Abstract
We investigated the effect of AGG interruptions on fragile X repeat instability upon transmission of fragile X intermediate and small premutation alleles with 45–69 CGG repeats. The FMR1 repeat structure was determined for 375 mothers, 48 fathers, and 538 offspring (457 maternal and 81 paternal transmissions) using a novel PCR assay to determine repeat length and AGG interruptions. The number of AGG interruptions and the length of uninterrupted CGG repeats at the 3′ end were correlated with repeat instability on transmission. Maternal alleles with no AGGs conferred the greatest risk for unstable transmissions. All nine full mutation expansions were inherited from maternal alleles with no AGGs. Furthermore, the magnitude of repeat expansion was larger for alleles lacking AGG interruptions. Transmissions from paternal alleles with no AGGs also exhibited greater instability than those with one or more AGGs. Our results demonstrate that characterization of the AGG structure within the FMR1 repeat allows more accurate risk estimates of repeat instability and expansion to full mutations for intermediate and small premutation alleles.
Keywords: fragile X, FMR1, trinucleotide repeat instability
INTRODUCTION
The fragile X syndrome (FXS, OMIM 300624), a common form of X-linked intellectual and developmental disability, occurs in approximately 1/4,000 males and 1/8,000 females [Crawford et al., 2001]. The mutation is an expansion of a CGG repeat in the 5′ untranslated region of the FMR1 gene [Kremer et al., 1991; Oberlé et al., 1991; Verkerk et al., 1991; Yu et al., 1991] that prevents expression of the FMR1 protein through methylation of the promoter region and results in the syndrome in affected individuals. The polymorphic CGG repeat region has been divided into four classes based on repeat length [Maddalena et al., 2001]: normal (6–44 repeats), intermediate (45–54 repeats), premutation (55–200 repeats) and full mutation (>200 repeats). Normal alleles are highly stable when passed from parent to child whereas some intermediate alleles may exhibit intergenerational instability. Premutation alleles are unstable and may expand to the full mutation in one generation. Although paternal premutation transmission to a daughter may be unstable, expansion to a full mutation occurs almost exclusively when a premutation allele is transmitted from mother to child, not from father to daughter. As documented in many studies, the risk for full mutation expansion in female transmissions of premutation alleles increases rapidly with increased repeat length [Fu et al., 1991; Nolin et al., 2003]. In families where full mutation expansions have occurred, large changes in repeat size are frequently observed in other transmissions within the family.
In the past, expanded alleles were ascertained from individuals affected with FXS. Thus, the alleles in these families were known to be unstable and capable of expanding to a full mutation. Recently, women in the United States and other countries have been screened for their fragile X carrier status, which has resulted in the identification of intermediate and premutation alleles with no known history of instability. Studies in Israel [Berkenstadt et al., 2007] reported a carrier frequency of ~1:150 for women with premutation alleles while in Canada a frequency of 1:259 has been observed [Rousseau et al., 1995]. In the United States estimates range from 1:151 to 1:382 [Cronister et al., 2005, 2008; Iong et al., 2011; Seltzer et al., 2012] with frequencies ~1:200 identified in recent studies. One U.S. prevalence study determined that 75% of newly identified premutation alleles have fewer than 70 repeats [Hantash et al., 2011]. While the prevalence of intermediate and small premutation alleles is high, prenatal studies of these maternal alleles have shown that many undergo little or no change in repeat size on transmission [Nolin et al., 2011]. The expansion risks for these newly identified alleles contrasts sharply with the risk estimates based on studies of families that include an affected individual. Thus, increased screening of women for their fragile X status has led to the identification of intermediate and small premutation alleles whose risk for expansion is unknown.
The number of AGG interruptions within the repeat region has been linked to repeat instability and risk of expansion to a full mutation. Eichler et al. [1994, 1996] examined the structure of the FMR1 repeat in the general population and families with FXS and observed interspersed AGGs within the repeat region in nearly all alleles in the general population. The most frequent allele pattern was two AGGs at positions 10 or 11, and 20 or 21 repeats. Unstable alleles in families with FXS contained no or few AGGs in the 5′ region of the repeat and long stretches of uninterrupted CGG sat the 3′ end of the repeat. The authors suggested that AGG interruptions that differentiate CGG repeat alleles are responsible for most of the variance in stability. This hypothesis has been difficult to test, however, because of the technical challenges in analyzing the AGG structures in the two X chromosomes in females.
In this study, we used a PCR assay based on triplet CGG repeat primed PCR [Chen et al., 2010; Nolin et al., 2011] to detect AGG interruptions in males and females with intermediate and small premutation alleles. We examined the association of the repeat instability on transmission with repeat length, AGG structure and 3′ uninterrupted repeat length in a large cohort of maternal and paternal alleles to derive better predictors for expansion of the repeat region. The results of our study have immediate implications in predicting risk of CGG repeat expansion based on AGG status as well as repeat length for intermediate and small premutation alleles.
MATERIALS AND METHODS
Subjects
The 377 families in the study with 45–69 CGG repeats in the FMR1 gene were ascertained as follows: 214 from population screening, 70 from an individual with developmental disabilities of unknown etiology, 48 with a history of FXS, three from individuals with neurological symptoms, one from an individual with premature ovarian insufficiency, and 41 for whom ascertainment was unknown. Purified genomic DNA was collected under Institutional Review Board approvals from four U.S. institutions (New York State Institute for Basic Research in Developmental Disabilities, Rush University Medical School, Emory University, and the UC Davis–MIND Institute).
PCR Protocol and Data Analysis
The AGG interruption pattern within the FMR1 repeat was determined at Asuragen (Austin, TX). Three PCR assays were performed to resolve the AGG status of each allele. The assays included a CGG repeat primed PCR [Tassone et al., 2008; Chen et al., 2010] and two additional PCR assays derived from a previously validated long-read PCR technique [Filipovic-Sadic et al., 2010] that resolved ambiguities in the repeat primed assay. Transmission data for the 377 families in this study were derived from CGG repeat length and AGG analysis of 971 DNA samples. The DNA samples were diluted to 20 ng/µl and a total of 120 ng of DNA was used across the PCR reactions. The PCR products, approximately 2,900 in total, were analyzed using a 3500×l Genetic Analyzer (Life Technologies, Carlsbad, CA). In general, samples were batched in groups of 94 samples and two controls per microtiter plate. Using three thermal cyclers and one 3500×l, the turnaround time for data acquisition was 24–36 hr per sample batch. Across 971 samples, genotypes were resolved for 928 in the first run, resulting in an initial pass rate of 95.6%. Only 1.6% (16/971) failed with all PCR assays. In cases where a failure was noted in any assay from the first run, interpretable data were produced for 88% (38/43) of those samples after repeat testing. Thus, 99.5% of all samples (966/971) were assigned a CGG repeat length and AGG genotype using only PCR-based data.
The specific AGG structure of each allele was deduced from the electropherogram data as previously described [Chen et al., 2010]. To evaluate the performance of this method for determining CGG repeat length and AGG genotyping, PCR results were compared to DNA sequencing using a separate set of 10 male and five female genomic DNA samples. All results were in agreement with DNA sequencing including additional comparisons to six cell line DNA samples, NA20232, NA11472, NA20230, CD00014, NA20231, and NA06892 (Coriell Cell Repositories, Hampton, NJ) corresponding to 46, 47, 54, 56, 76, and 93 CGG repeats, respectively. The AGG PCR genotyping was further verified by DNA sequencing and haplotype analysis in a separate sample cohort [Nolin et al., 2011].
Allele instability was defined as any measurable repeat change from parent to child. The CGG repeat primed PCR assay can identify a single repeat change [Chen et al., 2010]. The average repeat difference from 18 parent–child alleles that were determined to differ by exactly one CGG was 0.9 ± 0.2 CGG, compared to <0.3 CGG from multi-day and operator testing of five alleles ranging from 20 to 120 CGGs (54 replicates of each allele). Thus, the PCR method identified modest changes in repeat quantification and transmission stability. In addition, a reproducibility study was performed to evaluate the consistency of the CGG repeat number measured for five alleles ranging from 20 to 120 CGG. Across 28 independent PCR runs, the same number of CGG repeats and AGG interruptions was determined for each of the five alleles tested.
Statistical Methods
All analyses were stratified by parental origin of the transmission. Logistic regression was used to test which measures of repeat structure best predicted the risk for instability (any change in overall repeat length during transmission = 1 vs. no change = 0). The predictor variables included overall repeat length, the number of AGG interspersions in the parent and the length of 3′ uninterrupted CGG repeats. Similarly, linear regression was used to test for associations with the magnitude of instability, or the difference between the parental and offspring repeat lengths for that transmitted allele, and the predicted variables outlined above. ANOVA models were used to test for mean differences in magnitude of instability by number of AGG interspersions (0, 1, or >1 AGG interspersions). All analyses were run using SAS V9.2. All statistical models derived from the entire cohort were also tested using only the subset of subjects who were recruited from screening. The results were similar between sample sets; therefore, we report findings only for the larger dataset that includes all samples ascertained.
RESULTS
Maternal Transmissions
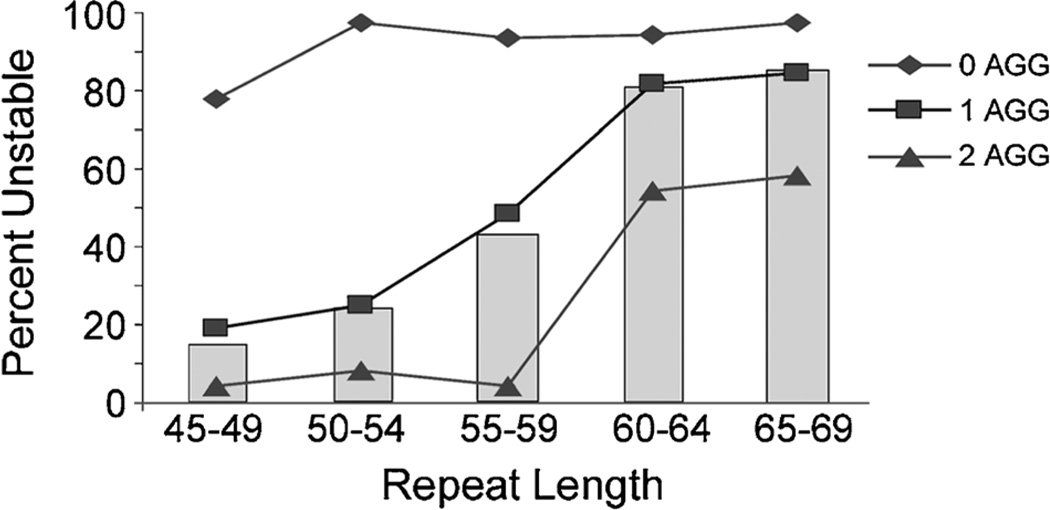
To determine the relationship of AGG structure to repeat instability, we characterized the number and allele-specific location of AGG interruptions within the FMR1 CGG repeat for 375 mothers with intermediate (45–54 repeats) and small premutation alleles (55–69 repeats) and examined the repeat length and AGG structure in their offspring. An unstable transmission was defined as a change of one or more repeats from parent to child. Table I summarizes the AGG structures and instability for 457 maternal transmissions. Of the 457 transmissions, 103 (23%) contained no AGGs, 180 (39%) included one, 159 (35%) included two, 13 (3%) included three, and two (0.4%) included four. In Table II, the transmissions are separated into groups of five repeats by maternal repeat size. Nine full mutation expansions occurred among the 457 maternal transmissions: eight were from families with a history of FXS and one was ascertained from a screening population. All of these full mutations expanded from maternal alleles with no AGGs. The smallest maternal allele expanding to a full mutation had 59 repeats. Two other maternal alleles with 60 and 64 repeats, and six with 65–69 repeats expanded to full mutations. Among the size categories, nearly all alleles with no AGGs (97% [100/103]) were unstable on transmission (Table I).
TABLE I.
AGG Structure and Percent Unstable Transmissions
| No. AGGs | Maternal no. unstable/total |
% | Paternal no. unstable/total |
% |
|---|---|---|---|---|
| 0 | 100/103 | 97 | 33/35 | 94 |
| 1 | 85/180 | 47 | 10/21 | 48 |
| 2 | 31/159 | 19 | 7/23 | 30 |
| 3 | 1/13 | 8 | 0/1 | 0 |
| 4 | 0/2 | 0 | 0/1 | 0 |
TABLE II.
The Effect of Maternal Repeat Size and Number of AGGs on Unstable Transmissions and Full Mutation Expansions
| Maternal repeat size |
No. AGGs |
Total transmissions |
Unstable transmissions |
% | No. full mutation expansion |
% |
|---|---|---|---|---|---|---|
| 45–49 | 0 | 5 | 4 | 80 | 0 | |
| 1 | 30 | 6 | 20 | 0 | ||
| 2 | 43 | 2 | 5 | 0 | ||
| 3 | 3 | 0 | 0 | |||
| 4 | 1 | 0 | 0 | |||
| Subtotal | 82 | 12 | 15 | |||
| 50–54 | 0 | 7 | 7 | 100 | 0 | |
| 1 | 38 | 10 | 26 | 0 | ||
| 2 | 34 | 3 | 9 | 0 | ||
| 3 | 2 | 0 | 0 | |||
| 4 | 1 | 0 | 0 | |||
| Subtotal | 82 | 20 | 24 | |||
| 55–59 | 0 | 23 | 22 | 96 | 1 | 4 |
| 1 | 72 | 37 | 51 | 0 | ||
| 2 | 40 | 2 | 5 | 0 | ||
| 3 | 5 | 0 | 0 | |||
| 4 | 0 | 0 | 0 | |||
| Subtotal | 140 | 61 | 44 | |||
| 60–64 | 0 | 35 | 34 | 97 | 2 | 6 |
| 1 | 25 | 21 | 84 | 0 | ||
| 2 | 27 | 15 | 56 | 0 | ||
| 3 | 1 | 1 | 100 | 0 | ||
| 4 | 0 | 0 | 0 | |||
| Subtotal | 88 | 71 | 81 | |||
| 65–69 | 0 | 33 | 33 | 100 | 6 | 18 |
| 1 | 15 | 12 | 80 | 0 | ||
| 2 | 15 | 9 | 60 | 0 | ||
| 3 | 2 | 0 | 0 | |||
| 4 | 0 | 0 | 0 | |||
| Subtotal | 65 | 54 | 83 | |||
| Total | 457 | 218 | 47 | 9 | 2 |
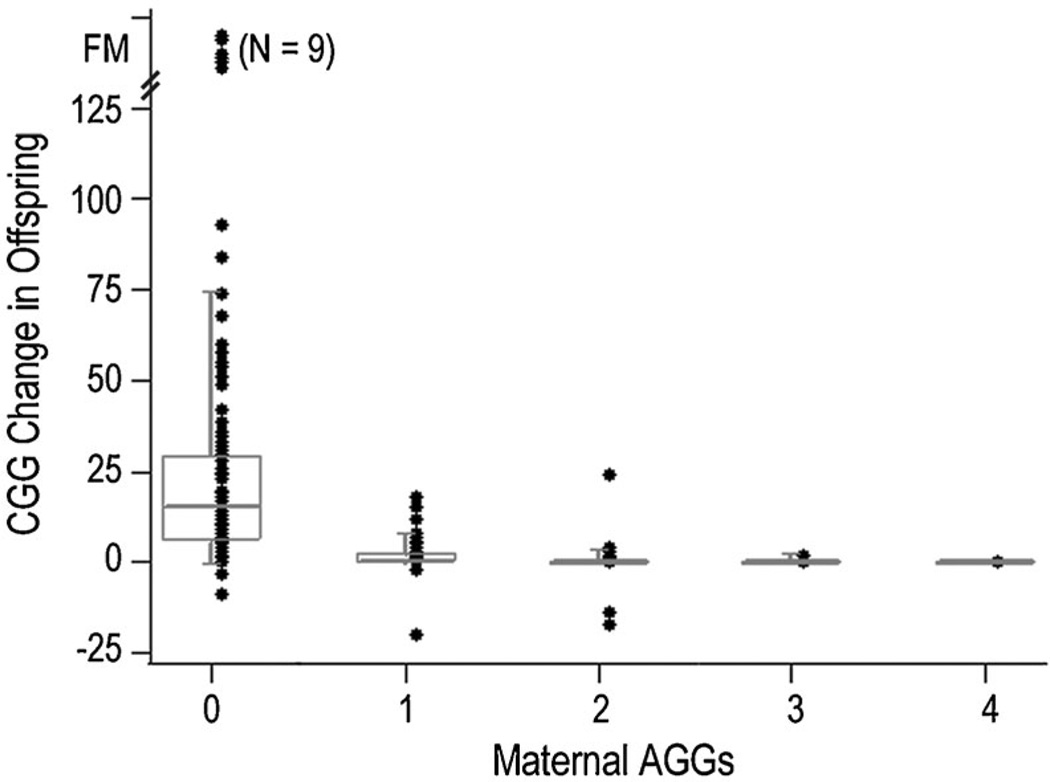
The number of AGGs had a substantial impact on the risk and the magnitude of repeat change from mother to child. The risk of instability associated with the number of AGGs compared to total maternal repeat length alone is shown in Figure 1. For example, maternal alleles with 55–59 repeats had a 42% overall risk of instability. However, for those with no AGG the risk of instability was 96%, for one AGG the risk was 51%, and for two AGGs the risk was only 5%, corresponding to a 19-fold range of risk based on AGG status alone. Moreover, as shown in Figure 2, the number of AGGs impacted both the occurrence of instability and the magnitude of repeat change. The largest range of change and all full mutation expansions were observed in alleles lacking AGGs. Based on logistic regression with instability as the outcome measure, the absence of AGGs within the repeat (0 AGG vs. ≥1 AGGs) was found to confer the greatest risk for unstable transmissions (OR = 67.51 [20.96 –217.42]; P < 0.0001). Alleles with two or more AGGs exhibited greater stability than did those with one or no AGGs (<2 AGGs = 1 vs. ≥2 AGGs = 0; OR = 0.12 (0.08–0.19); P < 0.0001). Both the range and the median of the repeat change magnitude were greatest for alleles with no AGGs and least for those with two AGGs (Table III; F2,454 = 99.14; P < 0.0001).
FIG. 1.
Percentage of unstable maternal transmissions by repeat length and the number of AGG interruptions. The gray columns represent the unstable transmissions for each repeat size category using total repeat length.
FIG. 2.
The change in repeat length from mother to offspring as a function of the number of AGGs. Full mutation expansions are indicated by FM on the Y axis. Box plots represent the 25th to the 75th percentiles of the repeat length change.
TABLE III.
Range of Repeat Change in Unstable Transmissions Excluding Full Mutation Expansions
| Maternal repeat size |
No. AGGs |
No. unstable/total transmissions |
Range of repeat change |
Median repeat change |
|---|---|---|---|---|
| 45–49 | 0 | 4/5 | 1–5 | 1.5 |
| 1 | 6/30 | 1–2 | 1 | |
| 2 | 2/43 | 1–2 | 1.5 | |
| 50–54 | 0 | 7/7 | 1–31 | 5 |
| 1 | 10/38a | 1–20 | 2 | |
| 2 | 3/34 | 1–4 | 3 | |
| 55–59 | 0 | 21/22b | 1–55 | 10 |
| 1 | 37/72c | 1–5 | 2 | |
| 2 | 2/40 | 1 | 1 | |
| 60–64 | 0 | 32/33b | 4–93 | 16 |
| 1 | 21/25 | 1–7 | 4 | |
| 2 | 15/27a | 1–24 | 2 | |
| 65–69 | 0 | 27/27a,b | 1–60 | 19 |
| 1 | 12/15 | 2–18 | 6 | |
| 2 | 9/15 | 1–4 | 2 |
Including two contractions.
Excluding full mutations.
Including one contraction.
We also considered the contribution of the uninterrupted repeat size at the 3′ end following any AGG interruptions to predict allele instability (Table IV). As expected, the longest uninterrupted CGGs were associated with the greatest risk for instability; the OR for instability was 1.23 (1.18–1.28, P < 0.0001). The effect of the length of the 3′ uninterrupted repeat on the magnitude of repeat change was also significant (R2 = 0.23; F1,455 = 135.99, P < 0.0001). All alleles with 61–69 uninterrupted CGGs (61/61) and 88% (80/91) of those with 49–60 CGGs were unstable. Transmissions of maternal alleles with 39–48 uninterrupted CGGs were highly variable with 44% (66/149) unstable making this the most difficult group for predicting allele instability. Adding the number of AGG interspersions into this model did not increase the ability to predict instability for this specific group of alleles (P = 0.23). Alleles with fewer than 39 uninterrupted CGGs were stably transmitted in 93% (145/156) of cases.
TABLE IV.
3′ CGG Uninterrupted Repeats and Percent Unstable Transmissions
| 3′ uninterrupted CGG repeats |
No. unstable/total maternal transmissions |
% | No. unstable/total paternal transmissions |
% |
|---|---|---|---|---|
| <25 | 0/10 | 0 | 0/1 | 0 |
| 25–32 | 2/59 | 3 | 1/9 | 11 |
| 33–38 | 9/87 | 10 | 6/19 | 32 |
| 39–48 | 66/149 | 44 | 7/13 | 54 |
| 49–60 | 80/91 | 88 | 17/20 | 85 |
| 61–69 | 61/61 | 100 | 19/19 | 100 |
We examined the two repeat structure characteristics together to determine their influence on the magnitude of instability because the instability appeared to increase among alleles with no AGG compared with those with one, even though the length of the 3′ uninterrupted CGGs was the same. For example, the magnitude of expansion for alleles with 59 CGG and no AGG, (CGG)59, was greater than that for alleles with one AGG and the same uninterrupted repeat length, (CGG)9AGG(CGG)59 (Table V). Using regression analysis, we first examined the parameters separately. Compared to repeat length alone, the proportion of explained variance in the magnitude of change increased 1.6-fold using maternal AGG number and 1.9-fold using maternal 3′ uninterrupted repeat length. There was a similar increase in the variance explained when we combined maternal repeat length or 3′ uninterrupted CGGs with the number of AGGs (Table VI). Attempting more complicated or higher-order models with both variables included did not improve our ability to predict the change in repeat size. Thus, although we observed patterns that suggested a combined effect of AGG number and 3′ uninterrupted CGGs, we did not detect a statistically significant effect. This result is not surprising since the instability observed in smaller intermediate alleles was modest compared to the larger premutation alleles.
TABLE V.
Comparison of Repeat Size in Offspring with 59 Uninterrupted CGGs in Maternal Alleles
| Maternal repeat structurea |
Offspring repeat size |
Repeat size increase |
|---|---|---|
| 59 | 61 | 2 |
| 59 | 69 | 10 |
| 59 | 82 | 23 |
| 59 | 85 | 26 |
| 59 | 98 | 39 |
| 59 | 114 | 114 |
| 59 | >200 | >140 |
| 9 + 59 | 69 | 0 |
| 9 + 59 | 72 | 3 |
| 9 + 59 | 72 | 3 |
| 9 + 59 | 81 | 12 |
| 9 + 59 | 87 | 18 |
Notation for repeat structure: the number represents the number of CGG repeats, + represents the AGG interruption within that series of CGG repeats.
TABLE VI.
Outcome of Regression Models That Examine the Effect of Repeat Structure Features on the Magnitude of Repeat Instability
| Maternal transmission N = 457 | Paternal transmissions N = 81 | |||||||
|---|---|---|---|---|---|---|---|---|
| Parental predictor variable |
β coeff. | Standardized β coeff. |
P-value | R2 | β coeff. | Standardized β |
P-value | R2 |
| Repeat length | 1.14 | 0.34 | <0.0001 | 0.12 | 0.34 | 0.27 | 0.0151 | 0.06 |
| No. of AGGs | −11.49 | −0.44 | <0.0001 | 0.19 | −3.80 | −0.41 | 0.0001 | 0.16 |
| 3′ uninterrupted repeat | 0.84 | 0.48 | <0.0001 | 0.23 | 0.25 | 0.40 | 0.0002 | 0.15 |
| No. of AGGs + repeat length | −9.62 0.74 |
−0.37 0.22 |
<0.0001 | 0.23 | −3.42 0.10 |
−0.37 0.08 |
0.0006 | 0.15 |
| No. of AGGs + 3′ uninterrupted repeata | −3.29 0.65 |
−0.13 0.38 |
<0.0001 | 0.23 | −2.28 0.11 |
0.30 0.44 |
0. 0006 | 0.15 |
The beta coefficient (β coeff.) for each predictor variable and P-value and R2 for the overall model are provided.
The equation was a linear fit to y = β1x + B0 for single terms and y = (β1x1 + β2x2) + B0 for two term equations. All models were significant thus the change in R2, or the variance explained by the different models, was emphasized.
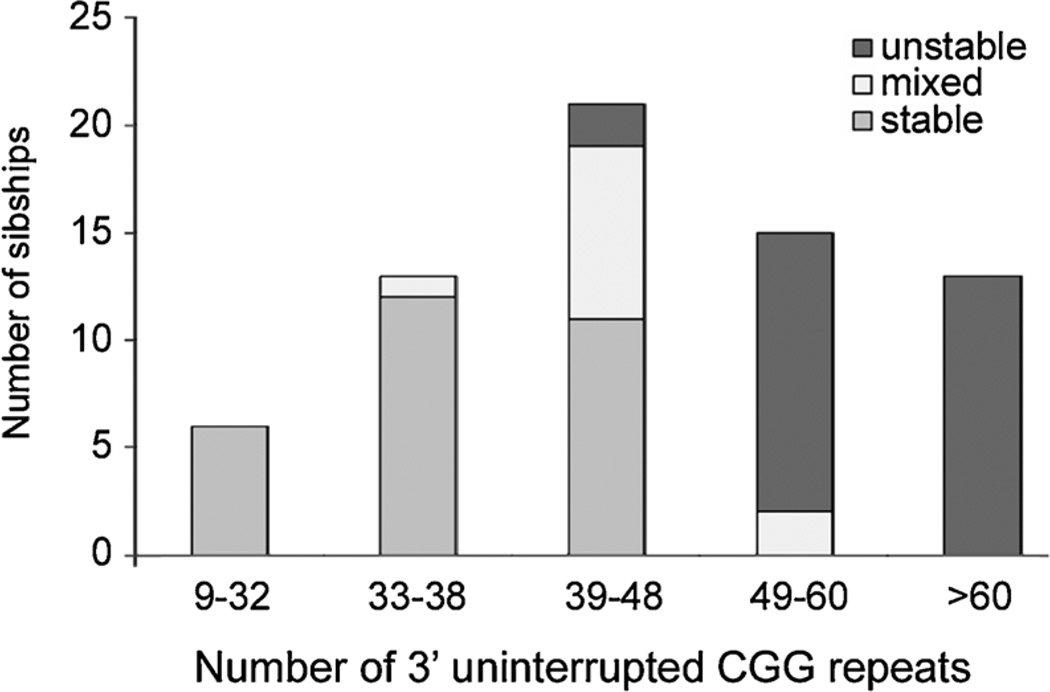
The same patterns of repeat instability were observed among families with more than one offspring (Fig. 3). There were a total of 151 offspring among 68 sibships. Transmissions within a sibship were scored as all stable, all unstable or mixed (both stable and unstable) and grouped by the number of 3′ uninterrupted CGG repeats. As in the previous analysis, transmissions from mothers with 39–48 uninterrupted repeats were the least predictable group with regard to instability.
FIG. 3.
Stability of transmissions within sibships compared to the maternal 3′ uninterrupted CGGs. The X axis indicates the length of the maternal 3′ uninterrupted CGGs. The Y axis indicates the number of sibships. Transmissions within sibships are shown as all stable (medium gray), all unstable (dark gray), or mixed (light gray) with both stable and unstable transmissions.
Contractions to smaller repeat sizes were observed in seven (1.5%) maternal transmissions. These contractions had a median decrease of nine repeats and ranged from a loss of 2 to 20 repeats. The maternal sizes were 54, 54, 59, 62, 64, 69, and 69 repeats. The first three had one AGG, the next two had two AGGs, and the last two had no AGG interruptions. All the maternal alleles included 42 or more 3′ uninterrupted repeats. The contraction of one maternal allele from 62 repeats with two AGGs to 45 repeats in the child resulted in the loss of the two AGG interruptions. This was the only loss of AGGs observed among the 457 maternal transmissions. No loss of AGGs was identified concurrent with expansions of repeat length from mother to child.
Paternal Transmissions
The 81 paternal transmissions from 48 fathers are summarized in Table VII with 50/81 (81%) unstable. While the number of transmissions was fewer than the maternal set, the same patterns of instability were present in this range of 45–69 repeats. Specifically, alleles with no AGG were least stable (33/35, 94%;OR = 28.13 [5.99 –132.23]; P < 0.0001) and those with two or more were most stable (7/25, 28%; OR = 0.12 [0.04–0.34]; P < 0.0001). The magnitude of change was also greatest for alleles with no AGG and for those with larger repeats. The largest expansions occurred in two fathers each with 60 repeats and no AGG. One had a daughter with 114 repeats and the other, one with 109 repeats. A comparison of the uninterrupted CGG repeats at the 3′ end following any AGG interruptions in paternal transmissions showed a pattern similar to maternal transmissions of increasing instability from smaller to larger sizes. All alleles with >60 uninterrupted repeats (19/19) were unstable and 85% of alleles with 49–60 uninterrupted CGGs (17/20) were unstable. Alleles with 39–48 repeats had variable instability (54% [7/13]). Three (3.7%) contractions were observed among the 81 paternal transmissions resulting in a loss of one or two repeats and a median of two repeats without a loss of AGGs. These contractions occurred in paternal alleles with 63, 65, and 66 repeats (two, one, and zero AGGs, respectively). Similar to maternal transmissions, we were able to increase the variance explained for the magnitude of instability by 2.6-fold using either the number of paternal AGGs or the uninterrupted 3′ repeat compared to the traditional method of using repeat size alone (Table VI). Using both predictor variables in a multivariate model did not improve our ability to predict the instability (Table VI).
TABLE VII.
Effect of Paternal Repeat Size and Number of AGGs on Unstable Transmissions
| Paternal repeat size |
No. AGGs |
No. unstable/total transmissions |
% | Range of repeat change |
Median repeat change |
|---|---|---|---|---|---|
| 45–49 | 0 | 1/1 | 100 | 3 | 3 |
| 1 | 1/8 | 13 | 2 | 2 | |
| 2 | 0/5 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | |
| 4 | 0/1 | 0 | 0 | 0 | |
| 50–54 | 0 | 2/2 | 100 | 2–3 | 2.5 |
| 1 | 2/3 | 67 | 1–2 | 1.5 | |
| 2 | 1/4 | 25 | 1 | 1 | |
| 3 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | |
| 55–59 | 0 | 6/7 | 86 | 3–13 | 4.5 |
| 1 | 3/5 | 60 | 2–4 | 2 | |
| 2 | 5/11 | 45 | 1–2 | 1 | |
| 3 | 0/1 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | |
| 60–64 | 0 | 14/15 | 93 | 2–54 | 10 |
| 1 | 1/1 | 100 | 7 | 7 | |
| 2 | 1/3a | 33 | 2 | 2 | |
| 3 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | |
| 65–69 | 0 | 10/10a | 100 | 1–11 | 4 |
| 1 | 3/4a | 75 | 1–11 | 3 | |
| 2 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | |
| Total | 50/81 | 81 |
Including one contraction.
DISCUSSION
Our study demonstrates that AGG analysis identifies maternal FMR1 alleles with 45–69 repeats that are at greatest risk for instability as well as for expansion to the full mutation. In recent years, fragile X carrier screening of pregnant women and women evaluated for infertility has been performed at many centers. Normal alleles aside, these studies identified primarily intermediate or small premutation alleles from 45 to 69 repeats. Two studies [Cronister et al., 2008; Seltzer et al., 2012] identified 1.9–2.8% of women with no family history of fragile X as carriers of intermediate alleles. In addition, 0.4–0.7% of women with no family history were found to carry premutation alleles with more than 54 repeats. While many of the alleles will be stably transmitted or increase by a small number of repeats, a few will be highly unstable. To date, the major problem in screening women has been our inability to predict the risk of instability and expansion to full mutation associated with newly identified alleles since repeat size alone does not accurately predict instability for intermediate and small premutation alleles.
Here, we have shown that either AGG structure or 3′ uninterrupted CGG length are better predictors of instability compared with overall repeat length, the parameter that is currently used in clinical settings. For example, irrespective of overall repeat length, the greatest risk factor for an unstable transmission was the absence of AGGs within the FMR1 repeat. In our study, 100 of 103 maternal transmissions with no AGGs were unstable. Maternal repeats with no AGGs also exhibited the greatest magnitude of repeat instability as compared to alleles with one or two AGGs. More importantly, the nine maternal alleles that expanded to full mutations all contained no AGGs indicating that premutation maternal alleles without AGGs are at greatest risk for full mutation expansion in a single transmission.
We considered different models to predict risk by including: AGG status alone, the 3′ uninterrupted repeat length alone and combinations of 3′ CGG or total repeat length and AGG status. The length of the 3′ uninterrupted repeat was the single-factor variable that best predicted the magnitude of change in repeat length for alleles of 45–69 repeats. However, in a recent study that reported expansion risks for premutation alleles, the combination of total length and number of AGG interruptions yielded the best predictive model for risk of expansion to a full mutation upon transmission, particularly for alleles with fewer than 100 repeats [Yrigollen et al., 2012]. Our current study substantially improves upon those results by analyzing a significantly larger cohort in the small premutation range that better represents the allele sizes and expansion risks in the general population. Eichler et al. [1994] suggested that alleles with >34 uninterrupted CGG repeats at the 3′ end are at risk for instability. Analysis of 3′ CGG length in our dataset showed that all maternal alleles with >60 uninterrupted repeats at the 3′ end were unstable. Setting a practical threshold for instability below this length is more difficult as alleles with 39 ndash;48 3′ uninterruped repeats were variable with respect to instability. For clinical purposes, a threshold for instability at 34 3′ uninterrupted CGG repeats may be too low. The difficulty of making predictions for alleles with 39–48 3′-uninterrupted repeats may be due either to insufficient data or to underlying DNA structural alternatives that occur within this allele range.
We also observed that the combination of repeat length with the number of AGGs had a similar predictive effect on instability. In comparing alleles with equivalent 3′-uninterrupted repeat lengths, the risks and magnitude of expansion are roughly equivalent regardless of which model is used. However, as shown in Table V, in the 55–59 repeat class, repeat expansion is highly dependent on the number of AGGs. Thus, a model that includes either overall repeat length or 3′ uninterrupted CGG length along with AGG number may be more predictive over the entire range of premutation alleles from 55 to 200. This is supported by other studies that indicate better prediction when both repeat length and AGG number are included [Nolin et al., 1999; Crawford et al., 2000; Yrigollen et al., 2012].
A greater percentage (81%) of paternal than maternal (47%) transmissions were unstable, consistent with studies indicating greater instability of normal or intermediate paternal alleles [Sullivan et al., 2002; Nolin et al., 2011]. Nevertheless, a comparison of maternal and paternal transmissions here suggests that many of the same patterns of instability are present in both. Alleles with no AGGs are most likely to be unstable for maternal (100/103) and for paternal (33/35) transmissions. In addition, the greatest magnitude of change is also observed in both maternal and paternal transmissions with no AGGs. Conversely, fewer unstable transmissions are observed for alleles with two AGGs for both maternal (31/159) and paternal (7/23) transmissions.
The only loss of an AGG interruption in 81 paternal or 457 maternal transmissions occurred in a repeat contraction from 62 to 45 suggesting a loss of AGGs is a rare event. There has been a single report of a loss of AGGs in a father who carried 52 repeats with two AGGs and transmitted an allele with 56 repeats and no AGGs to his daughter [Fernandez-Carvajal et al., 2009]. Our AGG analysis showed an increase in the proportion of alleles with no AGGs from the smaller to the larger maternal repeat categories. We also showed that alleles with no AGGs had a greater frequency and magnitude of instability on transmission than those with one or two AGG interruptions. These patterns and the rarity of an observed loss of an interrupting AGG sequence suggest that the enrichment of alleles with no AGGs in the larger repeat categories results primarily from the gradual addition of CGG repeats. We suggest that a major mutational pathway for increases in repeat size occurs in alleles with no AGG interruptions rather than expansions accompanied by a loss of AGGs.
In this study, we show a 2–3-fold increase in our ability to predict magnitude of instability with AGG/CGG repeat structure information relative to using repeat length alone. However, only about 23% and 15% of the variance in the magnitude of the change in repeat length in maternal and paternal transmissions, respectively, was explained by these features. Thus, other factors must contribute to instability and expansion to a full mutation. A recent study indicated that the inclusion of AGG interruption information has a major impact on the accuracy of risk predictions for CGG repeat expansion from larger premutation alleles to a full mutation [Yrigollen et al., 2012]. The authors noted that their observations were derived from a limited number of unique haplotypes and suggested the essential need to refine models of repeat expansion from larger premutation alleles greater than 69 CGG using a larger cohort and different populations.
The identification of AGG interruptions will allow more accurate risk estimations in counseling women who carry FMR1 intermediate and small premutation alleles. Our study demonstrates that alleles without AGGs are at greatest risk for instability and expansion to a full mutation. This observation has different practical implications for the various repeat sizes and will be most useful for women with alleles greater than 49 repeats.
For alleles with 45–49 repeats, prenatal testing or analysis of other family members is unnecessary even in the absence of any AGGs. Size increases are small and there is no apparent risk of full mutation expansion in a single transmission. Alleles with 50–54 repeats and no AGGs are likely to be unstable and may expand into premutation alleles. One maternal 54 repeat allele in our study did expand to 85 repeats in a child. However, this study and others suggest there is little, if any, risk of expansion to a full mutation. For women carrying alleles with greater than 54 repeats and no AGGs there is a clear risk for expansion to a full mutation in the next generation. Women with these alleles should be offered the option of fragile X prenatal testing. For other alleles in this repeat range with at least one AGG, a consideration of prenatal testing is more complicated. While our study suggests little or no full mutation expansion risk for these alleles, the numbers are insufficient to provide specific guidelines at this time. Decisions about prenatal diagnosis should be made on an individual basis for each woman taking into account her level of anxiety, infertility issues, advanced maternal age or other relevant factors.
ACKNOWLEDGMENTS
The authors thank the families whose support made this work possible. The authors thank Victor Kaytser and Lili Zhou for preparation of the samples from Rush University Medical Center. This work was supported in part by the New York State Institute for Basic Research in Developmental Disabilities, the NYS Office of People with Developmental Disabilities and by National Institutes of Health grants R01 HD29909, P30 HD24064 (S.L.S and E.A.), and HD02274 (F.T.), 2R44HD066953-02 (AGH, GJL and SS). Andrew Hadd, Gary Latham, and Sachin Sah have stock options in Asuragen, Inc.
REFERENCES
- Berkenstadt M, Ries-Levavi L, Cuckle H, Peleg L, Barkai G. Preconceptional and prenatal screening for fragile X syndrome: Experience with 40,000 tests. Prenat Diag. 2007;27:991–994. doi: 10.1002/pd.1815. [DOI] [PubMed] [Google Scholar]
- Chen L, Hadd A, Sah S, Filipovic-Sadic S, Krosting J, Sekinger E, Pan R, Hagerman PJ, Stenzel TT, Tassone F, Latham GJ. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. 2010;12:589–600. doi: 10.2353/jmoldx.2010.090227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Zhang F, Wilson B, Warren ST, Sherman SL. Fragile X CGG repeat structures among African-Americans: Identification of a novel factor responsibility for repeat instability. Hum Molec Genet. 2000;9:1759–1769. doi: 10.1093/hmg/9.12.1759. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: Human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronister A, DeMaio M, Mahoney MJ, Donnenfeld AE, Hallam S. Fragile X syndrome carrier screening in the prenatal genetic counseling setting. Genet Med. 2005;7:246–250. doi: 10.1097/01.gim.0000159898.90221.d3. [DOI] [PubMed] [Google Scholar]
- Cronister A, Teicher J, Rohlfs EM, Donnenfeld A, Hallam S. Prevalence and instability of fragile X alleles. Obstet Gynecol. 2008;111:596–601. doi: 10.1097/AOG.0b013e318163be0b. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Holden J, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Macpherson JN, Murray A, Jacobs PA, Chakravarti A, Nelson DL. Haplotype and intersperson analysis of the FMR1 CGG repeat indentifies two different mutational pathways for the origin of the fragile X syndrome. Hum Molec Genet. 1996;5:319–330. doi: 10.1093/hmg/5.3.319. [DOI] [PubMed] [Google Scholar]
- Fernandez-Carvajal I, Posadas BL, Pan R, Raske C, Hagerman P, Tassone F. Expansion of an FMR1 grey-zone allele to a full mutation in two generations. J Molec Diagn. 2009;11:306–311. doi: 10.2353/jmoldx.2009.080174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic-Sadic S, Sah S, Chen L, Krosting J, Sekinger E, Zhang W, Hagerman PJ, Stenzel TT, Hadd AG, Latham GJ, Tassone F. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr, Warren ST, Oostra BA, Nelson DL, Caskey CT. Variation of the CGG repeat at the fragile X site results in genetic instability: Resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Hantash FM, Goos DM, Crossley B, Anderson B, Zhang K, Sun W, Strom CM. FMR1 premutation carrier frequency in patients undergoing routine population-based carrier screening: Insights into the prevalence of fragile X syndrome, fragile X-associated tremor/ataxia syndrome, and fragile X-associated primary ovarian insufficiency in the United States. Genet Med. 2011;13:39–45. doi: 10.1097/GIM.0b013e3181fa9fad. [DOI] [PubMed] [Google Scholar]
- Iong KP, Tong T, Gane LW, Sorensen P, Berry-Kravis E, Nguyen D, Mu Y, Skinner D, Bailey D, Hagerman RJ, Tassone F. Newborn screening in fragile X syndrome: Prevalence and allele distribution of the FMR1 gene. Vancouver, Canada: Amer College Med Genet Meeting; 2011. [Google Scholar]
- Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- Maddalena A, Richards CS, McGinnis MJ, Brothman A, Desnick RJ, Grier RE, Hirsch B, Jacky P, McDowell GA, Popovich B, Watson M, Wolff DJ. Technical standards and guidelines for fragile X: The first of a series of disease-specific supplements to the standards and guidelines for clinical genetics laboratories of the American College of Medical Genetics. Genet Med. 2001;3:200–205. doi: 10.1097/00125817-200105000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolin SL, Houck GE, Gargano AD, Blumstein H, Dobkin CS, Brown WT. FMR1 CGG-repeat instability in single sperm and lymphocytes of fragile-X premutation males. Am J Hum Genet. 1999;65:680–688. doi: 10.1086/302543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolin SL, Brown WT, Glicksman A, Houck GE, Gargano AD, Sullivan A, Biancalana V, Bröndum-Nielsen K, Hjalgrim H, Holinski-Feder E, Kooy F, Longshore J, Macpherson J, Mandel JL, Matthijs G, Rousseau F, Steinbach P, Väisänen ML, von Koskull H, Sherman SL. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am J Hum Genet. 2003;72:454–464. doi: 10.1086/367713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolin SL, Glicksman A, Ding X, Ersalesi N, Brown WT, Sherman SL, Dobkin C. Fragile X analysis of 1112 prenatal samples from 1991 to 2010. Prenat Diagn. 2011;31:925–931. doi: 10.1002/pd.2815. [DOI] [PubMed] [Google Scholar]
- Oberlé I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boué J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Rouillard P, Morel ML, Khandjian EW, Morgan K. Prevalence of carriers of premutation-size alleles of the FMR1 gene and implications for the population genetics of the fragile X syndrome. Am J Hum Genet. 1995;57:1006–1018. [PMC free article] [PubMed] [Google Scholar]
- Seltzer MM, Baker MW, Hong J, Maenner M, Greenberg J, Mandel D. Prevalence of CGG expansions of the FMR1 gene in a US population-based sample. Am J Med Genet Part B Neuropsychiatr Genet. 2012;159B:589–597. doi: 10.1002/ajmg.b.32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AK, Crawford DC, Scott EH, Leslie ML, Sherman SL. Paternally transmitted FMR1 alleles are less stable than maternally transmitted alleles in the common and intermediate size range. Am J Hum Genet. 2002;70:1532–1544. doi: 10.1086/340846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. A rapid polymerase chain reaction-based screening method for indentification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Duhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BE, van Ommen GJB, Blonden LAJ, Riggins G, Chastain HJL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostra BA, Warren ST. Identification of a gene FMR-1 containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:6–15. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Yrigollen CM, Durbin-Johnson B, Gane L, Nelson DL, Hagerman R, Hagerman PJ, Tassone F. AGG interuptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet Med. 2012;14:729–736. doi: 10.1038/gim.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Fragile X genotype characterized by an unstable region of DNA. Science. 1991;252:1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]