Abstract
Background: In colorectal cancer, there are significant differences between synchronous and metachronous distant metastases. However in recent studies, synchronous and metachronous metastases were always lumped together, neglecting their clinical and molecular differences. The mechanism of the latency of metachronous metastases is still unclear. We conducted this study to reveal the relationship between EGFR pathways and metachronous metastases, and try to find efficient predictors. Methods: PCRs and pyrosequencing were used to detect KRAS, BRAF, PIK3CA and PTEN mutations in primary tumor tissues in a total of 281 patients from 2002 to 2008. Patients were identified into three groups: no-metastases group, synchronous-metastases group and metachronous-metastases group. Clinical and survival data were collected from a prospective database. Results: KRAS codon 13 mutation was an independent predictor only for metachronous distant metastases (OR = 11.857, P < 0.001), but not for synchronous metastases. Male gender (OR = 2.233, P = 0.024), primary tumor located at rectum (OR = 0.404, P = 0.041), and primary pN2 stage (OR = 3.361, P = 0.01) were also independent predictors for metachronous distant metastases. Different SNPs in KRAS worked significantly different in determining synchronous or metachronous metastases. BRAF mutation (Univariate, OR = 11.5, P = 0.039) and > 200 ng/ml preoperative CEA (Univariate, OR = 41, P = 0.011) potentially predicted metastases within 6 months after primary tumor resection. After metachronous metastases, radical resection (HR = 0.280, P = 0.002) was the most important protective factor for long-term survival. Conclusion: There were significant clinical and molecular differences between synchronous and metachronous metastases. As an independent predictor, KRAS codon 13 mutation might be the key to explain the mechanism of colorectal cancer metachronous distant metastases. Together with clinical characteristics, it could aid in the early detection of metachronous metastases.
Keywords: Colorectal cancer, metachronous metastases, predictor, KRAS mutation
Introduction
Colorectal cancer are common throughout the world [1]. And distant metastases are the major cause of death in colorectal cancer patients. About 20% patients present with synchronous distant metastases at the time of diagnosis or within 6 months after primary tumor resections. Another 30% will develop clinically detectable metastases afterwards, metachronous distant metastases [2,3]. Compared with synchronous metastases, patients with metachronous metastases have better prognosis after metastases resections [4-6]. However, metachronous metastases are more difficult to detect. Lack of clinical symptoms makes metachronous metastases easily ignored. Such a long latency before the occurrence of metastases also makes follow-up hard, demanding high cost. Therefore, it’s necessary to find efficient predictors for metachronous distant metastases.
The epidermal growth factor receptor (EGFR) has been proved crucial in determining the development of colorectal cancer. It has also become a major molecular target for anticancer therapies. As a transmembrane tyrosine kinase receptor, it triggers two main signaling pathways, the RAS-RAF-MAPK pathway and the PI3K-PTEN-AKT pathway [7]. Mutations in KRAS, BRAF, PIK3CA or PTEN genes in the two pathways result in continuous activation of the downstream signal transduction, regardless of whether the EGFRs activated. Present studies have showed KRAS to be pivotal in predicting the efficacy of anti-EGFR therapy [8,9]. BRAF, PIK3CA and PTEN are also prognosis factors for colorectal cancer [10-14]. However in these studies reported, synchronous and metachronous metastases are lumped together, neglecting their clinical and molecular differences. Thus, it’s unable to explain the key mechanism of metachronous metastases: how could metachronous metastases have such a long latency?
Therefore, we conducted this study, including patients with no metastases, with synchronous metastases, and with metachronous metastases as three groups respectively. We aimed to investigate gene mutations in EGFR pathway (KRAS, BRAF, PIK3CA and PTEN) in these three different groups and to reveal the possible mechanisms of metachronous metastases. We also expected to find some predictors for metachronous distant metastases after primary tumor resections.
Patients and methods
Study population
Patients diagnosed with colorectal cancer during January 2002 to December 2008 were randomly identified from the colorectal cancer database of the General Surgery Department of Zhongshan Hospital, Fudan University (Shanghai, China). The inclusion criteria were as follows: colorectal carcinoma determined by pathological evidence; primary tumor resections (only R0 resections permitted); no chemotherapy, radiotherapy or interventional therapy before primary tumor resections; and no targeted therapy during the course of the disease. Distant metastases were defined as metastases to organs far from the primary tumor sites (such as the liver, lungs, bones, brain, adrenal glands, or other distant sites). Abdominal or retroperitoneal lymph node metastases, peritoneal dissemination or pelvic recurrence were not included as distant metastases. Adjuvant chemotherapy after primary tumor resection was permitted. Transcatheter arterial chemoembolization (TACE) and transcatheter arterial infusion (TAI) were permitted only after the occurrence of metastases.
Three groups were established in this study: the no-metastases group, the synchronous-metastases group and the metachronous-metastases group. Primary tumor recurrences were permitted in the no-metastases group. The synchronous-metastases group was defined as a diagnosis of distant metastases together with or within a six-month interval of the diagnosis of the primary colorectal cancer. The metachronous-metastases group was defined as diagnosis of distant metastases more than six months after primary tumor resection. If patients in the metachronous-metastases group had both primary recurrences and distant metastases, the metastases must occur together or before the primary recurrence. This study was approved by the institutional review board of Zhongshan Hospital, Fudan University. And the investigators obtained informed consent from each patient.
DNA extraction and mutation detection
DNA was extracted from formalin-fixed paraffin-embedded (FFPE) primary tumor samples using the GTpure DNA FFPE Tissue Kit (Gene Tech (Shanghai) Co. Ltd., Shanghai, China). The DNA concentration and purity were tested using spectrophotometry. DNA was amplified with specific primers for exons where “hot-spot” mutations were located. The mutation status of KRAS (exon 2), BRAF (exon 15), PIK3CA (exon 9 and 20), and PTEN (exon 5, 7 and 8) were investigated by polymerase chain reaction (PCR) amplification (Primers were designed and synthesized by Gene Tech (Shanghai) Co. Ltd., Shanghai, China). A total 50 μl PCR system contained: template DNA 50 ng, forward primer (10 mM) 0.5 μl, reverse primer (10 mM) 0.5 μl, dNTP (2.5 mM) 4 μl, Hotstart Taq (2.5 U/μl, DBI Bioscience, German) 1 μl, 10 × Hotstart PCR Buffer 5 μl, MgCl2 (25 mM) 4 μl.
PCRs for KRAS, BRAF and PIK3CA were run at 95°C 5 min for initial denaturation, then 56°C 20 sec, 72°C 30 sec, 95°C 20 sec for 45 cycles, and 72°C 5 min for last elongation. Primers used:
For KRAS exon 2 (110 bp)
Forward 5’-TGTAAAACGACGGCCAGTTTATAAGGCCTGCTGAAAATGACTGAA-3’.
Reverse 5’-TGAATTAGCTGTATCGTCAAGGCACT-3’.
For BRAF exon 15 (120 bp)
Forward 5’-TGTAAAACGACGGCCAGTGAAGACCTCACAGTAAAAATAGGTGA-3’.
Reverse 5’-CCACAAAATGGATCCAGACA-3’.
For PIK3CA exon 9 (346 bp)
Forward 5’-TGTAAAACGACGGCCAGTATTATGTCTTAGATTGGTTC-3’.
Reverse 5’-AATCTCCATTTTAGCACT-3’.
For PIK3CA exon 20 (388 bp)
Forward 5’-TGTAAAACGACGGCCAGTGGAATGCCAGAACTACAA-3’.
Reverse 5’-AGTGCTATCAAACCCTGT-3’.
PCRs for PTEN was run at 95°C 5 min for initial denaturation, then 56°C 30 sec, 72°C 1 min, 95°C 30 sec for 45 cycles, and 72°C 5 min for last elongation. Primers used:
For PTEN exon 5 to 8 (638 bp)
Forward 5’-TGTAAAACGACGGCCAGTATCAAACCCTTTTGTGAAGA-3’.
Reverse 5’-TCTATACTGCAAATGCTATC-3’.
All forward primers were M13-tagged (5’-TGTAAAACGACGGCCAGT-3’) to receive a more specific PCR product during the sequencing reaction. The subsequent pyrosequencing was conducted using M13/UC forward sequencing primer (5’-TGTAAAACGACGGCCAGT-3’) in Pyro-Mark ID system (PSQ 96 MA, Biotage AB, Sweden).
Clinical data collection
This investigation was performed as a retrospective analysis. Contrast CTs/MRIs were used to clarify whether there were distant metastases before the primary tumor resections. The pathological tumor stage was documented according to the AJCC TNM classification (version 7, 2010). Follow-up principles were based on the Chinese guidelines for the diagnosis and comprehensive treatment of hepatic metastasis of colorectal cancer [15]: history, physical, carcinoembryonic antigen (CEA) and abdominal ultrasound every 3 months for 2 years, then every 6 months for 3 to 5 years, then every year after 5 years; chest/abdominal/pelvic CT scan every 6 months for 2 years, then every year after 2 years; colonoscopy 6 months after the primary tumor resection, then every year for 5 years. Once distant metastases were confirmed, the previous examinations were backtracked to identify the time at which the metastases first appeared. The data of metastases-free survival time and overall survival time were collected.
Statistical methods
For categorical parameters, correlation test and univariate analyses were conducted using two-sided Pearson’s χ2 tests or Fisher’s exact tests for samples with expected frequency < 5. For multivariate analyses of distant metastases, logistic regression was used. Odds ratios (ORs) were calculated to represent the weights of factors; an OR < 1 represents a protective factor, and an OR > 1 represents a risk factor. All summary statistics on survival data were calculated according to the Kaplan-Meier method and compared by the medians of the log-rank test. The median follow-up time was calculated using the reverse Kaplan-Meier method [16]. Cox regression was used to adjust the survival data. SPSS software (version 16.0; SPSS, Chicago, IL) was used for statistical analyses. For the correlation analyses, a P value of < 0.01 was considered to be significant. For other situations, a P value of < 0.05 was considered to be significant.
Results
Patients follow-up
A total of 281 patients were finally included in this study: 96 patients in no-metastases group, 92 patients in synchronous-metastases group, and 93 patients in metachronous-metastases group. The median follow-up time of all patients was 84 months (interquartile range, IQR = (78-89)). In no-metastases group, the median follow-up time was 86 months (IQR = (80-92)), and 80 patients (83%) had a survival time of more than 60 months. In synchronous-metastases group, the median follow-up time was 78 months (IQR = (73-89)). In metachronous-metastases group, the median follow-up time was 87 months (IQR = (71-103)). In no-metastases group, nine patients (9.4%) had local recurrences. In metachronous-metastases group, 79 (84.9%) patients had first metastases to the liver, 12 (12.9%) patients had first metastases to the lungs, and 2 (2.2%) patients had first metastases to the bones.
Mutation detection
KRAS, BRAF, PIK3CA and PTEN mutations were detected for all 281 patients. The mutation statuses are listed in Table 1. In RAS-RAF-MAPK signaling pathway, there was no overlap between the KRAS and BRAF mutations, with significant correlation (P < 0.001). In KRAS mutations, there was also no overlap between codon 12 and 13 mutations, with significant correlation (P < 0.001). It seemed that in RAS-RAF-MAPK pathway, gene mutations were mutally exclusive. However in PI3K-PTEN-AKT pathway, both PIK3CA and PTEN mutations could be detected in a same sample, seemed independent from each other, with no significant correlation (P = 0.952). The activation of two signaling pathways also seemed independent. KRAS/BRAF and PIK3CA/PTEN mutations could occur in one sample, with no significant correlation (P = 0.762).
Table 1.
Results of mutation detection
| No metastases | Synchronous metastases | Metachronous metastases | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Number | % | Number | % | Number | % | |
| Total patients (n) | 96 | - | 92 | - | 93 | - |
| All WT | 46 | 47.9 | 27 | 29.3 | 30 | 32.3 |
| Total KRAS MT | 28 | 29.2 | 43 | 46.7 | 44 | 47.3 |
| Total KRAS 12 MT | 24 | 25.0 | 36 | 39.1 | 23 | 24.7 |
| Total KRAS 13 MT | 4 | 4.2 | 7 | 7.6 | 21 | 22.6 |
| Total BRAF MT | 4 | 4.2 | 12 | 13.0 | 2 | 2.2 |
| RAS-RAF-MAPK MT | 32 | 33.3 | 55 | 59.8 | 46 | 49.5 |
| Total PIK3CA MT | 13 | 13.5 | 20 | 21.7 | 15 | 16.1 |
| Total PTEN MT | 15 | 15.6 | 14 | 15.2 | 17 | 18.3 |
| PI3K-PTEN-AKT MT | 26 | 27.1 | 33 | 35.9 | 28 | 30.1 |
| Only KRAS 12 MT | 19 | 19.8 | 20 | 21.7 | 15 | 16.1 |
| Only KRAS 13 MT | 2 | 2.1 | 4 | 4.3 | 18 | 19.4 |
| Only BRAF MT | 3 | 3.1 | 11 | 12.0 | 2 | 2.2 |
| Only PIK3CA MT | 7 | 7.3 | 5 | 5.4 | 7 | 7.5 |
| Only PTEN MT | 10 | 10.4 | 5 | 5.4 | 7 | 7.5 |
| Only KRAS + PIK3CA MT | 3 | 3.1 | 12 | 13.0 | 4 | 4.3 |
| Only KRAS 12 + PIK3CA MT | 2 | 2.1 | 8 | 8.7 | 2 | 2.2 |
| Only KRAS 13 + PIK3CA MT | 1 | 1.0 | 2 | 2.2 | 2 | 2.2 |
| Only KRAS + PTEN MT | 3 | 3.1 | 6 | 6.5 | 6 | 6.5 |
| Only KRAS 12 + PTEN MT | 2 | 2.1 | 6 | 6.5 | 6 | 6.5 |
| Only KRAS 13 + PTEN MT | 1 | 1.0 | 0 | 0.0 | 0 | 0.0 |
| KRAS + PIK3CA + PTEN MT | 1 | 1.0 | 2 | 2.2 | 1 | 1.1 |
| Only BRAF + PIK3CA MT | 1 | 1.0 | 1 | 1.1 | 0 | 0.0 |
| Only BRAF + PTEN MT | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Only PIK3CA + PTEN MT | 1 | 1.0 | 0 | 0.0 | 3 | 3.2 |
WT: wild type; MT: mutant type; Total: patients with at least one gene mutation; Only: patients without other gene mutations. RAS-RAF-MAPK MT: patients with any of KRAS or BRAF mutation. PI3K-PTEN-AKT MT: patients with any of PIK3CA or PTEN mutation.
Correlation between mutation status and clinicopathological characteristics
Our analyses identified 3 significant correlations (P < 0.01) between mutation status and clinicopathological characteristics, as follows: “primary tumor location” significantly associated with PIK3CA mutations (P = 0.009), and presence of “distant metastases” associated with KRAS (P < 0.001) and BRAF (P = 0.006) mutations. The other 9 potential correlation factors (P < 0.10) were also as follows: “age” associated with PIK3CA mutation; “primary tumor location” associated with BRAF and PTEN mutations; “primary pT stage” associated with PTEN mutations; “primary pN stage” associated with KRAS and BRAF mutations; “primary histological type” associated with BRAF and PIK3CA mutations; and “CEA before primary tumor resection” associated with KRAS mutations. Details are provided in Table 2.
Table 2.
Correlation between gene mutations and clinicopathological factors
| KRAS | BRAF | PIK3CA | PTEN | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| WT | 12 MT | 13 MT | P value | WT | MT | P value | WT | MT | P value | WT | MT | P value | |
| Total patients (n) | 166 | 83 | 32 | 263 | 18 | 233 | 48 | 235 | 46 | ||||
| Age (year) | 0.140 | 0.958 | 0.090 | 0.117 | |||||||||
| < 55 | 68 | 22 | 8 | 92 | 6 | 86 | 12 | 76 | 22 | ||||
| 55-69 | 60 | 39 | 14 | 106 | 7 | 87 | 26 | 97 | 16 | ||||
| > 69 | 38 | 22 | 10 | 65 | 5 | 60 | 10 | 62 | 8 | ||||
| Sex | 0.278 | 0.942 | 0.677 | 0.615 | |||||||||
| Male | 109 | 46 | 19 | 163 | 11 | 143 | 31 | 144 | 30 | ||||
| Female | 57 | 37 | 13 | 100 | 7 | 90 | 17 | 91 | 16 | ||||
| Primary tumor location | 0.357 | 0.016 | 0.009 | 0.054 | |||||||||
| Right-sided | 51 | 28 | 10 | 78 | 11 | 66 | 23 | 77 | 12 | ||||
| Left-sided | 50 | 16 | 11 | 73 | 4 | 63 | 14 | 69 | 8 | ||||
| Rectum | 65 | 39 | 11 | 112 | 3 | 104 | 11 | 89 | 26 | ||||
| Primary pT Stage | 0.591 | 0.384 | 0.659 | 0.073 | |||||||||
| 1-2 | 22 | 12 | 2 | 35 | 1 | 28 | 8 | 30 | 6 | ||||
| 3 | 48 | 23 | 13 | 80 | 4 | 71 | 13 | 64 | 20 | ||||
| 4 | 96 | 48 | 17 | 148 | 13 | 134 | 27 | 141 | 20 | ||||
| Primary pN Stage | 0.053 | 0.091 | 0.460 | 0.731 | |||||||||
| 0 | 61 | 39 | 9 | 101 | 8 | 88 | 21 | 93 | 16 | ||||
| 1 | 52 | 24 | 17 | 91 | 2 | 76 | 17 | 78 | 15 | ||||
| 2 | 53 | 20 | 6 | 71 | 8 | 69 | 10 | 64 | 15 | ||||
| Primary differentiation | 0.498 | 0.252 | 0.759 | 0.288 | |||||||||
| Well to moderate | 103 | 50 | 23 | 167 | 9 | 145 | 31 | 144 | 32 | ||||
| Poor | 63 | 33 | 9 | 96 | 9 | 88 | 17 | 91 | 14 | ||||
| Primary histological type | 0.376 | 0.084 | 0.078 | 0.326 | |||||||||
| Non-mucinous | 140 | 64 | 26 | 218 | 12 | 195 | 35 | 190 | 40 | ||||
| Mucinous | 26 | 19 | 6 | 45 | 6 | 38 | 13 | 45 | 6 | ||||
| Pre-primary resection CEA | 0.076 | 0.194 | 0.562 | 0.954 | |||||||||
| < 5 ng/ml | 81 | 24 | 14 | 115 | 4 | 99 | 20 | 101 | 18 | ||||
| 5-200 ng/ml | 59 | 38 | 12 | 98 | 11 | 92 | 17 | 90 | 19 | ||||
| > 200 ng/ml | 11 | 13 | 3 | 26 | 1 | 23 | 4 | 22 | 5 | ||||
| Unknown | 15 | 8 | 3 | 24 | 2 | 19 | 7 | 22 | 4 | ||||
| Distant metastases | < 0.001 | 0.006 | 0.314 | 0.829 | |||||||||
| No | 68 | 24 | 4 | 92 | 4 | 83 | 13 | 81 | 15 | ||||
| Synchronous | 49 | 36 | 7 | 80 | 12 | 72 | 20 | 78 | 14 | ||||
| Metachronous | 49 | 23 | 21 | 91 | 2 | 78 | 15 | 76 | 17 | ||||
Pearson χ2 test was used in this analysis. WT: wild type; MT: mutant type; CEA: carcinoembryonic antigen. P value of < 0.01 was considered significant.
Analyses of the factors relevant to distant metastases
Univariate and multivariate analyses were conducted to find predictors for distant metastases. The results showed that for synchronous distant metastases, male gender (OR = 2.457, P = 0.038), primary pN2 stage (OR = 4.579, P = 0.006), BRAF mutations (OR = 4.419, P = 0.047) and > 5 ng/ml CEA before primary tumor resection (CEA = 5-200, OR = 4.789, P < 0.001; CEA > 200, OR = 80.799, P < 0.001) were independent risk factors; age > 69 (OR = 0.187, P = 0.003) was an independent protective factor. For metachronous distant metastases, male gender (OR = 2.233, P = 0.024), primary pN2 stage (OR = 3.361, P = 0.01) and KRAS codon 13 mutations (OR = 11.857, P < 0.001) were independent risk factors; the primary tumor located at rectum (OR = 0.404, P = 0.041) was an independent protective factor. The univariate analyses considered KRAS codon 12 mutation as a risk factor for synchronous metastases (OR = 2.082, P = 0.023). However after the multivariate correction, it was only potentially significant (OR = 2.271, P = 0.086). For PIK3CA and PTEN mutations, no statistically significant differences were detected in univariate or multivariate analyses.
At the same time, there were four factors playing significantly different rolls in predicting synchronous or metachronous metastases: age > 69 (P = 0.032), BRAF mutation status (P = 0.019), and > 5 ng/ml CEA before primary tumor resection (P < 0.001) were independent factors for synchronous metastases but not for metachronous metastases, and KRAS codon 13 mutations (P = 0.049) were independent factors for metachronous metastases but not for synchronous metastases. Details are provided in Table 3.
Table 3.
Univariate and multivariate analyses of clinicopathological factors and gene mutations in distant metastases
| Metastases status | No vs. Synchronous | No vs. Metachronous | Synchronous vs. Metachronous | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||
| No | Syn. | Meta. | Univariate | Multivariate | Univariate | Multivariate | Univariate | Multivariate | |||||||
|
|
|
|
|
|
|
||||||||||
| OR | P value | OR | P value | OR | P value | OR | P value | OR | P value | OR | P value | ||||
| Total patients (n) | 96 | 92 | 93 | ||||||||||||
| Sex | |||||||||||||||
| Male | 51 | 63 | 60 | 1.917 | 0.032 | 2.457 | 0.038 | 1.604 | 0.113 | 2.233 | 0.024 | 0.837 | 0.568 | 0.757 | 0.512 |
| Female | 45 | 29 | 33 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| Age | |||||||||||||||
| < 55 | 31 | 37 | 30 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| 55-69 | 34 | 40 | 39 | 0.986 | 0.966 | 1.030 | 0.951 | 1.185 | 0.625 | 1.151 | 0.735 | 1.203 | 0.580 | 1.534 | 0.351 |
| > 69 | 31 | 15 | 24 | 0.405 | 0.023 | 0.187 | 0.003 | 0.800 | 0.550 | 0.627 | 0.340 | 1.973 | 0.098 | 3.585 | 0.032 |
| Primary tumor location | |||||||||||||||
| Right-sided | 24 | 34 | 31 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| Left-sided | 26 | 27 | 24 | 0.733 | 0.417 | 0.671 | 0.446 | 0.715 | 0.392 | 0.556 | 0.202 | 0.975 | 0.946 | 0.922 | 0.879 |
| Rectum | 46 | 31 | 38 | 0.476 | 0.036 | 0.400 | 0.070 | 0.640 | 0.201 | 0.404 | 0.041 | 1.344 | 0.393 | 1.467 | 0.430 |
| Primary pT Stage | |||||||||||||||
| 1/2 | 16 | 11 | 9 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| 3 | 31 | 18 | 35 | 0.845 | 0.731 | 0.673 | 0.561 | 2.007 | 0.150 | 1.402 | 0.562 | 2.377 | 0.106 | 4.478 | 0.046 |
| 4 | 49 | 63 | 49 | 1.870 | 0.151 | 1.161 | 0.811 | 1.778 | 0.214 | 1.243 | 0.692 | 0.951 | 0.917 | 1.240 | 0.751 |
| Primary pN Stage | |||||||||||||||
| 0 | 45 | 30 | 34 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| 1 | 35 | 26 | 32 | 1.114 | 0.757 | 1.338 | 0.551 | 1.210 | 0.568 | 1.369 | 0.439 | 1.086 | 0.821 | 1.142 | 0.792 |
| 2 | 16 | 36 | 27 | 3.375 | 0.001 | 4.579 | 0.006 | 2.233 | 0.039 | 3.361 | 0.010 | 0.662 | 0.248 | 0.863 | 0.773 |
| Primary differentiation | |||||||||||||||
| G1-G2 | 64 | 53 | 59 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| G3-G4 | 32 | 39 | 34 | 1.472 | 0.201 | 1.336 | 0.335 | 1.153 | 0.642 | 1.042 | 0.912 | 0.783 | 0.417 | 0.967 | 0.934 |
| Primary histological type | |||||||||||||||
| Non-mucinous | 76 | 74 | 80 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| Mucinous | 20 | 18 | 13 | 0.924 | 0.829 | 0.738 | 0.532 | 0.618 | 0.217 | 0.396 | 0.056 | 0.668 | 0.311 | 0.639 | 0.412 |
| KRAS | |||||||||||||||
| WT | 68 | 49 | 49 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| Codon 12 MT | 24 | 36 | 23 | 2.082 | 0.023 | 2.271 | 0.086 | 1.330 | 0.411 | 2.278 | 0.054 | 0.639 | 0.181 | 0.709 | 0.472 |
| Codon 13 MT | 4 | 7 | 21 | 2.429 | 0.175 | 2.085 | 0.407 | 7.286 | 0.001 | 11.857 | < 0.001 | 3.000 | 0.022 | 3.764 | 0.049 |
| BRAF | |||||||||||||||
| WT | 92 | 80 | 91 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| MT | 4 | 12 | 2 | 3.450 | 0.038 | 4.419 | 0.047 | 0.505 | 0.437 | 0.758 | 0.785 | 0.147 | 0.014 | 0.099 | 0.019 |
| PIK3CA | |||||||||||||||
| WT | 83 | 72 | 78 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| MT | 13 | 20 | 15 | 1.774 | 0.143 | 1.977 | 0.204 | 1.228 | 0.617 | 1.195 | 0.720 | 0.692 | 0.331 | 0.458 | 0.133 |
| PTEN | |||||||||||||||
| WT | 81 | 78 | 76 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| MT | 15 | 14 | 17 | 0.969 | 0.938 | 0.960 | 0.944 | 1.208 | 0.627 | 1.219 | 0.667 | 1.246 | 0.578 | 1.764 | 0.304 |
| Pre-primary resection CEA | |||||||||||||||
| < 5 ng/ml | 51 | 18 | 50 | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - | 1 | - |
| 5-200 ng/ml | 31 | 45 | 33 | 4.113 | < 0.001 | 4.789 | < 0.001 | 1.086 | 0.797 | 1.046 | 0.902 | 0.264 | < 0.001 | 0.164 | < 0.001 |
| > 200 ng/ml | 1 | 25 | 1 | 70.833 | < 0.001 | 80.799 | < 0.001 | 1.020 | 0.989 | 0.756 | 0.862 | 0.140 | < 0.001 | 0.005 | < 0.001 |
| Unknow | 13 | 4 | 9 | 0.872 | 0.829 | 0.772 | 0.735 | 0.706 | 0.466 | 0.834 | 0.749 | 0.810 | 0.750 | 0.619 | 0.547 |
| Adjuvant CT | |||||||||||||||
| Oral Fu | 11 | - | 8 | - | - | - | - | 1 | - | 1 | - | - | - | - | - |
| FOLFOX | 72 | - | 71 | - | - | - | - | 1.356 | 0.538 | 1.274 | 0.711 | - | - | - | - |
| XELOX | 7 | - | 11 | - | - | - | - | 2.161 | 0.251 | 1.875 | 0.440 | - | - | - | - |
| Unknow | 6 | - | 3 | - | - | - | - | 0.688 | 0.658 | 0.531 | 0.551 | - | - | - | - |
Logistic regression model was used in the analyses. Syn.: synchronous; Meta.: metachronous; OR: odds ratio; WT: wild type; MT: mutant type; CEA: carcino-embryonic antigen; Adjuvant CT: adjuvant chemotherapy after primary tumor resection; Fu: fluorouracil; FOLFOX: fluorouracil, leucovorin, and oxaliplatin; XELOX: capecitabine and oxaliplatin.
We next conducted subgroup analyses for single nucleotide polymorphisms (SNPs) in KRAS exon 2. The 7 common mutation sites are listed in Table 4. The results of multivariate analyses showed that one KRAS codon 12 mutation (c.34G > T, p.G12C) was an independent risk factor for synchronous metastases (OR = 11.667, P = 0.026) but not for metachronous metastases (i.e., there was a significant difference between synchronous and metachronous metastases, P = 0.043). Another KRAS codon 12 mutation (c.35G > T, p.G12V) was a potential risk factor only for metachronous metastases (OR = 3.489, P = 0.062) but not for synchronous metastases (i.e., there was a significant difference between synchronous and metachronous metastases, P = 0.049). Different SNPs in KRAS played different roles in determining synchronous or metachronous metastases.
Table 4.
Multivariate analyses of common single nucleotide polymorphisms in KRAS mutations
| KRAS gene type | Metastases status | No vs. Syn. | No vs. meta. | Syn. vs. meta. | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| No | Syn. | Meta. | OR | P value | OR | P value | OR | P value | |
| Total patients (n) | 96 | 92 | 93 | ||||||
| Wild type | 68 | 49 | 49 | 1 | - | 1 | - | 1 | - |
| c.35G > A, p.G12D | 14 | 18 | 9 | 2.006 | 0.235 | 1.147 | 0.798 | 0.548 | 0.331 |
| c.35G > T, p.G12V | 7 | 6 | 9 | 0.343 | 0.280 | 3.489 | 0.062 | 6.424 | 0.049 |
| c.34G > T, p.G12C | 2 | 8 | 1 | 11.667 | 0.026 | 2.675 | 0.483 | 0.057 | 0.043 |
| c.35G > C, p.G12A | 0 | 1 | 2 | NE | 1.000 | NE | 0.999 | 1.382 | 0.838 |
| c.34G > A, p.G12S | 0 | 2 | 2 | NE | 0.999 | NE | 0.999 | 1.243 | 0.869 |
| c.34G > C, p.G12R | 1 | 1 | 0 | 5.655 | 0.407 | NE | 1.000 | NE | 1.000 |
| c.38G > A, p.G13D | 4 | 7 | 21 | 1.822 | 0.510 | 13.686 | < 0.001 | 3.930 | 0.047 |
Multivariate analysis included all other clinical and pathological characteristics in Table 3, and logistic regression was used. Duplicate results were omitted in this table. OR: odds ratio; Syn.: synchronous; Meta.: metachronous; NE: not evaluable.
Moreover, in synchronous-metastases group, 6 patients (6.5%) had distant metastases within 6 months after primary tumor resections, not at first diagnosis. The univariate pairwise comparison showed there were no significant difference between metastases within 6 months and metastases detected at first diagnosis. However, significantly more patients with BRAF mutations (P = 0.018) and > 200 ng/ml CEA before primary tumor resection (P = 0.011) were detected having metastases within 6 months rather than metachronous metastases. It seemed that metastases within 6 months were more in line with synchronous metastases detected at first diagnosis rather than metachronous metastases. The comparison between metastases within 6 months and no-metastases also showed that these short-term metastases could be predicted by BRAF mutations (OR = 11.5, P = 0.039) and > 200 ng/ml CEA before primary tumor resection (OR = 41, P = 0.011). Additional details are provided in Table 5.
Table 5.
Univariate analyses of synchronous metastases within 6 months after primary tumor resection
| Metastases status | P1 | P2 | P3 | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| No | Syn. | Meta. | |||||
|
| |||||||
| Within 6 months | First diagnosis | ||||||
| Total patients (n) | 96 | 6 | 86 | 93 | |||
| Sex | 0.684 | 0.722 | 0.738 | ||||
| Male | 51 | 4 | 59 | 60 | |||
| Female | 45 | 2 | 27 | 33 | |||
| Age | 0.368 | 0.392 | 0.567 | ||||
| < 55 | 31 | 1 | 36 | 30 | |||
| 55-69 | 34 | 4 | 36 | 39 | |||
| > 69 | 31 | 1 | 14 | 24 | |||
| Primary tumor location | 0.548 | 0.872 | 0.759 | ||||
| Right-sided | 24 | 3 | 31 | 31 | |||
| Left-sided | 26 | 1 | 26 | 24 | |||
| Rectum | 46 | 2 | 29 | 38 | |||
| Primary pT Stage | 0.853 | 0.823 | 0.554 | ||||
| 1/2 | 16 | 1 | 10 | 9 | |||
| 3 | 31 | 1 | 17 | 35 | |||
| 4 | 49 | 4 | 59 | 49 | |||
| Primary pN Stage | 0.473 | 1.000 | 1.000 | ||||
| 0 | 45 | 2 | 28 | 34 | |||
| 1 | 35 | 2 | 24 | 32 | |||
| 2 | 16 | 2 | 34 | 27 | |||
| Primary differentiation | 1.000 | 0.700 | 1.000 | ||||
| G1-G2 | 64 | 4 | 49 | 59 | |||
| G3-G4 | 32 | 2 | 37 | 34 | |||
| Primary histological type | 0.126 | 0.087 | 0.052 | ||||
| Non-mucinous | 76 | 3 | 71 | 80 | |||
| Mucinous | 20 | 3 | 15 | 13 | |||
| KRAS | 0.730 | 0.699 | 1.000 | ||||
| Wild type | 68 | 4 | 45 | 49 | |||
| Codon 12 Mutant type | 24 | 2 | 34 | 23 | |||
| Codon 13 Mutant type | 4 | 0 | 7 | 21 | |||
| BRAF | 0.039 | 0.174 | 0.018 | ||||
| Wild type | 92 | 4 | 76 | 91 | |||
| Mutant type | 4 | 2 | 10 | 2 | |||
| PIK3CA | 0.829 | 0.755 | 0.972 | ||||
| Wild type | 83 | 5 | 67 | 78 | |||
| Mutant type | 13 | 1 | 19 | 15 | |||
| PTEN | 0.946 | 0.919 | 0.921 | ||||
| Wild type | 81 | 5 | 73 | 76 | |||
| Mutant type | 15 | 1 | 13 | 17 | |||
| Pre-primary resection CEA | 0.011 | 1.000 | 0.011 | ||||
| ≤ 200 ng/ml | 82 | 4 | 59 | 83 | |||
| > 200 ng/ml | 1 | 2 | 23 | 1 | |||
| Unknown | 13 | 0 | 4 | 9 | |||
P1: synchronous metastases within 6 months after primary tumor resection vs. no metastases. P2: synchronous metastases within 6 months after primary tumor resection vs. synchronous metastases at first diagnosis. P3: synchronous metastases within 6 months after primary tumor resection vs. metachronous metastases. Syn.: synchronous; Meta.: metachronous Within 6 months: metastases were diagnosed within 6 months after primary tumor resections. First diagnosis: metastases were detected before or during the primary tumor resection. Fisher’s exact test was used for samples with expected frequency < 5. CEA: carcinoembryonic antigen.
KRAS mutation and long-term survival
The latency of metachronous distant metastases is showed in Figure 1. Half of all metachronous metastases occurred within 16 months, 75% occurred within 28 months, and 95% within 56 months. For patients with metachronous metastases, KRAS codon 13 mutations potentially resulted in shorter latency than KRAS wild type (median, 13 vs. 18 months, P = 0.066). But KRAS codon 12 mutations had no significant effect on the latency (Wild type vs. codon 12 mutation, 18 vs. 16 months, P = 0.775; codon 12 mutation vs. codon 13 mutation, 16 vs. 13 months, P = 0.277). Additional details are provided in Figure 2.
Figure 1.
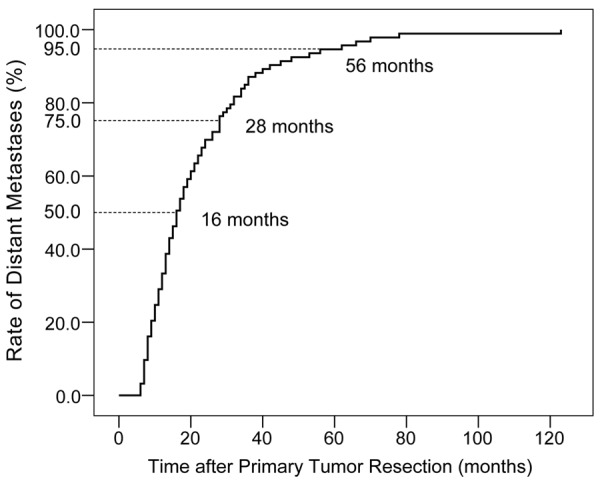
Timing of metachronous distant metastases. 50% of metachronous metastases occurred within 16 months, 75% occurred within 28 months, and 95% within 56 months after primary tumor resection.
Figure 2.
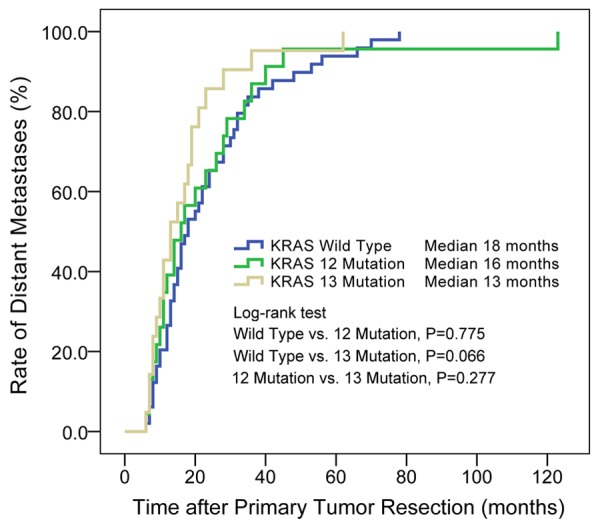
KRAS codon 12 and codon 13 mutations affected the latency of metachronous distant metastases after primary tumor resection. There were potential significant difference between patients with KRAS wild type and codon 13 mutation (P = 0.066). Median: median of the latency to metachronous distant metastases.
After resections of metastases, patients origionally diagnosed with metachronous metastases had longer overall survival time than synchronous metastases (median, 43 vs. 23 months, P = 0.050, details in Figure 3). The overall survival after occurrence of metachronous metastases is also showed in Figure 4. The survival curve showed no significant differences between patients with wild-type KRAS, codon 12 mutations and codon 13 mutations. Multivariate Cox regression in Table 6 showed that radical resection of metachronous metastases was a significant protective factor for long time survival (HR = 0.280, P = 0.002). Chemotherapy and TACE/TAI had similar effects, but were both far less effective than radical surgery. The survival curve in Figure 5 also shows that radical resection of metastases resulted in a subsequent median survival of 43 months, significantly longer than chemotherapy alone (median 6 months, P < 0.001) or chemotherapy plus TACE/TAI (median 5 months, P < 0.001).
Figure 3.
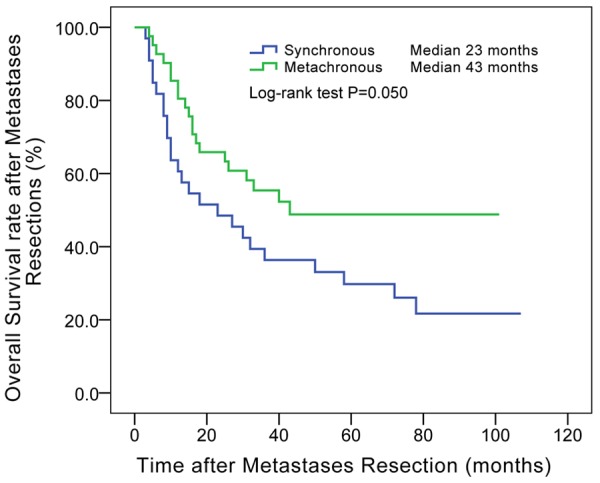
Overall survival after metastases resections between synchronous and metachronous metastases. Patients with metachronous metastases had longer survival time than patients with synchronous metastases after resections of metastases (median, 43 vs. 23 months, P = 0.050). Median: median of overall survival after resection of distant metastases.
Figure 4.
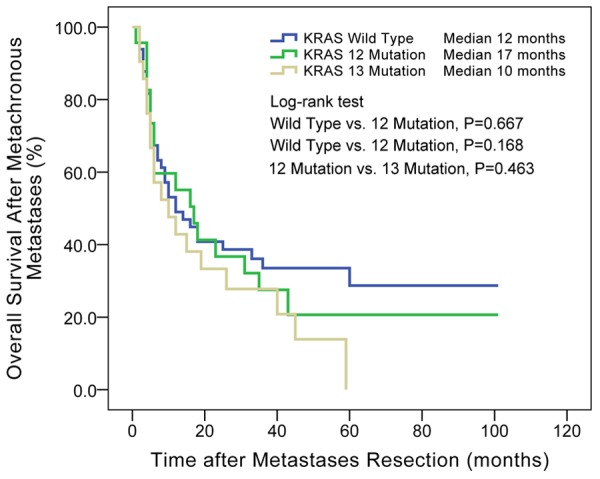
KRAS codon 12 and codon 13 mutations affected the overall survival after the occurrence of metachronous distant metastases. There was no significant difference among the three groups. Median: median of overall survival after occurrence of metachronous distant metastases.
Table 6.
Multivariate analyses of overall survival after metachronous metastases
| Number | HR | P value | |
|---|---|---|---|
| Total patients (n) | 93 | ||
| Sex | |||
| Male | 60 | 0.873 | 0.623 |
| Female | 33 | 1 | - |
| Age at metastases | |||
| < 55 | 26 | 1 | - |
| 55-69 | 38 | 1.130 | 0.718 |
| > 69 | 29 | 0.976 | 0.945 |
| Primary differentiation | |||
| G1-G2 | 59 | 1 | - |
| G3-G4 | 34 | 1.017 | 0.955 |
| Primary histological type | |||
| Non-mucinous | 80 | 1 | - |
| Mucinous | 13 | 0.806 | 0.626 |
| KRAS | |||
| Wild type | 49 | 1 | - |
| Codon 12 Mutant type | 23 | 1.046 | 0.901 |
| Codon 13 Mutant type | 21 | 1.150 | 0.704 |
| BRAF | |||
| Wild type | 91 | 1 | - |
| Mutant type | 2 | 2.255 | 0.381 |
| PIK3CA | |||
| Wild type | 78 | 1 | - |
| Mutant type | 15 | 0.818 | 0.674 |
| PTEN | |||
| Wild type | 76 | 1 | - |
| Mutant type | 17 | 0.518 | 0.101 |
| CEA at diagnosis of metastases | |||
| < 5 ng/ml | 25 | 1 | - |
| 5-200 ng/ml | 47 | 0.682 | 0.346 |
| > 200 ng/ml | 11 | 0.935 | 0.902 |
| Unknown | 10 | 1.074 | 0.897 |
| Treatment of metastases | |||
| Chemotherapy only | 21 | 1 | - |
| Chemotherapy + TACE/TAI | 31 | 1.479 | 0.284 |
| Radical surgery | 41 | 0.280 | 0.002 |
Gene type was detected based on the primary tumor. Age at metastases was based on the time metastases diagnosed. Cox-regression model was used in the multivariate analysis. HR: hazard ratio; CEA: carcinoembryonic antigen; TACE: transcatheter arterial chemoembolization; TAI: Transcatheter arterial infusion.
Figure 5.
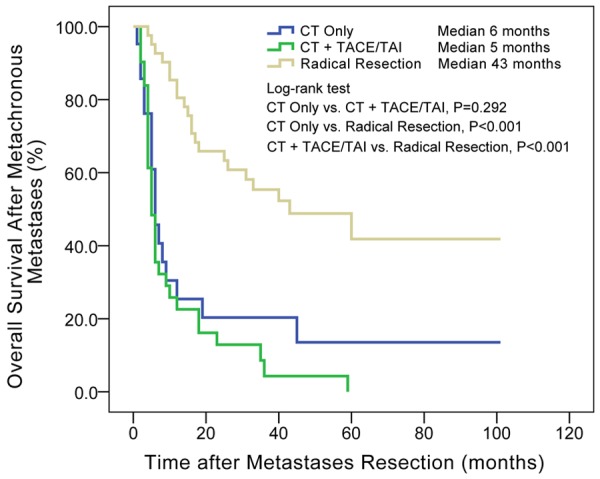
Different therapies affected overall survival after the occurrence of metachronous distant metastases. Radical resections of metastases had significant advantage over chemotherapy and TACE/TAI. Median: median of overall survival after occurrence of metachronous distant metastases; TACE: transcatheter arterial chemoembolization; TAI: transcatheter arterial infusion.
Discussion
In this study, we selected 3 groups of patients: patients without distant metastases, with synchronous distant metastases and with metachronous distant metastases. The detection and analysis of genotype and clinicopathological characteristics showed that age, sex, primary N stage, BRAF mutations and CEA levels before primary tumor resection were independent factors for synchronous metastases; sex, primary tumor location, primary N stage and KRAS codon 13 mutations were independent factors for metachronous distant metastases. The metastases occurred within 6 months after primary tumor resections seemed more in line with synchronous metastases detected at first diagnosis rather than metachronous metastases, and were potentially predicted by BRAF mutations and > 200 ng/ml CEA before primary tumor resection. Different SNPs of KRAS mutations played different roles in determining the timing of metastases. Moreover, we found that compared to KRAS wild type, KRAS codon 13 mutations potentially resulted in shorter latency of metastases. After the occurrence of metachronous distant metastases, treatment was the most important factor in determining the long-term survival of the patient.
The KRAS oncogene has a well-established role in tumor growth and regulation, and plays an important role in individualized molecular treatment of colorectal cancer. Since the multicenter “RASCAL” and “RASCAL II” studies [14,17], numerous studies have confirmed KRAS mutation as prognostic factor of colorectal cancer [18]. Along with the application of targeted therapy, KRAS mutation was also proved predictor for ineffective anti-EGFR treatment [8,9]. However, the difference between KRAS codon 12 and codon 13 mutations are still controversial. Experimental studies have demonstrated a reduced transforming activity of the codon 13 mutation compared with the codon 12 mutation in vitro systems [19-21]. Compared with KRAS codon 12 mutant cell lines, KRAS codon 13 mutation showed decreased anchorage-independent growth and higher levels of apoptosis, which suggested lower malignancy and better prognosis. Furthermore, recent computational molecular dynamics simulations demonstrated that KRAS codon 13 mutation had similar behavior as wild-type KRAS [22]. Patients who harbored KRAS 13 mutations might therefore benefit from treatment with anti-EGFR antibodies [23-25]. But clinical studies have opposite conclusions. Samowitz et al [26] retrospectively analyzed 1413 patients with colon cancer, showed that KRAS codon 13 mutation was associated with more risk (HR = 1.4, 95% CI = [0.95, 2.0]) than codon 12 mutation (HR = 1.0, 95% CI = [0.8, 1.2]) in survival. Bazan et al [27] demonstrated that KRAS codon 13 mutation was independently related to risk of relapse (HR = 1.79, P < 0.05) and death (HR = 1.93, P < 0.05), but not KRAS codon 12 mutation. Some other clinical studies also confirmed that KRAS codon 13 mutations were associated with more distant metastases and poorer prognosis than codon 12 mutations [27-30].
These contradictions between experimental and clinical studies might be explained by our study: KRAS codon 12 mutations were potential risk factor for both synchronous and metachronous metastases; but KRAS 13 mutations were risk factor only for metachronous metastases. Present experimental studies were always based on cell lines with short observation period, which was conducive to the expression of synchronous metastases, not associated with KRAS codon 13 mutations. However in clinical studies, observation period was long enough to fully reveal the traits of metachronous metastases, significantly associated with KRAS codon 13 mutations. Thus, the contradictions came from neglecting the differences between synchronous and metachronous metastases. It would be better making a distinction between synchronous and metachronous metastases in future researches.
The specific KRAS codon 13 mutation could also partially explain the long latency from primary tumor resection to occurrence of metachronous metastases. Different from KRAS codon 12 mutation, codon 13 mutation was more similar to wild type in molecular structure and function [22]. This meant a reduced activity of RAS-RAF-MAPK signaling pathway [19-21]. It could be suspected that the effect of KRAS codon 13 mutations was more moderate than codon 12 mutations, requiring a long-term accumulating process for the occurrence of detectable metastases. Moreover, KRAS codon 13 mutations were associated with lower levels of tumor-infiltrating mature dendritic cells [31]. This change of microenvironment might also help tumor cells hide for a long time, avoid being detected or destroyed by the immune system.
In addition, different SNPs in the KRAS gene had different effects on prognosis. In our study, one KRAS codon 12 mutation type (c.34G > T, p.G12C) showed more predictive capability of synchronous metastases, but another KRAS codon 12 mutation type (c.35G > T, p.G12V) tended to predict metachronous metastases. The other common KRAS codon 12 mutation type (c.35G > A, p.G12D) seemed meaningless in predicting metastases. This suggested that change of a single amino acid at a same site may result in different outcomes. Not only KRAS codon 13 mutations, but also different SNPs of KRAS codon 12 mutations should be paid more attention. As KRAS codon 13 mutations have been considered benefiting from anti-EGFR antibodies [24,25,32], specific SNPs in KRAS codon 12 should also be tested. And in individualized treatment, classification based on protein function might be better than simply based on the mutation site.
We also analyzed synchronous distant metastases occurred within 6 months after primary tumor resections. For these short-term metastases, the results of univariate analyses showed conformity with the synchronous metastases detected at first diagnosis, and could be predicted by BRAF mutations and > 200 ng/ml CEA before primary tumor resection. CEA is a traditional prognostic marker for colorectal cancer. BRAF mutation was also consistently associated with a worse prognosis in patients with metastatic disease in both retrospective clinical series and therapeutic trials [33,34]. For newly diagnosed patients with the two high-risk factors, if synchronous metastases were not detected, more detailed preoperative examinations should be carried out to reduce the risk of missed diagnosis.
For patients with metachronous distant metastases, resection of metastases was the major factor for long-term survival. Compared with surgery, chemotherapy or chemotherapy combined with TACE/TAI did not obtain satisfactory prognosis. Moreover, early detection was important for the radical resection of metastases. The independent factors in our study may help identify patients with high-risk of metachronous metastases in a simple way. The median metastatic time was approximately 16 months for all KRAS genotypes and earlier (approximately 13 months) for codon 13 mutations. During this period, intensive follow-ups could be conducted for high-risk patients, which may aid in early detection and treatment of metachronous distant metastases.
Acknowledgements
This study was supported by funds as follows: National Natural Science Foundation of China (81272390, 81372315), Shanghai Science and Technology Committee Talent Program (12XD1401900) and Outstanding Academic Leaders Project of Health System in Shanghai (XBR2011031).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.van der Wal GE, Gouw AS, Kamps JA, Moorlag HE, Bulthuis ML, Molema G, de Jong KP. Angiogenesis in synchronous and metachronous colorectal liver metastases: the liver as a permissive soil. Ann Surg. 2012;255:86–94. doi: 10.1097/SLA.0b013e318238346a. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Nordlinger B, Adam R, Kohne CH, Pozzo C, Poston G, Ychou M, Rougier P. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42:2212–2221. doi: 10.1016/j.ejca.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 4.de Jong MC, Pulitano C, Ribero D, Strub J, Mentha G, Schulick RD, Choti MA, Aldrighetti L, Capussotti L, Pawlik TM. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg. 2009;250:440–448. doi: 10.1097/SLA.0b013e3181b4539b. [DOI] [PubMed] [Google Scholar]
- 5.Tsai MS, Su YH, Ho MC, Liang JT, Chen TP, Lai HS, Lee PH. Clinicopathological features and prognosis in resectable synchronous and metachronous colorectal liver metastasis. Ann Surg Oncol. 2007;14:786–794. doi: 10.1245/s10434-006-9215-5. [DOI] [PubMed] [Google Scholar]
- 6.Fong Y. Surgical therapy of hepatic colorectal metastasis. CA Cancer J Clin. 1999;49:231–255. doi: 10.3322/canjclin.49.4.231. [DOI] [PubMed] [Google Scholar]
- 7.Haglund K, Rusten TE, Stenmark H. Aberrant receptor signaling and trafficking as mechanisms in oncogenesis. Crit Rev Oncog. 2007;13:39–74. doi: 10.1615/critrevoncog.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- 8.Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S, Schlichting M, Zubel A, Celik I, Rougier P, Ciardiello F. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J. Clin. Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 9.Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, Celik I, Schlichting M, Koralewski P. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535–1546. doi: 10.1093/annonc/mdq632. [DOI] [PubMed] [Google Scholar]
- 10.Zhu K, Yan H, Wang R, Zhu H, Meng X, Xu X, Dou X, Chen D. Mutations of KRAS and PIK3CA as independent predictors of distant metastases in colorectal cancer. Med Oncol. 2014;31:16. doi: 10.1007/s12032-014-0016-6. [DOI] [PubMed] [Google Scholar]
- 11.Eklof V, Wikberg ML, Edin S, Dahlin AM, Jonsson BA, Oberg A, Rutegard J, Palmqvist R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108:2153–2163. doi: 10.1038/bjc.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atreya CE, Sangale Z, Xu N, Matli MR, Tikishvili E, Welbourn W, Stone S, Shokat KM, Warren RS. PTEN expression is consistent in colorectal cancer primaries and metastases and associates with patient survival. Cancer Med. 2013;2:496–506. doi: 10.1002/cam4.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J. Clin. Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 14.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Qin X, Wang J, Zhang S, Zhong Y, Ren L, Wei Y, Zeng S, Wan D, Zheng S. Chinese guidelines for the diagnosis and comprehensive treatment of hepatic metastasis of colorectal cancer. J Cancer Res Clin Oncol. 2011;137:1379–1396. doi: 10.1007/s00432-011-0999-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17:343–346. doi: 10.1016/0197-2456(96)00075-x. [DOI] [PubMed] [Google Scholar]
- 17.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O’Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrington AK, Heinrich EL, Lee W, Duldulao M, Patel S, Sanchez J, Garcia-Aguilar J, Kim J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int J Mol Sci. 2012;13:12153–12168. doi: 10.3390/ijms131012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750–6756. [PubMed] [Google Scholar]
- 20.Ward RL, Todd AV, Santiago F, O’Connor T, Hawkins NJ. Activation of the K-ras oncogene in colorectal neoplasms is associated with decreased apoptosis. Cancer. 1997;79:1106–1113. [PubMed] [Google Scholar]
- 21.Bos JL, Toksoz D, Marshall CJ, Verlaan-de Vries M, Veeneman GH, van der Eb AJ, van Boom JH, Janssen JW, Steenvoorden AC. Amino-acid substitutions at codon 13 of the N-ras oncogene in human acute myeloid leukaemia. Nature. 1985;315:726–730. doi: 10.1038/315726a0. [DOI] [PubMed] [Google Scholar]
- 22.Chen CC, Er TK, Liu YY, Hwang JK, Barrio MJ, Rodrigo M, Garcia-Toro E, Herreros-Villanueva M. Computational analysis of KRAS mutations: implications for different effects on the KRAS p. G12D and p.G13D mutations. PLoS One. 2013;8:e55793. doi: 10.1371/journal.pone.0055793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar SS, Price TJ, Mohyieldin O, Borg M, Townsend A, Hardingham JE. KRAS G13D Mutation and Sensitivity to Cetuximab or Panitumumab in a Colorectal Cancer Cell Line Model. Gastrointest Cancer Res. 2014;7:23–26. [PMC free article] [PubMed] [Google Scholar]
- 24.Mao C, Huang YF, Yang ZY, Zheng DY, Chen JZ, Tang JL. KRAS p. G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: a systematic review and meta-analysis. Cancer. 2013;119:714–721. doi: 10.1002/cncr.27804. [DOI] [PubMed] [Google Scholar]
- 25.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 26.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193–1197. [PubMed] [Google Scholar]
- 27.Bazan V, Migliavacca M, Zanna I, Tubiolo C, Grassi N, Latteri MA, La Farina M, Albanese I, Dardanoni G, Salerno S, Tomasino RM, Labianca R, Gebbia N, Russo A. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol. 2002;13:1438–1446. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 28.Roszkowski K, Zurawski B, Jozwicki W, Basta P, Lewandowska MA. Impact of Specific KRAS Mutation in Exon 2 on Clinical Outcome of Chemotherapy- and Radiotherapy-Treated Colorectal Adenocarcinoma Patients. Mol Diagn Ther. 2014;18:559–566. doi: 10.1007/s40291-014-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modest DP, Stintzing S, Laubender RP, Neumann J, Jung A, Giessen C, Haas M, Aubele P, Schulz C, Boeck S, Stemmler HJ, Kirchner T, Heinemann V. Clinical characterization of patients with metastatic colorectal cancer depending on the KRAS status. Anticancer Drugs. 2011;22:913–918. doi: 10.1097/CAD.0b013e3283493160. [DOI] [PubMed] [Google Scholar]
- 30.Pajkos G, Kiss I, Sandor J, Ember I, Kishazi P. The prognostic value of the presence of mutations at the codons 12, 13, 61 of K-ras oncogene in colorectal cancer. Anticancer Res. 2000;20:1695–1701. [PubMed] [Google Scholar]
- 31.Kocian P, Sedivcova M, Drgac J, Cerna K, Hoch J, Kodet R, Bartunkova J, Spisek R, Fialova A. Tumor-infiltrating lymphocytes and dendritic cells in human colorectal cancer: their relationship to KRAS mutational status and disease recurrence. Hum Immunol. 2011;72:1022–1028. doi: 10.1016/j.humimm.2011.07.312. [DOI] [PubMed] [Google Scholar]
- 32.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O’Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S. Association of KRAS p. G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 33.Yaeger R, Cercek A, Chou JF, Sylvester BE, Kemeny NE, Hechtman JF, Ladanyi M, Rosen N, Weiser MR, Capanu M, Solit DB, D’Angelica MI, Vakiani E, Saltz LB. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer. 2014;120:2316–2324. doi: 10.1002/cncr.28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]


