Abstract
The prognosis of radioresistant colorectal cancer (CRC) is generally poor. Abnormal expression of microRNAs (miRNAs) is involved in the radiosensitivity of various tumor cells as these RNAs regulate biological signaling pathways. However, radioresistance-associated miRNAs in CRC have not yet been identified. In this study, we filtered out HCT116 and CCL-244 from seven CRC cell lines that showed the highest difference in radiosensitivity in a clonogenic assay. MiRNA sequencing identified 33 differentially expressed miRNAs (13 up-regulated and 20 down-regulated) in CCL-244 and 37 in HCT116 (20 up-regulated and 17 down-regulated) cells. MiR-100 was significantly down-regulated in CCL-244 cells after X-ray irradiation but not in HCT116 cells. Quantitative real-time PCR showed that the expression of miR-100 in CRC tissues was significantly lower than that in normal tissues. Thus, miR-100 seems to be involved in the radioresistance of CCL-244 cells. MiR-100 up-regulation sensitized CCL-244 cells to X-ray irradiation, which probably led to apoptosis and DNA double-strand breaks in these. In conclusion, to our knowledge, this is the first study to show that miR-100 may play an important role in regulating the radiosensitivity of CRC, and it may act as a new clinical target for CRC radiotherapy.
Keywords: Colorectal cancer, miRNA profiling, miR-100, radioresistance, radiosensitivity
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. Statistically, the United States ranks third in terms of both the incidence and mortality of CRC, although the death rates associated with CRC have declined [1]. In China, CRC is the third most malignant tumor. Despite improvements in surgical techniques, therapeutic levels, and treatment efficacy, the overall prognosis of CRC is not optimistic, with the 5-year survival rate still hovering at around 50%. National Comprehensive Cancer Network guidelines on rectal treatment indicate that T3-stage or N (+) preoperative as well as postoperative rectal cancer should be treated with neoadjuvant chemoradiotherapy [2]. Radiation therapy is as important as surgery and chemotherapy for the treatment of CRC. However, radioresistance leading to tumor recurrence and consequently, a poor prognosis, is still a serious concern. Therefore, overcoming radiation resistance, enhancing radiation sensitivity of CRC, and improving the efficacy of radiotherapy have important practical significance in the treatment of CRC.
MicroRNAs (miRNAs) are a group of small non-coding RNAs, about 22 nucleotides (nt) in length. MiRNAs act as guide molecules by base pairing with one or more specific sequences of mRNAs that recruit argonaute-containing RNA-induced silencing complex (RISC) at the post-transcriptional level. RISC combines with the target mRNA at the 3’ untranslated region (3’ UTR), and the miRNA base pairs with the target mRNA, causing cleavage of the target mRNA and inhibition of protein expression [3].
Accumulating evidence has indicated that miRNAs are implicated in cell radiosensitivity. Josson et al found that miR-521 modulated the expression of the DNA repair protein CSA and sensitized prostate cancer cells to radiation treatment. Transient overexpression of miR-181a significantly sensitized malignant glioma cells to radiation treatment concurrent with down-regulation of the Bcl-2 protein [4]. A previous study also reported that miR-125b plays a role in the radioresistance of oral squamous cell carcinoma, probably through intercellular adhesion molecule-2 signalling [5]. Multiple miRNAs are involved in the radioresistance and radiosensitivity of lung cancer; the up-regulation of let-7g, let-7c, and miR-210 could induce the resistance of lung cancer to radiotherapy or chemotherapy. Further, Liu et al found that miR-21 expression promoted radioresistance in non-small cell lung cancer cells by targeting PTEN [6-9], while the upregulation of let-7a and let-7b could enhance the radiation sensitivity of lung cancer. They also found that miR-449a enhanced the radiosensitivity of lung cancer by increasing irradiation-induced DNA damage and apoptosis and altering cell cycle distribution [10]. The expression levels of miR-125b, miR-137, and miR-145 are reportedly closely related to the radiosensitivity of CRC [11-13]. Thus, miRNAs seem to be involved in tumor radiosensitivity via various mechanisms.
To our knowledge, miRNA profiles in response to irradiation and their impact on radioresistance in CRC have not yet been reported. This study aimed to investigate the role of miRNAs in the radiosensitivity of CRC.
Materials and methods
Cell culture
Human CRC-derived cell lines HT29, LoVo, Hce8693, and CaCo2 were resuscitated and resuspended in RPMI 1640 medium (HyClone, Logan, UT, USA), while HCT116, CCL-244, and SW-480 were resuspended in high-glucose DMEM (Hyclone) supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone), 100 U/mL penicillin G, and 100 mg/mL streptomycin (Hyclone). The cells were then plated in 25-cm2 culture bottles and incubated in a 5% CO2-humidified atmosphere at 37°C. The media were changed every 3 days, and the cells were trypsinized using trypsin-edetic acid when they reached 80%-90% confluence.
Colony formation assay
The cells were inoculated on 6-well plates. After 24 h, they were exposed to X-rays with different irradiation doses of 0, 2, 4, 6 and 8 Gy. After 14 days of incubation at 37°C, the colonies were stained with Giemsa, and a minimum of 50 viable cells were counted. This process was repeated three times. The surviving fraction was calculated as a ratio of the number of colonies formed divided by the total number of cells plated times the plating efficiency. The colony formation assay was also conducted with transiently transfected cells.
miRNA sequencing
HCT116 and CCL-244 cells were inoculated in 6-well plates and exposed to 8-Gy X-ray irradiation or sham irradiation. After 48 h, these samples were lysed using Trizol reagent for total RNA collection. Subsequently, miRNA sequencing was performed using the Illumina sequencing platform single-end sequencing mode to conduct high-throughput sequencing for the four samples (Genenergy, Shanghai, China); each sample had three parallel groups. Differences between the groups were determined using the DESeq statistical tests of R software (genetic screening criteria: P ≤ 0.05 and fold difference in expression ≥ 2). Cluster 3.0 and TreeView software programs were used to analyze the differences in gene cluster analysis [14]. Differential expression of miRNA between irradiated and nonirradiated cells was analyzed after the sequencing was proofread using the genome and miRBase databases.
Collection of clinical samples
A total of 30 CRC specimens were collected from the Second Affiliated Hospital of Soochow University after primary resection of the primary lesions. Fresh tumor tissues of the colon at the same pathological stage and at a distance of 8 cm from the border of the tumor tissues were collected as the control samples. All resected tissue samples were stored in the freezer at -80°C. The diagnosis of CRC was based on World Health Organization criteria. Tumor differentiation was defined according to Edmondson-Steiner grading system, while tumor staging was determined according to the sixth edition of the tumor-node-metastasis classification of the International Union Against Cancer. This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Soochow University. Written informed consent was obtained from all participants. None of the patients had undergone any treatment before surgery.
Transient transfection
CCL-244 cells were plated in 6-well plates (3 × 105 cells/well) and transfected with 100-nM miR-100 mimics (GenePharma, Shanghai, China) using Lipofect (PolyPlus, Illkirch, France) according to the manufacturer’s instructions. Fifty nanomolar of miRNA mimics (GenePharma) were used as control miRNA. After transfection for 5 h, the serum-free medium was replaced by a medium containing 10% FBS. After transfection for 48 h, the cells were harvested for further experiments. Quantitative real-time PCR was used to determine the efficiency of transfection.
RNA preparation and quantitative real-time PCR
Total RNA was extracted from the oncocytes in the sections using the TRIzol® one-step extraction method according to the manufacturer’s instructions (Invitrogen, San Diego, CA, USA). Total RNA was collected according to the manufacturer’s instructions of the RecoverAll™ Total Nucleic Acid Isolation kit (Applied Biosystems, San Diego, CA, USA). Light absorption values were read at 230, 260, and 280 nm using spectrophotometry to determine the purity and density of the RNA. Real-time PCR was then performed according to the manufacturer’s (Hairpin-itTM miRNAs qPCR Quantitation Kit) and instrument instructions (185-5096 CFX96 real time PCR detection system; Bio-Rad, Hercules, California, USA). The SYBR and U6 genes were used for detection of gene amplification and normalization of each sample, respectively. MiR-100 was amplified using RT primer 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACAAG-3’ and Reverse Primer 5’-ATCCAGTGCAGGGTCCGAGG-3’; forward primer 5’-GCGGCGGAACCCGTAGATCCGAAC-3’. Real-time PCR was also used to confirm the expression of miR-100 after transfection and to verify its consistency with sequencing results.
Cell proliferation assay
After transfection, CCL-244 cells were seeded in 96-well plates at 4000 cells/well and cultured in a CO2 incubator. After 24 h, the cells were subjected to 8-Gy irradiation. After 48 more hours, 10 µl of CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well, and the plates were placed in a CO2 incubator for 2 h. The optical density was determined at 450 nm. Each group was set up in triplicate.
Cell apoptosis assays
Following conventional digestion, cells in the logarithmic growth phase were used to prepare a single cell suspension, 2 × 105 cells/ml was seeded in 6-well plates, and two vice-holes were set. Cell adhesion was performed after transfection. The cells were subjected to X-ray irradiation after 24 h. After 48 h, all cells were collected and centrifuged at 2,000 rpm for 5 min; the supernatant was discarded after washing with PBS and centrifugation was repeated twice. The cells were then stained with fluorescein FITC-conjugated Annexin V and PI (KeyGen, Nanjing, China) and analyzed by flow cytometry (Beckman Coulter, Brea, CA, USA). For test results, the ratio of early apoptosis cells was only to be counted.
DNA double-strand break assay
DSBs represent an important ionizing radiation-induced lesion. The rapid phosphorylation of histone H2AX at serine 139 is a sensitive marker for DNA DSBs induced by ionizing radiation, which can later be detected by immunofluorescence.
After transfection, the CCL-244 cells were fixed in 4% paraformaldehyde, permeabilized with 1% Triton X-100 for 10 min at room temperature, and then blocked with dilute 1% BSA (Solarbio, Beijing, China) for 1 h. The cells were incubated at 4°C overnight with the primary antibody γ-H2AX (Epitomics, Burlingame, CA, USA), which was diluted 1:1000 with 1% BSA. PBS was used to wash away the primary antibody. The cells were incubated with the secondary antibody at 37°C for 1 h, following which they were placed on coverslips and counterstained with 4-6-diamidino-2-phenylindole (Invitrogen) to mark the nuclei. Immunofluorescence staining was examined using a fluorescence microscope.
Western blot
Lysates from the CCL-244 cells were prepared after transfection and irradiation by incubating the cells in lysis buffer (125 mM Tris-HCl, pH 6.8, 2% SDS, 10% v/v, glycerol) at 4°C for 30 min. Protein concentration was determined using the BCA protein assay (Bio-Rad Laboratories). Next, 40 μg of cell lysates was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following SDS-PAGE, the proteins were transferred onto a PVDF membrane (Millipore, Temecula, CA, USA) that was blocked with 10% dry milk in TBST. The membrane was then probed with primary antibodies against Bcl-2, NF-κB, Caspase-3, and β-actin (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C for 12 h.
Statistical analysis
Data were expressed as mean ± standard error of the mean of at least three independent experiments. Standard error bars were included for all data points. Student’s t-test was used to analyze the data when only two groups were compared, while one-way analysis of variance was used in case of more than two groups. The DESeq statistical tests of Rsoftware (genetic screening criteria: P ≤ 0.05 and fold difference in expression ≥ 2) was used to analyze the differences between the two groups of sequencing results. Statistical analysis was performed using the Statistical Package for the Social Sciences Version 17.0 (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant. GraphPad Prism 5 software (San Diego, CA, USA) was used to draw survival curves with data from the colony formation assays.
Results
Selection of experimental CRC cell lines by comparing the radiosensitivities of different CRC cancer cells
In order to select the appropriate cell lines for the experiment, a colony formation assay was used to detect the radiosensitivities of seven CRC cell lines, namely, HCT116, CCL-244, HT-29, LOVO, Hce-8693, CaCO2, and SW480. We found that the ascending order of these seven CRC cell lines in terms of radiosensitivity was CCL-244, SW480, HT29, LOVO, CaCO2, Hce8693, and HCT116 (Figure 1A). The mean lethal dose of ionizing radiation for these cells was represented by the values of the parameter D0. D0 values reflect the radiosensitivities of different cells; the lower the D0 values of cells, the more sensitive they are to radiation. The D0 values of each CRC cell line were consistent with the results of clonogenic assay (Figure 1B). As this study focused on the radiotolerance of CRC, the CCL-244 and HCT116 cell lines were selected for subsequent experiments.
Figure 1.
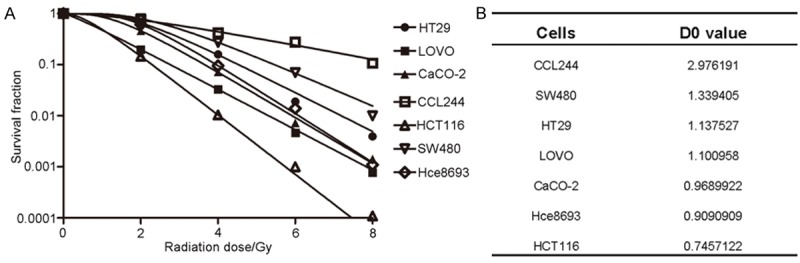
Comparison of the radiosensitivities of the seven CRC cell lines to X-ray irradiation. A. Cells were exposed to X-rays at different irradiation doses: 0, 2, 4, 6, and 8 Gy. After 14 days, the surviving fraction was calculated as a ratio of the number of colonies formed divided by the total number of cells plated times the plating efficiency. B. D0 values for each type of colorectal cancer cell line.
MiRNA profiling in CRC cells in response to X-ray irradiation
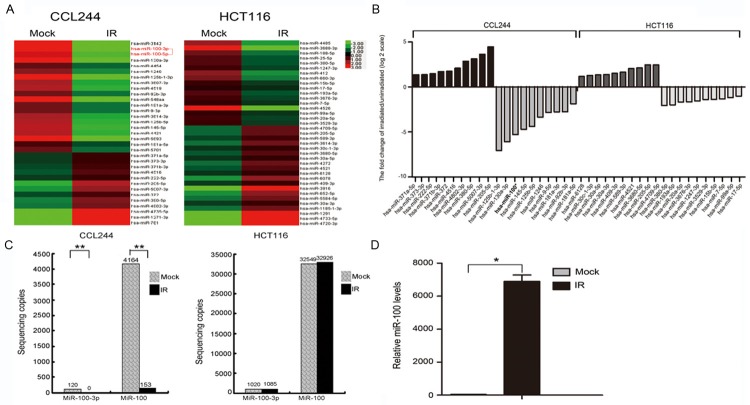
In order to identify the miRNAs implicated in CRC radiosensitivity, miRNA sequencing was used to monitor their expression in HCT116 and CCL-244 cells 48 h after radiation treatment. The results showed that radiation affected the expression of various miRNAs in CRC cells. A total of 33 miRNAs showed dysregulation of expression in CCL-244, with 13 miRNAs being up-regulated and 20 being down-regulated. In HCT116 cells, 37 miRNAs showed dysregulation of expression (20 miRNAs were up-regulated and 17 were down-regulated; Figure 2A). Table 1 and Figure 2B list the top 20 miRNAs that showed considerable up-regulation/down-regulation after irradiation. For this study, we selected miR-100 as the target miRNA, as it was significantly differentially expressed before and after irradiation in CCL-244 cells. In addition, the difference in this expression was statistically significant before and after irradiation in CCL-244 cells (P = 8.38 × 10-11) but not HCT116 cells (P = 0.11, Figure 2C). This dysregulation of miR-100 in cells with different radiosensitivities was noteworthy. Quantitative real-time PCR was used to confirm the sequencing results (P = 0.035, Figure 2D).
Figure 2.
miRNA sequencing results of colon cancer cells before and after X-ray irradiation. A. miRNA expression profiles of CCL-244 and HCT116 cells before and after X-ray irradiation determined by miRNA sequencing; the heatmap was generated with miRNAs whose expressions were down- or up-regulated more than 2-fold compared to controls. B. Ten miRNAs were differentially expressed in CCL-244 or HCT116 cells after exposure to 8-Gy X-ray irradiation compared with nonirradiated control cells. The irradiated/nonirradiated expression ratio was expressed as Log2. The bars indicate the means for this experiment. C. Expression of miR-100 in CCL-244 cells before and after exposure to 8-Gy X-ray irradiation (**P < 0.01); expression of miR-100 in HCT116 cells before and after exposure to 8-Gy X-ray irradiation (P > 0.05). D. Quantitative real-time PCR to confirm the expression of miR-100 after transfection and to examine consistency with sequencing results (*P < 0.05).
Table 1.
Summary of significantly differentially expressed miRNAs in irradiated CCL244 and HCT116 compared with no irradiated CCL244 and HCT116 respectively
| Cells | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| CCL244 | HCT116 | ||||||||||
|
|
|
||||||||||
| Upregulated miRNAs | Downregulated miRNAs | Upregulated miRNAs | Downregulated miRNAs | ||||||||
|
|
|
|
|
||||||||
| miRNA ID | Fold Changea | P-value | miRNA ID | Fold Changea | P-value | miRNA ID | Fold Changea | P-value | miRNA ID | Fold Changea | P-value |
| hsa-miR-371a-5p | 2.549911 | 0.021014 | hsa-miR-125b-1-3p | 0.007523 | 3.43×10^-5 | hsa-miR-6128 | 2.247243 | 0.047018 | hsa-miR-380-5p | 0.239406 | 0.006182 |
| hsa-miR-373-3p | 2.573817 | 0.023362 | hsa-miR-130a-3p | 0.014726 | 7.47×10^-4 | hsa-miR-30c-1-3p | 2.422787 | 0.0399391 | hsa-miR-193a-5p | 0.250288 | 0.010183 |
| hsa-miR-222-5p | 2.768475 | 0.035264 | hsa-miR-100* | 0.025431 | 8.3×10^-11 | hsa-miR-30a-5p | 2.539796 | 0.0128789 | hsa-miR-20a-5p | 0.313987 | 7.52×10^-5 |
| hsa-miR-371b-3p | 3.271834 | 0.011606 | hsa-miR-145-5p | 0.038029 | 8.81×10^-4 | hsa-miR-30a-3p | 2.604347 | 0.0447689 | hsa-miR-3676-3p | 0.318393 | 0.006701 |
| hsa-miR-372 | 3.350868 | 0.009543 | hsa-miR-125b-5p | 0.048064 | 3.21×10^-4 | hsa-miR-409-3p | 2.87287 | 0.0413885 | hsa-miR-1247-3p | 0.339676 | 0.038251 |
| hsa-miR-4516 | 4.251587 | 0.036335 | hsa-miR-1246 | 0.097547 | 7.35×10^-3 | hsa-miR-589-3p | 3.120256 | 0.0196415 | hsa-miR-3529-3p | 0.379937 | 0.007554 |
| hsa-miR-4802-3p | 7.267247 | 0.026147 | hsa-miR-9-5p | 0.140838 | 1.06×10^-2 | hsa-miR-4521 | 4.12975 | 0.0214253 | hsa-miR-15b-5p | 0.385549 | 0.005794 |
| hsa-miR-380-5p | 8.766837 | 0.001643 | hsa-miR-181a-3p | 0.146507 | 2.94×10^-2 | hsa-miR-3680-5p | 4.380038 | 0.0032954 | hsa-miR-7-5p | 0.398485 | 0.025984 |
| hsa-miR-5007-3p | 12.45814 | 0.016728 | hsa-miR-92b-3p | 0.147947 | 4.25×10^-4 | hsa-miR-205-5p | 5.506333 | 0.0433714 | hsa-miR-99a-5p | 0.439371 | 0.010586 |
| hsa-miR-205-5p | 22.1478 | 0.002933 | hsa-miR-181a-5p | 0.27454 | 2.74×10^-2 | hsa-miR-4709-5p | 5.506333 | 0.0100935 | hsa-miR-17-5p | 0.492995 | 0.017785 |
cancer tissues vs adjacent normal tissues;
hsa-miR-100.
MiR-100 was significantly down-regulated in CRC tissues compared with matched noncancerous colorectal tissues
Quantitative real-time PCR was used to detect miR-100 expression in CRC tissues and matched noncancerous tissues from 30 CRC patients. Table 2 shows the clinical characteristics of these patients. PCR showed lower expression of miR-100 in tumor tissues than in normal tissues (P < 0.01, Figure 3). Therefore, dysregulation of miR-100 expression may be associated with certain biological behaviors of CRC.
Table 2.
Clinical characteristic of the CRC patients
| Clinical parameters | Numbers |
|---|---|
| Cases | 30 |
| Age | |
| < 65 | 14 |
| ≥ 65 | 16 |
| Tumor size | |
| Small size (< 5 cm) | 20 |
| Large size ( ≥ 5 cm) | 10 |
| Gender | |
| Male | 15 |
| Female | 15 |
| Invasion levels | |
| Mucosa | 2 |
| Submucosa | 4 |
| Muscle | 7 |
| Serosa | 17 |
| TNM stage | |
| Stage 1-2 | 21 |
| Stage 3-4 | 9 |
| Lymph node metastasis | |
| Positive | 17 |
| Negative | 13 |
| Grade of tumors | |
| Low grade | 5 |
| Intermediate grade | 23 |
| High grade | 2 |
| Vascular invasion | |
| Positive | 14 |
| Negative | 16 |
| Perineural invasion | |
| Positive | 7 |
| Negative | 23 |
Figure 3.
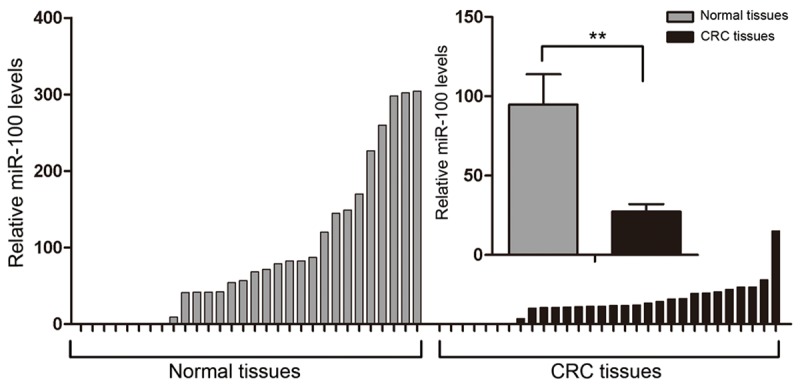
Different expression of miR-100 in CRC tissues compared with matched noncancerous colorectal tissues. Quantitative real-time PCR was performed to compare the expression of miR-100 between 30 CRC tissues and matched noncancerous colorectal tissues. U6 RNA was used as an internal control. The mean ΔCt of miR-100 in the 30 CRC tissues was compared with that in the matched noncancerous colorectal tissues (**P < 0.01). The mean and standard deviation of expression levels relative to U6 expression levels are shown and normalized to the expression in the normal tissue of each matched pair.
MiR-100 modulated the radiosensitivity of CCL-244 cells
The results of miRNA sequencing showed down-regulation of miR-100 in CCL-244 cells in response to radiation, while miR-100 showed no such response in HCT116 cells. We surmised that miR-100 could modulate the radiation response of CCL-244 cells. MiR-100 mimics were transiently transfected into CCL-244 cells to up-regulate miR-100, whereas negative control miR-100 mimics were transiently transfected as control. Quantitative real-time PCR confirmed the transfer of miR-100 mimics and miRNA negative control into the CCL-244 cells, as expected (P = 0.04, Figure 4A). After transfection, CCL-244 cells were exposed to 8-Gy X-ray irradiation. After 48 h, CCK-8 and colony formation assays were performed. The results of the CCK-8 assay showed that the survival rate of CCL-244 cells significantly decreased after miR-100 up-regulation as compared to the mock transfected cells (P = 0.029) and negative control miR-100 mimics (P < 0.01, Figure 4B). The results of the colony formation assay showed that the colony survival fraction significantly reduced after miR-100 up-regulation (Figure 4C). Taken together, the results suggest that miR-100 plays an important role in the radiosensitivity of CCL-244 cells.
Figure 4.
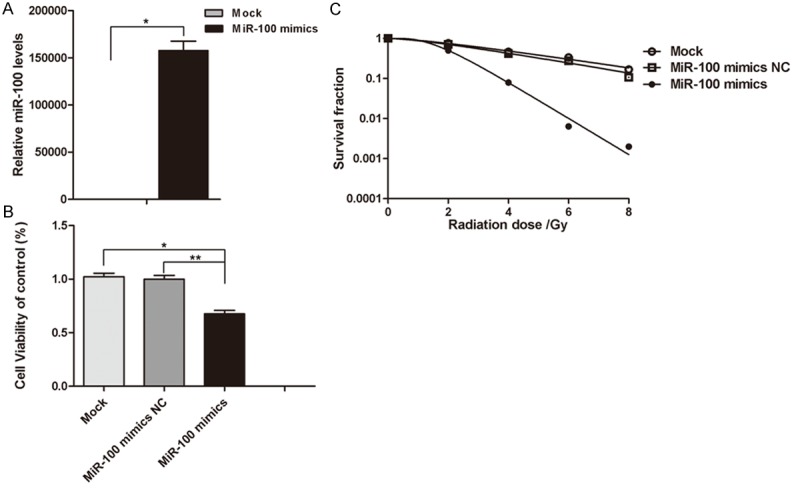
miR-100 regulated the radiosensitivity of CRC cells CCL-244. A. Quantitative real-time PCR was used to confirm the transfection efficiency. U6 RNA was used as an internal control. The mean ΔCt of miR-100 in CCL-244 cells treated with miR-100 mimics and negative control miR-100 mimics (*P < 0.05). B. CCL-244 cells treated with miR-100 mimics and negative control miR-100 mimics. After 24 h, the cells were subjected to 8-Gy irradiation. After 48 h, CCK-8 was used to detect cell viabilities (miR-100 mimics compared with miR-100 negative control, *P < 0.05 or untransfected, **P < 0.01). These results are representative of at least three separate experiments. C. CCL-244 cells treated with miR-100 mimics and negative control miR-100 mimics. After 24 h, the cells were subjected to 0, 2, 4, 6 and 8 Gy of irradiation. After 14 days, the colony formation rate in different treatment groups was examined.
MiR-100 promoted X-ray-induced apoptosis of CCL-244 cells and regulated the expression of apoptosis-related proteins
Apoptosis is one of the most important impairments occurring as a result of radiation therapy in tumors. In order to investigate whether miR-100 promotes the radiosensitivity of CCL-244 cells by inducing apoptosis, the cells were transfected with miR-100 mimic or negative control miRNA mimics as control. After incubation for 48 h, the cells were exposed to different doses of irradiation (0 or 8 Gy) and subjected to Annexin V/PI double staining and FACS analysis 48 h later, including nontransfected cells. The results showed that miR-100 not only significantly increased X-ray-induced apoptosis in CCL-244 cells as compared to nontransfected cells (P = 0.006) and negative control miR-100 mimics (P = 0.021) but also considerably improved the apoptosis rate of nonirradiated CCL-244 cells as compared to nontransfected cells (P = 0.046) and negative control miR-100 mimics (P = 0.035) (Figure 5A). Further, the expression of apoptosis-related molecules such as P53, Bcl-2, NF-κB, and caspase-3 was evaluated by western blot analysis. At 48 h after transfection, the cells were subjected to 8-Gy radiation and collected 48 h after irradiation. As shown in Figure 5B, miR-100 increased the expressions of pro-apoptotic proteins P53 and Caspase-3, and decreased the expressions of anti-apoptotic proteins Bcl-2 and NF-κB regardless of irradiation. Taken together, these results demonstrated that miR-100 attenuated X-ray irradiation-induced apoptosis in CCL-244 cells by regulating the expression of apoptosis-related proteins.
Figure 5.
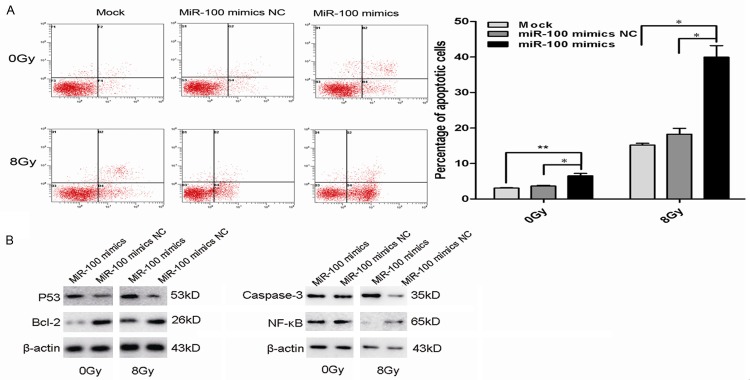
miR-100 promoted X-ray-induced apoptosis of CCL-244 cells. A. Flow cytometry assay to determine the apoptosis of CCL-244 cells transfected with miR-100 mimics or negative control miR-100 mimics and non-transfected cells. Percentage of apoptotic cells in non-transfected and transfected cells subjected to 0 and 8-Gy irradiation. Data are means ± SEM of three independent experiments. miR-100 significantly increased the X-ray-induced apoptosis of CCL-244 cells compared with non-transfected (**P < 0.01) and negative control miR-100 mimics (*P < 0.05) as well as the apoptosis of nonirradiated CCL-244 cells compared with non-transfected (*P < 0.05) and negative control miR-100 mimics (*P < 0.05). B. Western blot analysis of apoptotic marker proteins p53, Caspase-3, Bcl-2 or NF-κB in CCL-244 cells with miR-100 in transfected and non-transfected cells before and after exposure to 8-Gy X-ray irradiation. β-Actin was used as an internal control.
MiR-100 increased the number of γ-H2AX foci
It is known that irradiation causes DNA DSBs, which subsequently bring about the phosphorylation of γ-H2AX at serine 139 (γ-H2AX). At 48 h after the transfection of miR-100 mimics, the cells were subjected to 4-Gy irradiation, and γ-H2AX foci were detected at different time points. As shown in Figure 6A, before ionizing radiation, the γ-H2AX foci levels in each group were extremely low. About 30 min following 4-Gy irradiation, a rapid induction of γ-H2AX foci was observed, with up to more than 60 foci per cell (Figure 6A and 6B). The average number of foci then began to gradually decrease until 16 hours after irradiation. However, compared to the nontransfected and negative control miR-100 mimic groups, the group transfected with miR-100 mimics showed a significantly higher number of foci. After 2 h, the average number of foci in the group transfected with miR-100 mimics was 37, and that in the negative control miR-100 mimics group and the nontransfected group was 20 (P = 0.012) and 17 (P = 0.002), respectively (Figure 6B). After 8 and 16 h, the number of foci in the group transfected with miR-100 mimics was significantly greater than that in the other two groups (Figure 6B). These results suggest that miR-100 modulates the sensitization of CCL-244 cells to irradiation by augmenting irradiation-induced DSBs.
Figure 6.
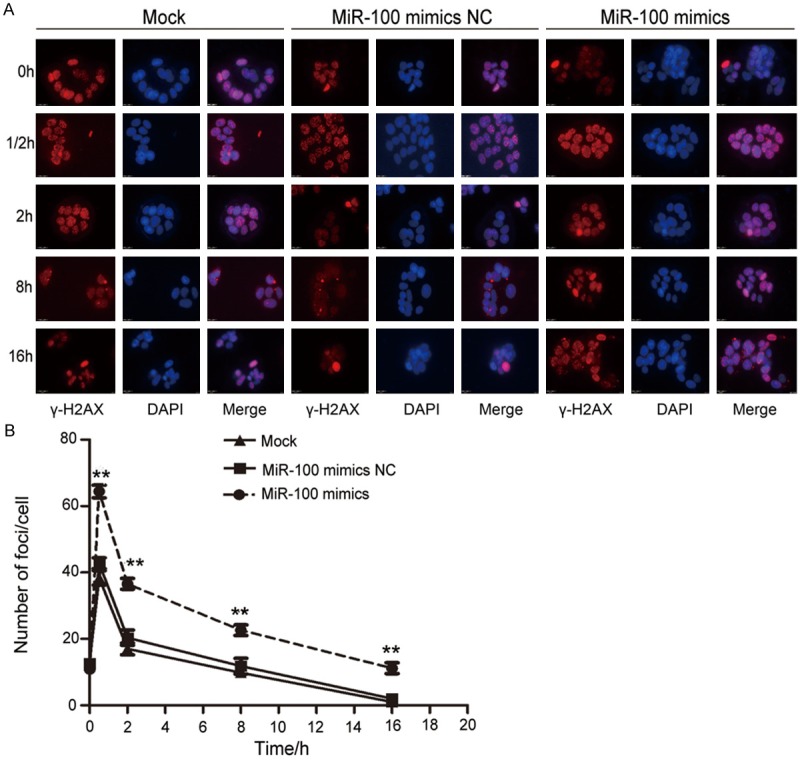
miR-100 increased γ-H2AX foci caused by X-ray irradiation and retarded DNA double strand breaks repair. A. Representative images of γ-H2AX foci for miR-100 mimics, miR-100 negative control, and non-transfected treated groups exposed to 4-Gy X-ray irradiation at different time points. B. Representative images of γ-H2AX foci in different groups of cells after 4-Gy X-ray irradiation. CCL-244 cells were stained for γ-H2AX at the indicated times and the mean number of γ-H2AX foci per cell (foci/cell) were then counted (**P < 0.01).
Discussion
Currently, radiotherapy is one of the principal treatment modalities for CRC. However, the radiation resistance of tumors has become an important concern among clinicians. Variable susceptibility of cells to radiotherapy is one of the main reasons for radioresistance. miRNAs have been reported to modulate radiosensitivity and have the potential to improve the efficacy of radiotherapy [15]. In this study, HCT116 and CCL-244 were identified as the most and least radiosensitive cell lines, respectively after the radiosensitivities of seven common CRC cell lines was compared. A microarray analysis was then performed to examine the miRNA expression profiles of CCL-244 and HCT116 cell lines before and after X-ray irradiation. MiR-100 was identified as a critical miRNA that was significantly down-regulated after irradiation in CCL-244 cells, suggesting its potential to regulate the radiosensitivity of these cells. Some previous studies have shown that miR-100 could significantly increase the sensitivity of CCL-244 cells to radiation, a finding consistent with our results. Studies have also shown that this increase in radiosensitivity could probably be brought about via radiation-induced apoptosis. In addition, we found that the expression of miR-100 in tumor tissues was significantly lower than that in normal tissues.
Many studies have reported that radiation alters miRNA expression profiles in tumor cells in glioblastoma, lung cancer, prostate cancer, and non-Hodgkin’s lymphoma. However, there are limited reports regarding CRC. Shin et al found that miRNAs are significantly influenced by irradiation in HCT116 human colon carcinoma cells [16]; these altered miRNAs may be related to cell radiosensitivity, as suggested by some previous studies on gliomas, prostate and lung cancers, and oral squamous cell carcinoma. Yang et al found that miR-210 regulates radiosensitivity in hypoxic human hepatoma by targeting the mitochondrion-associated, 3 (AIFM3) [17]. Further, miR-31 modulates the response of esophageal adenocarcinoma to irradiation, possibly via regulation of DNA repair [18]. Thus, the targets through which different miRNAs affect the radiosensitivity of tumor cells differ widely. Zhang et al [19] revealed that miR-221 and miR-222 regulate the radioresistance of gastric carcinoma cells by targeting PTEN. MiRNA-324-3p regulates the radioresistance of nasopharyngeal carcinoma by directly targeting WNT2B [20]. Further, one study reported that miRNAs may mediate signaling pathways in response to radiation [21].
Most of the current individual studies have not utilized the high-throughput screening method to reveal the miRNA profile. The present study is novel because we used advanced high-throughput sequencing to select miRNAs highly correlated with CRC radiosensitivity as the research focus.
We selected relatively insensitive colon cancer cells CCL-244 to simulate the radiation tolerance of CRC. We found that the difference in miR-100 expression was statistically significant (P < 0.05) before and after irradiation by detecting the expression profiles of miRNAs in CCL-244 cells. Studies have reported the involvement of miR-100 in carcinogenesis, progression, and prognosis of many kinds of cancers, such as prostate cancer; hepatocellular carcinoma; renal cell carcinoma; pancreatic, bladder, lung, breast, and cervical cancers; gliomas; and acute myeloid leukemia [22-37]. However, the impacts of miR-100 on the development of CRC and the related mechanisms have rarely been reported. Boni et al found that the single-nucleotide polymorphism rs1834306, located in the pri-miR-100 gene, was significantly correlated with a longer time to progression [38]. Thus, the clinical importance of miR-100 reiterates our purpose behind focusing on this miRNA.
Apoptosis may be one of the mechanisms of ionizing radiation-induced cell death [39], and it is believed that miRNAs regulate radiation-induced apoptosis. Wu et al showed that miR-148b increases the radiosensitivity of non-Hodgkin’s lymphoma cells, probably by promoting radiation-induced apoptosis [15]. Similarly, Wang et al found that miR-185 enhances radiation-induced apoptosis in gastric cancer 786-O cells by repressing the apoptosis-related ATR (ATM- and Rad3-related) pathway [40]. Some studies have shown that miRNA-21 contributes to the radioresistance of tumors by blocking the expression of critical apoptosis-related genes such as caspase-3 and PARP-1 [41]. Further, abnormal regulation of apoptosis signaling pathways occurs often in tumor cells, leading to radioresistance, which induces apoptosis. Therefore, an increasing number of cancer biologists are shifting their attention to the apoptosis mechanism in tumor cells [42]. Various signaling molecules are involved in apoptosis signaling pathways, which include the promotion of apoptosis by P53, Bax, and caspase-3 and the inhibition of apoptosis by Bcl-2 and NF-κB [43-47]. In this study, we examined apoptosis in CCL-244 cells in order to reveal the role played by miR-100 in the radiosensitivity of CRC. We found that CCL-244 cells transfected with miR-100 had a significantly higher percentage of apoptotic cells than the negative control-transfected group (P < 0.05). In addition, miR-100 increased the expressions of the pro-apoptotic proteins P53 and caspase-3 and decreased the expressions of the anti-apoptotic proteins Bcl-2 and NF-κB irrespective of radiation. Collectively, these data suggest that miR-100 increases radiosensitivity by promoting apoptosis.
The induction of DSBs is also an important mechanism of radiation-induced cell damage [48]. Radiation-induced γ-H2AX foci are representative of DSBs. In this study, we found that up-regulation of miR-100 significantly increased the number of γ-H2AX foci detected by immunofluorescence analysis in a time-dependent manner. These results confirm the notion that miR-100 plays an important role in the regulation of radiosensitivity of CCL-244 cells in CRC.
Conclusion
Our results demonstrated that up-regulation of miR-100 significantly increased the radiosensitivity of CCL-244 cells in CRC. The involvement of miR-100 in radiation-induced apoptosis and DNA DSBs of CRC cells may be the primary mechanism involved in the radiosensitivity of CRC. In addition, our clinical sample test results showed that miR-100 expression was significantly lower in tumor tissue than in normal tissue, a finding that may be useful to study the functional mechanism(s) underlying the oncogenic role of miR-100. MiR-100 may serve as a clinical target for reducing radiation resistance. However, further studies in vivo are warranted to improve our understanding of miR-100 regulation mechanisms involved in the radiosensitivity of CRC.
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (Grant No: 81172348, 81301933 and 81472917), Health Research Projects in Jiangsu Province (H201313), the projects of Suzhou Technology Bureau (SYSD2013090, SS0834) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure of conflict of interest
The authors have no conflict of interest.
Abbreviations
- miRNA
microRNA
- DSB
DNA double-strand break
- NCCN
National Comprehensive Cancer Network
- RISC
RNA-induced silencing complex
- 3’ UTR
3’Untranslated Region
- ICAM2
intercellular adhesion molecule-2
- OSCC
oral squamous cell carcinoma
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Engstrom PF, Arnoletti JP, Benson AB 3rd, Chen YJ, Choti MA, Cooper HS, Covey A, Dilawari RA, Early DS, Enzinger PC, Fakih MG, Fleshman J Jr, Fuchs C, Grem JL, Kiel K, Knol JA, Leong LA, Lin E, Mulcahy MF, Rao S, Ryan DP, Saltz L, Shibata D, Skibber JM, Sofocleous C, Thomas J, Venook AP, Willett C National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: rectal cancer. J Natl Compr Canc Netw. 2009;7:838–81. doi: 10.6004/jnccn.2009.0057. [DOI] [PubMed] [Google Scholar]
- 3.Xu X, Yang X, Xing C, Zhang S, Cao J. miRNA: The nemesis of gastric cancer (Review) Oncol Lett. 2013;6:631–41. doi: 10.3892/ol.2013.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23:997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- 5.Shiiba M, Shinozuka K, Saito K, Fushimi K, Kasamatsu A, Ogawara K, Uzawa K, Ito H, Takiguchi Y, Tanzawa H. MicroRNA-125b regulates proliferation and radioresistance of oral squamous cell carcinoma. Br J Cancer. 2013;108:1817–21. doi: 10.1038/bjc.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 7.Cui SY, Huang JY, Chen YT, Song HZ, Feng B, Huang GC, Wang R, Chen LB, De W. Let-7c Governs the Acquisition of Chemo- or Radioresistance and Epithelial-to-Mesenchymal Transition Phenotypes in Docetaxel-Resistant Lung Adenocarcinoma. Mol Cancer Res. 2013;11:699–713. doi: 10.1158/1541-7786.MCR-13-0019-T. [DOI] [PubMed] [Google Scholar]
- 8.Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noël A, Pouysségur J, Barbry P, Mazure NM, Mari B. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013;4:e544. doi: 10.1038/cddis.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol Cell Biochem. 2013;372:35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 10.Liu YJ, Lin YF, Chen YF, Luo EC, Sher YP, Tsai MH, Chuang EY, Lai LC. MicroRNA-449a enhances radiosensitivity in CL1-0 lung adenocarcinoma cells. PLoS One. 2013;8:e62383. doi: 10.1371/journal.pone.0062383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Rocca G, Badin M, Shi B, Xu SQ, Deangelis T, Sepp-Lorenzinoi L, Baserga R. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol. 2009;220:485–91. doi: 10.1002/jcp.21796. [DOI] [PubMed] [Google Scholar]
- 12.Manne U, Shanmugam C, Bovell L, Katkoori VR, Bumpers HL. miRNAs as biomarkers for management of patients with colorectal cancer. Biomark Med. 2010;4:761–70. doi: 10.2217/bmm.10.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alison MR, Lin WR, Lim SM, Nicholson LJ. Cancer stem cells: in the line of fire. Cancer Treat Rev. 2012;38:589–98. doi: 10.1016/j.ctrv.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Liu GL, Liu SH, Wang CX, Xu YL, Ying Y, Mao P. MicroRNA-148b enhances the radiosensitivity of non-Hodgkin’s Lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res. 2012;53:516–25. doi: 10.1093/jrr/rrs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin S, Cha HJ, Lee EM, Jung JH, Lee SJ, Park IC, Jin YW, An S. MicroRNAs are significantly influenced by p53 and radiation in HCT116 human colon carcinoma cells. Int J Oncol. 2009;34:1645–52. doi: 10.3892/ijo_00000295. [DOI] [PubMed] [Google Scholar]
- 17.Yang W, Sun T, Cao J, Liu F, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318:944–54. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Lynam-Lennon N, Reynolds JV, Marignol L, Sheils OM, Pidgeon GP, Maher SG. MicroRNA-31 modulates tumour sensitivity to radiation in oesophageal adenocarcinoma. J Mol Med (Berl) 2012;90:1449–58. doi: 10.1007/s00109-012-0924-x. [DOI] [PubMed] [Google Scholar]
- 19.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. MicroRNA-221 and microRNA- 222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li G, Liu Y, Su Z, Ren S, Zhu G, Tian Y, Qiu Y. MicroRNA-324-3p regulates nasopharyngeal carcinoma radioresistance by directly targeting WNT2B. Eur J Cancer. 2013;49:2596–607. doi: 10.1016/j.ejca.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in MicroRNA expression patterns in human fibroblasts after low-LET radiation. J Cell Biochem. 2008;105:824–34. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- 22.Leite KR, Sousa-Canavez JM, Reis ST, Tomiyama AH, Camara-Lopes LH, Sanudo A, Antunes AA, Srougi M. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol Oncol. 2011;29:265–9. doi: 10.1016/j.urolonc.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Leite KR, Tomiyama A, Reis ST, Sousa-Canavez JM, Sanudo A, DallOglio MF, Camara-Lopes LH, Srougi M. MicroRNA-100 expression is independently related to biochemical recurrence of prostate cancer. J Urol. 2011;185:1118–22. doi: 10.1016/j.juro.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Giangreco AA, Vaishnav A, Wagner D, Finelli A, Fleshner N, Van der Kwast T, Vieth R, Nonn L. Tumor suppressor microRNAs, miR-100 and -125b, are regulated by 1,25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Prev Res (Phila) 2013;6:483–94. doi: 10.1158/1940-6207.CAPR-12-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leite KR, Morais DR, Reis ST, Viana N, Moura C, Florez MG, Silva IA, Dip N, Srougi M. MicroRNA 100: a context dependent miRNA in prostate cancer. Clinics (Sao Paulo) 2013;68:797–802. doi: 10.6061/clinics/2013(06)12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen P, Zhao X, Ma L. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Mol Cell Biochem. 2013;383:49–58. doi: 10.1007/s11010-013-1753-0. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Chen L, Meng J, Chen M, Zhuang L, Zhang L. Overexpression of microRNA-100 predicts an unfavorable prognosis in renal cell carcinoma. Int Urol Nephrol. 2013;45:373–9. doi: 10.1007/s11255-012-0374-y. [DOI] [PubMed] [Google Scholar]
- 28.Huang JS, Egger ME, Grizzle WE, McNally LR. MicroRNA-100 regulates IGF1-receptor expression in metastatic pancreatic cancer cells. Biotech Histochem. 2013;88:397–402. doi: 10.3109/10520295.2012.762460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C, Zeng Q, Xu W, Jiao L, Chen Y, Zhang Z, Wu C, Jin T, Pan A, Wei R, Yang B, Sun Y. miRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTOR. Mol Cancer Ther. 2013;12:207–19. doi: 10.1158/1535-7163.MCT-12-0273. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira JC, Brassesco MS, Morales AG, Pezuk JA, Fedatto PF, da Silva GN, Scrideli CA, Tone LG. MicroRNA-100 acts as a tumor suppressor in human bladder carcinoma 5637 cells. Asian Pac J Cancer Prev. 2011;12:3001–4. [PubMed] [Google Scholar]
- 31.Wang S, Xue S, Dai Y, Yang J, Chen Z, Fang X, Zhou W, Wu W, Li Q. Reduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancer. Diagn Pathol. 2012;7:159. doi: 10.1186/1746-1596-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Lu KH, Liu ZL, Sun M, De W, Wang ZX. MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1. BMC Cancer. 2012;12:519. doi: 10.1186/1471-2407-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebeshuber CA, Martinez J. miR-100 suppresses IGF2 and inhibits breast tumorigenesis by interfering with proliferation and survival signaling. Oncogene. 2013;32:3306–10. doi: 10.1038/onc.2012.372. [DOI] [PubMed] [Google Scholar]
- 34.Li BH, Zhou JS, Ye F, Cheng XD, Zhou CY, Lu WG, Xie X. Reduced miR-100 expression in cervical cancer and precursors and its carcinogenic effect through targeting PLK1 protein. Eur J Cancer. 2011;47:2166–74. doi: 10.1016/j.ejca.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 35.Ng WL, Yan D, Zhang X, Mo YY, Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 2010;9:1170–5. doi: 10.1016/j.dnarep.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 36.Bai J, Guo A, Hong Z, Kuai W. Upregulation of microRNA-100 predicts poor prognosis in patients with pediatric acute myeloid leukemia. Onco Targets Ther. 2012;5:213–9. doi: 10.2147/OTT.S36017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng YS, Zhang H, Zhang XJ, Feng DD, Luo XQ, Zeng CW, Lin KY, Zhou H, Qu LH, Zhang P, Chen YQ. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene. 2012;31:80–92. doi: 10.1038/onc.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boni V, Zarate R, Villa JC, Bandrés E, Gomez MA, Maiello E, Garcia-Foncillas J, Aranda E. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J. 2011;11:429–36. doi: 10.1038/tpj.2010.58. [DOI] [PubMed] [Google Scholar]
- 39.Shinomiya N. New concepts in radiation-induced apoptosis: ‘premitotic apoptosis’ and ‘postmitotic apoptosis’. J Cell Mol Med. 2001;5:240–53. doi: 10.1111/j.1582-4934.2001.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, He J, Su F, Ding N, Hu W, Yao B, Wang W, Zhou G. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013;4:e699. doi: 10.1038/cddis.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang XC, Wang W, Zhang ZB, Zhao J, Tan XG, Luo JC. Overexpression of miRNA-21 promotes radiation-resistance of non-small cell lung cancer. Radiat Oncol. 2013;8:146. doi: 10.1186/1748-717X-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulze-Bergkamen H, Krammer PH. Apoptosis in cancer--implications for therapy. Semin Oncol. 2004;31:90–119. doi: 10.1053/j.seminoncol.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–9. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 44.Kitada S, Krajewski S, Miyashita T, Krajewska M, Reed JC. Gamma-radiation induces upregulation of Bax protein and apoptosis in radiosensitive cells in vivo. Oncogene. 1996;12:187–92. [PubMed] [Google Scholar]
- 45.Waterhouse NJ, Finucane DM, Green DR, Elce JS, Kumar S, Alnemri ES, Litwack G, Khanna K, Lavin MF, Watters DJ. Calpain activation is upstream of caspases in radiation-induced apoptosis. Cell Death Differ. 1998;5:1051–61. doi: 10.1038/sj.cdd.4400425. [DOI] [PubMed] [Google Scholar]
- 46.Mirkovic N, Voehringer DW, Story MD, McConkey DJ, McDonnell TJ, Meyn RE. Resistance to radiation-induced apoptosis in Bcl-2-expressing cells is reversed by depleting cellular thiols. Oncogene. 1997;15:1461–70. doi: 10.1038/sj.onc.1201310. [DOI] [PubMed] [Google Scholar]
- 47.Raffoul JJ, Wang Y, Kucuk O, Forman JD, Sarkar FH, Hillman GG. Genistein inhibits radiation-induced activation of NF-kappaB in prostate cancer cells promoting apoptosis and G2/M cell cycle arrest. BMC Cancer. 2006;6:107. doi: 10.1186/1471-2407-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modesti M, Kanaar R. DNA repair: spot(light)s on chromatin. Curr Biol. 2001;11:R229–32. doi: 10.1016/s0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]