SUMMARY
Biological circuits can be controlled by two general schemes: environmental sensing or autonomous programs. For viruses such as HIV, the prevailing hypothesis is that latent infection is controlled by cellular state (i.e. environment) with latency simply an epiphenomenon of infected cells transitioning from an activated to resting state. However, we find HIV expression persists despite the activated-to-resting cellular transition. Mathematical modeling indicates that HIV’s Tat positive-feedback circuitry enables this persistence and strongly controls latency. To overcome the inherent crosstalk between viral circuitry and cellular activation, and directly test this hypothesis, we synthetically decouple viral dependence on cellular environment from viral transcription. These circuits enable control of viral transcription without cellular activation and show that Tat feedback is sufficient to regulate latency independent of cellular activation. Overall, synthetic reconstruction demonstrates that a largely autonomous, viral-encoded program underlies HIV latency—potentially explaining why cell-targeted latency-reversing agents exhibit incomplete penetrance.
Keywords: fate specification, synthetic biology, positive feedback, cellular decision-making, gene-regulatory circuit, viral persistence, HIV latency, single-cell analysis
INTRODUCTION
Diverse biological systems, both natural and engineered, face the challenge of surviving in variant and unpredictable environmental conditions. One strategy is to sense surrounding conditions and respond with environment-specific developmental programs—there is a 1:1 correspondence between explicit sensor-actuators and the extremely reduced form of this scheme where sensing and actuation are so tightly coupled that environment entirely actuates the program (Bull and Vogt, 1979). An alternate strategy foregoes environmental sensing and actuation, instead relying on autonomous programs (Knedler, 1947), for example programs that intrinsically generate heterogeneity in phenotypes and allow probabilistic ‘bet hedging’ (Cohen, 1966). For many systems, such as bacteriophage-λ it is unclear if environmental sensor-actuator schemes or autonomous programs are employed (Arkin et al., 1998; St-Pierre and Endy, 2008; Zeng et al., 2010). The ensuing debates carry evolutionary significance since sensor-actuator regulation can be driven by crosstalk from coincidental signals and hence tied to unrelated epiphenomena, whereas autonomous circuits are invariably subjected to direct natural selection pressures. In other words, if a phenotype is controlled by sensor-actuator regulation it can be an ‘epiphenomenon’, but if autonomously regulated, the phenotype is invariably evolutionary hardwired and directly selected for.
For HIV, the debate is clinically relevant; it remains unclear whether the host-cell environment or autonomous viral circuitry controls proviral latency, a long-lived viral dormancy state that is the chief barrier to curative therapy (Richman et al., 2009; Weinberger and Weinberger, 2013). Upon infecting CD4+ T lymphocytes, HIV either actively replicates to rapidly produce progeny virions or can enter a long-lived quiescent state (proviral latency), from which it subsequently reactivates. These latently infected cells form a viral reservoir forcing patients to remain on lifelong suppressive therapy. The prevailing view (Coffin and Swanstrom, 2013; Siliciano and Greene, 2011) holds that proviral latency results from HIV transcription being controlled by the host-cell activation state (i.e. environment) since relaxation of activated lymphocytes, to a resting-memory state, is correlated with increased epigenetic silencing of the HIV promoter and increased cytoplasmic sequestration of transcription factors that activate HIV transcription (Pearson et al., 2008; Tyagi et al., 2010). In this model, HIV infects activated T cells, which allow active viral replication, and if these cells ‘relax’ to resting-memory T cells, which generally restrict HIV infection, viral latency ensues (Fig. 1, left).
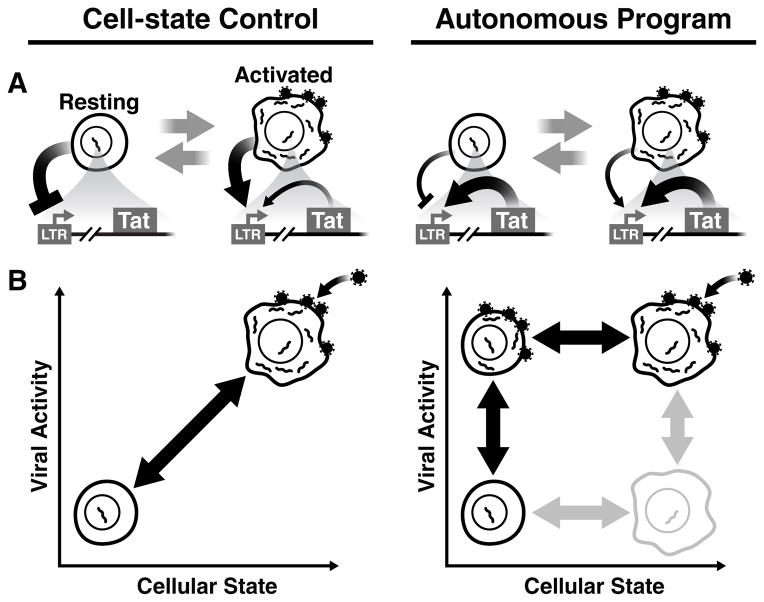
Fig. 1. Two models of HIV latency regulation: Cell-state control vs. Autonomous programming.
(A) Left: the prevailing hypothesis of HIV proviral latency regulation. As CD4+ T cells relax from an activated state (permissive to infection) to a resting-memory state, the host-cell environment silences HIV gene expression restricting Tat transactivation of the LTR. Right: The alternate hypothesis that HIV Tat positive feedback is robust to changes of the host-cell environment and operates autonomously despite changes in cell state. The overlapping nature of cellular and viral regulatory circuits confounds testing between these hypotheses (i.e. the LTR actuates Tat feedback but doubles as a sensor of the host-cell environment). (B) If cell state and viral circuitry can be orthogonalized (i.e. decoupled), the influence of cellular state on viral latency can be analyzed via an orthogonal 2D graphical correlation. Left: If cellular state dominates regulation of viral latency, resting cells would inhibit viral circuitry while active cells would induce viral gene expression generating a strong correlation between cell state and viral activity. Right: If an autonomous latency circuit regulates latency, both latent and active viral expression could be generated in either resting cells or activated T cells, producing little correlation between cell state and viral activity.
In contrast to the cellular control hypothesis, there is circumstantial evidence for an alternate model where latency is controlled by viral gene-regulatory circuitry (Ho et al., 2013; Jeeninga et al., 2008; Weinberger et al., 2005) without strict dependence on cellular state (Fig. 1, right). HIV encodes a transcriptional master circuit that is driven by the HIV Tat protein, which amplifies expression from the viral promoter within the HIV long terminal repeat (LTR), establishing positive feedback. Critically, minimal Tat positive-feedback circuits can recapitulate latency and stochastic fluctuations between a transcriptionally on and off state in the Tat circuit are sufficient to drive a phenotypic bifurcation between active and latent expression, even in non-resting cells (Weinberger et al., 2005). However, there is also evidence that cellular factors modulate stochastic HIV expression to drive latency (Burnett et al., 2009), confounding the hypothesis that latency is controlled by an autonomous viral circuit.
Here, we test between the cell-state and autonomous-circuit hypotheses for latency establishment. If latency is regulated by host-cell state, viral expression should be tightly correlated with cell state, whereas if the latency circuit is hardwired to function autonomously, then cellular state would be uncorrelated with viral expression and tuning viral circuitry, independent of cell state, would be sufficient to control HIV latency (Fig. 1B). Surprisingly, we find that viral expression is robust to cellular activation state in primary T cells, and mathematical models indicate this autonomy results from intrinsic properties of the HIV Tat positive-feedback circuit. However, directly testing circuit autonomy to cell state is confounded by overlap between cellular and viral networks—the same transcription factors that alter cellular activation also activate the HIV LTR, triggering Tat positive feedback (Karn, 2011). To circumvent this overlap, we synthetically reconstruct the Tat circuit to decouple viral dependence on the cellular environment from viral transcriptional regulation (i.e. decouple viral sensing and actuation). The refactored circuits chemically modulate viral expression independent of cellular activation levels and show that Tat circuitry is sufficient to overcome cell-driven silencing of HIV transcription during cellular relaxation from active to resting. Overall, the results argue that the Tat circuit is hardwired to establish latency largely autonomous of cellular state.
RESULTS
Donor-derived primary T lymphocytes maintain robust HIV expression during cellular relaxation from activated to resting
To test the prevailing ‘epiphenomenon’ hypothesis of HIV latency establishment, we αCD3/CD28 pre-activated donor-derived primary human CD4+ T lymphocytes (to achieve a CD25+CD69+ phenotype), infected them with full-length HIV-1 virus, and then removed activation stimuli, allowing infected cells to relax to a resting (CD25−CD69−) state (Fig. 2A). The virus used (HIV-d2GFP) encodes a short-lived two-hour half-life GFP (d2GFP) reporter to enable rapid detection of viral transcriptional silencing and is env-mutated (i.e. single-replication round) to avoid confounding the data with expansion of the infected-cell population. Infected cells were sampled periodically over two weeks for cellular-activation status (as quantified by CD25 and CD69) alongside viral-GFP expression.
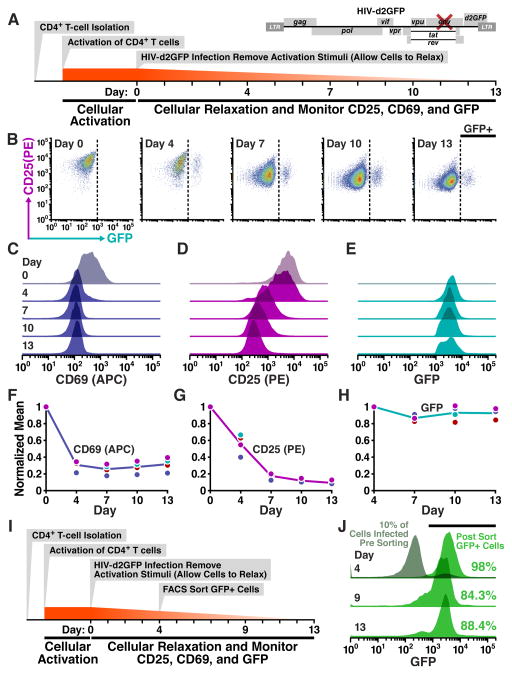
Fig. 2. HIV expression is autonomous to changes in cellular state: transitioning of primary T lymphocytes from activated to resting does not silence HIV expression.
(A) Schematic of activation, infection, and long-term observations of relaxing primary CD4+T cells with full-length HIV-d2GFP. Donor-derived primary cells were activated with αCD3/CD28 beads in the presence of rIL-2 for 3 days following which beads were removed and the cells were infected. At indicated time points, cells were collected for flow cytometry based measurement of CD25/CD69 levels and GFP expression. Data shown (in B-E) are representative of duplicate infections performed with cells from two donors. (B) Flow cytometry time course of CD25 and GFP levels taken on indicated days post infection. Dotted line indicates gating for productively infected cells (GFP+). (C–E) Histograms of cellular activation levels CD25 (C) and CD69 (D) of the entire population alongside GFP expression from productively infected cells (cells in GFP+ gate in B) over the course of 13 days post infection (17 days post cellular activation). (F–H) Cellular activation levels and GFP levels for all replicates over the experimental time course. Each dot indicates the time point from an independent infection and represents the geometric mean of the distribution as seen in C-E. Solid line connects the mean of the replicates. CD25 and CD69 normalized to day 0 (maximal); GFP normalized to day 4 when viral activity is first observed. (I) Schematic of FACS based isolation of productively infected cells. 4 days post infection, GFP+ cells were isolated and cultured (repeated for two donors). (J) Histograms of isolated GFP+ cells over time. Numbers indicate the proportion of cells that fall within the gate for positive GFP expression (marked by horizontal black bar). Day 4: Grey histogram shows the infected population prior to FACS-based separation. Viral titer was calibrated to achieve 10% infection (fraction of grey histogram that is GFP+ at day 4). Histogram in green (for Days 4, 9 and 13) shows the GFP expression in the isolated productively infected cells (post sort). All data shown above are from donor 1. See Fig. S1 for results from donor 2 and CD25 expression decline during the experiment.
Surprisingly, viral expression appears remarkably robust during the cellular transition from activated to resting (Fig. 2B,C–H). Despite drastic decline in cellular activation both in CD25 (Fig. 2D,G) and CD69 (Fig. 2C,F), viral activity (quantified by GFP expression of productively infected cells) remained relatively unchanged (Fig. 2B,E,H). The resilience of viral gene expression despite cellular relaxation is not due to differential relaxation of productively infected cells compared to the overall population as productively infected cells relax at the same rate as the overall population (Fig. S1).
Since human primary cells represent a mixed co-culture (i.e. infected and uninfected subsets of cells), which may obfuscate the interpretation of results (Jordan et al., 2003), we also performed a refined version of the experiment by isolating HIV-infected cells through FACS sorting and tracking this purified population of infected lymphocytes as cells relaxed to resting (Fig. 2I). As before, even after two weeks of culture ~90% of cells maintain high-level viral expression (Fig. 2J) despite cellular relaxation to resting (Fig. S1). Collectively, these two experiments show that despite a ten-fold decline in CD4+ T cell activation levels, the impact on viral gene expression is minimal, suggesting that viral circuitry is largely autonomous to cellular state.
Computational analysis predicts that Tat feedback circuitry can autonomously generate active and latent infection across a broad range of cellular-activation states
To investigate how viral transcription remains robust despite cell-state changes, we employ a simplified computational model of HIV transcriptional regulation (Fig. 3A) based on previous studies (Weinberger et al., 2008). This model builds off the standard two-state model of transcription (Kepler and Elston, 2001; Paulsson, 2004) and allows the LTR promoter to stochastically toggle between a transcriptionally non-permissive state (LTROFF) and a transcriptionally permissive state (LTRON) at rates koff and kon, respectively. In the LTRON state, Tat protein can transactivate the promoter, enhancing transcriptional elongation at a rate ktransact. These parameters (koff, kon, and ktransact) have been quantified by single-cell analysis (Dar et al., 2012; Singh et al., 2010; Weinberger et al., 2008) and measurements at thousands of HIV integration sites across the human genome show kon to be the predominant parameter that alters LTR activity in the regime required for latency (Dar et al., 2012), i.e. the weak expression regime. Potent cell-state activators, such as tumor necrosis factor alpha (TNFα) which acts through the same pathway as αCD3/CD28 activation, maximally stimulate LTR activity by increasing kon by 1.5- to two-fold (Dar et al., 2014; Dar et al., 2012; Jordan et al., 2001).
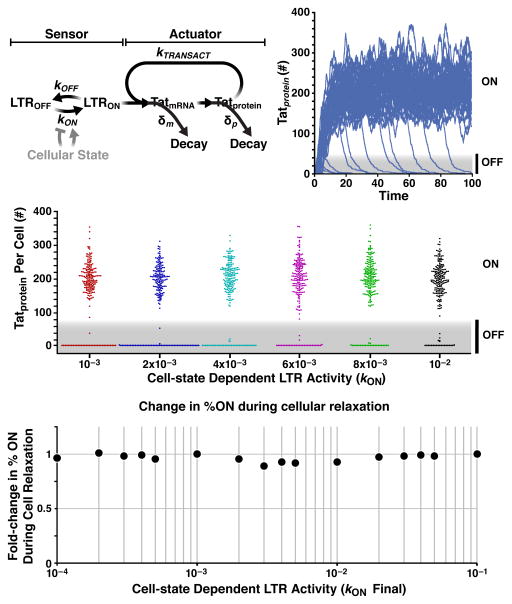
Fig. 3. Computational analysis predicts that Tat positive-feedback circuitry underlays HIV autonomy to cell state.
(A) Schematic of a simplified model of the Tat-feedback circuit. The LTR promoter can toggle between a state where transcriptional elongation is stalled (LTROFF) and a state where elongation proceeds (LTRON) at rates koff and kon, respectively, (Dar et al., 2012; Singh et al., 2010; Singh et al., 2012) and Tat protein transactivates the promoter by enhancing transcriptional elongation at a rate ktransact (Razooky and Weinberger, 2011). Tat protein and mRNA decay at rates δm and δp, respectively. (B) Stochastic Monte-Carlo simulations (“Gillespie” algorithm) of Tat protein levels (in arbitrary number of molecules) in individual cells over time (from reaction scheme in panel A). Each trajectory represents an individual cell; 100 single-cell trajectories shown (initial conditions for all species equal zero at time t=0, except LTRON = 1); see Extended Experimental Procedures for reaction rates. (C) Bee-Swarm plots of circuit activity (Tat levels at t=200) over a range of kon values. Each data point represents a single-cell trajectory, (200 trajectories shown per kon value). The width of the collection of cells (dots) having zero level of Tat (bottom of each kon value simulated) shows that high values of kon do generate less frequent latency (smaller number of dots). Compare for example the spread of red dots (kon=10−3) and black dots (kon=10−2) at 0 (D) Fold change in percentage of trajectories in ON state for two-fold reductions in kon. Circuit activity (%ON) is largely robust to reductions in LTR activity (i.e. kon), over three orders of magnitude. Phase-plane analysis (i.e. sensitivity analysis) from a closed-form analytical solution shows this behavior is robust across the physiological parameter regime (ktransact > kon). See also Fig. S2 and Table S1.
To determine if relaxation of activated T cells (i.e. decreases in kon) can drive LTR-Tat circuit shutoff and latency, we simulated infection of activated T cells and examined how tuning kon alters the fraction of trajectories in the ON state; i.e. initial conditions were LTRON = 1, and all other molecular species = 0 (see Table S1) thereby allowing efficient Tat turn-on in activated cells with subsequent stochastic circuit shutoff. The simplified model recapitulates previous results showing a phenotypic bifurcation in Tat levels (Weinberger et al., 2005), with a fraction of trajectories remaining ON and a fraction turning OFF (Fig. 3B) for any given kon across a broad range of values (Fig. 3C). Indeed, for LTR activities within three orders of magnitude (Fig. S2), any trajectory can maintain either an ON or OFF state purely by altering the level of Tat without a change in basal LTR activity. Thus, the model predicts that at a given cellular-activation state (kon value), circuit activity could be toggled ON and OFF simply by supplying Tat alone (e.g. in trans) without activating the LTR or changing the cellular-activation state (e.g. via TNFα). Moreover, the ON fraction can also be altered by changing Tat abundance—and hence feedback strength—through Tat half-life modulation (Fig. S2).
Next, we directly examined how decreases in kon influenced circuit activity. For all two-fold decreases in kon (over three orders of magnitude), there is > 90% robustness in the percentage of trajectories in the ON state (Fig. 3D). Two-fold decreases in LTR activity were examined because removal of cell-state activators (e.g. TNFα), result in 1.5- to two-fold reductions in LTR activity (Dar et al., 2012; Jordan et al., 2001), but comparable circuit robustness was observed for all 4-fold and even 1-Log reductions in kon (Fig. S2). In fact, the simplified nature of the computational model allows derivation of an analytical “closed-form” solution for the fraction of ON trajectories as a function of time for all parameters (see Supplemental Experimental Procedures), thereby enabling phase-plane analysis of the ON fraction as a function of kon and ktransact (Fig. S2). This phase-plane sensitivity analysis demonstrates that—throughout the physiological parameter regime of ktransact > kon (Dar et al., 2012; Molle et al., 2007)—even if an infected cell lives far longer than the in vivo lifetime of 40 h (Perelson et al., 1996) kon modulation cannot substantially alter the ON fraction. To be completely sure that these results were not a peculiarity of the specific model used, we also examined an alternate positive-feedback model topology (Weinberger et al., 2005)—which encodes substantially more molecular detail but is experimentally validated—and we observed similar circuit robustness to decreases in kon (Fig. S2). Analytical solution shows that this robustness results from the strong positive feedback (ktransact > kon), since changes in kon produce small corrections. Notably, despite the circuit’s robustness to cellular relaxation (kon decreases), high values of kon do generate less frequent latency in both the simplified model (Fig. 3C) and the complex models (Weinberger et al., 2005). In fact, the analytical solution quantifies how increases in kon (e.g. via NFκB stimulation) reactivate the circuit from a latent state (Eq. [12] Supplemental Experimental Procedures).
Overall, the results demonstrate robustness of LTR-Tat circuit activity to cellular relaxation (i.e. reductions in kon), consistent with primary-cell observations (Fig. 2), but, critically, also show sensitivity of latency to changes in Tat abundance or changes in Tat half-life. Below, we experimentally test these computational predictions: (i) that LTR-Tat circuit activity between latent and active can be toggled by Tat levels alone (i.e. independent of cellular-activation state), (ii) that Tat is more effective at activation from latency than cell-state modifiers, and, (iii) that cellular relaxation to resting does not silence Tat positive-feedback circuitry.
A minimal synthetic circuit shows that viral reactivation from latency can be toggled independent of cellular activation
To test if HIV gene-regulatory circuitry can control proviral latency without changes in cellular-activation state, we developed synthetic circuits where viral expression could be toggled independent of cell state. The synthetic circuits are based upon a minimal model of the HIV latency circuit and encode a transcriptional positive-feedback loop where HIV Tat amplifies expression from the HIV LTR promoter (Jordan et al., 2001; Weinberger et al., 2005). The minimal LTR-Tat circuit is sufficient to recapitulate latent gene expression; stimulation with cell-state modifiers reactivates proviral expression from a non-expressive ‘OFF’ state to a high-level ‘ON’ state.
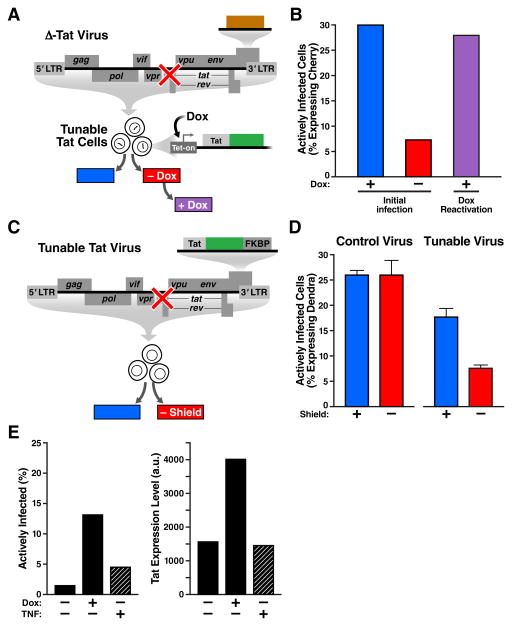
The minimalist synthetic toggle circuit encodes Tat fused to a controllable-proteolysis tag, FKBP (Banaszynski et al., 2006), under the control of the HIV LTR (Fig. 4A). FKBP degradation is reversibly inhibited by a small molecule, Shield-1, allowing Tat half-life to be rapidly tuned. The Tat-FKBP fusion was also tagged with a photo-switchable fluorescent protein, Dendra-2 (Gurskaya et al., 2006), which allows for light-based pulse-chase experiments (Zhang et al., 2007) to measure Tat half-life destabilization in single cells (Fig. S3). In this minimal LTR-Tat-Dendra-FKBP viral vector, Tat half-life is reduced to 2.5 hours in the absence of Shield-1 (a ~3.3-fold reduction from its native half-life) but returns to its native 8-hour half-life (Weinberger and Shenk, 2007) in the presence of 1 μM Shield-1.
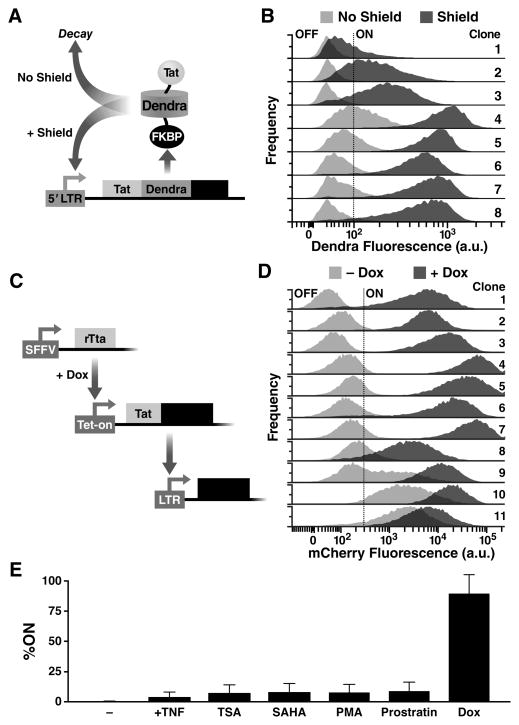
Fig. 4. Synthetic tuning of Tat circuit activity is sufficient to control latent HIV expression in the absence of cellular activation.
(A) Schematic of the minimal LTR-Tat-Dendra-FKBP lentiviral circuit. In the absence of Shield-1, the Tat-Dendra-FKBP fusion protein is rapidly degraded, diminishing positive feedback. When Shield-1 is added, FKBP-mediated proteolysis is blocked, allowing Tat levels to increase and enabling strong Tat positive feedback. (B) Flow cytometry histograms of eight isoclonal populations of Jurkat cells infected with LTR-Tat-Dendra-FKBP in the absence of Shield-1 (light gray histograms) or the presence of 1 μM Shield-1 (dark gray histograms). Gating of the Dendra-positive region (right of black-dashed line) was set relative to naïve, un-transduced Jurkat cells. See also fig. S3 and fig. S4. (C) Schematic of the synthetic system (left) and flow cytometry data of the LTR expression in cells transduced with the synthetic circuit (right). The synthetic circuit is composed of an rTta activator constitutively expressed from an SFFV promoter. In the presence of Dox, rTta protein activates the Tet-On promoter to drive expression of the Tat-Dendra fusion protein. Tat transactivates expression from the HIV-1 LTR promoter and LTR activity is measured by mCherry expression. (D) LTR mCherry expression is shown for 11 representative isoclonal populations in the absence of Dox (light gray histograms) or after Dox addition (dark gray histograms). (E) Flow cytometry analysis of a library containing 33 distinct LTR clonal integration sites subjected to Dox and a panel of standard cell-state modifiers: TNFα, phorbol myristate acetate (PMA), PMA-ionomycin, suberanilohydroxamic acid (SAHA/vorinostat), trichostatin A (TSA), or prostratin. Error bars show standard error.
Simulations predict that changes in Tat half-life should be sufficient to toggle HIV positive feedback between ‘ON’ and ‘OFF’ at a majority of viral integration sites (Fig. S2). As predicted, altering the Tat half-life by addition or removal of Shield-1 was sufficient to toggle between latent and active expression across an array of integration sites (Fig. 4B). The observed reactivation is not due to pleiotropic effects of Shield-1 since Tat-Dendra fusion proteins lacking FKBP are insensitive to Shield-1 (Fig. S3). Moreover, the increased expression levels cannot simply be due to an increase in the half-life of the reporter (Dendra-2) as the expression increases are substantially greater than the 3.3-fold increase in half-life caused by Shield-1 (Fig. S3). To be completely sure that reporter half-life changes were not accounting for the increased expression, we also decoupled the fluorescent reporter half-life from the Tat half-life by creating a polycistronic system where the reporter protein and Tat are transcriptionally fused but not translationally fused (Fig. S4). The polycistronic system corroborates the finding that Tat positive feedback is sufficient to control viral switching from an inexpressive ‘OFF’ to expressive ‘ON’ state (Fig. S4). Thus, in both the translational and transcriptional fusions, Shield-1 toggles the circuit between ON and OFF. These data indicate that tuning Tat positive feedback is sufficient to toggle HIV gene expression between a quiescent state and an actively expressing state, and that viral expression can be activated without activating cell state.
Tat induction alone is more efficient than cell-state activation for reactivating latent clones
One caveat of using tunable-proteolysis systems to toggle the Tat circuit is that a minimal level of Tat protein must be present in the off state—i.e. modulating protein half-life when protein concentration is zero has no effect. Thus, the Tat-FKBP approach is unable to test if Tat can reactivate latent cells that are fully silenced. To circumvent this obstacle and test if Tat induction is sufficient to reactivate completely silenced LTR’s, we developed a set of open-loop circuits, based on the Tet-On system (Gossen and Bujard, 1992), that induce Tat expression de novo. These systems allow tight induction of Tat expression upon Doxycycline (Dox) addition. To examine the effects of Tat induction on HIV gene-expression, these circuits were incorporated into cells that encoded an HIV LTR promoter driving the mCherry fluorescent reporter (Fig. 4C) and a library containing 33 distinct LTR clonal integration sites was examined.
The Tet-On circuits show that Tat by itself is sufficient to toggle cells between OFF and ON and to control the mean levels of LTR expression despite the large clonal variation (Fig. 4D). Importantly, a number of clones (‘Clones 1-3’) exhibit no detectable LTR expression in the absence of Tat induction—the conventional threshold for latency. But, inducing Tat expression is sufficient to fully reactivate these clones without the need for any cell-state activation signals.
Next, to test the effects of cell-state activation, Tet-inducible isoclonal populations were exposed to an array of standard cell-state modifiers. These agents are potent activators of T lymphocytes (Pazin et al., 1996) and also of the LTR (Jordan et al., 2001; Karn, 2011). For example, TNFα strongly activates T-cell state by stimulating nuclear localization of the nuclear factor of activated T-cells (NFAT) and by stimulating recruitment of the p50-RelA heterodimer to promoters containing NF-κB binding sites (Karin and Lin, 2002). If cell-state activation were the dominant factor controlling latency, then cell-state modulators should strongly reactivate latent mCherry expression in the Tet-inducible system. Strikingly, cell-state activation alone only slightly increases LTR expression, and the percentage of cells in the ON state, across the library of 33 distinct integration sites (Fig. 4E). In contrast, induction of Tat (by Dox) drastically increases the percentage of cells in the ‘ON’ state to near 100% (Fig. 4E). This dramatic difference between direct Tat induction versus cell-state modifiers demonstrates that ktransact > kon for the HIV circuitry and indicates that Tat-mediated transactivation is far stronger an effect than the switching of the LTR to an ‘ON’ state through cell-state modifications. Collectively, these data (Fig. 4E) indicate that activating cell-state alone is not sufficient to control HIV transcription. These results in no way exclude a role for cellular state in HIV reactivation in vivo. Rather, the sufficiency of Tat-mediated viral reactivation without cell-state modification emphasizes the autonomy of the HIV Tat circuit.
Refactoring of full-length replicating HIV indicates that latency establishment and reactivation depend on viral-circuit activity, and are largely independent of cellular activation
We next tested if viral circuitry could control latency in full-length replicating virus. First, we developed a decoupled system where Tat expression is controlled by the cells (via Tet-On) completely independently of the virus. The engineered cells, termed “Inducible-Tat cells”, contain a stable integrated Tet-inducible Tat-Dendra cassette and provide in trans complementation of Tat for a reengineered Tat-deleted full-length virus, the ΔTat-Cherry virus. The ΔTat-Cherry virus was constructed from a full-length HIV molecular clone containing a Tat deletion (Huang et al., 1994) and encodes an mCherry fluorescent reporter within nef (Fig. 5A). In these Inducible-Tat cells, viral gene expression can be toggled on even if initial Tat levels are zero and virus replicates only in the presence of Dox and, as with conventional strains, virus is inhibited by HIV protease inhibitors (Fig. S5). Inducing Tat expression in these cells during infection with ΔTat-Cherry virus shows a ~400% increase in active infection compared to non-induced ΔTat Cherry-infected cells (Fig. 5B), indicating that absence of Dox drives the virus to enter latency in agreement with findings that Tat protein can inhibit establishment of latency (Donahue et al., 2012). Strikingly, subsequent induction of Tat expression by Dox, fully reactivates latent virus to levels observed in the initial infection with Dox (Fig. 5B). Further, Dox was far more effective in reactivating latent virus than any of the standard cell-state modifiers: TNFα, PMA, PMA-ionomycin, SAHA/vorinostat, TSA, or prostratin (Fig. S5). Hence, latent provirus can be reactivated by Tat induction alone, without altering cellular-activation state, demonstrating that Tat is sufficient to control latent reactivation in full-length HIV.
Fig. 5. Tat feedback circuitry is sufficient to control active-versus-latent infection in full-length viruses.
(A) Schematic of experiment: A Jurkat cell-line where Tat-Dendra is expressed only in the presence of Dox, “Inducible-Tat Cells”, was infected with full-length ΔTat-Cherry virus in the presence (+) or absence (−) of Dox to score for latency and to score reactivation. Dox- infections were subsequently induced by Dox. (B) Percent of cells actively infected (actively expressing mCherry) two days post infection. 30% of cells were actively infected in the presence of Dox (blue), while only 7% of cells were actively infected in the absence of Dox (red). Upon subsequent Dox incubation of the Dox- infection, 28% of cells reactivated to active infection (purple), indicating virtually all latent cells can be reactivated with Tat induction. (C) Experiment schematic: CEM T cells were infected with either full-length Tat-FKBP Virus or Control Virus in the presence or absence of Shield-1. (D) Percent of cells actively infected (actively expressing Dendra) two days post infection. For the Control Virus infection, 25.8 ± 1.0% of cells exhibit active infection in the presence of 1 μM Shield-1 (blue), while 26.0 ± 2.7% exhibit active infection in the absence of Shield-1 (red). For the Tat-FKBP Virus infection, 17.5 ± 1.7% of cells exhibit active infection in the presence of 1 μM Shield-1 (blue), while 7.5 ± 1.0% of cells exhibit active infection in the absence of Shield-1 (red). Infections were performed in triplicate. Error bars = ±1 standard deviation. Control Virus infection and Tat-FKBP Virus infection are independent experiments (infection titers of the two are different). (E) Comparison of viral circuit versus cell-state activation by quantifying the percentage of delta-Tat virus infections that enter the active state. In the absence of TNFα or Dox, 2% of cells generate active HIV replication. Dox addition increases active infections to ~13%, while TNFα generates 4% actively infected cells. The same can be seen by plotting Tat expression level (Dendra). Again, TNFα by itself leaves expression level unchanged over that in absence of treatment. Addition of Dox leads to > 2-fold increase in expression. Also see Fig. S5 for the experiment repeated with Dox and a panel of cell-state modifiers.
Next, to check if Tat induction in cis (i.e. within the positive-feedback loop) could also control latency in full-length virus, we reengineered the ΔTat-Cherry virus to encode either the Tat-Dendra-FKBP cassette, referred to as “Tat-FKBP Virus” (Fig. 5C), or a control Tat-Dendra cassette, referred to as “Tat-Dendra Control Virus” or simply “Control Virus” (Fig. S5). As previously established in these nef-reporter viruses, actively replicating infections express reporter, while latent infections are quantified by absence of reporter expression (Jordan et al., 2003; Pearson et al., 2008). In Control-HIV infections, Shield-1 has no measureable affect on active-vs-latent infection (Fig. 5D). In striking contrast, in Tat-FKBP virus infections, modulating Tat positive-feedback strength with Shield-1 alters the percentage of actively infected cells by 141%, i.e. >2 fold (Fig. 5D). The reduction in actively infected cells is not due to reduced input virus since equivalent titers of virus (i.e. MOIs) were used in the presence and absence of Shield-1 and the lack of measureable difference in infection in Control-HIV infections indicates that Shield-1 is not inducing abortive infections and that hypothetical pleiotropic effects of Shield-1 cannot explain the difference in active-versus-latent infection. Overall, these results show that modulating viral feedback strength is sufficient to control the establishment of active-versus-latent infection in full-length replicating virus.
Tat induction is >300% more effective than cellular activation for reactivating full-length latent HIV
To directly compare the effects of tuning viral circuitry to altering cellular-activation state, Inducible Tet-Tat-Dendra cells were infected with ΔTat virus in the presence of Dox or TNFα (Fig. 5E). Modifying cellular activity with TNFα, in the absence of Tat induction, leads to a 1.5-fold change in the percentage of active infections (from 2% to 4% active infection) whereas Tat induction drastically increases, by >300%, the proportion of infections that are active (Fig. 5E). Similar results were seen in reactivating latent cells post infection (Fig. S5): inducible Tet-Tat-Dendra cells were infected with ΔTat virus and 3 days post infection were treated with either Dox or standard cell-state modulators (as well as combination of the two). Tat induction through Dox was significantly more effective at reactivation than the cell-state modifiers. Thus, as seen with the minimal-synthetic circuits (Fig. 4), perturbing viral circuitry provides substantially more potent reactivation of latency than targeting cell-state alone.
Tat circuitry is sufficient to autonomously regulate viral expression during the activated-to-resting transition in human primary T lymphocytes
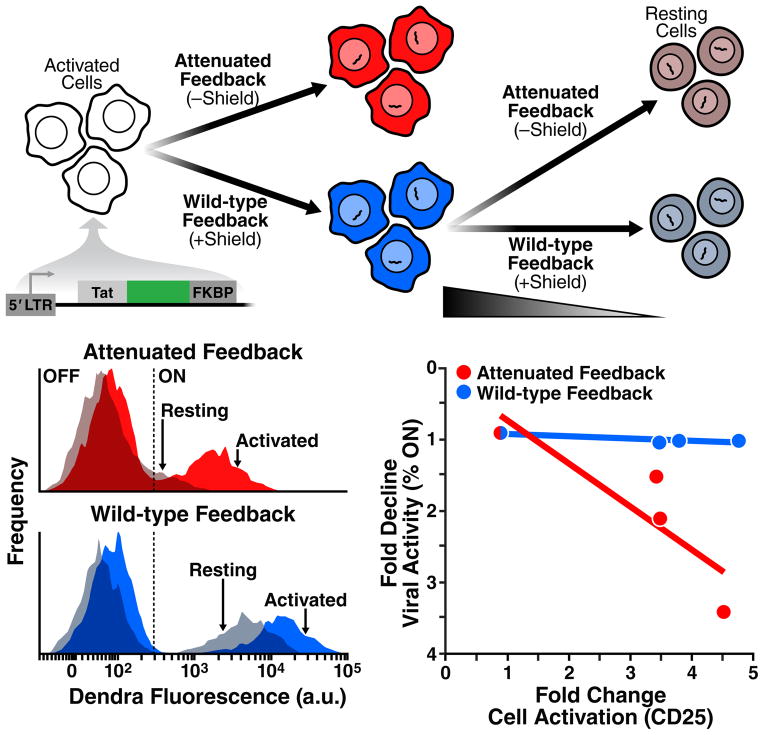
As a final test, we directly examined the model prediction that Tat circuitry alone is sufficient to explain the resilience of HIV transcription to cellular silencing during cellular relaxation from activated to resting (Fig. 3D). Activated primary CD4+ T cells were transduced with LTR-Tat-Dendra-FKBP virus and allowed to relax from an active to a resting-memory state while Tat positive-feedback strength was either maintained or attenuated by removing Shield-1 (Fig. 6A).
Fig. 6. Tat feedback circuitry is sufficient to autonomously regulate viral expression during the activated-to-resting transition in primary T cells.
(A) Experiment schematic: donor-derived primary CD4+ T lymphocytes were activated and infected with LTR-Tat-Dendra-FKBP in either the presence of Shield-1 (blue; wild-type feedback) or without Shield, (red; attenuated feedback) and cells were allowed to relax back to resting (as measured by CD25 surface expression) in presence/absence of Shield-1 (i.e. under wild-type/attenuated feedback). (B) Flow cytometry analysis of viral expression (Dendra fluorescence) in primary CD4+ T lymphocytes during transition from activated to resting in absence of Shield-1 (Attenuated feedback; upper panel) or presence of Shield-1 (Wild-type feedback; lower panel); activated lymphocytes shown as opaque histograms, resting lymphocytes shown as translucent histograms. (C) Plot of the fold change in the number of active infections for varying cellular state (fold change cell activation as measured by CD25 surface expression, see also Fig. S6.). If feedback strength is wild type (blue data points; blue trend line), the fold change in viral activity is uncorrelated with changing cell state. In the presence of attenuated feedback, the percentage of active infections is dependent on cell-state. Each data point is normalized against the percent of active infections in the lowest cell-state activation data point.
When Tat positive feedback is attenuated (by absence of Shield-1) as lymphocytes relax from activated to memory, significant silencing of HIV gene expression occurs (Fig. 6B, red histograms). However, when Tat positive-feedback strength is maintained at wild-type levels (via Shield-1 addition), only a slight shift in HIV gene expression occurs as lymphocytes transition from active to memory (Fig. 6B, blue histograms). Quantifying the relaxation of cellular-activation alongside viral latency reveals a remarkable relationship: if Tat feedback is attenuated, the cellular-activation state tightly controls entry to latency by significantly reducing the percentage of cells in active infection (Fig. 6C, red); however, when Tat feedback is active (the case in Fig. 2), the cellular activation state has no bearing on entrance into latency as the percentage of cells in active infection remains constant (Fig. 6C, blue)—i.e. the intact feedback circuit allows viral gene expression to act completely independent of cellular-activation state. Thus, active Tat feedback appears to buffer HIV from global transcriptional silencing as primary lymphocytes transition from active to resting memory.
DISCUSSION
Beginning with observations that HIV gene expression is largely autonomous to cellular relaxation (Fig. 2), computationally guided synthetic reconstruction revealed Tat positive feedback as the core mechanism underlying viral autonomy (Figs. 3–5). Strikingly, Tat feedback alone is sufficient to overcome cell-driven silencing of HIV transcription during cellular relaxation from active to resting in primary T cells (Fig. 6). These findings are consistent with patient-cell latent-reactivation experiments showing that direct addition of Tat activates viral expression and reverses latency in resting CD4+ T cells without requiring cellular activation (Lassen et al., 2006; Lin et al., 2003). Thus, in patient cells, Tat-mediated positive feedback also appears to regulate latency independent of cell state.
The data herein cannot discount one variant of the cell-state hypothesis which proposes that latency is established when HIV infects relaxing cells which are at an activation level just above a first threshold required for HIV infection and integration, but below a second threshold required to sustain active Tat expression and viral replication. Nevertheless, while the presence of two thresholds is plausible, the second (Tat activation) threshold being higher than the first (infection) threshold is not consistent with existing data. For example, although global activation of primary CD4+ T cells is required for efficient infection, HIV can be reactivated from latency in primary cells without global activating the cells (Xing et al., 2012). Similarly, the reactivation of HIV in resting T cells using Tat protein (Lassen et al., 2006; Lin et al., 2003), indicates that extremely low levels of cellular activation (i.e. in resting/quiescent cells) are still amenable to robust viral expression. Thus, since resting cells are at an activation level non-permissive to infection (Pan et al., 2013), but are sufficiently activated for Tat to function, the putative Tat-activation threshold is lower than the infection threshold and the two-threshold scenario appears unlikely.
If cellular relaxation does not lead to the establishment of HIV latency, how is HIV latency established? Previous studies demonstrated the intrinsic ability of the Tat positive-feedback circuit to rapidly and stochastically establish latency (Weinberger et al., 2005), consistent with recent primate studies showing that latency is rapidly established within the first three days of infection (Whitney et al., 2014), and with cell-culture models showing latency establishment immediately upon infection (Calvanese et al., 2013; Dahabieh et al., 2013). Given that resting CD4+ T lymphocytes are highly resistant to direct HIV infection (Pan et al., 2013), the rapid establishment of latency is difficult to reconcile with the cell-state epiphenomenon theory; productively infected cells live < 2 day in vivo (Perelson et al., 1997) while the process of T-cell transitioning from active to memory is a slow and low-probability process (Youngblood et al., 2013) occurring during and after vigorous expansion of effector lymphocytes that only begins weeks after infection (Kuroda et al., 1999). The alternate model examined here (Fig. 3), where intrinsic (stochastic) viral circuitry autonomously regulates HIV latency, also provides a mechanistic basis for recent observations in patient cells (Ho et al., 2013) showing that: (i) a significant fraction of latent proviruses are not induced even if cells are reactivated from a resting-memory state; and (ii) a second identical cellular stimulation (of already activated cells) induces additional latent proviruses to reactivate. These results indicate that viral reactivation is probabilistic. While particularly puzzling for the cellular-control hypothesis, probabilistic reactivation is consistent with HIV latency being regulated by an autonomous viral-encoded circuit influenced by stochastic gene-expression fluctuations, which provides rationale for targeting viral gene-expression circuitry to reactivate latent HIV (Dar et al., 2014).
To be completely clear, the viral-encoded latency model does not exclude a role for cellular state in regulating HIV proviral latency. In fact, the Tat-feedback model predicts that latency establishment is sharply reduced at higher cellular activation levels (Fig. 3C) and that cellular activation probabilistically reactivates latent virus (Eq. 12 in Supplemental Information). Experimentally, cellular activation clearly rescues attenuated feedback (Fig. 6B). Similarly, the ability of Tat expression to reactivate latent virus independent of cellular activation (Figs. 4,5) does not imply that in vivo latent reactivation occurs absent cellular activation. Rather, the results herein demonstrate—contrary to prevailing dogma—that there is also an underlying viral program that autonomously regulates proviral latency.
A viral-encoded latency program naturally raises questions on the evolutionary origin and function of HIV latency. While sensor-actuator circuitry would have been consistent with either the epiphenomenon hypothesis or evolutionary hardwiring, an autonomous regulatory circuit is invariably hardwired and must be selectively maintained—especially in a rapidly evolving virus under strong selection. So, how would latency be beneficial in the natural history of lentiviral infection? In a companion paper (Rouzine et al., 2015), we propose that latency may provide a fitness advantage by acting as a viral ‘bet-hedging’ strategy to enhance net viral transmission probability. An associated aspect is the decision-making architecture behind latency: Tat positive feedback maintains strong expression levels robust to cellular perturbations, while large stochastic fluctuations exhibited by the LTR promoter enable the system to probabilistically switch (Dar et al., 2012). Notably, this architecture has been theoretically proposed to be an unreliable environmental sensor in fluctuating environments (Brandman et al., 2005), suggesting that HIV’s circuit architecture is precisely the opposite configuration that would be required for a reliable environmental sensor – a reliable sensor would respond faithfully to environmental changes – and similar stochastic positive-feedback circuitry has been proposed for autonomous decision-making in other biological systems (Jilkine et al., 2011). Overall, viral evolution appears to have selected for circuitry that both maintains remarkable autonomy from environmental cues and simultaneously drives probabilistic on-off decision-making.
EXPERIMENTAL PROCEDURES
Primary-cell isolation and cell-culture conditions
Primary CD4+ T cells were isolated from peripheral blood obtained from Stanford Blood Bank (Palo Alto, CA) using RosetteSep™ Human CD4+ T Cell Enrichment Cocktail from STEMCELL™ Technologies and Ficoll as described (Terry et al., 2009). Once isolated, cells were either cultured as described (Terry et al., 2009) or frozen in 10% DMSO, 90% culture media at a density of 107 per mL. For infections, primary CD4+ T cells were pre-activated for 2–3 days with αCD3/CD28 beads (Dynabeads™, Life Technologies) as per manufacturer instructions. Cell activation was measured by flow cytometry with anti-CD25-PE-conjugated antibody and anti-CD69-APC-conjugated antibody from BD Biosciences™. Primary CD4+ T lymphocytes, Jurkat T Lymphocytes, and CEMs were all cultured in RPMI 1640 (supplemented with L-glutamine, 10% fetal bovine serum, and 1% penicillin-streptomycin) in a humidified environment at 37°C and 5% CO2. Jurkats and CEM were maintained by passage between 2×105 and 2×106 cells/mL. Primary cell media was supplemented with 20 U/ml r-IL2 (Peprotech™, 200-02).
Computational Modeling
A simplified two-state model of Tat positive feedback was constructed from experimental data of LTR toggling (Dar et al., 2014; Dar et al., 2012; Singh et al., 2010; Singh et al., 2012) and simulated using the Gillespie algorithm (Gillespie, 1977) to test how altering LTR basal transcription rate or Tat protein stability would affect the activity of the circuit. The chemical reaction scheme and parameters used are described in Table S1. The outputs from simulations are the different molecular species in arbitrary numbers. Stochastic simulations were run in Mathematica™ using the xSSA package (http://www.wolfram.com/mathematica/ and http://www.xlr8r.info/SSA/). Initial conditions for all species were set to 0, except for LTRON, which was set to 1, and simulations were run to time=200 (arbitrary time units); 500 simulation runs were conducted for each parameter set. See Extended Experimental Procedures for further details and explanation of simulations for the more complex model (Fig. S2).
Recombinant virus production and infections
Lentivirus was packaged in 293T cells and isolated as described (Dull et al., 1998; Weinberger et al., 2005). HIV-d2GFP (Jordan et al., 2003) was packaged with dual-tropic env-encoding plasmid pSVIII-92HT593.1 (NIH AIDS Reagents Program). Before infecting, primary cells, activation beads were removed and cells were mixed with appropriate amount of virus (to get <10% infection) in 100μl media and spinoculated at 32°C for 2 hours at 1000 × g.
To generate the isoclonal populations with engineered viral circuits, lentivirus was added to Jurkat T Lymphocytes at a low MOI to ensure a single integrated copy of proviral DNA in infected cells. Cells were stimulated with tumor necrosis factor alpha (TNFα) and Shield-1 for 18 hours before sorting for Dendra expressing cells. Isoclonal and polyclonal populations were created as described (Weinberger et al., 2005). Sorting and analysis of cells infected was performed on a FACSAria II™. The same procedure was followed to create the LTR-Tat-Dendra and LTR-mCherry-IRES-Tat-FKBP cell lines. Inducible-Tat cells were generated by transducing Jurkat cells with Tet-Tat-Dendra and SFFV-rTta lentivirus at high MOI. The cells were incubated in Dox for 24 h and then FACS sorted for Dendra+ cells to create a polyclonal population. To create the Tet-Tat-Dendra + LTR-mCherry cells, the polyclonal population was infected with LTR-mCherry lentivirus at a low MOI. Before sorting for mCherry+ and Dendra+ cells, Dox was added at 500ng/mL for 24 h, and single cells were FACS sorted and expanded to isolate isoclonal populations. The same procedure was followed for the Tet-Tat-Dendra-FKBP + LTR-mCherry populations, however, 24hrs before the sort 1uM Shield-1 and 500ng/mL Dox was added to the culture. All Inducible-Tat or Control-HIV infection experiments were performed by incubating 5 × 105 CEM cells in the same titer of Inducible-Tat or the same titer of Control-HIV in the presence or absence of Shield-1 and taking a flow cytometry time point after 48 h. Δ-Tat mCherry infections were carried out using 105 – 106 Inducible-Tat (Jurkat) cells in the presence or absence of 500ng/mL doxycycline.
Flow Cytometry and analysis
Flow cytometry data was collected on a BD FACSCalibur DxP8, BD LSR II, or HTFC Intellicyt™ for stably transduced lines and primary cells, and on a BD FACSAria II for replication competent virus assays and sorting. All flow cytometry experiments on replication competent virus were performed in BSL3 conditions (safety information available upon request). Flow cytometry data was analyzed in FlowJo™ (Treestar, Ashland, Oregon) and using customized MATLAB® code.
Supplementary Material
Acknowledgments
We are grateful to M. Thompson, O. Weiner, J. Toettcher, J. Miranda, A. Weinberger, M. Ott, E. Verdin, A. Frankel, M. Simpson, and G. Süel, for comments, helpful discussions, and providing reagents. BSR was supported by an NSF graduate research fellowship (GRFP, grant 1144247). This work was supported by NIH award R01-AI109593 and in part by the Center for Synthetic and Systems Biology at UCSF (P50GM081879), the NIH Delaney CARE Collaboratory of AIDS Researchers for a Cure (U19AI096113), and UCSF-GIVI CFAR (P30AI027763 & S10RR028962-01). LSW acknowledges support from NIH Director’s New Innovator Award Program (DP2-OD006677), the Alfred P. Sloan Foundation, and the Pew Scholar’s in the Biomedical Sciences.
Abbreviations
- HIV-1
human immunodeficiency virus type 1
- LTR
long terminal repeat
- FKBP
FK506-binding protein
- IRES
Internal Ribosome Entry Sequence
- TNFα
tumor necrosis factor α
- FACS
Fluorescence Assisted Cell Sorting
Footnotes
AUTHOR CONTRIBUTIONS
B.S.K., A.P., L.S.W., designed research and wrote the paper. B.S.K., A.P., K.A., executed the experiments and analyzed the data. I.R., L.S.W. performed mathematical analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Vogt RC. Temperature-dependent sex determination in turtles. Science. 1979;206:1186–1188. doi: 10.1126/science.505003. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Miller-Jensen K, Shah PS, Arkin AP, Schaffer DV. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog. 2009;5:e1000260. doi: 10.1371/journal.ppat.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V, Chavez L, Laurent T, Ding S, Verdin E. Dual-color HIV reporters trace a population of latently infected cells and enable their purification. Virology. 2013;446:283–292. doi: 10.1016/j.virol.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J, Swanstrom R. HIV pathogenesis: dynamics and genetics of viral populations and infected cells. Cold Spring Harb Perspect Med. 2013;3:a012526. doi: 10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. Optimizing reproduction in a randomly varying environment. J Theor Biol. 1966;12:119–129. doi: 10.1016/0022-5193(66)90188-3. [DOI] [PubMed] [Google Scholar]
- Dahabieh MS, Ooms M, Simon V, Sadowski I. A doubly fluorescent HIV-1 reporter shows that the majority of integrated HIV-1 is latent shortly after infection. J Virol. 2013;87:4716–4727. doi: 10.1128/JVI.03478-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar RD, Hosmane NN, Arkin MR, Siliciano RF, Weinberger LS. Screening for noise in gene expression identifies drug synergies. Science. 2014;344:1392–1396. doi: 10.1126/science.1250220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17454–17459. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue DA, Kuhl BD, Sloan RD, Wainberg MA. The viral protein Tat can inhibit the establishment of HIV-1 latency. J Virol. 2012;86:3253–3263. doi: 10.1128/JVI.06648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DT. Exact Stochastic Simulation of Coupled Chemical-Reactions. J Phys Chem-Us. 1977;81:2340–2361. [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-tored photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LM, Joshi A, Willey R, Orenstein J, Jeang KT. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. Embo J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, Westerhout EM, van Gerven ML, Berkhout B. HIV-1 latency in actively dividing human T cell lines. Retrovirology. 2008;5:37. doi: 10.1186/1742-4690-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilkine A, Angenent SB, Wu LF, Altschuler SJ. A density-dependent switch drives stochastic clustering and polarization of signaling molecules. PLoS Comput Biol. 2011;7:e1002271. doi: 10.1371/journal.pcbi.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. Embo J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- Karn J. The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit. Curr Opin HIV AIDS. 2011;6:4–11. doi: 10.1097/COH.0b013e328340ffbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, Elston TC. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys J. 2001;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knedler JW. Masterworks of science; digests of 13 great classics. Garden City, N.Y: Doubleday & Company, inc; 1947. [Google Scholar]
- Kuroda MJ, Schmitz JE, Charini WA, Nickerson CE, Lifton MA, Lord CI, Forman MA, Letvin NL. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Irwin D, Kanazawa S, Huang L, Romeo J, Yen TS, Peterlin BM. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: effects of Tat on the host and reservoir. J Virol. 2003;77:8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, Marcello A, Basyuk E. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology. 2007;4:36. doi: 10.1186/1742-4690-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Baldauf HM, Keppler OT, Fackler OT. Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 2013;23:876–885. doi: 10.1038/cr.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, Jones KA. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. Journal of virology. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Razooky BS, Weinberger LS. Mapping the architecture of the HIV-1 Tat circuit: A decision-making circuit that lacks bistability and exploits stochastic noise. Methods. 2011;53:68–77. doi: 10.1016/j.ymeth.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Rouzine I, Weinberger A, Weinberger LS. An evolutionary role for HIV latency in enhancing viral transmission. 2015 doi: 10.1016/j.cell.2015.02.017. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. HIV Latency. Cold Spring Harb Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Razooky B, Cox CD, Simpson ML, Weinberger LS. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys J. 2010;98:L32–34. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Razooky BS, Dar RD, Weinberger LS. Dynamics of protein noise can distinguish between alternate sources of gene-expression variability. Mol Syst Biol. 2012;8:607. doi: 10.1038/msb.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Endy D. Determination of cell fate selection during phage lambda infection. Proc Natl Acad Sci U S A. 2008;105:20705–20710. doi: 10.1073/pnas.0808831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry VH, Johnston IC, Spina CA. CD44 microbeads accelerate HIV-1 infection in T cells. Virology. 2009;388:294–304. doi: 10.1016/j.virol.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M, Pearson RJ, Karn J. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. Journal of virology. 2010;84:6425–6437. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AD, Weinberger LS. Stochastic fate selection in HIV-infected patients. Cell. 2013;155:497–499. doi: 10.1016/j.cell.2013.09.039. [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Shenk T. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol. 2007;5:e9. doi: 10.1371/journal.pbio.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JB, Hill A, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014 doi: 10.1038/nature13594. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Bhat S, Shroff NS, Zhang H, Lopez JA, Margolick JB, Liu JO, Siliciano RF. Novel structurally related compounds reactivate latent HIV-1 in a bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Antimicrob Chemother. 2012;67:398–403. doi: 10.1093/jac/dkr496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B, Hale JS, Ahmed R. T-cell memory differentiation: insights from transcriptional signatures and epigenetics. Immunology. 2013;139:277–284. doi: 10.1111/imm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Skinner SO, Zong C, Sippy J, Feiss M, Golding I. Decision making at a subcellular level determines the outcome of bacteriophage infection. Cell. 2010;141:682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gurskaya NG, Merzlyak EM, Staroverov DB, Mudrik NN, Samarkina ON, Vinokurov LM, Lukyanov S, Lukyanov KA. Method for real-time monitoring of protein degradation at the single cell level. Biotechniques. 2007;42:446, 448, 450. doi: 10.2144/000112453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.