Abstract
The c-Jun/AP-1 transcription factor controls key cellular behaviours, including proliferation and apoptosis, in response to JNK and Ras/MAPK signalling. While the JNK pathway has been well characterised, the mechanism of activation by Ras was elusive. Here we identify the uncharacterised ubiquitin ligase Trim7 as a critical component of AP-1 activation via Ras. We found that MSK1 directly phosphorylates Trim7 in response to direct activation by the Ras–Raf–MEK–ERK pathway, and this modification stimulates Trim7 E3 ubiquitin ligase activity. Trim7 mediates Lys63-linked ubiquitination of the AP-1 coactivator RACO-1, leading to RACO-1 protein stabilisation. Consequently, Trim7 depletion reduces RACO-1 levels and AP-1-dependent gene expression. Moreover, transgenic overexpression of Trim7 increases lung tumour burden in a Ras-driven cancer model, and knockdown of Trim7 in established xenografts reduces tumour growth. Thus, phosphorylation-ubiquitination crosstalk between MSK1, Trim7 and RACO-1 completes the long sought-after mechanism linking growth factor signalling and AP-1 activation.
Introduction
The Ras signalling pathway regulates a large and diverse array of cellular decisions, including cell proliferation. Around 30% of all human tumours harbour activating mutations in Ras or its downstream kinases, which contribute towards several aspects of the malignant phenotype such as deregulated growth, apoptosis and invasiveness1-3. Oncogenic Ras induces constitutive activation of many effectors that are normally activated by growth factor stimulation, driving cell growth and proliferation1. Among these effects is the upregulation of c-Jun, a member of the AP-1 transcriptional activator family, which controls transcription of cell cycle regulator genes including cyclinD1 and cdc24-7. In line with this, c-Jun−/− mouse embryonic fibroblasts (MEFs) display severe proliferation defects and deficiency in cell cycle re-entry after serum withdrawal4, 8, 9. c-Jun also has been demonstrated to be crucial for Ras-driven transformation, as c-Jun knock-out MEFs were refractory to the effects of oncogenic Ras10. c-Jun responds to growth factor stimulation via ERK as well as cellular stress via the JNK pathway, and mediates diverse cellular responses ranging from proliferation, migration and differentiation to tumourigenesis and cellular apoptosis4, 11, 12. The activation of c-Jun via JNK has been well characterised13, 14; however, the molecular mechanism connecting c-Jun and active Ras–Raf-MEK–ERK signalling, potentially crucial to its role in tumourigenesis, remains incomplete.
We previously described a novel c-Jun coactivator, RING domain AP-1 co-activator 1 (RACO-1), a RING domain-containing E3 ubiquitin ligase, which is stabilised by growth factor signalling. RACO-1 stability is regulated by a ubiquitin switch between Lys48 (K48) and Lys63 (K63)-linked ubiquitination, controlled by active MEK15 and by PRMT1 mediated arginine methylation16. Methylation of two arginine residues in the N terminus of RACO-1 (R98, 109) stabilises RACO-1 in a dimeric conformation and is a prerequisite for all known RACO-1 functions16.
RACO-1 depletion reduces cellular proliferation and downregulates several growth-associated AP-1 target genes, such as cdc2, cyclinD1 and hb-egf. On the other hand, transgenic overexpression of RACO-1 augments intestinal tumour formation triggered by aberrant Wnt signalling and cooperates with oncogenic Ras in colon epithelial hyperproliferation15. These data indicate that RACO-1 forms part of the link between Ras and c-Jun in tumourigenesis. However, the molecular players inducing the ubiquitin switch downstream of MEK to stabilise RACO-1 were not known.
In this study, we identify a previously uncharacterised ubiquitin ligase, Tripartite Motif-containing 7 (Trim7), which upon Ras–Raf-MEK–ERK pathway activation, is phosphorylated and activated by MSK1. Trim7 in turn ubiquitinates and stabilises RACO-1, leading to increased c-Jun transcription. These findings delineate the complete pathway by which growth factor signalling stimulates c-Jun function, and provide further evidence for the importance of phosphorylation-ubiquitination crosstalk in fundamental aspects of cell signalling. Underlining the importance of this pathway in vivo, we find that Trim7 cooperates with oncogenic Ras to drive lung tumourigenesis.
Results
Identification of Trim7 as a RACO-1 interacting protein
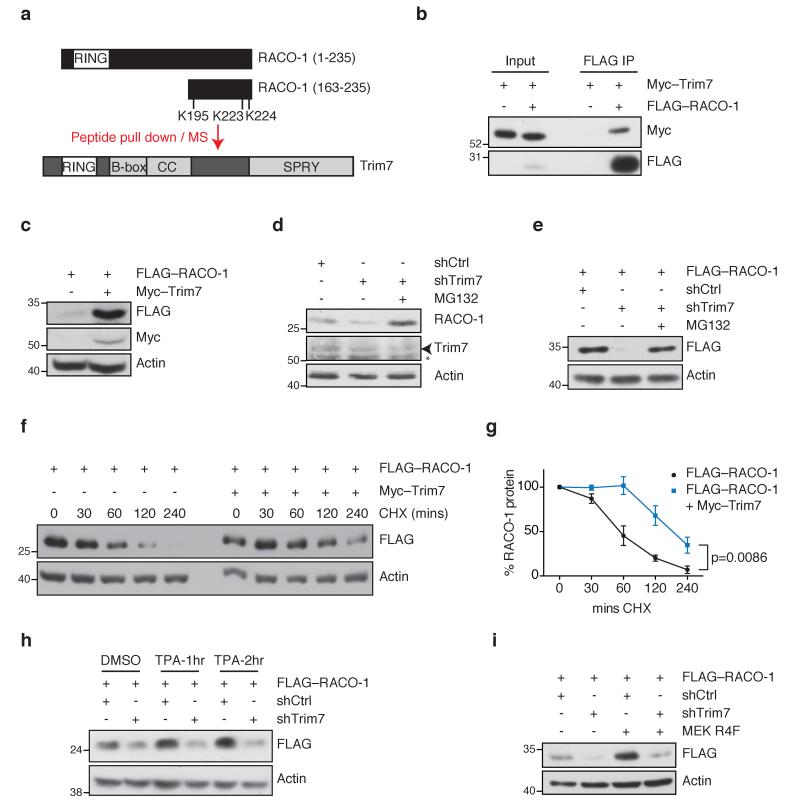
The c-Jun coactivator RACO-1 is stabilised by growth factor signalling via MEK, and is therefore a component of the uncharacterised pathway linking Ras signalling with c-Jun/AP-1 transcription15. To identify other components of this pathway, we used a biotinylated peptide (163-235) corresponding to the RACO-1 C-terminus, encompassing the ubiquitinated residues K195, K223 and K224, in order to find protein interactors likely to affect RACO-1 stability (Fig. 1a). RACO-1 (163-235) or the scrambled peptide was incubated with HeLa cell extract, followed by affinity capture using streptavidin beads. Using mass spectrometry, we identified a previously uncharacterised ubiquitin ligase, Trim7, interacting with RACO-1. The TRIM family of proteins is identified by tripartite motifs, which consist of a RING domain, followed by a B-Box and a coiled coil domain. The C-terminal end varies between family members and in the case of Trim7, contains a C-terminal B30.2 (SPRY) domain17 (Fig. 1a). Flag-tagged RACO-1 co-immunoprecipitated with Myc-tagged Trim7, confirming the interaction (Fig. 1b). Fine mapping with different deletion constructs encompassing RACO-1 showed that Trim7 interacts specifically with the very C-terminus of RACO-1 (Supplementary Fig. 1a,b).
Figure 1. Trim7 regulates RACO-1 stability in a MEK-dependent manner.
(a) Scheme of mass spectrometry experiment to identify RACO-1 interactors, showing domain structures of RACO-1 and Trim7. RING (zinc finger), B-box, coiled-coil (CC), and SPRY (protein-protein interaction) domains are shown. (b) FLAG–RACO-1 immunoprecipitation from cells transfected with Myc–Trim7 and blotted for Myc and FLAG. (c) Western blots showing the effect of Myc–Trim7 overexpression on FLAG–RACO-1 levels. (d) Western blot showing the effect of Trim7 depletion on endogenous RACO-1 levels in human lung H727 cells, in the presence and absence of proteasome inhibitor (MG132). Arrowhead indicates Trim7 band; asterisk indicates a non-specific band. (e) Western blots showing the effect of Trim7 depletion on FLAG–RACO-1 levels, in the presence and absence of MG132. (f) Western blots showing the degradation of FLAG–RACO-1 over time in the presence of cycloheximide (CHX), in control cells and cells overexpressing Myc–Trim7. (g) Quantification of (f) and 2 other similar CHX time course experiments showing RACO-1 stability. Error bars indicate s.e.m. P=0.0086 (Student’s t test comparing RACO-1 levels at 240 mins). (h) Western blot showing RACO-1 levels in control and Trim7-depleted cells after treatment with DMSO (vehicle) or TPA to stimulate MEK signalling. (i) Western blots showing the effect of Trim7 depletion by shRNA in the presence or absence of constitutively active MEK (MEKR4F). Actin is shown as loading control. Experiments were replicated at least three times.
Next, we investigated the effect of Trim7 on RACO-1 stability. RACO-1 protein is highly unstable in unstimulated conditions, since it is subject to autoubiquitination with K48-linked ubiquitin via its own RING domain and degradation by the proteasome15. However, overexpression of Myc–Trim7 in 293T cells greatly increased RACO-1 protein levels (Fig. 1c). Conversely, knocking down Trim7 decreased levels of endogenous RACO-1 in the H727 human lung adenocarcinoma cell line. This decrease occurred through proteasome-mediated degradation, as inhibiting the proteasome restored RACO-1 protein levels (Fig. 1d). Similar results were obtained with ectopically expressed FLAG-tagged RACO-1, and with two independent trim7 siRNAs (Fig. 1e and Supplementary Fig. 1c). Moreover, Trim7 knockdown in H727 cells had no significant effect on RACO-1 mRNA measured by quantitative RT-PCR (Supplementary Fig. 1d), excluding an effect of Trim7 on RACO-1 transcription. To determine whether Trim7 affects RACO-1 protein stability, we performed a time course experiment to monitor FLAG-RACO-1 degradation in the presence of cycloheximide to inhibit protein synthesis. Overexpression of Myc–Trim7 significantly slowed the degradation of RACO-1 (Fig. 1 f,g) while knockdown of Trim7 accelerated degradation of the protein (Supplementary Fig. 1e,f). Together with the results above, these findings demonstrate that Trim7 stabilises RACO-1 protein.
Our previous results showed that RACO-1 stability is regulated by MEK-ERK signalling and the effect of RACO-1 on c-Jun is independent of JNK phosphorylation15. In line with this, stimulation of JNK by anisomycin does not increase RACO-1 levels (Supplementary Fig. 1g). To test whether Trim7-mediated stabilisation of RACO-1 is under the control of the MEK-ERK pathway, Trim7 was knocked down by shRNA and the cells were treated with phorbol ester (TPA), which activates the MEK-ERK pathway. TPA treatment increased RACO-1 protein levels, but failed to do so in Trim7-depleted samples (Fig. 1h). As TPA is known to activate other signalling pathways in addition to MEK-ERK signalling, we used a constitutively active MEK1 (MEKR4F) to activate the MEK-ERK pathway specifically. MEKR4F increased RACO-1 protein levels in control cells but this effect was significantly reduced when Trim7 was knocked down (Fig. 1i). These results imply that RACO-1 stabilisation by MEK-ERK signalling requires Trim7.
Trim7 directly ubiquitinates RACO-1 using K63-linked chains
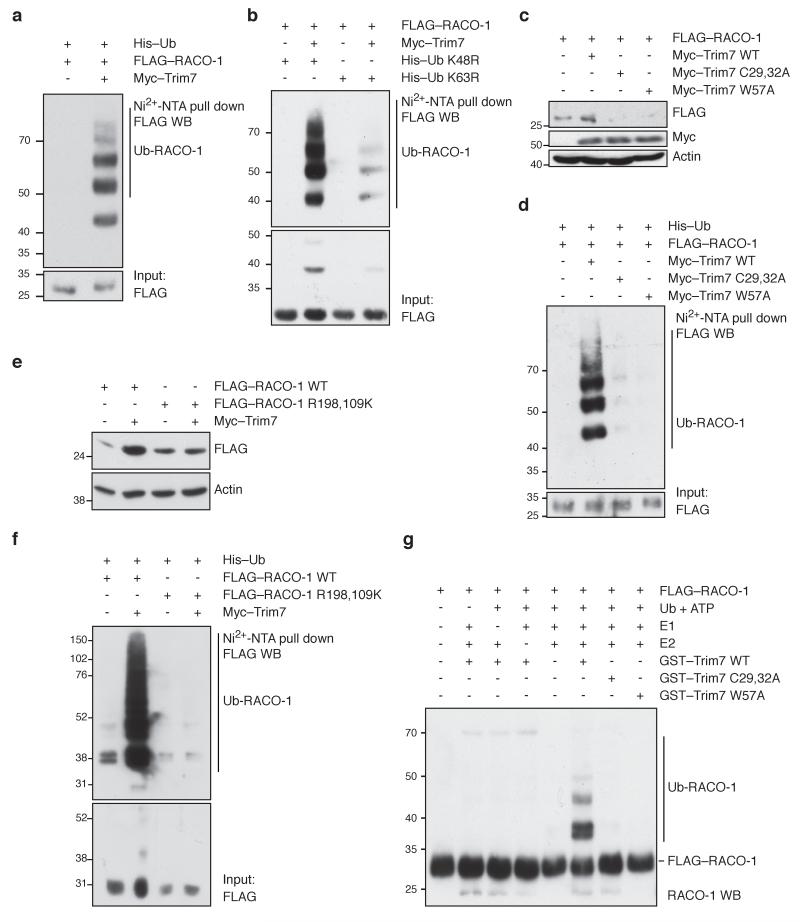
To investigate the mechanistic basis of RACO-1 stabilisation by Trim7, 293T cells were transfected with His-ubiquitin and RACO-1 with or without Trim7. In vivo ubiquitination assay using Ni2+-NTA pull down of ubiquitinated proteins showed that Trim7 stimulates RACO-1 ubiquitination (Fig. 2a). Next, we used two ubiquitin mutants, K48R and K63R, to identify the chain linkage of ubiquitinated RACO-1. Trim7 stimulated efficient ubiquitination of RACO-1 using K48R ubiquitin but ubiquitination was significantly reduced when K63R mutant ubiquitin was used. Thus, Trim7 promotes RACO-1 ubiquitination with K63-linked ubiquitin chains (Fig. 2b). Trim7 is predicted to be a ubiquitin ligase, as it contains a RING domain, but this has not been experimentally validated. To test whether the RING domain of Trim7 is important for RACO-1 ubiquitination, we mutated Cys29 and Cys32 to alanine (C29,32A), which destroys the integrity of the RING domain. In addition, we mutated a conserved tryptophan residue (Trp57) to alanine (W57A), which is important for the interaction of the RING domain with ubiquitin-conjugating enzymes (E2s)18. Both the mutant Trim7 proteins failed to stabilise RACO-1 (Fig. 2c). In vivo ubiquitination assay using wild-type (WT) or RING mutant Trim7 also demonstrated the inability of the Trim7 mutants to ubiquitinate RACO-1 (Fig. 2d). Interestingly, overexpression of mutant Trim7 reduced FLAG–RACO-1 protein below control levels (Fig. 2c), suggesting that the mutant has a dominant negative effect over the endogenous Trim7. In support of this idea, FLAG-tagged Trim7 was able to pull down Myc-tagged Trim7 by coimmunoprecipitation (Supplementary Fig. 2a), suggesting that, in common with other RING E3 ligases, Trim7 may act as a homodimer.
Figure 2. Trim7 ubiquitinates RACO-1 through K63 linkage.
(a) Ubiquitin pulldown using Ni2+-NTA resin to precipitate ubiquitinated proteins from cells transfected with FLAG–RACO-1 and Myc–Trim7 as indicated. (b) Ubiquitin pulldown as above using K48R or K63R mutant ubiquitin, which cannot form K48- or K63-linked ubiquitin chains, respectively. (c) Western blots showing FLAG–RACO-1 levels in Trim7-depleted cells reconstituted with wild-type, RING domain mutant (C29,32A), or E2 interaction mutant (W57A) Trim7. (d) Ubiquitin pulldown from cells expressing the above Trim7 mutants. (e) Western blots showing FLAG–RACO-1 and FLAG–RACO-1 R198,109K levels in the presence or absence of Trim7 overexpression. The R198,109K mutation disrupts the PRMT1 methylation sites and prevents RACO-1 dimerisation. (f) Ubiquitin pulldown from cells expressing FLAG–RACO-1 or FLAG–RACO-1 R198,109K and Myc–Trim7 as indicated. (g) In vitro ubiquitination assay using the indicated components together with immunoprecipitated FLAG–RACO-1 and purified recombinant GST–Trim7 wild-type (WT), GST–Trim7 C29,32A or GST–Trim7 W57A. Experiments were replicated at least three times.
Previously we have shown that active RACO-1 is a homodimer and PRMT1 mediated methylation of RACO-1 is a pre-requisite for efficient dimer formation16. In order to identify whether Trim7 preferentially ubiquitinates dimeric RACO-1, we used a mutant RACO-1 lacking two arginines which are targets for PRMT1-mediated methylation (RACO-1 R98,109K) that is therefore compromised in dimerisation ability. Overexpression of Trim7 did not stabilise RACO-1 R98,109K (Fig. 2e), and in vivo ubiquitination assay revealed that Trim7 was unable to stimulate ubiquitination of this mutant protein (Fig. 2f), suggesting that dimeric RACO-1 is the optimal substrate for Trim7.
Next, we wanted to test whether Trim7 directly ubiquitinates RACO-1. Ectopically expressed FLAG–RACO-1 was immunoprecipitated from 293T cells and used as a substrate for in vitro ubiquitination assay (Fig. 2g). Recombinant GST-Trim7 ubiquitinated RACO-1 only when all the purified components of the ubiquitination cascade (ubiquitin, ATP, E1, E2 and E3 ligases) were present. Moreover, the C29,32A and W57A mutants of Trim7 did not ubiquitinate RACO-1 (Fig. 2g), implying that RACO-1 ubiquitination requires a functional Trim7 RING domain. Taken together, these data suggest that Trim7 directly ubiquitinates RACO-1.
RACO-1 stabilisation by MEK requires MSK1 activity
We next investigated the mechanism of Trim7 regulation by growth factor signalling. The requirement of Trim7 for RACO-1 stabilisation by MEKR4F (Fig. 1g) suggested that the Trim7 regulatory activity is downstream of MEK. Knocking down ERK1/2 in 293T cells reduced RACO-1 levels, suggesting that Trim7 is under the control of ERK activity (Supplementary Fig. 2b). To test whether ERK directly phosphorylates Trim7, we carried out an in vitro kinase assay. Recombinant ERK efficiently phosphorylated Elk1, a canonical ERK substrate, but did not phosphorylate Trim7 (Supplementary Fig. 2c).
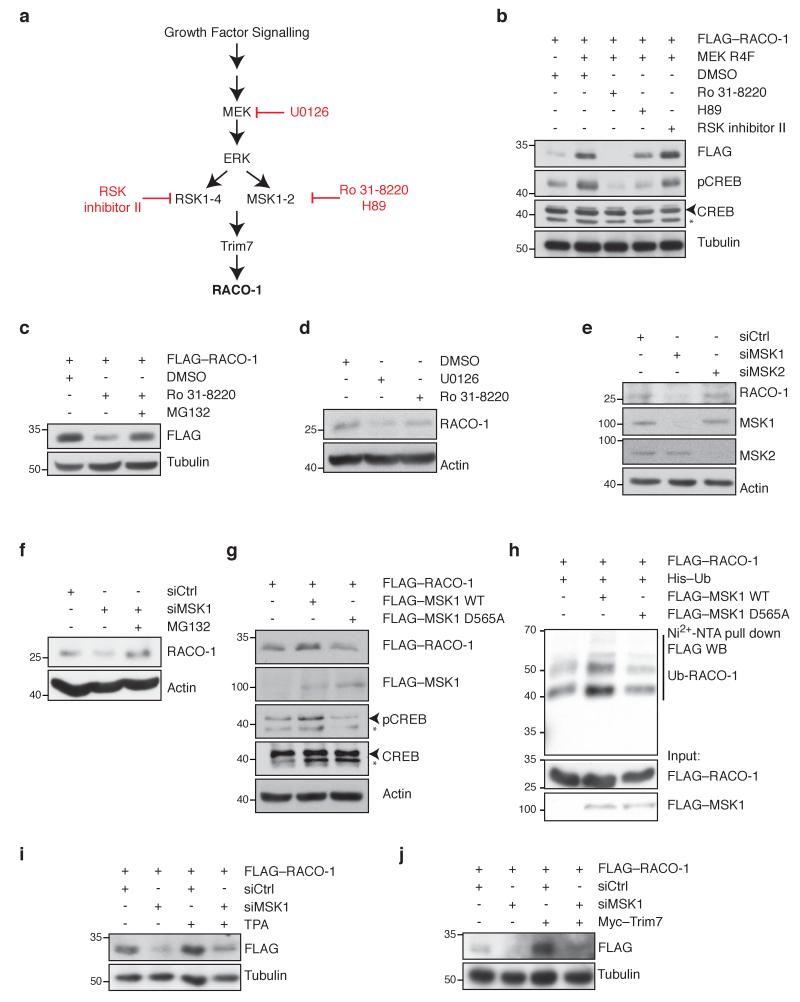
Mitogen and stress activated protein kinase (MSK) and 90kDa ribosomal S6 kinase (RSK) kinases mediate signal transduction downstream of MAPK signalling. Both the kinases are directly phosphorylated and activated by ERK kinases19, 20. In addition, MSK1 has been shown to induce immediate early gene expression upon Ras activation, via the nucleosomal response21-23. We therefore tested the effect of two MSK inhibitors, H-89 and Ro-31-8220, as well as RSK inhibitor II, on RACO-1 stability (Fig. 3a). Both MSK inhibitors destabilised RACO-1, while RSK inhibitor II had no effect on RACO-1 protein levels, suggesting that MSK is probably responsible for transducing the signals from active MEK-ERK signalling towards RACO-1 (Fig. 3b). The destabilisation of RACO-1 by Ro-31-8220 was proteasome dependent, as adding proteasome inhibitor reverted this effect (Fig. 3c). Inhibition of the MEK-ERK pathway in H727 cells with the MEK inhibitor U0126 and the MSK inhibitor Ro-31-8220 decreased endogenous RACO-1 protein levels to a similar extent (Fig. 3d).
Figure 3. RACO-1 stabilisation by MEK requires MSK1.
(a) Simplified scheme of MEK signalling pathway showing possible downstream effectors of Trim7 and RACO-1 regulation. Kinase inhibitors are shown in red. (b) Western blots showing the effect of the indicated kinase inhibitors on MEKR4F-induced FLAG–RACO-1 protein levels. Phosphorylated CREB (pCREB) is shown as a control for MSK inhibition. Tubulin is shown as loading control. Arrowhead indicates CREB band; asterisk indicates a non-specific band. (c) Western blots showing the effect of the MSK inhibitor Ro 31-8220 on FLAG–RACO-1 levels in the presence and absence of MG132. (d) Western blot showing the effect of the indicated inhibitors on endogenous RACO-1 levels in H727 cells. (e) siRNA-mediated depletion of MSK1 and MSK2 in H727 cells, showing the effect on endogenous RACO-1 levels. (f) Western blots showing the effect of MSK1 depletion on endogenous RACO-1 levels in H727 cells, in the presence and absence of MG132. (g) Western blot showing FLAG–RACO-1 levels in cells transfected with MSK1 or the catalytically impaired MSK1 mutant D565A. pCREB is shown as a control for MSK1 activity. Arrowhead indicates CREB/pCREB band; asterisk indicates a non-specific band. (h) Ubiquitin pulldown from cells expressing MSK1 or D565A mutant MSK1, showing ubiquitinated FLAG–RACO-1. (i) Western blots showing the effect of MSK1 depletion by siRNA on FLAG–RACO-1 levels in the presence or absence of TPA. (j) Western blots showing the effect of MSK1 depletion by siRNA on FLAG–RACO-1 levels in the presence or absence of Myc–Trim7 overexpression. Experiments were replicated at least three times.
The MSK protein kinase family consists of 2 members, MSK1 and MSK2. To determine whether MSK1 or MSK2 is responsible for RACO-1 stabilisation, we depleted each of these proteins individually by siRNA. Depletion of MSK1, but not MSK2, reduced endogenous RACO-1 protein levels, and this effect was rescued by proteasome inhibitor, suggesting that MSK1 is required for the stabilisation of endogenous RACO-1 (Fig. 3e,f). In support of this, overexpression of wild-type MSK1 increased RACO-1 protein levels, but a catalytically dead MSK1 protein (MSK1 D565A20) did not (Fig. 3g). In vivo ubiquitination assay also confirmed this conclusion, as overexpression of wild-type MSK1 increased RACO-1 ubiquitination, while MSK1 D565A had no effect (Fig. 3h). MSK1 was required for RACO-1 protein stabilisation by TPA (Fig. 3i). Similarly, MSK1 was also required for efficient RACO-1 stabilisation by Trim7 overexpression, suggesting that RACO-1 stabilisation by MEK-ERK signalling requires MSK1 (Fig. 3j).
MSK1 directly phosphorylates Trim7
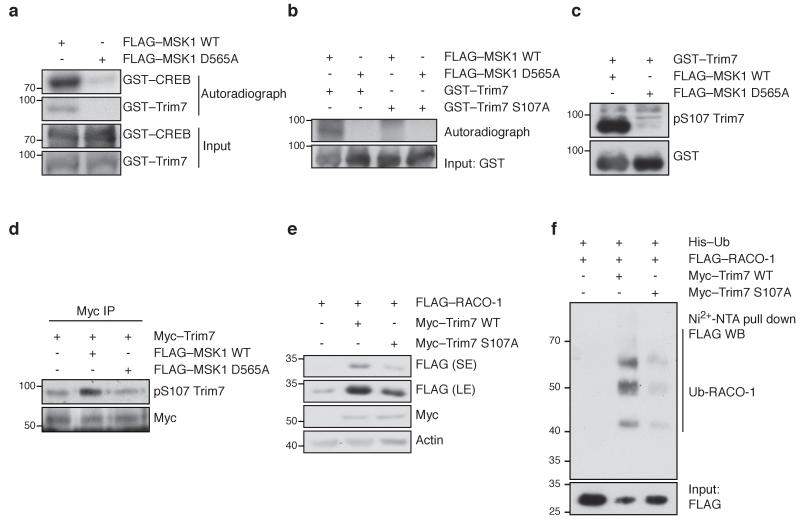
To investigate the mechanism of MSK1-mediated signal transduction resulting in RACO-1 stabilisation, we tested whether MSK1 directly phosphorylates Trim7. Wild-type MSK1 and the kinase-dead MSK1 D565A mutant were overexpressed in 293T cells, immunoprecipitated and used for in vitro kinase assay using radioactive γ32P-ATP. Wild-type MSK1 specifically phosphorylated recombinant GST-Trim7 while MSK1 D565A did not, suggesting that MSK1 directly phosphorylates Trim7 (Fig. 4a). MSK kinases phosphorylate serine residues within the consensus sequence R/KxRxxS or RRxS. Analysis of the Trim7 amino acid sequence revealed a consensus phosphorylation site, Ser107, for the MSK kinases (Supplementary Fig. 2d). Mutation of the S107 site on Trim7 abolished this phosphorylation (Fig. 4b), confirming S107 as the MSK1 phosphorylation site on Trim7.
Figure 4. MSK1 directly phosphorylates Trim7 and regulates RACO-1 stability.
(a) In vitro kinase assay using recombinant GST–Trim7 as a substrate for phosphorylation by FLAG-MSK1. GST–CREB and MSK1 D565A are shown as positive and negative controls, respectively. (b) In vitro kinase assay using recombinant GST–Trim7 or mutant GST–Trim7 S107A as a substrate for phosphorylation by FLAG-MSK1. (c) Western blot showing Trim7 phosphorylation at S107, using pS107 Trim7 antibody. (d) Myc–Trim7 immunoprecipitation from cells transfected with FLAG–MSK1 or FLAG–MSK1 D565A, showing Trim7 phosphorylation at S107. (e) Western blot showing FLAG–RACO-1 levels in cells transfected with Myc–Trim7 or mutant Myc–Trim7 S107A. SE, short exposure; LE, long exposure. (f) Ubiquitin pulldown from cells expressing Myc–Trim7 or mutant Myc–Trim7 S107A, showing ubiquitinated FLAG–RACO-1. Note that the amount of lysate was adjusted to account for the stabilisation of RACO-1 in Trim7-overexpressing cells, and so input levels do not reflect the amount of RACO-1 protein in cells. Experiments were replicated at least three times.
We generated an antibody specifically recognising phosphorylated serine 107 of Trim7 to further investigate Trim7 regulation. The pS107-TRIM7 antibody detected Trim7 phosphorylated with wild-type MSK1, but not the unphosphorylated Trim7, demonstrating the specificity of the antibody (Fig. 4c). Ectopic MSK1 expression increased Trim7 phosphorylation at S107, validating that this modification occurs in vivo (Fig. 4d). Mutation of Trim7 S107 to alanine inhibited the stabilisation of RACO-1, suggesting that phosphorylation of this site is required for stimulation of Trim7 activity (Fig. 4e). Consistent with this, Trim7 S107A was less able to ubiquitinate RACO-1 (Fig. 4f). To determine whether phosphorylation of Trim7 S107A affected its interaction with RACO-1, we performed co-immunoprecipitation experiments. Neither activation nor inhibition of MEK signalling (by TPA or U0126 respectively) nor mutation of the S107 phosphorylation site on Trim7 affected its interaction with RACO-1 (Supplementary Fig. 3a,b), suggesting that MEK-induced phosphorylation of Trim7 promotes its E3 ligase activity by increasing the efficiency of ubiquitin transfer rather than enhancing the interaction with RACO-1. Taken together, our data suggest that upon activation of the Ras-Raf-MEK-ERK pathway, MSK1 phosphorylates Trim7 on serine 107 and stimulates its E3 ligase activity towards RACO-1, resulting in RACO-1 stabilisation.
Trim7 promotes oncogenic Ras-mediated lung adenocarcinoma
Adenocarcinoma, a form of non-small cell lung cancer, is the most common type of lung cancer in humans. K-Ras mutations are commonly found in non-small cell lung cancers and are believed to play a key role in this malignancy. Data from The Cancer Genome Atlas indicates that Trim7 is altered in 5% of human lung adenocarcinomas, most frequently by overexpression (Supplementary Fig. 4a). Moreover, amplified or overexpressed RACO-1, present in 8% of these tumours, does not overlap with Trim7 alteration, and there is also a tendency towards mutual exclusivity with K-Ras mutations (Supplementary Fig. 4a)24, 25, in support of K-Ras, Trim7 and RACO-1 being in the same pathway. Thus we used a Ras-driven lung cancer model26, 27 to test the significance of the Trim7/RACO-1 axis in tumourigenesis. In this mouse model, Cre-mediated recombination induces the expression of oncogenic K-RasG12D 27. The transgene expression was activated in the lung by tracheal intubation of mice with Cre-expressing adenovirus (Adeno-Cre)26.
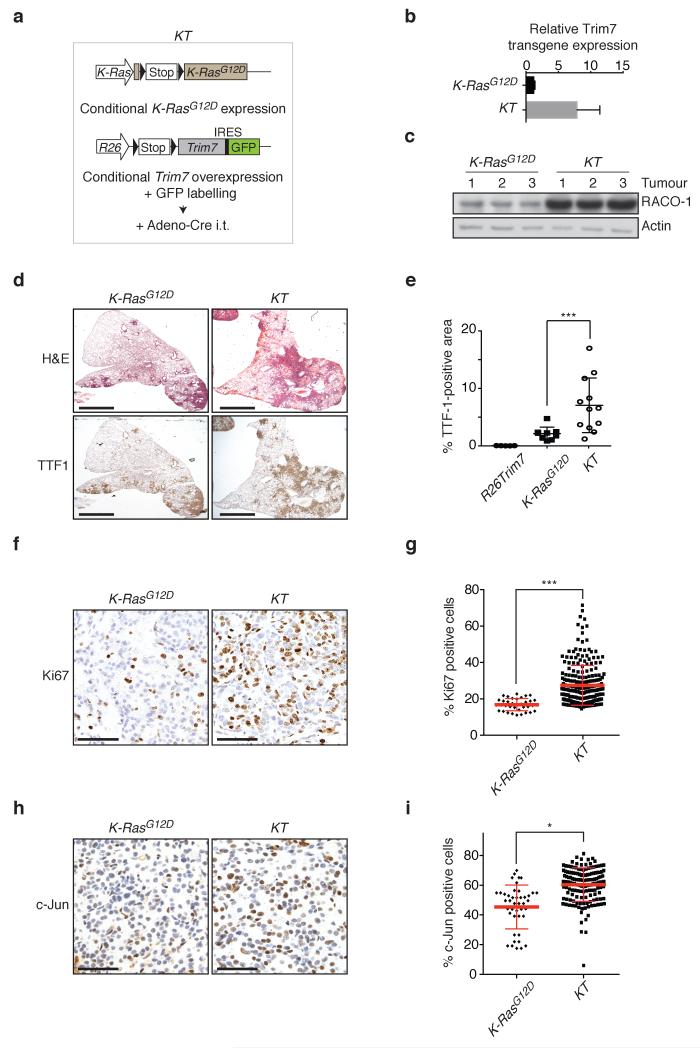
For transgenic overexpression of Trim7, we targeted a Cre-inducible human Trim7 transgene to the Rosa26 locus. The expression of the transgene is dependent on Cre recombinase-mediated excision of a STOP cassette (Rosa26-loxSTOPlox-Trim7-IRES-GFP or R26Trim7). The “KT” model combined inducible K-RasG12D expression with transgenic Trim7 and GFP expression from the Rosa26 promoter (LSL-K-RasG12D; R26-LSL-Trim7-IRES-GFP; plus Adeno-Cre) (Fig. 5a). Total lung extracts from either K-RasG12D or KT mice were used to isolate RNA and protein. Quantitative RT-PCR showed the expression of the Trim7 transgene in the lungs of KT mice (Fig. 5b). Western blot analysis of the proteins showed a significant stabilisation of RACO-1 in KT mice compared to K-RasG12D mice (Fig. 5c).
Figure 5. RACO-1 and Trim7 cooperate with K-RasG12D to drive lung tumourigenesis.
(a) Scheme of KT mouse model. i.t., intratracheal. (b) Quantitative PCR showing expression of the Trim7 transgene in KT mouse lung tissue. Error bars indicate s.d. n=3 tumours per genotype. (c) Western blot showing RACO-1 protein levels in KT mouse lung tissue. (d) Haematoxylin and Eosin (H&E) stain and TTF1 immunohistochemistry on whole lung sections from KT and K-RasG12D mice 12 weeks after Cre induction (1.25X magnification). Scale bar indicates 2mm. (e) Quantification of the percentage of total lung area stained with TTF-1 in mice of the indicated genotypes. Each data point represents a whole lung section from one mouse. n= 5 R26Trim7, 9 K-RasG12D, and 12 KT mice. (f, g) Staining (40X magnification)(f) and quantification (g) of Ki67-positive cells in lung tumours from mice of the indicated genotypes. Each data point represents 1 tumour. All tumours from one lung section per mouse were analysed (5 mice per genotype). n=36 K-RasG12D and 243 KT tumours. (h, i) Staining (40X magnification) (h) and quantification (i) of c-Jun-positive cells in lung tumours from mice of the indicated genotypes. Each data point represents 1 tumour. All tumours from one lung section per mouse were analysed (5 mice per genotype). n=43 K-RasG12D and 151 KT tumours. *p=0.02, **p<0.01, ***p<0.001 (Student’s t-test). Error bars in (e), (g), and (i) indicate mean ±s.e.m. Scale bars in (f) and (h) represent 100μm.
As expected, expression of oncogenic K-RasG12D alone resulted in formation of hyperplastic lesions and adenomas in the lungs that stain positive for TTF-1 (Fig. 5d). TTF-1 is frequently overexpressed in lung adenocarcinoma, and routinely used as a marker for these tumours 28, 29. Quantification of the TTF1-positive transformed area of the lungs from K-RasG12D and KT mice showed a significant increase in tumour burden in KT mice (Fig. 5d,e), whereas transgenic overexpression of Trim7 alone did not result in any TTF-1-positive lesions (Fig. 5e).
Detailed analysis of the tumours was carried out by immunohistochemical staining of the lung. Tumours from both mouse genotypes were positive for TTF1 and surfactant protein C (SpC), implying that they are adenocarcinomas (Supplementary Fig. 4b). KT tumours had significantly increased cellular proliferation compared with K-RasG12D tumours, as judged by quantification of Ki67-positive cells (Fig. 5f,g). Quantification of the percentage of c-Jun-positive cells per tumour showed a mild but significant increase in KT mice compared with K-RasG12D mice (Fig. 5h,i). Taken together, these data demonstrate that Trim7 cooperates with oncogenic Ras in lung adenocarcinoma formation.
Trim7 depletion restrains tumour cell proliferation
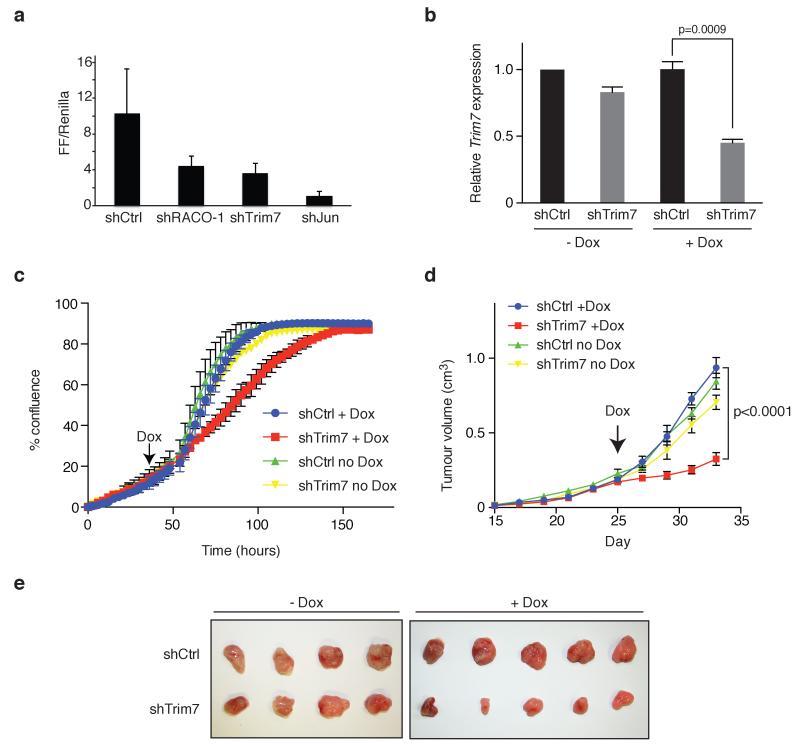
RACO-1 acts as a transcriptional coactivator of c-Jun and reduced RACO-1 level leads to a reduction in c-Jun transcriptional activity15, 16. A luciferase assay using a stably integrated chromosomal AP-1 reporter construct (collagenase-luciferase) showed a similar decrease in gene expression when either Trim7 or RACO-1 were depleted with shRNA, consistent with Trim7 regulating the transcription of c-Jun/AP-1 target genes via RACO-1 (Fig. 6a).
Figure 6. Trim7 depletion in established tumours reduces tumour growth.
(a) Luciferase assay in HeLa cells containing a stably integrated chromosomal collagenase-luciferase reporter as a read-out for AP-1 transcription. Error bars indicate s.d. (b) Quantitative PCR showing the level of Trim7 mRNA expression in H727 cells transfected with doxycycline (Dox)-inducible shRNA constructs. Error bars indicate s.d. (c) Time course showing proliferation of cells in (b) in vitro with and without addition of Dox to induce expression of the indicated shRNA. % confluence was measured automatically by Incucyte time lapse microscopy and used as a read out for cell proliferation. Error bars indicate s.d. (d) Time course showing growth of xenograft tumours from NOD/SCID mice following subcutaneous injection with H727 cells stably carrying Dox-inducible shRNA. Tumours were allowed to establish for 25 days and then animals were treated daily with Dox to induce Trim7 knockdown in the xenograft. The growth of similar xenografts in mice not treated with Dox is shown as a control. Tumours were measured with calipers every 2 days. n=5 mice (+Dox) and 4 mice (−Dox). Error bars indicate s.e.m. p<0.0001 at 33 days (Student’s t test). (e) Xenograft tumours from the experiment in (d) shown at experimental endpoint.
In human lung adenocarcinoma, oncogenic Ras mutations are frequently associated with mutations in other tumour suppressors such as p5330. Therefore, to test the effects of Trim7 depletion, we used primary murine lung adenocarcinoma cells derived from a K-RasG12D; p53ΔL/ΔL (KP) mouse line, which closely resembles human lung adenocarcinoma (Supplementary Fig. 5a). We generated KP cells stably expressing two different shRNAs against Trim7. Trim7 depletion suppressed the proliferation of KP cells, and the reduction in proliferation correlated with the extent of Trim7 knockdown in the two cell lines (Supplementary Fig. 5b,c). Trim7 depletion in KP cells with the most efficient knockdown construct (shTrim7-2) resulted in a reduced level of RACO-1 (Supplementary Fig. 5d) and quantitative RT-PCR analysis revealed that c-Jun target genes were downregulated in Trim7 knockdown cells (Supplementary Fig. 5e), supporting the observation that Trim7 regulates c-Jun transactivation function and cellular proliferation. To test the in vivo effect of Trim7 knockdown on KP cells, we subcutaneously injected shControl cells in the left flank and shTrim7-2 cells in the right flank of NOD-SCID mice. Trim7 knockdown resulted in significantly smaller xenograft tumours after 20 days (Supplementary Fig. 5f,g).
To further test the effect of Trim7 depletion in a clinically relevant scenario, we decided to knock down Trim7 in an already established tumour. To achieve this we generated H727 cell lines expressing a doxycycline-inducible shTrim7 or shControl construct (Fig. 6b). We first measured the proliferation of these cell lines in vitro. Cells were plated and allowed to grow for 36 hours without doxycycline, at which stage the proliferation of both cell lines was comparable. After 36 hours cells were treated with 2μg ml−1 doxycycline and their proliferation was monitored for a further 120 hours. Upon doxycycline treatment, shTrim7-expressing cells showed a significant decrease in proliferation (Fig. 6c).
Next we carried out a xenograft experiment with these cell lines. After sub-cutaneous injection of untreated shControl and shTrim7 cells into NOD-SCID mice as before, xenografts were allowed to grow to a palpable tumour mass, and then mice were given doxycycline every day to induce and maintain the expression of the shRNA. In control mice that did not receive doxycycline treatment, tumours grew equally (Fig. 6d,e). In the cohort that received doxycycline treatment, shTrim7-expressing tumours grew significantly slower than the control tumours (Fig. 6d,e). This experiment demonstrates that depletion of Trim7 in an established tumour can slow tumour cell proliferation. We conclude that Trim7 functions to stabilise RACO-1 and stimulate c-Jun-mediated gene expression and cellular proliferation, and is required for efficient tumour growth driven by oncogenic Ras.
Discussion
More than 25 years ago AP-1 was identified as an activity that bound to DNA elements in the promoters of the metallothionein and collagenase genes31, 32. The tumour promoter 12-O-tetradecanoyl phorbol 13-acetate (TPA) dramatically increases transcription of these genes in an AP-1-dependent manner. The classical Jun/Fos AP-1-binding site, TGACTCA, was identified as the element that mediates TPA induction of the metallothionein and collagenase genes, and is therefore known as the TPA response element. TPA strongly activates the MEK/ERK MAP kinase pathway, but is only a weak activator of JNK signalling and c-Jun N-terminal phosphorylation33. However, while JNK-mediated c-Jun phosphorylation is a well-established means of stimulating c-Jun/AP-1 activity, the precise molecular connection between c-Jun and growth factor signalling that is independent of N-terminal phosphorylation has been enigmatic.
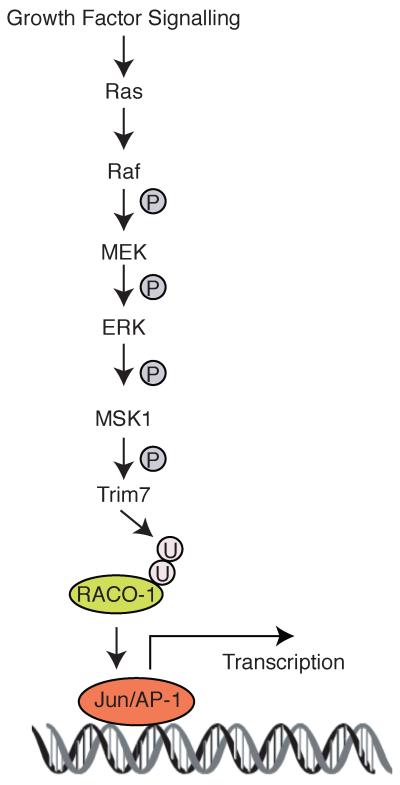
Our previous work demonstrated that RACO-1 functions as a molecular link between Ras signalling and c-Jun activity15. RACO-1 acts as a transcriptional coactivator of c-Jun and stimulates expression of target genes required for cellular proliferation, such as cyclinD1, cdc2, and hb-egf. RACO-1 stability plays a central role in the regulation of Ras-c-Jun crosstalk. Upon activation of growth factor signalling, the ubiquitination of RACO-1 shifts from degradative K48-linked autoubiquitination to a non-degradative K63-linked ubiquitination, thereby increasing RACO-1 protein levels and AP-1 activity. In addition, PRMT1-mediated methylation is required for RACO-1 to form a stable dimer which is responsive to MEK-ERK signalling and can interact with c-Jun. Here we have identified the E3 ubiquitin ligase Trim7 as another important regulator of Ras-c-Jun cross-talk, responsible for MEK-driven stabilisation of RACO-1 (Fig. 7).
Figure 7. Schematic model of the pathway linking Ras with c-Jun/AP-1.
A phosphorylation/ubiquitination cascade via MSK1, Trim7 and RACO-1 links growth factor signalling to c-Jun/AP-1 activation.
Trim family proteins are involved in a broad range of biological processes, and their alterations are associated with diverse pathological conditions, such as developmental disorders, neurodegenerative diseases, viral infections and cancer34-36. Trim7 was first identified as glycogenin interacting protein (GNIP) and shown to be important in glycogen biosynthesis37. Although ubiquitin ligase activity has been predicted for Trim7 due to the presence of the RING domain, no experimental validation has been carried out. Our data suggest that Trim7 has ubiquitin ligase activity towards RACO-1. Upon MEK/ERK activation, the ERK downstream kinase MSK1 is activated through phosphorylation, and active MSK1 phosphorylates Trim7, thereby stimulating its activity towards RACO-1. As with many other enzymes, activation by phosphorylation is common among E3 ubiquitin ligases; for example, the regulation of c-Jun turnover through JNK-dependent phosphorylation of the E3 ligase Itch38, 39. The cross-talk between different post-translational modifications has become a recurring theme in biology40, and many signalling pathways, including the Ras–Raf-MEK–ERK pathway, are subject to multiple protein modifications including farnesylation3, phosphorylation3 ubiquitination41, 42, sumoylation43 and methylation 44, 45, among others. This enables the pathway to be responsive to different signal inputs with tighter multi-layered regulation. Our study adds to the growing body of data showing that phosphorylation and ubiquitination not only cooperate via priming or competing modifications on the same protein, but also contribute as integral components of signal transmission cascades40, 41, 46.
A significant amount of research has been carried out to develop inhibitors of the Ras signalling pathway for therapeutic intervention in human malignancies. It is becoming increasingly apparent that monotherapy targeting a single kinase is only transiently effective, as the cells become resistant to the inhibitor via various mechanisms, as in the case of the B-Raf inhibitor vemurafenib47-51. Combined inhibition of B-Raf and MEK has shown promising results and has now been approved for treatment of B-Raf-V600E metastatic melanoma. However the long-term efficacy of this treatment has been limited by emergence of resistance through mutation in MEK and copy number gain in BRAF-V600E52. Targeting effectors downstream of ERK is a promising approach, which could improve efficacy and reduce toxicity 53, 54. Our data show that knocking down Trim7 in primary KP lung tumour cells reduced cell proliferation, and significantly decreased tumour growth in xenograft experiments. Moreover, depletion of Trim7 in an established xenograft tumour also reduced tumour growth, supporting the idea that therapeutic intervention in the MSK1/Trim7/RACO-1 signalling pathway is potentially promising.
Methods
Cell lines and antibodies
Human embryonic kidney (HEK) 293T cells, human lung adenocarcinoma H727 cells and primary murine lung adenocarcinoma cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS). HEK293T and H727 cells were obtained from Cancer Research UK Cell Services. Polyclonal RACO-1 (HPA030098) and Trim7 (HPA039213) antibodies were from Sigma. Antibodies against p-ERK1/2 (9106), total ERK1/2 (9102), pCREB (9191), total CREB (9197), MSK1 (3489) and MSK2 (3679) were from Cell Signaling Technology; Jun (610326) from BD transduction laboratories; Flag (A8592) and HA (H 6908) from Sigma; β-actin (ab49900), GFP (ab290) from Abcam; and GST (sc-459) from Santa Cruz. Mouse monoclonal antibody against Myc tag was generated in-house. Anti-FLAG HRP antibody was used at 1:5000 dilution and anti- β-actin HRP antibody was used at 1:30,000 dilution. All other antibodies were used at 1:1000 dilution for western blots. Uncropped scans of western blots are shown in Supplementary Fig. 6.
cDNA constructs and transfections
RACO-1 cDNA was cloned in BamHI-EcoRI sites of pCMV2B to generate FLAG-RACO-1, and in HindIII-BamHI sites of pEGFP(C3) to generate GFP-RACO-1. Deletion mutants and point mutants of RACO-1 were generated in the FLAG-RACO-1 construct and verified by sequencing.15. Human Trim7 isoform 1 cDNA in pDONR vector (Invitrogen) was obtained from Genecopoeia and transferred to N-terminal Myc and GST-tagged pDEST vector (Invitrogen) by LR recombination, as per manufacturer’s instructions. Site-directed mutagenesis of Trim7 was carried out on Trim7/pDONR and then moved to Myc- or GST-tagged pDEST vector by LR recombination. His-tagged ubiquitin K48R and ubiquitin K63R constructs were a gift from Andreas Hock (University of Marburg, Germany), and MEK1R4F was a gift from Melanie Cobb (The University of Texas Southwestern Medical Center). Flag-tagged MSK1, MSK1 D565A and GST-tagged CREB were obtained from the MRC Protein Phosphorylation and Ubiquitylation Unit, Dundee. All mutagenic constructs generated were confirmed by sequence analysis. HEK293T and H727 cells were transfected with Polyethyleneimine (PEI).
RNAi constructs and Lentivirus production
siRNA sequences for human Trim7, MSK1 and MSK2 knockdown were designed using Dharmacon siDESIGN centre. Oligonucleotide sequences (sense strand) were: siTrim7 (1): 5′-CGGAAAAGAAGGAGAGCAA-3′; siTrim7 3′-UTR (2): 5′-GCAUCACGGCUCUCCAGCA-3′; siMSK1 (1): 5′-GCAGAAGAGUUCACAGAAA-3′; siMSK1 (2): 5′-GGAAAUAACAGCUCUGAAA-3′; siMSK2 (1): 5′-CGGCCGAGAUCAUGUGCAA-3′; siMSK2 (2): 5′-GCCGAGAUCAUGUGCAAAA-3′. RISC free siRNA from Dharmacon was used as a control. shRNA sequence for human RACO-1 (5′-tgTGATGGACCGTAGGAAGAAttcaagagaTTCTTCCTACGGTCCATCAttttttc-3′) was cloned into the IRES-GFP lentiviral construct pLentiLox 3.716. shRNA for human c-Jun (SHCLND-NM_002228) and non-targeting shRNA (SHC016) were obtained from Sigma. siTrim7(1) was cloned into pLentiLox 3.7. Hairpin sequence (shown in upper case characters) was: shTrim7- 5′-tg CGGAAAAGAAGGAGAGCAA ttcaagaga TTGCTCTCCTTCTTTTCCG ttttttc-3′. shRNA against luciferase (5′-tg CGTACGCGGAATACTTCGA ttcaagaga TCGAAGTATTCCGCGTACG ttttttc-3′) was cloned in pLentiLox 3.7 and used as a control. GIPZ shRNA constructs for mouse Trim7 (shTrim7-1, clone V3LMM_428696: GAACTTCTTCAGCAATCCT and shTrim7-2, clone V3LMM_428699: TGTTGAGTTGCATTGCCCA) and TRIPZ Dox-inducible shRNA constructs for human Trim7 (shTrim7, clone V3THS_351926: TGAGCTGTCTCCTGGATCT) were obtained from Thermo Scientific. For lentiviral production, low passage HEK293T were transfected with the respective shRNA construct and the accessory plasmids VSV-G and Δ8.2 using PEI. After 48 hrs, medium containing the infectious virus was centrifuged, filter-sterilised, supplemented with 8 μg ml−1 polybrene and used to infect KP or H727 cells for 48 hrs. Cells were allowed to recover for another 48 hrs before adding puromycin for selection of cells stably expressing the shRNA. Puromycin resistant cells were further sorted for high GFP (in the case of GIPZ constructs) or high RFP (in the case of TripZ constructs) expression by flow cytometry.
Reporter assay
Stable HeLa clones bearing an array of amplified Collagenase-Luciferase gene units were obtained by blasticidin selection after cotransfection of equimolar amounts of −517/+63 Collagenase-Luc55, 56 and pΔBN-AR1, which promotes an amplification of cotransfected plasmids57. Cells were plated in 24 wells and transfected with 250 ng shRNA constructs with 50 ng Ub-Renilla as an internal control. After 48 hrs the cells were lysed and luciferase activity was measured using the Dual Luciferase reporter assay (Promega), as per manufacturer’s instructions.
qRT-PCR on cell lines
RNA was extracted from cells using the Qiagen RNAeasy mini kit, DNase treated (Ambion) and cDNA synthesised using Superscript III (Invitrogen) according to the manufacturer’s instructions. QPCR was performed using a ABIPrism 7500HT sequence detection system with SYBR Green incorporation. QPCR primers were: mouse Trim7, forward: 5′-cagctcaactgtgggcact-3′, reverse: 5′-gcatgtcctccactgcatag-3′; mouse c-Jun, forward: 5′-tgaaagctgtgtcccctgtc-3′, reverse: 5′-atcacagcacatgccacttc-3′; mouse cyclin D1, forward: 5′-gtgcgtgcagaaggagattgt-3′, reverse: 5′-ctcacagacctccagcatcca-3′; mouse HB–EGF, forward: 5′-tgctgccgtcggtgatg-3′, reverse: 5′-caccaacgcggacaacact-3′; mouse cdc2, forward: 5′-ggacgagaacggcttggat-3′, reverse: 5′-attcgtttggcaggatcatagac-3′; mouse CD44, forward: 5′-ctcctggcactggctctga-3′, reverse: 5′-ctgcccacaccttctcctactatt-3′; mouse β-actin, forward: 5′-atgctccccgggctgtat-3′, reverse: 5′-cataggagtccttctgacccattc-3′, human Trim7, forward: 5′-ttctccagcttctgggtagc-3′, reverse: 5′-cgctgagcagggagtagc-3′; human c-Jun, forward:. 5′-ccaaaggatagtgcgatgttt-3′, reverse: 5′-ctgtccctctccactgcaac-3′; human CyclinD1, forward: 5′-agctcctgtgctgcgaagtggaaac-3′, reverse: 5′-agtgttcaatgaaatcgtgcggggt-3′; human cdc2, forward: 5′-tggatctgaagaaatacttggattcta-3′, reverse: 5′-caatcccctgtaggatttgg-3′; human CD44, forward: 5′-tgccgctttgcaggtgat-3′, reverse: 5′-ggcctccgtccgagaga-3′; human MSK1, forward: 5′-tgctgacagattttggtctga-3′, reverse: 5′-atagttccacaaaaggaatatgctc-3′; human MSK2, forward: 5′-cacgaggtgcatcacgac-3′, reverse: 5′-gctgaagtgccgcttctt-3′; human Actin, forward: 5′-tggatcagcaagcaggagtat-3′, reverse: 5′-gcatttgcggtggacgat-3′. All primer pairs generated a single PCR product, as determined by dissociation curve analysis.
Immunoprecipitations
HEK293T cells expressing Myc-Trim7 with or without FLAG-RACO-1 were lysed with Buffer A (0.2% NP 40, 150 mM NaCl, 20 mM Tris pH7.5, 500 μM EDTA, protease inhibitor cocktail (Sigma), 100 μM PMSF, 1 mM Na3VO4, 50 mM NaF, 1 mM β-glycerophosphate, 1mM DTT) and subjected to Flag immunoprecipitation with anti-Flag agarose beads (A2220, Sigma). For coIP of GFP–RACO-1 with Myc–Trim7, 293T cells expressing Myc–Trim7 and wt or deletion mutant GFP-RACO-1 were lysed in buffer A and then subjected to Myc immunoprecipitation with anti-Myc agarose beads (A7470, Sigma).
In vivo and in vitro ubiquitination assays
For in vivo ubiquitination assays, His–Ub was affinity purified with Ni2+-NTA-agarose beads58. Briefly, FLAG–RACO-1 and His-Ub with or without Myc–Trim7 were transfected in 293T cells. Cells were lysed in buffer B (100 mM Sodium phosphate pH 8.0, 5M Guanidinium Hydrochloride) containing 10 mM imidazole. Ubiquitinated proteins were precipitated using Ni2+-NTA agarose beads (Qiagen). Beads were washed thrice in buffer B containing 20 mM imidazole, then thrice in buffer containing 1 M guanidinium hydrochloride and finally thrice in buffer C (25 mM Tris pH 6.5, 20 mM imidazole). Proteins were eluted from the beads by addition of 250 mM imidazole and subjected to western blotting.
For in vitro ubiquitination, FLAG-RACO-1 was immunoprecipitated from Trim7-depleted HEK293T cells using anti-FLAG M2 agarose beads (Sigma), washed with buffer A containing 1M NaCl, then with buffer A containing no NaCl and finally with buffer A containing 150 mM NaCl. Immunoprecipitated protein was eluted by 100ng ml−1 3X-FLAG peptide (Sigma). Eluted FLAG RACO-1 was incubated with 50 nM UBE1, 500 nM UbcH5a, 5 μM HA–Ubiquitin and GST–Trim7 wt or mutants in buffer E (50 mM Tris Cl pH 8.0, 10 mM MgCl2, 2 mM DTT and 4mM ATP) for 2 hours at 30°C. Reactions were stopped by addition of SDS loading dye and subjected to western blotting with RACO-1 antibody as above.
Protein purification
GST–Trim7 wt or mutant expression vectors were transformed into BL21 (DE3) bacteria and a single colony expanded. Cultures were grown to an optical density of 0.6 and protein expression was induced by the addition of 1mM IPTG for 4 h at 37°C. Pelleted bacteria were lysed by sonication in Buffer D (20 mM Tris pH 7.5, 150 mM NaCl, 0.5% Triton-X100, 100 μM PMSF, 5 mM β-mercaptoethanol, 10% glycerol). Fusion proteins were isolated by affinity purification using Glutathione Sepharose 4B beads (Amersham), washed 4 times with Buffer D containing 2 mM glutathione, and then the protein was eluted by 20 mM glutathione. Purified protein was dialysed against buffer D to remove glutathione before storage.
The construct encoding the mouse C-terminal activation domain of Elk-1 (GST-(His)6-Elk-1 residues 309-429) was inserted into the BamHI-NotI cloning sites of pET-41a59 and expressed in E. coli strain BL21(DE3) (Novagen) as an N-terminal GST-(His)6 fusion protein. For the purification of the bacterially expressed GST-(His)6-Elk-1 309-429 construct, cells were lysed in buffer E (50mM Tris pH 8.0, 300mM NaCl, 5mM DTT) with complete protease inhibitor cocktail (Roche Diagnostics). The cleared lysate was applied to glutathione sepharose resin (GE Healthcare Life Sciences), washed extensively first with buffer E and finally with buffer F (50mM Tris pH 8.0, 150mM NaCl, 5mM DTT). The GST-(His)6 tag was cleaved overnight by incubating the protein with 1:50 (w/w) GST-3C protease on the resin. Cleaved protein was collected and further purified on a Superdex 75 HR10/30 size exclusion column (GE Healthcare Life Sciences) in buffer G (25mM Tris pH 7.5, 150mM NaCl).
Constitutively active ERK2 was expressed in bacteria as an N-terminal (His)6 fusion protein and purified using the protocol of Cobb and colleagues60. Briefly, expression of recombinant ERK2 was induced with 0.25 mM isopropyl-β-D-thiogalactopyranoside at 30°C for 12-14 h. Cells were lysed in buffer H (50 mM sodium phosphate, pH 8.0, 0.3 M NaCl) with complete protease inhibitor cocktail (Roche Diagnostics). The lysate was clarified at 20,000 g for 30 min at 4°C. The resulting supernatant was applied to Ni2+-NTA resin equilibrated in buffer H, washed with 20 volumes of buffer I (20 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 10 mM imidazole) and eluted with 0.25 M imidazole, pH 7.0, in buffer I. Eluted protein was dialyzed overnight against buffer J (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM dithiothreitol, 1 mM EGTA) and applied to a Mono Q column (GE Healthcare) equilibrated in the same buffer, and eluted with 500 mM NaCl in buffer I. Fractions containing purified ERK2 were pooled, dialyzed against buffer I containing 20% glycerol and protease inhibitors, frozen in liquid nitrogen, and stored at −80°C in small aliquots.
In vitro kinase assay
For ERK kinase activity, GST or GST–Trim7 or Elk1 was incubated with ERK2 in buffer K (50 mM Tris-Cl pH 8.0, 150 mM NaCl, 10 mM MgCl2, 1mM EDTA and 2mM DTT) containing 0.2 mM ATP and 20 μCi γ32P ATP for 1 hr at 30 °C. The reaction was stopped by adding SDS loading dye and analysed on SDS PAGE. The gel was dried and bands were visualised by autoradiography.
For MSK1 kinase reaction, FLAG–MSK1 WT or FLAG–MSK1 D565A were ectopically expressed in 293T cells, immunoprecipitated by anti-FLAG M2 agarose beads (Sigma), washed and eluted using 100 μg ml−1 3X FLAG peptide (Sigma). Eluted MSK1 or MSK1 D565A was incubated with GST–Trim7 or GST–CREB in buffer H containing 20uM ATP and 20 μCi γ32P ATP for 1 hr at 30 °C. The reactions were analysed as described above.
Ethical considerations
Experiments using mice (Mus musculus) were carried out with the approval of the London Research Institute’s Ethical Review Committee according to the UK Animals (Scientific Procedures) Act 1986. For the lung cancer model, animals were culled 12 weeks after intubation, before tumours outwardly affect the health of the animal. For xenograft experiments, subcutaneous tumours were allowed to reach a maximum of 1.4cm in length before the experiment was terminated.
Generation of transgenic mice
The conditional ROSA26-Trim7-IRES-eGFP-pA+ targeting vector (using human cDNA sequence for Trim7 isoform 1) was generated through LR reactions between the pDONR-Trim7 and the Gateway compatible pROSA26-DV1 plasmids61. The ROSA26-Trim7 conditional targeting vector was linearised with Pvu1 and electroporated into the G4 ES cell line62 and ES clones were selected by G418 resistance and screened by PCR analysis61. Correctly targeted ROSA26-Trim7 ES cell clones were aggregated with wild-type Swiss diploid host embryos and strong male chimeras were generated that transmitted the conditional ROSA26-Trim7-IRES-eGFP-pA+ transgene to F1 generation offspring61. These mice were then bred with the K-RasG12D mouse model26, 27 to generate KT mice.
Adenoviral infection
Adeno-Cre virus (Ad5CMVCre) was purchased from the Gene Transfer Vector Core, University of Iowa. Intratracheal intubation26 was carried out on animals at 8-12 weeks of age using a dose of 2.5 × 10^7 viral particles per mouse. Briefly, the adenovirus was prepared in MEM and 10mM calcium chloride was added and allowed to precipitate at room temperature. The mouse was anaesthetised and the virus preparation was administered via a catheter inserted into the trachea. Both male and female transgenic mice were used. A sample size of at least 5 mice per genotype was used to allow statistical analysis to be performed, with no sample exclusion criteria. Mice were culled 12 weeks after intubation for analysis.
Immunohistochemistry and qRT–PCR on transgenic mouse tissue
Mice were killed by cervical dislocation and lungs were perfused with PBS and then 10% NBF. Lung tissue was incubated in 10% NBF overnight and washed with 70% ethanol. Paraffin-embedded 4 μM sections were stained with the following antibodies: TTF-1 (ab40880, Abcam), GFP (ab290, Abcam), Surfactant Protein C (AB3786, Millipore), c-Jun (610326, BD Biosciences), Ki67 (M7248, DAKO). All the antibodies were used at 1:100 dilution for immunohistochemistry. TTF-1 was used as a marker of lung adenocarcinoma to differentiate transformed from untransformed lung tissue. ImageJ software was used to quantify the TTF1-positive transformed lung area. Lungs from 5 R26Trim7 mice, 9 K-RasG12D mice, and 12 KT mice were analysed. Each data point in Figure 5e represents one mouse.
For Ki67 or c-Jun staining, the entire lung section was imaged using a Nanozoomer (Hamamatsu) slide scanner. The tumour areas were outlined and Ki67- or c-Jun-positive and negative nuclei were quantified automatically by APPsolute Digital Pathology software (VisioPharm) using a standard threshold staining intensity. For each tumour, the number of Ki67- or c-Jun-positive cells was calculated and represented as percentage of total number of cells. 5 mice of each genotype were used; in total, 36 K-RasG12D and 243 KT tumours were analysed for Ki67 staining and 43 K-RasG12D and 151 KT tumours were analysed for c-Jun staining. Each data point in Figure 5g and Figure 5i represents one tumour.
For RNA and protein isolation, lungs were perfused with PBS and minced and then resuspended in DMEM containing collagenase type III and incubated for 30 min at 37°C in a shaking incubator. Cells were centrifuged for 5 min at 300 ×g and then passed through a 70 μm cell strainer and then used for either protein preparation or RNA isolation as described above.
Xenograft experiments
Female NOD/SCID mice were subcutaneously injected at 8 weeks of age with shControl cells (stably transfected with shRNA construct as described above) on the left flank and shTrim7 cells on the right. A sample size of at least 4 mice per cohort was used to allow statistical analysis to be performed, with no sample exclusion criteria. For the “+Dox” cohort, 5 mice received a daily dose of 5mg ml−1 aqueous solution of doxycycline by oral gavage starting from day 25 and continuing to the end of the experiment. For the “no Dox” cohort, 4 mice did not receive any treatment as a control. Randomization and blinding were not used. Subcutaneous tumour volumes were determined every 2 days by measuring the external dimensions of the tumour with calipers. Two perpendicular diameters were measured in each case and the longer designated “length” (L) and the shorter “width” (W). The tumour volume (V) was calculated using the following equation, taken from Sanceau et al63:
The experiment was terminated once the largest xenograft tumour reached 1.4cm in length, whereupon the mice were culled and the tumours excised and photographed.
Mass spectrometry
Biotinylated RACO-1(163-235) and scrambled peptides (2 mg) were incubated with Dynabeads M280 streptavidin in PBS at 4 °C overnight. Peptide bound beads were washed and resuspended in buffer A. HeLa cells were lysed in buffer A and then incubated with Dynabead-bound peptides for 2 h at 4 °C. The beads were washed three times with buffer A containing 300 mM NaCl. For LC-tandem mass spectrometry (MS-MS), polyacrylamide gel slices (1 to 2 mm) containing the purified proteins were prepared for analysis using the Janus liquid handling system (Perkin Elmer, United Kingdom). Briefly, the excised protein gel pieces were placed in wells of a 96-well microtiter plate, destained with 50% (vol/vol) acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. Alkylated proteins were digested with 6 ng μl–1 trypsin (Promega, United Kingdom) overnight at 37°C. The resulting peptides were extracted in 1% (vol/vol) formic acid, 2% (vol/vol) acetonitrile and analysed by nanoscale capillary LC-MS-MS using a nanoAcquity UPLC (Waters, United Kingdom) to deliver a flow of 300 nl min−1. A C18 Symmetry 5-μm, 180-μm by 20-mm μ-precolumn (Waters, United Kingdom) trapped the peptides prior to separation on a C18 BEH130 1.7-μm, 75-μm by 250-mm analytical UPLC column (Waters, United Kingdom). The peptides were eluted with a gradient of acetonitrile. The analytical column outlet was interfaced with a Triversa NanoMate microfluidic chip for mass spectrometric analysis (Advion, United Kingdom). Mass spectrometric information was obtained using an orthogonal-acceleration quadrupole-time of flight mass spectrometer (Synapt HDMS; Waters, United Kingdom). Data-dependent analysis was carried out, for which automatic MS-MS data on the 8 most intense, multiply charged precursor ions in the m/z range of 400 to 1,500 were acquired. MS-MS data were acquired over the m/z range of 50 to 1,995. LC-MS-MS data were then searched against the UniProt KB (release 15.5) protein database using the Mascot search engine program (Matrix Science, United Kingdom)64.
Supplementary Material
Acknowledgements
We are grateful to the LRI Biological Resource Unit, Experimental Histopathology, Peptide Chemistry, High Throughput Screening and Protein Analysis and Proteomics laboratories for technical help. A. Hock, M. Cobb, and the MRC Protein Phosphorylation and Ubiquitylation Unit kindly provided reagents. NOD-SCID mice were kindly provided by D. Bonnet. We thank C. Davies for critical reading of the manuscript and C. Cremona for assistance with manuscript preparation. A.C. was funded by AICR grant 12-0149. M.E.D. was supported by a DFG fellowship (DI 1679/1-1). The London Research Institute is funded by Cancer Research UK.
Footnotes
Conflict of Interest statement
There are no conflicts of interest to disclose.
References
- 1.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nature reviews. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 3.Downward J. Targeting RAS signalling pathways in cancer therapy. Nature reviews. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 4.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 5.Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 6.Ryseck RP, Hirai SI, Yaniv M, Bravo R. Transcriptional activation of c-jun during the G0/G1 transition in mouse fibroblasts. Nature. 1988;334:535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- 7.Wada T, et al. MKK7 couples stress signalling to G2/M cell-cycle progression and cellular senescence. Nature cell biology. 2004;6:215–226. doi: 10.1038/ncb1098. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RS, van Lingen B, Papaioannou VE, Spiegelmann BM. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 9.Schreiber M, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes & development. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Molecular and cellular biology. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 12.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nature reviews. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 13.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 14.Aguilera C, et al. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231–235. doi: 10.1038/nature09607. [DOI] [PubMed] [Google Scholar]
- 15.Davies CC, et al. Identification of a co-activator that links growth factor signalling to c-Jun/AP-1 activation. Nature cell biology. 2010;12:963–972. doi: 10.1038/ncb2098. [DOI] [PubMed] [Google Scholar]
- 16.Davies CC, Chakraborty A, Diefenbacher ME, Skehel M, s A. Arginine methylation of the c-Jun coactivator RACO-1 is required for c-Jun/AP-1 activation. The EMBO journal. 2013;32:1556–1567. doi: 10.1038/emboj.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 18.Joazeiro CA, et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 19.Dalby KN, Morrice N, Caudwell FB, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. The Journal of biological chemistry. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 20.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. The EMBO journal. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HG, et al. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer research. 2008;68:2538–2547. doi: 10.1158/0008-5472.CAN-07-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang S, Iversen L, Kragballe K, Arthur JS, Johansen C. Mice lacking MSK1 and MSK2 show reduced skin tumor development in a two-stage chemical carcinogenesis model. Cancer investigation. 2011;29:240–245. doi: 10.3109/07357907.2010.550594. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Cadahia B, et al. Role of MSK1 in the malignant phenotype of Ras-transformed mouse fibroblasts. The Journal of biological chemistry. 2011;286:42–49. doi: 10.1074/jbc.M110.156687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwei KA, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 31.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 32.Angel P, et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 33.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. The EMBO journal. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nature reviews. Immunology. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. BioEssays: news and reviews in molecular, cellular and developmental biology. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 36.Hatakeyama S. TRIM proteins and cancer. Nature reviews. Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 37.Skurat AV, Dietrich AD, Zhai L, Roach PJ. GNIP, a novel protein that binds and activates glycogenin, the self-glucosylating initiator of glycogen biosynthesis. The Journal of biological chemistry. 2002;277:19331–19338. doi: 10.1074/jbc.M201190200. [DOI] [PubMed] [Google Scholar]
- 38.Gao M, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- 39.Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Molecular cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen LK, Kolch W, Kholodenko BN. When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling. Cell communication and signaling: CCS. 2013;11:52. doi: 10.1186/1478-811X-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laine A, Ronai Z. Ubiquitin chains in the ladder of MAPK signaling. Science’s STKE: signal transduction knowledge environment. 2005;2005:re5. doi: 10.1126/stke.2812005re5. [DOI] [PubMed] [Google Scholar]
- 43.Kubota Y, O’Grady P, Saito H, Takekawa M. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nature cell biology. 2011;13:282–291. doi: 10.1038/ncb2169. [DOI] [PubMed] [Google Scholar]
- 44.Hsu JM, et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nature cell biology. 2011;13:174–181. doi: 10.1038/ncb2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andreu-Perez P, et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Science signaling. 2011;4:ra58. doi: 10.1126/scisignal.2001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao M, Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Molecular cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 51.Lito P, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villanueva J, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell reports. 2013;4:1090–1099. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 54.Nissan MH, Rosen N, Solit DB. ERK pathway inhibitors: how low should we go? Cancer discovery. 2013;3:719–721. doi: 10.1158/2159-8290.CD-13-0245. [DOI] [PubMed] [Google Scholar]
- 55.Schneikert J, et al. Androgen receptor-Ets protein interaction is a novel mechanism for steroid hormone-mediated down-modulation of matrix metalloproteinase expression. The Journal of biological chemistry. 1996;271:23907–23913. doi: 10.1074/jbc.271.39.23907. [DOI] [PubMed] [Google Scholar]
- 56.Diefenbacher ME, et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. The Journal of clinical investigation. 2014 doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu N, Hashizume T, Shingaki K, Kawamoto JK. Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer research. 2003;63:5281–5290. [PubMed] [Google Scholar]
- 58.Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 60.Khokhlatchev A, et al. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. The Journal of biological chemistry. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- 61.Nyabi O, et al. Efficient mouse transgenesis using Gateway-compatible ROSA26 locus targeting vectors and F1 hybrid ES cells. Nucleic acids research. 2009;37:e55. doi: 10.1093/nar/gkp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.George SH, et al. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4455–4460. doi: 10.1073/pnas.0609277104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanceau J, Poupon MF, Delattre O, Sastre-Garau X, Wietzerbin J. Strong inhibition of Ewing tumor xenograft growth by combination of human interferon-alpha or interferon-beta with ifosfamide. Oncogene. 2002;21:7700–7709. doi: 10.1038/sj.onc.1205881. [DOI] [PubMed] [Google Scholar]
- 64.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.