF2K Ser366, catalyzed by p90RSK downstream of Ras-Erk to inhibit eEF2K (Wang et al., 2001), was retained in transformed cells under ND (Figure 1C).
These results point to eEF2K inhibition and deregulated eEF2 activity in transformed cells under ND. We therefore examined contributions of Ras-Erk-p90RSK and AMPK to this process, as both regulate eEF2K (Proud, 2007). Chemical inhibitors of p90RSK (PD184352 and BI-D1870; data not shown) or dominant-negative p90RSK (DN-RSK; Figure S1G) each reduced eEF2K-Ser366 phosphorylation and markedly enhanced eEF2 phosphorylation in EN and RasV12 cells under ND. Next, we ectopically activated AMPK under ND using AICAR and SMER28, chemical inducers of AMPK, or constitutively active AMPK. Each strongly induced eEF2 phosphorylation without changing eEF2K mRNA (data not shown) or protein levels (Figures S2A and S2B). Therefore, eEF2K activity is deregulated in transformed cells under ND by sustained Ras-Erk-p90RSK activity and reduced AMPK activation. The former likely occurs through constitutively high Ras activity in RasV12 or EN-transformed cells (Figure 1C). To explore the basis of defective AMPK activation, we measured AMP:ATP and ADP:ATP ratios, known physiological inducers of AMPK (Hardie, 2011), because ATP levels were elevated in transformed cells (Figure S1F). Whereas both ratios were elevated in control cells under ND and correlated with AMPK induction (Figure 1C), ratios were virtually unchanged in transformed cells (Figures 1D and S2C). Therefore EN- and RasV12-transformed cells unexpectedly exhibit a defect in the energy response to ND by maintaining high ATP and failing to raise AMP:ATP and ADP:ATP ratios. To confirm that sustained ATP levels inhibit AMPK activation, we blocked ATP production with 2-deoxyglucose (2-DG) or rotenone. Both agents, but not an Akt inhibitor, induced AMPK activity under ND and markedly increased eEF2 phosphorylation (Figure 1E). Together, this indicates that transformation sensitizes fibroblasts to ND in association with defects in AMPK and p90RSK regulation of eEF2.
Adaptation of Transformed Cells to Chronic ND Correlates with Increased AMPK-eEF2K Pathway Activation
We next hypothesized that rare transformed cells might adapt and acquire resistance to prolonged ND. Therefore, transformed cells were subjected to repeated cycles of prolonged ND followed by nutrient resupplementation (Figure 1F and Extended Experimental Procedures). Indeed, after several weeks, stable populations of “selected” EN or RasV12 cells, designated EN selected (EN-S) and RasV12 selected (RasV12-S), respectively, emerged with the capacity to survive under acute ND (Figure 1F), which was retained after multiple passages (tested out to ten serial passages). Importantly, unselected EN- and RasV12-transformed cells subjected to the same adaptation protocol but with glucose and amino acids (EN-S+Glc+AA and RasV12-S+Glc+AA), were equally sensitive to ND as their parental counterparts (Figure S2D), indicating that adaptive reprogramming requires chronic exposure to ND. Notably, EN-S and RasV12-S cells retained soft-agar-colony-forming activity (data not shown) and tumorigenicity (Figure S2E). As observed for unselected cells, EN-S and RasV12-S cells rapidly shut down mTORC1 signaling during acute ND and exhibited similar levels of Akt activation and autophagic flux compared to unselected counterparts (Figures 1G and S2F). In contrast, they displayed increased AMPK and ACC phosphorylation under ND, suggesting that, like non-transformed cells, selected cells had reacquired the ability to activate AMPK under ND (Figure 1G). This correlated with massively increased eEF2 phosphorylation, which was not due to altered eEF2K transcript or protein levels (Figures 1G and S2G). This confirms a correlation between eEF2 phosphorylation and survival capacity under acute ND. This was not due to inhibition of the Ras-Erk-p90RSK pathway, as eEF2K Ser366 phosphorylation was preserved in selected cells under ND (Figure 1G). However, blocking AMPK activity with dominant-negative AMPK (DN-AMPK) markedly reduced eEF2 phosphorylation in selected cells (Figure 1H) and significantly increased ND-induced cell death (Figure 1I), highlighting the importance of AMPK reinduction in resistance to ND. Mechanistically, this was not due to changes in LKB1 expression (Figure 1G), which directly activates AMPK (Hardie, 2011). However, AMP:ATP and ADP:ATP ratios were both markedly increased in EN-S and RasV12-S cells under ND compared to parental EN and RasV12 cells (Figures 1D and S2C). Therefore, unlike nonselected tumor cells, selected cells were able to raise AMP:ATP and ADP:ATP ratios when nutrient depleted, strongly suggesting that adaptation of EN and RasV12 cells to ND is linked to reactivation of an AMPK-eEF2K axis.
Translation Elongation Is Deregulated in Transformed Fibroblasts under ND and Restricts Cell Survival
We next asked whether eEF2 deregulation impacts protein synthesis under ND, predicting that high eEF2 activity would sustain protein synthesis under ND, thereby sensitizing nonselected tumor cells to apoptosis. However, rates of global protein synthesis were equivalently reduced under ND in both transformed and nontransformed cells using [35S]-methionine/cysteine incorporation (Figure 2A) and pulse-labeling with L-azidohomoalanine (AHA) (Somasekharan et al., 2012) (Figure S3A). This is consistent with the observed block in mTORC1 signaling under ND (Figure 1C) and argues against a major role for sustained global protein synthesis in hypersensitivity of nonselected transformed cells to ND. It is noteworthy that a low level of protein synthesis was still observed up to 6 hr under ND (Figure S3B).
Figure 2. Translation Elongation Activity Is Sustained in Transformed Cells under ND and Reduces Their Survival.
(A) Protein synthesis levels in NIH 3T3 MSCV, EN, and RasV12 cells deprived of nutrients for 5 min or 1, 3, or 6 hr determined by [35S]-methionine/cysteine(Met/Cys) incorporation. Results are expressed as a percentage of [35S]-Met/Cys incorporation/mg protein relative to MSCV cells at 5 min (n = 2).
(B) Polysome profiles for NIH 3T3 MSCV, EN, and RasV12 cells grown in complete media or under ND for 70 min as described in Extended Experimental Procedures. P/S indicates ratio of polysomal to subpolysomal (40S, 60S, and 80S) fractions.
(C) Ribosome half-transit times for NIH 3T3 MSCV, EN, and RasV12 cells grown in complete media or under ND for 70 min. [35S]-Met/Cys incorporation into all polypeptides (postmitochondrial supernatant, PMS) and into polypeptides released from ribosomes (postribosomal supernatant, PRS) was obtained by linear regression analysis. Representative results from three different experiments are shown.
(D and E) siRNA-mediated knockdown of eEF2 in NIH 3T3 EN and RasV12 cells. Cells were transiently transfected with 12.5 nM of control (CTRL) or eEF2-directed siRNAs, grown in complete media for 48 hr, and placed under ND for 48 hr. Lysates were either analyzed by immunoblotting (D) or assayed for caspase-3 activity (n = 3) (E).
(F and G) siRNA-mediated knockdown of eEF2 in NIH 3T3 EN-S/MSCV, EN-S/DN-AMPK, RasV12-S/MSCV, and RasV12-S/DN-AMPK cells. Cells were transfected, treated, and analyzed as in (D) and (E).
Where shown, data are reported as means ± SD with indicated significance (**p < 0.005). See also Figure S3.
We next examined the effects of ND on translation initiation versus elongation by analyzing polysome profiles from sucrose gradients. Ribosomal 40S-, 60S-, and 80S-mRNA complexes represent translationally inactive complexes (subpolysomal fractions), whereas polyribosome (polysome) fractions correspond to translationally active complexes. ND markedly increased 60S fractions in both control and transformed cells, indicating a global block in translation initiation (Figure 2B). This is consistent with loss of 4EBP1 phosphorylation (Figure 1C), which blocks cap-dependent translation initiation, the major rate-limiting step of mRNA translation (Mathews et al., 2007). Whereas ratios of polysomal to subpolysomal fractions (P/S) were not significantly affected by ND in nontransformed cells (4% ± 0.2% reduction), P/S ratios were dramatically decreased in EN and RasV12 cells (82% ± 2% and 91% ± 3%, respectively) under ND (Figure 2B). Because initiation is blocked, this strongly suggests that, in transformed cells, ribosomes are continuously running off transcripts (i.e., there is minimal ribosome stalling) under ND, leading to polysomal disaggregation due to sustained translation elongation, thus decreasing P/S ratios (Mathews et al., 2007). Furthermore, under ND, eEF2 remained in an unphosphorylated state in mouse embryonic fibroblasts (MEFs) lacking eEF2K (eEF2K−/−), indicating that translation elongation was maintained in eEF2K−/− cells under ND, in contrast to wild-type eEF2K+/+ cells in which eEF2 was strongly phosphorylated (Figure S3C). ND correlated with accumulation of 60S fractions in both eEF2K+/+ and eEF2K−/− cells, which is consistent with blocked translation initiation (Figure S3D). However, the P/S ratio was more dramatically reduced by ND in eEF2K−/− cells (89% ± 2%) compared to eEF2K+/+ cells (49% ± 1%) (Figure S3D). This indicates that retained eEF2 activity leads to sustained translation elongation and polysome disaggregation under ND.
To conclusively demonstrate differential translation elongation in control versus transformed cells, we measured ribosome half-transit times under ND. These refer to the time required for a ribosome to translate an average-sized mRNA and are the inverse of the elongation rate (Fan and Penman, 1970). Under ND, ribosome half-transit times were ~2-fold longer in nontransformed cells, which is indicative of slower elongation rates (Figure 2C). In contrast, half-transit times were only slightly increased by ND in EN and RasV12 cells (i.e., ~1.25-fold and ~1.2-fold, respectively) (Figure 2C). This was not due to differences in intracellular amino acid levels between control and transformed cells, which were decreased equivalently during ND (Figure S3E). These data support a model whereby, in nonselected tumor cells under ND, retained eEF2 activity leads to sustained translation elongation. To link this to cell death under ND, we specifically impaired translation elongation in EN and RasV12 cells by eEF2 knockdown (Figure 2D). Indeed, ND-induced cell death was substantially reduced in transformed cells after eEF2 knockdown compared to small interfering RNA (siRNA) controls (Figure 2E). This further supports a role for deregulated eEF2 activity in hypersensitivity of transformed cells to ND. In addition, EN-S/DN-AMPK and RasV12-S/DN-AMPK cells, which are highly susceptible to ND (Figure 1I), were rescued from cell death by eEF2 knockdown, suggesting that AMPK promotes survival of adapted transformed cells by inhibiting translation elongation (Figures 2F and 2G). Therefore, sustained translation elongation renders transformed cells hypersensitive to ND.
eEF2K Is Critical for Cell Survival under ND
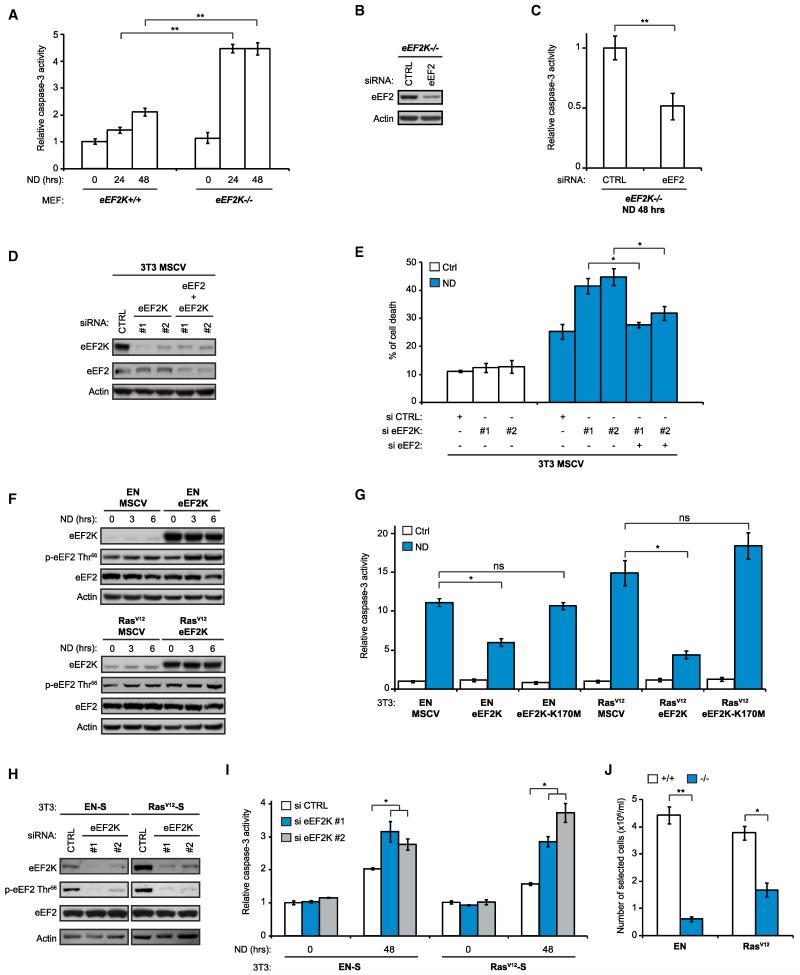
Because translation elongation is tightly controlled by eEF2K (Carlberg et al., 1990), we next examined the impact of eEF2K expression on cell survival under ND. Similar to nonselected tumor cells, eEF2K−/− MEFs were highly sensitive to ND compared with eEF2K+/+ MEFs and underwent apoptotic cell death under ND (Figures 3A and S4A). In contrast to previous reports (Wu et al., 2006), this was not associated with defects in autophagy (Figures S4B and S4C). To establish whether eEF2 is linked to survival responses under ND, we performed eEF2 knockdown in eEF2K−/− cells, which harbor high eEF2 activity under ND (Figure S3C). This markedly increased cell survival compared to siRNA controls (Figures 3B and 3C). Moreover, eEF2K knockdown in NIH 3T3 cells with two independent eEF2K siRNAs strongly sensitized cells to ND (Figures 3D and 3E). However, coknockdown of eEF2 completely rescued cells from cell death (Figures 3D and 3E), confirming that eEF2K mediates survival under ND through its control of eEF2. Conversely, stable overexpression of eEF2K in EN- and RasV12-transformed NIH 3T3 cells led to increased eEF2 phosphorylation and cell survival under ND (Figures 3F and 3G). This was not observed with kinase-dead eEF2K, implying that eEF2 phosphorylation and inhibition of translation elongation are critical for survival under ND (Figures 3G and S4D).
Figure 3. eEF2K Is Critical for the Adaptation of Fibroblasts to ND.
(A) Caspase-3 activity assays for eEF2K+/+ and eEF2K−/− MEFs grown in complete media or under ND for the indicated times (n = 3).
(B and C) siRNA-mediated knockdown of eEF2 in eEF2K−/− MEFs. Cells were transiently transfected with 12.5 nM of control (CTRL) or eEF2-directed siRNAs, grown in complete media for 48 hr, and placed under ND for 48 hr. Cell lysates were either analyzed by immunoblotting (B) or assayed for caspase-3 activity (n = 3) (C).
(D and E) siRNA-mediated knockdown of eEF2K and eEF2 in NIH 3T3 MSCV cells by transient transfection with 37.5 nM of control (siCTRL) or 25 nM of eEF2K siRNAs (si eEF2K#1 and eEF2K#2) combined with 12.5 nM of control or eEF2 siRNAs. Cells were treated as described in (B) and (C). Lysates were analyzed by immunoblotting (D) or assayed for cell death (n = 3) (E).
(F) Immunoblot analysis of NIH 3T3 EN/MSCV, EN/eEF2K, RasV12/MSCV, and RasV12/eEF2K grown in complete media or under ND for the indicated times.
(G) Caspase-3 activity assays for NIH 3T3 EN/MSCV, EN/eEF2K, EN/eEF2K-K170M (kinase-dead), RasV12/MSCV, RasV12/eEF2K, and RasV12/eEF2K-K170M cells grown in complete media (Ctrl) or under ND for 48 hr (n = 3).
(H and I) siRNA-mediated knockdown of eEF2K in NIH 3T3 EN-S and RasV12-S cells. Cells were transiently transfected with 25 nM of control (siCTRL) or eEF2K (si eEF2K#1 and eEF2K#2) siRNAs and grown in complete media for 72 hr. Cells were placed under ND either for 3 hr and analyzed by immunoblotting (H) or for 72 hr and assayed for caspase-3 activity (n = 3) (I).
(J) Cell adaptation assays for eEF2K+/+ and eEF2K−/− MEFs expressing EN or RasV12. Cells were subjected to the adaptation protocol described in the Extended Experimental Procedures. Results are expressed as the number of cells alive at the completion of the experiment (n = 3).
Where shown, data are reported as means ± SD with indicated significance (*p < 0.05 and **p < 0.01; ns, nonsignificant). See also Figure S4.
To directly test the role of eEF2K in the adaptive response of transformed fibroblasts to ND, we performed eEF2K knockdown in EN-S and RasV12-S cells (which have high phospho-eEF2 levels; Figure 1G) using two independent siRNAs. This blocked eEF2 phosphorylation (Figure 3H) and rendered cells significantly more susceptible to ND compared to siRNA control cells (Figure 3I). Finally, eEF2K−/− MEFs transformed with EN or RasV12 exhibited similar growth in vitro under ambient conditions as their eEF2K+/+ counterparts (Figures S4E and S4F) and were similarly tumorigenic in athymic mouse (Figure S4G). However, when these cells were subjected to the above adaptation protocol (Figure 1F), transformed eEF2K−/− MEFs had a strikingly reduced capacity to adapt to repeated ND compared to transformed eEF2K+/+ MEFs (Figure 3J). These findings clearly establish eEF2K as a critical survival factor under ND and in the adaptive response of transformed fibroblasts to nutrient stress.
eEF2K Is Critical for Survival of Human Tumor Cells under Nutrient Stress
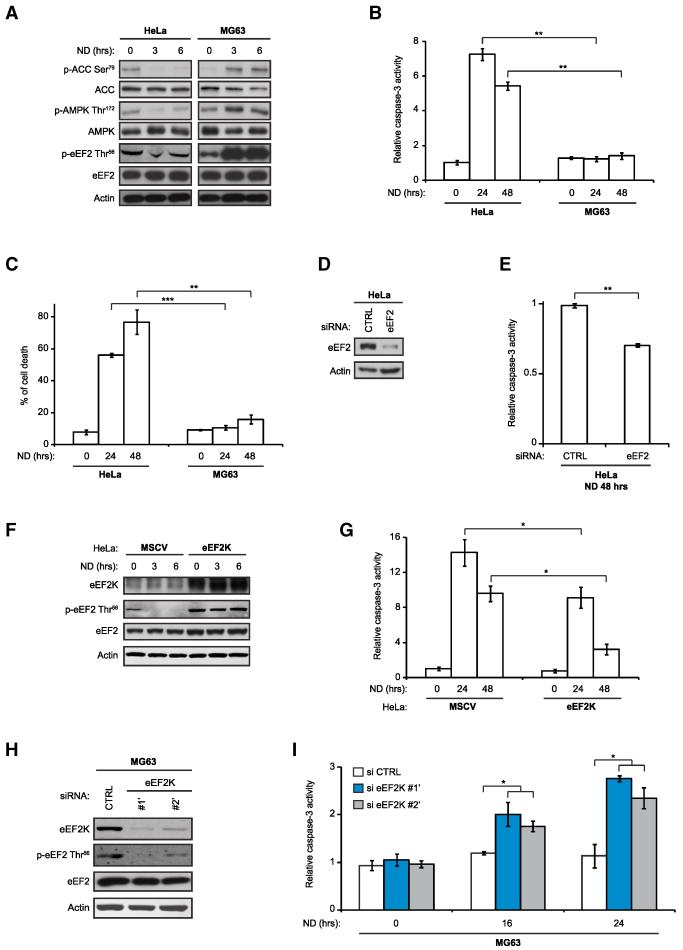
To extend these findings to human cells, we screened tumor cell lines for eEF2 phosphorylation under ND. This identified two lines with widely divergent levels: HeLa cells showed minimal eEF2 phosphorylation, whereas MG63 osteosarcoma cells exhibited high eEF2 phosphorylation under ND (Figure 4A). HeLa cells exhibited extremely low AMPK activity under ND as reported (Liang et al., 2007), whereas, in MG63 cells, AMPK activity was high (Figure 4A). This was tightly associated with survival: HeLa cells were highly sensitive to ND, whereas MG63 cells were almost completely resistant to cell death under these conditions (Figures 4B and 4C), with HeLa cells being particularly sensitive to glucose withdrawal (Figure S5A). To link this to eEF2 and translation elongation, we knocked down eEF2 in HeLa cells, which significantly reduced cell death under ND compared to siRNA controls (Figures 4D and 4E). Moreover, stable eEF2K overexpression increased eEF2 phosphorylation and cell survival in HeLa cells under ND compared to parental cells (Figures 4F, 4G, and S5B). This strongly argues that hypersensitivity of HeLa cells is related to reduced eEF2K activity and sustained translation elongation. Knockdown of eEF2K, which reduced eEF2 phosphorylation compared to controls (Figure 4H), strongly increased sensitivity of MG63 cells to ND (Figures 4I and S5C). This was phenocopied by knockdown of AMPK-subunits α1/α2, which sensitized MG63 cells to ND-induced cell death, supporting the importance of an AMPK-eEF2K axis in protecting human tumor cells from ND (Figures S5D and S5E). Overall, these results demonstrate that eEF2K is critical for survival of human tumor cells under ND.
Figure 4. eEF2K Facilitates Survival of Human Tumor Cells in Response to ND.
(A) Immunoblot analysis of HeLa and MG63 cells grown in complete media or under ND for the indicated times.
(B and C) Caspase-3 activity assays (B) or cell death assays (C) for HeLa and MG63 cells grown in complete media or under ND for the indicated times (n = 3).
(D and E) siRNA-mediated knockdown of eEF2 in HeLa cells. Cells were transiently transfected with 12.5 nM of control (CTRL) or eEF2-directed siRNAs, grown in complete media for 48 hr, and placed under ND for 48 hr. Cell lysates were either analyzed by immunoblotting (D) or assayed for caspase-3 activity (n = 3) (E).
(F and G) Immunoblot analysis (F) or caspase-3 activity assays (n = 3) (G) of HeLa MSCV and HeLa eEF2K cells grown in complete media or under ND for the indicated times.
(H and I) siRNA-mediated knockdown of eEF2K in MG63 cells. Cells were transiently transfected with 25 nM of control (siCTRL) or eEF2K (si eEF2K#1′ and eEF2K#2′) siRNAs and grown in complete media for 72 hr. Cells were placed under ND either for 3 hr and analyzed by immunoblotting (H) or for 72 hr and assayed for caspase-3 activity (n = 3) (I). Where shown, data are reported as means ± SD with indicated significance (*p < 0.05, **p < 0.01, and ***p < 0.005). See also Figure S5.
eEF2K Protects Tumors against Caloric-Restriction-Induced Cell Death In Vivo
To determine the relevance of these findings in vivo, EN- and RasV12-transformed NIH 3T3 cells overexpressing eEF2K or MSCV vector control were subcutaneously implanted in nu/nu immunocompromised mice. Mice were either fed a standard ad libitum (AL) diet or, prior to tumor development, caloric restriction (CR) corresponding to 60% caloric intake of the AL diet. As described (Kalaany and Sabatini, 2009), CR led to significant reductions of blood IGF1, insulin, and glucose levels (Figures S6A–S6C). Tumor sizes in EN and RasV12 vector control mice were each severely reduced (by ~50%) by CR compared to AL (compare MSCV bars in Figures 5A and 5B). Although eEF2K-overexpressing tumors were smaller than control tumors under AL diets, likely due to reduced translation elongation and therefore overall growth, their sizes were virtually unaffected by CR (Figures 5A, 5B, and S6D). Notably, EN/eEF2K and RasV12/eEF2K tumors eventually grew to larger sizes than controls when mice were reverted to an AL diet following CR (Figures S6E and S6F), highlighting the survival advantage of high eEF2K expression in tumors. Morphologic examination revealed massive necrosis within EN and RasV12 vector control tumors after CR compared to AL diets. In dramatic contrast, EN/eEF2K and RasV12/eEF2K tumors from CR animals exhibited no apparent necrosis (Figures 5C and S6G). In EN and RasV12 control tumors, CR massively increased apoptosis as measured by IHC of cleaved caspase-3, which was not observed in EN/eEF2K and RasV12/eEF2K tumors (Figures 5D and S6H), further accentuating the protective effect of eEF2K in vivo. We then assessed eEF2K activity in AL versus CR tumors by analyzing phospho-eEF2 levels by IHC or immunoblotting. Phosphorylation of eEF2 was poorly induced by CR in EN and RasV12 control tumors (see MSCV panels in Figures 5E, 5F, S6I, and S6J), which correlated with sustained eEF2K inhibition under CR as indicated by retention of p90RSK-mediated eEF2K Ser366-phosphorylation (Figure S6K). In contrast, eEF2 phosphorylation was strongly induced in EN/eEF2K and RasV12/eEF2K tumors under either AL or CR and was more pronounced under CR (see eEF2K panels in Figures 5E, 5F, S6I, and S6J). This was confirmed under CR using eEF2K−/− and +/+ MEFs transformed with RasV12 (Figure S6L). Although tumor sizes were reduced by CR in RasV12/eEF2K+/+ MEF tumors, effects were more significant in RasV12/eEF2K−/− MEF tumors (Figure 5G). CR again led to significantly increased necrosis and apoptosis in RasV12/eEF2K−/− as compared to RasV12/eEF2K+/+ tumors (Figures 5H and 5I), indicating that genetic loss of eEF2K further increases sensitivity of tumors to CR. These results are consistent with a model whereby eEF2K activity protects tumors from CR-induced cell death in vivo.
Figure 5. eEF2K Promotes Resistance of Tumors to Caloric Restriction-Induced Cell Death In Vivo.
(A and B) Tumor volumes of NIH 3T3 RasV12/MSCV and RasV12/eEF2K (A) or EN/MSCV and EN/eEF2K (B) xenografts implanted subcutaneously in nu/nu mice. Mice were fed either AL or placed on CR diets (n = 10 mice/group).
(C) Hematoxylin and eosin staining (H&E) of tumor xenografts from (A). Black arrows indicate regions of necrosis. Results are expressed as percentages of necrotic/total tumor areas (n = 3 mice per group).
(D) Immunohistochemical (IHC) staining of tumor xenografts from (A) with antibodies against cleaved caspase-3. Graphs indicate percent of total cells that are positive for cleaved caspase-3 (n = 3 mice per group).
(E) IHC staining of tumor xenografts from (A) with anti-phospho-eEF2 antibodies.
(F) Immunoblot analysis of tumor lysates of tumor xenografts from (A).
(G) Tumor volumes of eEF2K+/+ RasV12 and eEF2K−/− RasV12 xenografts implanted subcutaneously in nu/nu mice. Mice were fed either AL or placed on CR diets (n = 10 mice per group).
(H) H&E staining of tumor xenografts from (G). Black arrows indicate areas of necrosis. Results are expressed as percentages of necrotic/total tumor areas (n = 3 mice per group).
(I) IHC staining of tumor xenografts from (G) with antibodies to cleaved caspase-3. Graphs indicate the percent of total cells positive for cleaved caspase-3 (n = 3 mice per group).
Where shown, data are reported as means ± SEM with indicated significance (*p < 0.05 and **p < 0.01; ns, nonsignificant). See also Figure S6.
Expression of eEF2K Is Associated with Poor Prognosis in Medulloblastoma and Glioblastoma Multiforme
To probe these findings in primary human tumors, we focused on medulloblastoma (MB), the most common malignant brain tumor of childhood, and glioblastoma multiforme (GBM), the most highly malignant adult brain tumor, because eEF2K expression data were publically available for these tumors (Northcott et al., 2011). In MB, high eEF2K transcript levels correlated with the most aggressive and metastatic subtype of MB, namely group 3 tumors (Northcott et al., 2011). Gene expression profiles from five independent MB cohorts (total n = 286) demonstrated highly significant upregulation of eEF2K in group 3 relative to non-group-3 tumors (Figures 6A and 6B). Furthermore, high eEF2K expression correlated with significantly decreased overall survival in all disease variants (p = 0.00003; Figure 6C), as well as in aggressive non-WNT/non-SHH subtypes (p = 0.002; Figure 6D). EEF2K expression was significantly upregulated in GBM compared to normal human brain tissue (~2.5-fold; Figure 6E). Among GBM subtypes, eEF2K expression was specifically increased in classical and mesenchymal subtypes (Figure 6F), both of which are associated with poor overall and progression-free survival (Verhaak et al., 2010). EEF2K expression strongly correlated with decreased overall survival across all human glioma subtypes (astrocytoma, oligodendroglioma, mixed glioma, and GBM) (p = 0.0001; Figure 6G), as well as in GBM itself (p = 0.001; Figure 6H). Together, these findings highlight eEF2K expression as a prognostic biomarker in aggressive human brain tumors.
Figure 6. eEF2K Transcript Levels Are Associated with Poor Prognosis in Medulloblastoma and Glioblastoma Multiforme.
(A and B) Expression levels of eEF2K in the Toronto MB cohort (A) or in the European MB data set (B) (see Extended Experimental Procedures). FC and CB indicate fetal and adult cerebellum, respectively; MBs indicate the total cohort, and WNT, SHH, group 3, and group 4 indicate the specific disease subgroups. p values are shown below each panel and were generated using a Mann-Whitney U test.
(C and D) Kaplan-Meier estimates of overall survival for MB patients from all subgroups (C) or from non-WNT/non-SHH subgroups (D) classified by eEF2K mRNA expression levels. The number of patients at risk is indicated for time increments of 24 months. p values were calculated using a log rank test.
(E and F) Expression of eEF2K mRNA in normal human brain (NHB; n = 10) compared to GBM (n = 25) (E) or in specific disease subtypes of GBM (proneural, neural, classical, and mesenchymal) (F). p values were determined by ANOVA with a Bonferroni post hoc test.
(G and H) Kaplan-Meier estimates of overall survival for all glioma patients (G) or GBM patients (H) classified by eEF2K mRNA expression levels. Survival is indicated for time increments of 2,000 (G) or 500 days (H). p values were calculated using a log rank test.
(I and J) IHC staining for phospho-eEF2 in primary tumors (I, 1–2) and corresponding metastases (I, 3–4) from a mouse model of MB (Wu et al., 2012) or in human primary MB (J). Black arrows indicate the tumor regions, and white triangles indicate adjacent normal cerebellar tissue.
(K) siRNA-mediated knockdown of eEF2K in d283 MB cells. Cells were transiently transfected with 25 nM of control (siCTRL) or eEF2K siRNAs (si eEF2K#1′ and eEF2K#2′), grown in complete media for 72 hr, and harvested or placed under ND for 24 hr. Cell lysates were assayed for caspase-3 activity. Error bars indicate SD for n = 3 (*p < 0.05).
See also Figure S7.
We next evaluated eEF2K activity in MB tumor samples by IHC for phospho-eEF2. In a mouse model of MB (Wu et al., 2012), both primary tumors (Figure 6I, panels 1 and 2) and matching metastases (Figure 6I, panels 3 and 4) showed extensive tumor-specific eEF2 phosphorylation (i.e., inactivation), whereas adjacent normal cerebellar tissue was negative. Similar findings were observed in primary human MB tumors (n = 7) (Figure 6J). Thus, eEF2K activity is strongly induced in MB tissues, both in primary tumor and metastatic compartments, compared to normal cerebellar tissue. Finally, we tested the potential involvement of eEF2K in the response of MB cells to ND. The group 3 human MB d283 cell line was highly resistant to ND (Figures S7A and S7B). However, d283 cells transfected with eEF2K siRNAs exhibited significantly higher cell death under ND compared to controls (Figures 6K and S7C). This suggests that eEF2K also promotes survival of MB cells under ND. Together, these findings indicate that eEF2K expression is predictive of outcome in MB and GBM and that eEF2K activity protects MB cells from ND.
The eEF2K Ortholog efk-1 Is Critical for Survival of C. elegans under ND
To test whether the protective role of eEF2K is conserved, we examined physiological responses to ND in vivo using the nematode worm Caenorhabditis elegans. In the absence of nutrients, newly hatched first larval stage (L1) C. elegans worms enter a dormant state termed “L1 diapause,” in which development is halted but survival is sustained for 2 to 3 weeks (Johnson et al., 1984), thus providing a useful model to study survival mechanisms under ND independently of concomitant developmental programs. We assessed a deletion allele of the C. elegans eEF2K ortholog, efk-1(ok3609), in survival of starved L1 larvae to determine whether there is a conserved role for efk-1 in managing ND. Whereas the deletion of efk-1 had only a minor effect on lifespans of animals grown under nutrient-rich conditions, survival of efk-1(ok3609) was substantially reduced under ND compared with wild-type (wt) (N2) worms (Figures 7A and 7B). Indeed, the mean lifespan of N2 animals in the absence of nutrients was 13.1 days, whereas efk-1 mutant worms had a mean lifespan of only 7.4 days (p < 0.00058; Figure 7B). Moreover, efk-1 transcripts were markedly induced by ND in wt L1 larvae (Figure 7C), as were eEF2K levels across various mouse and human cell lines subjected to ND (Figure 7D). These findings provide compelling evidence that the eEF2K ortholog efk-1 is a component of the physiological response of C. elegans to ND and, thus, that the role of eEF2K in this pathway is highly conserved.
Figure 7. The eEF2K Ortholog efk-1 Supports Survival of C. elegans under ND In Vivo.
(A) Lifespans of C. elegans WT (N2) and efk-1 (ok3609) homozygous deletion mutants under ambient growth conditions.
(B) Lifespans of N2 and efk-1 (ok3609) mutants at the L1 stage of development for the indicated days in the absence of food. Error bars indicate SEM for n = 142–150.
(C) Quantitative RT-PCR for efk-1 mRNA levels in N2 worms held at the L1 stage for 24 hr in the absence of food (starved) or then placed on food (OP50 bacteria) for 3 hr (fed). Transcript levels were normalized to γ-tubulin (tbg-1) levels. Error bars indicate SD for n = 3 (*p < 0.05).
(D) Quantitative RT-PCR for eEF2K transcripts in the indicated mammalian cell lines grown in complete media (Ctrl) or under ND for 24 hr. Transcript levels were normalized to β-actin (actb). Results are shown as relative levels in ND versus control conditions for each cell line. Error bars indicate SD for n = 2 (*p < 0.05).
(E) Model for the proposed role of eEF2K/efk-1 in adaptation to ND in normal and tumor tissues. Gray arrows and bars indicate release from regulatory effects of upstream pathways.
DISCUSSION
efk-1/eEF2K Is a Component of the Stress Response to ND
Nutrient deprivation activates multiple transcriptional cascades to reprogram metabolism when nutrients are limited, such as induction of AMPK subunit α-2 (prkaa-2/aak-2) (Baugh et al., 2009; Jagoe et al., 2002). Here, we report that mammalian eEF2K and its C. elegans ortholog efk-1 are ND-responsive genes and are components of a nutrient stress adaptive program. In mice, eEF2K expression is upregulated when neonates are deprived of transplacental nutrient supply (Sakagami et al., 2002). Along with our findings, this suggests that the efk-1/eEF2K system may have evolved to preserve cell survival during physiological ND (Figure 7E). Given that eEF2K blocks mRNA translation elongation, this may reflect the need to block translation elongation along with initiation for an optimal adaptive response. Indeed, 4EBP1 expression increases under ND (Jagoe et al., 2002) to support adaptation to metabolic stress, including ND and hypoxia (Braunstein et al., 2007; Teleman et al., 2005).
eEF2K Blocks Translation Elongation to Protect Cells from ND
Inhibiting global mRNA translation increases resistance to ND in vivo (Pan et al., 2007; Teleman et al., 2005). Indeed, mTORC1 is inactivated under ND to preserve energy balance and amino acids (Choo et al., 2010; Teleman et al., 2005). Our findings suggest that mTORC1 inhibition alone may be insufficient to fully protect cells from ND and that eEF2K activation is also critical. The latter relies on inhibition of the eEF2K substrate, eEF2, supporting the notion that both translation initiation and elongation must be inhibited to promote survival (Figure 7E). Unexpectedly, inhibition of translation elongation was not linked to preservation of ATP nor to altered ROS or autophagic flux. Moreover, inhibition of elongation did not affect global protein synthesis rates in nutrient-deprived cells, as overall protein synthesis was already strongly reduced due to the block in translation initiation. Nevertheless, eEF2K activation does appear to promote stalling of ribosomes on transcripts, secondary to blocked translation elongation. This suggests that, in the absence of eEF2K activation, polypeptides still elongate under ND and some transcripts may even bypass the block in initiation through alternative initiation mechanisms (cap-independent or uORF-based processes), thus supporting their continued translation under nutrient stress. Therefore, eEF2K may block synthesis of specific subgroups of proteins instead of affecting overall synthesis. This is particularly relevant if the latter include proapoptotic factors or anabolic drivers whose inhibition is critical for adaptation to ND. In keeping with this notion, 4E-BP, which is protective under ND (Teleman et al., 2005), blocks synthesis of specific subsets of proteins involved in proliferation and polyamine biosynthesis (Dowling et al., 2010).
eEF2K Is Hijacked by Tumor Cells for Adaptation to ND
During tumor development, transformed cells must adapt to metabolic stress such as ND because anabolic processes typically driven by oncogenic pathways may have deleterious effects on the cell when nutrients are scarce (Jones and Thompson, 2009). Our in vitro studies indicate that hypersensitivity of RasV12- or EN-transformed cells to ND is due to a block in eEF2K activation through a combination of sustained Ras-Erk-p90RSK signaling and dampened AMPK induction. Given that the Ras-Erk pathway is overactive in ~25% of human tumors, eEF2K may be inhibited in diverse tumors, therefore restricting their capacity to survive when nutrients are limited. However, we observed that RasV12- or EN-transformed cells can adapt to nutrient stress, even with sustained Ras-Erk activity. This is in agreement with previous studies reporting that, although oncogenic Ras primarily sensitizes fibroblasts to reduced nutrients (Chiaradonna et al., 2006), it can also support adaptation to ND by increasing Glut1 and glucose uptake in tumor cells (Yun et al., 2009). We propose an additional mechanism for adaptation in which reactivation of an AMPK-eEF2K axis occurs via the ability of these cells, unexpectedly, to raise their AMP:ATP and ADP:ATP ratios. In contrast, defective AMPK activation in nonadapted cells results from sustained ATP levels, thus blocking the increase in AMP:ATP and ADP:ATP ratios. This suggests that nonadapted tumor cells may maintain specific anabolic processes under ND that sustain ATP.
It is tempting to speculate that eEF2K-mediated adaptation to ND mimics the process occurring in primary tumors subjected to repeated cycles of metabolic stress. The mechanism of ND-induced eEF2K activation in vitro appears to involve, at least in part, activation of an AMPK-eEF2K pathway by modulation of cellular AMP:ATP and ADP:ATP ratios. In vivo, defects in eEF2K activity rendered tumor xenografts sensitive to CR as reported for other models (Kalaany and Sabatini, 2009). Although reduction of glucose levels under CR is modest compared to the severe conditions imposed in vitro, this suggests that eEF2K-deficient tumors may be sensitive to even moderate reductions in glucose in vivo. In addition, a major effect of CR is to reduce plasma levels of insulin and IGF1 (Kalaany and Sabatini, 2009). Because eEF2K is negatively regulated by these factors (Wang et al., 2001), in vivo necrosis in eEF2K-deficient tumors under CR may reflect enhanced sensitivity of tumors to decreased plasma levels of insulin and IGF1. We speculate that the combined effect of compromised plasma nutrient availability, including modestly reduced glucose levels, coupled with decreased insulin and IGF1 levels, collaborate to render eEF2K-deficient tumors sensitive to CR.
In summary, our data reinforce the importance of AMPK for management of nutrient stress in tumors, as AMPK supports adaptation to ND by activating eEF2K and blocking translation elongation, in addition to inhibiting ACC and fatty acid synthesis as reported (Jeon et al., 2012). Thus, eEF2K emerges as a critical prosurvival factor that can be exploited by tumors to support adaptation to metabolic stress. The development of therapeutic strategies to target the eEF2K kinase in tumors, especially aggressive human brain tumors, warrants further investigation. Such strategies may be particularly relevant if combined with CR mimetics such as 2-DG, angiogenesis inhibitors, or resveratrol, as eEF2K may selectively protect tumors under CR rather than under nonstress conditions. In conclusion, inhibition of mRNA translation elongation represents an important mechanism for tumor cells to adapt to ND, highlighting previously unrecognized possibilities for therapeutic targeting of metabolically adapted tumor cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Xenografts
NIH 3T3, MG63, HeLa, and d283 cells were purchased from ATCC. NIH 3T3 cells stably expressing EN or K-RasV12 were as described (Ng et al., 2012). Immortalized WT and eEF2K-deficient MEFs were gifts from Dr. Alexey Ryazanov (University of Medicine and Dentistry of New Jersey). ND was performed with ~50% subconfluent cultures in Hanks buffered saline solution (HBSS)-HEPES (CaCl2·2H2O, 0.185 g/l; MgSO4·7H2O, 0.2 g/l; KCl, 0.4 g/l; KH2PO4 [anhydrous], 0.06 g/l; NaHCO3, 0.35 g/l; NaCl, 8 g/l; Na2HPO4·7H2O, 0.09 g/l; 20 mM HEPES, pH 7.4) containing no glucose, as for the indicated times, and resupplemented with glucose (4.5 g/l; Fisher) and/or 1× MEM essential and nonessential amino acid solutions (Invitrogen) and glutamine (2 mM; Invitrogen) as indicated. For tumor xenografts, cells were injected subcutaneously (5 × 105 cells per site) into flanks of nu/nu mice (Harlan Laboratories). CR experiments were carried out as described (Kalaany and Sabatini, 2009); mice in the CR group received 60% of the daily food intake of ad libitum counterparts. Nine days postinjection, tumor tissues were either snap frozen in liquid N2 or formalin fixed for IHC.
Protein Synthesis Rate
For [35S]-methionine-cysteine incorporation, cells were pulse labeled with 10 μCi [35S]-methionine and cysteine mix per ml (EasyTag EXPRESS, Perkin Elmer) for 5 min in HBSS-HEPES as described (Ng et al., 2012). For AHA incorporation assays, cells were incubated in methionine-free DMEM containing 10% CS for 1 hr and pulse labeled with 50 μM L-AHA for 45 min in methionine-free DMEM containing 10% CS or in HBSS-HEPES. Cells were either processed as described in Somasekharan et al. (2012) or fixed, permeabilized, and labeled using a Click-iT Cell Reaction Buffer Kit (Invitrogen) for the Alexa Fluor 488 azide probe (Invitrogen) and sorted by FACS (BD Bioscience).
C. elegans Studies
C. elegans strains were cultured at 20°C on nematode growth media (NGM) plates using standard protocols (Brenner, 1974). WT Bristol N2 strain worms and efk-1(ok3609) deletion mutants were obtained from the Caenorhabditis Genetics Centre (CGC) of the National Institutes of Health Center for Research Resources.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Tang, M. Robertson, and C. Chow for technical assistance and B. Fonseca (McGill University), S. Aparicio, S. Mullaly, T. Ng (UBC), J.A. Pospisilik, and R. Teperino (Max Planck Institute, Freiburg) for helpful discussions. This work was supported by the Canadian Cancer Society Research Institute and the British Columbia Cancer Foundation (to P.H.S.) through generous donations from Team Finn and other riders in the Ride to Conquer Cancer and by the National Institutes of Health (CA159859), the Terry Fox Research Institute, and the Pediatric Brain Tumor Foundation (to M.D.T.).
Footnotes
Supplemental Information includes Extended Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.04.055.
REFERENCES
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995;312:163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- Carlberg U, Nilsson A, Nygård O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- Caro-Maldonado A, Muñoz-Pinedo C. Dying for something to eat: how cells respond to starvation. The Open Cell Signaling Journal. 2011;3:42–51. [Google Scholar]
- Chiaradonna F, Sacco E, Manzoni R, Giorgio M, Vanoni M, Alberghina L. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25:5391–5404. doi: 10.1038/sj.onc.1209528. [DOI] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol. Cell. 2010;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Penman S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: response to food deprivation. FASEB J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE, Mitchell DH, Kline S, Kemal R, Foy J. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 1984;28:23–40. doi: 10.1016/0047-6374(84)90150-7. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–731. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat. Genet. 1998;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB. Origin and principles of translational control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 1–40. [Google Scholar]
- Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, Moretti M, De Smaele E, Beg AA, Tergaonkar V, et al. NF-κB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat. Cell Biol. 2011;13:1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JA, Chang SH, Dvorak AM, Dvorak HF. Why are tumour blood vessels abnormal and why is it important to know? Br. J. Cancer. 2009;100:865–869. doi: 10.1038/sj.bjc.6604929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TL, Leprivier G, Robertson MD, Chow C, Martin MJ, Laderoute KR, Davicioni E, Triche TJ, Sorensen PH. The AMPK stress response pathway mediates anoikis resistance through inhibition of mTOR and suppression of protein synthesis. Cell Death Differ. 2012;19:501–510. doi: 10.1038/cdd.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature. 1988;334:170–173. doi: 10.1038/334170a0. [DOI] [PubMed] [Google Scholar]
- Sakagami H, Nishimura H, Saito R, Kondo H. Transient upregulation of elongation factor-2 kinase (Ca2+/calmodulin-dependent protein kinase III) messenger RNA in developing mouse brain. Neurosci. Lett. 2002;330:41–44. doi: 10.1016/s0304-3940(02)00715-2. [DOI] [PubMed] [Google Scholar]
- Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc. Natl. Acad. Sci. USA. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan SP, Stoynov N, Rotblat B, Leprivier G, Galpin JD, Ahern CA, Foster LJ, Sorensen PH. Identification and quantification of newly synthesized proteins translationally regulated by YB-1 using a novel Click-SILAC approach. J. Proteomics. 2012;77:e1–e10. doi: 10.1016/j.jprot.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yang JM, Jin S, Zhang H, Hait WN. Elongation factor-2 kinase regulates autophagy in human glioblastoma cells. Cancer Res. 2006;66:3015–3023. doi: 10.1158/0008-5472.CAN-05-1554. [DOI] [PubMed] [Google Scholar]
- Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, Croul S, Bouffet E, Fults DW, Eberhart CG, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.